Abstract
How BAK and BAX induce mitochondrial outer membrane (MOM) permeabilization (MOMP) during apoptosis is incompletely understood. Here we have used molecular dynamics simulations, surface plasmon resonance, and assays for membrane permeabilization in vitro and in vivo to assess the structure and function of selected BAK subdomains and their derivatives. Results of these studies demonstrate that BAK helical regions α5 and α6 bind the MOM lipid cardiolipin. While individual peptides corresponding to these helical regions lack the full biological activity of BAK, tandem peptides corresponding to α4–α5, α5–α6, or α6–α7/8 can localize exogenous proteins to mitochondria, permeabilize liposomes composed of MOM lipids, and cause MOMP in the absence of the remainder of the BAK protein. Importantly, the ability of these tandem helices to induce MOMP under cell-free conditions is diminished by mutations that disrupt the U-shaped helix-turn-helix structure of the tandem peptides or decrease their lipid binding. Likewise, BAK-induced apoptosis in intact cells is diminished by CLS1 gene interruption, which decreases mitochondrial cardiolipin content, or by BAK mutations that disrupt the U-shaped tandem peptide structure or diminish lipid binding. Collectively, these results suggest that BAK structural rearrangements during apoptosis might mobilize helices involved in specific protein-lipid interactions that are critical for MOMP.
Subject terms: Biochemistry, Physiology
Introduction
The pro-apoptotic BCL2 family members BAK and BAX play critical roles in cellular life and death decisions [1–4]. These two proteins are present as inactive monomers in healthy cells, with BAK tethered to the mitochondrial outer membrane (MOM) and BAX in the cytoplasm [5]. Upon activation by BH3-only proteins or release from anti-apoptotic BCL2 family members [6–11], BAK and BAX directly mediate MOM permeabilization (MOMP), thereby releasing cytochrome c and other mitochondrial intermembrane proteins to trigger caspase activation [12–14]. Despite extensive study, the structural and functional changes that accompany BAK or BAX activation and lead to MOMP remain incompletely understood [15, 16].
Earlier studies demonstrated that BAK and BAX monomers are globular proteins consisting of a central hydrophobic helix (α5) surrounded by eight additional helices [17, 18]. Monomeric BAK consists of five long helices (α1, α2, α3, α4, and α6) that form a circle around α5 and two short helices (α7 and α8) that join the rest of the protein to α9, which is thought to span the MOM [17]. The major structural difference between monomeric BAK and BAX is the orientation of helix α9, which extends away from the remainder of the globular BAK protein but is buried in a hydrophobic groove formed by helices α3, α4, and α5 of BAX [18]. These structural differences are thought to explain why BAK is constitutively localized to the cytoplasmic surface of the MOM and BAX resides in cytoplasm until activation is triggered [5, 19]. Importantly, however, BAK does not need to be tethered to the MOM to permeabilize it. Under cell-free conditions, BAK lacking α9 can permeabilize liposomes or the MOM upon activation [8, 20–22].
A major unresolved question is how activated BAK and BAX cause MOMP. At least four models that emphasize different aspects of BAX/BAK biology have been proposed: i) insertion of BAX hydrophobic regions into the MOM to form a discrete channel [23, 24], ii) collapse of BAX or BAK hydrophobic regions consisting of helices α5 and α6 onto the MOM surface to disrupt lipid–lipid interactions [25], iii) formation of a clamp-like structure consisting of α6 peptides from two different BAX monomers in an activated dimer that pulls lipids away from a central hole to rent the MOM [26, 27], and iv) aggregation of BAK dimers to form disordered clusters that rupture the MOM [28].
Several structural studies have provided further insight into BAK activation and action. First, the structure of BAK bound to the BID BH3 domain revealed that ligation of the BAK BH3 binding groove is associated with a structural rearrangement that separates the α6–α8 segment from the α2–α5 core [29]. Second, crystal structures of BAK α2–α5 dimers fused to enhanced green fluorescent protein (EGFP) [29] or alone [PBD accession 4U2V, see also ref. 30] revealed the presence of symmetric BH3-in-groove dimers of core regions, consistent with earlier biochemical analysis [31]. More recently, a detergent-induced BAK α2-α8 dimer was reported to contain a BH3-in groove dimer of core subdomains with extended α6-α8 regions [22]. Although this detergent-induced dimer was able to bind to and permeabilize the MOM, the mechanistic basis for the MOMP and the role of the α6-α8 region in this process were not examined.
Here we focus on the functional properties of a fragment containing four helices in the C-terminal half of BAK (α4, α5, α6, and the region containing α7 and α8, which we abbreviate “α7/8”). Our binding studies show that two of these BAK helices can bind cardiolipin (CL), a mitochondrial phospholipid that has previously been detected at contact sites between the mitochondrial inner and outer membranes [32] and has been implicated in binding of truncated BID to the MOM [33, 34]. Moreover, subdomains containing two of these helices in tandem, i.e., α4–α5, α5–α6, or α6–α7/8, retain critical features of activated BAK, including strong MOM binding, permeabilization of CL-containing membranes, release of cytochrome c, and cell killing. Conversely, depletion of CL inhibits BAK-mediated membrane permeabilization under cell-free conditions and in intact cells, highlighting the potential importance of BAK/CL interactions to MOMP.
Results
BAK auto-activation in intact cells
Our previous studies indicated that purified, recombinant BAK is able to bind to itself, leading to multimerization and activation of its membrane permeabilizing activity at micromolar concentrations in vitro [35]. To further assess the ability of BAK to undergo autoactivation, we interrupted the genes encoding BAX, BAK, NOXA, PUMA, BIM, BID, HRK, and BMF in the human T cell leukemia line Jurkat and then expressed the human BAK cDNA in a doxycycline-inducible manner (Fig. S1a). As BAK increased, the percentage of BAK present in oligomers also increased (Fig. S1b). This oligomerization occurred in the absence of pro-apoptotic stimuli or expression of known direct activators and was accompanied by increased binding to MCL1 and BCLXL (Fig. S1c), release of cytochrome c (cyto c) from mitochondria (Fig. S1d) and binding of annexin V (Fig. S1e), suggesting BAK activation, MOMP, and induction of apoptosis. Combined with our previous results obtained under cell-free conditions, these observations provide the starting point for examining MOMP by various BAK constructs in vitro and in vivo.
BAKΔΤΜ-induced MOMP parallels MOM binding
To further study MOMP, we removed BAK α9 to generate a protein that is soluble in aqueous buffers in the unactivated state [8, 20, 21] and replaced the two endogenous cysteines with serines to yield the cysteine-less protein BAK2SΔTM. Like wildtype BAK without α9 [35], BAK2SΔTM released cyto c from isolated mitochondria whether reducing agent was present or not (Fig. 1a). We then introduced cysteines in neighboring positions on adjacent helices and assessed the impact of cross-linking on ability of BAK2SΔTM to bind and permeabilize mitochondria from Bax−/−/Bak−/− MEFs.
Fig. 1. Activation-associated binding of BAKΔTM to the MOM requires movement of multiple helices.
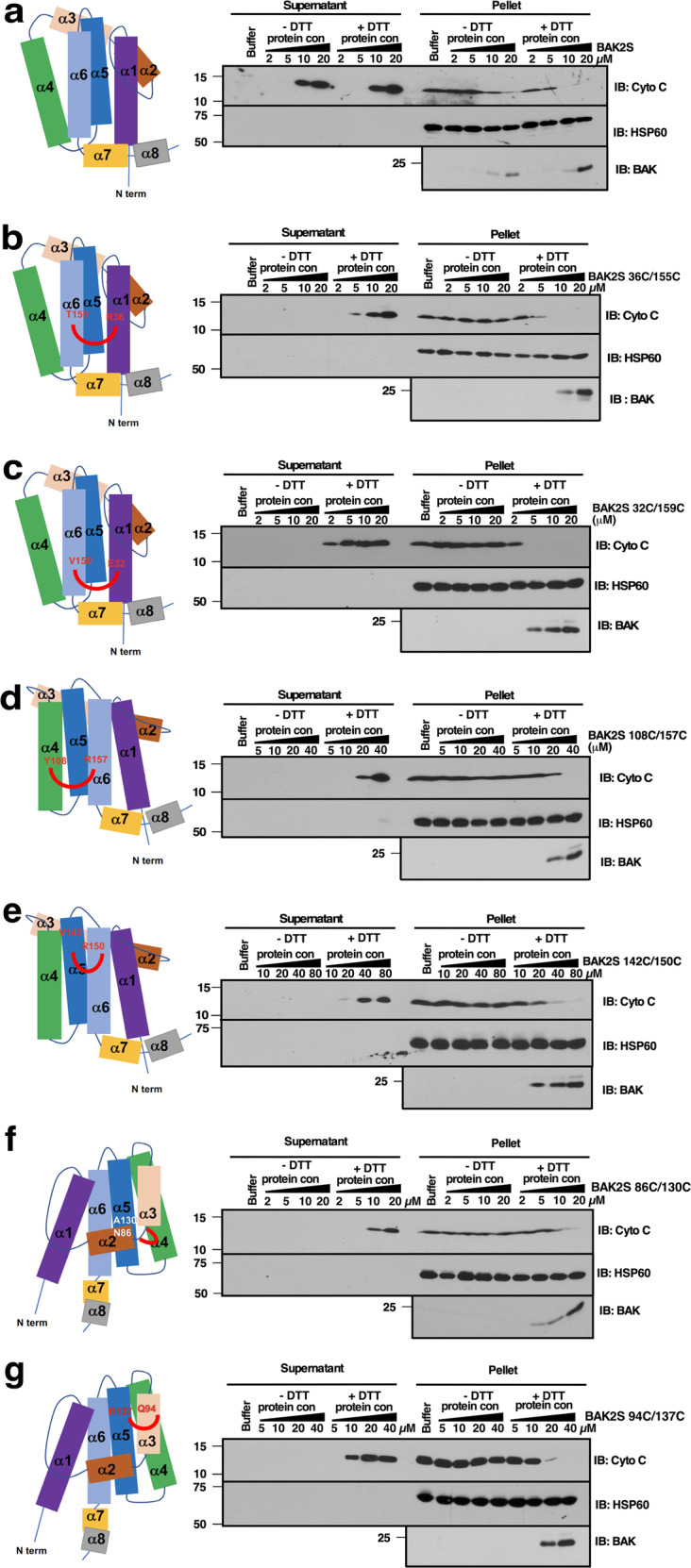
Mitochondria from Bax−/−Bak−/− MEFs were incubated at 25 °C for 90 min with different BAKΔTM (BAK 1-186) mutant proteins (a BAK2SΔTM; b BAK2SΔTM 36C/155C; c BAK2SΔTM 32C/159C; d BAK2SΔTM 108C/157C; e BAK2SΔTM 142C/150C; f BAK2SΔTM 86C/130C; g BAK2SΔTM 94C/137C) at the indicated concentrations under nonreducing (− DTT) or reducing conditions (+ DTT, pre-incubated with DTT for 2 h). The supernatants and pellets were then subjected to SDS-PAGE and immunoblotting. Left panels, diagrams of the BAKΔTM structure showing locations of amino acids (red or white lettering) that were substituted with cysteines so crosslinks (depicted by red curves) could be introduced under nonreducing conditions. See also Fig. S2.
