Abstract
As the relationship between human genes and various malfunctions and diseases becomes revealed at an ever-increasing pace, the need arises for the development of rapid genetic screening methods for diagnostic purposes. Genetic diseases show great diversity. Some are caused by a few characteristic localised mutations, while others arise from a large number of variations. Hence, it is unlikely that a single, general diagnostic method that applies to all cases will ever exist. Instead, a combination of methods is frequently applied. Here we propose the use of a dramatic colour change that a cyanine dye, 3,3′-diethylthiadicarbocyanine, displays upon binding to DNA–PNA duplexes. This method could become an inexpensive, fast and simple genetic screening test by visual inspection, with no need for complicated equipment. Our results demonstrate that this diagnostic method may be sufficiently sensitive to discriminate between even a fully complementary and a single mutation DNA sequence.
INTRODUCTION
The number of genes associated with disease states has increased markedly with the sequencing of the human genome. Some genes that cause disease show only a few specific mutations (e.g. sickle cell anaemia), whereas others have a large number of possible variations (e.g. cystic fibrosis, with over 800 mutant variants documented so far; 1). As a consequence, the methods for genetic screening may need to vary from one case to another. Among the more complete standardised tests on the market is a test for cystic fibrosis (Genzyme Genetics CF87), which can detect 87 of the more than 800 cystic fibrosis mutations. To screen for the remaining 90% of cystic fibrosis mutations or to screen for mutations in another disease-causing gene that does not have a standardised test, combinations of different screening methods are usually used. The most general way of screening for mutations is via methods such as single-strand conformational polymorphism (2) or direct sequencing (3) of the unknown sequence, but these approaches are extremely time-consuming and expensive. When only a limited number of sequence mutations cause a certain genetic disease, allele-specific amplification (4), oligonucleotide ligation assay (5) and multiplex allele-specific diagnostic assay (MASDA) (6) can be used. These methods have the ability to simultaneously screen for up to ∼100 known mutations (MASDA being the most efficient) (6) and have, for that reason, helped to increase the throughput. Nevertheless, the need for expensive equipment makes some of these methods inaccessible to less specialised laboratories.
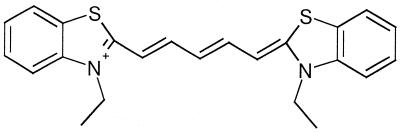
For a genetic screening method to be useful it needs to be capable of detecting even a single base-pair mutation. It is of course also a great advantage if the method is fast and straightforward. It should require as uncomplicated and as inexpensive equipment as possible to be useful in less equipped laboratories. To meet these needs, we present a colourimetric method that screens for genetic mutations. It draws on the differences in stability between fully complementary and mutated hybrid duplexes formed between DNA and a peptide nucleic acid (PNA). These differences in duplex stability have earlier been suggested for usage in genetic screening by making use of capillary electrophoresis (7). Our method uses the following: a single-stranded (ss)DNA sequence of the part of the gene to be examined; a ssPNA oligomer that is complementary to the non-mutated form of the DNA sequence; a cyanine dye, 3,3′-diethylthiadicarbocyanine iodide [DiSc2(5)] (Fig. 1) that changes colour from blue to purple upon binding to DNA–PNA duplexes (8). It is this colour change that is used to visualise formation/dissociation of the hybrid duplex. The colour change for the fully complementary DNA–PNA duplex occurs at a higher temperature than that for duplexes involving DNA mutations, owing to the greater stability of the former. Colour changes on DNA binding of cyanine dyes, especially to hairpin structures (9) and to DNA triplexes (10), have been studied previously, but not as a function of temperature. The method presented here should be applicable to genetic diseases corresponding to only a few mutations in a gene sequence as well as to more complicated screenings.
Figure 1.
Chemical structure of the dye DiSc2(5), 3,3′-diethylthiadicarbocyanine.
MATERIALS AND METHODS
Materials
The reverse phase HPLC purified PNA oligomer, complementary to a 15 nt sequence of the cystic fibrosis regulator gene, was purchased from Applied Biosystems (Framingham, MA). The cyanine dye DiSc2(5) was obtained from Sigma-Aldrich (Milwaukee, WI). The ssDNA sequences (wild-type DNA, 1MU DNA, 2MU DNA and ΔF508) were all synthesised at the PAN Facility at Stanford University (Stanford, CA).
