Abstract
The 1986 Chernobyl nuclear disaster initiated a series of catastrophic events resulting in long-term and widespread environmental contamination. We characterize the genetic structure of 302 dogs representing three free-roaming dog populations living within the power plant itself, as well as those 15 to 45 kilometers from the disaster site. Genome-wide profiles from Chernobyl, purebred and free-breeding dogs, worldwide reveal that the individuals from the power plant and Chernobyl City are genetically distinct, with the former displaying increased intrapopulation genetic similarity and differentiation. Analysis of shared ancestral genome segments highlights differences in the extent and timing of western breed introgression. Kinship analysis reveals 15 families, with the largest spanning all collection sites within the radioactive exclusion zone, reflecting migration of dogs between the power plant and Chernobyl City. This study presents the first characterization of a domestic species in Chernobyl, establishing their importance for genetic studies into the effects of exposure to long-term, low-dose ionizing radiation.
Characterization of free-breeding dogs in the Chernobyl Exclusion Zone establishes a mechanism for genetic mapping studies.
INTRODUCTION
In April of 1986, the world’s largest nuclear disaster to date occurred in the Chernobyl Nuclear Power Plant (CNPP) in a part of Ukraine that was formerly within the Soviet Union, permanently altering the ecological landscape of the region. The steam explosion inside reactor four and fires that subsequently burned for 10 days released vast quantities of cesium-137, iodine-131, and other radionuclides that spread via weather patterns across Ukraine, Belarus, Russia, other parts of Europe (1–3), and even North America (4). The 2600-km2 area extending around the power plant, which is now known as the Chernobyl Exclusion Zone (CEZ), was most profoundly affected by the radioactive cloud, generating an ecological catastrophe of massive proportions.
The abundance of wildlife populations within the CEZ was substantially reduced following the accident (5), and although some species appear to have recovered, likely due to a lack of human disturbance, many have not (6). One of the greatest concerns is that continued environmental pollution, including radiation and heavy metal poisoning, may raise or lower genetic species diversity depending on directional selection, bottleneck events, or alteration of migration patterns (7). Increased genetic diversity via elevated mutation rates may be more likely in highly mutagenetic environments, such as that of Chernobyl (8), or other radioactive places on Earth (9). Conversely, a reduction in the mating population from the initial effects of the disaster, including high doses of radiation and fires, may markedly reduce genetic diversity. To date, no population genetic studies of Chernobyl organisms have included large-bodied mammals, such as canines. Thus, nonhuman mammals in the CEZ are greatly understudied, despite their potential to offer powerful insights into the history and survival of life in this hostile environment.
The domestic dog presents an interesting case in this regard, as little is known about the origin of the free-roaming dog populations in the Chernobyl region or how canine populations survived after the explosion. One claim is that the current populations are descended from pets left behind by individuals evacuated from cities such as Pripyat (10), which was once home to approximately 50,000 people. After the evacuation of people from the CEZ, the Ukrainian Ministry of Internal Affairs initiated the culling of abandoned pets to prevent the potential spread of radioactive contamination (10). However, some dogs were believed to have evaded hunters, escaping beyond the CNPP, and were subsequently fed and cared for by camps of Chernobyl clean-up workers (10) and, more recently, by tourists to the CEZ. It remains unclear the degree to which dog populations have expanded from their original 1986 founders, how many distinct populations remain, how diverse these populations are, or if they are bounded by geographic constraints. Further, the role of modern purebred versus free-breeding dogs in reconstituting the Chernobyl dog populations remains unknown. This division is of interest given that the population structure of free-breeding dogs, which encompasses the majority of domestic dogs in the world today, is distinct from that of purebred dogs. Specifically, purebred dogs, by definition, have closed breeding pools such that only members of a specified breed can be included in an established breeding program, often leading to limited genetic variation within any given breed (11–14).
In this study, we demonstrate that there are two locally distinct populations in this region, those from Chernobyl City, located 15 km from the CNPP, and dogs living in the CNPP. Within the CNPP itself, we describe discrete populations, including multiple nuclear families with connections to families in Chernobyl City. Further, by identifying differences in genetic diversity and breed ancestry, we characterize underlying genomic differences between the two primary populations within the CEZ, enabling a robust comparison of Chernobyl dogs to other free-breeding dogs throughout Ukraine and surrounding countries. These findings constitute a critical first step in the characterization of a unique target population that holds great promise for investigations into the effects of continuous environmental radiation exposure on a large-bodied mammalian species. Classification of discrete geographically defined populations, ancestry, and the existence of family structures within and among populations are areas of inquiry needed to design studies aimed at finding critical genetic variants that have accumulated for more than 30 years in this hostile, contaminated environment.
RESULTS
Sample collection includes dogs from distinct locations in the CEZ
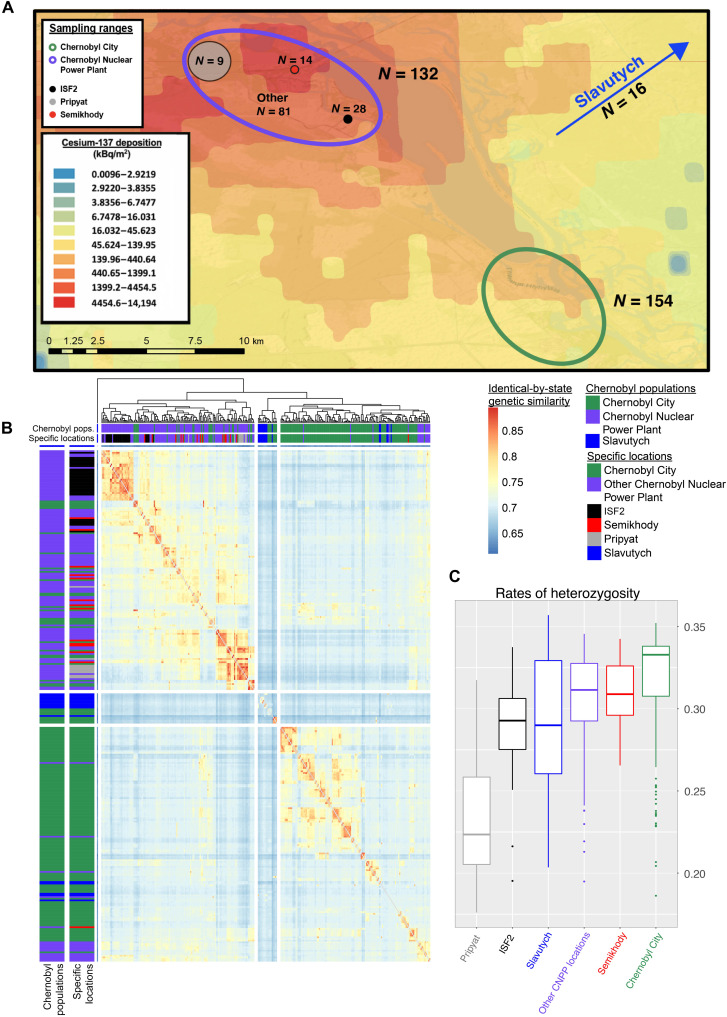
The Chernobyl Dog Research Initiative was formed in June 2017 in response to a substantial increase in the feral dog population size, which was estimated at one time to exceed 800 individuals. In 2017 to 2019, three clinics providing veterinary care for free-roaming dogs in and around the CEZ were established strategically to sample the greatest geographic diversity of dogs. During this period, blood samples from 302 dogs were obtained and preserved for subsequent studies, including those herein. Samples from 132 dogs were collected from the first clinic location, situated inside the industrial areas of the CNPP. Dogs now occupying the CNPP region are often fed by power plant workers and live in the power plant itself, including the Semikhody train station and the interim used fuel storage facilities (ISF2), which stores spent nuclear fuel. We also sampled dogs from areas directly adjacent to the power plant in the heavily wooded region around the city of Pripyat, located approximately 3.5 km from the CNPP. Pripyat remains uninhabited today due to high contamination levels, although workers and tourists visit the CNPP daily. Fourteen dogs were sampled from the Semikhody train station, 28 from ISF2, and 9 from Pripyat (Fig. 1A and fig. S1, A and B). The remainder of this set of samples were collected from dogs found at distinct locations throughout the CNPP.
Fig. 1. Map and analyses of genetic relationships between dogs from Slavutych, Chernobyl City, and the CNPP.
(A) Sampling ranges of dogs within the CEZ. Map background shows the levels of cesium-137 deposition as a proxy for contamination (1). (B) Hierarchically clustered heatmap based on identical-by-state genetic similarity between individuals. (C) Average rates of heterozygosity of individuals separated by sampling location.
In addition to the CNPP-dwelling dogs, an additional large population occupies Chernobyl City, a residential area approximately 15 km from the CNPP, which was largely abandoned after the disaster, leaving only a small number (<500) of people. A second clinic location was set up in Chernobyl City itself where blood samples from 154 dogs were collected. After the nuclear disaster, displaced CNPP workers and their families were moved to the city of Slavutych, a comparatively less contaminated area, 45 km across the Belarusian border where the majority still reside. One of the 2019 clinic locations was in the city of Slavutych, where 16 samples were collected (Fig. 1A and fig. S1, A and B). See Materials and Methods for details.
Chernobyl dog population genetic structure is shaped by geographic location
To understand the genetic relationships within and among the Chernobyl dog populations sampled above, we first constructed a hierarchically clustered heatmap based on identical-by-state (IBS) genetic similarities using 129,497 genome-wide single-nucleotide polymorphisms (SNPs) from the 302 samples (Fig. 1B and Materials and Methods). In addition, we applied F3 statistical analysis to the aforementioned dataset to measure admixture between Chernobyl populations (fig. S1D and Materials and Methods). We first sought to determine whether the dog populations in the CNPP, Chernobyl City, and Slavutych are genetically differentiated and, if so, to quantify differences in relatedness. We found, first, that there are three genetically independent populations, such that the majority of individuals cluster in the heatmap based on capture location, with dogs from each location more closely related to each other than to those from other locations. However, there is some overlap, as 27 dogs sampled in Chernobyl City cluster with dogs sampled in the CNPP. In addition, 16 dogs from the CNPP and 5 from Slavutych cluster with the dogs from Chernobyl City. The dogs from Slavutych split into their own cluster, along with 10 Chernobyl City dogs (Fig. 1B). We see the lowest F3 values (F3 = 0.62 to 0.65) between samples from Slavutych and any of the other Chernobyl populations (fig. S1D).
