Abstract
The hexanucleotide GGGGCC repeat expansion in the intronic region of C9orf72 is the most common cause of Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The repeat expansion-generated toxic RNAs and dipeptide repeats (DPRs) including poly-GR, have been extensively studied in neurodegeneration. Moreover, haploinsufficiency has been implicated as a disease mechanism but how C9orf72 deficiency contributes to neurodegeneration remains unclear. Here, we show that C9orf72 deficiency exacerbates poly-GR-induced neurodegeneration by attenuating non-homologous end joining (NHEJ) repair. We demonstrate that C9orf72 localizes to the nucleus and is rapidly recruited to sites of DNA damage. C9orf72 deficiency resulted in impaired NHEJ repair through attenuated DNA-PK complex assembly and DNA damage response (DDR) signaling. In mouse models, we found that C9orf72 deficiency exacerbated poly-GR-induced neuronal loss, glial activation, and neuromuscular deficits. Furthermore, DNA damage accumulated in C9orf72-deficient neurons that expressed poly-GR, resulting in excessive activation of PARP-1. PARP-1 inhibition rescued neuronal death in cultured neurons treated with poly-GR peptides. Together, our results support a pathological mechanism where C9orf72 haploinsufficiency synergizes with poly-GR-induced DNA double-strand breaks to exacerbate the accumulation of DNA damage and PARP-1 overactivation in C9orf72 ALS/FTD patients.
Subject terms: Chromatin, Neuroscience
Introduction
The GGGGCC repeat expansion in the noncoding region of the C9orf72 gene is the most common cause of Amyotrophic lateral sclerosis (ALS)/ frontotemporal dementia (FTD) [1, 2]. The pathological mechanisms of C9orf72 repeat expansions include gain of toxicity and loss of function [3]. Gain of toxicity from repeated RNAs and dipeptide repeats (DPRs) in neurodegeneration has been extensively studied. Repeat-containing RNAs can form RNA foci, which may sequester RNA-binding proteins and induce DNA damage in neurons [4–7]. In addition, the repeated RNAs can be translated to generate five distinct DPRs via a repeat-associated non-ATG (RAN) translation mechanism [8–10]. Among these DPR proteins, the arginine-rich ones (poly-GR and poly-PR) are most toxic when ectopically expressed in Drosophila, zebrafish, and mice [3, 11, 12]. Poly-GR and poly-PR interact with RNA-binding proteins and ribosomes to attenuate mRNA translation and nucleocytoplasmic transport [13–15]. Furthermore, the poly-GR and poly-PR attenuate mitochondrial function and elevate cellular oxidative stress to induce DNA damage [12, 16–19]. Increased DNA damage in neurons may activate p53 to drive neurodegeneration [20–22].
A growing body of evidence demonstrates that reduced C9orf72 function also plays a role in neurodegeneration. The reduction of C9orf72 transcript and protein level has been demonstrated in C9orf72 ALS/FTD patients [1, 4, 8, 23–25]. Haploinsufficiency of C9orf72 leads to neurodegeneration in C9orf72 ALS iPSC-derived motor neurons [26]. Furthermore, C9orf72 deficiency exacerbates motor deficits in mice containing repeat expansions [27, 28]. However, the mechanism by which C9orf72 deficiency leads to neurodegeneration remains unclear due to a limited understanding of C9orf72 function. C9orf72 is a DENN-like protein that might regulate autophagic flux and control endosome trafficking [29–33]. C9orf72-deficient mice display deficits in immune system, likely through dysfunction of autophagy [34–37]. Moreover, C9orf72 regulates the assembly of oxidative phosphorylation complex I [38]. Thus, identifying novel C9orf72 functions will likely contribute to a better understanding of C9orf72 ALS/FTD pathology.
In this study, we discovered that, in response to DNA damage, C9orf72 localized to the nucleus and regulated non-homologous end joining (NHEJ) repair by affecting DNA-PK complex assembly. Loss of C9orf72 function exacerbated neurodegeneration and neuromuscular junction deficits in mouse models. Furthermore, accumulated DNA damage in C9orf72-deficient neurons with poly-GR expression led to neuron loss through PARP-1 overactivation. These observations reveal an unexpected function of C9orf72 in the nucleus and provide evidence how reduced C9orf72 functions in NHEJ repair might contribute to DNA damage accumulation and neurodegeneration in C9orf72 ALS/FTD patients.
Results
Accumulation of nuclear C9orf72 at DNA damage sites
To identify potential novel functions for C9orf72, we first sought to identify protein interactors. We immunoprecipitated C9orf72 from HEK293T cells transfected with Flag-C9orf72, and subjected the immunoprecipitated proteins to mass spectrometric analysis. Several C9orf72-binding proteins were identified (Supplementary Table 1), including known C9orf72 interactors SMCR8 and WDR41 [29, 32, 39]. Unexpectedly, among the top-30 abundant binding proteins, 13 proteins localized to the nucleus (Fig. 1a). We were particularly interested in DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (Supplementary Table 1), a key kinase in the NHEJ repair pathway. Thus, we hypothesized that C9orf72 might function in the nucleus to participate in DNA damage repair.
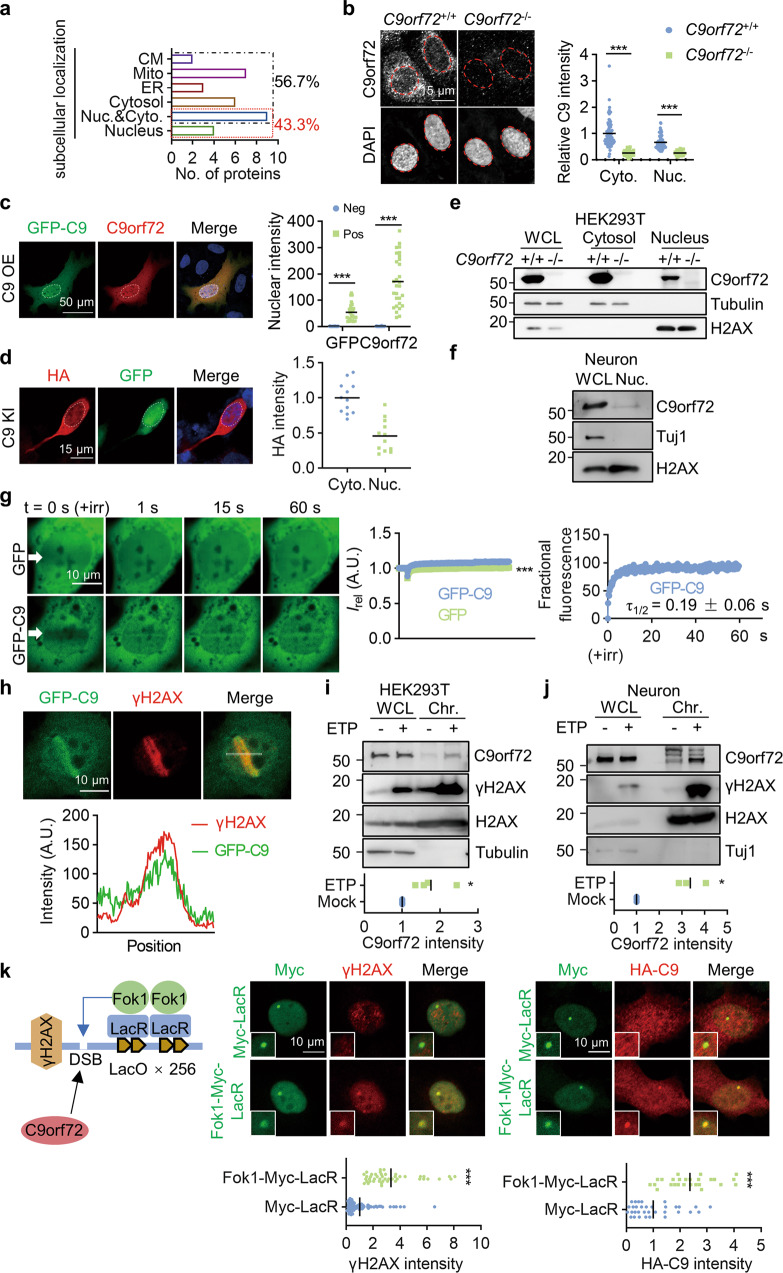
Fig. 1. Accumulation of nuclear C9orf72 at DSB sites.
