Abstract
During the past 60 years, perceptions about the origins of mental illness have shifted toward a biomedical model, depicting depression as a biological disorder caused by genetic abnormalities and/or chemical imbalances. Despite benevolent intentions to reduce stigma, biogenetic messages promote prognostic pessimism, reduce feelings of agency, and alter treatment preferences, motivations, and expectations. However, no research has examined how these messages influence neural markers of ruminative activity or decision-making, a gap this study sought to fill. In this pre-registered, clinical trial (NCT03998748), 49 participants with current or past depressive experiences completed a sham saliva test and were randomly assigned to receive feedback that they either have (gene-present; n = 24) or do not have (gene-absent; n = 25) a genetic predisposition to depression. Before and after receiving the feedback, resting-state activity and neural correlates of cognitive control (error-related negativity [ERN] and error positivity [Pe]) were measured using high-density electroencephalogram (EEG). Participants also completed self-report measures of beliefs about the malleability and prognosis of depression and treatment motivation. Contrary to hypotheses, biogenetic feedback did not alter perceptions or beliefs about depression, nor did it alter EEG markers of self-directed rumination nor neurophysiological correlates of cognitive control. Explanations of these null findings are discussed in the context of prior studies.
Supplementary Information
The online version contains supplementary material available at 10.3758/s13415-023-01073-9.
Keywords: Chemical imbalance, Etiological beliefs, Error positivity, Error-related negativity, Depression
Introduction
Trends in public perceptions of mental health have evolved over time, with a gradual shift toward biological explanations of psychopathology—explanations that mental health problems are caused primarily by genetic and chemical disruptions (Haslam & Kvaale, 2015; Pescosolido et al., 2010). This change in narrative came with an unfortunate cost. A predominant focus on a highly simplistic, biological model overshadows psychosocial factors that profoundly impact mental health, particularly depression. Although the biomedical model might reflect an attempt to remove blame associated with mental illness (Lee et al., 2016), it also leads to a reduced sense of agency and control (Lebowitz & Appelbaum, 2019). This has profound ramifications for patient motivation—a potent predictor of treatment engagement and efficacy (Miller, 1985)—which relies on a sense of personal agency and expectancy for success (Kelly et al., 1991). Indeed, among people with depression, biological explanations increase prognostic pessimism, decrease self-efficacy, and promote negative expectancies about treatment success (Deacon & Baird, 2009; Kemp et al., 2014; Lebowitz et al., 2021; Schroder et al., 2020).
The theory of genetic essentialism could explain the negative effects of biogenetic messaging on patient perceptions. Based in biological determinism, this is the false belief that genes represent the fundamental essence of a trait and fully determine one’s fate (including health outcomes; Dar-Nimrod & Heine, 2011). Essentialism therefore promotes a skewed perception of permanency and intractability that precludes hope for remediation. In this context, biogenetic causal attribution is a significant predictor of longer expected symptom duration among those with symptomatic depression (Lebowitz et al., 2013). Furthermore, people who are led to believe that they have a genetic predisposition for depression retrospectively report higher depressive symptoms in the prior 2 weeks (Lebowitz & Ahn, 2017) and show reduced confidence in mood-regulation abilities (Lebowitz & Ahn, 2018) compared with those who are told that they do not have this predisposition. Furthermore, interventions that emphasize the malleability of genetics and neurochemistry reduce prognostic pessimism and decrease hopelessness (Lebowitz et al., 2013), both of which could last several weeks (Lebowitz & Ahn, 2015). This suggests that the perception of one’s traits as being changeable could protect against prognostic pessimism and poor outcomes. Indeed, growth mindsets—beliefs in the malleability of human attributes—are associated with positive health outcomes; specifically, reduced psychological distress (e.g., depression, anxiety, stress), elevated treatment value (i.e., expectations that treatment will be valuable and effective), and active coping (Burnette et al., 2020). By contrast, fixed mindsets—beliefs that human attributes are immutable—are associated with anxiety, avoidance, and helplessness (Schroder, 2021).
Research pertaining to mindsets, or implicit beliefs about the malleability of self-attributes, sheds some light on potential mechanisms that might be affected by genetic feedback. Briefly, mindset theory suggests that individuals differ on how much they believe certain attributes, such as intelligence and anxiety, can change. Mindset induction studies show dissociable information-processing patterns among fixed- and growth-minded individuals. Inducing a fixed mindset enhances attention to responses in a cognitive control task (Eriksen flanker task; Eriksen & Eriksen, 1974) but impairs adaptive brain-behavior correlations following error commission (Schroder et al., 2014). By contrast, inducing a growth mindset enhances attention toward task-relevant stimuli and improves post-error performance (Schroder et al., 2014). Thus, beliefs about the malleability of traits are thought to dictate how people perceive performance errors, effort, and ability (Dweck, 1999).
Prior research in the field of biogenetic messaging is severely limited due to a single modality of measurement: self-report. Indeed, the impact of biogenetic messaging on neural correlates of cognition remains unexplored. Recent studies using event-related potentials (ERPs) suggest mindsets differ in terms of their neurophysiological reactions to mistakes (see Schroder, 2021 for review). The error positivity (Pe)—an ERP signal that denotes conscious attention allocation to an error (Falkenstein et al., 2000)—has been linked to the emotional significance of errors (Falkenstein et al., 2000; Ridderinkhof et al., 2009). A growth mindset has been associated with a greater Pe amplitude (Moser et al., 2011; Schroder et al., 2017), suggesting these individuals are particularly cognizant of having made a mistake. However, mindsets that are experimentally induced show different effects—inducing a fixed mindset by emphasizing the importance of genetics led to a greater Pe amplitude. However, the Pe was strongly correlated with improved post-error performance (i.e., faster responses after errors) in the growth (but not fixed) mindset group (Schroder et al., 2014). This highlights a greater efficiency and ability for those induced with a growth mindset to rebound after making a mistake, likely due to the recruitment of cognitive control resources after error commission. Based on the mindset literature, it is conceivable that similar neurocognitive mechanisms underlie fixed mindedness and biogenetic causal attributions of depression—both of which are characterized by the perception that self-attributes are immutable (Deacon & Baird, 2009; Dweck et al., 1995; Haslam et al., 2006).
In this pre-registered clinical trial (NCT03998748), we examined the effects of biogenetic feedback on subjective perceptions of depression, cognitive control, as well as neural correlates of rumination-like brain activity using resting-state EEG in individuals with past or current MDD symptoms. Using a previously validated sham saliva test (Lebowitz & Ahn, 2017), we randomly assigned participants to receive either a positive (gene-present) or negative (gene-absent) genetic test result. Based on the literature outlined above, we hypothesized that those who believe they are genetically vulnerable to depression would (1) endorse reduced perceived control over their emotions, (2) perceive their traits and emotions to be less malleable, (3) show increased prognostic pessimism, and (4) endorse pharmacotherapy as more effective than psychotherapy. In addition, this study was the first to examine neural consequences associated with genetic feedback; based on mindset literature we expected that the gene-present condition would be associated with (5) stronger activity in default-mode network (DMN) regions, which have been linked to rumination and depression (Pizzagalli, 2011) and (6) a larger Pe, perhaps reflecting increased attention allocation to errors (similar to the findings of the fixed mindset induction from Schroder et al., 2014). As a secondary objective, we explored whether genetic feedback altered the error-related negativity (ERN), a neural correlate of automatic, unconscious detection of response conflict (Falkenstein et al., 2000; Gehring et al., 1993). This study was expected to inform our understanding of how people with depression engage in cognitive control after receiving biogenetic messages that they are likely to encounter in the media, at school, or in a healthcare setting.
Method
Pre-registration
The study design, recruitment strategy, inclusion criteria, and several hypotheses were pre-registered at an online depository for clinical trials (https://clinicaltrials.gov/ct2/show/study/NCT03998748). In that registration, we specified two primary EEG hypotheses regarding genetic manipulation: increased default mode network activation, and increased error positivity. We pre-registered self-report hypotheses that the genetic feedback condition would (1) have poorer perceived control over emotions, (2) have an expectation that depression would last for a longer amount of time, (3) endorse a preference for medication over psychotherapy, and (4) view medication as more effective than psychotherapy. Although we listed all measures in the pre-registration, we did not specify in the registration website the particular hypotheses, so other outcomes detailed here should be considered exploratory.
Participants
Forty-nine adults (21 females; Mage = 23.10, range 18-44) were recruited from the larger Boston community and randomly assigned to receive either positive (gene-present; n = 24) or negative (gene-absent; n = 25) genetic feedback for depression using a sham saliva test. The original clinical trials registration specified a recruitment goal of 80 (which was determined by budgetary feasibility and not a power analysis). However, due to the COVID-19 pandemic, personnel changes, and budgetary constraints, this sample size was not achievable, so a new target sample size was set (N = 50). Participants were recruited by McLean Hospital’s Center for Depression, Anxiety & Stress Research. All participants had current or past MDD symptoms as determined by the Mini International Neuropsychiatric Interview (MINI; Sheehan et al., 1998), which was administered by a PhD-level clinical psychologist or a Master of Social Work interviewer.
Inclusion criteria consisted of the following: age 18 to 45 years; score ≥14 on the Beck Depression Inventory-II (BDI-II; Beck et al., 1996) or a history of 2 weeks of anhedonia and/or low mood; right handedness (Chapman & Chapman, 1987); normal or corrected-to-normal vision; fluency in written and spoken English; and absence of any psychotropic medications or psychotherapy for at least 2 weeks. Participants were excluded if they had any lifetime history of mania, bipolar disorder, or psychosis and if they had experience with psychiatric genotyping. Finally, participants were excluded if they tested positive for any current drug use as assessed by a urine drug test immediately before the procedure. Participants were compensated $100.00 USD for completing the study in addition to compensation for travel. All participants provided written, informed consent before study procedures, and the Mass General Brigham Institutional Review Board approved the study.