We next separate dogs from ISF2, Semikhody, and Pripyat from other locations within the CNPP and investigate relatedness within these groups. We also measure admixture between these groups and populations in Chernobyl City or Slavutych. Individuals from ISF2 appear to be the most genetically similar and form a tight cluster in the heatmap, with elevated IBS values. Dogs from Pripyat also appear genetically similar relative to other dogs from the CNPP. Dogs from the Semikhody train station do not cluster together in the heatmap and are intermingled with dogs from multiple locations within the CNPP. One of the Semikhody dogs groups with Chernobyl City dogs (Fig. 1B). We see greater admixture between ISF2, Semikhody, Pripyat, and dogs sampled from other locations within the CNPP area (F3 = 0.094 to 0.141) than we do between dogs from any of the CNPP populations and those from Chernobyl City (F3 = 0.072 to 0.079) (fig. S1D). Overall, this suggests that there is gene flow between locations within and closest to the CNPP including between ISF2, Pripyat, Semikhody, and other locations within the CNPP. There is only minimal gene flow between CNPP populations and those from Chernobyl City.
We next measured the genome-wide heterozygosity per individual to characterize the level of genetic diversity within each population (Fig. 1C). We observe that the dogs from other locations within the CNPP, excluding Pripyat, ISF2, and Semikhody, have significantly lower rates of heterozygosity than those from Chernobyl City (mean = 0.32 and 0.34, respectively; P = 3.093 × 10−6). Moreover, Pripyat dogs have significantly lower rates of heterozygosity compared to dogs from other locations in the CNPP (mean = 0.26 and 0.32, respectively; P = 6 × 10−3), suggesting that dogs from this area may have experienced increased inbreeding or that the population initiated from a small number of founders. In contrast, dogs from the Semikhody train station and ISF2 show similar rates of heterozygosity to other dogs from the CNPP (P = 0.07 and 0.09, respectively). We observed that dogs sampled from Slavutych have slightly lower median rates of heterozygosity compared to dogs from both the CNPP and Chernobyl City, although Slavutych dogs have the widest distribution of values (0.23 to 0.38). Overall, dogs from Chernobyl City have the highest average rates of heterozygosity (0.34), suggesting that this group is, comparatively, the most outbred (Fig. 1C).
Chernobyl dogs are genetically distinct from other free-breeding and purebred dog populations
To test genetic relatedness between dogs from Chernobyl, purebred dogs, and free-breeding dogs from several adjacent countries, we performed principal components analysis (PCA) using the quality pruned dataset (Fig. 2, A to C, and Materials and Methods). Locations closest to and within the CNPP, including Pripyat, ISF2, and the Semikhody train station, showed extensive genetic similarity and gene flow between groups (Fig. 1B and fig. S1D). Therefore, samples from these locations are collectively labeled as CNPP in the PCA and haplotype sharing analyses. The PCA included 232 free-breeding dogs from 12 countries that were split into four geographic regions: Eastern Europe, central Asia, eastern Asia, and the Middle East (fig. S2). In addition, 49 free-breeding dogs from the Ukrainian city of Vinnytsia, which is located approximately 350 km southwest of Chernobyl, were also included. To provide a comparison to purebred populations, we used genetic data from 1324 dogs from 162 breeds recognized by the Fédération Cynologique International, which are largely of western European descent (15). The 162 breeds were organized into clades of related breeds based on a bootstrapped phylogenetic tree of only purebred dogs, and the clades were named according to their general function or breed type for ease of comparison (e.g., shepherds and related breeds, U.K. flock guardians, and related breeds) (see Materials and Methods and fig. S3).
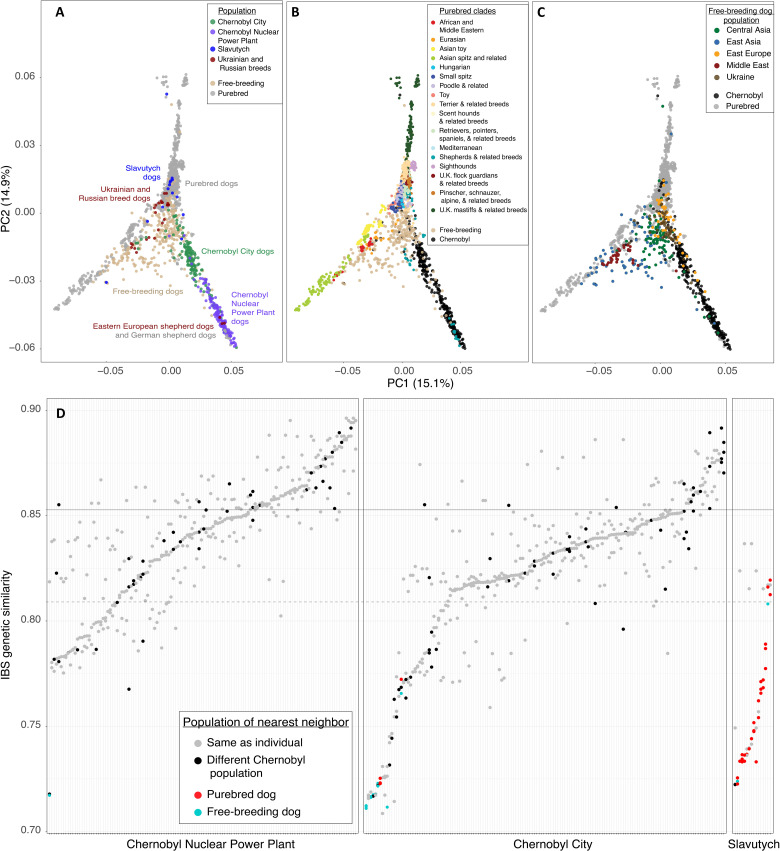
Fig. 2. Genetic differentiation of Chernobyl dog populations.
(A) PCA including purebred, Ukrainian/Russian breed, free-breeding, Slavutych, and Chernobyl dogs. Samples are colored by population. (B) PCA with purebred dog samples colored by clade. (C) PCA with village dog samples colored by geographic region. A full list of individuals and their classifications can be found in table S1. (D) Nearest genetic neighbor analysis showing distribution of relatedness of each individual from the CNPP, Chernobyl City, and Slavutych to the three most closely related individuals in the entire dataset of Chernobyl, purebred, and other free-breeding dogs. The black dotted horizontal line represents the first quartile, and the black solid horizontal line represents the third quartile.
We found that principal component 1 (PC1) (15.1% variance) corresponds to a portion of the diversity present among purebred dogs and free-breeding dogs related to geographic origin (Fig. 2B). For example, the Asian spitz and related breeds separate from European-origin breed dogs across PC1 and further separate into breed clusters along the same axis (Fig. 2B). Free-breeding dogs are also distributed along PC1 and separate primarily based on geographic location and genetic relatedness to purebred dogs from the same region (Fig. 2C and fig. S2). PC2 (14.9% variance) corresponds to diversity within the Chernobyl populations, in addition to that of breed dogs, demonstrating that dogs sampled in the CNPP separate from those sampled in Chernobyl City (Fig. 2A). Separation of the CNPP and Chernobyl City populations from purebred dogs and free-breeding dogs from other geographical regions suggests that dogs living in Chernobyl are genetically distinct from those outside of the region. Variance along PC2 is similar within the CNPP and Chernobyl City populations, and there is some overlap between dogs from each sampling location. The German shepherd dogs and Eastern European shepherds cluster with dogs from the CNPP and separate from the rest of the purebred dogs in the dataset (Fig. 2A). Dogs from Slavutych do not cluster as a single population, instead grouping according to genetic relatedness with breed dogs (Fig. 2, A and D).
We next examined genetic relationships among Chernobyl dogs in the context of other dog populations to determine which, if any, individuals have nonnative ancestry. We used IBS genetic distance metrics to identify the three nearest genetic neighbors for each Chernobyl dog (Fig. 2D). We examined the IBS distances beyond a dog’s own population by including individuals from the CNPP, Chernobyl City, Slavutych, purebred dogs, and free-breeding dogs from several geographic regions, as previously described, to determine whether dogs migrated between Chernobyl populations and to assess whether they are closely related to nonlocal dogs (Materials and Methods). We found that 39 dogs from Chernobyl City and 35 dogs from the CNPP have at least one nearest genetic neighbor from a different population within the CEZ. One dog from the CNPP and seven dogs from Chernobyl City have at least one nearest genetic neighbor from a free-breeding dog population outside of Ukraine and are less similar to each other than to their other nearest genetic neighbors, indicating that these individuals are unlikely to have local ancestry. In addition, one dog from Chernobyl City has purebred Siberian huskies as its nearest genetic neighbors, and another has a single German shepherd dog as one of its nearest genetic neighbors (Fig. 2D). None of the Slavutych dogs have all three of their nearest genetic neighbors from Slavutych, indicating that dogs from this location do not make up a closed breeding population. Most of the dogs from Slavutych have different types of purebred dogs as their nearest genetic neighbors, suggesting that the majority do not have local ancestry (Fig. 2D).
Kinship analysis reveals complex family relationships among Chernobyl dog populations
IBS analysis revealed several closely related groups of dogs from the CNPP and Chernobyl City (Fig. 1B). To investigate potential family relationships, both within and between populations, kinship estimates were calculated for each pair of dogs. When plotted, the distribution of estimates revealed a peak overlapping 0.25, which indicates several first-order (sibling or parent-offspring) family relationships (Fig. 3A). However, when these estimates were plotted against the proportion of markers showing zero IBS in an effort to distinguish parent-offspring relationships from those of siblings, it was difficult to determine a cutoff point for siblings versus half-siblings (Fig. 3A). By comparison, parent-offspring relationships were easily distinguishable from all other relationships as almost no markers showed zero IBS between these pairs. Therefore, we focused our subsequent kinship analysis exclusively on parent-offspring relationships.
