Abstract
Background
Infection with SARS-CoV-2 may perturb normal microbiota, leading to secondary infections that can complicate the viral disease. The aim of this study was to probe the alteration of nasopharyngeal (NP) microbiota in the context of SARS-CoV-2 infection and obesity and to identify other respiratory pathogens among COVID-19 cases that may affect patients’ health.
Methods
A total of 107 NP swabs, including 22 from control subjects and 85 from COVID-19 patients, were processed for 6S amplicon sequencing. The respiratory pathogens causing secondary infections were identified by RT-PCR assay, using a kit that contained specific primers and probes combinations to amplify 33 known respiratory pathogens.
Results
No significant (p > 0.05) difference was observed in the alpha and beta diversity analysis, but specific taxa differed significantly between the control and COVID-19 patient groups. Genera of Sphingomonas, Kurthia, Microbacterium, Methylobacterium, Brevibacillus, Bacillus, Acinetobacter, Lactococcus, and Haemophilus was significantly abundant (p < 0.05) in COVID-19 patients compared with a healthy control group. Staphylococcus was found in relatively high abundance (35.7 %) in the COVID-19 patient groups, mainly those treated with antibiotics. A relatively high percentage of Streptococcus was detected in COVID-19 patient groups with obesity or other comorbidities. Respiratory pathogens, including Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Salmonella species, along with Pneumocystis jirovecii fungal species were detected by RT-PCR mainly in the COVID-19 patients. Klebsiella pneumoniae was commonly found in most of the samples from the control and COVID-19 patients. Four COVID-19 patients had viral coinfections with human adenovirus, human rhinovirus, enterovirus, and human parainfluenza virus 1.
Conclusions
Overall, no substantial difference was observed in the predominant NP bacterial community, but specific taxa were significantly changed between the healthy control and COVID-19 patients. Comparatively, an increased number of respiratory pathogens were identified in COVID-19 patients, and NP colonization by K. pneumoniae was probably occurring in the local population.
Keywords: COVID-19, Nasopharyngeal microbiome, 16S amplicon, Secondary infection, Saudi Arabia
Introduction
The world is in the midst of a pandemic era. Several viral outbreaks have occurred in the recent past, including Ebola, influenza, Zika, SARS-CoV-1, and MERS-CoV. The outbreak of COVID-19, caused by SARS-CoV-2, started toward the end of 2019. As of November 2022, over 600 million people have been infected, with a huge loss of life worldwide (WHO: https://covid19.who.int/). The emergence of new variants, such as Omicron and Delta, is an ongoing threat [1]. The disease symptoms include fever, cough, sore throat, breathlessness, fatigue, and malaise, among others [2]. The infection is mild in the majority of cases, but in some cases, pneumonia, acute respiratory distress syndrome, and sepsis can develop, with multi-organ failure and death as potential outcomes [2]. The severity and negative outcomes of COVID-19 are related to several risk factors, such as age, and comorbidities [3]. Furthermore, perturbation of normal respiratory microbes may result in secondary infections, which increases the risk of complications associated with respiratory viral infection and thus contributes to an overall increase in death and morbidity [4], [5].
The microbiome of the upper respiratory system works as a gatekeeper for respiratory health by inhibiting both commensal and opportunistic pathogens, and it also plays a vital role in host immune response [6], [7]. Previous studies demonstrated that respiratory viruses influence the microbiota of the respiratory tract [7], [8]. As a result, colonization of the nasopharynx by opportunistic pathogens such as Streptococcus, Acinetobacter, Moraxella, Corynebacterium, and Staphylococcus can increase and ultimately contribute to the severity of the disease [8], [9], [10]. In an analysis of 22 studies, it was reported that, respectively, 7 % and 14 % of hospitalized and intensive care unit patients with COVID-19 had a bacterial co-infection [11]. Moreover, studies carried out in the United States, China, and Italy found that a majority of COVID-19 patients with severe disease had at least one comorbidity such as diabetes, obesity, hypertension, cardiovascular disease, and chronic kidney disease [2], [3], [12]. A meta-analysis of 14 studies found that obesity is a predictor of mortality in patients with COVID-19, which is consistent with a UK study of COVID-19 patients in hospitals [13], [14]. Previous studies have demonstrated that obesity is associated with altered gut microbiota [15]. However, the impact of obesity and other comorbidities on the respiratory microbiome in COVID-19 patients has rarely been examined.
A range of observations have been reported about the nasopharyngeal (NP) microbiome in SARS-CoV-2 positive and negative samples, from no differences in those with mild COVID-19 to taxa-specific differences reported in various cities and patient populations [8], [9], [16]. Ventero et al. reported differences in the NP microbiome of COVID-19 patients compared with control subjects and identified operational taxonomic units (OTUs), mostly belonging to the Bacteroidetes and Firmicutes, specifically in patients with positive SARS-CoV-2 tests [17]. Differences in Shannon and Chao1 indexes have also been reported [18]. In contrast, several studies found no differences in either bacterial richness, diversity, or composition of the NP microbiome between COVID-19 patients and control subjects [16], [19]. However, according to some studies, interactions between microbes and the immune system are species-specific, indicating that even small differences in the diversity and composition of the microbiota can have significant effects on an individual’s health [6], [20].
The conflicting results on the NP microbiome composition associated with COVID-19 could be attributable to differences in SARS-CoV-2 variants, respiratory tract sampling location, severity of disease, comorbidities, variation in the individual respiratory microbiome, and environmental factors based on the geographic location of a study [3], [21]. Most studies on the respiratory microbiome association with COVID-19 were reported from China, and data from other countries are needed, as ethnicity is known to be a significant factor in the diversity of the microbiome [22]. The aim of the current study was to probe the alteration of NP microbiota in patients with COVID-19 compared with healthy subjects and to identify secondary microbial infections among COVID-19 patients that may have an impact on their health. Furthermore, we particularly focused on obesity as a comorbidity. Obesity is a major health issue in Saudi Arabia, with a prevalence of more than 25 % in the adult population. We identified the alteration of the NP microbiome with obesity, a major risk factor for COVID-19-associated complications.