a Diagram showing subcellular localization of proteins identified as potential C9orf72 binding proteins by IP-MASS analysis. CM, cell membrane; Mito, mitochondria; ER, endoplasmic reticulum. b, c C9orf72 localized to the nucleus and cytosol in cells. b The endogenous C9orf72 was stained with anti-C9orf72 antibody (GTX632041) in WT and C9orf72 KO U2OS cells. Red circles indicate the nuclei (left). Quantitative analysis of C9orf72 intensity in the cytosol and nucleus (right). Data are mean (n = 63-96 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). c The C9orf72 KO U2OS cells were transfected with GFP-C9orf72. The localization of GFP-C9orf72 was revealed by anti-GFP and anti-C9orf72 (GTX632041) antibodies. White circles indicate the nuclei (left). Quantitative analysis of C9orf72 intensity in the nucleus (right). Data are mean (n = 29 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). d The HA tag was inserted into endogenous C9orf72 gene in HEK293T cells by CRISPR-Cas9. The localization of C9orf72 was revealed by anti-HA antibody, and cells transfected with CRISPR construct were indicated by GFP. White circles indicate the nuclei (left). Quantitative analysis of HA intensity (right). Data are mean (n = 12 cells from 3 independent experiments). e, f Nuclear localization of endogenous C9orf72 was revealed by subcellular fraction analysis in HEK293T cells (e) and neurons (f) (n = 3 independent experiments). g Accumulation of GFP-C9orf72 protein at sites of DNA DSB. The U2OS cells were transfected with the constructs encoding GFP or GFP-C9orf72. Two days after transfection, cells were stained with Hoechst and subjected to laser-generated DNA lesion by a 405 nm laser (+irr). Time-lapse images of the transfected cells were taken by a confocal microscope. White arrows indicate the damaged regions of interest (ROI) (left). Quantitative analysis of relative fluorescence intensity (Irel) as a function of time at lesioned ROIs for U2OS cells transfected with GFP (green) or GFP-C9orf72 (blue) (middle) (right). Data are mean (n = 12–16 cells from 3 independent experiments; Two-way ANOVA; ***, p < 0.001). h GFP-C9orf72 co-localized with γH2AX in U2OS cells after DNA DSB induction. U2OS cells transfected with GFP-C9orf72 were fixed immediately after laser-induced DNA damage and stained with anti-γH2AX antibody. The fluorescence-intensity profile of GFP-C9orf72 (green) and γH2AX (red) were obtained along the white line. i, j Increased C9orf72 in the chromatin fraction after DNA damage. HEK293T cells (i) and neurons (j) were treated with ETP (10 μM) for 4 h. The whole cell lysate (WCL) and chromatin fraction (Chr.) were isolated for immunoblotting analysis. Quantitative analysis of C9orf72 intensity in the chromatin fraction. Data are mean (n = 3-4 independent experiments; paired t test; *, p < 0.05). k Schematic diagram of the LacO/LacR (Lac operator/Lac repressor) system to analyze recruitment of C9orf72 on chromatin after DNA DSB (left). U2OS LacO cells contain Lac operator array in the genome to tether LacR protein at LacO array. U2OS LacO cells were transfected with Fok1-LacR to tether the nuclease Fok1 at LacO array to generate DNA DSB. U2OS LacO cells expressing Fok1-Myc-LacR or Myc-LacR, together with HA-C9orf72 were stained with anti-γH2AX (middle) (n = 45-88 cells from 3 independent experiments) or anti-HA (right) (n = 26-33 cells from 3 independent experiments) antibodies. Quantitative analysis of γH2AX intensity or HA-C9orf72 intensity. Data are mean (unpaired t-test; ***, p < 0.001).
To test this hypothesis, we first sought to determine whether C9orf72 localized to the nucleus. Because uncertainty in the specificity of C9orf72 antibodies in immunofluorescence staining, the nuclear localization of C9orf72 is controversial [40]. We therefore performed immunofluorescence staining in WT and C9orf72 knockout (KO) U2OS cells using a C9orf72 antibody (GTX632041) [40]. The GTX632041 immunofluorescence staining revealed punctate signal mainly in the cytosol. The signal in the nucleus was much weaker and was dramatically reduced in C9orf72 KO cells (Fig. 1b). In contrast, the GTX632041 fluorescent signal was diffuse in the cytosol and nucleus in C9orf72 KO cells transfected with GFP-C9orf72 (Fig. 1c), similar to what was observed in HA-tagged endogenous C9orf72 cells (Fig. 1d), and 2×HA-tagged endogenous C9orf72 cells [41]. We performed subcellular fraction analysis, which also revealed that C9orf72 was present in the cytosolic and nuclear fractions in HEK293T cells and neurons (Fig. 1e-f). Together, these results indicate that C9orf72 can localize to the nucleus.
To determine whether C9orf72 participates in DNA damage repair, we examined the recruitment of C9orf72 to sites of DNA damage. We transfected GFP or GFP-C9orf72 into U2OS cells and used laser microirradiation to generate subnuclear DNA double-strand breaks (DSBs) within the nucleus of Hoechst-stained living cells. GFP-C9orf72 accumulated at laser-lesioned regions almost immediately after microirradiation (τ1/2 = 0.19 ± 0.06 s) (Fig. 1g). Moreover, GFP-C9orf72 co-localized with the DNA DSB marker γH2AX (H2AX phosphorylation on S139) after laser microirradiation (Fig. 1h). In addition, C9orf72 was enriched in the chromatin fraction in HEK293T cells and neurons treated with the topoisomerase II inhibitor Etoposide (ETP) to induce DNA DSB [42] (Fig. 1i-j). We also used the LacR/LacO system to generate DNA DSB by tethering the nuclease (Myc-tagged Fok1-LacR) at an integrated LacO array in U2OS cells [43–45]. Tethering of Fok1-LacR at the LacO array led to DNA DSB revealed by γH2AX staining (Fig. 1k). Consistently, HA-C9orf72 was recruited to the DNA damage sites induced by Fok1-LacR (Fig. 1k). Together, these results indicate that C9orf72 is rapidly recruited to DNA DSB sites and suggest that it may play a role in DNA damage repair.
Attenuated NHEJ repair in C9orf72-deficient neurons
Accumulation of DNA damage, particularly DNA DSB, has been linked to various neurodegenerative diseases [12, 16, 46]. Two main pathways are involved in DSB repair: NHEJ that repairs DSB by directly ligating the broken ends, and homologous recombination (HR) that repairs DSB by using a homologous DNA template and selectively occurs in the S and G2 phases in cell cycles [47]. Therefore, NHEJ is the primary pathway of DNA DSB repair in neurons.
To define the role of C9orf72 in DNA damage repair, we examined DNA damage in cultured neurons by using a single-cell gel electrophoresis assay (comet assay). C9orf72 KO neurons displayed longer tail moments than WT neurons even without treating with a DNA-damaging agent (H2O2) (Fig. 2a), suggesting an increased amount of DNA damage in C9orf72 KO neurons. In the presence of H2O2, both WT and KO neurons showed increased tail moments (Fig. 2a). After H2O2 withdrawal and recovery for 24 h, the tail moments in WT neurons were significantly reduced, whereas C9orf72 KO neurons continued to have long tail moments (Fig. 2a), suggesting that C9orf72 KO neurons are deficient in DNA damage repair. ETP can induce DNA DSB and thus increase γH2AX. We compared the kinetics of the ETP-induced γH2AX signal between WT and C9orf72 KO HEK293T cells. The γH2AX levels in C9orf72 KO cells were significantly lower than that of WT cells after ETP treatment (Fig. 2b). Consistently, immunofluorescence analysis revealed that the intensity of γH2AX in C9orf72 KO neurons was significantly lower than that in WT neurons after ETP treatment (Fig. 2c). We also observed a reduction in γH2AX staining in C9orf72 KO neurons derived from human neural progenitor cells (NPCs) after DNA DSB induction (Fig. S1a-b). Interestingly, despite the increased levels of DNA damage (Fig. 2a), the γH2AX level, which serves as not only a marker of DNA DSB but also as a signaling molecule in DNA DSB repair, was reduced in C9orf72 KO neurons after DNA DSB induction (Fig. 2b-c). These results indicate that C9orf72 may be involved in the initiation of the cellular DNA damage response (DDR) and H2AX phosphorylation. A similar phenotype was also observed in SIRT1- and FUS-deficient neurons [48, 49].