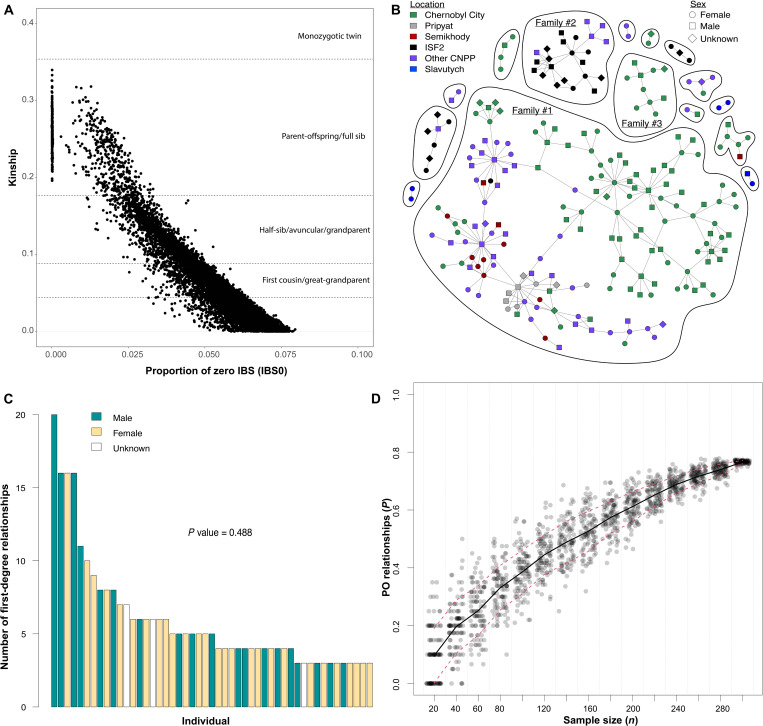
Fig. 3. Identification and analysis of family structure in the Chernobyl dog populations.
(A) Scatterplot showing pairwise kinship relationships among dogs from the Chernobyl populations. Proportion of zero identity-by-state below 0.0004 differentiates parent-offspring relationships. (B) Networks of parent-offspring relationships. (C) Distribution of the number of first-degree relationships for individuals with more than two first-degree family relationships colored to indicate sex. P value corresponds to a Wilcoxon rank sum test used to determine whether there is a significant difference in the number of first-degree family relationships between males and females. (D) Resampling of the Chernobyl dog population to determine the impact of sample size on the discovery of parent-offspring (PO) pairs. The y axis represents the proportion of individuals within the resampled population belonging to a parent-offspring pair.
Once parent-offspring relationships were identified, single linkage clustering was used to define families (Fig. 3B). Notably, 77% of dogs (233 of 302) have a parent or offspring within the dataset, and a total of 15 discrete families were defined. The largest family group, termed Family #1, is composed of more than 162 individuals, with multiple connections between dogs from different capture locations (Fig. 3B). This suggests that there is gene flow between populations. The seven smallest family groups, defined as having only two individuals, are each composed of a single parent and single offspring. Most dogs belong to one of three large families, each containing more than 10 members (Fig. 3B, labeled Families #1 to #3). Within the complex genealogy of Family #1, subgroups of closely related individuals are connected to one or two other subgroups from different locations by a single pair of individuals. All nine dogs from Pripyat belong to a complicated family network of parent-offspring relationships within Family #1. Ten of the 14 dogs from the Semikhody train station belong to the same family network and are connected by a single dog to the family group from Pripyat within Family #1.
The kinship analysis shows that genetic relationships between free breeding dogs in Chernobyl are complex, with many instances of breeding across multiple generations, making it difficult to infer kinship beyond the first degree. We were, however, able to investigate the sex distribution of dogs with more than two parent-offspring connections to test for potential popular sire effects (Fig. 3C). Male dogs typically mate continuously, while females only mate when they are in heat; thus, successful males will sire a disproportionate number of offspring, leading to overrepresentation in the gene pool. Our analysis was restricted to dogs connected by more than two parent-offspring relationships to ensure that those analyzed are a parent to at least one other dog, as directionality of inheritance could not be inferred from the observed relationships. Our results showed no significant difference between the distribution of males and females (P = 0.488) indicating, unexpectedly, that there is no obvious popular sire effect (Fig. 3C). This could be the result of sampling bias, as females were more often captured for surgery and sampling with their offspring, while males do not provide care for offspring after birth, thus yielding more complete family groups for females than males. Overall, 60 females from Chernobyl City, 54 from the CNPP, and 5 from Slavutych had at least one parent or offspring in the dataset. An additional 52 males from Chernobyl City, 39 from the CNPP, and 1 from Slavutych also had at least one parent or offspring in the dataset. We were unable to confirm the sex of 7 dogs from Chernobyl City and 15 from the CNPP.
To determine the potential relationship between sample size and kinship structure in the CEZ populations, we randomly subsampled Chernobyl dogs at various sample sizes and counted the total number of parent-offspring relationships. Our results showed that the observed extent of family structure is related to the number of individuals sampled (Fig. 3D). At lower levels of sampling, many family relationships would have been missed. For example, if we had only sampled 100 individuals, then approximately 40% of dogs would have been identified as members of a family. However, sampling of more than 300 individuals revealed that 77% of dogs were identified as family members, indicating the value of sampling large numbers of individuals for inferring family structure. Because 77% of dogs were observed in parent-offspring relationships, the analysis is approaching saturation and it is unlikely that additional sampling would reveal further family structure. Moreover, after capture, dogs are either spayed or neutered, which is expected to slow the growth of dog families in the CEZ.
Chernobyl dog populations differ in levels of purebred dog haplotype sharing
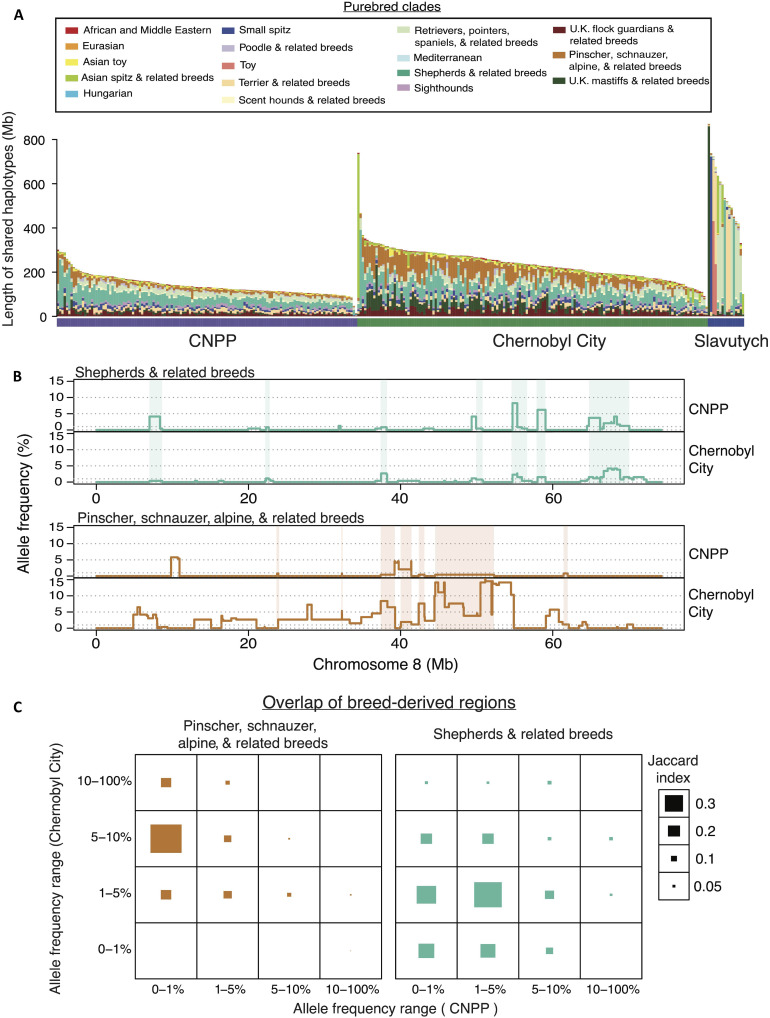
To identify purebred contributions to the CNPP and Chernobyl City populations or shared ancestry with particular breed types, we determined the total length of shared haplotypes in identical-by-descent (IBD) segments between each Chernobyl dog and each of 1324 breed dogs, which themselves represent 162 individual breeds (Fig. 4A and Materials and Methods). On the basis of the average total length of IBD segment sharing between each breed dog and Chernobyl dog, we found that individuals from both Chernobyl City and CNPP have the greatest level of haplotype sharing with German shepherd dogs, followed by Eastern European shepherds and other shepherd-related breeds, which is consistent with the PCA results (Figs. 2A and 4A). Dogs from Chernobyl City also have elevated sharing with the boxer and rottweiler breeds. Of all Chernobyl dog populations, dogs from Slavutych show the greatest haplotype sharing with purebred dogs, particularly with the Labrador retriever, boxer, and Yorkshire terrier (Fig. 4A).
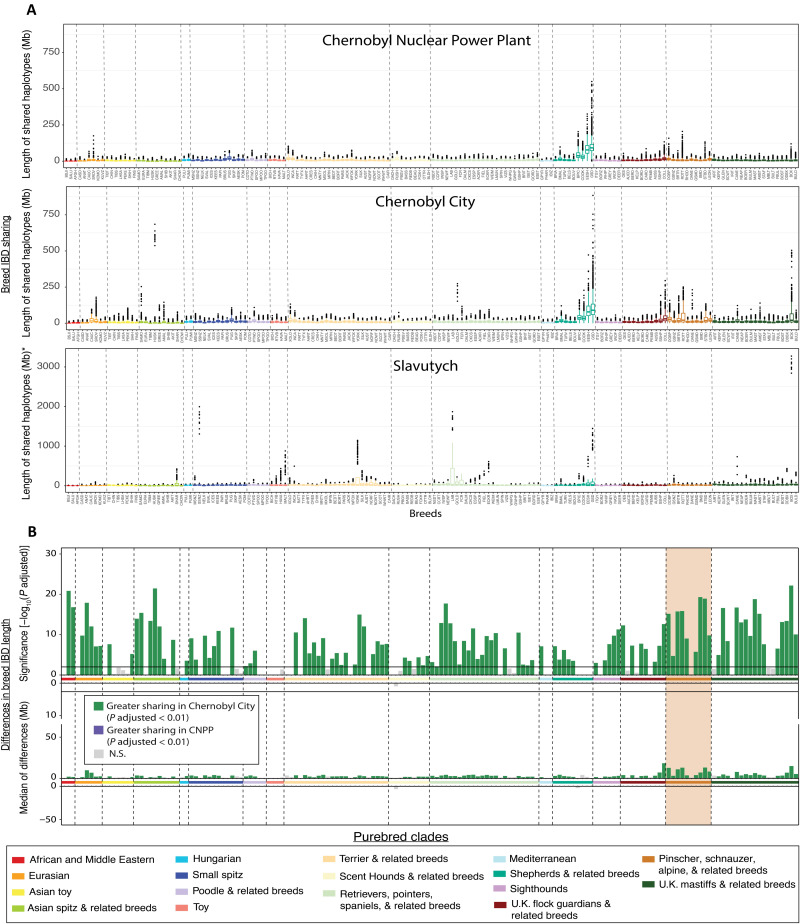
Fig. 4. Differences in breed ancestry between Chernobyl populations.
