Abstract
Alternative precursor messenger RNA splicing is instrumental in expanding the proteome of higher eukaryotes, and changes in 3′ splice site (3'ss) usage contribute to human disease. We demonstrate by small interfering RNA–mediated knockdowns, followed by RNA sequencing, that many proteins first recruited to human C* spliceosomes, which catalyze step 2 of splicing, regulate alternative splicing, including the selection of alternatively spliced NAGNAG 3′ss. Cryo–electron microscopy and protein cross-linking reveal the molecular architecture of these proteins in C* spliceosomes, providing mechanistic and structural insights into how they influence 3'ss usage. They further elucidate the path of the 3′ region of the intron, allowing a structure-based model for how the C* spliceosome potentially scans for the proximal 3′ss. By combining biochemical and structural approaches with genome-wide functional analyses, our studies reveal widespread regulation of alternative 3′ss usage after step 1 of splicing and the likely mechanisms whereby C* proteins influence NAGNAG 3′ss choices.
Cryo-EM and global functional analyses reveal how human C* spliceosomal proteins regulate alternative 3' splice site choices.
INTRODUCTION
The structure and composition of the spliceosome changes continuously throughout the precursor messenger RNA (pre-mRNA) splicing process, leading to the formation of distinct spliceosomal complexes, including the C, C*, and P complexes that are formed during splicing catalysis (1–3). Pre-mRNA splicing is catalyzed by the U2 and U6 small nuclear RNAs (snRNAs) that fold into a catalytically active three-dimensional (3D) structure that coordinates the two catalytic Mg2+ ions required for splicing catalysis (4). During the first catalytic step of splicing (denoted branching), the 2′-OH group of the branch site adenosine (BS-A) carries out a nucleophilic attack at the 5′ splice site (ss). This generates the spliceosomal C complex, which contains the intermediates of the splicing reaction, namely, the cleaved 5′ exon and the intron lariat-3′ exon, in which the 5′ end of the intron is ligated to the 2′-OH of the BS-A forming a branched intron. During step 2, which is catalyzed by the C* complex, the 3′-OH group of the 5′ exon attacks the 3′ss, leading to intron excision and ligation of the 5′ and 3′ exons.
The transition from a spliceosomal C complex into an activated C* complex is catalyzed by the RNA helicase PRP16, which facilitates ribonucleoprotein (RNP) rearrangements that lead to the movement of the branched intron structure and the branched U2/BS helix (henceforth denoted the branched helix) away from the catalytic center (5–8). This allows the 3′ exon to dock near the cleaved 5′ exon and for the recruitment of step 2 factors such as SLU7, PRP18, and the DEAH-box helicase PRP22. Catalysis of step 2 of pre-mRNA splicing converts the C* complex into the spliceosomal P (postcatalytic) complex, which contains the spliced mRNA and the excised intron-lariat. PRP22 facilitates step 2 in an adenosine triphosphate (ATP)–independent manner, at least in yeast (9). Its ATP-dependent helicase activity is subsequently required for proofreading exon ligation and for the subsequent release of the spliced mRNA from the P complex (10–12).
Cryo–electron microscopy (cryo-EM) of human (h) C* and P complexes (7, 13, 14) revealed that, as in yeast, the 3′ss AG dinucleotide is docked by noncanonical base pairing interactions between the guanine of the 3′ss and the 5′ss guanine (G+1) at the 5′ end of the intron, and between the adenine of the 3′ss and the BS-A. Thus, stabilization of the nucleotides comprising the branched intron structure is important for stable docking of the 3′ss AG. The guanine base of the 3′ss also stacks against A45 of the U6 snRNA, which forms two hydrogen bonds with the second intron nucleotide U+2 (7, 13, 14). Docking of the 3′ss AG to nucleotides of the branched intron structure creates an RNA loop between the BS-A and the 3′ss. In higher eukaryotes, this loop is composed mainly of the polypyrimidine tract (PPT). The distance between the BS-A and 3′ss is typically short [i.e., ca 15 to 35 nucleotides (nt)], but in some instances, it is more than 100 nt (15). However, except for the position of a few nucleotides downstream of the BS-A, the path of this proposed RNA loop, which likely is important for stable 3′ss docking, could not be discerned in previous cryo-EM studies.
Given the limited RNA-RNA interactions involving the 3′ss, stable docking of the latter for catalytic step 2 must be aided by spliceosomal proteins. In both yeast and humans, the interaction of the 3′ss with the branched intron structure and U6 before step 2 is stabilized in part by the PRP8 α-finger and the β-hairpin of the PRP8 ribonuclease H-like domain (PRP8RH) (7, 13, 14, 16, 17). The latter, together with PRP17, SLU7, CDC5L, and the metazoan-specific proteins PRKRIP1 and CACTIN that are absent in Saccharomyces cerevisiae, tethers the branched helix to its new position in C* (5–7). The spatial organization of regions of the metazoan-specific proteins FAM32A, PRKRIP1, CACTIN, SDE2, and NKAP in hC* and/or hP suggests that they help to stabilize the RNP conformation of the spliceosome that promotes exon ligation (7, 13). Numerous additional proteins that are absent in S. cerevisiae spliceosomes are recruited to human C/C* complexes (18). However, for many of these proteins, it is not clear whether they are recruited first during C or C* complex formation, and little or nothing is known about their spatial organization and function in the spliceosome, including whether they may also help to facilitate the docking of the 3′ss in a manner conducive for step 2 of splicing.
The majority of pre-mRNAs are alternatively spliced in higher eukaryotes, generating multiple distinct mRNAs and proteins from a single pre-mRNA species (19). A common type of alternative splicing in mammals is the use of an alternative 5′ or 3′ss, which affects the length of the exon by altering its 5′ or 3′ end (20). There is a preferred directionality in alternative 3′ss selection, with proximal sites (relative to the BS-A) preferentially chosen over more distal ones in many, but not all, cases (21–23). This led to the proposal that the 3′ss is selected by linear scanning for the first AG downstream of the BS/PPT (21–24). However, the mechanism of this potential scanning process and the factors that are involved have remained enigmatic. As a strict linear search is not compatible with several experimental observations, additional or alternative modes of 3′ss selection, including direct competition between closely spaced 3′ss, must also exist (22, 23).
Competing 3′ss can be located far apart or directly adjacent to one another, as is the case for NAGNAG (N, any nucleotide, A, adenosine, G, guanosine) alternative splicing (25). Widespread tissue-specific alternative NAGNAG splicing has been documented, confirming that this form of alternative splicing is biologically important (26, 27). Regulated NAGNAG 3′ss are found in thousands of human genes and represent the second most common form of alternative splicing in which the reading frame is preserved (27). However, the mechanisms regulating whether the distal or proximal AG is used and at what stage of splicing this regulation occurs have remained elusive since the discovery of widespread NAGNAG splicing (28). Because of their extreme proximity, classical mechanisms governing 3′ss choice, such as the binding of regulatory factors closer to one site or the other, cannot apply to NAGNAG alternative splicing. The nature of the intron sequences upstream of a NAGNAG site appears to modulate usage of the proximal or distal site (27), but as NAGNAG sites are spliced in a tissue-specific manner, trans-acting regulatory factors must also be involved (27, 28). Spliceosomal proteins involved in the docking of the 3′ss, including those recruited after the first step of the splicing reaction has occurred, are potential candidates for regulating the choice of competing, adjacent NAGNAG 3′ss, by fine-tuning the RNP structure of the spliceosome in the vicinity of the 3′ss directly before step 2.
Here, we unambiguously identify numerous metazoan-specific proteins that are first recruited to human spliceosomes at the C* complex stage. We subsequently perform comprehensive structural and global functional analyses of human C* proteins. Small interfering RNA (siRNA)–mediated knockdowns of 13 of the C* proteins, followed by RNA sequencing (RNA-seq), demonstrated that many of them play a role in alternative splicing, particularly in the selection of alternatively spliced NAGNAG 3′ss. Cryo-EM coupled with protein cross-linking allowed us to map the previously unknown location of several of the latter proteins in hC*. The 3D structural organization of C* proteins that modulate 3′ss selection reveals how, by stabilizing the RNP conformation of the C* complex, they likely promote docking of the 3′ss into the C* active site and further provides mechanistic insights into how they control NAGNAG 3′ss selection. We could also map the path of the PPT loop between the BS-A and the docked 3′ss and elucidate how it is stabilized in a protein pocket formed in part by C* proteins. This allowed us to generate a model for how the PPT loop is involved in a potential scanning mechanism that leads to the preferential selection of the proximal 3′ss. By combining extensive RNA-seq analyses of multiple C* protein knockdowns with cryo-EM, our data provide new insights into the roles of metazoan-specific spliceosomal proteins in alternative 3′ss selection and reveals widespread regulation of alternative splicing after step 1, at the C* complex stage.
RESULTS
Identification of proteins enriched in the hC* complex
Identifying the precise stage when proteins are recruited to the spliceosome is important for defining their potential functions and also facilitates the identification and placement of spliceosomal proteins located in less well-resolved regions of cryo-EM spliceosome structures. To identify proteins first recruited during C* complex formation, we affinity-purified human spliceosomal complexes, formed on either PM5 or MINX/MINXGG pre-mRNA (fig. S1A), that were stalled either directly before PRP16 action (i.e., at the C stage) or directly before the second catalytic step of splicing (i.e., at the C* stage). Both PM5 and MINXGG C* complexes contained predominantly pre-mRNA splicing intermediates and U2, U5, and U6 snRNA (fig. S1B). Mass spectrometry (table S1) and immunoblotting (fig. S1C) revealed that a large number of proteins not found in S. cerevisiae spliceosomes are predominantly recruited during the C-to-C* complex transition—henceforth denoted C* proteins—although several are still present in the hP complex (13) (see also below). These include, among others, FAM32A, SDE2, CACTIN, PRKRIP1, NKAP, TLS1, CXORF56, FAM50A, PPIL3, PPIG, ESS2, and NOSIP, the RNA helicases DDX41 and DHX35, and GPATCH1, a potential DHX35 activating protein (fig. S1D). As these proteins are recruited directly before step 2, they likely function at the C* or a later stage of splicing and potentially regulate alternative 3′ss selection after catalytic step 1. Consistent with their designation as C* complexes, C-specific proteins, as well as the pre-C* protein FAM192A (7), are underrepresented or absent in PM5 and MINXGG complexes assembled in the presence of wild-type (WT) PRP16 (fig. S1C and table S1).