Materials and methods
Sampling
A total of 140 NP swabs were initially included in this cross-sectional study and were collected between February and June 2022. Based on the exclusion and inclusion criteria of the study, 22 NP swabs from control subjects and 85 NP swabs from COVID-19 patients (Table S1) were processed for NP microbiome analysis. Patients younger than 18 years, those who were pregnant, or those with incomplete metadata information mentioned in Table S1were excluded. Participants with negative SARS-CoV-2 test results were divided into two groups: the healthy control group (HC) without any comorbidities, and the control group with obesity (COb) of ≥ 30 BMI but did not have any other comorbid conditions (Table S1). The COVID-19 patients were divided into four groups: patients with no comorbidities (PNCo), patients with obesity only (POb), patients with other comorbidities (PCo), and patients who used antibiotics within 2 months prior to sampling (PAb).
Nucleic acid extraction and qPCR screening of SARS-CoV-2
In a negative-pressure BSL-2 laboratory, NP samples were processed in a Class II biosafety cabinet. Total nucleic acid was extracted from the samples using the ExiPrep™ 96 Viral DNA/RNA kit (BiONEER, Seoul, South Korea) using the BiONEER extractor system with a 200-µL sample volume and a 50-µL elution volume. Real-time PCR kit of PowerChek SARS-CoV-2 (Kogenebiotech, Seoul, Korea) was used to detect the virus by mixing 5 µL of extracted nucleic acid with 11 µL of RT-PCR Premix and 4 µL of Primer/Probe Mix to make a 20-µL reaction volume. All the samples were tested for both the E gene specific for Sarbecovirus and the RdRp gene specific for SARS-CoV-2. Both probes were labeled with the fluorophore fluorescein. An internal control and a specific labeled probe for the internal control were included in the reaction mixture. Thermal cycling was performed at 50 °C for 30 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min using the LightCycler® 480 Instrument II (Roche, Germany).
S amplicon sequencing and bioinformatic analysis
DNA from the NP swab samples was extracted following the protocol of the ExiPrep™ bacteria genomic DNA kit (BiONEER). A previously described method was used for an amplicon sequence targeting the V3-V4 variable region using the universal primers 341 F and 785 R [23]. Briefly, using the Qubit system (Invitrogen, USA), the DNA concentration was determined. Limited PCR cycles were used to join sequencing adapters and dual-index barcodes. Following purification with Agencourt AMPure beads (Agencourt, USA), the Nextera XT kit was used to normalize the libraries. A single flow cell was used to sequence the samples via the MiSeq system (Illumina, Inc., USA).
Pair-end FASTQ files were assembled using PANDAseq [24]. Sequence reads were filtered, including primer and barcode region cleaning, and chimaeras and singleton reads were deleted. High-quality sequence reads above Q20 were grouped into OTUs with ≥97 % sequence similarity, using the EzBiocloud Microbiome Taxonomic Profile pipeline [25]. Two negative controls were included in this study to identify and address contamination from exogenous sources including reagents, which was previously described as being of particular relevance in low biomass samples [26]. The Kolmogorov-Smirnov D test was used to determine the normality of the data. Alpha and beta diversity analysis was performed using the MicrobiomeAnalyst pipeline [27]. The significance of beta diversity among groups was evaluated with nonmetric multidimensional scaling (NMDS) based on permutational MANOVA. A linear discriminant analysis effect size (LEfSe), Wilcoxon rank sum test, and Pearson r correlation analysis were used to identify differences in taxa abundance between patients and healthy control groups and to identify positive and negative correlations of specific taxa with each group.
RT-PCR screening of respiratory pathogens
The total nucleic acid extracted from COVID-19 patients and control groups, along with a negative control, was also tested by the FTD® Respiratory Pathogens 33 kit (SIEMENS Healthineers, Germany). In the FTD assay, 33 respiratory pathogens are simultaneously amplified using primer-probe mixtures in one-step multiplex RT-PCR, including 18 viruses and their types, such as influenza and parainfluenza viruses, coronaviruses, rhinovirus, adenovirus, Pneumocystis jirovecii (fungi); bacterial pathogens including Mycoplasma pneumoniae, Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Moraxella catarrhalis, and Klebsiella pneumoniae; and an internal control. Assay procedures for FTD-33 were followed according to the manufacturer’s guidelines (SIEMENS Healthineers). For the PCR assays, 10 µL of extracted nucleic acid samples was mixed with 20 µL of a master mix containing 12.5 µL of buffer, 1.5 µL of the primer-probe mix, and 1 µL of enzyme. Multiplex real-time RT-PCR was performed with the following thermal conditions: 50 °C for 15 min, 94 °C for 1 min, 40 cycles of 94 °C for 8 s, and 60 °C for 1 min. A sigmoidal curve with a CT value of< 40 was considered positive for a pathogen.
Results
The mean age of patients was 44.2 ± 17.6 years, and that of control subjects was 38.4 ± 8.0 years (Table S1). Regarding health conditions, 40 % of patients had COVID-19 without any comorbidities, 16.5 % had obesity only, and 28.2 % of the patients had other comorbidities, such as diabetes, hypertension, chronic cardiovascular disease, and chronic kidney disease. Antibiotics had been used by 15.3 % of patients. COVID-19 patients had mild to moderate symptoms, mainly cough (70.6 %), fever (63.5 %), sore throat (41.2 %), shortness of breath (27.1 %), headache (23.5 %), and runny nose (22.4 %), and chest x-ray/computed tomography findings (ground-glass opacity, pleural effusion, or consolidation) were noted in 11.8 % patients (Table S1).