(A) IBD haplotype sharing with purebred dogs. Each point represents the average length of shared haplotypes of individuals from each population of interest with each breed. *Y-axis scale of the Slavutych plot is different from that of the CNPP and Chernobyl City. The x axis shows abbreviations for each breed comparison defined in table S1. (B) Differences between CNPP and Chernobyl City IBD haplotype sharing with breeds. Each vertical bar represents the −log10(P value) calculated for pairwise comparisons of average sharing between each Chernobyl population and each breed. Vertical bars flipped above the zero line indicate greater average IBD haplotype sharing between dogs from that breed and dogs from Chernobyl City. Breeds are ordered on the x axis according to their placement on a bootstrapped phylogenetic tree found in fig. S2. Breeds are colored according to clade. N.S., not significant.
To characterize differences in western breed contributions to Chernobyl City versus CNPP populations, we used a Wilcoxon signed-rank test, assigning P values to pairwise comparisons of shared haplotypes between each of the two Chernobyl populations and each breed (Fig. 4B). We calculated the average total length of IBD haplotypes shared between each individual Chernobyl dog and each purebred dog (Materials and Methods). We then compared breed averages between CNPP and Chernobyl City. Dogs from Slavutych were omitted from this analysis because of variability in breed composition between members of the population.
Compared to CNPP dogs, we found that dogs from Chernobyl City have significantly greater haplotype sharing with 111 of 162 pure breeds (P adjusted < 0.01). Chernobyl City dogs show the greatest differences in median sharing with breeds in a clade consisting of pinscher, schnauzer, alpine, and related breeds, suggesting that sharing of haplotypes associated with this group likely contributes to differentiation of the Chernobyl City and CNPP populations. On average, Chernobyl City dogs share 7.7 Mb more of their genome with the above pinscher clade than do the CNPP dogs (Fig. 4, A and B). Differences in median IBD haplotype sharing shows that dogs from the CNPP have slightly greater sharing with the Russian hound and berger Picard, although these differences are not statistically significant [P adjusted(Russian hound) = 0.1; P adjusted(berger Picard) = 1] (Fig. 4B).
Identification of breed-derived genomic regions within the Chernobyl dog populations reveals independent admixture events
To further investigate modern breed contributions to Chernobyl dog populations, we assigned purebred dog “clade labels” to genomic segments based on shared haplotypes between Chernobyl dog and purebred dog chromosomes within each clade (see Materials and Methods). Characterizing individual genomes in this manner made it possible to localize and track the genomic contributions of breed-derived haplotypes within the Chernobyl populations, allowing us to differentiate more recently introgressed breed-derived haplotypes from those that are likely ancestral. Ancestral haplotypes that have persisted in Chernobyl populations provide a target for future studies aimed at measuring the accumulation of genetic variation in the presence of continued radioactive contamination. Direct comparisons between these breed-derived haplotypes versus those same haplotype segments in purebred populations that were not exposed to radiation may reveal the magnitude of genomic scarring caused by long-term multigenerational radiation exposure.
Overall, the individual composition of breed-derived haplotypes is consistent with observations from analysis of shared IBD segments described above (Fig. 4A). For example, shepherd-related IBD segments are found frequently in individuals from both populations, while segments from the pinscher clade are more frequently represented in the Chernobyl City dogs (Fig. 5A). Alternatively, individual dogs from Slavutych carry breed-derived haplotypes from only one or two clades rather than breed-derived haplotypes from all highly represented clades in the population (Fig. 5A). Therefore, Slavutych dogs maintain large signatures of purebred ancestry, which is distinct from that observed in the CNPP and Chernobyl City populations. The latter reflect mixed ancestry and resemble free-breeding dog populations. This is consistent with the veterinary clinic notes from sample collection in Slavutych where both owned and unowned free-breeding dogs and residents’ pets were sampled.
Fig. 5. The genomic landscape of breed-derived haplotypes within the dogs of Chernobyl.
(A) Individual composition of breed-derived haplotypes shown as total length of shared haplotypes between Chernobyl dogs and purebred populations. (B) Example of the genomic distribution and haplotype frequency of clade labeled regions along a single chromosome for each population. Breed-derived haplotypes shared between both populations are highlighted to indicate overlap. (C) Overall sharing of breed-derived IBD segments across populations. Sharing is measured as the Jaccard index for overlapping segments of breed-derived IBD across both populations. IBD segments were categorized according to their frequency within each population. Frequency ranges are displayed along the x and y axes.
Next, we determined whether breed-derived haplotypes contributed to shared ancestry between the CNPP and Chernobyl City dogs. We used population haplotype frequencies to distinguish recent from ancestral breed contributions. Higher allele frequencies likely indicate ancestral contributions, as those haplotypes have had longer to accumulate within the population. We therefore tested whether high-frequency breed-derived haplotypes in the genomes of dogs from either population overlapped. We focused, specifically, on the shepherd and pinscher clade haplotypes, as they are the most frequent in both populations (Figs. 4A and 5B and fig. S4). Results show that pinscher clade haplotypes observed at high frequencies (>5%) in the Chernobyl City population are observed at low frequencies (<1%) in the CNPP population. For example, 39% of pinscher clade haplotypes existing at a frequency of 5 to 10% in Chernobyl City dogs occur at <1% frequency in the CNPP dogs (Fig. 5C). Consistent with above results, the pinscher clade haplotypes are largely distinct between the two populations (Fig. 4B), with migration between Chernobyl City and the CNPP explaining minor overlap (Figs. 3B and 5C). Therefore, high-frequency pincher clade haplotypes, which make up 20% of Chernobyl City chromosomal regions, likely had limited radiation exposure, as these haplotypes are mainly restricted to the Chernobyl City population, which exists in a less contaminated area of the CEZ than the CNPP (Fig. 1A).
Conversely, shepherd clade haplotypes are more evenly shared between Chernobyl City and the CNPP. While there were few haplotypes at ≥5% frequency shared between the two populations, high-frequency haplotypes in one population tend to exist at moderate frequency in the other. For example, 36% of shepherd clade haplotypes, which occur at a frequency of 5 to 10% in Chernobyl City dogs exist at a frequency of 1 to 5% in the CNPP population. Yet, 52% of shepherd clade haplotypes, which are present at a frequency of 5 to 10% in the CNPP, exist at a frequency of 1 to 5% in the Chernobyl City population (Fig. 5C). Parent-offspring relationships spanning both locations indicate that migration plays a role in shaping the landscape of shared haplotypes between populations (Figs. 2B and 3B), as is observed with the pincher clade haplotypes. However, sharing of shepherd clade haplotypes across both populations is maintained at elevated frequencies, with the overlap between the two populations suggesting shared ancestry in addition to recent admixture. Our examination of breed-derived segment lengths across haplotype frequencies also supports both recent purebred admixture and shared shepherd-related ancestry in the Chernobyl City and CNPP populations (fig. S5). Thus, the high-frequency shepherd haplotypes, which make up 9% of CNPP chromosomal regions, are high priority targets for measuring genomic scarring of long-term multigenerational exposure to environmental radiation.
Comparisons to free-breeding dog populations outside Chernobyl reveal patterns of breed haplotype sharing consistent with geographic origin
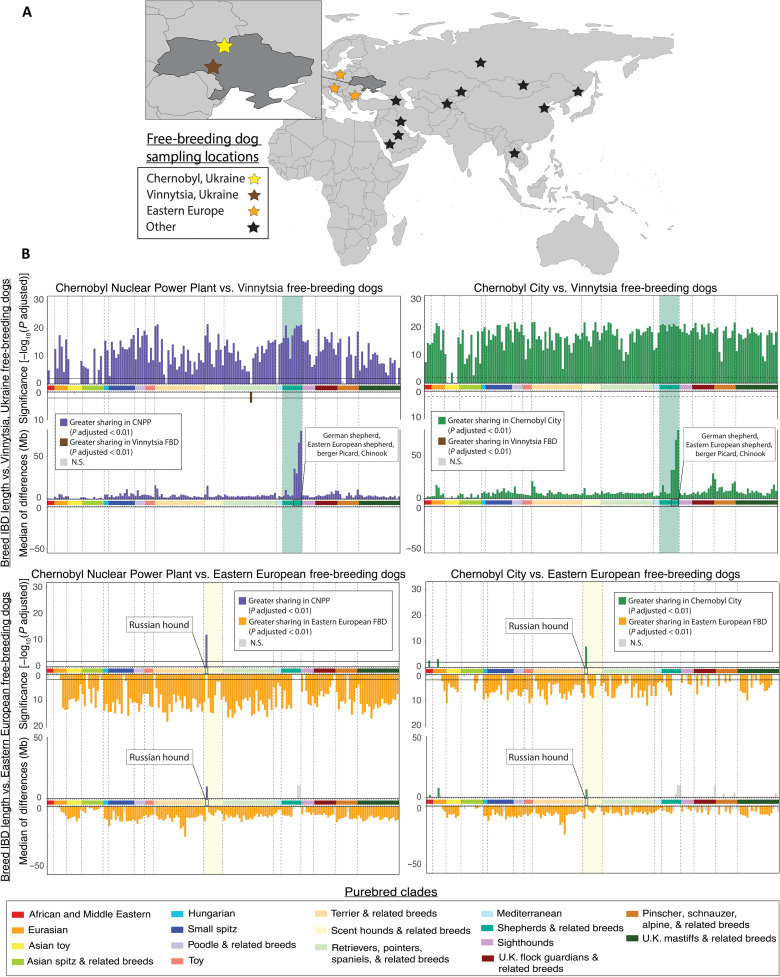
Breed-based interrogation of Chernobyl dog ancestry indicates minimal haplotype sharing with modern purebred dogs, suggesting a genetic composition similar to that of other free-breeding populations. We next sought to characterize the ancestry of Chernobyl dogs as it relates to free-breeding dog populations outside of Chernobyl, using the same methodology as shown in Fig. 4B. We thus compared the breed composition of Chernobyl populations to that of other free-breeding dog populations, starting with free-breeding dogs from Vinnytsia, Ukraine (Fig. 6A). We found that both Chernobyl populations have significantly greater haplotype sharing with most established breeds than do the free-breeding dogs from Vinnytsia (Fig. 6B).