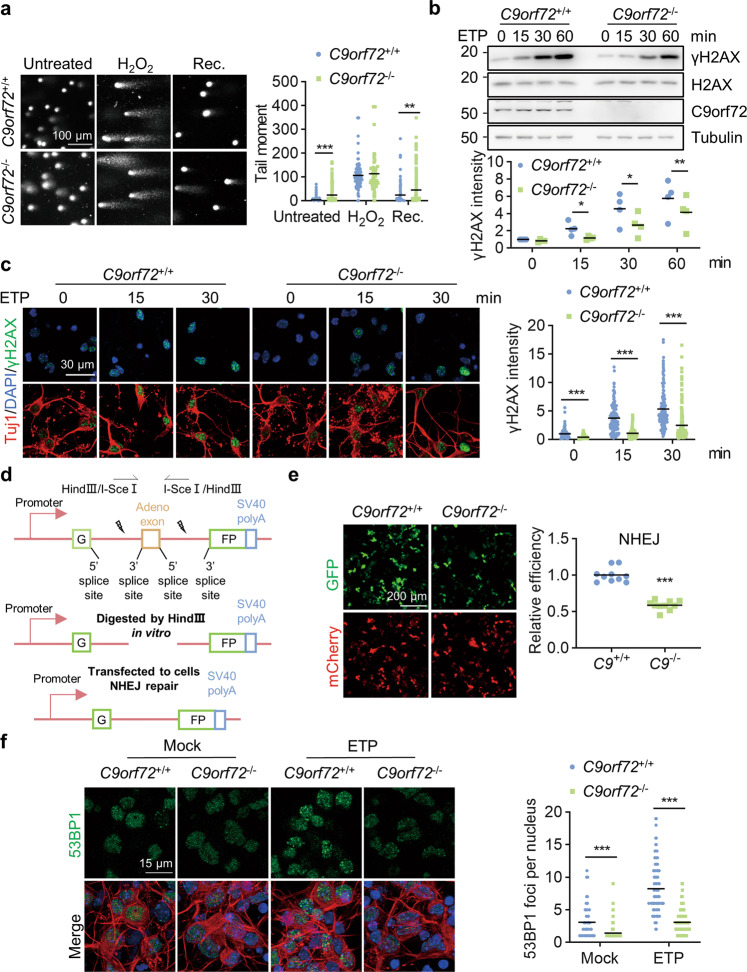
Fig. 2. C9orf72 deficiency leads to attenuated NHEJ repair.
a Impaired DNA damage repair in C9orf72−/− neurons. Primary cultured cortical neurons from C9orf72+/+ and C9orf72−/− mice were treated with 200 μM H2O2 for 3 h (H2O2). After withdrawal of H2O2, the neurons were cultured for 24 h to repair DNA damage (Rec.). DNA damage in neurons was assessed by a comet assay. Quantitative analysis of tail moment. Data are mean (n = 67–237 cells from 3 independent experiments; unpaired t-test; **, p < 0.01; ***, p < 0.001). b Reduced γH2AX level in C9orf72 KO cells compared with WT cells after ETP treatment. HEK293T cells with indicated genotypes were treated with 10 μM ETP for the indicated time points. The γH2AX level was revealed by blotting with anti-γH2AX antibody. Quantitative analysis of γH2AX intensity. Data are mean (n = 4 independent experiments; paired t-test; *, p < 0.05; **, p < 0.01). c Decreased γH2AX level in C9orf72−/− neurons compared with WT neurons after ETP treatment. Primary cultured cortical neurons from C9orf72+/+ and C9orf72−/− mice were treated with 10 μM ETP for the indicated time points, fixed and stained with anti-γH2AX antibody (green) and anti-Tuj1 antibody (red). Quantitative analysis of γH2AX intensity. Data are mean (n = 145–200 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). d Diagram of NHEJ reporter construct. The NHEJ report construct contains a GFP gene with two HindIII sites in the intron region. The DSB was induced by digestion with HindIII in vitro. e Attenuated NHEJ repair capacity in C9orf72 KO cells. DNA DSB was generated by HindIII digestion in the NHEJ reporter construct. Indicated genotypes HEK293T cells were transfected with cleaved reporter construct. The GFP-positive cells were counted as successful NHEJ repair and mCherry was used for transfection control. Quantitative analysis of relative NHEJ efficiency. Data are mean (n = 10 images of 3 independent experiments; unpaired t-test; ***, p < 0.001). f Reduced number of 53BP1 foci in C9orf72−/− neurons. Primary cultured cortical neurons with indicated genotypes were treated with 10 μM ETP for 30 min, fixed, and stained with anti-53BP1 (green) and anti-Tuj1 (red) antibodies. Quantitative analysis of 53BP1 foci in the nucleus. Data are mean (n = 112–150 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001).
To determine whether C9orf72 functions in NHEJ repair, we performed a NHEJ reporter assay, using a construct consisting of a GFP gene with an artificially engineered intron and two HindIII restriction sites for DSB induction (Fig. 2d) [50]. The HEK293T cells were transfected with the cleaved NHEJ reporter construct together with an mCherry construct as a transfection control. The NHEJ-mediated repair was largely reduced in cells treated with the DNA-PKcs inhibitor NU7026 (Fig. S2a). We next transfected the cleaved NHEJ reporter into WT and C9orf72 KO HEK293T cells. The percentage of GFP-positive cells was greatly reduced in C9orf72 KO cells (Fig. 2e), suggesting that C9orf72 deficiency causes marked impairment in NHEJ-mediated DSB repair. Moreover, the ATM inhibitor KU55933 did not alter the NHEJ repair efficiency in C9orf72 KO cells (Fig. S2b). The alt-NHEJ repair efficiency and HR repair efficiency were comparable between WT and C9orf72 KO cells (Fig. S2c–f). To further confirm that C9orf72 participates in NHEJ-mediated DNA repair, we examined the immunofluorescent signal of 53BP1 in neurons after DSB induction. DNA DSB-induced 53BP1 foci formation was significantly reduced in C9orf72 KO neurons compared to WT neurons (Fig. 2f). These results indicate that C9orf72 deficiency leads to attenuated NHEJ-mediated DNA repair in neurons.
C9orf72 functions in DNA-PK assembly
NHEJ-mediated DNA repair requires many proteins, including DNA-dependent protein kinase (DNA-PK) complex, the end processing complex, and the Ligase 4/XRCC4 complex [51]. DNA-PKcs together with the Ku70/80 heterodimer comprise the DNA-PK complex, and initiate the NHEJ process after DNA DSB [52]. Moreover, DNA-PKcs belongs to the atypical phosphatidylinositol-3 kinase-related kinase (PIKK) family which controls the initiation of DDR signaling and H2AX phosphorylation [53, 54]. In response to DNA DSB, DNA-PKcs undergoes autophosphorylation, and phosphorylation at T2609 and S2056 indicates NHEJ pathway activation [55].