Post–step 1 regulation of alternative 3′ss selection by C* proteins
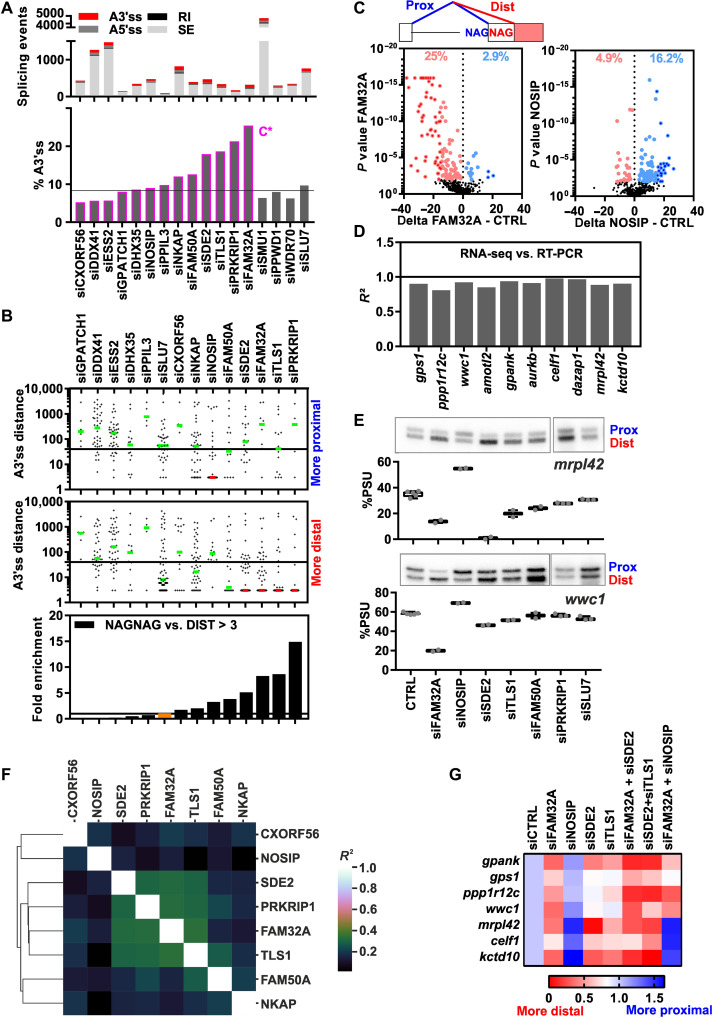
To investigate the function of C* proteins in splicing, we analyzed the effects of siRNA-mediated knockdown of 13 C* proteins in HeLa cells by RNA-seq (table S2 and data files S1 and S2). Efficient knockdown of each protein was confirmed by RNA-seq and by immunoblotting (fig. S2, A and B). All C* protein knockdowns led to changes in global alternative splicing patterns but to various degrees (Fig. 1A, fig. S3A, and data files S3 to S6). Knockdown of most C* proteins led to a clear increase in the percent of altered splicing events that involved alternative 3′ splice site (A3′ss) selection, compared to control knockdowns of proteins specific for B or C spliceosomal complexes, with the strongest increase observed upon knockdown of NKAP, FAM50A, SDE2, TLS1, PRKRIP1, and FAM32A (Fig. 1A and fig. S3A). Searches for common features of the effected A3′ss revealed a broad distribution of upstream intron or downstream exon length (fig. S3B). Notably, knockdown of eight of the analyzed C* proteins had significant effects on the selection of A3′ss separated by exactly three nucleotides (i.e., NAGNAG 3′ss). Alternative NAGNAG splicing comprised ~25 to 75% of the A3′ss affected by knockdown of CXORF56, NOSIP, FAM50A, NKAP, SDE2, PRKRIP1, TLS1, or FAM32A (Fig. 1B and fig. S3C). RNA-seq data showed that knockdown of individual C* proteins significantly altered 3′ss usage for ~15 to 30% of all quantified NAGNAG sites (Fig. 1C, fig. S3D, and data S7). These RNA-seq results were validated by reverse transcription polymerase chain reaction (RT-PCR), both qualitatively (100% validation rate) and quantitatively [R2 for the percent proximal site usage (%PSU) of RNA-seq versus RT-PCR close to 1], for 10 selected NAGNAG introns (Fig. 1, D and E, and fig. S3E). Notably, knockdown of FAM32A, SDE2, PRKRIP1, TLS1, NKAP, and FAM50A promoted usage of the distal (downstream) A3′ss in most cases, whereas NOSIP knockdown led to the preferred usage of the proximal A3′ss (Fig. 1, B and C, and fig. S3,C and D; see also mrpl42 in Fig. 1E, top). While NOSIP knockdown most strongly increased the %PSU of NAGNAG introns where the distal AG is preferentially used (%PSU = 0 to 50), the remaining C* proteins most strongly reduced the PSU of NAGNAG introns with a basal %PSU between 50 and 90 (i.e., where the proximal site is most often selected) (fig. S4, A and B). A pairwise correlation matrix revealed overlapping targets especially for SDE2, PRKRIP1, TLS1, and FAM32A (Fig. 1F), which was also validated for selected NAGNAG introns by RT-PCR (i.e., gpank1 in fig. S3E). A Venn diagram showing the overlap of FAM32A, SDE2, TLS1, and PRKRIP1 target genes revealed that more than 50% of the genes targeted by one of these factors are also regulated by at least a second factor (fig. S4C and table S3). Coregulated genes showed a significant Gene Ontology (GO) term enrichment for positive regulation of double-strand break repair [fold enrichment: 24.09; false discovery rate (FDR): 3.17 × 10−2], pointing toward a potential biological function of C* factor–regulated NAGNAG splicing. We also found targets that are predominately affected by the knockdown of a single C* protein (e.g., wwc1; Fig. 1E, bottom), indicating some degree of target specificity.
Fig. 1. C* complex proteins regulate NAGNAG 3'ss selection.
(A) Effects of C* protein knockdown on splicing as determined by RNA-seq. Top: rMATS-derived altered splicing changes for skipped exons (SE), retained introns (RI), A5′ss, and A3′ss for the indicated knockdowns. Bottom: %A3′ss of all alternative splicing events affected by each knockdown. Black line, fraction of A3′ss in all targets quantified by rMATS. (B) Scatter dot blot of distances between A3′ss. For each C* protein (indicated above), targets are separated into knockdown-induced proximal 3′ss usage (top) or distal 3′ss usage (middle). Green lines, median distance between A3′ss. Red line, median of three (>50% with a distance of exactly three). Black line, median 3′ss distance among all A3′ss quantified by rMATS. Bottom: Normalized ratio between alternatively spliced NAGNAG 3′ss and A3′ss with a distance larger than 3 nt (DIST > 3). The fraction of NAGNAGs among all A3′ss quantified by rMATS is set to 1 (black line). SLU7 is shown for comparison. (C) Volcano blots summarizing the global effect of FAM32A (left) or NOSIP (right) knockdown on NAGNAG 3′ss selection. See fig. S3D for further details. (D) Correlation coefficients for 10 tested NAGNAG splicing events, comparing RNA-seq and RT-PCR–derived %PSU values. (E) Validation RT-PCRs for NAGNAG alternative splicing of mrpl42 (top) and wwc1 (bottom) upon C* protein knockdown (indicated below). See fig. S3E for further details. (F) Overlap of alternatively spliced NAGNAG sites affected by individual C* protein knockdowns. For each of the eight knockdowns, rMATS-derived P values were correlated with all knockdowns in a pairwise manner. Right: Correlation scores, where darker colors indicate a lower percent of overlapping NAGNAG targets. (G) Cross-talk of C* complex factors during NAGNAG 3′ss selection. Mean PCR-derived %PSU values are shown for seven targets relative to the siCTRL upon single and double C* protein knockdown.
To investigate whether C* proteins cooperate during NAGNAG 3'ss selection, we performed double knockdowns (Fig. 1G and fig. S4, D to F). Additive effects (i.e., enhanced usage of the distal 3′ss) were observed with FAM32A/SDE2 and SDE2/TLS1 knockdowns for several NAGNAG introns such as gpank1 and wwc1 (Fig. 1G and fig. S4F). Knockdown of NOSIP antagonized the effects of the FAM32A knockdown, indicating that NOSIP acts in a dominant manner in promoting usage of the distal site (i.e., mrpl42 and celf1; Fig. 1G and fig. S4F). Together, our data indicate that several C* proteins regulate A3′ss choices and, for many NAGNAG alternative splicing events, play an important role in ensuring the preferred usage of the proximal 3′ss. They additionally reveal widespread regulation of alternative splicing that occurs after the first transesterification reaction has already occurred.
Overall structure of the PM5 C* complex
To elucidate the spatial organization of C* proteins and the potential mechanisms whereby they modulate 3′ss selection, we performed single-particle cryo-EM and determined the structure of the human PM5 C* complex (fig. S5, A to E, and table S4). The major part of the PM5 C* complex (colored region in fig. S5C) could be resolved at a resolution of 2.7 to 4 Å and at 3 Å or better in regions of its catalytic RNP core. By fitting published structures of spliceosome components and/or structural domains newly predicted by AlphaFold (29, 30) into the EM density map (table S5), together with protein cross-linking coupled with mass spectrometry (CXMS) (data S8), we generated a 3D model of the hC* complex (Fig. 2). Using this combinatorial structural approach, we could map regions/domains of several C* proteins including DDX41, FAM50A, CXORF56, TLS1, PPIL3, ESS2, and NOSIP, as well as additional regions of NKAP and SDE2, thus providing numerous new insights into the 3D structure of the hC* complex.
Fig. 2. 3D cryo-EM model of the hC* complex.
Two different views of the molecular architecture of hC* complexes formed on PM5 pre-mRNA. Bottom: Summary of all modeled proteins and RNAs with color code. Black dot, proteins localized in PM5 hC* that were depleted by siRNA-mediated knockdown experiments.
Formation of a tandem helicase module by DDX41 and PRP22
The conserved RNA helicase PRP22 is docked via its C-terminal (CT) domain to PRP8 and SKIP in PM5 C* as in previously reported hC* and P complexes (Fig. 2 and fig. S6, A and B) (5–7, 13, 14). However, we could also map additional regions of PRP22 that were either incorrectly localized (e.g., amino acids 511 to 567) or not localized at all in previous human cryo-EM structures (fig. S6, A to C). The latter includes the CT tail of PRP22, which follows a very similar path as in the yeast P complex (31), reaching out to the α-finger of PRP8 in the catalytic core (see below) and thereby establishing a direct connection between the PRP22 helicase domain and the catalytic center. In contrast to previous studies, our data indicate that PRP22 amino acids 511 to 567 are located on the opposite side of the PRP22 helicase domain and also exhibit a more elongated path (fig. S6, A to C). DDX41 RNA helicase, a multifunctional protein that is involved in pre-mRNA splicing and is linked to various cancers, including myelodysplastic syndromes and acute myeloid leukemia (32, 33), could be localized close to the RecA2 domain of PRP22 (Fig. 2 and fig. S6B). Thus, DDX41 and PRP22 form a tandem helicase module, raising the possibility that DDX41 may collaborate with PRP22 during the second step of splicing and/or in displacing the mRNA from the spliceosome. DDX41 is located far away from the hC* catalytic center (Fig. 2), which might explain the limited effect that its knockdown had on NAGNAG site selection (fig. S3C) and, together with its strong impact on skipped exons (Fig. 1A), points to a more general role in controlling alternative splicing.
Mimicry of the 3′ss and 3′ exon by PM5 nucleotides downstream of the BS-A
The catalytic U2/U6 RNA center and nucleotides of the 5′ exon and of the PM5 intron comprising the branched helix are well defined in PM5 C* (fig. S7, A and B). Unexpectedly, although PM5 lacks a 3′ss AG downstream of the BS-A (fig. S1A), a dinucleotide (most likely an AC, see also below) located ~15 nt downstream of the BS-A is docked to the latter and to G+1 at the 5′ end of the intron (Fig. 3, A and B). Furthermore, U6-A45 also stacks with the last base of this PM5 dinucleotide. Thus, the latter, which we refer to as the PM5 3′ss mimic, is docked to the catalytic center in a similar manner to a bona fide 3′ss AG (Fig. 3, A and B). The last nucleotide of the 3′ss mimic is still covalently bound to the first nucleotide of the 3′ exon mimic (Fig. 3B). Likewise, the 3′ terminal nucleotide of the 5′ exon (G-1), which is located close to the active site, no longer exhibits a flipped-out conformation, as observed in the hC complex (fig. S7C) (8) but is not ligated to any other PM5 nucleotide (Fig. 3B). Thus, exon ligation has not taken place, confirming that the complex formed on PM5 has not progressed to the P complex stage. In summary, by using the PM5 pre-mRNA construct, we could isolate hC* complexes in which a dinucleotide that mimics the 3′ss is stably docked to the catalytic center, just before the second catalytic step.