Clinical measures
To assess baseline (pre-saliva test) mood states, participants completed a series of self-report inventories:
Beck Depression Inventory-II (BDI-II, Beck et al., 1996)
The BDI-II is a well-validated measure of depressive symptom severity. It consists of 21 statements of depression. Participants rate their severity of each symptom pertaining to the last 2 weeks by using a 0 (minimally symptomatic) to 3 (maximally symptomatic) scale. Higher BDI-II scores denote greater depression severity. BDI-II categories define depression severity as follows: minimal (0-13), mild (14-19), moderate (20-28), severe (29-63). The BDI-II had excellent internal reliability in this sample (α = 0.94).
Quick Inventory of Depressive Symptomatology-16 (QIDS-16; Rush et al., 2003)
The QIDS-16 is a commonly used 16-item measure of depressive symptoms, and responses are given on a 0-3 response scale. In addition to the total score (α = 0.88), the QIDS-16 has three additional scores, pertaining to sleep changes, weight changes, and psychomotor changes, which are determined by the highest single-item score among a subset of responses for these domains.
Beck Hopelessness Scale (BHS; Beck et al., 1974)
The BHS is a widely used measure of hopelessness and consists of 20 true/false items. The BHS in this study provided adequate internal reliability (α = 0.90).
Ruminative Response Scale (RRS; Treynor et al., 2003)
The RRS is a 22-item measure of different rumination styles, which are rated on a 1 (almost never) to 4 (almost always) Likert-type scale. It consists of three subscales, which each showed adequate internal reliability in the current study: brooding (α = 0.82), reflection (α = 0.77), and depression (α = 0.92).
Mood and Anxiety Symptom Questionnaire (MASQ; Watson et al., 1995)
The short adaptation of the MASQ is a widely used 39-item measure of dimensions of anxiety and depression over the last week. Participants rate on a 1 (Not at all) to 5 (Extremely) Likert-type scale about their symptoms of anxiety (e.g., “felt tense or high strung”) and depression (e.g., “felt like I was having a lot of fun” [reverse-scored]). For the current study, we evaluated the anhedonic depression subscale (α = 0.94) and its constituent components, positive affect (α = 0.95), and loss of interest (α = 0.87).
Positive and Negative Affect Schedule (PANAS; Watson et al., 1988)
The PANAS is a well-validated measure of trait positive and negative affect. Participants respond on a 1 to 4 Likert-type scale to a variety of descriptors of how they generally feel. In this study, both positive (α = 0.90) and negative (α = 0.90) subscales showed adequate internal reliability.
Perceived Stress Scale (PSS; Cohen et al., 1983)
The PSS is a 14-item measure of the subjective experience of stress over the past month. Participants respond to a set of items (e.g., “In the last month, how often have you been upset because of something that happened unexpectedly?”) on a 0 (“Never”) to 4 (“Very often”) Likert-type scale. Items on the PSS were found to be adequately reliable in this study (α = 0.87).
To assess the impact of biogenetic messaging on perceptions about depression, participants completed an additional series of self-report inventories following the saliva test:
Negative Mood Regulation Scale (NMR; Catanzaro & Mearns, 1990)
The original 30-item NMR is a measure of beliefs about the capacity to regulate emotions, behaviors, and thoughts. The full NMR scale was administered, but we used the modified scoring outlined by Lebowitz and Ahn (2018), which consists of 17 items, in an attempt to replicate their findings. The total score was internally reliable (α = 0.80). The cognitive subscale showed adequate, albeit low, internal reliability (α = 0.65), and the general subscale showed adequate reliability (α = 0.72); however, the behavioral subscale showed unacceptable internal reliability (α = 0.40).
Implicit Theories Questionnaires (ITQ; Schroder et al., 2015)
We assessed malleability beliefs for five different self-attributes: depression, anxiety, intelligence, emotion, and personality. Each of these attributes was measured with four items (except for personality, which was measured with 3 items). Participants are shown a series of fixed-minded statements (e.g., “To be honest, you cannot really do much to change how anxious you are”) and rate their agreement on a 1 (Strongly Disagree) to 6 (Strongly Agree) Likert-type scale. Items are reverse-scored such that higher scores indicate greater growth mindset endorsement. In the current study, all subscales were reliable: depression (α = 0.91), anxiety (α = 0.98), intelligence (α = 0.94), emotion (α = 0.78), and personality (α = 0.91).
Perceptions of Depression Scale (PDS; Deacon & Baird, 2009)
The PDS is a scale that was developed to assess different attitudes about depression, including stigma (e.g., “To what extent would you feel personally responsible for having developed depression?”), prognosis (e.g., “To what extent would you believe you could eventually recover from your depression?”), and treatment (e.g., “How effective would you expect medication to be in treating your depression?”). Items are rated on a 0 (Not at all) to 4 (Extremely) Likert-type scale. All three subscales displayed adequate internal reliability: stigma (α = 0.80), prognosis (α = 0.75), and treatment (α = 0.75). Higher scores indicate higher stigma endorsement, better perceived prognosis, and more optimistic attitudes about treatment, respectively.
Future depression scale (Lebowitz & Ahn, 2017)
The Future Depression Scale consisted of two items: “What do you think the odds are (from 0% to 100%) that you will experience an episode of Major Depression at some point in the future?” and “What do you think the odds are (from 0% to 100%) that your child or children will suffer from Major Depression at some point? (If you do not currently have children, please answer this question imagining that you have one or more children at some point in the future.)”
Credibility and Expectancy Questionnaire (CEQ; Devilly & Borkovec, 2000)
The CEQ is a 6-item measure assessing treatment credibility and expectancy. We administered the CEQ two times, once pertaining to medication and once pertaining to therapy (the order of the CEQ scales was counterbalanced across participants). All scores were internally reliable (therapy credibility: α = 0.81; medication credibility: α = 0.75; therapy expectancy: α = 0.88; medication expectancy: α = 0.92). Finally, participants responded to one item to assess the likelihood of initiating either therapy or medication within the next year on a 0 (Not at all likely) to 5 (Extremely likely) Likert-type scale.
Hypothetical Treatment Choice (HTQ, Cochran et al., 2008)
The HTQ is a single-item assessment of participant preferences for treatment. The item is as follows: “If you struggle with or if you were to struggle with mental health problems (e.g., anxiety, depression) and had a choice between no treatment, individual therapy, or medication to help you with your mental health problems, which would you choose?”
Beliefs about the Causes of Depression Scale (BCD; adapted from France et al., 2007)
The BCD in this study consisted of four items corresponding to different etiological beliefs about depression. Participants were asked, “How likely is it that depression might be caused by the following” and given four possible choices: “Recent or ongoing stressful experiences”; “Difficult childhood experiences”; “Chemical imbalance”; and “Genetic/inherited problems.” Items were rated on a 1 (Very unlikely) to 6 (Very likely) Likert-type scale. Each item was evaluated independently, so no internal reliability information is available.
Following study completion (immediately before debriefing), participants completed the Experiment Questionnaire/Manipulation Check, which asked what they believed the study was intended to measure and to rate their understanding of, agreement with, and perceived credibility of the saliva test on a scale of 1 (“Not at all”) to 6 (“Very much”). The latter was used to evaluate whether the saliva test deception was successful; specifically, scores >3 on this measure denoted credibility.
Flanker task
For the Flanker task (Eriksen & Eriksen, 1974), participants were seated approximately 70 cm from a computer monitor in the EEG booth. Participants were instructed to indicate (as quickly and accurately as possible) the direction of a target (center) arrow that was flanked by congruent (<<<<<) or incongruent (<<><<) arrows.
In the current study, two versions of the Flanker task were used. In the first version of the task (PsychoPy v3.0), which was completed by 21 subjects, the flanking arrows were presented alone (100 ms) and were then joined by the central arrow (50 ms) for a total stimulus duration of 150 ms. Participants completed five blocks of 70 trials (46 congruent, 24 incongruent), for a total of 350 trials (230 congruent, 120 incongruent). Due to higher-than-expected error rates on this task version, trial timing was slightly modified to improve accuracy. In the modified version (E-Prime v2.0), which was completed by the remaining 28 subjects, the flanking arrows were presented simultaneously with the central arrow (200 ms). Stimulus presentation was followed by a blank screen (950 ms) with an intertrial interval (ITI) that varied randomly from 1,150 ms to 1,650 ms. For this task, participants completed 12 blocks of 20 trials (10 congruent, 10 incongruent), for a total of 240 trials (120 congruent; 120 incongruent). To elicit a sufficient number of errors, participants were told after each block to respond faster if they achieved >75% accuracy and to slow down if their accuracy was <75%. Task differences did not emerge in any of our analyses; thus, we combined data from both tasks in these analyses. By design of the change in task, the overall accuracy for the first task (T1: M = 75.87%, SD = 13.77; T2: M = 69.59%, SD = 15.73%) was significantly lower than the second task (T1: M = 89.22%, SD = 6.50; T2: 89.67%, SD = 3.98) (t(42)s = 4.23 and 6.04, ps < 0.001, respectively). The number of errors on the first task (T1: M = 115, SD = 8.74; T2: 109.70, SD = 15.08) also was higher than the second task (T1: M = 8.33, SD = 4.68; T2: M = 5.79. SD = 2.77) (t(42)s = 51.63 and 33.17, ps < 0.001, respectively). Importantly, the Pe and ERN have been found to be reliable with a minimum of six error trials (Olvet & Hajcak, 2009a, 2009b), and ERP reliability information is presented in Table S1 in the Supplemental Material. The proportion of participants from each feedback condition did not differ between task types [χ2(1) = 0.55, p = 0.46].