Fig. 6. Breed haplotype sharing of Chernobyl dogs compared to European free-breeding dogs.
(A) Map of approximate free-breeding dog (FBD) sampling locations. See fig. S4 for breed haplotype comparisons to free-breeding dog populations outside of Eastern Europe. (B) Differences in breed IBD haplotype sharing between each Chernobyl population and free-breeding dogs from Vinnytsia, Ukraine. In the plots of significance, each vertical bar represents the −log10(P adjusted value) calculated for pairwise comparisons of average sharing with each breed between one of the Chernobyl populations and free-breeding dogs from Vinnytsia or Eastern Europe. Purple or green vertical bars flipped up on each plot indicate increased haplotype sharing of a breed with one of the Chernobyl populations. Brown or orange vertical bars flipped down indicate increased sharing with Vinnytsia or Eastern European free-breeding dogs, respectively. The black solid line on the plot of significance indicates the threshold for significance at a 99% confidence interval for each comparison. Each vertical bar in the second set of plots represents the median of differences in average breed haplotype sharing between one of the Chernobyl populations and free-breeding dogs from Vinnytsia or Eastern Europe. Plots on the left side of the figure compare average breed haplotype sharing of CNPP dogs to average breed sharing of Vinnytsia or Eastern European free-breeding dogs, while plots on the right side of the figure compare Chernobyl City dogs with Vinnytsia or Eastern European free-breeding dogs. Breeds on the x axis of each plot are ordered according to their placement on a bootstrapped phylogenetic tree found in fig. S3 and grouped by genetic similarity into clades. Horizontal bars are colored according to the purebred clade.
To contextualize the differences in ancestry between the two Chernobyl populations and the free-breeding dogs from Vinnytsia, we examined differences in breed haplotype sharing between Chernobyl dogs and free-breeding dog populations from other countries in Eastern Europe. We found that haplotype sharing with breeds in the Chernobyl populations is less than that observed in free-breeding dogs from elsewhere in Eastern Europe, except for sharing with the Russian hound, a breed with origins in a nearby geographic region (Fig. 6B). Differences in sharing with shepherd-related breeds, including the German shepherd dog, Eastern European shepherd, berger Picard, and Chinook are not significant between Chernobyl populations and Eastern European free-breeding dogs (P adjusted = 1). This suggests that the previously observed shared ancestry between Chernobyl dogs and shepherd-related breeds is potentially a feature of Eastern European free-breeding dog populations in general and not necessarily unique to the Chernobyl populations (Fig. 6B and fig. S2). By comparison, shared ancestry with the Russian hound appears to be unique to free-breeding populations in Ukraine, including dogs from Chernobyl, and is not observed in free-breeding populations elsewhere in Eastern Europe (e.g., Poland, Bulgaria, and Slovenia) (Fig. 6B). The significance of Russian hound ancestry among CNPP dogs, compared to that of Eastern European, central Asian, east Asian, and Middle Eastern dogs (P adjusted = 7.4 × 10−13, 3.2 × 10−24, 3.69 × 10−27, and 6.1 × 10−17, respectively), is similar to the level of significance observed between east Asian free-breeding dogs or Middle Eastern free-breeding dogs with pure breeds that originate in those geographic regions (i.e., purebred Saluki dogs and Middle Eastern free-breeding dogs, P adjusted =1.7 × 10−17; Fig. 6B and fig. S2). Together, our results indicate that the CNPP and Chernobyl City populations exhibit genetic compositions consistent with global free-breeding dog populations that are, in large part, reproductively isolated from purebred populations but which continue to share an ancestral relationship with breeds that historically derive from the same geographic locations.
DISCUSSION
In this study, we characterize the genetic composition of free-breeding dogs living within and around the site of the 1986 Chernobyl nuclear disaster. Previous studies have shown that the two largest nuclear disasters in history, occurring at the CNPP in 1986 and the Fukushima Daiichi Nuclear Power Plant in 2011, both led to massive ecological consequences for wildlife and domestic animals (16–18). However, far more radioactivity was released at Chernobyl than Fukushima, including approximately six times more cesium-137, a long-lived radionuclide with a half-life of over 30 years (19). We present the first genetic analysis of domestic dogs affected by a nuclear disaster, establishing that populations of semi-feral dogs have likely populated the Chernobyl nuclear disaster site in the decades since the accident. Some dogs are living in, and breeding around, highly contaminated areas such as the Chernobyl New Safe Confinement structure, built to contain radioactivity from the damaged reactor, and storage areas for spent nuclear fuel (ISF2). Hence, the dogs of Chernobyl are of immense scientific relevance for understanding the impact of harsh environmental conditions on wildlife and humans alike, particularly the genetic health effects of exposure to long-term, low-dose ionizing radiation and other contaminants (20, 21), i.e., their adaptation to harsh living conditions makes them an ideal system in which to identify mutational signatures resulting from historical and ongoing radiation exposures (22).
As a first step toward a broader study of the genetic effects of long-term radionuclide contamination on free-living dogs, we performed genetic analyses of three populations living at varying distances from the disaster site: Slavutych (45 km), Chernobyl City (15 km), and the CNPP itself, which includes dogs from specific locations within the CNPP including the Semikhody train station, ISF2, and Pripyat (Fig. 1A and fig. S1A). Our study design aimed to capture population differences as they align with variation in radiation exposure based on approximate distance from the disaster site. Previous analyses on small mammals have shown a positive association between average internal dose and proximity to the reactor (23). Although radioactivity measurements vary based on the method and the radionuclide measured, recent estimates show that cesium-137 deposition in different parts of the power plant ranges from 10 to 400 times higher than in Chernobyl City (Fig. 1A) (1).
Before the effects of radiation on the whole genomes of this population can be isolated from other influencing factors, the demography and history of the population itself need to be understood in order that a rigorous framework for experimental design can be constructed, as we do herein. Despite the evidence of clear genetic differentiation between dog populations in Chernobyl City, the CNPP, and Slavutych relative to dog populations elsewhere (Fig. 2A), our genome-wide SNP analysis of kinship and F3 statistical analysis identifies signatures of admixture between these three major populations (Fig. 3B and fig. S1D). We observe extensive relatedness and discrete family structures in which first-degree relationships are evident, particularly within the CNPP (Figs. 1B and 3B). For example, we find the most substantial signatures of admixture between locations that are geographically closest to one another (<5 km apart), including Pripyat, ISF2, the Semikhody train station, and other locations within the CNPP (fig. S1D). This was perhaps expected given the mobility of free-breeding dogs. Unexpectedly, however, our kinship analysis, which was aimed at identifying first-order relationships between dogs, revealed that even within the relatively small area of ISF2 (approximately 0.2 km2), three discrete family groups exist (Fig. 3B).
Consistent with previous studies, our findings highlight the tendency of semi-feral dogs, much like their wild canid ancestors, to form packs of related individuals (24). However, our findings also reveal that within this region, small family groups or packs of free-roaming dogs coexist in close proximity to each other, a phenomenon at odds with the generally territorial nature of domestic dog’s closest ancestor, the gray wolf (25). Free-roaming dogs in urban areas tend to adapt their territoriality and day-to-day movement in response to humans in the region; generally, their home range consists of a small core, where they sleep, and a buffer zone, where they search for food (26–28). The combination of observed behaviors in the Chernobyl dogs and their complex family structures suggests that the Chernobyl dog populations violate the assumption of random mating that is inherent to many population genetic models. When increasing the specificity of sampling location alone, for instance, considering only dogs from the CNPP or Chernobyl City themselves, the observation of complex family structure remains.
To further characterize the suitability of the Chernobyl dog population as a system for studies aimed at identifying genomic loci associated with long-term radiation exposure, we ascertained the degree to which Chernobyl dog populations are genetically isolated and whether their genomic structure follows patterns observed in other free-breeding dog populations. Genomic differences between purebred and free-breeding dogs stem from the fact that established breeds are largely defined by artificial selection, unlike free-breeding and village dog populations. In addition, free-breeding dog populations tend to have varying degrees of regionally specific native ancestry, maintained through isolation from purebred populations (12–14, 29).
Similarity to other free-breeding dog populations, versus purebred dog populations, is indicative of the Chernobyl dogs’ origin in the CEZ region. For example, elevated haplotype sharing with purebred populations might suggest that the original population has been largely replaced by modern pet dogs, leading to intrinsically lower genomic variation from which to distinguish mutations related to radiation exposure. However, this would also make the Chernobyl dogs less than ideal candidates for future genomic studies into cumulative DNA damage and for finding genetic variants associated with population survival and propagation. It is formally possible that some of the early genetic scars present in dogs living in the region immediately after the explosion that have been lost in modern populations are now replaced by large signatures of purebred ancestry. However, we demonstrate that this is unlikely. Our examination of dogs from Ukraine and neighboring countries in Eastern Europe revealed that both the Chernobyl City and CNPP populations have a similar genetic structure to free-breeding dog populations, reflecting a history of admixture, indicating that dogs have existed in the Chernobyl region for a long period of time, potentially since the disaster, or even earlier. Genetic differentiation from other purebred and free-breeding dogs suggests that the Chernobyl populations have a unique genomic signature, supporting their utility in further genomic studies (Figs. 4 to 6).
Even at the individual level, ancestry can vary within free-breeding dog populations, with some dogs reflecting native populations more strongly than others (14, 29). Individually, most dogs from the CNPP and Chernobyl City have minimal levels of breed-characteristic contributions from purebred clades, although when considered as a whole we found differences in ancestral relationships, such that dogs from Chernobyl City likely have ancestral contributions from breeds not detected in the CNPP dogs (Figs. 5, A to C and 6B). We also found recent contributions from the U.K. mastiffs and related breeds as well as the U.K. flock guardian clade, as defined by phylogenetic analysis (fig. S3), in the Chernobyl City population but not in the CNPP (Fig. 4B). Unlike the CNPP, which is an industrial area without permanent inhabitants, Chernobyl City is a historically residential area. Although it was evacuated at the time of the disaster, residents have begun moving back to the area, potentially bringing with them pet dogs responsible for the recent modern breed contributions to Chernobyl City. In addition, safety barriers erected to contain radioactivity and to maintain internal security within the CNPP likely contribute to reproductive isolation and act as a barrier to gene flow, leading to higher inbreeding in the power plant dogs, further accentuating the effect of the disaster on local dog populations.