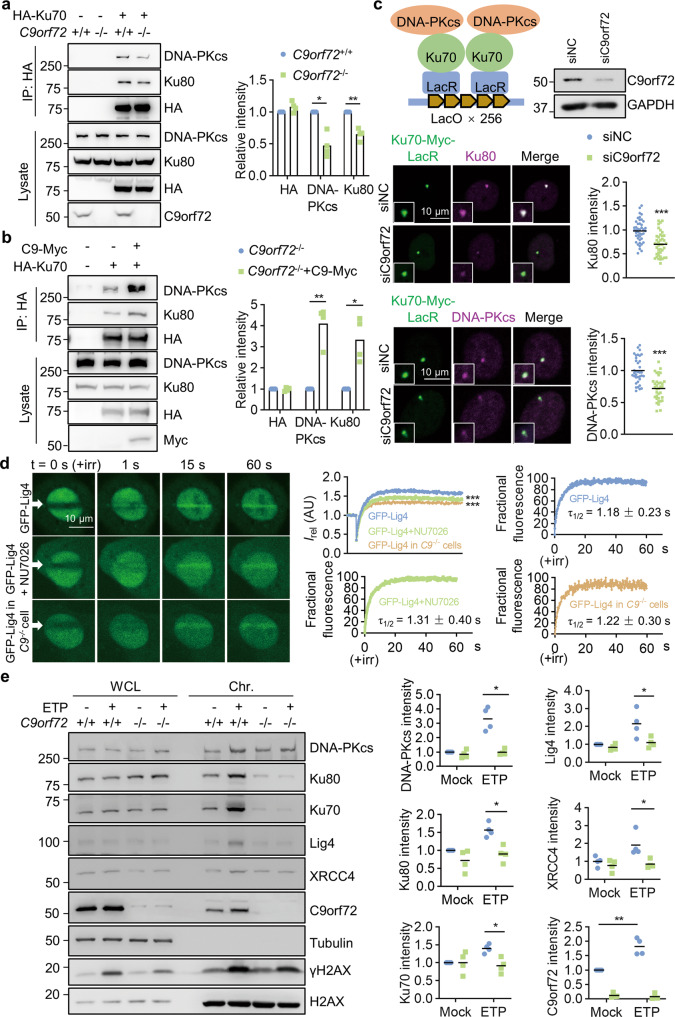
We examined DNA-PKcs phosphorylation kinetics in WT and C9orf72 KO HEK293T cells following ETP treatment. DNA-PKcs phosphorylation levels were significantly reduced in C9orf72 KO cells compared to WT cells after ETP treatment (Fig. 3a). Consistently, immunofluorescence staining revealed that DNA-PKcs phosphorylation was significantly lower in C9orf72 KO neurons than in WT neurons (Fig. 3b), suggesting that C9orf72 participates in DNA-PKcs activation. Notably, the reduced γH2AX levels in C9orf72 KO cells following DNA DSB (Fig. 2b, c) may result from attenuated DNA-PKcs activation [56]. We thus tested whether C9orf72 regulates DNA-PKcs phosphorylation through interaction with the DNA-PK complex. We performed immunoprecipitation in HEK293T cells transfected with Flag-C9orf72 and detected co-immunoprecipitated Ku70, Ku80, and DNA-PKcs (Fig. 3c, d). Interestingly, the C9orf72 and DNA-PK complex interaction was increased after DNA DSB induction (Fig. 3c, d). Moreover, we used the LacR/LacO system to tether Myc-tagged Ku70-LacR or Ku80-LacR fusion proteins at the integrated LacO array. HA-C9orf72 co-localized with Myc-tagged Ku70 or Ku80 at nuclear foci (Fig. 3e), indicating that C9orf72 interacts with Ku70/Ku80 on chromatin.
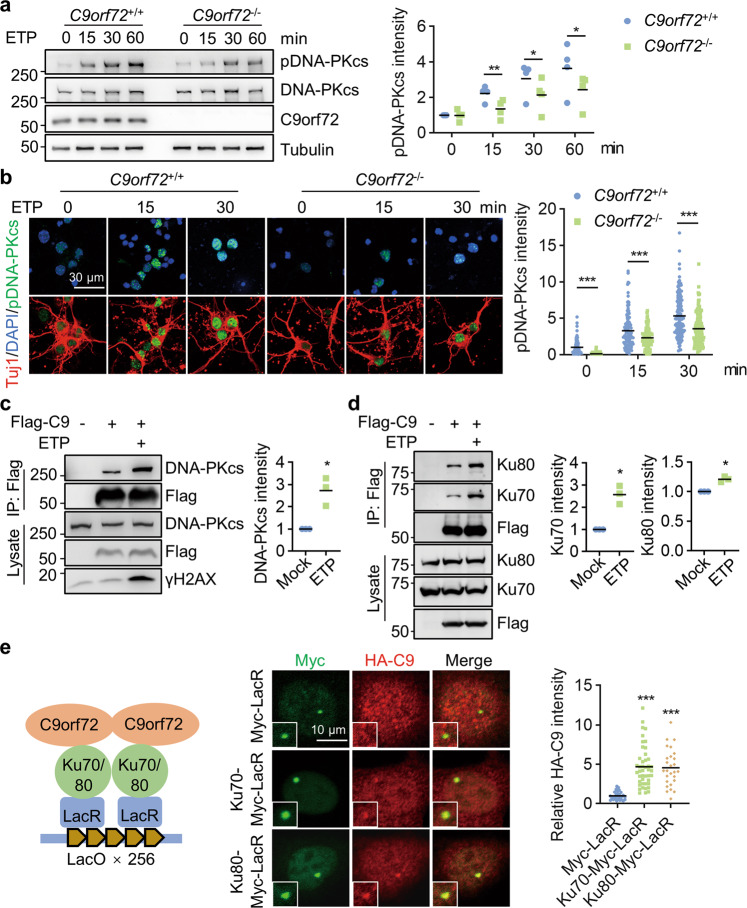
Fig. 3. C9orf72 binds with DNA-PK complex to regulate DNA-PKcs phosphorylation.
a Impaired induction of DNA-PKcs phosphorylation in C9orf72-deficient cells after DNA DSB. HEK293T cells were treated with 10 μM ETP for the indicated time. Cell lysates were blotted for indicated antibodies. Quantitative analysis of pDNA-PKcs intensity. Data are mean (n = 4 independent experiments; paired t-test; *, p < 0.05; **, p < 0.01). b Impaired induction of DNA-PKcs phosphorylation in C9orf72−/− neurons treated with ETP. Primary cultured cortical neurons from C9orf72+/+ and C9orf72−/− mice were treated with 10 μM ETP for the indicated time, fixed, and stained with anti-pDNA-PKcs antibody (green) and anti-Tuj1 antibody (red). Quantitative analysis of pDNA-PKcs intensity. Data are mean (n = 115–150 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). c, d Increased interaction between C9orf72 and DNA-PK complex after DNA damage. HEK293T cells were transfected with Flag-C9orf72 and treated with ETP. The immunoprecipitated complex was probed with anti-DNA-PKcs antibody (c), and anti-Ku70, and anti-K80 antibodies (d). Quantitative analysis of intensities. Data are mean (n = 3 independent experiments; paired t-test; *, p < 0.05). e Schematic diagram of the LacO/LacR system to analyze C9orf72 interaction with Ku70 and Ku80 on chromatin (left). U2OS LacO cells were transfected with HA-C9orf72 and Ku70-Myc-LacR or Ku80-Myc-LacR, and stained with anti-Myc (green), and anti-HA (red) antibodies. Quantitative analysis of HA-C9orf72 intensity. Data are mean (n = 27–44 cells from 3 independent experiments; one-way ANOVA; ***, p < 0.001).
To investigate whether C9orf72 binds to DNA-PK complex components directly, we purified Flag-C9orf72 protein and performed pull-down assay. The results revealed that Flag-C9orf72 did not interact with purified GFP-Ku70 or GFP-Ku80 (Fig. S3a, b). In contrast, Flag-C9orf72 protein interacted with DNA-PKcs in the cell lysate (Fig. 4a). In addition, HA-C9orf72 interacted with purified DNA-PKcs KIP domain (Fig. 4b), but not the FAT domain (Fig. S3c) in the pull-down assay. Moreover, DNA-PKcs deficiency attenuated C9orf72 and Ku70/80 interaction in co-immunoprecipitation experiment (Fig. 4c). However, C9orf72 and DNA-PKcs interaction was not altered in Ku70-deficient cells (Fig. 4d). We also found that the C-terminal domain of C9orf72 was involved in binding DNA-PKcs (Fig. 4 e, f). Together, these results suggest that C9orf72 may interact with DNA-PKcs directly to associate with DNA-PK complex.