Saliva test
Based on a previously established protocol (Lebowitz & Ahn, 2017, 2018), participants completed a sham saliva test. Extensive pilot testing with volunteer employees of the hospital before data collection led to the final saliva test protocol that allowed for clear instructions and research assistant blinding to the condition. The experimenter, who was blind to condition assignment, informed participants that they would complete a saliva test and provided context. Participants were then provided with the saliva testing kit (Appendix Fig. 5), which was kept in a brown paper lunch bag and included a small container of dextrose-dissolved mouthwash and a glucose test strip that participants were told gauged salivary levels of 5-Hydroxyindoleacetic acid, a metabolite of serotonin. They also were given a laptop computer, which included information about the “genetic” test, which stated that their saliva would be tested for 5-Hydroxyindoleacetic acid, and low levels were associated with increased genetic risk for MDD. Then, participants were shown instructions on how to self-administer the saliva test. Following Lebowitz and Ahn (2017), participants were instructed to rinse their mouth with the mouthwash for 7 seconds (to “eliminate impurities from your saliva and increase saliva production”) and insert the test strip under their tongue for 10 seconds. Unbeknownst to participants, the test strip was sensitive to glucose (a component of the mouthwash), causing the strip to turn brown for all participants. Participants were randomly assigned to receive computer feedback indicating that the brown test strip signifies that they either (a) have a genetic vulnerability to depression [gene-present condition] or (b) do not have a genetic vulnerability to depression [gene-absent condition]. In reality, the genetic/nongenetic feedback given to participants had been predetermined by another research assistant at the center who was uninvolved in the study and kept a password-protected file separate from all study staff. At no point in the study did study staff learn about the experimental condition for any participant until data collection was complete. The saliva test was administered immediately after the first flanker task was completed (Time 1).
Fig. 5.

Sham saliva test kit. The kit consisted of a container of mouthwash mixed with glucose, a test strip (sensitive to dextrose) and a sanitary wipe, all of which were enclosed in a plastic container inside of a brown bag. Participants were given the entire kit and asked to follow instructions on the computer to complete the saliva test and learn the results.
Psychophysiological recording and data reduction
Continuous EEG recordings were conducted in an acoustically and electrically shielded room using a high-density (96-channel), EEG data collection system (Brain Products GmbH). Participants were instructed to reduce movement as much as possible during the recordings. During data collection, Channel 1 (Cz) was used as the online reference channel. Offline processing using BrainVision Analyzer 2.0 (Brain Products) was performed to eliminate EEG artifacts. Large muscle activity and EEG activity during breaks was manually removed by visual inspection followed by band-pass filtering with cutoffs of 0.1 and 30 Hz, 24 dB/oct roll-off. Independent component analysis (ICA) removed blinks, eye movements, and electrocardiogram. Corrupted channels were interpolated using spline interpolation. Electrode recordings were re-referenced to the average activity of all electrodes.
For resting state analyses, EEG data were recorded during four minutes of eyes-closed trials immediately before and 10 min after the onset of genetic feedback. Resting state source-localization was estimated using standardized low resolution brain electromagnetic tomography (sLORETA), which calculates localization based on images of standardized current density (Grech et al., 2008). Current source density distribution was estimated on a 6,239-voxel grid at 5-mm spatial resolution. Given the exploratory nature of this study, we used whole-brain analyses to evaluate whether the genetic feedback had an impact on resting-state EEG activity (specifically with regards to the DMN).
For analyses of the ERPs during the flanker task, response-locked data were segmented into individual epochs beginning 1,500 ms before response onset and continuing 1,500 ms after the response. Epochs were rejected as artifactual if any of the following criteria were met: (a) voltage step exceeding 50 μV in 200-ms time intervals, (b) a voltage difference of more than 150 μV within a trial, or (c) a maximum voltage difference of less than 0.5 μV within a 100-ms interval. Epochs from responses with reaction times (RTs) <150 ms or >3 SDs from the intra-individually computed RT distribution were excluded.
For resting EEG analyses, we explored group differences (gene-present vs. gene-absent) at each time point (pre- and post-feedback) across all frequency bands. For analyses of the Flanker task, the Pe was the primary ERP of interest, and we evaluated the ERN as a secondary objective. For the Pe, a parietally distributed component, the average voltage potentials from Channels 34 and 35 were taken to approximate the Pz electrode, as the EEG cap did not have a true Pz electrode. The Pe was then quantified by calculating the average amplitude within the 200- to 400-ms post-response window on error trials, and the correct-trial counterpart was calculated in the same window on correct trials. The ERN was quantified as the average voltage at Channel 2 (FCz) in the 0- to 100-ms post-response window on error trials, and its correct-trial counterpart, the correct-response negativity (CRN), was measured in the same time window after correct trials. Participants with fewer than six error trials were excluded from the behavioral and ERP analyses (Olvet & Hajcak, 2009a, 2009b).
Procedure
Individuals who were interested in the study completed an online eligibility survey (Research Electronic Data Capture; REDCap; Vanderbilt University), which included detailed questions about mental health history. Eligible participants were invited to the laboratory, provided informed consent, and were asked to complete a urine drug screen. Following the screen, they completed the MINI (~1 h) and the first series of self-report questionnaires on a laptop (~30 min). Afterwards, the experimenter fitted the EEG cap and gelled each electrode onto the scalp (~45 min). Participants completed the Flanker task twice (10 min x 2) and a resting state EEG recording was performed before each task (8 min x 2; 4 min eyes-closed trials). The saliva test (10 min) was administered between the first (Time 1) and second (Time 2) task sessions. To maintain consistency, the second round of testing did not resume until exactly 10 min had passed from the onset of saliva testing (even if participants had finished early). Following the tasks, participants completed the second series of self-report questionnaires on the laptop (~30 min). The EEG cap was removed and participants were thoroughly debriefed (5-10 min). In accordance with previously established protocols (Lebowitz & Ahn, 2017), the experimenter revealed the nature of the deception, explained why it was necessary, and outlined the concept of random assignment. Before departing, participants completed a short quiz consisting of items that asked whether genetic testing occurred. If they did not respond accurately, the debriefing procedure was repeated until full comprehension was achieved. Care was taken such that all participants knew there was no genetic testing done in the testing session before leaving the facility. In total, each study session took approximately 4 h to complete.
Data analysis
For the demographics survey, three participants from the gene-absent and one participant from the gene-present condition did not complete income-related questions. Ethnicity and marital status questions were not completed by one participant in the gene-absent and gene-present conditions, respectively. Furthermore, one participant in the gene-present group did not complete the BDI-II. All post-saliva test self-report measures were completed by the full sample. Finally, two participants from each feedback condition were excluded from behavioral and ERP analyses on account of having fewer than six errors in the Flanker task (Olvet & Hajcak, 2009a, 2009b) (final sample sizes: n = 23 gene-absent; n = 22 gene-present).
All self-report measures were analyzed using either a chi-square (χ2) test for categorical variables (e.g., sex, race/ethnicity, BDI-II category) or an independent samples t-test for continuous variables (e.g., age, BDI-II score). For the CEQ, a two-way mixed-model analysis of variance (ANOVA) was conducted with Condition (gene-absent, gene-present) as the between-subjects factor and Treatment Credibility/Expectancy (therapy, medication) as the within-subjects factor. For the Flanker task, accuracy (% correct) and RT (ms; correct trials only) were recorded for each trial type (congruent, incongruent) as primary indices of cognitive control. To assess Flanker interference effects, three-way, mixed-model ANOVAs were performed for accuracy and RT using Time (Time 1 [pre-saliva test], Time 2 [post-saliva test]), and Trial Type (congruent, incongruent) as within-subjects factors and Condition (gene-absent, gene-present) as the between-subjects factor. For ERP analyses, three-way Time*Condition*Accuracy (correct, incorrect) ANOVAs were performed for the Pe and ERN, respectively. Given that some participants completed a modified version of the Flanker task, we used Task Type (Task 1 [PsychoPy], Task 2 [E-Prime]) as an additional between-subjects factor for the behavioral and ERP analyses. These analyses were conducted in addition to analyses of the full sample.
Pairwise comparisons were reported for significant interactions using Bonferroni multiple-significance-test corrections. The Bonferroni correction was applied across all tests in the current study. Whenever necessary, Greenhouse Geisser corrections were used for violations of sphericity in mixed-model ANOVAs. Finally, Cohen’s d was reported for effect size.
Results
Demographic characteristics
Demographic information is summarized in Table 1. The gene-absent and gene-present groups did not differ by age [t(47) = 0.30, p = 0.77] [Cohen’s d = 0.09], sex at birth [χ2(1) = 0.98, p = 0.32] [Phi(φ) = 0.14], income [χ2(5) = 0.85, p = 0.97] [φ = 0.13], ethnicity/race [χ2s(1) < 2.00, ps > 0.16] [φ = 0.04], or marital status [χ2(1) = 0.36, p = 0.55] [φ = 0.09]. The groups did not differ by highest level of completed schooling [χ2(7) = 5.48, p = 0.24] [φ = 0.33]; however, the gene-absent group reported a higher number of years spent in school [t(45) = 2.11, p = 0.04] [d = 0.62]. Importantly, results did not change when education (number of years) was used as a covariate for each analysis.
Table 1.