Fig. 3. An AC dinucleotide that mimics a 3′ss is docked in the PM5 hC* active site via interactions with the BS-A, 5′ss, and U6-A45.
(A) Schematic of the docking of a 3′ss mimic (A169 and C170) in PM5 C* via interactions with the BS-A, G+1, and U+2 of the intron, and U6-A45 in comparison with the docking of the 3′ss in the hP complex [Protein Data Bank (PDB) 6QDV]. Base pairing and stacking interactions are indicated by shading. (B) Close-up of the 3′ss dinucleotides and neighboring nucleotides in the catalytic core of PM5 hC* (this study) and hP (PDB 6QDV) complexes, and their fit to the respective cryo-EM densities.
Regulation of 3′ss selection via direct interaction of FAM32A with nucleotides in the C* catalytic core
Knockdown of FAM32A had the strongest effect on alternative 3′ss usage (Fig. 1A). In PM5 C*, FAM32A runs along the interface between the PRP8 endonuclease (En) and N-terminal domains (NTDs) (fig S7D). Its CT tail extends into the C* catalytic core, with FAM32A residues K107 and S109 interacting with the backbone of the last three nucleotides at the 3′ end of the 5′ exon (fig. S7, D and E). Knockdown of FAM32A may thus destabilize the 3′ end of the 5′ exon in C*, including the 3′OH of the cleaved 5′ exon that acts as the nucleophile for step 2. In hC*, G-1 of the 5′ exon is still base-paired to U40 of U5 loop 1, while in hP, the base of G-1 has turned away from U5 loop 1 by about 3 Å (fig. S7C), and its new position is stabilized by FAM32A residues K107 and S109, which appear to be more stably bound in hP compared to hC* (fig. S7C). This suggests that another role of FAM32A may be to help drive the step 2 reaction forward by removing G-1 from the catalytic center. T111 of FAM32A is located in the vicinity of the 3′ss dinucleotides in PM5 C* (fig. S7E) and may thus directly influence 3′ss docking, whereas V108 is positioned close to PRP8NTD (fig. S7F).
Regulation of 3′ss usage by C* proteins via tethering the branched helix and/or PRP8RH
In PM5 C*, the branched intron structure and branched U2/BS helix, which have moved out of the catalytic center, are tethered to their new position by the repositioned PRP8RH domain that directly contacts the BS and also U6 nucleotides of the 5′ss/U6 ACAGA helix, as observed in hC* and hP complexes (fig. S8A). PRKRIP1 and the β-sandwich domain (BSD) (amino acids 637 to 758) of CACTIN, along with the conserved splicing factor PRP17, are docked to PRP8RH and cooperate in stabilizing the new position of the branched helix, which is important for stable docking of the 3′ss (fig. S8, B and C). Several PRKRIP1 amino acids, including G73, A74, and S76, interact with intron nucleotides directly flanking the BS-A (fig. S8D). Moreover, SLU7 also interacts with PRP8RH, and the NKAP329–358 domain docks to the linker between PRP8RH and PRP8En, which will also reduce the flexibility of PRP8RH (fig. S8, C and E). Thus, several C* proteins, such as CACTIN, PRKRIP1, and NKAP, aid PRP8RH in stabilizing/organizing RNA nucleotides in the C* catalytic center. Knockdown of these C* proteins could, therefore, destabilize PRP8RH and/or the branched intron structure and branched helix in hC*. This, in turn, may weaken the interaction between the 3′ss nucleotides and the BS-A and/or G+1 of the intron. As PRP8RH also tethers the U6/5′ss helix, the enhanced flexibility of the latter may also weaken the base-stacking interactions between the preferentially selected 3′ss AG dinucleotide and U6-A45. Destabilization of these interactions could enhance the usage of more distally located A3′ss, including regulated NAGNAG sites.
Path of the PPT loop and its stabilization by C* proteins
In the PM5 C* complex, nucleotides between the BS-A and the docked 3′ss mimic form a loop—henceforth termed the PPT loop—whose stability/configuration likely plays a decisive role in the positioning/stable docking of the 3′ss in the C* catalytic center. Three nucleotides (C153, C154, and U155) directly downstream of the BS-A, as well as one upstream of the 3′ss mimic, can be readily localized in the PM5 C* map (fig. S9A). The remaining PPT nucleotides between the BS-A and 3′ss mimic are less well resolved, but the apparent path of this PPT loop could be traced in the PM5 C* EM density in a continuous manner (fig. S9A). The loop in PM5 C* appears to be composed of 15 nt, which is consistent with the recognition of the AC dinucleotide at PM5 positions 169 to 170 as the 3′ss (Fig. 4A). In addition to residues of the RH, RT-finger 1, and α-finger domains of PRP8, several C* proteins also form part of the pocket that cradles the intron loop and thus help to tether the position of the latter (Fig. 4, B to E). For example, the 5′ region of the PPT loop directly downstream of the branched helix appears to be initially stabilized by the interface formed by PRP8RH and a loop (amino acids 68 to 78) of PRKRIP1 (Fig. 4, B and C). It then runs upward in a narrow pocket formed by the basic inner surface of an α-helical protrusion (amino acids 657 to 663) of CACTINBSD and of a CT region of PRP22 (amino acids 1214 to 1220) that is docked to the PRP8FINGER1 (Fig. 4, A to C). The top part of the PPT loop is sandwiched between the former two domains, with W655 and adjacent amino acids of CACTINBSD appearing to interact with one or more of the nucleotides of the loop (fig. S9B). The NKAP329–359 domain, which bridges the N-terminal and CT parts of SLU7 in PM5 C* and hP, is located on the 3′ side of the PPT loop (Fig. 4, B and D, and fig. S9B). A long α helix (amino acids 360 to 413) of NKAP that, aided by CXMS, we can localize in C* extends from the NKAP329–359 domain at the intron loop pocket toward PRP22, docking at its other end to PRP22RecA2 (Fig. 5A and figs. S6C, S9B, and S10A). The base of the NKAP α helix, together with the NKAP329–359 domain, appears to interact with nucleotides in the 3′ region of the PM5 PPT loop, while also interacting with SLU7 (Fig. 4, D and E, and fig. S9B). Lastly, the 3′ end of the PPT loop interacts with the α-finger of PRP8 (Fig. 4, B to D), which also stabilizes the docking of the 3′ss with the branched intron structure.
Fig. 4. Path of the PPT loop in hC*.
(A) Schematic of RNAs in the RNP core of the PM5 hC* complexes. Yellow balls, positions of the catalytic Mg2+ ions M1 and M2, as well as structural Mg2+ ions M3-M5. (B) Overview of the path of the PPT loop and 5′ end of the 3′ exon mimic (3′ exon*). (C and D) Tight fit of the 5′ region (C) and 3′ region (D) of the PPT loop, with space filling models of proteins forming the PPT loop pocket. (E) Electrostatic surface potential of proteins forming the PPT loop pocket, where blue indicates a positive charge and red indicates a negative charge. (F) Extended positive surface of CACTIN. Dashed line, the potential path for nucleotides of introns with a longer distance between the BS-A and 3′ss.
Fig. 5. Spatial organization of the C* proteins FAM50A, CXORF56, TLS1, SDE2, ESS2, and NOSIP in hC*.
(A) FAM50A, CXORF56, and TLS1 interact with the β-sandwich domain of CACTIN, and FAM50A bridges BRR2 and PRP22. Bottom: Schematic of the domain structures of CACTIN, CXORF56, FAM50A, and TLS1, as predicted primarily by AlphaFold. Light green boxes, predicted domains not modeled in PM5 C*. RS, rich in serine-arginine dipeptides; H, helix; HD, helical domain; BSD; β-sandwich domain; GD, globular domain. (B) SDE2 stabilizes SYF2 and the U2/U6 helix II in hC*. Bottom: Schematic of the domain structures of SDE2 and SYF2. Color code as in (A). Helices that could be localized in C* are indicated by an asterisk. Abbreviations as in (A). SAP: SAF-A/B, Acinus, and PIAS domain. Red box, helical bundle formed by SDE2, SYF1, and CDC5L. (C) Localization of NOSIP and ESS2 in hC*. Bottom: Schematic of the domain structures of NOSIP and ESS2. H, helix; UB, U-box domain. Color code as in (A).
The RNA strand downstream of the 3′ss mimic follows a very similar path as the 3′ exon in the yeast P complex and extends to PRP22, where it is bound by its RecA domains (fig. S9, C and D). Initially, the RNA makes a ~180° turn around the PRP8 α-finger, and ~14 nts downstream of the 3′ss are located in the positively charged RNA exit channel (also called the 3′ exon channel) composed of the PRP8 Thumb, RT-Finger1, and the 1400 stalk domain of the linker (fig. S9, C and D). This channel is additionally flanked by the PRP22 CT tail and by SLU7 and is enclosed at the 5′ end by the PRP8 α-finger (Fig. 4, B and C, and fig. S9D). The ensuing ca 9 nt of the 3′ exon mimic are then guided by the PRP22 CT domain toward the PRP22 RecA domains (fig. S9D). While short PPT loops like that in PM5 C* can be accommodated in the narrow pocket described above, additional nucleotides of longer loops may protrude from the opening at the top of the pocket and run along the extended basic surface of the CACTINBSD (Fig. 4F).
Together, our structure suggests that the C* proteins PRKRIP1, CACTIN, and NKAP, in cooperation with several PRP8 domains and PRP22, may also directly influence the conformation/stability of the PPT loop during step 2 of splicing and, as a consequence, influence 3′ss docking/selection. Knockdown of these C* proteins is expected to alter the architecture of the PPT binding pocket, particularly its basic protein surfaces, and lead to a less constrained PPT loop and potentially a less stably docked 3′ss AG. In the case of regulated NAGNAG sites, these changes in the RNP environment of the PPT loop likely lead to increased flexibility, which may lead to the enhanced usage of the distal 3′ss that we observed in our knockdown experiments.
