The COVID-19 pandemic and discovery of SARS-CoV-2-like viruses in Rhinolophus bats has reinforced the need to identify wildlife sources of zoonotic pathogens and to forecast where and when spillover to humans is likely to occur. Although we have long recognised that most emerging infectious diseases, especially viral infections, originate in wildlife, the global virome remains poorly characterised.1 Growing quantities of host–virus association data and advancements in statistical modelling are now facilitating our ability to predict probable wildlife hosts and prioritise field sampling to uncover novel virus diversity.2 How best to follow such species-level predictions for downstream insights most relevant to the risk of zoonotic spillover is an outstanding challenge.
Zoonotic spillover requires a pathogen to overcome many hierarchical barriers, spanning from circulation in wildlife to human exposure and susceptibility.3 Thus, characterising how zoonotic pathogens persist in wildlife and identifying the conditions that shape the prevalence of infection and viral shedding over space and time is essential for predicting where and when spillover is likely to occur.4 Immunology has a key role in deciphering these infection dynamics because the immune response dictates susceptibility, tolerance of infection, and pathogen shedding. Identifying the within-host processes governing infection dynamics in wildlife is crucial for forecasting the spatial and temporal distribution of infectious hosts. For example, pathogens that confer lifelong immunity in their host will generate distinct epidemiological patterns in wildlife compared with pathogens that induce short-lived immunity or those with cycles of chronic and acute infection.5 Ultimately, understanding such immunological mechanisms is necessary to develop accurate mathematical models of pathogen transmission, which in turn are important predictive tools.6
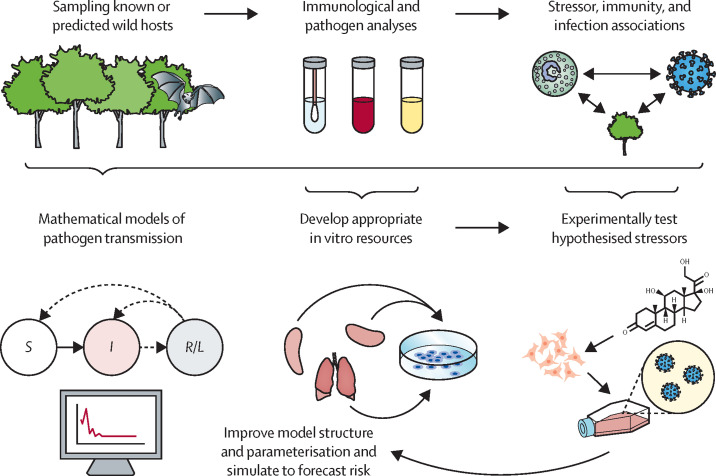
We suggest that greater integration of wildlife field studies and mechanistic molecular investigations in the laboratory is central to achieving and enhancing these predictive goals (figure ). Bats are one such host taxon in which this combination of approaches will be especially fruitful. Various zoonotic viruses circulate in wild bats, with experimental infections establishing select species as competent hosts for Hendra and Nipah viruses, Marburg virus, and rabies virus, among others.5, 7 The persistence of viruses in bats that often cause lethal disease in other mammals stems from a combination of host metapopulation dynamics and several within-host mechanisms, including low probabilities of lethal infection, high tolerance, and reactivation from chronic infection.5 Characterising the immune state of wild bats has been important to lend initial support for such mechanisms. Given the remote nature of field studies and the small sample volumes that can be collected non-lethally, field studies have largely relied on simple immunological tools. However, increasingly available annotated genomes are making shotgun techniques, such as transcriptomics and proteomics, now feasible for wild bat samples.8
Figure.
Proposed iterative pipeline for coupling wildlife field studies and mechanistic molecular investigations in the laboratory
Field sampling of known or predicted hosts can uncover novel diversity of zoonotic pathogens; establish associations between stressors, immunity, and infection; and enable initial mathematical models of transmission dynamics. Results of field studies can guide the development of appropriate in-vitro resources and, in turn, enable mechanistic experiments of stressors and pathogen shedding under controlled conditions. Such results could be used to refine mathematical models to better predict where and when zoonotic hosts are infectious and therefore more likely to transmit to humans. Figure created with BioRender.com.
Field studies have identified important sources of immunological variation in wild bats that could relate to viral susceptibility, replication, and shedding. Reproduction and migration both modulate bat immunity9 with plausible effects on virus dynamics, including reactivation of chronic infection.7 Analyses of Australian flying foxes also suggest nutritional stress as a driver of bat–virus interactions, with pulses of Hendra virus shedding occurring in winter after food shortages, especially in urban habitats.4 More broadly, field studies have suggested crucial roles for both intrinsic (eg, reproduction and migration) and extrinsic (eg, food scarcity and land conversion) stressors on shaping bat immunity and within-host dynamics of viral infection.7
Parallel to and independently of these field studies, laboratory investigations have shed light on how viruses replicate in bat cells and the associated antiviral responses generated upon infection. Early studies developed cell lines for select species,10 providing key tools to explore how viruses interact with the cellular processes of bats. Over 14 years, primary and immortalised cells of bats and, as of 2020, organoids have been used to mechanistically investigate how antiviral processes have evolved differently in bats to control virus replication and inflammation better than human cells do.11 The diversity of bats—over 1450 species—has made performing molecular studies across this mammalian order challenging because reagents do not necessarily cross-react. As with field studies, annotated genomes have begun to facilitate novel in‑vitro techniques in bats, such as bulk and single-cell transcriptomics of infected cells.12 However, such molecular resources have largely been developed from relatively accessible bat species, and cell lines have been derived from tissues that are also comparatively easy to manipulate. The development of relevant resources derived from bat species that carry viruses of zoonotic potential is now needed.