Nasopharyngeal bacterial community analysis
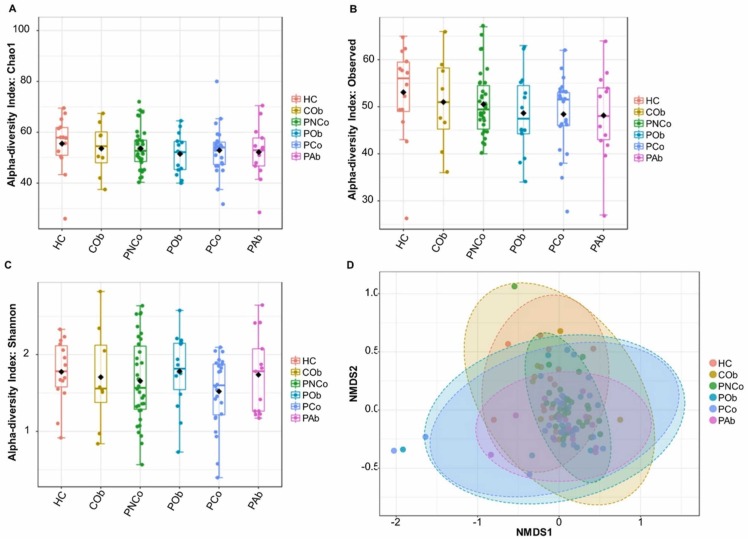
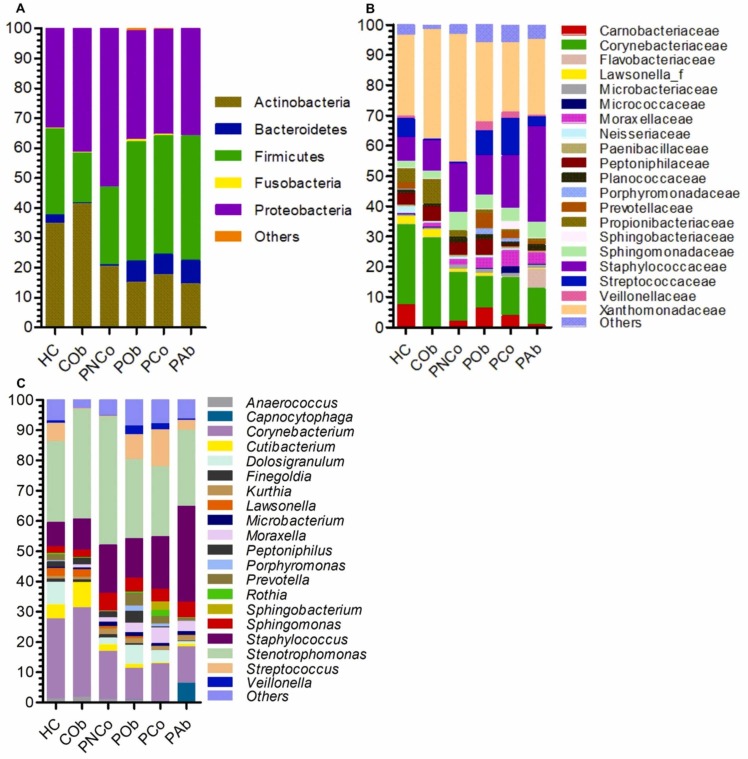
A total of 6465,308 reads were obtained from 16 S amplicon sequencing and had high quality (≥Q20 score). An average of 59,863 reads per sample was obtained. A slight decrease was found in Chao1 and observed species in the COVID-19 patient groups, but the difference in the alpha and beta diversity analysis between control and COVID-19 patient groups was not significant (p > 0.05) ( Fig. 1A-D). A total 22 phyla and candidate phyla were identified in the NP samples. Proteobacteria were predominantly found in all the groups, followed by Actinobacteria, Firmicutes, and Bacteroidetes ( Fig. 2A). Phylum Actinobacteria was found to be positively correlated (p = 0.02) with the control groups. A total of 182 families were identified from the NP samples. Xanthomonadaceae, Corynebacteriaceae, and Staphylococcaceae were predominantly found in the studied groups, but the relative abundance varied among the control and patient groups (Fig. 2B). A total of 826 OTUs were retrieved at two or more counts. Among them, 424 were classified at the genus level. Twenty genera were commonly found in> 50 % of the samples, representing more than 70 % relative abundance of NP bacteria in the studied groups. Fifty-two genera were commonly found in ≥ 20 % of samples, representing more than 90 % relative abundance of NP bacteria in each group. Stenotrophomonas bacteria were predominantly found in all the groups followed by Corynebacterium, Staphylococcus, Sphingomonas, Streptococcus, Cutibacterium, and Moraxella (Fig. 2C). Corynebacterium was found at relatively high abundance in the control groups (28.4 % ± 1.3 %) compared with COVID-19 patient groups (13.4 % ± 2 %). Staphylococcus was found at a relatively high abundance in the COVID-19 patient groups, mainly those treated with antibiotics (PAb = 35.7 %). Streptococcus was found at a relatively high abundance in the COVID-19 patient groups with obesity (POb = 8.9 %) or other comorbidities (PCo = 13.5 %). Several genera with pathogenic species, such as Acinetobacter, Microbacterium, Stenotrophomonas, Streptococcus, and Pseudomonas, were detected in the studied groups, but their relative abundance was not significantly different between control and COVID-19 patient groups.
Fig. 1.
Alpha and beta diversity analysis of nasopharyngeal bacterial community from the control and COVID-19 patients groups. (A) Chao1 estimate of richness, (B) observed species, (C) Shannon index, and (D) beta diversity analysis using Non-metric Multidimensional Scaling (NMDS). HC, healthy control group; COb, control with obesity; PNCo, COVID-19 patients with no comorbidities; POb, COVID-19 patients with obesity only; PCo, COVID-19 patients with other comorbidities; PAb, COVID-19 patients with antibiotic use.
Fig. 2.
Nasopharyngeal bacterial community taxonomy. (A) The bar charts show the average relative percentage abundance of (A) phyla, (B) families, and (C) genera among the studied sample groups. HC, healthy control group; COb, control with obesity; PNCo, COVID-19 patients with no comorbidities; POb, COVID-19 patients with obesity only; PCo, COVID-19 patients with other comorbidities; PAb, COVID-19 patients with antibiotic use.