Indirect modulation of 3′ss selection by TLS1, CXORF56, and FAM50A via their interactions with CACTIN
Mutations in several C* proteins, such as NKAP, FAM50A, and CXORF56, whose genes are located on the X chromosome, are linked to intellectual disabilities (34–36). Guided by CXMS and AlphaFold structural predictions, we could map regions of the newly identified C* proteins FAM50A and CXORF56, as well as TLS1, close to the CACTINBSD. For example, the globular domain (GD) in the CT part of FAM50A (amino acids 197 to 271) could be mapped close to the α-helical protuberance of CACTINBSD that forms part of the PPT loop pocket (Fig. 5A and fig. S10B). In addition, the elongated β-sandwich domain (amino acids 28 to 120) of CXORF56 could be tentatively placed directly adjacent to the CACTINBSD, although the orientation of this domain could not be unambiguously determined (Fig. 5A and fig. S10B). TLS1 meanders throughout the C* complex, with its N-terminal region docking to BRR2 (Fig. 5A and fig. S10, C and D) (37). Two predicted α helices in its CT part can be located between CACTINBSD and FAM50AGD (TLS1 amino acids 159 to 172) or between CACTINBSD and CXORF56BSD (TLS1 amino acids 179 to 208) (Fig. 5A and fig. S10B). Thus, TLS1 tethers these domains to each other, consistent with cross-links identified between all four of these proteins (fig. S10B and data file S8). Regions of TLS1, CXORF56, and FAM50A can also be fit into similar, unassigned regions of the hP complex EM density (fig. S11, A to E), indicating that these proteins remain bound after the C*-to-P transition. As described above, CACTIN likely promotes 3′ss docking/selection by stabilizing the branched intron structure and branched helix and by affecting the conformation and/or stability of the PPT loop. The close proximity of TLS1, FAM50A, and CXORF56 to the CACTINBSD raises the possibility that they may indirectly affect 3′ss selection by stabilizing the CACTINBSD.
Additional domains of FAM50A, CXORF56, and SLU7 extend from the RNP core to peripheral regions of C* (fig. S10, C and D). The C terminus of SLU7 extends to the BRR2 N-terminal helicase cassette (BRR2NC) and PRP8Jab1 domains and, together with PPIL3, appears to stabilize the position of BRR2/PRP8Jab1 in C* (Fig. 5A and fig. S10A). The peripherally located 3′ domain of U2 snRNP is tethered via ISY1 to the N-terminal region of SYF1 (fig. S12, A and B). The extensive contacts among the various C* proteins and their additional interactions with other spliceosomal proteins, not only in the RNP core, but also at the periphery of hC*, is consistent with the idea that the C* proteins play an important role in stabilizing the entire C* RNP conformation.
Stabilization of SYF2 and the U2/U6 helix II in C* by SDE2
We could also localize SDE2 in the hC* complex. Like in hP, SDE2 helix 109 to 125 forms a helical bundle with the CT helix (amino acids 736 to 749) of SYF1 and CDC5L helix 11 (H11) (amino acids 275 to 281) (Fig. 5B), both of which can first be discerned at the C* stage. SDE2 was previously proposed to facilitate CACTIN binding and thus, indirectly, also to help stabilize the branched helix (13). In C*, SDE2 appears to mainly stabilize and/or promote the formation of several SYF2 α helices that are absent in hC (8), but in PM5 C* are located at the base and tip of U2/U6 helix II (Fig. 5B and fig. S12A). Two N-terminal SYF2 helices and a C-terminally located SDE2 α helix, which can first be discerned in hC*, also latch on to neighboring half a tetratricopeptide (HAT) repeats of SYF1, tethering the SYF1/SYF3 basket structure to U2/U6 helix II (Fig. 5 and fig. S12A). Together, this results in an improved resolution of the EM density in hC* at the base of helix II and at the U2 linker (nucleotides 12 to 19) connecting helix II to U2/U6 helix Ia, suggesting that both RNA regions are more stable in C*. This in turn could potentially also affect the positioning or stability of U2/U6 helix I and thus the catalytic U2/U6 RNA center, which, in turn, may promote 3′ss docking. Thus, the knockdown of SDE2 could destabilize SYF2 and U2/U6 helices II and Ia and, in this way, potentially alter the base pairing and/or stacking interactions with the preferentially used 3′ss AG, leading to enhanced usage of a more distal 3′ss. Alternatively, or in addition, it could also indirectly alter 3′ss selection by destabilizing CACTIN and, as a consequence, the branched helix and/or PPT loop–binding pocket.
Interaction with PRP8 and the potential modulation of its conformation by NOSIP
NOSIP belongs to the family of E3 ubiquitin ligases. It has two U-box domains, one at its C terminus and one located centrally that is split into two parts that are separated by 104 amino acids, a subset of which (amino acids 67 to 123) are predicted to form a long α helix (Fig. 5C). Both U box domains are located in a cleft between PRP8RT and SNU114 (Fig. 5C), near the pivot point where PRP8Large moves with respect to PRP8NTD during the B-to-Bact-to-C complex transitions. The NOSIP α helix comprised of amino acids 67 to 123 protrudes from the central U-box domain and, based on cross-linking, further extends to the interface between the SYF3 and SYF1 HAT repeats (Fig. 5C and fig. S12, C and D), connecting PRP8RT with the SYF3/SYF1 basket structure. This elongated α helix of NOSIP and its CT U-box can also be fit into unassigned EM density of the human P complex at similar positions (fig. S11F), indicating that NOSIP remains bound and is organized in a similar manner in hP. We can additionally trace the path of the remaining stretch of NOSIP residues (i.e., amino acids 117 to 234) that links the C terminus of the α helix to NOSIP’s CT U box (Fig. 5C and fig. S12, C and D). As NOSIP binds PRP8, its absence may lead to changes in the local conformation of PRP8, including potentially changes in the positioning of its RH domain and/or α-finger, which could directly affect selection of the 3′ss AG. NOSIP appears to promote a PRP8 conformation that favors usage of distal 3′ss AGs, as knockdown of NOSIP, unlike the other tested C* proteins, leads to the preferred usage of the proximal NAGNAG site. ESS2, a protein linked to the autosomal dominant DiGeorge syndrome (38), is also located near NOSIP. Guided by cross-links (data S8), we could map two α-helical regions (amino acids 32 to 97 and 140 to 177) of ESS2, which contact the CDC5L Myb domain and interact with the N-terminal part of the long α helix of SKIP that binds PRP22 (Fig. 5C and fig. S12, C and D). As SKIP appears to interact with PRP22 only at the late stage of the C-to-C* transformation (7), ESS2 may play a role in stabilizing the connection between PRP22 and the spliceosomal core via SKIP.
Identification of FAM32A, TLS1, and PRKRIP1 amino acids required for regulation of NAGNAG 3'ss choice
To dissect the function in NAGNAG alternative splicing of regions of selected C* proteins that are located in structurally important positions, we performed knockdown-rescue experiments in human embryonic kidney (HEK) 293 cells, focusing initially on the evolutionary conserved CT region of FAM32A. As in HeLa cells, RT-PCRs confirmed the preferential usage of the distal NAGNAG site after FAM32A knockdown in HEK293 cells for all investigated targets (Fig. 6A and fig. S13A), arguing for a cell type–independent function in NAGNAG 3'ss selection. Moreover, FAM32A not only promotes the selection of the proximal alternative NAGNAG site but also when the A3′ss are separated by a distance of <100 nt (fig. S13B). Knockdown of FAM32A did not affect splicing efficiency (figs. S13, C and D), indicating that, in vivo, FAM32A is primarily involved in regulating alternative splice site choices. Overexpression of an siRNA-resistant, WT version of FAM32A reverted knockdown %PSU values to control levels, ruling out potential siRNA-mediated off target effects. Deletion of solely the CT lysine residue (K112), which contacts the BS-A in hP (13), but could not be clearly localized in PM5 C*, had no effect on the rescue efficiency (Fig. 6A and fig. S13A). In contrast, overexpression of FAM32A variants lacking the last 7 or 17 amino acids failed to restore usage of the proximal NAGNAG site to control levels after FAM32A knockdown (Fig. 6A and fig. S13A), indicating an important role for one or more of the seven CT-most residues of FAM32A in A3′ss selection. Even in the presence of endogenous FAM32A (i.e., without siRNA-mediated FAM32A knockdown), overexpression of CT deletion mutants of FAM32A (i.e., FAM32AΔ17 or FAM32AΔ7) promoted usage of the distal NAGNAG site (i.e., led to reduced %PSU values), demonstrating that they act in a dominant-negative manner (Fig. 6B and fig. S14A). RNA-seq of cells overexpressing FAM32AΔ17 or FAM32AΔ7 confirmed a global effect on NAGNAG selection, similar to the effect observed after FAM32A knockdown (Fig. 6C,, fig. S14B, and data file S9). Notably, there is a strong overlap between NAGNAG introns affected by knockdown of FAM32A and overexpression of FAM32AΔ17 (fig. S14C). We have also complemented the deletion studies by investigating the effects of alanine substitutions of selected CT amino acids of FAM32A on NAGNAG 3'ss selection. Overexpression of FAM32A mutants harboring single W110A, V108A, or T111A substitutions led to strongly reduced %PSU values or, in the case of S109A or K107A substitutions, to mild but significantly reduced values (Fig. 6B and fig. S14A). In PM5 C*, W110 of FAM32A stacks on K1570 of the PRP8En domain and V108 is embedded in a hydrophobic pocket (fig. S7F). These interactions may thus play a key role in stabilizing the position of neighboring residues in the FAM32A CT tail, including S109 and K107. As T111 is located close to the 3′ss nucleotides (i.e., A169 and C170), its substitution by alanine could disturb the stable docking of the proximal 3′ss and allow competition by the distal 3′ss.
Fig. 6. Identification of FAM32A, TLS1, and PRKRIP1 residues that regulate NAGNAG 3'ss choice.
(A) NAGNAG alternative splicing after FAM32A knockdown and rescue in HEK293 cells. Top: Sequence of FAM32A’s CT tail. Middle: Representative gel showing RT-PCR products for gpank1, after knockdown of FAM32A and transfection of a construct expressing siRNA-resistant versions of WT FAM32A or deletion mutants thereof, as indicated. Bottom: Quantification of independent RNA samples (n = 2 to 5). The line in each box depicts the median, and whiskers show the minimum to maximum values. All individual data points are shown. Statistical significance was determined by unpaired t tests and indicated by asterisks *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. Green, mutants inducing distal 3′ss usage. (B) FAM32A mutants act in a dominant-negative manner. HEK293 cells were transfected with plasmids encoding the indicated FAM32A mutants and their effect on gpank1 NAGNAG splicing assayed by RT-PCR (n > 3). Quantification as in (A). Significance is indicated relative to the empty vector. (C) Volcano plot illustrating the global impact of FAM32A CT deletion (Δ17) on NAGNAG A3′ss choice (relative to CTRL cells transfected with an empty vector), as determined by RNA-seq. Significant distal AG usage upon overexpression is highlighted red and proximal AG usage is blue (|Δ%PSU| > 15 in dark red/blue). Top: Fraction of NAGNAG 3'ss whose regulation is significantly altered by FAM32AΔ17. (D) TLS1 mutants lacking α helices 159 to 172 and/or 178 to 208 act in a dominant-negative manner on NAGNAG A3′ss selection. Top: Schematic of predicted domains in TLS1’s CT region. HEK293 cells were transfected with the indicated TLS1 variants and their effect on gpank1 NAGNAG splicing (n > 3) investigated by RT-PCR. Quantification as in (B). ns, not significant. (E) PRKRIP1 mutants lacking amino acids 72 to 76, 62 to 75, or 62 to 93 act in a dominant-negative manner on NAGNAG A3′ss selection. Top: Domain structure of PRKRIP1’s central region. Experiments performed as described in (D).