Introduction of cysteines at two positions along α6 and corresponding positions along α1 [36] yielded BAK2SΔTM 36C/155C and BAK2SΔTM 32C/159C (left panels, Fig. 1b, c). When dithiothreitol was added to reduce spontaneously formed cross-links, these BAK constructs became stably bound to mitochondria (bottom panels, Fig. 1b, c) and released cyto c (right panels, Fig. 1b, c) despite the absence of the classical transmembrane domain. In contrast, in the absence of reducing agent these BAK2SΔTM derivatives exhibited enhanced mobility, consistent with a more compact structure (Fig. S2), and failed to stably bind to mitochondria or induce MOMP. BAK2SΔTM also failed to stably bind mitochondria or induce MOMP under nonreducing conditions when juxtaposed cysteines were used to tether α6 to α4 or α5 (Fig. 1d, e) or α5 to α2 or α3 (Fig. 1f, g). These observations raised the possibility that structural rearrangements accompanying BAK activation might be important for BAK/MOM interactions that are required for MOMP. This possibility prompted us to examine BAK/MOM interactions in greater detail.
Multiple BAK helices are capable of interacting with the MOM
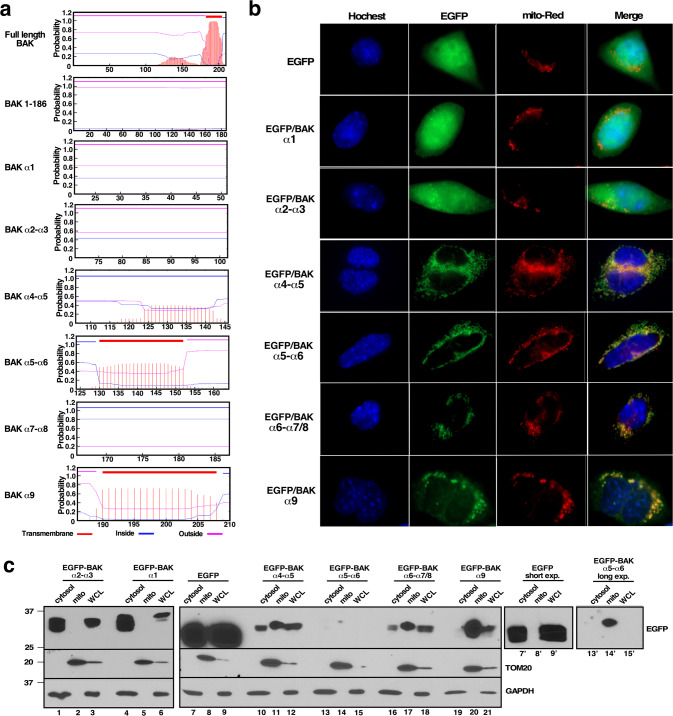
Previous studies have focused extensively on BAK α5 [26, 27] and BAK α5-α6 [25] as potential MOM binding regions. Our in silico analysis suggested that the potential lipid binding region of BAK might be more extensive (Fig. 2a). In addition to the established transmembrane domain BAK α9 (amino acids 187–211), a second region encompassing amino acids 115 to 178 also appeared positive in this analysis (Fig. 2a, top panel). In particular, regions of BAK α4-α5 (amino acids 105–146) and α5–α6 (amino acids 124–164) were also predicted to interact with membranes (Fig. 2a, 5th and 6th panels). In contrast, other helices such as α1 and α2–α3 were not predicted to be membrane-inserting regions (Fig. 2a).
Fig. 2. BAK helical fragments other than α9 target mitochondria.
a Full length BAK, BAKΔTM (1-186), and BAK helices α1 (aa 21–51), α2–α3 (aa 70–101), α4–α5 (aa 105–146), α5–α6 (aa 124–164), α7–α8 (aa 165–186) were analyzed using the TMHMM program (www.cbs.dtu.dk/services/TMHMM/), and the probability of each particular 20 aa sequence being associated with a membrane as a transmembrane sequence was calculated. BAK α9 (aa 187-210), which is known to form a transmembrane domain [60], served as a positive control in these calculations. b After cDNAs encoding the indicated BAK helices fused to the C-terminus of EGFP were transfected into Bax−/−Bak−/− MEFs, cells were incubated in the presence of Q-VD-OPh and bortezomib, which was added to stabilize the fusion protein as illustrated in Figs. S7 and S8. After 24 h, samples were analyzed by confocal microscopy for colocalization of EGFP and MitoTracker Red. c cDNAs encoding the indicated BAK peptides fused to the C-terminus of EGFP were transfected into BAX−/−BAK−/− HCT116 cells. After a 24-h incubation in the presence of Q-VD-OPh and bortezomib, cytosol and mitochondria (mito) were separated and analyzed along with whole cell lysates (WCL). Gels were loaded with 50 µg WCL or cytosolic protein and 20 µg mitochondrial protein. Blots that were exposed and probed simultaneously are shown as two panels because the number of lanes exceeds the capacity to run on a single gel. Lanes 7′-9′, shorter exposure of EGFP blot shown in lanes 7–9. Lanes 13′-15′, longer exposure of EGFP blot shown in lanes 13–15.
To test these predictions, we expressed cDNAs encoding helices α1, α2-α3, α4-α5, α5-α6, or α6-α7/8 fused to the C-terminus of EGFP in Bax−/−/Bak−/− MEFs. Fluorescence microscopy demonstrated that α4-α5, α5-α6 or α6-α7/8 targeted EGFP to a compartment that colocalized with Mitotracker red (Figs. 2b and S3). Moreover, the association was stable enough to permit recovery of a substantial portion of each fusion protein with mitochondria during cell fractionation (Fig. 2c, lanes 11, 14′ and 17).