Differentially abundant taxa between healthy control and COVID-19 patients
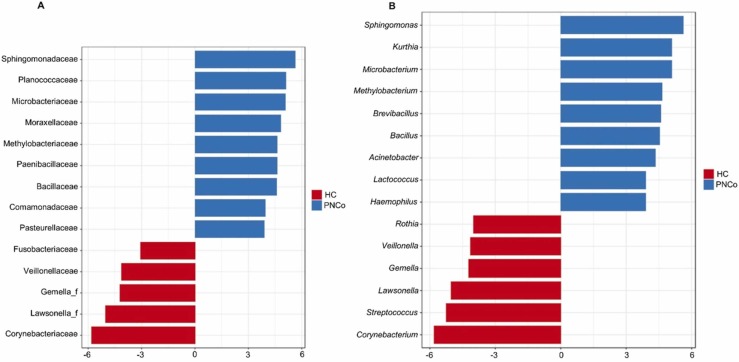
LEfSe identified bacterial taxa that were differentially abundant between the healthy control and COVID-19 patients without comorbidities group (PNCo). Differentially increased abundance of Actinobacteria and Fusobacteria was found in HC compared with PNCo, whereas Proteobacteria were abundantly found in the PNCo group. Nine families of Sphingomonadaceae, Planococcaceae, Microbacteriaceae, Moraxellaceae, Methylobacteriaceae, Paenibacillaceae, Bacillaceae, Comamonadaceae, and Pasteurellaceae were found at significantly (p < 0.05) high abundance in PNCo compared with the HC group ( Fig. 3A). Corynebacteriaceae, Lawsonella, Gemella, Veillonellaceae, and Fusobacteriaceae were significantly higher in the HC group (Fig. 3A). The LEfSe identified 15 genera that were differentially abundant (p < 0.05) between the HC and PNCo groups (Fig. 3B). Genera associated with PNCo were Sphingomonas, Kurthia, Microbacterium, Methylobacterium, Brevibacillus, Bacillus, Acinetobacter, Lactococcus, and Haemophilus. In the HC group, Corynebacterium, Streptococcus, Lawsonella, Gemella, Veillonella, and Rothia were differently more (p < 0.05) abundant compared with PNCo group (Fig. 3B).
Fig. 3.
Analysis of nasopharyngeal bacteria community using linear discriminant analysis of effect size (LEfSe). Analysis identified significantly (p < 0.05) different in relative abundance of specific (A) families, and (B) genera in the nasopharyngeal microbiome between SARS-CoV2 negative healthy controls (HC) and COVID-19 patients with no comorbidities (PNCo) groups.
Obesity influence on NP bacterial community with COVID-19
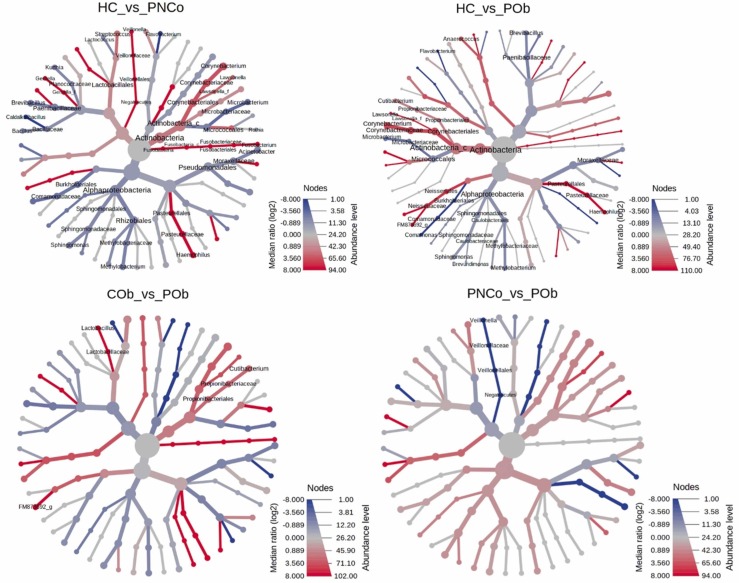
Pairwise comparison using the Wilcoxon rank sum test identified eighteen genera that were significantly different in relative abundance between HC and PNCo groups, and in comparison, 13 genera were significantly different between the HC and POb groups ( Fig. 4). Genera of Flavobacterium, Corynebacterium, Haemophilus, Sphingomonas, Lawsonella, Methylobacterium, Microbacterium, and Brevibacillus; were commonly found and significantly (p < 0.05) different in relative abundance in the two COVID-19 patient groups PNCo and POb compared with the HC group (Fig. 4). In addition, genera of Anaerococcus, Brevundimonas, Comamonas, and Cutibacterium were significantly different in relative abundance between the HC and POb groups. When the control (COb) and COVID-19 patient groups with obesity (POb) were compared, Cutibacterium and Lactobacillus were found to be significantly (p < 0.05) different in relative abundance. Veillonella was significantly (p = 0.04) different in comparison between the PNCo and POb groups (Fig. 4). Only Comamonas was significantly (p = 0.002) different in relative abundance between the control HC and COb groups.
Fig. 4.
Heat tree visualization of taxonomic differences using Wilcoxon Rank Sum test. A heat tree illustrates the genera differences between SARS-CoV2 negative control and COVID-19 patient groups. The color gradient and the size of the node, edge, and label are based on the log2 ratio of median abundance. HC, healthy control group; COb, control with obesity; PNCo, COVID-19 patients with no comorbidities; POb, COVID-19 patients with obesity only.
RT-PCR analysis of respiratory pathogens
Seven bacterial pathogens, including K. pneumoniae, S. aureus, S. pneumoniae, H. influenzae, L. pneumophila/L. longbeachae, M. catarrhalis, and Salmonella species, along with P. jirovecii fungal species and human adenovirus, enterovirus, and human parainfluenza virus 1, were detected mainly in the COVID-19 patients by RT-PCR ( Table 1). Among the 85 COVID-19 patients tested, at least one bacterial pathogen was detected in 79 samples, mainly K. pneumoniae (92.9 %). Four patients had mixed infection of bacteria and respiratory viruses (Table 1). Specifically, infection with K. pneumoniae as the only bacterial pathogen was detected in 50 patients. A dual bacterial infection was identified in 19 patients, who mainly tested positive for K. pneumonia and S. aureus (14 patients). Four patients had positive results for three distinct bacterial pathogens, and two patients had four distinct bacterial pathogens or including a fungal pathogen. We also found K. pneumonia predominantly in the control groups (20/22, 90.9 %) subjects. In addition, in the control group, three subjects were positive for S. aureus, two for M. catarrhalis, and one for P. jirovecii (Table 1). The detection of K. pneumoniae at high abundance in both patients and control groups suggested widespread NP colonization of K. pneumoniae in the local population (Table 1).