To gain more insight into the mechanism whereby TLS1 regulates NAGNAG alternative splicing, we assayed the effects of the overexpression of various TLS1 deletion mutants on the splicing of NAGNAG-containing introns, focusing on the two TLS1 α helices that appear to tether the GDs of FAM50A and CXORF56 with the CACTIN BSD (Fig. 5A). Overexpression in HEK293 cells of a TLS mutant lacking α helices 179 to 208 had a significant effect on NAGNAG site choice, leading to enhanced usage of the distal AG compared to the WT TLS1 protein, as assayed by RT-PCR of the NAGNAG-containing introns from gpank1 (Fig. 6D) and ppp1r12c (fig. S14D). The effect was even stronger when α helices 159 to 172 and adjacent residues were additionally deleted (TLS1 Δ140-289). In contrast, deletion of amino acids 208 to 289 comprising the TLS1 C terminus had little effect. Together, these data are consistent with the idea that both α helices contribute to TLS1 function during NAGNAG 3′ss selection.
Our structure suggests that PRKRIP1 residues 72 to 76 play a key role in stabilizing the position of the branched intron structure and the 5′ region of the PPT loop. Overexpression in HEK293 cells of a PRKRIP1 deletion mutant lacking amino acids 72 to 76 leads to enhanced usage of the distal AG compared to the WT protein (Fig. 6E and fig. S14E). This effect was not significantly enhanced by the deletion of additional PRKRIP1 loop nucleotides (Δ62-75) or the N-terminal 18 amino acids of the long PRKRIP1 α helix comprised of amino acids 76 to 142 (i.e., PRKRIP1 Δ62-93) that connects U2 snRNP to PRP8RH (Fig. 6E and fig. S14E). These results indicate that the effects of PRKRIP1 knockdown on NAGNAG 3′ss usage are mediated primarily by amino acids 72 to 76 and further suggest that alterations in the conformation and/or stability of the 5′ region of the PPT loop indeed play a role in the regulation of NAGNAG 3′ss usage. Alternatively, or in addition, deletion of PRKRIP1 residues 62 to 75, which are close to the BS-A and the G+1 and U+2 nt of the intron, may destabilize their interactions with the preferentially used 3′ss and enhance usage of an alternative 3′ss. In summary, the dominant-negative effects of several FAM32A, PRKRIP1, and TLS1 deletion mutants provide first mechanistic insights into key protein domains that stabilize a conformation of the C* complex that promotes the preferential usage of proximal NAGNAG sites.
Potential “scanning” mechanism leading to the preferential selection of the proximal 3′ss AG
Previous studies showing that proximal, as opposed to distal, 3′ss are often preferentially used led to spliceosome scanning models in which the 3′ss is selected by linear scanning for the first AG downstream of the BS/PPT (21–24). The previously unknown information provided by the PM5 C* structure, particularly the path of nucleotides between the BS-A and the 3′ss, and the molecular architecture of the protein pocket that binds these nucleotides, allows new insights into possible mechanisms that lead to the preferred usage of the proximal 3′ss AG in those instances where closely spaced, competing 3′ss use the same BS. In the hC complex, ca 14 nt directly downstream of the BS-A are bound in a channel formed by PRP8’s RT-Thumb, RT-Finger 1, and stalk 1400 (i.e., the RNA exit channel), and intron nucleotides further downstream are bound by the helicase PRP16 (Fig. 7A) (8, 39). The ATP-dependent action of PRP16 during the C-to-C* transition, which displaces numerous proteins, leads to a ca 2-nm movement of the branched intron structure away from the catalytic center, the repositioning of the PRP8 RH domain and α-finger, and the release of PRP16 from the 3′ region of the intron, allowing recruitment of PRP22, SLU7, and the C* proteins. We propose that due to the movement of the branched intron structure and branched helix, nucleotides directly downstream of the BS-A will be released from the RNA exit channel. This will allow the initial looping out of several PPT nucleotides, which will be further facilitated by stochastic movements (Fig. 7A). As the exit channel is closed at one end by the rearranged PRP8 α-finger, nucleotides of the PPT will pass by it and be guided close to the 3′ss docking nucleotides, namely, the BS-A, G+1 of the intron, and U6-A45 (Fig. 7B). At this scanning stage, the α-finger will not yet be stably clamped down on the RNA in order not to hinder its movement. The repositioning of the branched helix is expected to mobilize only a limited number of PPT nucleotides and “pull” them toward the emerging PPT pocket. The tight confines of the PPT loop pocket, together with the PRP8 α-finger, would limit stochastic movements of the PPT nucleotides to one dimension, and the basic surfaces of the PPT loop pocket would facilitate the subsequent emergence of additional PPT nucleotides from the exit channel. At the same time, it may potentially act as a backstop (pawl), ensuring that looped-out PPT nucleotides do not move back into this channel. The emergence of the first AG dinucleotide downstream from the exit channel would allow it to be stably bound, before a more distal site emerges, by base pairing/stacking interactions with nucleotides of the branched intron structure and U6-A45, and its position could then be additionally stabilized in C* by clamping down of the PRP8 α-finger. If the proximal-most 3′AG is not stably bound, or bound at a very slow rate, additional intron nucleotides that contain a distal 3′ss will be pulled past the α-finger until a downstream 3′ss dinucleotide is stably docked. While such a mechanism can be envisaged when the distance between the BS-A and competing 3′ss is relatively short (as is the case for most introns), different mechanisms, which may even involve an additional RNA helicase, are likely at play when this distance is longer. One potential candidate is the RNA helicase DHX35, which we could not unambiguously localize in PM5 C*, and whose role in splicing remains unknown.
Fig. 7. Proposed mechanism for preferential selection of the proximal 3′ss AG.
(A) Repositioning of the branched helix during the C-to-C* transition leads to the movement of the PPT and 3′ exon nucleotides, leading to the looping out of the PPT and docking of the 3′ss to nucleotides of the branched intron structure. Organization of the PPT, 3′ exon nucleotides, and branched helix in hC (top) versus PM5 hC*. The proposed path of the first 10 nt of the PPT shown for hC is based on the hC cryo-EM structure (8, 39), whereas that of the next 11 nt is based on the path of the intron in the yeast Ci complex (44). For simplicity, only the NTD, En, RT, α-finger (α), and stalk (St) domains of PRP8, and the helicases PRP16 or PRP22 are shown. (B) Schematic of the PPT loop–binding pocket and docking of the proximal (top) or, upon loss of FAM32A (as an example), of the distal (bottom) 3′ss AG of the celf1 pre-mRNA containing an alternatively spliced NAGNAG 3′ss.
DISCUSSION
In this study, we have combined biochemical and structural approaches with genome-wide functional analyses. Our studies not only provide evidence that NAGNAG splicing regulation occurs at the C* stage, a long-standing idea (28) that has remained speculative until now, but also provide insights into how this regulation is achieved by C* proteins. Specifically, the latter may stabilize the position of the docking site for the 3′ss, namely, the branched intron structure, the architecture of the PPT loop, and/or the structure of the PRP8 α-finger that directly contacts the backbone of the 3′ss. This regulation can be achieved either by proteins that contact these RNA elements or protein domains or by those located further away via their interaction with the aforementioned proteins. This contrasts with other forms of alternative splicing regulation where regulatory factors bind regulatory sequences and promote or hinder the assembly of the splicing machinery at one of the competing sites and where splice site selection predominantly takes place at early spliceosome assembly stages before catalytic step 1 (20). Regulation of alternative splicing after step 1 was previously reported for the inclusion/skipping of exon 3 of the Sex-lethal (SXL) pre-mRNA that is controlled by SPF45 and SXL (40). However, post–step 1 regulation by SPF45 and SXL has not been reported for any other pre-mRNA substrates including those with alternatively spliced NAGNAG 3'ss. Thus, our studies uncover splicing factors that globally regulate alternative splice site choices after step 1 has occurred.
Knockdown of most of the C* proteins analyzed shifted the preferred usage of the upstream NAGNAG 3′ss site to the downstream site, as in the case of FAM32A, PRKRIP1, FAM50A, SDE2, NKAP, and TLS1, whereas knockdown of NOSIP had the opposite effect. Thus, the former C* proteins appear to support the aforementioned 3′ss scanning mechanism that favors selection of the proximal NAG site. However, the changes observed after knockdown of a given protein were quantitative in nature and not all-or-none effects, and for some introns, knockdown of a given C* protein had the opposite effect on 3′ss usage or no significant effect at all. Some NAGNAG 3'ss choices have been shown to be influenced by the direct cis-regulatory environment (i.e., the RNA sequence directly adjacent or upstream of the first NAG) and also other features of the nucleotides comprising the PPT loop, suggesting that the organization of the latter plays an important role in 3'ss choice (27). On the basis of our structural studies, the length of the PPT or secondary structural elements, for example, could be envisioned to negatively affect the docking of the PPT loop in its tight cavity, which could potentially lead to enhanced usage of the distal site 3′ss. Our C* structure indicates, however, that the effects of such cis-acting RNA features are likely modulated by C* proteins, which form the PPT loop–binding pocket and play a key role in determining which alternative NAGNAG site is ultimately stably docked in the C* complex. Thus, the final outcome of NAGNAG 3'ss choice—whether the proximal or distal site is preferentially used—is likely dependent on the interplay between one or more C* protein and intron-specific, cis-regulatory features. In other cases, alternative NAGNAG splicing may be regulated by C* proteins in a manner that is largely independent of sequence and context. In the future, additional biochemical and cryo-EM studies may clarify the details of the RNP code governing the regulation of 3′ss choices and the roles of the individual C* proteins.
As proteins that both promote proximal usage or enhance distal usage are found in the same C* complex formed on a given pre-mRNA substrate, there must also be an interplay between the regulatory effects of these proteins. The results of double knockdowns of selected C* proteins suggest that they act incrementally or, in the case of NOSIP, in an overriding, antagonistic manner, consistent with the fact that several of them appear to affect 3′ss selection by different mechanisms, as described above. Approximately 25% of regulated NAGNAG sites undergo tissue-specific alternative splicing that is often conserved between species (27), demonstrating that NAGNAG alternative splicing is not purely stochastic but rather regulated and functionally important. Our data indicate that tissue-specific changes in the expression levels of one or more C* protein could lead to changes in the 3'ss that is preferentially used. In addition, changes in their posttranslational modification status, which could prevent their stable or productive recruitment to the C* complex, could also conceivably shift the preferentially used NAGNAG 3′ss from the proximal to distal site in the case of NKAP, FAM50A, SDE2, TLS1, PRKRIP1, and FAM32A or have the opposite effect with NOSIP. At least one C* protein (i.e., FAM32A) exhibits a homogeneous expression pattern at the RNA level across various tissues (41). However, it is currently unknown whether there are tissue-specific differences at the protein level, including changes in posttranslational modifications.
RNA-seq data have confirmed the presence of multiple alternatively spliced 3′ss (including a few NAGNAG sites) in S. cerevisiae (42), raising the question how 3′ss selection occurs in yeast in the absence of homologs of the hC* proteins. The aforementioned A3′ss are more homogeneous compared to those in human, with only small variations in BS-A to 3′ss distance (43). Thus, alternative 3'ss selection likely would not require a large number of regulatory proteins, as we show are present in the human spliceosome. The catalytic core and the mechanism of 3′ss AG recognition are conserved between S. cerevisiae and humans, and the path of the downstream exon region leading to PRP22 is very similar in the C* complex of both organisms. Aside from the absence of C* proteins, one major difference is the presence of Yju2 in yeast C*, which stabilizes the branched U2/BS helix (44), but dissociates from the human spliceosome during the C-to-C* transition (fig. S1). In the human C*/P complex, the absence of YJU2 was proposed to be compensated for by C* proteins (13). While there are no obvious yeast orthologs for the human proteins that cradle the PPT loop in hC* complexes, it will be interesting to see in the future whether yeast C* spliceosomes that are assembled on introns containing alternative 3′ss, may recruit one or more currently unknown, yeast-specific C* protein that would facilitate alternative 3′ss selection.