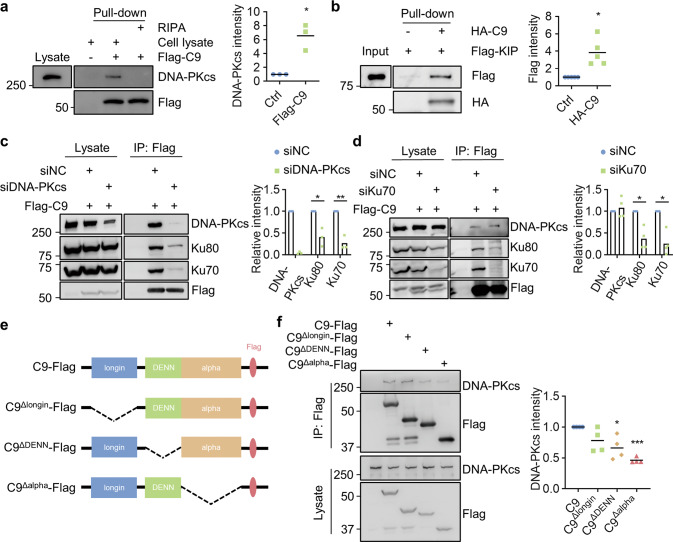
Fig. 4. C9orf72 directly interacts with DNA-PKcs.
a, b C9orf72 interacts with DNA-PKcs directly. a The purified Flag-C9orf72 protein pulled down DNA-PKcs in the cell lysate. Quantitative analysis of DNA-PKcs intensity. b HA-C9orf72 protein directly interacted with Flag-tagged KIP domain of DNA-PKcs in in-vitro pulldown assay. Data are mean (n = 3–5 independent experiments; paired t-test; *, p < 0.05). c, d DNA-PKcs deficiency impaired interaction between C9orf72 and Ku70, Ku80. HEK293T cells were transfected with Flag-C9orf72 and siDNA-PKcs (c), or siKu70 (d), and immunoprecipitated with anti-Flag antibody. The immunoprecipitated complex was probed with indicated antibodies. Quantitative analysis of intensities of Ku70, Ku80, and DNA-PKcs. Data are mean (n = 3 independent experiments; paired t-test; *, p < 0.05; **, p < 0.01). e, f The C terminus of C9orf72 is required for interaction with DNA-PKcs. e Schematic diagram of C9orf72 constructs. f HEK293T cells were transfected with C9orf72-Flag or indicated deletions. The immunoprecipitated complex was probed with DNA-PKcs antibody. Quantitative analysis of DNA-PKcs intensity. Data are mean (n = 4 independent experiments; paired t-test; *, p < 0.05; ***, p < 0.001).
To determine how C9orf72 regulates DNA-PKcs phosphorylation following DNA DSB, we suspected that C9orf72 may affect DNA-PK complex assembly. We found that the Ku70, Ku80, and DNA-PKcs interaction was significantly reduced in C9orf72 KO HEK293T cells (Fig. 5a). Furthermore, the interaction between Ku70, Ku80, and DNA-PKcs was markedly increased when C9orf72 was overexpressed in the KO cells (Fig. 5b), suggesting that C9orf72 promotes DNA-PK complex assembly. By using the LacR/LacO system, we observed Ku70 and Ku80 interaction revealed by a Myc-tagged Ku70-LacR fusion protein and Ku80 immunostaining. Consistent with the co-immunoprecipitation experiment, the intensity of Ku80 staining was significantly reduced in C9orf72-deficient cells (Fig. 5c). Similarly, the interaction between Ku70 and DNA-PKcs was also decreased in C9orf72-deficient cells (Fig. 5c). Together, these results provide evidence that C9orf72 contributes to DNA-PKcs activation by regulating DNA-PK complex assembly at DNA DSB sites.
Fig. 5. C9orf72 promotes DNA-PK complex assembly and recruitment of Ligase 4/XCCR4 complex.
a C9orf72 deficiency reduced DNA-PK complex assembly. HEK293T cells with indicated genotypes were transfected with HA-Ku70, and immunoprecipitated with anti-HA antibody. The immunoprecipitated complex was probed with indicated antibodies. Quantitative analysis of intensities of Ku70, Ku80 and DNA-PKcs. Data are mean (n = 4 independent experiments; paired t-test; *, p < 0.05; **, p < 0.01). b C9orf72 overexpression rescues DNA-PK complex assembly in C9orf72 KO cells. C9orf72 KO HEK293T cells were transfected with HA-Ku70 with or without C9orf72-Myc, and immunoprecipitated with anti-HA antibody. The immunoprecipitated complex was probed with indicated antibodies. Quantitative analysis of intensities of Ku70, Ku80, and DNA-PKcs. Data are mean (n = 4 independent experiments; paired t-test; *, p < 0.05; **, p < 0.01). c Schematic diagram and representative images of reduced Ku70 interaction with Ku80 or DNA-PKcs on chromatin in C9orf72-deficient cells. U2OS LacO cells were transfected with control siRNA or C9orf72 siRNA, and followed by transfection with Ku70-Myc-LacR in 12 h. The U2OS cells were stained with indicated antibodies. Quantitative analysis of Ku80 and DNA-PKcs intensities. Data are mean (n = 35–43 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). d Reduced GFP-Lig4 accumulation at laser-lesioned sites in C9orf72-deficient cells. The U2OS cells with indicated genotypes were transfected with GFP-Lig4. Two days after transfection, cells were stained with Hoechst and subjected to laser-generated DNA lesion by a 405 nm laser (+irr). Time-lapse images of the transfected cells were taken by a confocal microscope. White arrows indicate the damaged regions of interest (ROI). Data are mean (n = 3 independent experiments; Two-way ANOVA; ***, p < 0.001). Quantitative analysis of relative fluorescence intensity (Irel) as a function of time at lesioned ROIs for U2OS cells transfected with GFP-Lig4 (blue, WT cells, n = 11; green, NU7026-treated cells, n = 10; yellow, C9orf72−/− cells, n = 12). Modeling and regression analysis of the quantitative data. e C9orf72 deficiency reduced recruitment of NHEJ repair proteins on the chromatin after DNA damage. HEK293T cells with indicated genotypes were treated with ETP (5 μM) overnight and performed chromatin fraction assay. NHEJ proteins were probed by indicated antibodies in immunoblots. Quantitative analysis of intensities of indicated proteins. Data are mean (n = 4 independent experiments; paired t-test; *, p < 0.05; **, p < 0.01).
C9orf72 promotes recruitment of Ligase 4 to DNA damage sites
During NHEJ, the initial event is the binding of the Ku70/Ku80 complex to DNA broken ends and recruiting DNA-PKcs to form the DNA-PK complex. Then, the broken DNA ends are processed by several nucleases and polymerases. Finally, the Ligase 4/XRCC4 complex is recruited to DNA DSB sites and catalyzes the ligation of the DNA ends [51]. To determine whether recruitment of the Ligase 4/XRCC4 complex is altered in C9orf72-deficient cells, we monitored the localization kinetics of GFP-Lig4 in response to laser-induced DNA damage. Indeed, GFP-Lig4 was recruited to laser-lesioned regions after microirradiation (τ1/2 = 1.18 ± 0.23 s). The recruitment of Ligase 4 is dependent on DNA-PKcs activation, and inhibition of DNA-PKcs by NU7026 caused a significant reduction of the maximum intensity of GFP-Lig4 at laser-lesioned regions (Fig. 5d). The maximum intensity of GFP-Lig4 at laser-lesioned regions was largely reduced in C9orf72 KO cells compared to that of WT cells, whereas the kinetics of GFP-Lig4 recruitment was similar (WT τ1/2 = 1.18 ± 0.23 s versus KO τ1/2 = 1.22 ± 0.30 s) (Fig. 5d). We examined whether C9orf72 regulates DNA DSB-induced recruitment of the endogenous Ligase 4/XRCC4 complex to chromatin. Chromatin fraction analysis revealed that ETP treatment induced the accumulation of C9orf72, Ligase 4, and XRCC4 on chromatin in WT HEK293T cells. In contrast, DNA damage-induced accumulation of the Ligase 4/XRCC4 complex was significantly decreased in C9orf72 KO cells (Fig. 5e). These results indicate that C9orf72 promotes recruitment of the Ligase 4/XRCC4 complex to DNA damage sites.