Table 1.
Identification of multiple respiratory pathogens in the nasopharyngeal swab samples by RT-PCR.
| Pathogens | HC (n = 14) |
COb (n = 8) |
PNCo (n = 34) |
PCo (n = 24) |
POb (n = 14) |
PAb (n = 13) |
|---|---|---|---|---|---|---|
| K. pneumoniae | 11 | 3 | 24 | 12 | 9 | 5 |
|
K. pneumoniae S. aureus |
1 | 2 | 4 | 6 | 1 | 3 |
|
K. pneumoniae S. pneumoniae |
1 | |||||
|
K. pneumoniae H. influenzae |
1 | 1 | ||||
|
K. pneumoniae M. catarrhalis |
1 | 1 | 1 | 1 | ||
|
K. pneumoniae P. jirovecii |
1 | |||||
|
K. pneumoniae S. aureus M. catarrhalis |
1 | |||||
|
K. pneumoniae S. aureus Salmonella spp. |
1 | |||||
|
K. pneumoniae S. pneumoniae Salmonella spp. |
1 | |||||
|
K. pneumoniae S. pneumoniae H. influenzae |
1 | |||||
|
K. pneumoniae S. aureus M. catarrhalis H. influenzae |
1 | |||||
|
K. pneumoniae S. aureus L. pneumophila/ L. longbeachae P. jirovecii |
1 | |||||
|
K. pneumoniae Human adenovirus |
1 | |||||
|
K. pneumoniae Human rhinovirus |
1 | |||||
| K. pneumoniae, Enterovirus | 1 | |||||
|
K. pneumonia, S. aureus HPIV 1 |
1 |
HC, healthy control; COb, control with obesity; PNCo, COVID-19 patients with no comorbidities; POb, COVID-19 patients with obesity only; PCo, COVID-19 patients with other comorbidities; PAb, COVID-19 patients with antibiotic use; HPIV 1, human parainfluenza virus 1.
Discussion
A number of studies have examined the modification of respiratory microbiota with SARS-CoV-2 infection and assessed the complication associated with the disease [28], [29], [30]. The reported results varied from no difference in mild COVID-19 to specific taxa alterations in the NP microbiome that differed between studies and patient populations [16], [28], [30], [31], [32]. Our 16S amplicon data showed a slight decrease in bacterial diversity in the COVID-19 patients, but the difference was not statistically significant for alpha- and beta-diversity analysis between control (SARS-CoV-2 negative) and COVID-19 patients, which is in agreement with previous studies on the NP microbiome of COVID-19 patients [8], [16], [33]. Hernández-Terán et al. found differences in the abundance of specific phyla and genera between healthy control and mild symptomatic COVID-19 patients, but they observed no differences in alpha and beta diversity [28]. A study from India reported a substantial shift in the composition of the nasal microbiome in SARS-CoV-2-infected individuals in the PCoA and NMDS plots, but they did not observe any significant difference in the alpha diversity based on Shannon, Simpson, Cho1, and Abundance-based Coverage Estimator indexes [34]. In contrast, some studies showed an increase in COVID-19-associated respiratory tract microbiota diversity [32].
This study revealed that the abundance of specific taxa varied between control and COVID-19 patients, and it identified bacterial phyla of Proteobacteria predominantly in all groups, followed by Actinobacteria, Firmicutes, and Bacteroidetes. However, the relative abundance of Actinobacteria and Fusobacteria was significantly decreased in the COVID-19 patients compared with the healthy control group. Previous studies in COVID-19 individuals showed a similar general microbial composition from the NP site [16], [17]. However, the relative abundance of those dominant phyla varied among the studies [17], [28]. Consistent with our finding, Nardelli et al. reported a reduction in Fusobacteria and Actinobacteria at all taxonomic levels, and importantly of Corynebacterium in COVID-19 patients compared with healthy controls [33], [35]. Fusobacteria adhere to a wide range of human cell types and may play a role in modulating the host's inflammatory response [33]. Previously, Fusobacterium periodonticum was shown to have a negative correlation with severity of symptoms in COVID-19 patients, suggesting a potential protective role of Fusobacterium against SARS-CoV-2 [33]. Moreover, we found several genera were differentially abundant between HC and PNCo groups based on LEfSe; these genera included Sphingomonas, Kurthia, Microbacterium, Methylobacterium, Brevibacillus, Bacillus, Acinetobacter, Lactococcus, Haemophilus, Corynebacterium, Streptococcus, Lawsonella, Gemella, Veillonella, and Rothia. Previous studies reported differences in the respiratory microbiome between COVID-19 patients and healthy controls, but few changes are consistent across studies [21]. From the review of COVID-19 studies, Merenstein et al. found that the NP microbiome was dominated by Staphylococcus, Corynebacterium, Streptococcus, Prevotella, and Veillonella [21]. In another study, the nasal microbiome of noninfected individuals contained Streptococcus, Veillonella, Rothia, Prevotella, Haemophilus, Neisseria, Stenotrophomonas, Leptotrichia, Fusobacterium, Acinetobacter, Alloprevotella, and Megasphaera, which are known to form the upper respiratory microbial community and to modulate host responses against viral infections [8]. Gao and colleagues found higher levels of the genera Halomonas, Granulicatella, Leptotrichia, and Streptococcus in COVID-19 patients, and Neisseria, Prevotella, Alloprevotella, Fusobacterium, and Haemophilus were less prevalent [36]. A reduction in the relative abundance of Corynebacterium was detected in our COVID-19 patients. It is considered a commensal organism and may play a negative role in S. pneumoniae colonization, as previously reported [37]. Recent studies have linked the decreased abundance of Corynebacterium to the severity of COVID-19 symptoms like anosmia and loss of smell [35], [38]. Veillonella and Gemella, which were identified relatively abundant in the control group in this study, were previously reported as a commensal bacteria of NP and oral cavity [39], [40]. Overall, several studies identified taxa that were differentially abundant in COVID-19, but it was difficult to find any single taxa or microbiome metric that was consistently altered across multiple studies [21], [29], [31], [41]. Several factors might contribute to the observed differences in the taxa identified between COVID-19 patients and control individuals, such as the severity of the disease, subject inclusion criteria, variation in the individual respiratory microbiome, and environmental factors based on geographical locations, among others. In particular, the effects of comorbidities and antibiotic use in the context of COVID-19 have been neglected in many studies, hindering direct comparisons between them.