Colourimetric assay using only the human eye
Both the PNA oligomer and the different DNA sequences were dissolved in 5 mM phosphate buffer, pH 7.5. Concentrations were determined spectrophotometrically at 85°C. The concentration of PNA oligomer (ɛ260 nm = 164 900 M–1 cm–1) was set to ∼300 µM and the concentration of DNA sequence (wild-type DNA ɛ260 nm = 499 100 M–1 cm–1, 1MU DNA ɛ260 nm = 497 000 M–1 cm–1, 2MU DNA ɛ260 nm = 498 700 M–1 cm–1 and ΔF508 ɛ260 nm = 475 700 M–1 cm–1) was set to ∼100 µM. DiSc2(5) (ɛ652 nm = 260 000 M–1 cm–1) was dissolved in methanol and the concentration determined spectrophotometrically as ∼2 mM. An aliquot of 17 µl of 5 mM phosphate buffer, pH 7.5, was added to an Eppendorf tube followed by 1 µl of PNA and 1 µl of DNA. After vortexing and 5 min heating (95°C), the samples were left to cool and hybridise at room temperature for 15 min. Thereafter, ∼0.7 µl of the dye was added and the samples were heated (95°C) again for 1–2 min. During the cooling to room temperature the colour changes of the samples were observed and monitored with a camera.
Colourimetric genetic screening using a UV/Vis spectrophotometer
A Cary 4B spectrophotometer was used for the Tm measurements. The absorbance changes were measured at 646 nm, the ɛmax of the unbound dye in water. The concentration of PNA oligomer was 0.4 µM, DNA sequences 0.13 µM and DiSc2(5) 5 µM. The buffer used was 5 mM phosphate, pH 7.5. The temperature of the samples decreased continuously at a rate of 0.5°C min–1 from 85 to 15°C.
RESULTS AND DISCUSSION
Colourimetric genetic screening using only the human eye
To assess the potential of DiSc2(5) as an indicator for genetic screening, a ssDNA sequence containing the most common mutation of the cystic fibrosis transmembrane regulator (CFTR) gene, ΔF508 (11), and two ssDNA sequences containing one and two mismatches (1MU and 2MU DNA, respectively) were compared with the non-mutated ssDNA (wild-type DNA) when hybridising to a PNA fully complementary to the wild-type DNA (Table 1). The wild-type 50mer DNA originates from the region surrounding that part of the CFTR gene where the ΔF508 mutation, which causes ∼70% of cystic fibrosis cases (11), is situated. The two other DNA sequences (1MU and 2MU DNA), being from the same region of the gene as the wild-type DNA, are not known to cause cystic fibrosis in nature but are merely model sequences to test that DiSc2(5) can discriminate between fully matched and one and two mismatches.
Table 1. DNA and PNA sequences.