Demographic information
| Gene-absent n = 25 |
Gene-present n = 24 |
|
|---|---|---|
| Age, mean (SD) | 23.32 (5.52) | 22.88 (4.88) |
| Sex at birth, no. (%) | ||
|
Female Male |
9 (36) 16 (64) |
12 (50) 12 (50) |
| Income, no. (%) | ||
|
<$50,000 $50,000-$100,000 >$100,000 |
10 (40) 5 (20) 7 (28) |
8 (33) 6 (25) 9 (38) |
| Education (# years), mean (SD) | 14.85 (1.78)* | 13.77 (1.72) |
| Education (highest level), no. (%) | ||
|
High school Some college Junior college Four-year college Graduate/professional school |
5 (20) 6 (24) 1 (4) 11 (44) 2 (8) |
4 (17) 13 (54) 0 (0) 6 (25) 1 (4) |
| Ethnicity, no. (%) | ||
|
Hispanic/Latino Non-Hispanic/Latino |
4 (16) 20 (80) |
4 (17) 20 (83) |
| Race, no. (%) | ||
|
White Asian American Indian/Alaska Native Black/African American Native Hawaiian/other Pacific Islander Other |
13 (52) 7 (28) 2 (8) 2 (8) 0 (0) 4 (16) |
16 (67) 9 (38) 1 (4) 0 (0) 0 (0) 0 (0) |
| Marital status, No. (%) | ||
|
Never married Married Divorced |
22 (88) 2 (8) 0 (0) |
23 (96) 1 (4) 0 (0) |
Clinical characteristics
Clinical information is summarized in Table 2 (MINI) and Table 3 (self-report). As ascertained by the clinical interview (Table 2), the proportion of participants with a current or past major depressive episode, or those in remission, did not differ between conditions (χ2s < 0.42, ps > 0.52). Similarly, the number of lifetime major depressive episodes [t(40) = 1.25, p = 0.22] and the age of onset [t(35) = 0.45, p = 0.66] also did not differ between conditions. Furthermore, the proportion of participants with an anxiety disorder, substance use disorder, or eating disorder did not differ between conditions (χ2s < 3.07, ps > 0.08), nor did the severity (ts < 1.00, ps > 0.33) or age of onset (ts < 2.61, ps > 0.05) for these diagnoses. In terms of self-reported clinical characteristics (Table 3), the gene-absent and gene-present groups did not differ by BDI-II score [t(46) = 0.58, p = 0.57] or BDI-II categories [χ2(3) = 1.28, p = 0.73], nor did they differ by scores on the QIDS, BHS, RRS, MASQ, PANAS, or PSS [all ts(47) < 0.64, ps > 0.40].
Table 2.
Clinical characteristics of the sample from the MINI-5
| DSM-5 diagnosis | Gene-absent n = 25 |
Gene-present n = 24 |
|---|---|---|
| Major depressive disorder1 | ||
|
Current (%) Remission (%) Lifetime (%) No. of episodes, mean (SD) Age of onset, mean (SD) |
9 (36) 3 (12) 20 (80) 2.86 (2.96) 16.24 (4.41) |
8 (33) 2 (8) 20 (87) 5.32 (8.35) 15.65 (3.50) |
|
Anxiety disorder (%) Age of onset, mean (SD) |
9 (36) 13.00 (3.63) |
8 (33) 15.86 (8.71) |
|
Substance/alcohol use disorder (%) Severity, mean (SD) (/4) Age of onset, mean (SD) |
7 (28) 2.08 (0.80) 18.50 (0.58) |
6 (25) 2.17 (0.98) 18.40 (1.82) |
|
Eating disorder (%) Severity, mean (SD) (/4) |
2 (8) 1.67 (2.08) |
2 (8) 2.50 (1.73) |
These data were ascertained by a clinical interview using the Mini-International Neuropsychiatric Interview DSM-5 (MINI-5). 1Percentages do not add up to 100, because some participants with a current major depressive episode also had a past episode (i.e., current and lifetime). No participants in either condition met criteria for bipolar disorder, mania, or psychosis, as per exclusion criteria. Severity scores were calculated on a 4-point Likert-type scale. Anxiety disorder = panic disorder, agoraphobia, social anxiety disorder, obsessive compulsive disorder, generalized anxiety disorder. Eating disorder = bulimia, binge eating.
Table 3.
Self-reported clinical characteristics at baseline
| Measure | Gene-absent n = 25 |
Gene-present n = 24 |
|---|---|---|
| BDI-II, Mean (SD) | 19.72 (12.63) | 17.65 (12.01) |
| BDI-II Category, No. (%) | ||
| Minimal | 10 (40) | 10 (42) |
| Mild | 4 (16) | 4 (17) |
| Moderate | 5 (20) | 2 (8) |
| Severe | 6 (24) | 7 (29) |
| QIDS, Mean (SD) | ||
| Sleep | 2.00 (0.91) | 2.04 (0.86) |
| Weight | 1.36 (0.95) | 1.13 (0.99) |
| Psychomotor | 0.96 (0.75) | 1.13 (1.03) |
| Total | 10.08 (5.29) | 10.34 (5.96) |
| BHS, Mean (SD) | 8.28 (4.53) | 7.71 (5.84) |
| RRS, Mean (SD) | ||
| Brooding | 2.20 (0.79) | 2.07 (0.70) |
| Reflection | 2.06 (0.75) | 2.10 (0.54) |
| Depression | 2.39 (0.75) | 2.27 (0.59) |
| MASQ, Mean (SD) | ||
| Anhedonic Depression | 69.84 (15.15) | 68.83 (16.34) |
| Positive Affect | 51.24 (10.83) | 51.71 (10.80) |
| Loss of Interest | 18.60 (6.34) | 17.13 (6.76) |
| PANAS, Mean (SD) | ||
| Positive Affect | 2.41 (0.75) | 2.37 (0.82) |
| Negative Affect | 1.98 (0.79) | 1.94 (0.82) |
| PSS, Mean (SD) | 22.84 (6.65) | 21.63 (6.97) |
Note All clinical information was collected prior to the saliva test manipulation. One participant in the gene-present group did not complete the BDI-II. There were no statistically significant group differences on any baseline clinical scales. BDI-II Beck Depression Inventory-II; QIDS Quick Inventory of Depressive Symptomatology; BHS Beck Hopelessness Scale; RRS Ruminative Response Scale; MASQ Mood and Anxiety Symptom Questionnaire; PANAS Positive and Negative Affect Schedule; PSS Perceived Stress Scale
Effects of biogenetic messaging on self-reported perceptions of depression
Saliva test ratings
There were no group differences in ratings of understanding, credibility, or agreement with the saliva test [all ts(47) < 0.70, ps > 0.49]. There also was no correlation between credibility and BDI-II score in either feedback condition [rs < 0.08, ps > 0.71], suggesting depression severity was not related to skepticism about the legitimacy of the test.
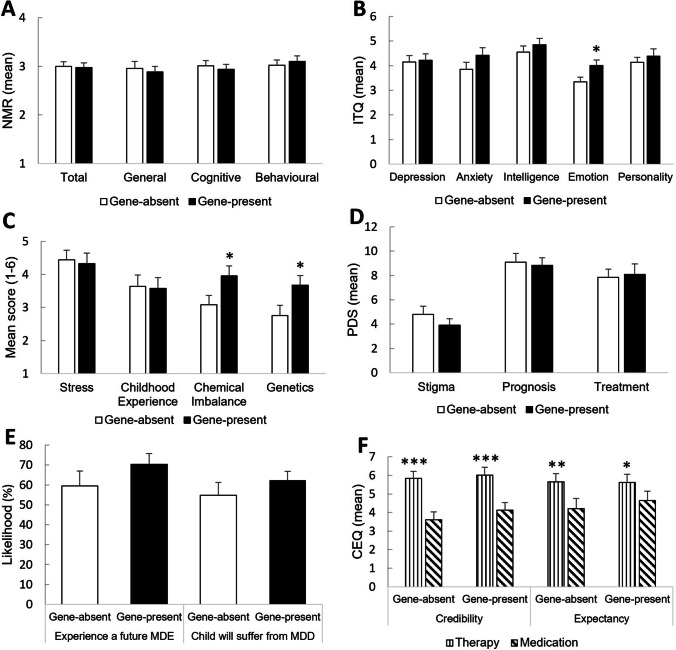
Emotion regulation and malleability beliefs
There were no group differences on mood regulation (NMR), or beliefs about intelligence, anxiety, and depression [ts(47) < 1.37, ps > 0.18]. Contrary to hypotheses, emotions were rated as more changeable in the gene-present compared to gene-absent condition [t(47) = 2.28, p = 0.03 (Cohen’s d = 0.65)].
Beliefs about the etiology of depression
As shown in Fig. 1C, there were no group differences in perceptions of environmental contributions to depression; specifically, the perception that depression is caused by recent or ongoing stress or difficult childhood experiences [ts(47) < 0.25, ps > 0.80]. As evidence of the manipulation’s effectiveness, the gene-present condition rated biological contributions as significantly more likely to cause depression than did the gene-absent group, with higher ratings for chemical imbalance [t(47) = 2.14, p = 0.04 (d = 0.61)] and genetics [t(47) = 2.11, p = 0.04 (d = 0.60)] as significant causes of depression. The gene-present group did not show lower scores on the PDS for stigma, prognosis or treatment [ts(47) < 1.02, ps > 0.31] (Fig. 1D). Relatedly, there were no group differences in perceptions about the future (Fig. 1E); specifically, self-reported likelihood of a future major depressive episode or that their children would eventually suffer from MDD [ts(47) < 1.15, ps > 0.26]. None of the nonsignificant effects were altered when excluding those who discredited the saliva test [ts < 1.19, ps > 0.25]. These data are inconsistent with our pre-registered hypothesis that the genetic condition would lead to poorer perceptions of the future.
Fig. 1.