The PM5 C* 3′ss mimic (an AC dinucleotide) is docked to the catalytic center in a similar manner to a bona fide 3′ss AG, consistent with PM5 C* being an on-path complex. Previous studies from our laboratory analyzing spliceosomes formed on the PM5 pre-mRNA substrate showed that purified PM5 C* complexes were active in bimolecular (trans) splicing assays, indicating that they can catalyze exon ligation in the presence of a bona fide 3′ss AG nucleotide (45). However, PM5 C* does not catalyze step 2 of canonical cis-splicing, suggesting that it may be a dead-end complex. In our hC* complex, the donor 3′ OH of G-1 of the 5′ exon coordinates the catalytic metal ion M1 at a distance of only ca 1.8 Å, but M1 is positioned ca 4.8 Å away from the phosphorous atom of the 5′ nucleotide of the 3′ exon mimic (U171), which may result from the 3′ss mimic containing a C instead of a G. This may, in turn, explain, at least in part, why catalytic step 2 does not occur with complexes formed on the PM5 pre-mRNA. Assuming that there is proofreading at this stage by PRP22, this could potentially lead to the PM5 C* complex being earmarked in the cell for the discard pathway. Together, our combined functional and structural studies identify transiently interacting, human spliceosomal proteins that regulate alternative NAGNAG 3′ss selection directly before step 2 and provide first insights into the potential mechanisms whereby they influence 3′ss usage.
MATERIALS AND METHODS
MS2 affinity purification of spliceosomes
HeLa S3 cells were obtained from the Helmholtz Zentrum für Infektionsforschung, Braunschweig, and tested negative for mycoplasma. HeLa nuclear extracts were prepared according to Dignam et al. (46) and dialyzed twice for 2.5 hours against 50 volumes of Roeder D buffer [20 mM Hepes-KOH (pH 7.9), 0.2 mM EDTA (pH 8.0), 1.5 mM MgCl2, 100 mM KCl, 10% (v/v) glycerol, 0.5 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride]. For purification of spliceosomal C complexes assembled on MINX or PM5 pre-mRNA, dialyzed nuclear extracts were preincubated for 10 min at 30°C with 1 μM of dominant-negative mutant of PRP16 protein (dnPRP16) (8). For both C and C* purifications, m7G(5′)ppp(5′)G-capped PM5, MINX, or MINX GG pre-mRNAs (5 nM) were preincubated with 20 nM MS2-MBP fusion protein for 30 min on ice before addition to the splicing reaction. Splicing reactions were carried out at 30°C with 40% (v/v) nuclear extract in splicing buffer [3 mM MgCl2, 65 mM KCl, 20 mM Hepes-KOH (pH 7.9), 2 mM ATP, and 20 mM creatine phosphate]. Splicing was carried out for 1.5 hours for purification of C complexes assembled on MINX and PM5 pre-mRNA, 3 hours for C* complexes assembled on PM5, and 1 hour for C* complexes assembled on MINX GG pre-mRNA. Splicing reactions were then chilled on ice, centrifuged for 30 min at 12,000 rpm to remove aggregates, and loaded onto an MBP Trap HP column (GE Healthcare) after the addition of 100 mM NaCl. The column was washed with G-150 buffer [20 mM Hepes-KOH (pH 7.9), 1.5 mM MgCl2, and 150 mM NaCl], and the complexes were eluted with G-150 buffer containing 1 mM maltose. Eluted complexes were loaded onto a 36-ml linear 5 to 20% (w/v) sucrose gradient prepared in G-150 buffer and centrifuged at 27,200 rpm for 9 hours at 4°C in a Surespin 630 (Thermo Fisher Scientific) rotor, and fractions were harvested from the bottom of the gradient. RNA from peak gradient fractions was separated on denaturing 4 to 12% NuPAGE gels (Life Technologies) and visualized by staining with SYBER Gold (Thermo Fischer Scientific).
Mass spectrometry
Purified spliceosomal C and C* complexes were denatured with 1% Rapigest SF surfactant (Waters) in 25 mM ammonia bicarbonate buffer, reduced with 10 mM DTT for 1 hour at 37°C, alkylated with 33 mM iodoacetamide for 1 hour at 25°C in the dark, diluted with 25 mM ammonia bicarbonate to decrease the RapiGest SF concentration to 0.1%, and in-solution–digested with trypsin (Promega) in a 1:30 ratio (w/w) for 20 hours at 37°C. The samples were acidified by trifluoroacetic acid and centrifuged to remove the hydrolytic products of RapiGest SF. The peptides were reverse phase–extracted from the supernatants using C18 micro-spin columns (Harvard Apparatus) and analyzed in Q Exactive HF-X and Q Exactive mass spectrometers coupled to Ultimate 3000 uHPLC (Thermo Fisher Scientific) under standard conditions. Proteins were identified by searching fragment spectra against UniProt (universal protein database) using Mascot as a search engine.
Cross-linking of PM5 C* complexes and cross-link identification
Purified spliceosomal PM5 C* complexes were cross-linked with 150 μM BS3 for 30 min at 20°C. After a buffer exchange to decrease the sucrose concentration to below 2% and a concentration step in an Amicon Ultracell-50 centrifugal filter unit with a 50-kDa cutoff (Merck Millipore), the BS3 cross-linked spliceosomes were subjected to 5 to 20% sucrose gradient centrifugation as described above. Spliceosomes from peak fractions were pelleted by ultracentrifugation in an S100-AT4 rotor (Thermo Fisher Scientific) and analyzed as described previously (47). Briefly, peptides generated after in-solution tryptic digestion were reverse phase–extracted and fractionated by gel filtration on a Superdex Peptide PC3.2/30 column (GE Healthcare). Fifty-microliter fractions corresponding to an elution volume of 1.2 to 1.8 ml were analyzed in triplicate on Thermo Fisher Scientific Q Exactive HF, Q Exactive HF-X, and Orbitrap Fusion Tribrid mass spectrometers. Protein-protein cross-links were identified by pLink 2.3.9 search engine (http://pfind.org/software/pLink) according to the recommendations of the developer (48). For simplicity, the cross-link score is represented as a negative value of the common logarithm of the original pLink score [i.e., score = −log10(pLink score)]. For the model building, a maximum distance of 30 Å between the Cα atoms of the cross-linked lysines was allowed.
Cryo-EM sample preparation
Purified spliceosomal PM5 C* complexes were in-batch cross-linked with 0.1% glutaraldehyde for 1 hour at 4°C and quenched with 100 mM aspartate (pH 8.0) on ice. The sample was buffer-exchanged and concentrated to 1 ml in an Amicon 50-kDa cutoff unit and purified further by a second sucrose density gradient centrifugation step as described above. Fractions were harvested from the bottom of the gradient. The peak fractions containing PM5 C* complexes were pooled, buffer-exchanged, and concentrated in an Amicon 50-kDa cutoff unit to decrease the sucrose below 0.1% and reach a protein concentration of 0.8 g/liter. UltrAUFoil Gold 200 mesh grids with R2/2 holey gold film (Quantifoil) were glow-discharged for 100 s at 15 mA. After applying a 4-μl sample and a wait time of 15 s, the grid was blotted for 3 s with a blot force of 5 and vitrified by plunging into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 4°C and 100% humidity.
Cryo-EM data acquisition
Cryo-EM data were acquired using a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV using Serial EM software (49). Micrographs were taken in energy-filtered transmission electron microscopy (EFTEM) mode with a slit width of 20 eV using a Quantum LS energy filter and a K3 direct electron detector (Gatan) at a nominal magnification of ×81,000 corresponding to a calibrated pixel size of 1.05 Å per pixel. Three exposures per hole were recorded in counting mode with a −0.5- to −3-μm defocus range. Each exposure was taken for 1.465 s with 40 movie frames at a dose rate of 33.295 e− per pixel per second, corresponding to a dose of 1.11 e−/Å2 per frame and resulting in a total dose of 44.24 e−/Å2. A total of 14,388 micrographs were collected.
Image processing
Motion correction, dose weighting, contrast transfer function (CTF) estimation, and particle picking were performed using Warp v.1.0.7 (50). Picked particles were extracted using a box size of 580 × 580 pixels and imported into cryoSPARC v.2.15. Three rounds of 2D classification were performed with 50 classes and 40 iterations. After removal of junk and redundant particles, 1,150,057 particles were retained and used for ab initio 3D model building with three classes. Approximately 52% of the particles contributed to the best model that resembled previously published C* and P complexes, while two other models apparently corresponded to contaminating, incomplete, or broken complexes and were not processed further. An initial refinement, with subsequent local and global CTF refinements, was carried out in cryoSPARC and yielded a ~2.7-Å EM density map using the gold standard Fourier shell correlation of 0.143. To faithfully visualize peripheral parts of the complex, a local-resolution filtering routine was applied. Attempts to improve the map using Relion v.3.1 were unsuccessful.
Model building
To generate a model of the human PM5 C* complex, we first rigid-body–fitted previously published protein and RNA structures from the human spliceosomal C* and P complexes using UCSF Chimera (51). To obtain a better fit into the EM density map, individual RNAs and proteins and domains thereof were subsequently refitted using UCSF Chimera and manually readjusted in Coot (52). After an initial round of real-space refinement in Phenix (53) and a manual optimization in Coot to improve the fit, the map was searched for unassigned elements. Guided by the composition of the PM5 C* complex (fig. S1 and table S1) and CXMS data (data S8), candidate proteins that were cross-linked to the already modeled parts of the complex were selected. Published experimental models or AlphaFold-predicted models (29, 30) of these candidates were examined and docked using UCSF Chimera. The individual models were manually checked and rebuilt in Coot if the resolution of the map allowed it. Using this approach, we were able to extend the models of several proteins including NKAP, PRP22, SDE2, and SYF2. Furthermore, it was possible to build partial models of ESS2 and NOSIP, as well as to rigid-body dock parts of CXORF56, PPIL3, and TLS1 not localized/modeled in previously published spliceosomal complexes. All AlphaFold models of various structural domains and helices of C* proteins (CXORF56, FAM50A, NKAP, NOSIP, SDE2, TLS1, and ESS2) that were fitted into our PM5 hC* density belong to the confident and very confident classes (as defined by the AlphaFold program). Although some single-stranded stretches (e.g., FAM50A amino acids 118 to 128 and 154 to 168 and NOSIP amino acids 121 to 158) belong to lower confidence classes, they fit well into our hC* EM density and, in some cases, even better into unassigned EM density of the previously published hP complex (see fig. S11). In addition, their location in PM5 C* is supported by protein cross-linking. For PRP22, AlphaFold-predicted models of some of the peripheral parts of its helicase domain (e.g., amino acids 107 to 154, 386 to 501, and 530 to 556) had a lower confidence score, but they fit well into the PM5 hC*, as well as hP densities, and were further supported by protein cross-linking data (see fig. S6). Several nucleotides of the intronic RNA (the PPT loop) were built de novo. The RNA fragment that mimics the 3′ exon was docked using a corresponding RNA element from the S. cerevisiae P complex structure and rebuilt in Coot. The model, excluding its parts located in the less well-resolved peripheral parts of the map, was iteratively refined in Phenix and inspected/adjusted in Coot. The model was validated in Phenix using a cryo-EM validation package (table S4). A summary of the appropriate existing atomic coordinate models used as templates and the procedures used to generate the model is provided in table S5. PyMOL (https://pymol.org/2/) and UCSF Chimera were used to generate the figures.
siRNA knockdowns
HeLa SS6 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin (100 μg/ml). For siRNA transfection (see table S6 for a list of siRNAs used), cells were grown in six-well plates in the absence of antibiotics. Transfections were carried out using Lipofectamine RNAiMAX transfection reagent (Thermo Fischer Scientific) according to the manufacturer’s protocol. AllStars Negative Control siRNA (Qiagen) was used as a control. Seventy-two hours after transfection, cells were washed with 1× PBS, transferred to Eppendorf tubes, and pelleted by centrifuging 5 min at 200g. RNA was obtained from the cell pellets using a Qiagen RNA isolation kit.