Loss of C9orf72 in mice expressing poly-GR exacerbates neurodegeneration
Previous studies have shown that ectopic expression of poly-GR increased DNA DSB in neurons [12, 16]. To investigate whether C9orf72 deficiency exacerbates DNA damage accumulation and neurodegeneration in neurons with toxic poly-GR expression, we used adeno-associated virus (AAV9) to deliver GFP (AAV-GFP), or GFP-poly-GR50 (AAV-GR50) [3, 12] to the nervous system of WT and C9orf72−/− mice. We administered the AAVs into the cerebroventricular region of mice at postnatal day 0 (P0). One month later, we mainly observed GFP signals in neurons, including NeuN-positive neurons in the cortex and choline acetyltransferase (ChAT)-positive motor neurons in the spinal cord (Fig. S4a, b).
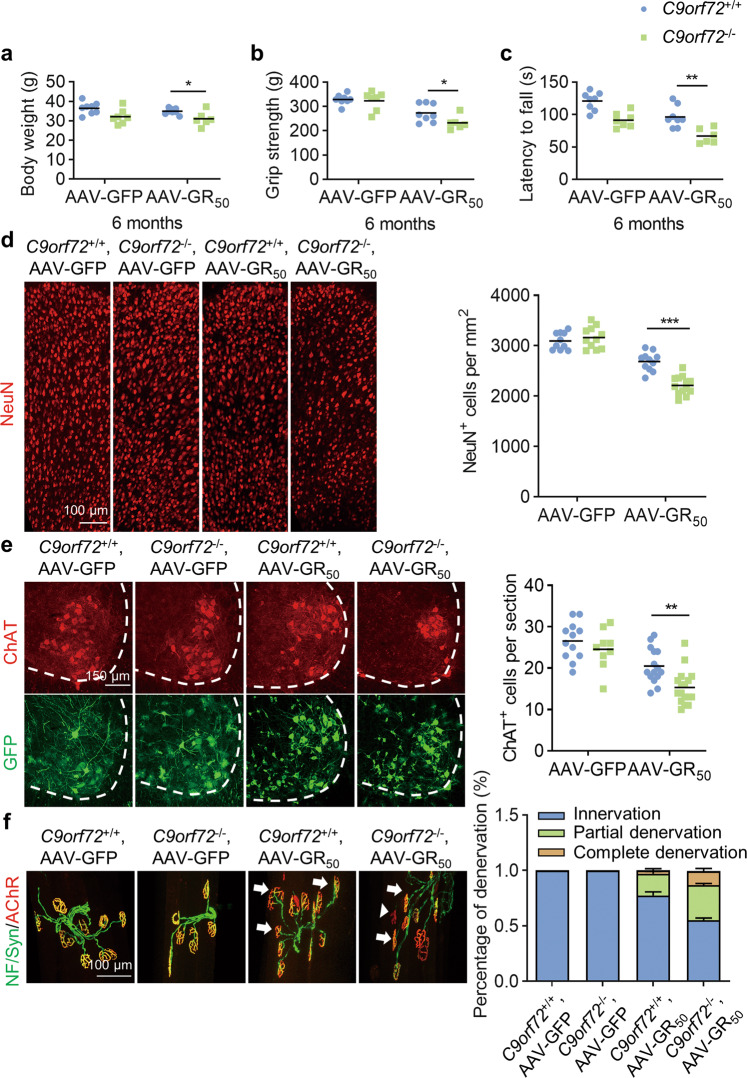
Six months after virus infection, the C9orf72−/− mice infected with AAV-GR50 displayed reduced body weight (Fig. 6a), decreased grip strength (Fig. 6b), and worsened rotarod experiment performance (Fig. 6c). The number of NeuN-positive neurons was significantly reduced in C9orf72+/+, AAV-GR50 mice compared to C9orf72+/+, AAV-GFP mice, and was further reduced in C9orf72−/−, AAV-GR50 mice (Fig. 6d). Consistently, poly-GR expression caused loss of ChAT-positive motor neurons in the lumbar spinal cord. Moreover, C9orf72 deficiency in poly-GR expression mice caused severe motor neuron degeneration (Fig. 6e). The intensity of GFAP and Iba1 was increased in the cortex and lumbar spinal cord in C9orf72−/−, AAV-GR50 mice (Fig. S5a–d), indicating increased activation of astrocytes and microglia in both cortex and spinal cord. Together, these results indicate that loss of C9orf72 exacerbates neuronal loss caused by the poly-GR.
Fig. 6. C9orf72 deficiency exacerbates motor deficits and neurodegeneration in poly-GR expression mice.
a Reduced body weight of C9orf72−/−, AAV-GR50 mice at 6 months of age. Data are mean (n = 6–8 mice; unpaired t-test; *, p < 0.05). b Reduced grip strength of four limbs in C9orf72−/− mice injected with AAV-GR50 at 6 months of age. Data are mean (n = 6–8 mice; unpaired t-test; *, p < 0.05). c Impaired motor performance on an accelerating rotarod test of C9orf72−/−, AAV-GR50 mice at 6 months of age. Data are mean (n = 6–8 mice; unpaired t-test; **, p < 0.01). d Decreased number of NeuN-positive cortical neurons in the motor cortex of C9orf72−/−, AAV-GR50 mice at 6 months of age. The brain slices from mice of indicated genotypes were stained with anti-NeuN antibody to label cortical neurons in the motor cortex. Data are mean (n = 10–13 sections of 3 mice; unpaired t-test; ***, p < 0.001). e Decreased number of ChAT-positive motor neurons in the spinal cord of C9orf72−/−, AAV-GR50 mice at 6 months of age. The lumbar spinal cord slices from mice of indicated genotypes were stained with anti-ChAT antibody to reveal motor neurons in the anterior horn. Data are mean (n = 9–17 sections of 3 mice; unpaired t-test; **, p < 0.01). f Increased neuromuscular junction denervation in C9orf72−/−, AAV-GR50 mice at 6 months of age. The tibialis anterior muscle fibers from mice of indicated genotypes were whole mount stained with CF555-BTX (red) to label AChR clusters and anti-NF/synapsin-1 antibodies (green) to label motor nerve terminals. Arrows, partially denervated NMJ; arrow heads, completely denervated NMJ (n = 9–21 images of 3 mice).
The withdrawal of motor nerve terminals from muscle fibers is an early event in ALS, and denervation of the neuromuscular junction (NMJ) contributes to motor deficits and muscle atrophy [57, 58]. To characterize NMJs, we dissected tibialis anterior (TA) muscle fibers and stained them with anti-neurofilament/synapsin-1 antibodies and CF555-conjugated α-bungarotoxin (CF555-BTX) to label pre-synaptic nerve terminals and post-synaptic acetylcholine receptors (AChR), respectively. AAV-GR50 mice showed increased partially-denervated and completely-denervated NMJs, and motor nerve denervation was much more severe in C9orf72−/−, AAV-GR50 mice (Fig. 6f). Thus, C9orf72 deficiency exacerbates neurodegenerative phenotypes of poly-GR expressing mice.