BAK tandem peptides release cyto c and induce cell death
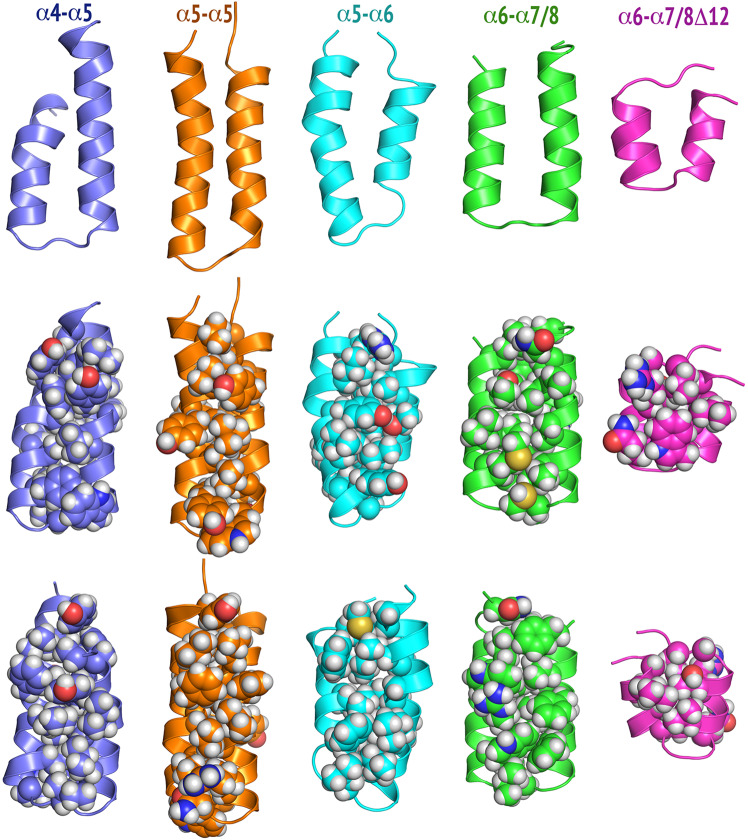
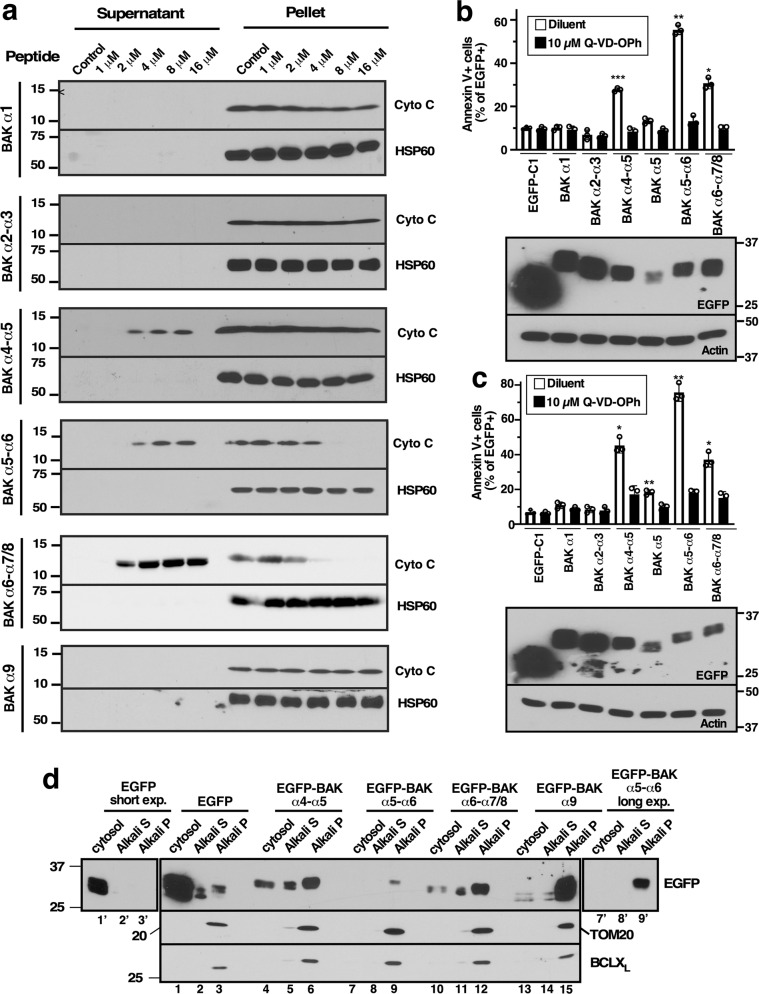
Further experiments examined the structure of individual tandem helices and their ability to initiate mitochondrial cyto c release. Synthetic BAK α4–α5, α5–α6, and α6–α7/8 employed in these studies exhibited substantial α-helicity when analyzed by circular dichroism spectroscopy (Fig. S4). In addition, molecular dynamics simulations indicated that the most populated or second most populated conformation predicted for each construct consists of two helices in a U-shaped helix-turn-helix configuration (Figs. 3 and S5, Table S1). These synthetic peptides induced dose-dependent cyto c release from Bak/Bax-deficient mitochondria (Figs. 4a and S6). Moreover, cDNAs encoding the same tandem helices fused to the C-terminus of EGFP induced caspase-dependent killing in BAX−/−/BAK−/− HCT116 cells and BAK−/− Jurkat cells (Figs. 4b, c, S7, S8). Introduction of an EGFP A207K mutation, which diminishes dimerization seen with wildtype EGFP [37], had no impact on killing (Fig. S9), suggesting that EGFP-induced dimerization is not driving the killing by these constructs. Importantly, the active EGFP fusion proteins became tightly associated with membranes and, at least in part, resisted release with 0.1 M sodium carbonate (Fig. 4d, lanes 6, 9’ and 12 vs. lane 3’). Collectively, these results suggested that multiple BAK tandem peptide regions might be able to bind to and permeabilize mitochondria, thereby inducing apoptosis.
Fig. 3. The first or second most populated conformations of tandem helix peptides and selected variants.
Top: Front view of the backbone conformation in the cartoon model. Middle: Front view of the backbone conformation in the cartoon model with side-chain conformations in the sphere model. Bottom: Back view of the backbone conformation in the cartoon model with side-chain conformations in the sphere model. The cartoon model shows two parallel helices (U-shaped conformation), while the sphere model shows the favorable interhelical side-chain interactions that contribute to the stability of the U-shaped structures. The H, N, and O atoms are colored in white, blue, and red, respectively. The C atoms in α4–α5, α5–α5, α5–α6, α6–α7/8, and α6–α7/8Δ12 are colored in slate, orange, cyan, green, and magenta, respectively. These populated conformations were identified from 20 distinct and independent molecular dynamics simulations with an aggregated simulation time of 37.92 microseconds for each peptide. Additional details are found in the Supplementary Methods, Supplementary Results and Discussion, Table S1 and Fig. S5.
Fig. 4. BAK tandem helices directly permeabilize the MOM and induce cell death.
a After mitochondria from Bax−/−Bak−/− MEFs were incubated with BAK peptides α1 (aa 21–51), α2-α3 (aa 70–101), α4-α5 (aa 105–146), α5-α6 (aa 124–164), α6–α7/8 (aa 149–186) at 25 °C for 90 min, the supernatants and pellets were subjected to SDS-PAGE and immunoblotting. Additional assays for mitochondrial cytochrome release c are shown in Figure S6. b, c After cDNAs encoding BAK peptides fused at their N termini to EGFP were transfected into BAX−/−BAK−/− HCT116 cells (b) or BAK−/− Jurkat cells (c), cells were incubated in the presence bortezomib without or with Q-VD-OPh for 24 h, stained with APC-conjugated Annexin V, and subjected to flow microfluorimetry. The percentage EGFP+ cells that were Annexin V+ is indicated. Lower panels in b and c, whole cell lysates subjected to immunoblotting. As indicated in the Materials and Methods, error bars in b and c as well as all subsequent figures represent mean ± sd of three independent experiments. *, **, and *** indicate p < 0.05, p < 0.01, and p < 0.001, respectively, in unpaired two-tailed t tests after correction for multiple comparisons by the method of Scheffé [59]. Asterisks in b and c reflect comparisons to EGFP-C1 transfected cells. See also Figs. S7–S9. d After BAX−/−BAK−/− HCT116 cells were transfected with the indicated plasmids, cells were incubated for 24 h in the presence of bortezomib and Q-VD-OPh. Following fractionation of cells into cytosol and mitochondria, mitochondrial fractions were further treated with 0.1 M sodium carbonate (pH 11.5) for 20 min and centrifuged to isolate alkali extractable (Alkali S) and alkali-insoluble (Alkali P) fractions, which were subjected to immunoblotting. Lanes 1′–3′, shorter exposure of EGFP blot for lanes 1–3. Lanes 7′–9′, longer exposure of EGFP blot for lanes 7–9.
Binding of BAK helices to MOM lipids
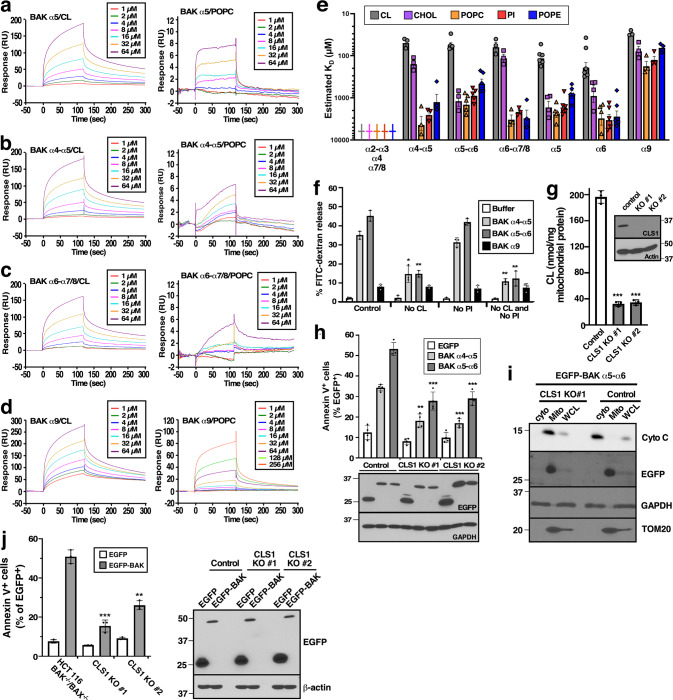
To assess the possibility that these BAK peptides might recognize a feature unique to the mitochondrial surface such as MOM lipids, we performed surface plasmon resonance (SPR) using peptides immobilized on sensor chips and MOM lipids in solution (Figs. 5a–e, S10, S11). In these studies, BAK α5 showed a preference for CL, a lipid reportedly enriched in the MOM [38, 39], over POPC or other membrane lipids (Fig. 5a, e). A similar binding preference was also observed for α6 and the tandem constructs α4–α5, α5–α6, and α6-α7/8 (Fig. 5b, c, e). In contrast, the known transmembrane domain α9 bound all lipids with limited selectivity; and synthetic α2–α3, α4, and α7/8 bound lipids poorly in these assays (Fig. 5d, e).