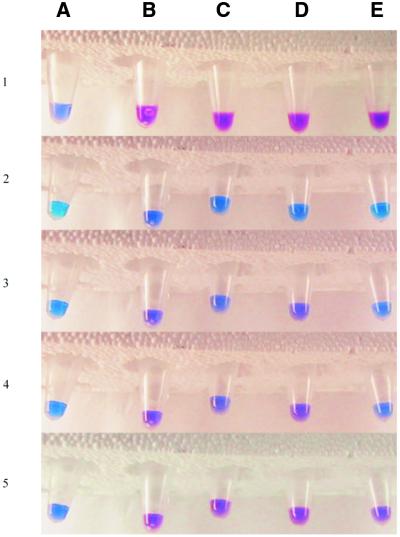
The samples containing PNA, DiSc2(5) and the different DNA strands, each in one Eppendorf tube, were heated to ∼95°C in a water bath and thereafter cooled to room temperature. A colour change from blue (free dye) to purple (dye bound to DNA–PNA duplex) occurs as hybridisation proceeds (Fig. 2). As expected, the DNA–PNA duplex involving wild-type DNA and the PNA fully complementary to the DNA sequence forms first because it is the most stable. Colour changes are then observed for the 1MU DNA, 2MU DNA and ΔF508 hybrids, in that order, which is the order of decreasing stability. Eppendorf tube A, containing only dye, remained blue throughout the experiment (Fig. 2), whereas the other samples all changed colour from dark blue to various purple tones. The top row of Figure 2 (row 1) was photographed before final heating of the samples. This shows the variation in colour between tubes containing dye with (B–E) and without (A) a DNA–PNA hybrid. As can be seen in row 2 in Figure 2, the sample that first started to turn purple was the one with wild-type DNA–PNA (B2). Similarly, tube D3, containing the 1MU DNA–PNA hybrid, had become purple, while the 2MU DNA–PNA sample in tube C3 had only just started to turn purple. In row 4 one can further see that the 2MU DNA–PNA sample (tube C4) had become more purple, while tube E4, containing ΔF508 and PNA, was still blue. Row 5 of Figure 2 shows the tubes at room temperature (after ∼1 min cooling) when all of the DNA–PNA hybrids had annealed and become purple. Our observations clearly indicate that the exceptionally large shift (>100 nm) in the wavelength maximum of DiSc2(5) upon binding to DNA–PNA duplexes can indeed be used to discriminate visually between fully matched and one and two mismatches in this system. We suggest that this behaviour is a general one. The assay could probably be further modified to suit different systems, e.g. by changing the salt concentration or the water:methanol ratio. The dye is more soluble in methanol than in water and the presence of methanol can be expected to decrease binding to the DNA–PNA duplex and therefore also the tendency to form aggregates.
Figure 2.
Colourimetric genetic screening by visual inspection. The Eppendorf tubes contain, from left to right: (A) dye only; (B) dye, PNA and wild-type DNA; (C) dye, PNA and 2MU DNA; (D) dye, PNA and 1MU DNA; (E) dye, PNA and ΔF508. Row 1 was taken after adding the dye to the hybridised samples and before the final heating. The rest of the rows (2–5) were all taken after the final heating to ∼95°C during cooling to room temperature. The rows of samples are taken in sequence with the last being row 5, i.e. the temperature decreases towards the bottom. The time elapsed after taking the samples out of the ∼95°C water bath until all of the DNA–PNA samples had turned purple was ∼1 min.
Colourimetric genetic screening using a UV/Vis spectrophotometer
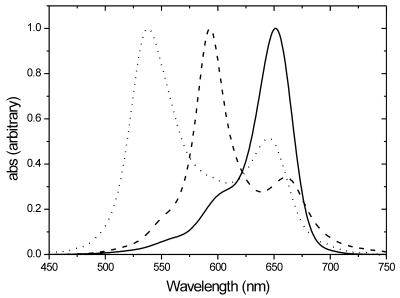
The large shift in wavelength maximum of DiSc2(5) upon binding to DNA–DNA (∼60 nm) as well as to DNA–PNA duplexes (∼115 nm), shown in Figure 3, was reported by Armitage and co-workers (8,12). The shift occurring when the dye binds to DNA–DNA duplexes is not large enough to see merely by watching the samples, but the extremely large shift when binding to DNA–PNA duplexes is sufficient to be observed just using your eyes. To verify the results obtained above using the naked eye, DNA–PNA duplex melting experiments were performed. When the samples are cooled from high temperature and the internal motion of the duplexes decreases, more dye binds to DNA–PNA duplexes and forms aggregates and the absorbance at the wavelength maximum of the free dye (646 nm) decreases. As can be seen from Table 2, the hybrid between ΔF508 and PNA shows the lowest dye aggregate Tm (estimated using the maximum of the derivative of the melting curves) followed by the 2MU DNA–PNA, 1MU DNA–PNA and wild-type DNA–PNA hybrids. This behaviour supports the results from the colourimetric genetic screening using only the human eye, in which the most stable duplex (highest Tm), that between wild-type DNA and PNA, turns purple before the other samples. The small differences in dye aggregate Tm, measuring the absorbance changes for DiSc2(5), compared to the large differences normally obtained when measuring duplex Tm at 260 nm for fully complementary DNA–PNA duplexes and DNA–PNA duplexes with one mismatch (ΔTm ≈ 10–15°C), have also been reported in a previous study (8). This kind of melting curve experiment could also be performed using purely DNA oligomers, as there are no problems for a spectrophotometer in distinguishing between bound and unbound dye.