By investigating the frequencies of breed-derived haplotype segments within the Chernobyl City and CNPP populations, we were able to place a relative timestamp on the introduction of breed dog haplotypes into these populations, demonstrating which breed contributions are likely due to ancestral sharing versus those representing recent introgression (Fig. 5). The duration with which these genomic segments have persisted in the dog populations corresponds to the length of time they were exposed to environmental radiation. For example, sharing with shepherd-related breeds is likely ancestral within both CEZ populations, and these haplotypes have likely existed in this population for the longest period of time. Conversely, sharing with pincher-related breeds is observed mostly among dogs from Chernobyl City, suggesting more recent introgression (Fig. 5C). The relative timing of breed-derived haplotype introduction into the CEZ populations enables a direct comparison of these haplotype segments to the same segments in purebred populations that were not exposed to radiation. Moreover, the relative difference in exposure levels and duration between the shepherd-related haplotypes in CNPP dogs versus the pinscher-related haplotypes in Chernobyl City dogs creates a gradient for measuring radiation scarring of the genome on these haplotype backgrounds, each of which make up >5% of the genome. These results thus define haplotype segments within the genome most likely to vary with the persistent presence of external radiation and heavy metal exposures (Fig. 5C).
Our findings highlight important aspects of the Chernobyl dogs’ history as it relates to their genetic background and population structure. Designing genome-wide association studies to find radiation “survival loci” using conventional methods will be difficult in this population. Extreme family relatedness, ambiguous population history, and elevated levels of inbreeding complicate the assignment of subpopulations to “case” or “control” status. The problem is further confounded by the differing influence of purebred dog contributions. Only by determining the ancestral makeup of each population, acknowledging differences in population history such as the timing of admixture with various breed types, comparing those data to that from surrounding free-breeding dog populations, and ultimately identifying both ancestral and recently emerged haplotypes can studies be appropriately designed, and loci of interest correctly identified.
The idea that the dogs now living in the greater Chernobyl area are descendants of the pets left behind by evacuees after the nuclear disaster remains uncertain (10). Our findings indicate that the CEZ populations share ancestry with shepherd-related breeds, perhaps suggesting that they descend from the same, likely small, founding population of dogs that remained after the disaster and subsequent culling. Evidence of genetic isolation within the CNPP population suggests that this group is most likely to represent the original dog population that inhabited the region before or immediately following the nuclear disaster (Figs. 1 and 2 and 4 to 6). However, any breed that is not present in our purebred dog dataset would not be revealed as a component of current CEZ dogs, and it is likely that at least some of the genetic composition of owned dogs in this region from the 1980s is missing from our purebred dataset. Thus, the extent to which CNPP ancestors were reproductively isolated pets versus owned and free-roaming pets or stray or semi-feral dogs before the nuclear disaster remains unclear.
The relative isolation and discrete genetic makeup of the CNPP population are key factors in the characterization of this population and design of all future mapping studies. There have been fewer genetic contributions from modern dogs into the CNPP population, suggesting that they have lived and reproduced in this environment for a longer period of time than dogs from Chernobyl City or Slavutych. Uniquely, each individual population in the Chernobyl region has experienced differential levels of contamination that are well recorded (1), offering additional advantages in experimental design. Our identification of shared genomic haplotypes and establishment of modern versus ancestral origins present a target for future genetic studies of radiation signatures. The Chernobyl dog population has great potential for informing environmental resource management studies in a resurging population. Its greatest potential, however, lies in understanding the biological underpinnings of animal and, ultimately, human survival in regions of high and continuous environmental assault.
MATERIALS AND METHODS
Sample collection
The radiation heatmap in Fig. 1A was created using ArcMap in ArcGIS (v10.6.1) with a map of cesium-137 deposition levels overlayed (1). Sample collection was organized by the Chernobyl Research Initiative at the University of South Carolina. Blood samples from the Chernobyl dogs were humanely collected alongside a transient spay, neuter, and vaccination clinic sponsored by animal welfare organizations including the International Society for Prevention of Cruelty to Animals and Clean Futures Fund. The annual spay/neuter/vaccination clinics were established as an alternative to culling free-roaming dogs, with the goal of preventing the spread of zoonotic diseases to increasing numbers of tourists in the region as well as workers associated with the construction of the Chernobyl New Safe Confinement facility. When free-roaming dogs in and around the CEZ received veterinary care, blood samples were obtained and preserved for subsequent studies, including those herein. Dogs were captured by veterinarians and qualified volunteers using humane chemical sedation and mechanical techniques, minimizing stress to animals as much as possible. Using both chemical and mechanical capture techniques reduced the effects of sampling bias by capture and permitted sampling of more fearful individuals from a distance. Following anesthetization for surgery, blood samples were collected using either a catheter or capillary tube, collecting blood exposed from the surgery by a licensed veterinarian or veterinary technician. No animals were euthanized for the purpose of this study, and the use of the word “capture” in this manuscript implies temporary capture for veterinary care. All such dogs captured for veterinary care were promptly and safely released back into their environment following surgical recovery. All procedures were conducted with the permission of the CNPP authorities under the supervision of licensed veterinarians and veterinary technicians. Data collected for this paper were gathered opportunistically while animals were being treated by the medical program and, hence, are exempt from Institutional Animal Care and Use Committee (IACUC) approval.
DNA preservation and extraction
Blood samples were collected into one of three types of tubes and processed at either the National Human Genome Research Institute (NHGRI) at the U.S. National Institutes of Health (NIH) IACUC protocol GFS-05-1 or Columbia University (CU). NHGRI blood samples collected in 2017 were preserved in RNAlater (Thermo Fisher Scientific, Rockville, MD), samples from 2018 were preserved in 90% ethanol solution, and 2019 samples were preserved in acid citrate dextrose anticoagulant tubes. After collection, samples were transported at room temperature to NHGRI in Bethesda, MD; anonymized; and entered into the NHGRI samples database. Samples collected in 2018 were extracted using a modified ammonium acetate precipitation protocol (30). DNA was extracted from both the 2017 and 2019 blood samples using a phenol chloroform extraction protocol (31). Each year’s collection produced increasingly higher DNA yields. Samples were aliquoted for long-term storage and stored at −80°C. Samples acquired by CU were preserved in PAXgene Blood DNA tubes (QIAGEN) for both the 2018 and 2019 sampling years. For those samples, DNA was isolated according to the manufacturer’s protocols using the Maxwell RSC Whole Blood DNA Kit and a Maxwell RSC instrument (Promega Corp., Madison, WI).
SNP genotyping
A total of 406 samples were genotyped using Illumina CanineHD 170k SNP arrays at NHGRI, thus maintaining consistency with previously analyzed datasets of purebred dogs (1296 dogs from 157 breeds) (15) and free-breeding dogs (232 from 12 countries) (13). Genotype calls were made with GenomeStudio (v2011.1) using genotyping module v1.9.4 (Illumina). One hundred nineteen samples (61 from the CNPP and 58 from Chernobyl City) were acquired by CU and genotyped by North Carolina State University for 714,000 loci using the Axiom Canine HD array (Thermo Fisher Applied Biosystems, Waltham, MA). Of these, three samples were lacking from the NHGRI dataset. These three were downsampled to match sites from the Illumina CanineHD 170k SNP array. Because of the larger sample numbers, we elected to use the entirety of the Illumina Canine HD dataset and the three downsampled samples from CU.
The purebred dog dataset included 1324 individuals from 162 breeds, six of which originated in either Russia or Ukraine (table S1). Of these, 1296 purebred dogs from 157 breeds had been previously genotyped using the Illumina CanineHD 170k SNP array (table S1) (15). An additional 28 dogs from six breeds originating in either Russia or Ukraine were downsampled from publicly available whole-genome sequence data (www.ncbi.nlm.nih.gov/bioproject/PRJNA648123) and merged with the dataset of purebred and Chernobyl dogs genotyped on the 170k array. In addition, a dataset of 281 free-breeding dogs from 13 countries were included in our analyses of population structure (table S1). Of these, 232 were from a publicly available dataset (13). All were genotyped using the Illumina CanineHD 170k SNP array. Forty-nine free-breeding dog samples from Vinnytsia, Ukraine were genotyped by the Ramaciotti Centre for Genomics, Sydney, Australia on the Axiom Canine HD array (Thermo Fisher Applied Biosystems, Waltham, MA) and downsampled to match sites from the Illumina CanineHD 170k SNP array (table S1).
All 281 free-breeding dog samples were merged with the purebred and Chernobyl dog datasets using PLINK (v1.9) software (32), after which we retained a dataset of 2012 dogs and 163,828 SNPs. Duplicate samples were identified using the --genome flag in PLINK. There was a clear separation of duplicate samples from unique samples whereby IBD proportions greater than 97% were consistent with duplicates, and the next highest IBD proportion was approximately 76%. One hundred one Chernobyl dog samples were found to be duplicates or triplicates of dogs already present in the dataset. This was expected, given that some individuals were sampled across multiple collection years. For individuals sampled multiple times, we maintained the best quality sample. In addition, we excluded four samples from unknown locations and one from a checkpoint outside the study location.
Before removing poor quality samples, the dataset was pruned for variants that were missing >10% of data, removing 34,331 SNPs and yielding a genotyping rate of 99.8%. Quality filtering was used to remove samples missing >10% of data (n = 1). Our final dataset included 132 dogs from the CNPP, 154 from Chernobyl City, 16 from Slavutych, 1324 purebred dogs, and 281 free-breeding dogs, each genotyped at 129,497 informative SNPs. SNP array data are available on Gene Expression Omnibus (GEO) accession ID GSE219090.