Fig. 5. Preferential binding of BAK peptides to MOM lipids is essential for MOMP.
a–d Representative SPR sensograms of immobilized BAK α5 (a), α4-α5 (b), α6-α7/8 (c), or α9 (d) interacting with a series of cardiolipin (CL, left) or POPC concentrations (right). e Equilibrium dissociation constants (KDs) of the indicated BAK single helices or tandem helices with MOM lipid components evaluated by SPR. Daggers indicate lack of detectable binding (KD > 10,000 µM) to α2-α3, α4, or α7/8 under the conditions of the assay. f After FITC-dextran-containing liposomes with different compositions [control, lacking CL (No CL), lacking PI (No PI), lacking both CL and PI (No CL and No PI)] were incubated with the indicated BAK peptides (10 µM) for 90 min at 37 °C, the percentage of FITC-dextran release was measured. g After knockout of CSL1 in BAX−/−BAK−/− HCT116 cells, two clones were assayed for mitochondrial CL content. Inset, whole cell lysates are subjected to immunoblotting. h CSL1−/− cells from panel g were transfected with the indicated EGFP-tagged plasmids, incubated for 24 h in the presence of bortezomib, stained with APC-Annexin V, and analyzed by flow microfluorimetry. The percentage of EGFP+ cells that were also Annexin V+ is indicated. i CLS1−/− clone #1 (which is also BAX−/−BAK−/−) was transfected with EGFP-BAK α5–α6 and incubated with bortezomib and Q-VD-Ph for 24 h. After whole cell lysates, mitochondria and cytosol were isolated, fractions were subjected to western blotting. j CSL1−/− cells from g were transfected with plasmid encoding full-length BAK fused to the C-terminus of EGFP, incubated for 24 h, stained with APC-Annexin V, and analyzed by flow microfluorimetry. The percentage of EGFP+ cells that were also Annexin V+ is indicated in the left panel. The right panel shows protein expression in samples where 10 µM Q-VD-OPh was added to prevent apoptosis-associated cell rupture. Error bars in e–h and j, mean ± sd of three independent experiments. Asterisks in g, h, and j reflect comparison to pooled cells transfected with Cas9 and nontargeting sgRNA (g), these same control cells transfected with the indicated BAK construct (h) or BAK−/−BAX−/− cells without CSLS1 gene interruption (j). See also Figs. S10–S13.
Based on these results, we examined the ability of the α4–α5 and α5–α6 peptides to permeabilize liposomes with different lipid compositions. Interestingly, the ability of α4-α5 or α5-α6 to permeabilize liposomes composed of MOM lipids was markedly diminished by omission of CL but not PI (Figs. 5f, S12).
To assess the role of CL in a cellular context, we interrupted CLS1, the gene encoding cardiolipin synthase, in BAX−/−/BAK−/− HCT116 cells. CLS1 depletion (inset, Fig. 5g) and the resulting 83% decrease in mitochondrial CL content (Fig. 5g) were accompanied by attenuated cytotoxicity of EGFP fused to BAK α4-α5 or α5-α6 (Fig. 5h). This diminished killing reflected decreased mitochondrial cyto c release despite trafficking of the EGFP-BAK fragment to mitochondria (Fig. 5i). Consistent with these results, the ability of full-length BAK to kill HCT116 or Jurkat cells was likewise diminished by CLS1 depletion (Figs. 5j, S13). Collectively, these observations raise the possibility that CL binding contributes to BAK-mediated MOMP.
Modifications to tandem helices modulate MOMP
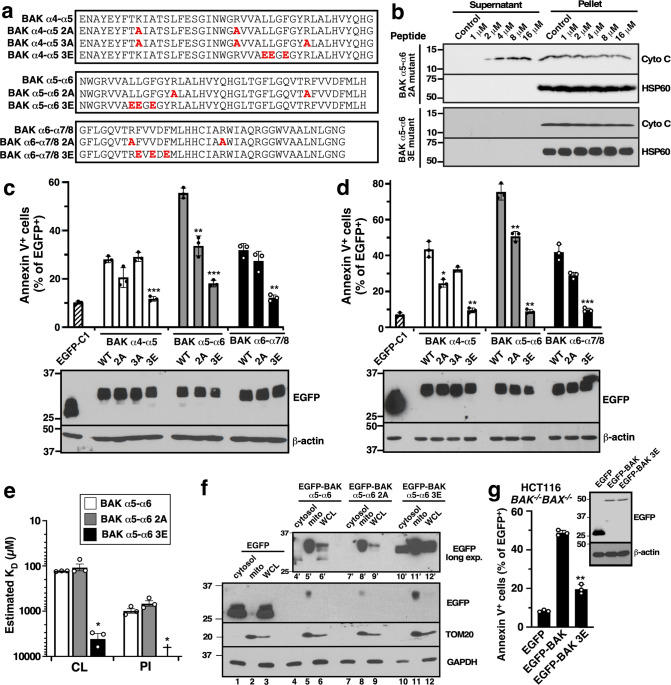
In an attempt to gain further insight, we mutated selected residues in the tandem helices (Fig. 6a) and examined the impact on cytochrome c release (Fig. 6b) and killing (Fig. 6c, d). Mutation of the cationic residues to neutral ones had a small but discernable impact on killing (e.g., the “2A” peptides), whereas mutation of the hydrophobic residues to anionic ones (the “3E” peptides) markedly diminished cytochrome c release and cell killing. These changes reflected distortion of the U-shaped conformations (Fig. S5), decreased affinity of the 3E peptide for CL and PI (Fig. 6e), and decreased association of the 3E peptide with mitochondria, as indicated by increased recovery of the fusion protein in cytosol (Fig. 6f, lane 10’ vs. lane 4’). Introduction of the same 3E mutation into α5 or α6 of the full-length BAK protein likewise diminished the ability to kill cells (Figs. 6g, S14) even though this BAK construct contained the α9 helix that tethers it to mitochondria.
Fig. 6. BAK peptide alterations modulate lipid binding and cytochrome c release.
a Sequences of peptide variants or EGFP-tagged peptide variants used in this figure. b After mitochondria from Bax−/−Bak−/− MEFs were incubated for 90 min at 25 °C with the indicated BAK peptides, supernatants and pellets were harvested for immunoblotting. c, d cDNAs encoding EGFP fused to the indicated BAK peptide variants were transfected into BAX−/−BAK−/− HCT116 cells (c) or BAK−/− Jurkat cells (d). After a 24-h incubation in the presence of bortezomib, cells were stained with APC-Annexin V and subjected to flow microfluorimetry. Lower panels in c and d, after aliquots of cells transfected with the indicated constructs were incubated with bortezomib + Q-VD-OPh (to prevent caspase-mediated cell rupture) for 24 h, whole cell lysates were subjected to SDS-PAGE. e KDs of cardiolipin (CL) or phosphatidylinositol (PI) binding to the indicated peptides by SPR. f cDNAs encoding the indicated BAK peptides fused to the C-terminus of EGFP were transfected into BAX−/−BAK−/− HCT116 cells. After a 24-h incubation in the presence of Q-VD-OPh and bortezomib, proteins from cytosol (50 µg) and mitochondria (mito, 20 µg) were separated and analyzed together with whole cell lysates (WCL, 50 µg) by immunoblotting. Lanes 4′-12′, longer exposure of EGFP blot shown in lanes 4–12. g 24 h after BAX−/−BAK−/− HCT116 cells were transfected with cDNA encoding the indicated full-length BAK protein fused to the C-terminus of EGFP, annexin V binding was assayed. Inset in g, transfected cells were incubated for 24 h in the presence of Q-VD-OPh before whole cell lysates were prepared. Asterisks reflect comparison to wildtype peptide sequence (e) or EGFP fused to the corresponding unmutated (WT) sequence (c, d, g).
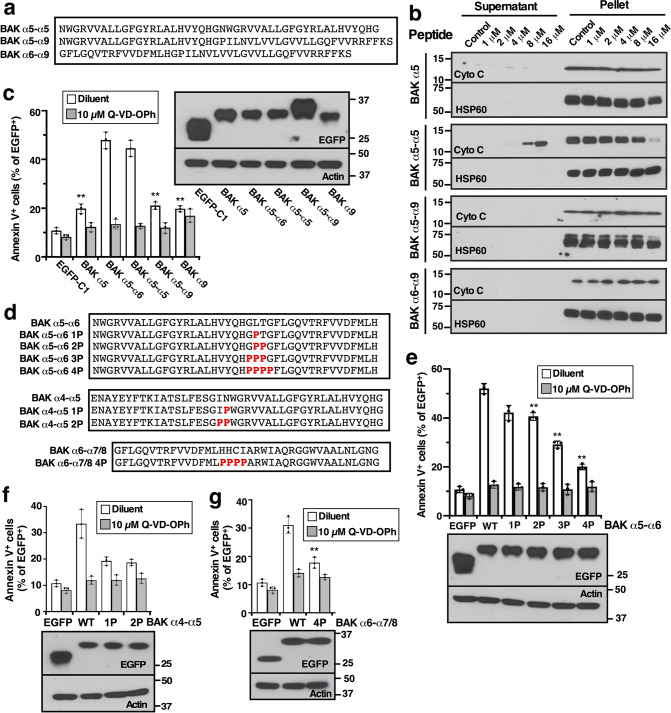
To further assess the requirements for MOMP, we generated constructs corresponding to the tandem helices BAK α5-α9 and BAK α6-α9 (Fig. 7a). These peptides failed to permeabilize mitochondria (Fig. 7b) or kill cells when fused to EGFP (Fig. 7c). Conversely, even though α5 as a single hydrophobic α helix did not induce killing, a BAK α5-α5 head-to-tail pair of helices was able to permeabilize mitochondria (Fig. 7b) and kill cells (Fig. 7c). Recognizing that α5-α5 readily adopts a U-shaped conformation whereas α5-α9 does not (Figs. 3 and S5), additional experiments examined the impact of altering the orientation of the α-helices. When prolines were substituted for amino acids in the joint between the antiparallel helices of α5-α6 (Fig. 7d), the propensity to adopt a U-shaped conformation decreased (Fig. S5) and the cytotoxicity of these constructs when introduced into BAX−/−/BAK−/− HCT116 cells at the C-terminus of EGFP diminished (Fig. 7e). Similar results were observed upon introduction of prolines into the corresponding locations in α4-α5 and α6-α7/8 (Figs. 7f, g and S5), suggesting that orientation of the tandem helices might be important for MOMP.