Clinical outcomes of COVID-19 are associated with the presence of comorbidities (type 2 diabetes, obesity, and cardiovascular diseases) that may also contribute to alteration of the NP microbiota [28], [42]. Previously, it was demonstrated that not only gut microbiota changed with obesity, but nasal and oral microbiota also varied in individuals with obesity relative to healthy individuals [15], [43]. Our study extends these findings by showing that an increased number of genera were significantly different in terms of relative abundance between healthy control and COVID-19 patients with obesity compared with control group with obesity. However, no significant difference was noticed in the NP bacterial community between healthy and obese subjects negative for SARS-CoV-2, which suggests that the NP microbiota alteration was enhanced in COVID-19 with obesity. Previously, obesity was associated with increased production of inflammatory molecules in COVID-19 patients, which may have influenced the NP microbiome among obese COVID-19 patients [13]. Fiazo et al. demonstrated reduced effectiveness of the COVID-19 vaccine in inducing protective humoral immunity among obese subjects [44].
It is well-known that commensal bacteria play an imperative role in host immunity; however, during dysbiosis, impaired host immunity, and breakdown of the local epithelial barrier in response to COVID-19, some of these commensal bacteria can become opportunistic pathogens that may lead to secondary infection [11], [45]. We identified 20 genera from 16 S amplicon sequencing, such as Acinetobacter, Microbacterium, Stenotrophomonas, Streptococcus, and Pseudomonas, which include pathogenic bacteria in the healthy control and COVID-19 patients; however, most of these bacteria had a relatively minor abundance. Moreover, respiratory bacterial pathogens of S. aureus, S. pneumoniae, H. influenzae, Legionella pneumophila/Legionella longbeachae, M. catarrhalis, and Salmonella species, along with P. jirovecii fungal species, were detected mainly in the COVID-19 patients based on the RT-PCR assays that were able to detect 33 respiratory pathogens. The pathobionts identified in COVID-19 patients are consistent with literature implicating some of these opportunistic pathogens in respiratory virus infection severity and could lead to secondary infections [8], [10], [28]. Vijay et al. isolated in high abundance of K. pneumoniae followed by Acinetobacter baumannii and found relatively high mortality among patients who developed secondary infections in admitted COVID-19 patients [46]. In this study, K. pneumoniae was commonly found in most of the samples from the control and COVID-19 patients. The detection of K. pneumoniae at a high abundance in both patient and control groups suggested NP colonization of K. pneumoniae in the local population. Consistent with our study, S. aureus, S. pneumoniae, H. influenzae, and M. catarrhalis, which commonly cause pneumonia, were frequently found in COVID-19 patients [47]. Secondary bacterial pneumonia caused by these pathogens can lead to severe and fatal diseases in COVID-19 patients with pre-existing comorbidities, as reported previously [47].
The prevalence of P. jirovecii fungal pathogen in our samples was low compared to previous studies that found 26–33 % fungal infection in ICU patients [48], [49]. However, it is important to pay close attention to fungus co-infection because of the high risk of mortality. Consistent with our finding, viruses other than SARS-CoV-2, such as influenza A/B, human metapneumovirus, Parainfluenza, Enterovirus, Rhinovirus, and Adenovirus, have also been detected in the respiratory tracts of COVID-19 patients, but they are rare [50]. Zhang et al., from metatranscriptomic analysis of the COVID-19 cohort, detected 16 different microorganisms, the most prevalent being human alpha-herpesvirus 1 and Candida albicans, along with eight other viral pathogens, including human influenza virus and respiratory syncytial virus [51]. Overall, several respiratory pathogens have been identified in infected individuals, which indicates that SARS-CoV-2 infection leads to the presence of microbial pathogens that may result in secondary infections. So, the potential for preventing and treating secondary infections is suggested in managing COVID-19. A relatively small sample size of the control groups compared to COVID-19 patients is a limitation of the study. However, the data was normalized and statistical analysis was performed to overcome this limitation including dividing the COVID-19 patients into groups based on clinical data of the patients.
Conclusion
This study provides insight into the systematic analysis of NP bacterial community composition in the Saudi Arabian population and alternation in the specific taxa with COVID-19 infection and obesity. Our data suggest that in contrast to high-level dysbiosis in NP bacterial communities with SAR-CoV-2 infection, specific taxa such as Lawsonella, Cutibacterium, Corynebacterium, Gemella, Haemophilus, Fusobacterium, Raoultella, and Veillonella were differentially abundant between healthy control and COVID-19 patients only. Obesity altered the abundance of several genera in COVID-19 patients compared with healthy control subjects. This study identified several pathogenic and opportunistic pathogenic bacteria, viruses, and fungi in COVID-19 patients. Together with other reports, these findings reinforce the risk of secondary infection in COVID-19 cases and point to the importance of screening patients for other respiratory pathogens to prescribe more precisely targeted antibiotics.
Ethical approval
Ethical approval of the study was obtained from the bioethics committee of the Center of Excellence in Genomic Medicine Research from Applied Medical Sciences (Ref: 03-CEGMR-Bioeth-2022).
Funding
This study was funded by Jameel Fund for Infectious Disease Research and Innovation in Saudi Arabia, grant number (8-7)2120191). The authors therefore acknowledge with thanks King Abdulaziz University and Community Jameel for their financial and technical support.
CRediT authorship contribution statement
Conceived and designed the experiments: MY, EIA, MA, AAS, AMK; Performed the experiments: HAA, TA, AMH, MMA, MEH; Collection and analysis of the data: DHB, BA, AMK, SAE, MME; Manuscript writing: MY, EIA, AAS, SAE, BA; Funding acquisition: MY, EIA, AMH; Samples collection: AAS, MA, DHB, BA, MEH. All authors had an opportunity to review the manuscript and approve its final submitted version.