Accumulation of DNA damage in C9orf72-deficient neurons with poly-GR expression
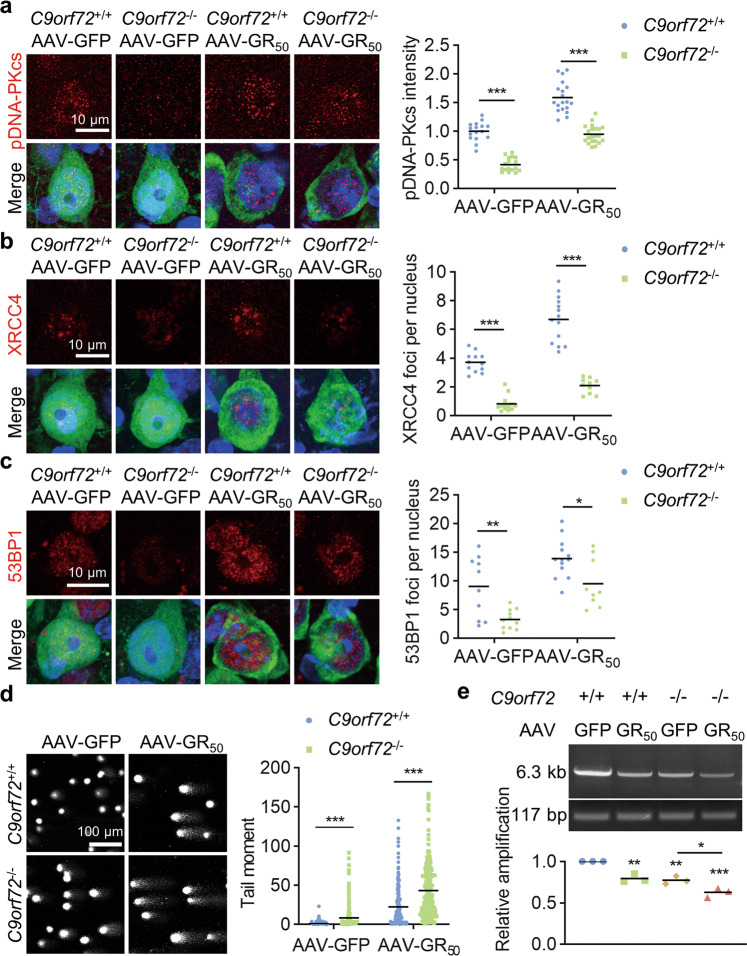
To determine whether NHEJ repair is attenuated in C9orf72-deficient neurons, we performed immunostaining in the brain sections. Consistent with previous results, pDNA-PKcs intensity was reduced in C9orf72−/− neurons compared to WT neurons (Fig. 7a). Moreover, the number of XRCC4 and 53BP1 foci was reduced in C9orf72−/− neurons in both AAV-GFP and AAV-GR50 mice (Fig. 7b, c). Impaired NHEJ repair in neurons may lead to accumulated DNA damage. We thus isolated cortical neurons from AAV-infected mice and performed comet assays. The poly-GR-expressing neurons displayed longer tail moments than the GFP-expressing neurons in WT mice (Fig. 7d). The C9orf72-deficient neurons with poly-GR expression displayed the longest tail moments (Fig. 7d). Similar to what was observed in AAV mice, neural progenitor cells derived from a C9ALS patient displayed attenuated NHEJ repair signal transduction (Fig. S6a-d) and impaired DNA repair efficiency (Fig. S6e). Moreover, we isolated genomic DNA from the cortex to perform long amplicon PCR (LA-PCR). We amplified a 6.3-kb amplicon derived from the Dntt gene and used a 117-bp amplicon in the mitochondrial DNA as a control. The long amplicon intensity was significantly reduced in the cortex from poly-GR-expressing mice and C9orf72 KO mice (Fig. 7e) and was further decreased in the cortex from C9orf72−/−, AAV-GR50 mice (Fig. 7e). Together, these results indicate that C9orf72 deficiency leads to impaired NHEJ repair and accumulated DNA damage in neurons with poly-GR expression.
Fig. 7. C9orf72 deficiency impairs NHEJ repair to induce DNA damage accumulation in poly-GR expression mice.
a–c C9orf72 deficiency attenuated NHEJ repair in neurons. The brain slices from mice of indicated genotypes were stained with anti-pDNA-PKcs (a), anti-XRCC4 (b), and anti-53BP1 (c) antibodies, respectively. Data are mean (n = 9–21 sections of 3 mice; unpaired t-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001). d Accumulated DNA damage in C9orf72−/−, AAV-GR50 mice. The cortical neurons were separated from cortex with indicated genotypes. DNA damage in neurons was assessed by the comet assay. Quantitative analysis of tail moment. Data are mean (n = 256 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). e Reduced genomic integrity in the cortex of C9orf72−/−, AAV-GR50 mice. The genomic DNA was purified from the cortex of indicated genotypes. A 6.3-kb amplicon in Dntt gene was amplified by long amplicon PCR. Quantitative analysis of the long amplicon. Data are mean (n = 3 independent experiments; Two-way ANOVA; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
Excessive activation of poly(ADP-ribose) polymerase-1 (PARP-1) in C9orf72-deficient neurons with poly-GR expression
Accumulated DNA damage in neurons may induce activation of poly(ADP-ribose) polymerase (PARP), producing PAR polymers. PARP-1 is the largest amount of PARP activity to produce PAR polymers in cells. Excessive activation of PARP-1 may lead to exhaustion of NAD+ and ATP pools, and eventually cell death [59, 60]. Moreover, PARP-1 overactivation has been implicated in many neurological disorders, including ALS [61–64].
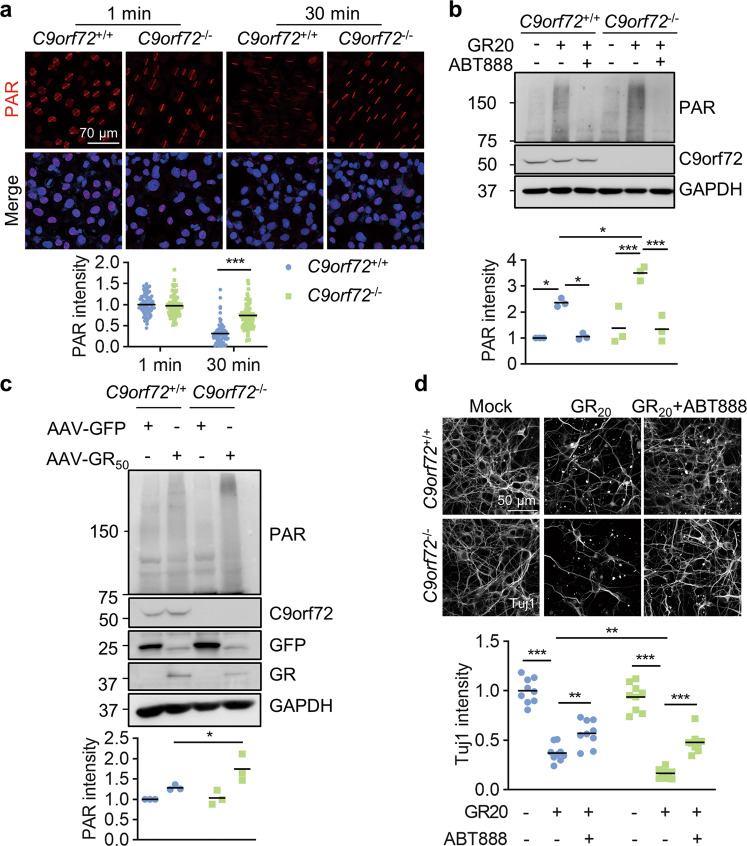
To determine whether C9orf72 participates in PARP-1 activation, we performed microirradiation experiment in U2OS cells. The immunostaining results revealed that the PAR staining in C9orf72 KO cells was comparable to WT cells at 1 min and remained at a high level even 30 min after microirradiation (Fig. 8a). This result suggests that C9orf72 deficiency leads to the constitutive activation of PARP-1 and PAR accumulation at DNA damage sites, likely due to attenuated DNA damage repair. We next analyzed PAR production in cortical neurons and immunoblotting revealed that PAR polymers were significantly increased in WT neurons treated with the poly-GR peptide. Moreover, poly-GR treatment induced more PAR production in C9orf72 KO neurons compared to WT neurons (Fig. 8b). Poly-GR-induced PAR production in neurons likely results from PARP activation, because PAR production was abolished in neurons by treatment with the PARP inhibitor ABT-888 (veliparib). Moreover, we observed elevated PAR production in the cortex of C9orf72−/−, AAV-GR50 mice (Fig. 8c). Treatment of poly-GR caused neuronal death and axon degeneration in WT neurons (Fig. 8d). Neuronal death in C9orf72 KO neurons was significantly increased compared to WT neurons after poly-GR treatment (Fig. 8d). Moreover, apoptosis was significantly increased in poly-GR-treated C9orf72 KO neurons; however, autophagy was similar between WT and C9orf72 KO neurons after poly-GR treatment (Fig. S7a, b). Importantly, treatment with the PARP inhibitor ABT-888 protected neurons from poly-GR-induced neurodegeneration in both WT and C9orf72 KO neurons (Fig. 8d). These results indicate that PARP-1 may be involved in poly-GR-induced neurodegeneration. Thus, DNA damage accumulation in C9orf72-deficient neurons with poly-GR expression leads to PARP-1 activation, thereby contributing to neurodegeneration.