Fig. 7. Effect of α9 or proline insertion on BAK tandem peptide-induced membrane permeabilization and cytotoxicity.
a, d Sequence of peptide variants or EGFP-tagged peptide variants used in this figure. b After mitochondria from Bax−/−Bak−/− MEFs were incubated for 90 min at 25 °C with the indicated BAK peptides, supernatants and pellets were harvested for immunoblotting. c, e–g cDNAs encoding EGFP fused to the indicated BAK peptide variants were transfected into BAX−/−BAK−/− HCT116 cells. After a 24-h incubation in the presence of bortezomib, cells were stained with APC-Annexin V and subjected to flow microfluorimetry. Inset in c and lower panels in e–g, after aliquots of cells transfected with the indicated constructs were incubated with bortezomib in the presence of Q-VD-OPh, whole cell lysates were subjected to SDS-PAGE. Asterisks reflect comparison to cells transfected with EGFP-BAK α5-α6 (c) or cells transfected with EGFP fused to the unmutated (WT) BAK peptide (e–g).
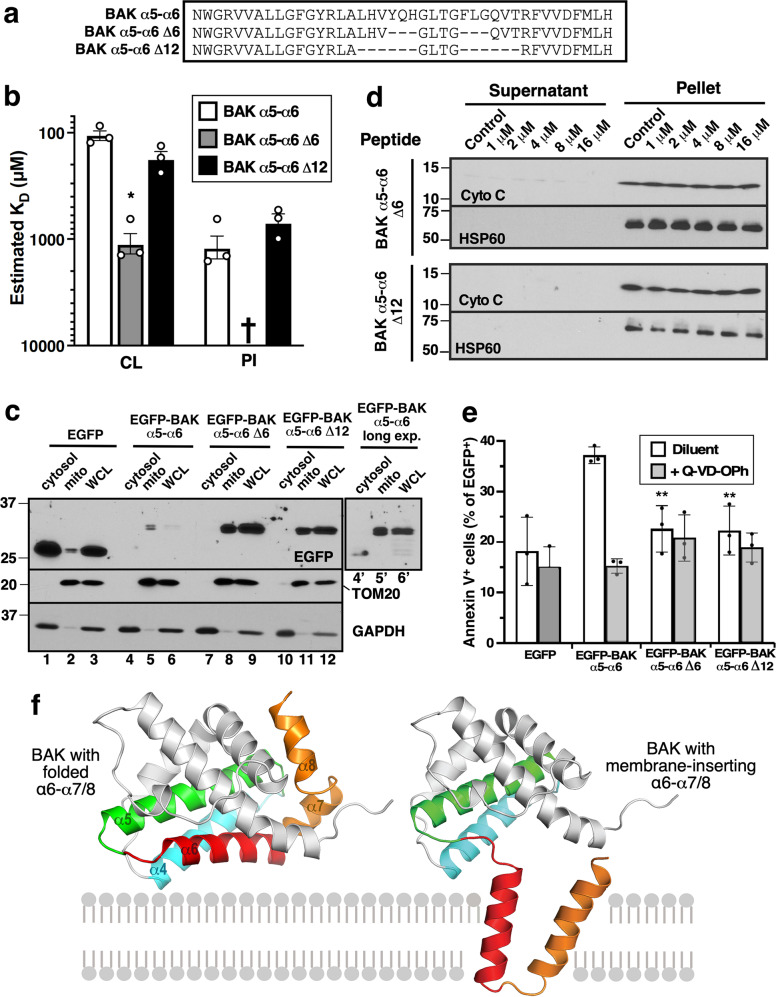
In a final set of experiments, we assessed the impact of shortening the tandem helices (Figs. 8a and S15a). Two of these constructs were particularly informative. The α5–α6 Δ12 construct, shortened by six amino acids on each helix, bound CL and PI with affinities similar to those of full-length α5–α6 peptide (Fig. 8b) and localized EGFP to mitochondria (Fig. 8c). In contrast to α5–α6, however, α5–α6 Δ12 exhibited diminished propensity to form a U-shaped structure (Table S1 and Fig. S5) and decreased ability to permeabilize mitochondria in vitro (Fig. 8d) or kill transfected cells (Fig. 8e). Conversely, α6–α7/8 Δ12, which retains the ability to form structures with two parallel helices, including a U-shaped structure (Table S1 and Fig. S5), but has diminished affinity for CL and PI (Fig. S15b) also exhibits diminished ability to permeabilize mitochondria (Fig. S15c) and diminished ability to kill cells (Fig. S15d). Collectively, results in Figs. 5–8 and S10–S15 suggest that CL binding is required for tight MOM association, whereas the folding of a pair of long helices back on themselves to form a U-shaped helix-turn-helix structure might play a role in subsequent MOMP.
Fig. 8. Effect of helix shortening on MOMP and cell killing.
a Sequences of hairpin shortened BAK α5-α6 peptide mutants or EGFP-tagged peptide mutants used in b–e. b KDs of CL or PI interacting with the indicated immobilized peptides as evaluated by SPR. † indicates KD > 10 mM. *, p < 0.05 relative to α5–α6. c After cDNAs encoding the indicated BAK α5–α6 helix shortened helixes fused to the C-terminus of EGFP were transfected into BAX−/−BAK−/− HCT116 cells, cultures were incubated for 24 h in the presence of Q-VD-OPh and bortezomib and subjected to fractionation. Aliquots containing 50 µg of protein from whole cell lysates (WCL) and 5 µg of protein from mitochondria (mito) were subjected to SDS-PAGE and immunoblotting. Lanes 4’-6’, longer exposure of EGFP blot is shown in lanes 4–6. d After mitochondria from Bax−/−Bak−/− MEFs were incubated for 90 min at 25 °C with the indicated hairpin shortened BAK α5-α6 peptides, supernatants and pellets were subjected to SDS-PAGE and immunoblotting. e cDNAs encoding EGFP-tagged BAK peptide mutants were transfected into BAX−/−BAK−/− HCT116 cells. After a 24-h incubation in the presence of bortezomib ± Q-VD-OPh, cells were stained with APC-Annexin V and subjected to flow microfluorimetry. Asterisks indicate comparison to cells transfected with EGFP-BAK α5-α6. f Model consistent with observations in the present work. See text for further discussion.
Discussion
Starting with the observation that BAK lacking the classical C-terminal transmembrane helix becomes stably bound to mitochondria during MOMP, we have examined the interaction of various BAK regions with lipids, liposomes, or mitochondria under cell-free conditions and in intact cells. The results of these studies provide potentially important new insight into the process of MOMP.
We initially examined the actions of BAK helical regions, both individually and in tandem pairs. While it has previously been reported that BAX α5 is able to permeabilize liposomes in vitro [40–42], our studies indicate that the BAK α5 peptide alone does not permeabilize mitochondria. In contrast, when BAK α5 is fused to α4, another α5, or α6, the resulting constructs target EGFP to mitochondria, permeabilize mitochondria under cell-free conditions and induce apoptosis in a cellular context (Figs. 2, 4, 7, and S3). BAK α6 has similar properties when fused to BAK α7/8, but not as a single peptide. These observations suggest that the minimal functional unit for induction of MOMP and cell killing is a tandem pair of BAK helical peptides.
Further studies using SPR demonstrated that α5 and α6 preferentially bind CL compared to other lipids (Fig. 5e). The potential importance of CL binding was highlighted by several observations. First, the ability of tandem peptides to permeabilize liposomes decreased in the absence of CL (Fig. 5f). Second, CLS1 knockout, which diminished mitochondrial CL, diminished the cytotoxicity of tandem BAK peptides in a cellular context (Fig. 5g, h). Further, CLS1 knockout diminished the ability of full-length BAK to release mitochondrial cyto c and trigger apoptosis in two different cellular contexts (Figs. 5j, S13c).
Although the possibility that MOMP involves CL, a lipid that is particularly abundant in the mitochondrial inner membrane [43], might seem counterintuitive, CL has also been detected in small amounts in highly purified MOMs from various cells [44, 45] and is concentrated at contact sites between the mitochondrial inner and outer membranes [32]. Moreover, previous studies have reported that CL contributes to apoptotic events at the MOM [46], including recruitment of tBID [33, 34], the ability of tBID to recruit BAX [47, 48], and enhanced activation of caspase 8 at the mitochondrial surface during death ligand-induced apoptosis [49]. These observations provide precedent for the present suggestion that interaction of BAK with CL in the MOM might contribute to MOMP. On the other hand, we also observed that CLS1 knockout did not completely eliminate BAK-induced apoptosis (Figs. 5j, S13c). While this might reflect incomplete CL depletion (Figs. 5g, S13a) due to alternative synthetic pathways, we cannot eliminate the possibility that BAK interactions with other MOM lipids also contribute to MOMP in a cellular context. Likewise, we cannot completely rule out the possibility that CSL1 knockout impairs MOMP through a generalized effect on MOM fluidity rather than disruption of specific BAK-lipid interactions. However, the fact that CL is concentrated mainly in the mitochondrial inner membrane and at intermembrane contact sites rather than throughout the MOM argues against this alternative explanation.