For overexpression experiments, a codon-optimized, siRNA-resistant version of FAM32A and PRKRIP1 with an N-terminal FLAG-tag, and an siRNA-resistant, CT FLAG-tagged version of TLS1 (37) were generated by PCR and cloned into the pTWIST-cytomegalovirus expression vector. HEK293 cells were cultured as described above for HeLa cells. Transfections were carried out using a Rotifect transfection reagent (Carl Roth) according to the manufacturer’s protocol. For rescue experiments, the respective siRNAs were transfected 2 hours after the overexpression constructs, and RNA was isolated after 48 hours. Overexpression was confirmed by Western blotting against the FLAG epitope. RNA was obtained using RNA Tri-flüssig (Bio&Sell) followed by deoxyribonuclease I (DNase I) digestion for 20 min at 37°C and extraction with ROTI-Aqua-P/C/I (Carl Roth), according to the manufacturer’s instructions.
Western blotting
For Western blot analysis of purified C and C* complexes, 100 fmol of each complex was separated on 4 to 12% NuPAGE gels and transferred to a Hybond P membrane. For Western blot analysis after siRNA knockdown, cells were pelleted as described above and suspended in 150 μl of HeLa lysis buffer [30 mM tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100 (v/v), 0.2% SDS (w/v), 1× cOmplete protease inhibitor (Roche), 1× PhosSTOP (Roche), and 5% glycerol (v/v)]. After brief vortexing, samples were incubated 15 min on ice and then sonicated in a Bioruptor for 3 min (30 s on, 30 s off) at maximum intensity in a water bath at 2°C. The protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Twenty micrograms of protein extract was separated by SDS–polyacrylamide gel electrophoresis and then transferred to Hybond P membranes (Protran, Whatman). The membranes were first blocked with 5% milk in 1× TBS-T buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.1% Tween 20] and then incubated with the corresponding antibodies (see table S7). The membranes were then washed with TBS-T and incubated with either horseradish peroxidase–conjugated goat anti-rabbit immunoglobulin G (IgG) (1:50,000; 111-035-144, Jackson Immunoresearch, USA) or goat anti-mouse IgG (1:10,000; 115-035-003, Jackson Immunoresearch, USA). After washing, membranes were immunostained using an enhanced chemiluminescence detection kit (GE Healthcare), and the signal was visualized using an Amersham Imager 680.
RNA-seq and data analysis
RNA-seq was performed in biological triplicates using DNase I–digested RNA samples for library preparation. Libraries were prepared using ribosomal RNA depletion method at BGI Genomics and sequenced using DNBSeq PE100 sequencing. Each knockdown was compared to a batch si control (siCTRL) triplicate, prepared, and sequenced on the same day (see table S2 for a summary of knockdowns and batch siCTRL samples). This yielded ~50 to 60 million paired-end 100-nt reads for each knockdown and control sample. Reads were aligned to the human hg38 genome, using STAR (v2.7.9a), yielding, on average, ~75% uniquely aligned reads. Alternative splicing changes were then calculated using rMATS (v3.1.0) and further filtered using a standard Python code. To obtain only high-confidence splicing targets, we compared each knockdown triplicate against the batch siCTRL and additionally against siCTRL of the first round of knockdowns (samples from the first round were additionally tested against siCTRL-B). For each knockdown versus control comparison, we applied the following filters: (i) P value < 0.01, (ii) |Δ%PSU| > 15, and (iii) >100 combined junction reads in the six tested knockdown and control samples. Only target events that passed these filters against both siCTRL batches were considered significantly changed (see data files S3 to S6). Similar filters were applied for a comparable Whippet analysis (P > 0.95). A3′ss distance, exon length (short variant), and intron length of regulated introns were derived using Python code. To correlate NAGNAG splicing (data S7) changes after each C* protein knockdown, we used the pandas corr function. Results were clustered using seaborn clustermap. Beforehand, a data frame comparing only P values for each C* protein against the same siCTRL (set no. 1) was normalized such that each P > 0.05 was set to 1. GO term enrichments were calculated using PANTHER v17.0 with only expressed genes as a background list. Whippet-derived gene level expressed as transcripts per million (TPM) values were used to confirm knockdowns of the investigated spliceosome factors (data S2). Mean TPM values of siRNA-treated samples were normalized to the mean TPM of the batch siCTRL samples and plotted as a heatmap in GraphPad Prism. To analyze global changes in gene expression, we compared triplicate siRNA samples against the batch siCTRL. Only genes with a (i) median TPM > 5 (across all RNA-seq samples), (ii) unpaired two-sided t test–derived P value < 0.001, (iii) |log2 fold change| > 0.5, and (iv) |ΔTPM| > 5 were considered to be differentially expressed.
Duplicate overexpression samples of FAM32A WT, Δ7, and Δ17 were compared with an empty vector CTRL. For library preparation, DNase I–digested RNA samples were purified using the polyA+ selection method at BGI Genomics and sequenced using DNBSeq PE150 sequencing. STAR (yielding ~95% uniquely aligned reads) and rMATS analyses were performed and filtered as described above (data file S9). For comparison of siRNA targets in HeLa cells and targets of the overexpressed, dominant-negative mutants in HEK293T cells, an overlap with NAGNAG coordinates was calculated for siFAM32A targets (for better comparison only against the batch siCTRL) and targets of the Δ17 mutant. Hypergeometric P values were calculated using all quantified NAGNAGs as population size. For sashimi blots of gapdh and ppp1r12c, merged bam files of siCTRL-B and siFAM32A were analyzed using the sashimi blot option of the integrative genomics viewer (IGV) . For ppp1r12c, only the NAGNAG intron is visualized. Within sashimi blots, junction read coverage minimum was set to >100 per track. Images were extracted as svg files and edited in Corel draw.
RT-PCR, siRNA knockdowns and rescue, and overexpression of mutant proteins
To investigate NAGNAG alternative splicing by RT-PCR, we optimized our established protocol for RNA extraction and RT-PCR (54). RNA was extracted using RNATri (Bio&Sell) followed by DNase I digestion, and 1 μg of RNA was used in a gene-specific RT reaction (combining up to four gene-specific reverse primers). PCR with a 32P-labeled forward primer was performed, and products were separated by denaturing 6% PAGE and quantified using PhosphorImager and ImageQuantTL software. During primer design, products were restricted to a length of 90 to 150 nt to optimize separation of the two NAGNAG products. To validate our rMATS-derived %PSU values, RT-PCRs targeting 10 NAGNAG introns (see table S8 for primer sequences) were performed using RNA samples from seven knockdowns and two batches of siCTRL. Mean rMATS-derived %PSU values were correlated against mean PCR-derived %PSU values. The high degree of reproducibility also enabled us to use RT-PCRs to compare single and double knockdowns.
Acknowledgments
We would like to thank HPC Service of ZEDAT, Freie Universität Berlin, for computing time and A. Neumann (Omiqa Bioinformatics) for help in establishing bioinformatics analyses. We are grateful to T. Conrad for HeLa cell production in a bioreactor, H. Kohansal for preparing HeLa nuclear extract, and U. Steuerwald, W. Lendeckel, M. Raabe, and R. Pflanz for technical assistance.
Funding: This work was supported by funding from the Max Planck Society to R.L. and by core funding from the Freie Universität Berlin to F.H.. Additional funding was provided by the Deutsche Forschungsgemeinschaft (grants HE5398/4-2 to F.H. and SFB860 to H.U.).