Mean (+SEM) post-feedback perceptions of depression malleability, etiology, and treatment. (A) No group differences emerged within the general, cognitive or behavioral subscales of the Negative Mood Regulation Scale (NMR). (B) For the Implicit Theories Questionnaire (ITQ), emotions were rated as more malleable in the gene-present compared with the gene-absent group; however, there were no group differences for depression, anxiety, intelligence, or personality. (C) For perceptions of depression etiology, the gene-present group rated biological, but not environmental, contributions as significantly more likely to cause depression than did the gene‐absent group. (D) For the Perceptions of Depression Scale (PDS), there were no group differences in perceptions of stigma, prognosis, or treatment. (E) In the Future Scale, there were no group differences in perceptions about the likelihood of experiencing a future major depressive episode (MDE) or that their children would suffer from major depressive disorder (MDD). (F) For the Credibility and Expectancy Questionnaire (CEQ), both feedback conditions rated therapy as more credible and effective than medication. Statistical significance: *p < 0.05; **p < 0.01; ***p < 0.001
Treatment ratings
Figure 1F shows perceptions of treatment credibility and expectancy as measured by the CEQ. For both credibility and expectancy, there was a main effect of Treatment, such that therapy was rated more favorably than medication across both feedback conditions [Fs > 13.46, ps < 0.001]. There were no main effects of Condition or Treatment*Condition interactions [Fs < 0.51, ps > 0.48], suggesting genetic feedback did not modulate perceptions of depression treatment. The final item on the CEQ, which asked likelihood of initiating treatment, was not different between experimental groups for therapy (t(47) = 0.56, p = 0.58) or medication (t(47) = 0.98, p = 0.33).
Relatedly, when asked to choose between therapy, medication or no treatment (HTQ Scale; data not shown graphically), the majority of participants in both conditions chose therapy (gene-absent: n = 21; gene-present: n = 14) over medication (gene-absent: n = 2; gene-present: n = 4) or no treatment (gene-absent: n = 2; gene-present: n = 6). There were no differences between conditions in treatment preference (χ2 = 4.05, df = 2, p = 0.13). No additional effects emerged when excluding those who discredited the saliva test [Fs < 0.36, ps > 0.55; χ2(2) = 4.62, p = 0.10]. Together, these data are inconsistent with our pre-registered hypotheses regarding differential treatment perceptions between the two groups.
Effects of biogenetic feedback on resting EEG
To test our pre-registered hypothesis regarding resting-state EEG, we used LORETA software to evaluate whether the genetic and control conditions differed in resting-state EEG power before versus after the intervention. A 2 x 2 independent and paired samples t-test design was implemented using the Statistics package within LORETA. The model was specified as such: Genetic (T2 – T1) minus Control (T2 – T1), with larger t values corresponding to greater current source density after the genetic feedback. Our pre-registered hypothesis was that receiving the genetic feedback would lead to more activity in default mode network regions. This hypothesis was not supported in an exploratory whole-brain analysis using the seven different EEG frequency bands for resting-state data (delta, theta, alpha1, alpha2, beta1, beta2, beta3). Specifically, no significant results were found for any frequency band, correcting for 6,000 comparisons (p < 0.05), as none of the comparisons met or exceeded the critical t-value of 4.64 (all ts < 3.90). No effects emerged when restricting analyses to those who rated the saliva test as credible (critical t = 5.36, all ts < 4.65).
Effects of biogenetic feedback on behavioral correlates of cognitive control
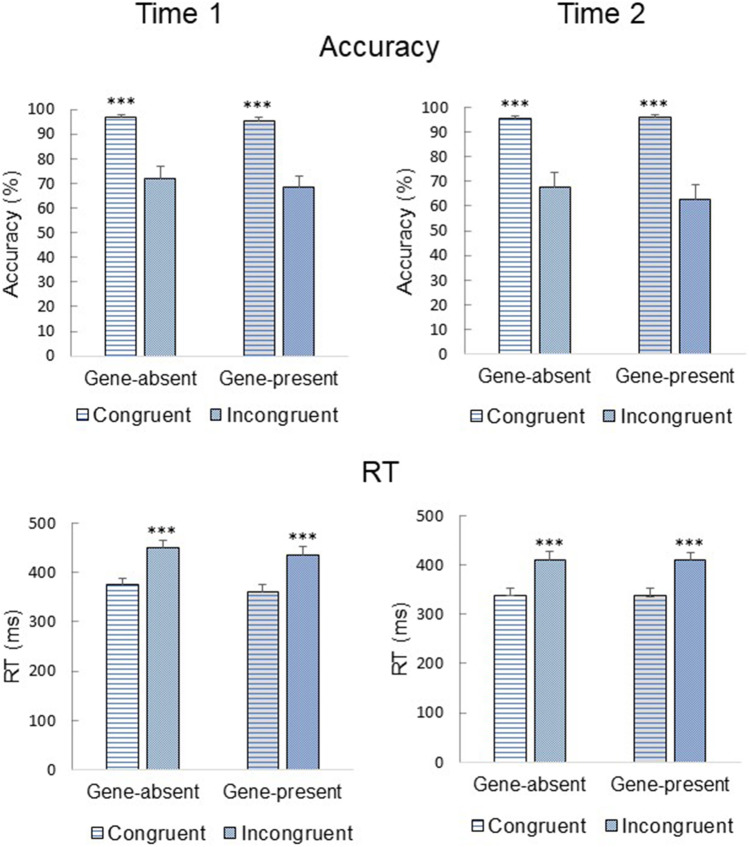
Accuracy and RT results are presented in Fig. 2. A main effect of Time emerged for both accuracy [F(1,42) = 7.76, p = 0.008] and RT [F(1,42) = 92.32, p < 0.001]. Overall, accuracy was higher at Time 1 compared with Time 2; however, RT was faster at Time 2 compared with Time 1. These data are consistent with a speed-accuracy tradeoff (faster RT and poorer accuracy) in Time 2, likely due to fatigue. A main effect of Trial Type also emerged for accuracy [F(1,42) = 74.83, p < 0.001] and RT [F(1,42) = 315.99, p < 0.001], such that incongruent accuracy was lower and RT was slower compared with congruent accuracy and RT. There were no main effects of Condition (ps > 0.56). These findings suggest that the task elicited the expected interference effects, but there was no effect of genetic feedback.
Fig. 2.
Mean (+SEM) accuracy and reaction time (RT) for congruent and incongruent trials in the Flanker task before (Time 1) and after (Time 2) receiving biogenetic feedback. For both the gene-absent and gene-present groups, accuracy was higher and RT was faster for congruent compared with incongruent trials. There were no group differences between the two time points. Statistical significance: ***p < 0.001
Effects of biogenetic feedback on electrophysiological correlates of cognitive control
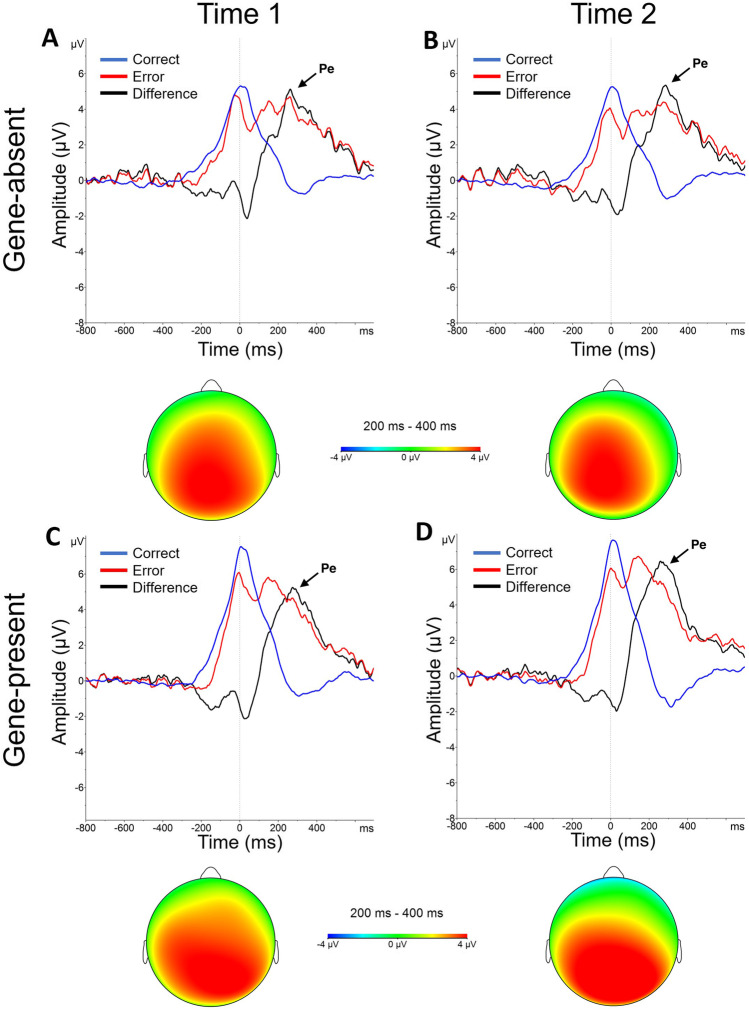
Pe
As shown in Fig. 3, a large positive deflection was observed in the 200-400 ms post-response window on error trials, consistent with the presence of a Pe. Indeed, a main effect of Accuracy emerged [F(1,42) = 122.89, p < 0.001], revealing a larger Pe for errors compared with correct responses. No additional main effects emerged (ps > 0.53). Critically, no interactions emerged (ps > 0.14), suggesting genetic feedback did not modulate the Pe. These findings are inconsistent with our pre-registered hypothesis that the Pe would be increased after the gene-present condition.
Fig. 3.