In an attempt to understand why single CL-binding BAK helices are not able induce MOMP, we examined various derivatives of BAK tandem helices. A common feature among the tandem helices that induced MOMP (α4–α5, α5–α5, α5–α6, and α6–α7/8) was their ability to form a U-shaped structure (Figs. 3, S5 and Table S1). Importantly, three different types of alterations that diminished this propensity to assume a U-shaped conformation (Fig. S5), i.e., substitution of a region that does not form the helix-helix interactions thought to stabilize this conformation (α5–α9, Fig. 7), introduction of prolines (Fig. 7), and shortening of the helices (α5–α6 Δ12, Fig. 8), concomitantly diminished the ability of the tandem peptides to permeabilize mitochondria and, when fused to EGFP, kill cells. The α5–α6 Δ12 construct is particularly informative because this tandem peptide retains the ability to bind CL and traffic to mitochondria but is unable to induce MOMP, suggesting the potential importance of a step involving a U-shaped tandem peptide with a membrane-spanning length after binding of BAK to MOM lipids.
Several models have previously been advanced to explain BAX- or BAK-induced MOMP. One suggests that α6 helices from two monomers of BAX form a clamp that pulls MOM lipids away from a central pore. This model is difficult to reconcile with the inability of BAK α5 or α6 alone to permeabilize liposomes or mitochondria. Another model suggests that aggregates of activated BAK permeabilize the MOM. In contrast, the present results suggest that α4–α5 and α5–α6 bound to EGFP A207K are acting as monomers (e.g., Fig. S9b), although we cannot rule out the possibility that α6–α7/8 acts as a multimer. A third model suggests that BAK α5 and α6 sit on the surface of the MOM and disrupt the lipid bilayer. This model is difficult to reconcile with the apparent need for tandem peptides to assume a U-shaped conformation in order to induce MOMP.
One possibility, depicted in Fig. 8f, is that tandem helices mobilized during BAK activation (e.g., α6–α7/8) bind CL and insert into the MOM as a hairpin-like structure during MOMP. This model is consistent with recent studies showing that a region consisting of α6-α8 separates from the BAK core during activation [22, 29] as well as earlier observations showing that BAK becomes more tightly associated with the MOM upon activation [5]. Membrane insertion of this hairpin-like structure during MOMP would be consistent with both the effect of the proline substitutions (Fig. 7d–g) and the impact of shortening the tandem α-helices (Fig. 8b–e). While the recently reported structure of the α2–α8 BAK dimer fails to show the hairpin-like moiety depicted in Fig. 8f, it is important to note that the crystal structure was determined in the absence of MOM lipids.
On the other hand, it is possible that the model shown in Fig. 8f represents an intermediate formed during a series of activation-associated BAK structural rearrangements. Based on the current results, we cannot rule out the possibility that α4 and/or α5 also participate in membrane binding and permeabilization in some way upon the insertion of α6-α7/8 into the MOM (e.g., Fig. S16). This possibility would be consistent with a recent report that the structure of BAK in association with lipid membranes is different from BAK in aqueous solution [50] as well as an earlier report that BAX α2–α5 is sufficient to induce apoptosis when targeted to mitochondria [51].
In summary, the present results highlight a critical role for CL in BAK action under cell-free conditions and in intact cells. In addition, they demonstrate that BAK α5 and α6 are capable of binding CL and, when present as U-shaped tandem helices, sufficient to bind to mitochondria, induce MOMP and kill cells. These results not only provide potential new insight into the proapoptotic action of BAK, but also provide a new series of peptides that, if targeted selectively to cancer cells, could potentially be explored for therapeutic efficacy in the absence of the portions of the BAX and BAK molecules that participate in sequestration by antiapoptotic BCL2 paralogs.
Methods and methods
Materials
Reagents were obtained as follows: Ni2+-NTA-agarose from Merck Biosciences (Burlington, MA); lipids and extruder from Avanti Polar Lipids (Alabaster, AL); CM5 biosensor chips from GE Healthcare (Chicago, IL); Q-VD-OPh from SM Biochemicals (Anaheim, CA); reduced and oxidized glutathione from Millipore-Sigma (St. Louis, MO); glutathione-agarose from Thermo Scientific (Waltham, MA); and FITC-labeled dextran 10 from Invitrogen (Carlsbad, CA). BAK peptides were generated by solid phase synthesis in the Mayo Clinic Proteomics Research Center (Rochester, MN). Antibodies to the following antigens were purchased from the indicated suppliers: Heat shock protein 60 (HSP60, cat # 4870S), glyceraldehyde phosphate dehydrogenase (GAPDH, 2118S) and GFP (2555S) from Cell Signaling Technology (Danvers, MA); cytochrome c (556433) from BD Biosciences (Franklin Lakes, NJ); BAK (06-536) from Millipore-Sigma (Burlington, MA); β-actin (sc-376421), TOM20 (sc-17764) and CLS1 (sc-514986) from Santa Cruz Biotechnology (Dallas, TX).
Cell culture
BAK−/− Jurkat cells were generated using CRISPR-Cas9 [52]. Cells were maintained at densities below 106 cells ml−1 in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (FBS), 100 units ml−1 penicillin G, 100 μg ml−1 streptomycin, and 2 mM glutamine. Previous studies have shown that these cells have extremely low levels of BAX [53]. BAX−/−BAK−/− HCT116 cells from Richard Youle [54] were propagated in McCoy’s 5A medium containing 10% heat-inactivated FBS along with penicillin, streptomycin, and glutamine. Bax−/−Bak−/− MEFs were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS, 250 μM L-asparagine, and 55 μM 2-mercaptoethanol. Prior to cryopreservation in multiple replicates, cell lines were authenticated by short tandem repeat profiling in the Mayo Clinic Cytogenetics Core, examined for absence of the pertinent proteins by immunoblotting and assayed for mycoplasma contamination. Fresh aliquots were thawed every 6–12 weeks.
Jurkat cells lacking activator BH3-only proteins as well as BAK and BAX (Jurkat 8KO cells) were generated from Jurkat 6KO cells (with interrupted genes encoding BAK, BAX, BIM, PUMA, NOXA, and BID) [55] by interruption of the BMF and HRK genes using oligonucleotides 5′-GGATGATGTGTTCCAACCAG-3′ and 5′-AGCTGCACCAGCGCACCATG-3′ as guide RNAs to human BMF coding nucleotides 33–52 (NCBI accession # NM_001003940) and HRK coding nucleotides 128-147 (NCBI accession # NM_003806), respectively. In brief, oligonucleotides were synthesized, annealed, and cloned into the BsmBI site of the lentiCRISPR-v2 plasmid (Addgene). Viruses were packaged in HEK293T cells using Lipofectamine 2000 with the packaging vector psPAX3, envelope vector pMD2.G, and lentiCRISPR-v2-BMF or-HRK. A week after transduction with the resulting viruses, cells were cloned by limiting dilution. Because BMF and HRK were not detectable by immunoblotting in the parental cells, clones were assayed for gene interruption by sequencing genomic DNA across the targeted region.
Full-length CRISPR/Cas9-resistant human BAK [55] was inserted into the BamHI and EcoRI sites of pRRL.TRE3G.Blast.IRES.rtTA3 plasmid. After packaging, the construct was transduced into cloned Jurkat 8KO cells, which were selected with 5 µg/ml blastocidin, cloned by limiting dilution, and assayed by immunoblotting to identify clones that expressed BAK in a doxycycline-inducible manner.
Mammalian expression plasmids, mutations, and transient transfection
cDNAs encoding the indicated fragments of human BAK (GenBank BC004431) were cloned into the Xho1 and BamH1 sites of pEGFP-C1 to yield constructs fused at their N termini to EGFP. All mutations were generated using a Qiagen QuickChange II site-direct mutagenesis kit (Agilent Technologies) and confirmed by Sanger sequencing. Empty pEGFP-C1 served as a control.
To ensure protein expression, log phase BAX−/−BAK−/− HCT116 cells or BAK−/− Jurkat cells growing in antibiotic-free medium were transiently transfected with the indicated plasmid using a BTX 830 square wave electroporator delivering a single pulse at 240 mV for 10 msec. Cells were treated with 50 nM bortezomib (BAX−/−BAK−/− HCT116) or 15 nM bortezomib (BAK−/− Jurkat) to induce fusion protein stabilization (Figs. S7 and S8) and incubated for 24 h before analysis for apoptosis using APC-coupled annexin V [35, 56]. All experiments included transfection with empty vector to assure that the bortezomib treatment per se did not induce apoptosis under the indicated conditions.
Fluorescence microscopy
After Bax−/−Bak−/− MEFs were transfected with the indicated EGFP-tagged plasmids, cells were grown in the presence of 10 μM Q-VD-OPh for 24 h and treated for 20 min with 20 nM MitoTracker red. The cells were then fixed with 4% paraformaldehyde, permeabilized with 0.15% (w/v) Triton X-100, and labeled with rabbit anti-GFP-antibody followed by Alexa Fluor 488-labeled secondary antibody (ThermoFisher). Images were recorded on a Zeiss Axio Observer fluorescence microscope (Carl Zeiss Inc, Thornwood, NY) using a Plan-Apochromat 63×/N.A. 1.40 lens. To assess colocalization, intensity maps of MitoTracker red and Alexa Fluor 488 were superimposed using Zeiss Zen software.
Cell fractionation
Cells were washed with calcium- and magnesium-free Dulbecco’s phosphate-buffered saline (PBS), incubated for 20 min in ice-cold hypotonic buffer [25 mM HEPES (pH 7.5), 5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM PMSF, 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 1 mM sodium vanadate, and 20 nM microcystin] and lysed with 35 strokes in a tight-fitting Dounce homogenizer. After cell lysis was confirmed by Trypan blue staining, lysates were sedimented at 800 × g for 15 min to remove the nuclei (pellet). The supernatant was carefully collected and spun at 4500 × g for 15 min to collect crude mitochondria (pellet) and cytosol (supernatant), which was spun a second time at 4500 × g to remove any unsedimented mitochondria.