Phylogenetic analysis
Pairwise IBS genetic similarity and distance matrices computed using PLINK were used to create a heatmap of genetic similarity using R (v4.0.2) and neighbor-joining (NJ) phylogenetic tree in PHYLIP (v3.696) (33). The heatmap included 132 CNPP, 154 Chernobyl City, and 16 Slavutych dogs (Fig. 1B). The heatmap was created, and the hierarchical clustering was performed using the pheatmap package in R (34). Pairwise IBS genetic similarities were calculated using PLINK with --distance ibs. The phylogenetic tree included only purebred dogs (fig. S3), and IBS genetic distances were calculated using PLINK with --distance 1-ibs to obtain a matrix of pairwise relatedness. The matrix was then used as input to create NJ phylogenetic trees using PHYLIP, which were then plotted with the ggtree package in R (35). Bootstrapping was performed using 100 distance matrices resampled with replacement from the original. The NJ tree, created using only purebred dogs (fig. S3), was used as the x-axis order of breeds for Figs. 4 and 6 and fig. S2. In addition, purebred dogs downsampled from published whole-genome sequence data were included in this tree to confirm accuracy of the dataset merge by checking for the correct placement of breeds on the tree.
Heterozygosity, F3 statistics, and nearest genetic neighbor analysis
Average rates of heterozygosity were calculated using PLINK with --het to obtain observed counts of autosomal homozygosity across all loci for each individual from the CNPP, Chernobyl City, or Slavutych. These were then used to calculate rates of heterozygosity per individual using the formula ((total # of sites − observed count of hom. sites)/total # of sites). Average rates of heterozygosity were calculated for Slavutych, Chernobyl City, Pripyat, ISF2, Semikhody, and other locations within the CNPP. To test for significant differences between average heterozygosity in each population, t tests were performed using R (v1.40.0).
F3 statistics were calculated between dogs from ISF2, Semikhody, Pripyat, other locations in the CNPP, Chernobyl City, and Slavutych using the ADMIXTOOLS package on R (v1.40.0) with the qp3Pop command (36). Middle East village dogs were used as an outgroup for all F3 statistics.
To detect outliers with nonlocal ancestry and investigate genetic relationships in Chernobyl dogs in relation to all other dogs in the dataset, we used the PLINK --neighbor flag, which creates a distribution of relatedness by ranking each individual’s genetic distance to its three most closely related neighbors with respect to all other individuals in the dataset. For this analysis, we plotted the raw IBS distance for each individual’s three nearest neighbors. Outliers in the distribution were determined to be those outside of the first and third quartiles of IBS distances.
Principal components analysis
We performed PCA using the combined dataset described above (132 dogs from the CNPP, 154 from Chernobyl City, 16 from Slavutych, 1324 purebred dogs, and 281 free-breeding dogs) using PLINK with --pca 50. PCA is sensitive to sample size and sample relatedness; however, when the dataset was reduced to exclude first-degree relationships, leaving 78 dogs from Chernobyl City and 63 dogs from the CNPP, trends in population structure and genetic differentiation remain the same (fig. S6). To reduce sample numbers for this PCA, we used PLINK with --king-cutoff 0.17 before running --pca 50.
Kinship analysis
First-degree family relationships were estimated using the KING (v2.2.7) software toolset for family inference (37) using the dataset of 132 CNPP, 154 Chernobyl City, and 16 Slavutych dogs. Relationships were estimated using the --kinship flag in KING to determine pairwise kinship coefficients between each pair of dogs. A zero IBS cutoff value of 0.0004 was used to distinguish parent-offspring relationships from sibling relationships. Parent-offspring relationships had kinship values between 0.1960 and 0.3398 (Fig. 3A). Sex of each dog was determined using the PLINK --check-sex function and cross-referenced with clinic notes made by veterinarians at the time of spay/neuter when possible. The role of sample size in finding parent-offspring pairs was determined using a resampling approach. Here, at each subsample size, 100 iterations of sampling without replacement of the Chernobyl dog population were performed, where, for each iteration, the total number of parent-offspring pairs was counted.
IBD haplotype sharing analyses
IBD genomic segments were identified with BEAGLE (v4.1) (38) using the following parameters: ibd = true window = 1000 overlap = 100 ibd = true ibdcm = 0.25 ibdtrim = 10. We determined the total length of shared haplotypes in IBD segments between each free-breeding and each purebred dog and then took the average of each individual’s sharing per breed (Fig. 4A). These values were then compared to determine statistical differences in sharing between free-breeding dog populations using a Wilcoxon signed-rank test, assigning P values to pairwise comparisons of shared haplotypes between each of the free-breeding populations and each breed (Figs. 4B and 6B).
Assignment of breed-derived genomic regions
Genomic segments were assigned as being breed-derived based on IBD haplotype sharing results produced by BEAGLE (v4.1) (38) using the following parameters: ibd = true window = 1000 overlap = 100 ibd = true ibdcm = 0.25 ibdtrim = 10. Breed assignments for genomic segments were only performed using haplotype sharing between Chernobyl dogs and purebred dogs. The only breeds considered in this analysis were those that had at least 10 members. IBD segments were initially assigned breed labels according to the following criteria: (i) Chernobyl dog genomic region displaying IBD sharing with >25% of chromosomes within a single breed and (ii) the genomic region displaying IBD sharing with <0.1% of chromosomes from dogs outside of that breed’s clade using clades of related breeds defined using a bootstrapped phylogenetic tree composed of only purebred dogs (fig. S3). Next, IBD breed segments from different breeds were combined according to their clade membership. For example, overlapping segments from two different breeds of the same clade were considered as a single breed-derived IBD segment, labeled according to the clade of both breeds. This approach ensured that no single haplotype would be assigned more than one clade label. Assignment of shared ancestry is based on haplotypes that are characteristic of a specific breed/clade, excluding haplotypes that are either common to all breeds, or shared among distantly related breeds. Furthermore, genomic ancestry assignments were examined by clade rather than individual breed, as extensive sharing of haplotypes between breeds within the same clade often makes it difficult to determine the precise breed origin of a given haplotype (15). Our stringent approach to breed haplotype assignment required that haplotypes be at a high frequency within a breed (>25% of chromosomes) and extremely rare outside the clade (<0.1% of chromosomes) to be labeled. Therefore, much of the genome cannot be assigned a breed/clade, especially if that haplotype is observed across other clades. All genomic interval operations were performed using the Genomic Ranges package (v1.40.0) in R (39).
Frequency of breed-derived alleles and population comparisons
For all subsequent analyses comparing Chernobyl City and CNPP populations, samples were filtered to remove individuals whose genetic profile was characteristic of the other population. Genetic profiles of samples were captured by PCA where the samples from each population mostly clustered according to their PC1 values (fig. S7). A total of 131 Chernobyl City samples with PC1 values > 0 were kept, as were 121 CNPP samples with PC1 values < 0. Allele frequencies of clade assignments were calculated by counting the number of clade segments that overlapped each marker. Haplotype sharing of clade IBD segments across populations was calculated by measuring the overlap between clade assigned markers in each population for the following allele frequency ranges: 0 to 1%, 1 to 5%, 5 to 10%, and 10 to 100%. Significance was determined by performing a Fisher’s exact test and corrected for multiple testing by calculating the false discovery rate using the P adjusted function in R. The test considered the number of markers that were assigned the same clade label in both populations, assigned a clade label in only one of the populations, and not assigned a clade label in either population. The size of the overlap was reported as the Jaccard index, which is calculated as the intersect of population clade labels divided by the union of population clade labels.
Acknowledgments
We thank L. Hixson and E. Kambarian from the Clean Futures Fund for organizing the canine wellness campaigns in Chernobyl, Ukraine. We also thank J. Chertok, K. Russell, N. Hank, C. Rulison, J. Hecla, and all volunteers who participated in the Dogs of Chernobyl animal wellness campaigns for assistance with field work, sample collection, and helpful suggestions. Veterinary clinics in Chernobyl were supported by Clean Futures Fund International+ and the Society for the Prevention of Cruelty to Animals International. We thank A. Lischyshyna and all the members of the Four Paws Ukraine team for the collection of dog samples from Vinnytsia, Ukraine.
Funding: This work is supported by the Intramural Program of the NHGRI (to G.J.S., R.M.B., E.V.D., H.G.P., and E.A.O.), University of South Carolina Office of Research (to G.J.S. and T.A.M.), University of South Carolina Honors College (to T.A.M.), Samuel Freeman Charitable Trust (to G.J.S. and T.A.M.), 2022 Flagship Project Funding NIH Intramural Sequencing Center (to E.A.O.), Clean Futures Fund International+ (to J.A.B.), NC State Genetic and Genomics Academy Graduate Fellowship (to M.D.), NC State Cancer Genomics Fund (to M.B.), and Polish National Agency for Academic Exchange (NAWA, Polish Returns Fellowship PPN/PPO/2018/1/00037) (to M.P.).
Author contributions: Conceptualization: T.A.M. Methodology: G.J.S., R.M.B., E.V.D., H.G.P., and E.A.O. Sample acquisition: G.J.S., J.A.B., M.P., W.B., I.C., .G.M, N.K., and T.A.M. Sample processing: G.J.S., H.G.P., R.T., and N.K. Investigation: G.J.S., R.M.B., M.D., M.B., E.A.O., and T.A.M. Visualization: G.J.S. and R.M.B. Supervision: M.B., E.A.O., T.A.M. Writing—original draft: G.J.S., R.M.B., and E.A.O. Writing—review and editing: M.D., E.V.D., M.B., E.A.O., and T.A.M.
Competing interests: The authors declare that they have no competing interest.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The SNP array data have been deposited in the database GEO (accession ID GSE219090). The dataset used in this study is also available at https://research.nhgri.nih.gov/dog_genome/data_release/index.shtml.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Legend for table S1
Other Supplementary Material for this : manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.N. Evangeliou, T. Hamburger, N. Talerko, S. Zibtsev, Y. Bondar, A. Stohl, Y. Balkanski, T. A. Mousseau, A. P. Moller, Reconstructing the Chernobyl Nuclear Power Plant (CNPP) accident 30 years after. A unique database of air concentration and deposition measurements over Europe. Environ. Pollut. 216, 408–418 (2016). [DOI] [PubMed] [Google Scholar]
- 2.International Atomic Energy Agency (IAEA), Environmental consequences of the Chornobyl accident and their remediation: Twenty years of experience (Radiological Assessment Reports Series, IAEA, 2006).
- 3.M. De Cort, G. Dubois, S. Fridman, M. Germenchuk, Y. Izrael, A. Janssens, A. Jones, G. Kelly, E. Kvasnikova, I. Matveenko, I. Nazarov, Y. Pokumeiko, V. Sitak, E. Stukin, L. Tabachny, Y. Tsaturov, S. Avdyushin, Atlas of Caesium Deposition on Europe after the Chernobyl Accident (Office for Official Publications of the European Communities, 2009).