Error positivity (Pe). Response-locked, grand‐averaged Pe waveforms at electrode site Pz for the (A-B) gene-absent and (C-D) gene-present conditions before (Time 1) and after (Time 2) biogenetic feedback. Pe was larger for errors than correct responses in both feedback conditions at each time point. Lines represent correct responses (blue), errors (red) and difference waves (error - correct) (black). The vertical line at time zero reflects response onset. Scalp topographies show difference waves for Pe amplitude in the 200-400 ms post-response time window
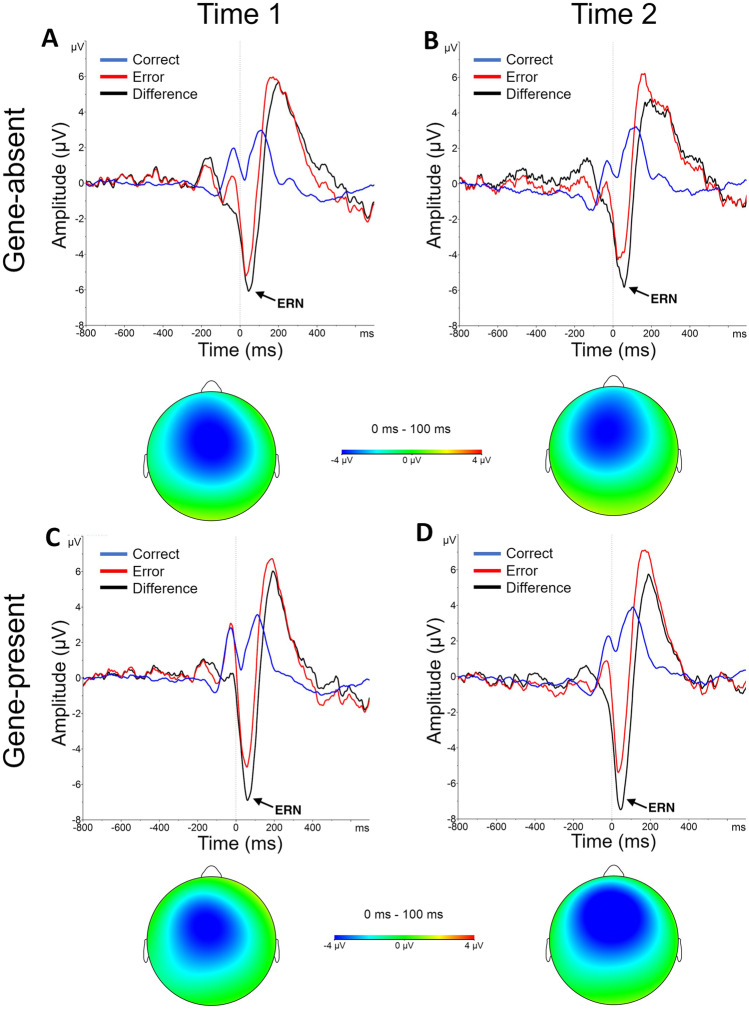
ERN
As shown in Fig. 4, a sharp negative deflection emerged in the 0-100 ms window on error trials, consistent with an ERN effect. Indeed, a main effect of Accuracy emerged [F(1,42) = 70.82, p < 0.001], revealing a larger (more negative) ERN amplitude for errors compared to correct responses, highlighting a robust ERN elicited by the task. However, no significant interactions emerged (ps > 0.32), suggesting genetic feedback did not modulate the ERN. No additional effects emerged when restricting analyses to those who rated the test as credible [Fs < 2.21, ps > 0.15].
Fig. 4.
Error-related negativity (ERN). Response‐locked, grand-averaged ERN waveforms at electrode site FCz for the (A-B) gene-absent and (C-D) gene-present conditions before (Time 1) and after (Time 2) biogenetic feedback. ERN was larger for errors than correct responses in both feedback conditions at each time point. Lines represent correct responses (blue), errors (red), and difference waves (error - correct) (black). The vertical line at time zero reflects response onset. Scalp topographies show difference waves for ERN amplitude in the 0-100 ms post-response time window
Additional Bayesian analyses for the self-report, behavioral, and ERP measures are presented in the Supplemental Material (Table S2). They demonstrate that there was moderate evidence for the null hypothesis in most cases.
Discussion
This study was designed to examine how receiving genetic feedback about depression impacted beliefs, expectations, and brain activity. Contrary to our hypotheses, personalized genetic feedback about depression had few immediate effects on participants’ beliefs about depression, treatment expectancies, or neural activity during a cognitive task. However, genetic feedback did increase the belief that depression has a biological origin (i.e., chemical imbalance or genetic predisposition) and unexpectedly increased participants’ growth mindset of emotions. We discuss possible explanations for the null findings and why they necessitate further research in this field.
We did not replicate a prior finding that biogenetic feedback promotes pessimism about the ability to alleviate or overcome depressive symptoms (as measured by the NMR; Kemp et al., 2014; Lebowitz & Ahn, 2018). Notably, the current study employed a similar saliva test as Lebowitz and Ahn (2018), but a key difference between the studies was the depression severity of the sample. In the current study, we recruited individuals with current and past MDD symptoms (allowing for BDI < 13 if past MDD was reported), resulting in a mixed sample of those with symptomatic and remitted depression. By contrast, Lebowitz and Ahn (2018) exclusively recruited individuals with symptomatic depression and a BDI-II > 13. Interestingly, effects of NMR disappeared when restricting analyses to low-scoring BDI-II participants (see Lebowitz & Ahn, 2018 supplemental materials), perhaps reflecting reduced sensitivity to genetic feedback among those with minimal depression. In theory, effects of genetic feedback on depression beliefs might be more salient among symptomatic individuals with significant current depression, but this was not supported by the current findings. In fact, additional post-hoc analyses revealed no effect of NMR when restricted to high BDI-II scorers or when using BDI-II as a covariate in these analyses (data not shown). A more plausible explanation for the discrepant findings might reflect a lack of statistical power. Indeed, some argue that samples smaller than 20 per condition are not powerful enough to detect effects in experimental psychology, contributing to Type II errors (Simmons et al., 2011). Although our sample sizes were sufficient in this context (n = 24-25 per condition), a G*Power analysis (Faul et al., 2007) revealed that a total sample of N = 190 (n = 95 per condition) would be required to achieve the small-medium effect size of 0.41 reported in Lebowitz and Ahn (2018). Thus, these discrepancies (as well as the inability to replicate the NMR effect) might reflect insufficient statistical power, perhaps underscoring a broader problem in psychological research (Simmons et al., 2011). Alternatively, the effects of biogenetic messaging may be small, consistent with meta-analytic review (Kvaale et al., 2013).
Interestingly, perceptions of self-stigma did not differ based on feedback type, contradicting a previous finding that biological explanations about depression etiology reduce self-stigma compared with cognitive-behavioral explanations (Lee et al., 2016). However, the finding that biogenetic feedback did not reduce self-stigma is consistent with other studies (Kemp et al., 2014; see Kvaale et al., 2013 for a meta-analysis) and suggests that biological explanations of depression do not always affect perceptions of stigma, as previously thought (for more, see Deacon & Baird, 2009). As a caveat, depression self-stigma can be influenced by a complex array of factors (e.g., symptom severity, race, coping abilities), as well as a protracted process of internalizing perceptions of public stigma (Kanter et al., 2008; Link & Phelan, 2001). Although one explanation for this null finding is that participants did not have enough time to process and internalize the newly acquired genetic information to have an impact on stigma, a previous study using a different saliva test feedback paradigm did find immediate post-feedback impacts on stigma (Kemp et al., 2014), suggesting this finding is nuanced and potentially sensitive to the precise manipulation.
Biogenetic feedback in the current study did not increase expectations or preferences for medication over therapy as previously shown (Kemp et al., 2014). Interestingly, both feedback conditions showed a strong preference for psychotherapy over medication, as well as a belief that therapy was more credible and effective than medication. Prior research has shown that treatments are considered more effective if the perceived etiology (e.g., low serotonin levels) is congruent with the treatment focus (e.g., selective serotonin reuptake inhibitors; Iselin & Addis, 2003). In the current study, a strong preexisting preference for therapy across both conditions may have superseded any immediate influence of genetic feedback on choice behavior. This is supported by the fact that only a small number of individuals in either condition reported a preference for medication (gene-absent: 8%, gene-present: 17%); in fact, a similar proportion of people reported a preference not to undergo any form of treatment as did a preference for medication. By contrast, most participants showed a preference for psychotherapy, which accords with a meta-analysis showing a threefold preference for psychotherapy in both treatment-seeking and nonseeking individuals (McHugh et al., 2013). Future studies should systematically evaluate and control for preexisting treatment expectancies, shedding light on whether preferences are acutely swayed by genetic feedback (Kemp et al., 2014) or if inherent biases are unyielding to these manipulations.
Unexpectedly, the gene-present condition was associated with greater endorsement of the growth mindset of emotions, suggesting participants who had just received positive genetic feedback about depression felt they could change their emotions more. This finding runs counter to most previous studies examining the impacts of biogenetic messages and beliefs on perceptions of emotional control. However, in the absence of effects on mindsets of anxiety, depression, and intelligence (and all other belief-related outcomes in this study), this finding is somewhat difficult to interpret. Previous findings suggest that the mindset of anxiety, over and above the mindset of emotion, predicts treatment outcomes, emotion regulation strategies, and symptoms of anxiety and depression (Schroder et al., 2015, 2019). Understanding the practical significance of this finding will be important for future studies to investigate.
The manipulation had no significant impact on either resting-state indices of rumination (e.g., default mode network areas) nor neural correlates of cognitive control. The current study is the first to examine neural consequences of genetic feedback, limiting our ability to interpret the null findings within the context of a broader literature. As one possibility, temporal restrictions may have impeded immediate neurophysiological effects of the feedback. Participants completed the flanker task approximately 10 minutes after learning their personalized results, which might have been insufficient to allow for internalization and impacts on cognitive control. However, several studies have shown rapid effects of mood/mindset induction paradigms on the Pe (Larson et al., 2013; Schroder et al., 2014) and ERN (Olvet & Hajcak, 2012). Alternatively, biogenetic messaging might not broadly affect all psychological attributes, warranting further research into domain-specific effects. The flanker task relies heavily on selective attention and executive control and may thus not target the emotional processing potentially impacted by genetic feedback.