Sodium carbonate extraction
Mitochondria were incubated in 0.1 M sodium carbonate (pH 11.5) at 25 °C for 20 min and sedimented at 14,000 × g for 15 min. Supernatants and pellets were then analyzed by immunoblotting.
CL downregulation
CLS1, the gene encoding Cardiolipin Synthase 1, was interrupted using CRISPR/Cas9 in BAK−/−/BAX−/− HCT116 cells. In brief, oligonucleotides (5′-GCGTGGCA- CGCGGCTCGTGG-3′) guiding to human CLS1 nucleotides 14–33 (Genbank accession number NM_19095) were synthesized, annealed, and cloned into the BsmBI site of the lentiCRISPR-v2 (Addgene) plasmid. Viruses were packaged in HEK293T cells by transfecting with the packaging vector psPAX3, envelope vector pMD2.G, and lentiCRISPR-v2-CLS1 14–33 using Lipofectamine 2000. Two days after transduction with virus, BAK−/−/BAX−/− HCT116 cells were selected with 3 μg ml−1 puromicin and cloned by limiting dilution. CLS1 knockout was verified by immunoblotting. Similar procedures were utilized to interrupt the CLS1 gene in Jurkat cells.
Measurement of CL content
After control or CLS1−/− cells were washed with PBS, mitochondria were isolated [8]. Aliquots containing 30 µg of mitochondrial protein (quantified using the bicinchoninic acid method) were assayed for CL using a BioVision fluorimetric CL detection kit (Biovision, Milpitas, CA). After a 5 min incubation with probe at 25 °C, samples and CL standards were examined on a Molecular Devices Spectramax plate reader with excitation and emission wavelengths of 340 and 380 nm, respectively.
Protein expression and purification
Plasmid encoding BAKΔTM (GenBank BC004431, residues 1-186) in pET29b(+) was a kind gift from Qian Liu and Kalle Gehring (McGill University, Montreal, Canada). BAK mutants were generated by site-directed mutagenesis (Agilent Technologies). All plasmids were subjected to Sanger sequencing to verify the described alteration and confirm that no additional mutations were present.
To obtain BAKΔTM (WT and mutants), plasmids were transformed into E. coli BL21 (DE3) by heat shock. After bacteria were grown to an optical density of 0.8, 1 mM IPTG was used to induce protein expression at 16 °C for 24 h. Sonication was performed on ice in TS buffer [150 mM NaCl containing 10 mM Tris-HCl (pH 7.4) and 1 mM PMSF]. After centrifugation, supernatants were applied to Ni2+-NTA-agarose columns, which were washed with 20 volumes of TS buffer followed by 10 volumes TS buffer containing 40 mM imidazole. Proteins were then eluted with TS buffer containing 200 mM imidazole.
Cyto c release
BAKΔTM (wt or mutant) was dialyzed against HK buffer (20 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM EGTA, pH 7.4). Before experiments, BAKΔTM was treated with either 5 mM DTT for 2 h to ensure a reduced condition or 5 mM oxidized GSH to ensure cysteine cross-linking. Mitochondria purified from Bax−/−Bak−/− MEFs [57] were incubated with protein or the indicated peptides at 25 °C for 90 min. After centrifugation (10,000 g, 15 min), supernatants and pellets were harvested for immunoblotting. To assess BAKΔTM stable association with mitochondria, pellets were washed with HK buffer before lysis in SDS sample buffer.
For cyto c release by synthetic BAK peptides or mutant BAK peptides, the peptides were dissolved in DMSO (BAKα5, BAKα9) or H2O (other peptides) and then diluted in HK buffer before experiments.
Preparation of FITC-dextran lipid vesicles
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-plamitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE), L-α-phosphatidyl-inositol (PI), cardiolipin (CL), and cholesterol (CHOL) at a weight ratio of 41:22:9:20:8 were dried as thin films in glass test tubes under nitrogen and then under vacuum for 16 h. The lipid composition for liposomes designated as “No CL,” “No PI,” and “No CL and PI” are 61:22:9:0:8, 50:22:0:20:8 and 70:22:0:0:8 for POPC:PE:PI:CL:CHOL, respectively. To encapsulate FITC-labeled dextran 10 (F-d10), 50 mg lipid in 1 ml of 20 mM HEPES, 150 KCl (pH 7.0) buffer was mixed with 50 mg F-d10, sonicated and extruded 15 times through a 400 nm polycarbonate membrane. Untrapped F-d10 was removed by gel filtration on Sephacryl S-300 HR (GE Healthcare). Phosphate was determined by colorimetric assay (Abcam, Cambridge, UK).
Liposome permeabilization assay [modified from 21]
Release of F-d10 from large unilamellar vesicles (LUVs) was monitored by fluorescence dequenching using a fluorimetric plate reader. After BAK peptides were added to LUVs (final lipid concentration 10 µg ml−1), 96 well plates were incubated at 37 °C, mixed and assayed (excitation 485 nm, emission 538 nm) every 20 s. F-d10 release was quantified by the equation ((Fsample−Fblank)/(FTriton−Fblank) × 100), where Fsample, Fblank, and FTriton are fluorescence of reagent-, buffer-, and 1% Triton-treated LUVs.
Surface plasmon resonance (SPR)
BAK peptides were immobilized on a CM5 chip using a Biacore T200 biosensor (GE Healthcare). Binding assays were performed at 25 °C using CHAPS buffer (20 mM HEPES, 150 mM NaCl, 1% CHAPS, pH 7.4). Lipids were dissolved in CHAPS buffer and injected at 30 µl/min for 120 s. Bound lipids were allowed to dissociate in CHAPS buffer at 30 µl/min for 10 min and then desorbed with 2 M MgCl2. Binding kinetics were derived using BIA evaluation software (Biacore, Uppsala, Sweden). For binding to CL, both KD and Rmax values for binding to CL were calculated using steady-state analysis [58]. For all the other lipids, the Rmax was estimated based on maximum CL binding (taking into account differences in lipid molecular weight) and the binding relative to the Rmax was then used to estimate the KD using steady state analysis with constant Rmax [58].
Analytical gel filtration
24 h after transfection with the indicated plasmids, cells were collected and lysed in buffer consisting of 20 mM HEPES (pH 7.4), 150 mM NaCl, 1% CHAPS, and protease inhibitor mix (Roche, cat #11836170001). After lysates were sedimented at 14000 × g for 30 min, 200 μl aliquots of supernatant were subjected to FPLC on a Superdex S200 size exclusion column and 500 µl fractions were collected. The fractions were then separated by SDS-PAGE and blotted with anti-GFP antibody. The molecular markers (Millipore Sigma) in the same buffer were also chromatographed on the same column.
Statistical analysis
Experiments examining binding affinities, permeabilization of mitochondria or liposomes, or cell killing were performed at least three times independently on separate days using different preparations of purified BAK protein or peptides and different SPR chips. Graphs were prepared in Prism 9.0.1 (GraphPad Software, San Diego, CA). Error bars in all figures represent mean ± sd of 3 independent experiments. *, **, and *** indicate p < 0.05, p < 0.01, and p < 0.001, respectively, in two-tailed unpaired t tests after correction for multiple comparisons by the method of Scheffé [59] using StatView (SAS Institute, Cary, NC).
Additional methods
Additional methods, including a description of molecular dynamics simulations and circular dichroism spectroscopy, as well as further results and discussion, are found in the Supplementary Material.
Supplementary information
Original blots for supplementary figures
Acknowledgements
We thank David Toft, Richard Youle, Qian Liu, and Kalle Gehring for gifts of reagents; Eric Roush (Cytiva) for advice regarding surface plasmon resonance analysis; Gregory Gores, Roderick Brown, and Husheng Ding for helpful discussions; and the two anonymous reviewers for insightful suggestions. We also acknowledge the computing resources provided by the University of Minnesota Supercomputing Institute and the Mayo Clinic high-performance computing facility at the University of Illinois Urbana-Champaign National Center for Supercomputing Applications.
Author contributions
Designed study: HD and SHK Conducted experiments: HD, KLP, KSF, XWM, AV, Y-PP. Analyzed data: HD, CC, MR-A, Y-PP, SHK. Wrote manuscript: HD, SHK. Edited and approved manuscript: HD, KLP, KSF, XWM, KLP, AV, CC, MR-A, Y-PP, SHK.
Funding
This work was supported in part by grants from the National Cancer Institute (R01 CA166741, R01 CA225996 and P30 CA015083).
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by P. Salomoni
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Haiming Dai, Scott H. Kaufmann.
Contributor Information
Haiming Dai, Email: Dai.Haiming@Mayo.edu.
Scott H. Kaufmann, Email: Kaufmann.Scott@Mayo.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01083-z.