- 4.R. Whitman, “Cesium-137 fallout in Indiana soil,” thesis, Ball State University (2017). [Google Scholar]
- 5.K. Beaugelin-Seiller, J. Garnier-Laplace, C. Della-Vedova, J. M. Metivier, H. Lepage, T. A. Mousseau, A. P. Moller, Dose reconstruction supports the interpretation of decreased abundance of mammals in the Chernobyl Exclusion Zone. Sci. Rep. 10, 14083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.T. G. Deryabina, S. V. Kuchmel, L. L. Nagorskaya, T. G. Hinton, J. C. Beasley, A. Lerebours, J. T. Smith, Long-term census data reveal abundant wildlife populations at Chernobyl. Curr. Biol. 25, R824–R826 (2015). [DOI] [PubMed] [Google Scholar]
- 7.N. M. van Straalen, M. J. T. N. Timmermans, Genetic variation in toxicant-stressed populations: An evaluation of the “Genetic Erosion” hypothesis. Hum. Ecol. Risk Assess. Int. J. 8, 983–1002 (2002). [Google Scholar]
- 8.A. P. Moller, T. A. Mousseau, Strong effects of ionizing radiation from Chernobyl on mutation rates. Sci. Rep. 5, 8363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. P. Moller, T. A. Mousseau, The effects of natural variation in background radioactivity on humans, animals and other organisms. Biol. Rev. Camb. Philos. Soc. 88, 226–254 (2013). [DOI] [PubMed] [Google Scholar]
- 10.A. Higginbotham, Midnight in Chernobyl (Simon & Schuster Inc., 2019).
- 11.B. M. Vonholdt, J. P. Pollinger, K. E. Lohmueller, E. Han, H. G. Parker, P. Quignon, J. D. Degenhardt, A. R. Boyko, D. A. Earl, A. Auton, A. Reynolds, K. Bryc, A. Brisbin, J. C. Knowles, D. S. Mosher, T. C. Spady, A. Elkahloun, E. Geffen, M. Pilot, W. Jedrzejewski, C. Greco, E. Randi, D. Bannasch, A. Wilton, J. Shearman, M. Musiani, M. Cargill, P. G. Jones, Z. Qian, W. Huang, Z. L. Ding, Y. P. Zhang, C. D. Bustamante, E. A. Ostrander, J. Novembre, R. K. Wayne, Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.L. M. Shannon, R. H. Boyko, M. Castelhano, E. Corey, J. J. Hayward, C. McLean, M. E. White, M. Abi Said, B. A. Anita, N. I. Bondjengo, J. Calero, A. Galov, M. Hedimbi, B. Imam, R. Khalap, D. Lally, A. Masta, K. C. Oliveira, L. Perez, J. Randall, N. M. Tam, F. J. Trujillo-Cornejo, C. Valeriano, N. B. Sutter, R. J. Todhunter, C. D. Bustamante, A. R. Boyko, Genetic structure in village dogs reveals a Central Asian domestication origin. Proc. Natl. Acad. Sci. U.S.A. 112, 13639–13644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. Pilot, T. Malewski, A. E. Moura, T. Grzybowski, K. Olenski, A. Rusc, S. Kaminski, F. Ruiz Fadel, D. S. Mills, A. N. Alagaili, O. B. Mohammed, G. Klys, I. M. Okhlopkov, E. Suchecka, W. Bogdanowicz, On the origin of mongrels: Evolutionary history of free-breeding dogs in Eurasia. Proc. Biol. Sci. 282, 20152189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. R. Boyko, R. H. Boyko, C. M. Boyko, H. G. Parker, M. Castelhano, L. Corey, J. D. Degenhardt, A. Auton, M. Hedimbi, R. Kityo, E. A. Ostrander, J. Schoenebeck, R. J. Todhunter, P. Jones, C. D. Bustamante, Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc. Natl. Acad. Sci. U.S.A. 106, 13903–13908 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.H. G. Parker, D. L. Dreger, M. Rimbault, B. W. Davis, A. B. Mullen, G. Carpintero-Ramirez, E. A. Ostrander, Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 19, 697–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. A. Mousseau, The biology of Chernobyl. Annu. Rev. Ecol. Evol. Syst. 52, 87–109 (2021). [Google Scholar]
- 17.T. A. Mousseau, A. P. Moller, Genetic and ecological studies of animals in Chernobyl and Fukushima. J. Hered. 105, 704–709 (2014). [DOI] [PubMed] [Google Scholar]
- 18.M. Nagasawa, K. Mogi, T. Kikusui, Continued distress among abandoned dogs in Fukushima. Sci. Rep. 2, 724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.T. Imanaka, in Low-Dose Radiation Effects on Animals and Ecosystems: Long-Term Study on the Fukushima Nuclear Accident, M. Fukumoto, Ed. (Springer, 2020), pp. 249–259. [Google Scholar]
- 20.P. Lehmann, Z. Boratynski, T. Mappes, T. A. Mousseau, A. P. Moller, Fitness costs of increased cataract frequency and cumulative radiation dose in natural mammalian populations from Chernobyl. Sci. Rep. 6, 19974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.I. Galván, A. Bonisoli-Alquati, S. Jenkinson, G. Ghanem, K. Wakamatsu, T. A. Mousseau, A. P. Møller, Chronic exposure to low-dose radiation at Chernobyl favours adaptation to oxidative stress in birds. Funct. Ecol. 28, 1387–1403 (2014). [Google Scholar]
- 22.L. M. Morton, D. M. Karyadi, C. Stewart, T. I. Bogdanova, E. T. Dawson, M. K. Steinberg, J. Dai, S. W. Hartley, S. J. Schonfeld, J. N. Sampson, Y. E. Maruvka, V. Kapoor, D. A. Ramsden, J. Carvajal-Garcia, C. M. Perou, J. S. Parker, M. Krznaric, M. Yeager, J. F. Boland, A. Hutchinson, B. D. Hicks, C. L. Dagnall, J. M. Gastier-Foster, J. Bowen, O. Lee, M. J. Machiela, E. K. Cahoon, A. V. Brenner, K. Mabuchi, V. Drozdovitch, S. Masiuk, M. Chepurny, L. Y. Zurnadzhy, M. Hatch, A. Berrington de Gonzalez, G. A. Thomas, M. D. Tronko, G. Getz, S. J. Chanock, Radiation-related genomic profile of papillary thyroid carcinoma after the Chernobyl accident. Science 372, eabg2538 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R. K. Chesser, D. W. Sugg, M. D. Lomakin, R. A. van den Bussche, J. A. DeWoody, C. H. Jagoe, C. E. Dallas, F. W. Whicker, M. H. Smith, S. P. Gaschak, I. V. Chizhevsky, V. V. Lyabik, E. G. Buntova, K. Holloman, R. J. Baker, Concentrations and dose rate estimates of 134137cesium and 90strontium in small mammals at chornobyl, Ukraine. Environ. Toxicol. Chem. 19, 305–312 (2000). [Google Scholar]
- 24.E. Natoli, R. Bonanni, S. Cafazzo, D. S. Mills, D. Pontier, M. Pilot, Genetic inference of the mating system of free-ranging domestic dogs. Behav. Ecol. 32, 646–656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L. D. Mech, L. Boitani, in Wolves: Behavior, Ecology, and Conservation, L. D. Mech, L. Boitani, Ed. (Univ. of Chicago Press, 2003). [Google Scholar]
- 26.M. A. Lewis, J. D. Murray, Modelling territoriality and wolf–deer interactions. Nature 366, 738–740 (1993). [Google Scholar]
- 27.C. Warembourg, E. Wera, T. Odoch, P. M. Bulu, M. Berger-Gonzalez, D. Alvarez, M. F. Abakar, F. Maximiano Sousa, L. Cunha Silva, G. Alobo, V. D. Bal, A. L. Lopez Hernandez, E. Madaye, M. S. Meo, A. Naminou, P. Roquel, S. Hartnack, S. Durr, Comparative study of free-Roaming domestic dog management and roaming behavior across four countries: Chad, Guatemala, Indonesia, and Uganda. Front. Vet. Sci. 8, 617900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. V. Ivanter, N. A. Sedova, Ecological monitoring of urban groups of stray dogs: An example of the city of petrozavodsk. Russ. J. Ecol. 39, 105–110 (2008). [Google Scholar]
- 29.S. K. Brown, N. C. Pedersen, S. Jafarishorijeh, D. L. Bannasch, K. D. Ahrens, J. T. Wu, M. Okon, B. N. Sacks, Phylogenetic distinctiveness of Middle Eastern and Southeast Asian village dog Y chromosomes illuminates dog origins. PLOS ONE 6, e28496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D. S. Richardson, F. L. Jury, K. Blaakmeer, J. Komdeur, T. Burke, Parentage assignment and extra-group paternity in a cooperative breeder: The Seychelles warbler (Acrocephalus sechellensis). Mol. Ecol. 10, 2263–2273 (2001). [DOI] [PubMed] [Google Scholar]
- 31.G. I. Bell, J. H. Karam, W. J. Rutter, Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc. Natl. Acad. Sci. U.S.A. 78, 5759–5763 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. C. Chang, C. C. Chow, L. C. Tellier, S. Vattikuti, S. M. Purcell, J. J. Lee, Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Felsenstein, PHYLIP (Phylogeny Inference Package) version 3.6 (Department of Genome Sciences, University of Washington, 2005);https://evolution.genetics.washington.edu/phylip.html.
- 34.R. Kolde, pheatmap: Pretty Heatmaps (2019);https://rdrr.io/cran/pheatmap/.
- 35.G. Yu, Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinformatics 69, e96 (2020). [DOI] [PubMed] [Google Scholar]
- 36.N. Patterson, P. Moorjani, Y. Luo, S. Mallick, N. Rohland, Y. Zhan, T. Genschoreck, T. Webster, D. Reich, Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. Manichaikul, J. C. Mychaleckyj, S. S. Rich, K. Daly, M. Sale, W. M. Chen, Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.B. L. Browning, S. R. Browning, Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Lawrence, W. Huber, H. Pages, P. Aboyoun, M. Carlson, R. Gentleman, M. T. Morgan, V. J. Carey, Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Legend for table S1
Table S1