The current study had several strengths, including the use of a previously validated sham saliva test, well-validated measures of resting-state EEG, cognitive control, and the use of clinical interviews, and self-report measures. Moreover, this was the first study ever to examine the neural correlates of genetic feedback, and our design and hypotheses were pre-registered prior to data collection. However, the findings should be interpreted in the context of important limitations. As outlined previously, our study was likely underpowered to detect the rather small effects of genetic manipulations (Kvaale et al., 2013). The small sample size also might be problematic given the number of tests that were conducted in the study; however, our preregistered hypotheses helped to narrow the focus of these tests and we used robust Bonferroni corrections to correct against false positives. Relatedly, concerns surrounding response priming precluded a within-subjects assessment of the self-report data. Specifically, we only measured post-feedback effects to maintain the integrity of our genetic manipulation. Furthermore, our sample was racially and ethnically homogenous and heavily represented by participants identifying as white (>50% per condition). This is relevant because, according to past research, minoritized patients are significantly less likely to endorse biological causal attributions for depression (Khalsa et al., 2011; Schnittker et al., 2000) and more likely to prefer therapy over medication (Givens et al., 2007). Second, we implemented two versions of the Flanker task, with half of our sample completing a more challenging version (Task 1). Statistically, our effects did not differ based on task type; however, it is possible that the more difficult task elicited changes in mood. Finally, experimentally manipulating genetic feedback is a relatively novel and underused method in this field, necessitating further insight into participants’ perceptions and beliefs about sham genetic tests. Interestingly, our results did not change when excluding those who discredited (i.e., did not believe) the genetic test. Recent findings lend support to this phenomenon. Specifically, treatment preferences did not differ between those who were and were not skeptical of a sham “depression screening test” if they were given a biological explanation for depression (Salem et al., 2019). Paradoxically, skeptics given a biopsychosocial explanation for depression were more likely to accept therapy compared to medication, but there were no differences in treatment preference among nonskeptics (Salem et al., 2019). As the authors note, this unexpected finding requires replication to further understand its meaning and practical significance (Salem et al., 2019). To be sure, the budding literature on sham genetic test results and its impact on beliefs, preferences, and decision-making is nuanced.
Conclusions
There are mounting concerns surrounding the recent shift in public narratives about biogenetic explanations of mental illness. Among those with depression, these messages have reportedly increased prognostic pessimism and reduced confidence in treatment outcomes. In the current study, biogenetic feedback did not alter beliefs or expectancies about depression, nor did it affect neural correlates of cognitive control. As this area of inquiry is relatively new, our findings should be interpreted within the context of study limitations. We advocate for future research to replicate prior findings and investigate domain-specific effects of genetic feedback on neurophysiology. Understanding how biogenetic messaging affects perceptions and expectancies about depression could inform clinical interventions that mitigate its potential negative consequences.
Supplementary Information
(DOCX 33 kb)
Acknowledgments
The authors thank Monica Landi and Elena Molokotos for help in conducting the clinical interviews, and Micah Breiger, Rachel Lobien and Sara Perlo for help in blinding the randomization procedure. We also thank Samantha Linton and Dan Dillon for their assistance with the behavioral data analysis.
Financial disclosures
During the past 3 years, Dr. Pizzagalli has received consulting fees from Albright Stonebridge Group, Boehringer Ingelheim, Compass Pathways, Engrail Therapeutics, Neumora Therapeutics (formerly BlackThorn Therapeutics), Neurocrine Biosciences, Neuroscience Software, Otsuka, Sunovion, and Takeda; he has received honoraria from the Psychonomic Society and American Psychological Association (for editorial work) and from Alkermes; he has received research funding from the Brain and Behavior Research Foundation, the Dana Foundation, Millennium Pharmaceuticals, NIMH, and Wellcome Leap; he has received stock options from Compass Pathways, Engrail Therapeutics, Neumora Therapeutics, and Neuroscience Software. No funding from these entities was used to support the current work, and all views expressed are solely those of the authors. Dr. Schroder has received consulting fees from Loom. All other authors have no conflicts of interest or relevant disclosures.
Appendix
Funding
Funding from this study came from the Andrew Merrill Memorial Fund from McLean Hospital awarded to HSS.
Footnotes
Open practices statement
This study design and hypotheses were pre-registered at https://clinicaltrials.gov/ct2/show/NCT03998748. Data are available on the Open Science Framework: https://osf.io/v6mpx/.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Beck A, Weissman A, Lester D, Trexler L. The measurement of pessimism: The hopelessness scale. Journal of Consulting and Clinical Psychology. 1974;42:861–865. doi: 10.1037/h0037562. [DOI] [PubMed] [Google Scholar]
- Beck, A., Steer, R., & Brown, G. (1996). Beck depression inventory-II. San Antonio.
- Burnette JL, Knouse LE, Vavra DT, O'Boyle E, Brooks MA. Growth mindsets and psychological distress: A meta-analysis. Clinical Psychology Review. 2020;77:1–13. doi: 10.1016/j.cpr.2020.101816. [DOI] [PubMed] [Google Scholar]
- Catanzaro SJ, Mearns J. Measuring generalized expectancies for negative mood regulation: Initial scale development and implications. Journal of Personality Assessment. 1990;54(3-4):546–563. doi: 10.1080/00223891.1990.9674019. [DOI] [PubMed] [Google Scholar]
- Chapman L, Chapman J. The measurement of handedness. Brain and Cognition. 1987;6:175–183. doi: 10.1016/0278-2626(87)90118-7. [DOI] [PubMed] [Google Scholar]
- Cochran BN, Pruitt L, Fukuda S, Zoellner LA, Feeny NC. Reasons underlying treatment preference: An exploratory study. Journal of Interpersonal Violence. 2008;23:275–291. doi: 10.1177/0886260507309836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. Journal of Health and Social Behavior. 1983;24:386–396. doi: 10.2307/2136404. [DOI] [PubMed] [Google Scholar]
- Dar-Nimrod I, Heine SJ. Genetic essentialism: On the deceptive determinism of DNA. Psychological Bulletin. 2011;137(5):800–818. doi: 10.1037/a0021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon BJ, Baird GL. The chemical imbalance explanation of depression: Reducing blame at what cost? Journal of Social and Clinical Psychology. 2009;28(4):415–435. doi: 10.1521/jscp.2009.28.4.415. [DOI] [Google Scholar]
- Devilly GJ, Borkovec TD. Psychometric properties of the credibility/expectancy questionnaire. Journal of behavior therapy and experimental psychiatry. 2000;31(2):73–86. doi: 10.1016/S0005-7916(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Dweck CS. Self-theories: Their role in motivation, personality, and development. Psychology press; 1999. [PubMed] [Google Scholar]
- Dweck CS, Chiu CY, Hong YY. Implicit theories and their role in judgments and reactions: A word from two perspectives. Psychological Inquiry. 1995;6(4):267–285. doi: 10.1207/s15327965pli0604_1. [DOI] [Google Scholar]
- Eriksen BA, Eriksen CW. Effects of noise letters upon the identification of a target letter in a nonsearch task. Perception & Psychophysics. 1974;16(1):143–149. doi: 10.3758/BF03203267. [DOI] [Google Scholar]
- Falkenstein M, Hoormann J, Christ S, Hohnsbein J. ERP components on reaction errors and their functional significance: A tutorial. Biological Psychology. 2000;51(2-3):87–107. doi: 10.1016/S0301-0511(99)00031-9. [DOI] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A. G* power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior Research Methods. 2007;39(2):175–191. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
- France CM, Lysaker PH, Robinson RP. The chemical imbalance explanation for depression: Origins, lay endorsement, and clinical implications. Professional Psychology: Research and Practice. 2007;38(4):411. doi: 10.1037/0735-7028.38.4.411. [DOI] [Google Scholar]
- Gehring WJ, Goss B, Coles MG, Meyer DE, Donchin E. A neural system for error detection and compensation. Psychological Science. 1993;4(6):385–390. doi: 10.1111/j.1467-9280.1993.tb00586.x. [DOI] [Google Scholar]
- Givens JL, Houston TK, Van Voorhees BW, Ford DE, Cooper LA. Ethnicity and preferences for depression treatment. General Hospital Psychiatry. 2007;29(3):182–191. doi: 10.1016/j.genhosppsych.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Grech R, Cassar T, Muscat J, Camilleri KP, Fabri SG, Zervakis M, Vanrumste B. Review on solving the inverse problem in EEG source analysis. Journal of Neuroengineering and Rehabilitation. 2008;5(1):1–33. doi: 10.1186/1743-0003-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam N, Kvaale EP. Biogenetic explanations of mental disorder: The mixed-blessings model. Current Directions in Psychological Science. 2015;24(5):399–404. doi: 10.1177/0963721415588082. [DOI] [Google Scholar]
- Haslam N, Bastian B, Bain P, Kashima Y. Psychological essentialism, implicit theories, and intergroup relations. Group Processes Intergroup Relations. 2006;9:63–76. doi: 10.1177/1368430206059861. [DOI] [Google Scholar]