Greater synergy between field and molecular studies will accelerate key insights into the relationships between reservoir hosts and zoonotic pathogens. Dialogue between approaches is crucial to inform study design, including, but not limited to, sample collection and preservation in the field and the establishment of relevant resources in the laboratory.7, 8 Field studies are necessary to inform the choice of host species for immunological resource development, and virus tropism data from such surveys (eg, assaying different sample types) are needed to guide the selection of tissues for cell-line and organoid generation for mechanistic studies of host–virus interactions.
More generally, developing an iterative pipeline between field studies and molecular investigations will enhance efforts to predict zoonotic risk across a range of study systems, including, but not limited to, bat viruses, and especially in the context of parallel statistical and mechanistic modelling efforts (figure). By describing patterns in reservoir host infection prevalence, seroprevalence, and immune measures, field studies can first suggest plausible within-host mechanisms of pathogen circulation5, 9 and identify probable intrinsic or extrinsic stressors that facilitate susceptibility to infection, pathogen replication, and shedding.4, 9 These data-driven hypotheses can then be experimentally interrogated through mechanistic molecular investigations,10, 11 given that appropriate in-vitro resources are available (ie, matching bat species and viruses to those in field studies). For example, stressors of a particular intensity could be mimicked at the cellular level by challenging physiologically relevant in-vitro models with surrogate stimulants and cortisols (eg, modifying dose and duration), assessing whether innate immune changes align or depart from differential expression or abundance patterns observed in field studies, and how such changes affect viral susceptibility and replication. Laboratory-confirmed mechanisms could then be built back into mathematical models to forecast infection prevalence in reservoir hosts, explore different transmission contexts (eg, anthropogenic change), and simulate interventions. To facilitate such a translational pipeline, we therefore encourageecologists, virologists, microbiologists, immunologists, and evolutionary biologists to foster such interdisciplinary collaborations around zoonotic diseases.
AB is a co-inventor of the Eptesicus fuscus kidney cell line (Efk3B), which is sold through Kerafast, Boston, MA, USA. DJB was supported by National Geographic (NGS-55503R-19), the National Institute of General Medical Sciences of the National Institutes of Health (P20GM134973), and the National Science Foundation (BII 2213854). AB was supported by the Natural Sciences and Engineering Research Council of Canada, Canadian Institutes of Health Research, Saskatchewan Health Research Foundation, and Coronavirus Variants Rapid Response Network. Vaccine and Infectious Disease Organization (VIDO) of the University of Saskatchewan receives operational funding for its CL3 facility (InterVac) from the Canada Foundation for Innovation through the Major Science Initiatives. VIDO also receives operational funding from the Government of Saskatchewan via Innovation Saskatchewan and the Ministry of Agriculture.
References
- 1.Carlson CJ, Zipfel CM, Garnier R, Bansal S. Global estimates of mammalian viral diversity accounting for host sharing. Nat Ecol Evol. 2019;3:1070–1075. doi: 10.1038/s41559-019-0910-6. [DOI] [PubMed] [Google Scholar]
- 2.Becker DJ, Albery GF, Sjodin AR, et al. Optimising predictive models to prioritise viral discovery in zoonotic reservoirs. Lancet Microbe. 2022;3:e625–e637. doi: 10.1016/S2666-5247(21)00245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plowright RK, Parrish CR, McCallum H, et al. Pathways to zoonotic spillover. Nat Rev Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker DJ, Eby P, Madden W, Peel AJ, Plowright RK. Ecological conditions predict the intensity of Hendra virus excretion over space and time from bat reservoir hosts. Ecol Lett. 2023;26:23–36. doi: 10.1111/ele.14007. [DOI] [PubMed] [Google Scholar]
- 5.Plowright RK, Peel AJ, Streicker DG, et al. Transmission or within-host dynamics driving pulses of zoonotic viruses in reservoir-host populations. PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glennon EE, Bruijning M, Lessler J, et al. Challenges in modeling the emergence of novel pathogens. Epidemics. 2021;37 doi: 10.1016/j.epidem.2021.100516. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez V, Banerjee A. Molecular, ecological, and behavioral drivers of the bat-virus relationship. iScience. 2022;25 doi: 10.1016/j.isci.2022.104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neely BA, Janech MG, Fenton MB, Simmons NB, Bland AM, Becker DJ. Surveying the vampire bat (Desmodus rotundus) serum proteome: a resource for identifying immunological proteins and detecting pathogens. J Proteome Res. 2021;20:2547–2559. doi: 10.1021/acs.jproteome.0c00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voigt CC, Fritze M, Lindecke O, Costantini D, Pētersons G, Czirják GÁ. The immune response of bats differs between pre-migration and migration seasons. Sci Rep. 2020;10 doi: 10.1038/s41598-020-74473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krähling V, Dolnik O, Kolesnikova L, et al. Establishment of fruit bat cells (Rousettus aegyptiacus) as a model system for the investigation of filoviral infection. PLoS Negl Trop Dis. 2010;4:e802. doi: 10.1371/journal.pntd.0000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan LLY, Gamage AM, Tan CW, et al. Generation of self-replicating airway organoids from the cave nectar bat Eonycteris spelaea as a model system for studying host-pathogen interactions in the bat airway epithelium. Emerg Microbes Infect. 2023;12 doi: 10.1080/22221751.2022.2148561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamage AM, Chan WOY, Zhu F, et al. Single-cell transcriptome analysis of the in vivo response to viral infection in the cave nectar bat Eonycteris spelaea. Immunity. 2022;55:2187–2205. doi: 10.1016/j.immuni.2022.10.008. [DOI] [PubMed] [Google Scholar]