References
- 1.Adams JM, Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25:27–36. doi: 10.1038/cdd.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–91. doi: 10.1016/j.ccell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Jeng PS, Inoue-Yamauchi A, Hsieh JJ, Cheng EH. BH3-dependent and independent activation of BAX and BAK in mitochondrial apoptosis. Curr Opin Physiol. 2018;3:71–81. doi: 10.1016/j.cophys.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonsson B, Montessuit S, Sanchez B, Martinou JC. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J Biol Chem. 2001;276:11615–23. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 6.Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–31. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–99. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai H, Smith A, Meng XW, Schneider PA, Pang Y-P, Kaufmann SH. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol. 2011;194:39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HC, Kanai M, Inoue-Yamauchi A, Tu HC, Huang Y, Ren D, et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat Cell Biol. 2015;17:1270–81. doi: 10.1038/ncb3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci USA. 2013;110:E986–995. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang K, O’Neill KL, Li J, Zhou W, Han N, Pang X, et al. BH3-only proteins target BCL-xL/MCL-1, not BAX/BAK, to initiate apoptosis. Cell Res. 2019;29:942–52. [DOI] [PMC free article] [PubMed]
- 12.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 13.Ekert PG, Vaux DL. The mitochondrial death squad—hardened killers or innocent bystanders? Curr Opin Cell Biol. 2005;17:626–30. doi: 10.1016/j.ceb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–83. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birkinshaw RW, Czabotar PE. The BCL-2 family of proteins and mitochondrial outer membrane permeabilisation. Semin Cell Dev Biol. 2017;72:152–62. doi: 10.1016/j.semcdb.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Hauseman ZJ, Harvey EP, Newman CE, Wales TE, Bucci JC, Mintseris J, et al. Homogeneous oligomers of pro-apoptotic BAX reveal structural determinants of mitochondrial membrane permeabilization. Mol Cell. 2020;79:68–83.e67. doi: 10.1016/j.molcel.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K. The X-ray structure of a BAK homodimer reveals an inhibitory zinc-binding site. Mol Cell. 2006;24:677–88. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–54. doi: 10.1016/S0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 19.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–92. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du H, Wolf J, Schafer B, Moldoveanu T, Chipuk JE, Kuwana T. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 2011;286:491–501. doi: 10.1074/jbc.M110.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh KJ, Singh P, Lee K, Foss K, Lee S, Park M, et al. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J Biol Chem. 2010;285:28924–37. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birkinshaw RW, Iyer S, Lio D, Luo CS, Brouwer JM, Miller MS, et al. Structure of detergent-activated BAK dimers derived from the inert monomer. Mol Cell. 2021;81:2123–34.e2125. doi: 10.1016/j.molcel.2021.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, et al. Inhibition of Bax channel-forming activity by Bcl-2. Science. 1997;277:370–2. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 24.Schendel SL, Xie Z, Montal MO, Matsuyama S, Montal M, Reed JC. Channel formation by antiapoptotic protein Bcl-2. Proc Natl Acad Sci USA. 1997;94:5113–8. doi: 10.1073/pnas.94.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westphal D, Dewson G, Menard M, Frederick P, Iyer S, Bartolo R, et al. Apoptotic pore formation is associated with in-plane insertion of Bak or Bax central helices into the mitochondrial outer membrane. Proc Natl Acad Sci USA. 2014;111:E4076–85. doi: 10.1073/pnas.1415142111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, Garcia-Saez AJ, Bordignon E. Structural model of active Bax at the membrane. Mol Cell. 2014;56:496–505. doi: 10.1016/j.molcel.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvador-Gallego R, Mund M, Cosentino K, Schneider J, Unsay J, Schraermeyer U, et al. Bax assembly into rings and arcs in apoptotic mitochondria is linked to membrane pores. EMBO J. 2016;35:389–401. doi: 10.15252/embj.201593384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uren RT, O’Hely M, Iyer S, Bartolo R, Shi MX, Brouwer JM, et al. Disordered clusters of Bak dimers rupture mitochondria during apoptosis. Elife 2017;6:e19944. [DOI] [PMC free article] [PubMed]
- 29.Brouwer JM, Westphal D, Dewson G, Robin AY, Uren RT, Bartolo R, et al. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol Cell. 2014;55:938–46. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Cowan AD, Smith NA, Sandow JJ, Kapp EA, Rustam YH, Murphy JM, et al. BAK core dimers bind lipids and can be bridged by them. Nat Struct Mol Biol. 2020;27:1024–31. doi: 10.1038/s41594-020-0494-5. [DOI] [PubMed] [Google Scholar]
- 31.Dewson G, Kratina T, Sim HW, Puthalakath H, Adams JM, Colman PM, et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell. 2008;30:369–80. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Simbeni R, Pon L, Zinser E, Paltauf F, Daum G. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J Biol Chem. 1991;266:10047–9. doi: 10.1016/S0021-9258(18)99184-2. [DOI] [PubMed] [Google Scholar]
- 33.Lutter M, Fang M, Luo X, Nishijima M, Xie X, Wang X. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat Cell Biol. 2000;2:754–61. doi: 10.1038/35036395. [DOI] [PubMed] [Google Scholar]
- 34.Raemy E, Montessuit S, Pierredon S, van Kampen AH, Vaz FM, Martinou JC. Cardiolipin or MTCH2 can serve as tBID receptors during apoptosis. Cell Death Differ. 2016;23:1165–74. doi: 10.1038/cdd.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai H, Ding H, Meng XW, Peterson KL, Schneider PA, Karp JE, et al. Constitutive BAK activation as a determinant of drug sensitivity in malignant lymphohematopoietic cells. Genes Dev. 2015;29:2140–52. doi: 10.1101/gad.267997.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol Cell. 2009;36:696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–6. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 38.Gohil VM, Greenberg ML. Mitochondrial membrane biogenesis: phospholipids and proteins go hand in hand. J Cell Biol. 2009;184:469–72. doi: 10.1083/jcb.200901127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Fuertes G, Garcia-Saez AJ, Esteban-Martin S, Gimenez D, Sanchez-Munoz OL, Schwille P, et al. Pores formed by Baxalpha5 relax to a smaller size and keep at equilibrium. Biophys J. 2010;99:2917–25. doi: 10.1016/j.bpj.2010.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Saez AJ, Coraiola M, Serra MD, Mingarro I, Muller P, Salgado J. Peptides corresponding to helices 5 and 6 of Bax can independently form large lipid pores. FEBS J. 2006;273:971–81. doi: 10.1111/j.1742-4658.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- 42.Qian S, Wang W, Yang L, Huang HW. Structure of transmembrane pore induced by Bax-derived peptide: evidence for lipidic pores. Proc Natl Acad Sci USA. 2008;105:17379–83. doi: 10.1073/pnas.0807764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krebs JJ, Hauser H, Carafoli E. Asymmetric distribution of phospholipids in the inner membrane of beef heart mitochondria. J Biol Chem. 1979;254:5308–16. doi: 10.1016/S0021-9258(18)50596-2. [DOI] [PubMed] [Google Scholar]
- 44.Hovius R, Lambrechts H, Nicolay K, de Kruijff B. Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim Biophys Acta. 1990;1021:217–26. doi: 10.1016/0005-2736(90)90036-N. [DOI] [PubMed] [Google Scholar]
- 45.de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325:108–16. doi: 10.1016/S0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- 46.Schug ZT, Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim Biophys Acta. 2009;1788:2022–31. doi: 10.1016/j.bbamem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalvez F, Pariselli F, Dupaigne P, Budihardjo I, Lutter M, Antonsson B, et al. tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 2005;12:614–26. doi: 10.1038/sj.cdd.4401571. [DOI] [PubMed] [Google Scholar]
- 48.Lucken-Ardjomande S, Montessuit S, Martinou JC. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 2008;15:929–37. doi: 10.1038/cdd.2008.9. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJ, et al. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol. 2008;183:681–96. doi: 10.1083/jcb.200803129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandow JJ, Tan IK, Huang AS, Masaldan S, Bernardini JP, Wardak AZ, et al. Dynamic reconfiguration of pro-apoptotic BAK on membranes. EMBO J. 2021;40:e107237. doi: 10.15252/embj.2020107237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.George NM, Evans JJ, Luo X. A three-helix homo-oligomerization domain containing BH3 and BH1 is responsible for the apoptotic activity of Bax. Genes Dev. 2007;21:1937–48. doi: 10.1101/gad.1553607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai H, Ding H, Peterson KL, Meng XW, Schneider PA, Knorr KLB, et al. Measurement of BH3-only protein tolerance. Cell Death Differ. 2018;25:282–93. doi: 10.1038/cdd.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai H, Meng XW, Lee S-H, Schneider PA, Kaufmann SH. Context-dependent Bcl-2/Bak interactions regulate lymphoid cell apoptosis. J Biol Chem. 2009;284:18311–22. doi: 10.1074/jbc.M109.004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C, Youle RJ. Predominant requirement of Bax for apoptosis in HCT116 cells is determined by Mcl-1’s inhibitory effect on Bak. Oncogene. 2012;31:3177–89. doi: 10.1038/onc.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye K, Meng WX, Sun H, Wu B, Chen M, Pang YP, et al. Characterization of an alternative BAK-binding site for BH3 peptides. Nat Commun. 2020;11:3301. doi: 10.1038/s41467-020-17074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SH, Meng XW, Flatten KS, Loegering DA, Kaufmann SH. Phosphatidylserine exposure during apoptosis reflects bidirectional trafficking between plasma membrane and cytoplasm. Cell Death Differ. 2013;20:64–76. doi: 10.1038/cdd.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goping IS, Gross A, Lavoie JN, Nguyen M, Jemmerson R, Roth K, et al. Regulated targeting of BAX to mitochondria. J Cell Biol. 1998;143:207–15. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biacore T200 software handbook. GE Healthcare Bio-Sciences AB. Uppsala, Sweden, 2010. p. 165–168.
- 59.Scheffe H. The analysis of variance. New York: John Wiley & Sons, Inc.; 1999.
- 60.Ferrer PE, Frederick P, Gulbis JM, Dewson G, Kluck RM. Translocation of a Bak C-terminus mutant from cytosol to mitochondria to mediate cytochrome C release: implications for Bak and Bax apoptotic function. PLoS One. 2012;7:e31510. doi: 10.1371/journal.pone.0031510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original blots for supplementary figures
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files.