Figure 3.
Isotropic absorption spectra of DiSc2(5) in methanol (solid curve), bound to a DNA–DNA duplex in 5 mM phosphate buffer, pH 7.5, (dashed curve) and bound to a DNA–PNA duplex in 5 mM phosphate buffer, pH 7.5 (dotted curve). The spectra are normalised to the same ɛmax.
Table 2. Cyanine aggregate melting temperature for the DNA–PNA duplexes.
| DNA–PNA duplex | Melting temperature (Tm) (°C) |
|---|---|
| Wild-type DNA–PNA | 59 |
| ΔF508–PNA | 52 |
| 2MU DNA–PNA | 54 |
| 1MU DNA–PNA | 57 |
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge Professor Jeffrey J. Wine for helpful discussions about cystic fibrosis and Dr Tommi Ratilainen for helpful assistance when taking the pictures of the assay.
REFERENCES
- 1.Wine J.J., Kuo,E., Hurlock,G. and Moss,R.B. (2001) Comprehensive mutation screening in a cystic fibrosis center. Pediatrics, 107, 280–286. [DOI] [PubMed] [Google Scholar]
- 2.Orita M., Iwahana,H., Kanazawa,H., Hayashi,K. and Sekiya,T. (1989) Detection of polymorphisms of human DNA by gel-electrophoresis as single-strand conformation polymorphisms. Proc. Natl Acad. Sci. USA, 86, 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton C.R., Graham,A., Heptinstall,L.E., Powell,S.J., Summers,C., Kaisheker,N., Smith,J.C. and Markham,A.F. (1989) Analysis of any point mutation in DNA—the amplification refractory mutation system (Arms). Nucleic Acids Res., 17, 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 6.Shuber A.P., Michalowsky,L.A., Nass,G.S., Skoletsky,J., Hire,L.M., Kotsopoulos,S.K., Phipps,M.F., Barberio,D.M. and Klinger,K.W. (1997) High throughput parallel analysis of hundreds of patient samples for more than 100 mutations in multiple disease genes. Hum. Mol. Genet., 6, 337–347. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson C., Jonsson,M., Nordén,B., Dulay,M.T., Zare,R.N., Noolandi,J., Nielsen,P.E., Tsui,L.C. and Zielenski,J. (1996) Screening for genetic mutations. Nature, 380, 207. [DOI] [PubMed] [Google Scholar]
- 8.Smith J.O., Olson,D.A. and Armitage,B.A. (1999) Molecular recognition of PNA-containing hybrids: spontaneous assembly of helical cyanine dye aggregates on PNA templates. J. Am. Chem. Soc., 121, 2686–2695. [Google Scholar]
- 9.Chen Q., Kuntz,I.D. and Shafer,R.H. (1996) Spectroscopic recognition of guanine dimeric hairpin quadruplexes by a carbocyanine dye. Proc. Natl Acad. Sci. USA, 93, 2635–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren J. and Chaires,J.B. (2000) Preferential binding of 3,3′-diethyloxadicarbocyanine to triplex DNA. J. Am. Chem. Soc., 122, 424–425. [Google Scholar]
- 11.Kerem B., Rommens,J.M., Buchanan,J.A., Markiewicz,D., Cox,T.K., Chakravati,A., Buchwald,M. and Tsui,L.C. (1989) Identification of the cystic fibrosis gene: genetic analysis. Science, 245, 1073–1080. [DOI] [PubMed] [Google Scholar]
- 12.Seifert J.L., Connor,R.E., Kushon,S.A., Wang,M. and Armitage,B.A. (1999) Spontaneous assembly of helical cyanine dye aggregates on DNA nanotemplates. J. Am. Chem. Soc., 121, 2987–2995. [Google Scholar]