Competing interests
None declared.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jiph.2023.03.001.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
The sequence data of this study is submitted to the European Nucleotide Archive under the project no. PRJEB59756.
References
- 1.Esper F.P., Adhikari T.M., Tu Z.J., Cheng Y.W., El-Haddad K., Farkas D.H., Bosler D., Rhoads D., Procop G.W., Ko J.S., et al. Alpha to omicron: disease severity and clinical outcomes of major SARS-CoV-2 variants. J Infect Dis. 2022 doi: 10.1093/infdis/jiac411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Di Napoli R.: Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls. Treasure Island (FL); 2022. [PubMed]
- 3.Cekmen N., Ersoy Z., Gunay Y.I., Ghavam A.A., Tufan M.Y.S., Sahin I.M. Evaluation of coronavirus diseases (COVID-19) in terms of epidemiological and clinical features, comorbidities, diagnostic methods, treatment, and mortality. J Educ Health Promot. 2022;11:236. doi: 10.4103/jehp.jehp_1328_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R., Soucy J.R., Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26(12):1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shafran N., Shafran I., Ben-Zvi H., Sofer S., Sheena L., Krause I., Shlomai A., Goldberg E., Sklan E.H. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Sci Rep. 2021;11(1):12703. doi: 10.1038/s41598-021-92220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Man W.H., de Steenhuijsen Piters W.A., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15(5):259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumpitsch C., Koskinen K., Schopf V., Moissl-Eichinger C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019;17(1):87. doi: 10.1186/s12915-019-0703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A., Karyakarte R., Joshi S., Das R., Jani K., Shouche Y., Sharma A. Nasopharyngeal microbiome reveals the prevalence of opportunistic pathogens in SARS-CoV-2 infected individuals and their association with host types. Microbes Infect. 2022;24(1) doi: 10.1016/j.micinf.2021.104880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edouard S., Million M., Bachar D., Dubourg G., Michelle C., Ninove L., Charrel R., Raoult D. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis. 2018;37(9):1725–1733. doi: 10.1007/s10096-018-3305-8. [DOI] [PubMed] [Google Scholar]
- 10.Brugger S.D., Bomar L., Lemon K.P. Commensal-pathogen interactions along the human nasal passages. PLoS Pathog. 2016;12(7) doi: 10.1371/journal.ppat.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., Li Q., Jiang C., Zhou Y., Liu S., et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81(2):e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwok S., Adam S., Ho J.H., Iqbal Z., Turkington P., Razvi S., Le Roux C.W., Soran H., Syed A.A. Obesity: a critical risk factor in the COVID-19 pandemic. Clin Obes. 2020;10(6) doi: 10.1111/cob.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Docherty A.B., Harrison E.M., Green C.A., Hardwick H.E., Pius R., Norman L., Holden K.A., Read J.M., Dondelinger F., Carson G., et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crovesy L., Masterson D., Rosado E.L. Profile of the gut microbiota of adults with obesity: a systematic review. Eur J Clin Nutr. 2020;74(9):1251–1262. doi: 10.1038/s41430-020-0607-6. [DOI] [PubMed] [Google Scholar]
- 16.De Maio F., Posteraro B., Ponziani F.R., Cattani P., Gasbarrini A., Sanguinetti M. Nasopharyngeal microbiota profiling of SARS-CoV-2 infected patients. Biol Proced Online. 2020;22:18. doi: 10.1186/s12575-020-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventero M.P., Cuadrat R.R.C., Vidal I., Andrade B.G.N., Molina-Pardines C., Haro-Moreno J.M., Coutinho F.H., Merino E., Regitano L.C.A., Silveira C.B., et al. Nasopharyngeal microbial communities of patients infected with SARS-CoV-2 that developed COVID-19. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.637430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rueca M., Fontana A., Bartolini B., Piselli P., Mazzarelli A., Copetti M., Binda E., Perri F., Gruber C.E.M., Nicastri E., et al. Investigation of nasal/oropharyngeal microbial community of COVID-19 patients by 16S rDNA sequencing. Int J Environ Res Public Health. 2021;18(4) doi: 10.3390/ijerph18042174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun T., Halevi S., Hadar R., Efroni G., Glick Saar E., Keller N., Amir A., Amit S., Haberman Y. SARS-CoV-2 does not have a strong effect on the nasopharyngeal microbial composition. Sci Rep. 2021;11(1):8922. doi: 10.1038/s41598-021-88536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budden K.F., Shukla S.D., Rehman S.F., Bowerman K.L., Keely S., Hugenholtz P., Armstrong-James D.P.H., Adcock I.M., Chotirmall S.H., Chung K.F., et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. 2019;7(10):907–920. doi: 10.1016/S2213-2600(18)30510-1. [DOI] [PubMed] [Google Scholar]
- 21.Merenstein C., Bushman F.D., Collman R.G. Alterations in the respiratory tract microbiome in COVID-19: current observations and potential significance. Microbiome. 2022;10(1):165. doi: 10.1186/s40168-022-01342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks A.W., Priya S., Blekhman R., Bordenstein S.R. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018;16(12) doi: 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasir M., Angelakis E., Bibi F., Azhar E.I., Bachar D., Lagier J.C., Gaborit B., Hassan A.M., Jiman-Fatani A.A., Alshali K.Z., et al. Comparison of the gut microbiota of people in France and Saudi Arabia. Nutr Diabetes. 2015;5 doi: 10.1038/nutd.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masella A.P., Bartram A.K., Truszkowski J.M., Brown D.G., Neufeld J.D. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinforma. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shetty S.A., van Beek J., Bijvank E., Groot J., Kuiling S., Bosch T., van Baarle D., Fuentes S. Associations and recovery dynamics of the nasopharyngeal microbiota during influenza-like illness in the aging population. Sci Rep. 2022;12(1):1915. doi: 10.1038/s41598-022-05618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong J., Liu P., Zhou G., Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez-Teran A., Mejia-Nepomuceno F., Herrera M.T., Barreto O., Garcia E., Castillejos M., Boukadida C., Matias-Florentino M., Rincon-Rubio A., Avila-Rios S., et al. Dysbiosis and structural disruption of the respiratory microbiota in COVID-19 patients with severe and fatal outcomes. Sci Rep. 2021;11(1):21297. doi: 10.1038/s41598-021-00851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu B., Yan Y., Dong L., Han L., Liu Y., Yu J., Chen J., Yi D., Zhang M., Deng X., et al. Integrated characterization of SARS-CoV-2 genome, microbiome, antibiotic resistance and host response from single throat swabs. Cell Disco. 