Author contributions: L.E.A., V.-Y.F., C.H., P.L., K.M., C.G., and M.P. performed the RNAi knockdown and rescue experiments. M.P. performed the bioinformatics analysis with help from P.Y. L.E.A. purified and biochemically characterized the MINX and PM5 C and C* complexes, with help from D.E.A. during the early stages of the project. O.D. and H.U. analyzed protein-protein cross-linking data. O.D. and C.D. collected EM data and performed EM data preprocessing. O.D. carried out subsequent EM data processing and refinement. O.D. and B.K. built the PM5 C* model. R.L., M.P., and F.H. designed the study and supervised the work. All authors were involved in data interpretation. The manuscript was written by R.L., M.P., F.H., O.D., and C.L.W., with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data from the siRNA knockdowns and overexpression have beendeposited to Gene Expression Omnibus (GEO) and are accessible underaccession number GSE198614. The atomic coordinate file has been depositedin the Protein Data Bank under accession ID 8C6J, and the cryo-EM mapshave been deposited in the Electron Microscopy Data Bank EMD-16452. Alldata needed to evaluate the conclusions in the paper are present in thepaper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Tables S1 to S8
Legends for data files S1 to S9
References
Other Supplementary Material for this : manuscript includes the following:
Data files S1 to S9
REFERENCES AND NOTES
- 1.B. Kastner, C. L. Will, H. Stark, R. Lührmann, Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb. Perspect. Biol. 11, a032417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. E. Wilkinson, C. Charenton, K. Nagai, RNA splicing by the spliceosome. Annu. Rev. Biochem. 89, 359–388 (2020). [DOI] [PubMed] [Google Scholar]
- 3.R. Wan, R. Bai, X. Zhan, Y. Shi, How is precursor messenger RNA spliced by the spliceosome? Annu. Rev. Biochem. 89, 333–358 (2020). [DOI] [PubMed] [Google Scholar]
- 4.S. M. Fica, N. Tuttle, T. Novak, N. S. Li, J. Lu, P. Koodathingal, Q. Dai, J. P. Staley, J. A. Piccirilli, RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.K. Bertram, D. E. Agafonov, W. T. Liu, O. Dybkov, C. L. Will, K. Hartmuth, H. Urlaub, B. Kastner, H. Stark, R. Lührmann, Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542, 318–323 (2017). [DOI] [PubMed] [Google Scholar]
- 6.X. Zhang, C. Yan, J. Hang, L. I. Finci, J. Lei, Y. Shi, An atomic structure of the human spliceosome. Cell 169, 918–929.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 7.X. Zhan, Y. Lu, X. Zhang, C. Yan, Y. Shi, Mechanism of exon ligation by human spliceosome. Mol. Cell 82, 2769–2778.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 8.K. Bertram, L. El Ayoubi, O. Dybkov, D. E. Agafonov, C. L. Will, K. Hartmuth, H. Urlaub, B. Kastner, H. Stark, R. Lührmann, Structural insights into the roles of metazoan-specific splicing factors in the human step 1 spliceosome. Mol. Cell 80, 127–139.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 9.B. Schwer, C. H. Gross, Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17, 2086–2094 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. Company, J. Arenas, J. Abelson, Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349, 487–493 (1991). [DOI] [PubMed] [Google Scholar]
- 11.R. M. Mayas, H. Maita, J. P. Staley, Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 13, 482–490 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Ohno, Y. Shimura, A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 10, 997–1007 (1996). [DOI] [PubMed] [Google Scholar]
- 13.S. M. Fica, C. Oubridge, M. E. Wilkinson, A. J. Newman, K. Nagai, A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 363, 710–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.X. Zhang, X. Zhan, C. Yan, W. Zhang, D. Liu, J. Lei, Y. Shi, Structures of the human spliceosomes before and after release of the ligated exon. Cell Res. 29, 274–285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D. A. Bitton, C. Rallis, D. C. Jeffares, G. C. Smith, Y. Y. Chen, S. Codlin, S. Marguerat, J. Bahler, LaSSO, a strategy for genome-wide mapping of intronic lariats and branch points using RNA-seq. Genome Res. 24, 1169–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. M. Fica, C. Oubridge, W. P. Galej, M. E. Wilkinson, X. C. Bai, A. J. Newman, K. Nagai, Structure of a spliceosome remodelled for exon ligation. Nature 542, 377–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C. Yan, R. Wan, R. Bai, G. Huang, Y. Shi, Structure of a yeast step II catalytically activated spliceosome. Science 355, 149–155 (2017). [DOI] [PubMed] [Google Scholar]
- 18.D. E. Agafonov, J. Deckert, E. Wolf, P. Odenwälder, S. Bessonov, C. L. Will, H. Urlaub, R. Lührmann, Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell. Biol. 31, 2667–2682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Q. Pan, O. Shai, L. J. Lee, B. J. Frey, B. J. Blencowe, Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Y. Lee, D. C. Rio, Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. W. Smith, E. B. Porro, J. G. Patton, B. Nadal-Ginard, Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342, 243–247 (1989). [DOI] [PubMed] [Google Scholar]
- 22.C. W. Smith, T. T. Chu, B. Nadal-Ginard, Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 13, 4939–4952 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Chen, K. Anderson, M. J. Moore, Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc. Natl. Acad. Sci. U.S.A. 97, 593–598 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. Anderson, M. J. Moore, Bimolecular exon ligation by the human spliceosome. Science 276, 1712–1716 (1997). [DOI] [PubMed] [Google Scholar]
- 25.M. Hiller, M. Platzer, Widespread and subtle: Alternative splicing at short-distance tandem sites. Trends Genet. 24, 246–255 (2008). [DOI] [PubMed] [Google Scholar]
- 26.M. Hiller, K. Huse, K. Szafranski, N. Jahn, J. Hampe, S. Schreiber, R. Backofen, M. Platzer, Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 36, 1255–1257 (2004). [DOI] [PubMed] [Google Scholar]
- 27.R. K. Bradley, J. Merkin, N. J. Lambert, C. B. Burge, Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLOS Biol. 10, e1001229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. Busch, K. J. Hertel, Extensive regulation of NAGNAG alternative splicing: New tricks for the spliceosome? Genome Biol. 13, 143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Zidek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Varadi, S. Anyango, M. Deshpande, S. Nair, C. Natassia, G. Yordanova, D. Yuan, O. Stroe, G. Wood, A. Laydon, A. Zidek, T. Green, K. Tunyasuvunakool, S. Petersen, J. Jumper, E. Clancy, R. Green, A. Vora, M. Lutfi, M. Figurnov, A. Cowie, N. Hobbs, P. Kohli, G. Kleywegt, E. Birney, D. Hassabis, S. Velankar, AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S. Liu, X. Li, L. Zhang, J. Jiang, R. C. Hill, Y. Cui, K. C. Hansen, Z. H. Zhou, R. Zhao, Structure of the yeast spliceosomal postcatalytic P complex. Science 358, 1278–1283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Y. Jiang, Y. Zhu, Z.-J. Liu, S. Ouyang, The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 8, 83–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. Z. Andreou, DDX41: A multifunctional DEAD-box protein involved in pre-mRNA splicing and innate immunity. Biol. Chem. 402, 645–651 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Y.-R. Lee, K. Khan, K. Armfield-Uhas, S. Srikanth, N. A. Thompson, M. Pardo, L. Yu, J. W. Norris, Y. Peng, K. W. Gripp, K. A. Aleck, C. Li, E. Spence, T.-I. Choi, S. J. Kwon, H.-M. Park, D. Yu, W. D. Heo, M. R. Mooney, S. M. Baig, I. M. Wentzensen, A. Telegrafi, K. McWalter, T. Moreland, C. Roadhouse, K. Ramsey, M. J. Lyons, C. Skinner, E. Alexov, N. Katsanis, R. E. Stevenson, J. S. Choudhary, D. J. Adams, C.-H. Kim, E. E. Davis, C. E. Schwartz, Mutations in FAM50A suggest that Armfield XLID syndrome is a spliceosomopathy. Nat. Commun. 11, 3698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. E. Rocha, T. R. D. Silveira, E. Sasaki, D. M. Sas, C. M. Lourenco, K. K. Kandaswamy, C. Beetz, A. Rolfs, P. Bauer, W. Reardon, A. M. Bertoli-Avella, Novel clinical and genetic insight into CXorf56-associated intellectual disability. Eur. J. Hum. Genet. 28, 367–372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. K. Fiordaliso, A. Iwata-Otsubo, A. L. Ritter, M. Quesnel-Vallieres, K. Fujiki, E. Nishi, M. Hancarova, N. Miyake, J. E. V. Morton, S. Lee, K. Hackmann, M. Bando, K. Masuda, R. Nakato, M. Arakawa, E. Bhoj, D. Li, H. Hakonarson, R. Takeda, M. Harr, B. Keena, E. H. Zackai, N. Okamoto, S. Mizuno, J. M. Ko, A. Valachova, D. Prchalova, M. Vlckova, T. Pippucci, C. Seiler, M. Choi, N. Matsumoto, N. Di Donato, Y. Barash, Z. Sedlacek, K. Shirahige, K. Izumi, Missense mutations in NKAP cause a disorder of transcriptional regulation characterized by marfanoid habitus and cognitive impairment. Am. J. Hum. Genet. 105, 987–995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. Bergfort, M. Preußner, B. Kuropka, I. A. Ilik, T. Hilal, G. Weber, C. Freund, T. Aktas, F. Heyd, M. C. Wahl, A multi-factor trafficking site on the spliceosome remodeling enzyme BRR2 recruits C9ORF78 to regulate alternative splicing. Nat. Commun. 13, 1132 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.W. Gong, B. S. Emanuel, N. Galili, D. H. Kim, B. Roe, D. A. Driscoll, M. L. Budarf, Structural and mutational analysis of a conserved gene (DGSI) from the minimal DiGeorge syndrome critical region. Hum. Mol. Genet. 6, 267–276 (1997). [DOI] [PubMed] [Google Scholar]
- 39.X. Zhan, C. Yan, X. Zhang, J. Lei, Y. Shi, Structure of a human catalytic step I spliceosome. Science 359, 537–545 (2018). [DOI] [PubMed] [Google Scholar]
- 40.M. J. Lallena, K. J. Chalmers, S. Llamazares, A. I. Lamond, J. Valcarcel, Splicing regulation at the second catalytic step by sex-lethal involves 3′ splice site recognition by SPF45. Cell 109, 285–296 (2002). [DOI] [PubMed] [Google Scholar]
- 41.J. Jo, S. Choi, J. Oh, S.-G. Lee, S. Y. Choi, K. K. Kim, C. Park, Conventionally used reference genes are not outstanding for normalization of gene expression in human cancer research. BMC Bioinformatics 20, 245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.K. Schreiber, G. Csaba, M. Haslbeck, R. Zimmer, Alternative splicing in next generation sequencing data of Saccharomyces cerevisiae. PLOS ONE 10, e0140487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. J. Taggart, C.-L. Lin, B. Shrestha, C. Heintzelman, S. Kim, W. G. Fairbrother, Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 27, 639–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. E. Wilkinson, S. M. Fica, W. P. Galej, K. Nagai, Structural basis for conformational equilibrium of the catalytic spliceosome. Mol. Cell 81, 1439–1452.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. Bessonov, M. Anokhina, C. L. Will, H. Urlaub, R. Lührmann, Isolation of an active step I spliceosome and composition of its RNP core. Nature 452, 846–850 (2008). [DOI] [PubMed] [Google Scholar]
- 46.J. D. Dignam, R. M. Lebovitz, R. G. Roeder, Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. Bertram, D. E. Agafonov, O. Dybkov, D. Haselbach, M. N. Leelaram, C. L. Will, H. Urlaub, B. Kastner, R. Lührmann, H. Stark, Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170, 701–713.e11 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Z.-L. Chen, J.-M. Meng, Y. Cao, J.-L. Yin, R.-Q. Fang, S.-B. Fan, C. Liu, W.-F. Zeng, Y.-H. Ding, D. Tan, L. Wu, W.-J. Zhou, H. Chi, R.-X. Sun, M.-Q. Dong, S.-M. He, A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. N. Mastronarde, Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 50.D. Tegunov, P. Cramer, Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 52.P. Emsley, K. Cowtan, Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 53.P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L.-W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, P. H. Zwart, PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Preußner, G. Goldammer, A. Neumann, T. Haltenhof, P. Rautenstrauch, M. Müller-McNicoll, F. Heyd, Body temperature cycles control rhythmic alternative splicing in mammals. Mol. Cell 67, 433–446.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 55.M. C. Wollerton, C. Gooding, E. J. Wagner, M. A. Garcia-Blanco, C. W. J. Smith, Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 (2004). [DOI] [PubMed] [Google Scholar]
- 56.D. S. Horowitz, A. R. Krainer, A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev. 11, 139–151 (1997). [DOI] [PubMed] [Google Scholar]
- 57.C. Townsend, M. N. Leelaram, D. E. Agafonov, O. Dybkov, C. L. Will, K. Bertram, H. Urlaub, B. Kastner, H. Stark, R. Lührmann, Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation. Science 370, eabc3753 (2020). [DOI] [PubMed] [Google Scholar]
- 58.F. Hamann, L. C. Zimmerningkat, R. A. Becker, T. B. Garbers, P. Neumann, J. S. Hub, R. Ficner, The structure of Prp2 bound to RNA and ADP-BeF3− reveals structural features important for RNA unwinding by DEAH-box ATPases. Acta Crystallogr. D Struct. Biol. 77, 496–509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. J. Tauchert, J.-B. Fourmann, R. Lührmann, R. Ficner, Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. eLife 6, e21510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. D. Friedman, D. Nimbalkar, F. W. Quelle, Erythropoietin receptors associate with a ubiquitin ligase, p33RUL, and require its activity for erythropoietin-induced proliferation. J. Biol. Chem. 278, 26851–26861 (2003). [DOI] [PubMed] [Google Scholar]
- 61.C. Charenton, M. E. Wilkinson, K. Nagai, Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364, 362–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.P. Fabrizio, B. Laggerbauer, J. Lauber, W. S. Lane, R. Lührmann, An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16, 4092–4106 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D. Ortlepp, B. Laggerbauer, S. Müllner, T. Achsel, B. Kirschbaum, R. Lührmann, The mammalian homologue of Prp16p is overexpressed in a cell line tolerant to leflunomide, a new immunoregulatory drug effective against rheumatoid arthritis. RNA 4, 1007–1018 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 to S8
Legends for data files S1 to S9
References
Data files S1 to S9