- Iselin MG, Addis ME. Effects of etiology on perceived helpfulness of treatments for depression. Cognitive Therapy and Research. 2003;27(2):205–222. doi: 10.1023/A:1023513310243. [DOI] [Google Scholar]
- Kanter JW, Rusch LC, Brondino MJ. Depression self-stigma: A new measure and preliminary findings. The Journal of Nervous and Mental Disease. 2008;196(9):663–670. doi: 10.1097/NMD.0b013e318183f8af. [DOI] [PubMed] [Google Scholar]
- Kelly RB, Zyzanski SJ, Alemagno SA. Prediction of motivation and behavior change following health promotion: Role of health beliefs, social support, and self-efficacy. Social Science & Medicine. 1991;32(3):311–320. doi: 10.1016/0277-9536(91)90109-P. [DOI] [PubMed] [Google Scholar]
- Kemp JJ, Lickel JJ, Deacon BJ. Effects of a chemical imbalance causal explanation on individuals' perceptions of their depressive symptoms. Behaviour Research and Therapy. 2014;56:47–52. doi: 10.1016/j.brat.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Khalsa SR, McCarthy KS, Sharpless BA, Barrett MS, Barber JP. Beliefs about the causes of depression and treatment preferences. Journal of Clinical Psychology. 2011;67(6):539–549. doi: 10.1002/jclp.20785. [DOI] [PubMed] [Google Scholar]
- Kvaale EP, Haslam N, Gottdiener WH. The ‘side effects’ of medicalization: A meta-analytic review of how biogenetic explanations affect stigma. Clinical Psychology Review. 2013;33(6):782–794. doi: 10.1016/j.cpr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Larson MJ, Steffen PR, Primosch M. The impact of a brief mindfulness meditation intervention on cognitive control and error-related performance monitoring. Frontiers in Human Neuroscience. 2013;7:308–320. doi: 10.3389/fnhum.2013.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz MS, Ahn WK. Emphasizing malleability in the biology of depression: Durable effects on perceived agency and prognostic pessimism. Behaviour Research and Therapy. 2015;71:125–130. doi: 10.1016/j.brat.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz MS, Ahn WK. Testing positive for a genetic predisposition to depression magnifies retrospective memory for depressive symptoms. Journal of Consulting and Clinical Psychology. 2017;85(11):1052–1063. doi: 10.1037/ccp0000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz MS, Ahn WK. Blue genes? Understanding and mitigating negative consequences of personalized information about genetic risk for depression. Journal of Genetic Counseling. 2018;27(1):204–216. doi: 10.1007/s10897-017-0140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz MS, Appelbaum PS. Biomedical explanations of psychopathology and their implications for attitudes and beliefs about mental disorders. Annual Review of Clinical Psychology. 2019;15:555–577. doi: 10.1146/annurev-clinpsy-050718-095416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz MS, Ahn WK, Nolen-Hoeksema S. Fixable or fate? Perceptions of the biology of depression. Journal of Consulting and Clinical Psychology. 2013;81(3):518–527. doi: 10.1037/a0031730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz, M. S., Dolev-Amit, T., & Zilcha-Mano, S. (2021). Relationships of biomedical beliefs about depression to treatment-related expectancies in a treatment-seeking sample. Psychotherapy [preprint accessed Jun 2021]. 10.1037/pst0000320. [DOI] [PMC free article] [PubMed]
- Lee AA, Farrell NR, McKibbin CL, Deacon BJ. Comparing treatment relevant etiological explanations for depression and social anxiety: Effects on self-stigmatizing attitudes. Journal of Social and Clinical Psychology. 2016;35(7):571–588. doi: 10.1521/jscp.2016.35.7.571. [DOI] [Google Scholar]
- Link BG, Phelan JC. Conceptualizing stigma. Annual review of Sociology. 2001;27(1):363–385. doi: 10.1146/annurev.soc.27.1.363. [DOI] [Google Scholar]
- McHugh RK, Whitton SW, Peckham AD, Welge JA, Otto MW. Patient preference for psychological vs pharmacologic treatment of psychiatric disorders: A meta-analytic review. The Journal of Clinical Psychiatry. 2013;74(6):595–602. doi: 10.4088/JCP.12r07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WR. Motivation for treatment: A review with special emphasis on alcoholism. Psychological Bulletin. 1985;98(1):84–107. doi: 10.1037/0033-2909.98.1.84. [DOI] [PubMed] [Google Scholar]
- Moser JS, Schroder HS, Heeter C, Moran TP, Lee YH. Mind your errors: Evidence for a neural mechanism linking growth mind-set to adaptive posterror adjustments. Psychological Science. 2011;22(12):1484–1489. doi: 10.1177/0956797611419520. [DOI] [PubMed] [Google Scholar]
- Olvet DM, Hajcak G. Reliability of error-related brain activity. Brain Research. 2009;1284:89–99. doi: 10.1016/j.brainres.2009.05.079. [DOI] [PubMed] [Google Scholar]
- Olvet DM, Hajcak G. The stability of error-related brain activity with increasing trials. Psychophysiology. 2009;46(5):957–961. doi: 10.1111/j.1469-8986.2009.00848.x. [DOI] [PubMed] [Google Scholar]
- Olvet DM, Hajcak G. The error-related negativity relates to sadness following mood induction among individuals with high neuroticism. Social Cognitive and Affective Neuroscience. 2012;7(3):289–295. doi: 10.1093/scan/nsr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescosolido BA, Martin JK, Long JS, Medina TR, Phelan JC, Link BG. “A disease like any other”? A decade of change in public reactions to schizophrenia, depression, and alcohol dependence. American Journal of Psychiatry. 2010;167(11):1321–1330. doi: 10.1176/appi.ajp.2010.09121743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli DA. Frontocingulate dysfunction in depression: Toward biomarkers of treatment response. Neuropsychopharmacology. 2011;36(1):183–206. doi: 10.1038/npp.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderinkhof KR, Ramautar JR, Wijnen JG. To PE or not to PE: A P3-like ERP component reflecting the processing of response errors. Psychophysiology. 2009;46(3):531–538. doi: 10.1111/j.1469-8986.2009.00790.x. [DOI] [PubMed] [Google Scholar]
- Rush, A. J., Trivedi, M. H., Ibrahim, H. M., Carmody, T. J., Arnow, B., Klein, D. N., ... & Keller, M. B. (2003). The 16-Item Quick Inventory of Depressive Symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): A psychometric evaluation in patients with chronic major depression Biological Psychiatry, 54(5), 573–583. [DOI] [PubMed]
- Salem T, Winer ES, Jordan DG, Dorr MM. Doubting the diagnosis but seeking a talking cure: An experimental investigation of causal explanations for depression and willingness to accept treatment. Cognitive Therapy and Research. 2019;43(6):971–985. doi: 10.1007/s10608-019-10027-w. [DOI] [Google Scholar]
- Schnittker J, Freese J, Powell B. Nature, nurture, neither, nor: Black-white differences in beliefs about the causes and appropriate treatment of mental illness. Social Forces. 2000;78(3):1101–1132. doi: 10.2307/3005943. [DOI] [Google Scholar]
- Schroder HS. Mindsets in the clinic: Applying mindset theory to clinical psychology. Clinical Psychology Review. 2021;101957:1–16. doi: 10.1016/j.cpr.2020.101957. [DOI] [PubMed] [Google Scholar]
- Schroder HS, Dawood S, Yalch MM, Donnellan MB, Moser JS. The role of implicit theories in mental health symptoms, emotion regulation, and hypothetical treatment choices in college students. Cognitive Therapy and Research. 2015;39(2):120–139. doi: 10.1007/s10608-014-9652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder HS, Duda JM, Christensen K, Beard C, Björgvinsson T. Stressors and chemical imbalances: Beliefs about the causes of depression in an acute psychiatric treatment sample. Journal of Affective Disorders. 2020;276:537–545. doi: 10.1016/j.jad.2020.07.061. [DOI] [PubMed] [Google Scholar]
- Schroder HS, Fisher ME, Lin Y, Lo SL, Danovitch JH, Moser JS. Neural evidence for enhanced attention to mistakes among school-aged children with a growth mindset. Developmental Cognitive Neuroscience. 2017;24:42–50. doi: 10.1016/j.dcn.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder HS, Kneeland ET, Silverman AL, Beard CB, Bjorgvinsson T. Beliefs about the malleability of anxiety and general emotions and their relation to treatment outcomes in acute psychiatric treatment. Cognitive Therapy and Research. 2019;43:312–323. doi: 10.1007/s10608-018-9985-7. [DOI] [Google Scholar]
- Schroder HS, Moran TP, Donnellan MB, Moser JS. Mindset induction effects on cognitive control: A neurobehavioral investigation. Biological Psychology. 2014;103:27–37. doi: 10.1016/j.biopsycho.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavas J, Weiller E, Dubnar GC. The Mini-international neuropsychiatric interview (M.I.N.I): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. The Journal of Clinical Psychiatry. 1998;59:22–33. [PubMed] [Google Scholar]
- Simmons JP, Nelson LD, Simonsohn U. False-positive psychology: Undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological Science. 2011;22(11):1359–1366. doi: 10.1177/0956797611417632. [DOI] [PubMed] [Google Scholar]
- Treynor W, Gonzalez R, Nolen-Hoeksema S. Rumination reconsidered: A psychometric analysis. Cognitive Therapy and Research. 2003;27(3):247–259. doi: 10.1023/A:1023910315561. [DOI] [Google Scholar]
- Watson D, Clark LA, Tellegen A. Development and validation of brief measures of positive and negative affect: The PANAS scales. Journal of Personality and Social Psychology. 1988;54:1063–1070. doi: 10.1037/0022-3514.54.6.1063. [DOI] [PubMed] [Google Scholar]
- Watson D, Weber K, Assenheimer JS, Clark LA, Strauss ME, McCormick RA. Testing a tripartite model: I. evaluating the convergent and discriminant validity of anxiety and depression symptom scales. Journal of Abnormal Psychology. 1995;104(1):3–14. doi: 10.1037/0021-843X.104.1.3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 33 kb)