2021;7(1):19. doi: 10.1038/s41421-021-00248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mostafa H.H., Fissel J.A., Fanelli B., Bergman Y., Gniazdowski V., Dadlani M., Carroll K.C., Colwell R.R., Simner P.J. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. mBio. 2020;11(6) doi: 10.1128/mBio.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosas-Salazar C., Kimura K.S., Shilts M.H., Strickland B.A., Freeman M.H., Wessinger B.C., Gupta V., Brown H.M., Rajagopala S.V., Turner J.H., et al. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J Allergy Clin Immunol. 2021;147(4):1226–1233. doi: 10.1016/j.jaci.2021.02.001. e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y., Jia Z., Shi J., Wang W., He K. The active lung microbiota landscape of COVID-19 patients through the metatranscriptome data analysis. Bioimpacts. 2022;12(2):139–146. doi: 10.34172/bi.2021.23378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nardelli C., Gentile I., Setaro M., Di Domenico C., Pinchera B., Buonomo A.R., Zappulo E., Scotto R., Scaglione G.L., Castaldo G., et al. Nasopharyngeal microbiome signature in COVID-19 positive patients: can we definitively get a role to fusobacterium periodonticum? Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.625581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar D., Pandit R., Sharma S., Raval J., Patel Z., Joshi M., Joshi C.G. Nasopharyngeal microbiome of COVID-19 patients revealed a distinct bacterial profile in deceased and recovered individuals. Micro Pathog. 2022;173(Pt A) doi: 10.1016/j.micpath.2022.105829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nardelli C., Scaglione G.L., Testa D., Setaro M., Russo F., Di Domenico C., et al. Nasal microbiome in COVID-19: a potential role of corynebacterium in anosmia. Curr Microbiol. 2022;80(1):53. doi: 10.1007/s00284-022-03106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao M., Wang H., Luo H., Sun Y., Wang L., Ding S., et al. Characterization of the human oropharyngeal microbiomes in SARS-CoV-2 infection and recovery patients. Adv Sci (Weinh) 2021;8(20) doi: 10.1002/advs.202102785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bomar L., Brugger S.D., Yost B.H., Davies S.S., Lemon K.P. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio. 2016;7(1):e01725–01715. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallo O., Locatello L.G., Mazzoni A., Novelli L., Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2021;14(2):305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knapp S., Brodal C., Peterson J., Qi F., Kreth J., Merritt J. Natural competence is common among clinical isolates of veillonella parvula and is useful for genetic manipulation of this key member of the oral microbiome. Front Cell Infect Microbiol. 2017;7:139. doi: 10.3389/fcimb.2017.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Liu S., Zhang Z., Lee X., Wu W., Huang Z., et al. Association between the nasopharyngeal microbiome and metabolome in patients with COVID-19. Synth Syst Biotechnol. 2021;6(3):135–143. doi: 10.1016/j.synbio.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaibani P., Viciani E., Bartoletti M., Lewis R.E., Tonetti T., Lombardo D., et al. The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci Rep. 2021;11(1):10103. doi: 10.1038/s41598-021-89516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao L., Zhang C., Dong J., Zhao L., Li Y., Sun J. Oral microbiome and SARS-CoV-2: beware of lung co-infection. Front Microbiol. 2020;11:1840. doi: 10.3389/fmicb.2020.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michalovich D., Rodriguez-Perez N., Smolinska S., Pirozynski M., Mayhew D., Uddin S., et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat Commun. 2019;10(1):5711. doi: 10.1038/s41467-019-13751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faizo A.A., Qashqari F.S., El-Kafrawy S.A., Barasheed O., Almashjary M.N., Alfelali M., et al. A potential association between obesity and reduced effectiveness of COVID-19 vaccine-induced neutralizing humoral immunity. J Med Virol. 2022 doi: 10.1002/jmv.28130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto S., Saito M., Tamura A., Prawisuda D., Mizutani T., Yotsuyanagi H. The human microbiome and COVID-19: a systematic review. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0253293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vijay S., Bansal N., Rao B.K., Veeraraghavan B., Rodrigues C., Wattal C., et al. Secondary infections in hospitalized COVID-19 patients: Indian experience. Infect Drug Resist. 2021;14:1893–1903. doi: 10.2147/IDR.S299774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginsburg A.S., Klugman K.P. COVID-19 pneumonia and the appropriate use of antibiotics. Lancet Glob Health. 2020;8(12):e1453–e1454. doi: 10.1016/S2214-109X(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao Q., Ma Y., Ling Y., Jin W., Su Y., Wang Q., et al. Evaluation of superinfection, antimicrobial usage, and airway microbiome with metagenomic sequencing in COVID-19 patients: a cohort study in Shanghai. J Microbiol Immunol Infect. 2021;54(5):808–815. doi: 10.1016/j.jmii.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koehler P., Cornely O.A., Bottiger B.W., Dusse F., Eichenauer D.A., Fuchs F., et al. COVID-19 associated pulmonary aspergillosis. Mycoses. 2020;63(6):528–534. doi: 10.1111/myc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Musuuza J.S., Watson L., Parmasad V., Putman-Buehler N., Christensen L., Safdar N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: a systematic review and meta-analysis. PLoS One. 2021;16(5) doi: 10.1371/journal.pone.0251170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H., Ai J.W., Yang W., Zhou X., He F., Xie S., et al. Metatranscriptomic characterization of coronavirus disease 2019 identified a host transcriptional classifier associated with immune signaling. Clin Infect Dis. 2021;73(3):376–385. doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The sequence data of this study is submitted to the European Nucleotide Archive under the project no. PRJEB59756.