Fig. 8. C9orf72 deficiency exacerbates poly-GR-induced neuronal death by PARP-1 overactivation.
a Accumulated PAR polymers at laser-lesioned regions in C9orf72 KO cells. WT and C9orf72 KO U2OS cells were subjected to laser-generated DNA lesion by a 405 nm laser. The cells were fixed at 1 min or 30 min after microirradiation, and stained with anti-PAR antibody (red). Quantitative analysis of PAR intensity. Data are mean (n = 87–99 cells from 3 independent experiments; unpaired t-test; ***, p < 0.001). b Increased PAR in C9orf72 KO neurons after GR20 treatment. Primary neurons were cultured for 24 h and treated with poly-GR peptide (10 μM), with or without 10 μM ABT888 for 24 h. Proteins were analyzed by immunoblot. Data are mean (n = 3 independent experiments; Two-way ANOVA; *, p < 0.05; ***, p < 0.001). c Increased PAR polymers in the cortex of C9orf72−/−, AAV-GR50 mice. Proteins were extracted from the cortex of indicated genotypes and PAR polymers were analyzed by immunoblot. Quantitative analysis of PAR intensity. Data are mean (n = 3 independent experiments; paired t-test; *, p < 0.05). d PARP inhibition rescues poly-GR-induced neurodegeneration. The cortical neurons were cultured for 48 h and treated with poly-GR peptide (10 μM), with or without 10 μM ABT888 for 48 h. The neurons were fixed and stained with anti-Tuj1 antibody to reveal neurites. Quantitative analysis of Tuj1 intensity. Data are mean (n = 3 independent experiments; Two-way ANOVA; **, p < 0.01; ***, p < 0.001).
Discussion
In this study, we discovered a novel function of C9orf72 in the nucleus to control NHEJ repair by regulating DNA-PK complex assembly and initiation of DDR signaling. We explored C9orf72 functions by IP-MS and found that C9orf72 interacted with DNA-PKcs, a critical kinase for DDR signaling in the NHEJ repair pathway (Supplemental table 1, Fig. 3c). The nuclear C9orf72 was recruited at DNA damage sites immediately after DNA damage induction (Fig. 1g–k). C9orf72 deficiency led to attenuated NHEJ repair and more DNA damage in neurons (Fig. 2). Mechanistically, C9orf72 deficiency caused impaired DNA-PK complex assembly and Ligase 4/XRCC4 recruitment to DNA damage sits (Fig. 5). Attenuated DNA repair capability induced PARP-1 hyperactivation and neurodegeneration (Fig. 8).
Poly-GR expression in neurons impairs mitochondrial function and thereby elevates cellular oxidative stress and DNA damage in neurons in vitro and in vivo [14, 16]. Moreover, the GGGGCC-containing RNA might act to form DNA-RNA hybrids (R-loops), resulting in DNA DSB [5]. Furthermore, increased DNA damage was observed in motor neurons [5] and iPSC-derived neurons from C9orf72 ALS/FTD patients [16]. Our results support a model where toxicity from GGGGCC repeat expansions causes DNA damage and reduced C9orf72 function in neurons compromises NHEJ repair capacity, thus leading to DNA DSB accumulation. Consequently, DNA damage in neurons may cause PARP-1 overactivation and PAR production. Indeed, the PAR polymers in the nucleus have been observed in motor neurons of ALS patients [64]. Thus, C9orf72 deficiency exacerbates DNA damage accumulation and neurodegeneration in DPR-expressing neurons, likely through DNA-damage-induced PARP-1 overactivation.
PARP-1 senses DNA damage and produces PAR by using oxidized NAD. The PAR polymers are linked covalently to nuclear proteins and recruit DNA repair proteins to DNA damage sites to facilitate DNA repair. PARP-1 inhibitors can induce cell death in BRCA-mutant tumor cells. However, PARP-1 inhibition in response to DNA damage can prevent neuronal death. Furthermore, DNA damage-induced PARP-1 overactivation has been implicated in many neurological disorders [62, 63, 65]. The following explanations may address how PARP-1 overactivation is involved in neuronal death. Firstly, PARP-1 overactivation may cause consumption of cellular NAD, inhibition of glycolysis pathway, and ATP deprivation [59, 60]. Secondly, PARP-1 overactivation and PAR accumulation may induce neuronal death called parthanatos which requires AIF and MIF translocation to the nucleus to fragment DNA [61, 66]. Finally, p53 transcriptional activity has been reported to play a central role in poly-PR- or poly-GR-induced neuronal death [20, 21]. Moreover, PARylation of p53 results in p53 accumulation in the nucleus and increasing p53 transcriptional activity [67, 68]. Thus, increased DNA damage in neurons may lead to excessive activation of PARP-1 and p53 PARylation, which causes constitutive activation of p53-mediated transcription.
Materials and methods
Immunostaining
Immunostaining was performed as described previously [69, 70]. Briefly, mice were anesthetized with isoflurane and perfused with PBS, and followed by 50 ml 4% paraformaldehyde (PFA). The brain, spinal cord and Tibialis anterior (TA) muscle were dissected and post-fixed in 4% PFA at 4 °C overnight. Brain sections were prepared by using the vibratome (Leica, VT1200S). While the lumbar spinal cord was dehydrated using graded sucrose buffer, embedded in OCT (Sakura), and quick-frozen in −80 °C refrigerator.
Sections were incubated in a blocking buffer (1% BSA, 0.5% Triton X-100 in PBS) for 1 h, and then incubated with primary antibodies at 4 °C overnight. After washing 3 times with PBS, sections were incubated with fluorescence-conjugated secondary antibodies (Life Technologies) at 37 °C for 45 min. All primary and secondary antibodies were diluted in the immunostaining buffer (0.1% Triton X-100, 1% BSA in PBS). Tibialis anterior (TA) muscle was dissected into muscle fibers and stained whole-mount with antibodies against neurofilament and synapsin-1 (Cell signaling Technology) and CF555-conjugated α-bungarotoxin (Biotium).
The primary antibodies used in immunostaining were following: anti-NeuN (Cell Signaling Technology, Cat# 24307, RRID: AB_2651140), anti-DNA-PK (phospho S2056) (Abcam, Cat# ab124918, RRID: AB_11001004), anti-GFAP (UC Davis/NIH NeuroMab Facility, Cat# 75-240, RRID: AB_10672299), anti-Iba1 (Abcam, Cat# ab5076, RRID: AB_2224402), Anti-Choline Acetyltransferase (Millipore, Cat# AB144P, RRID: AB_2079751), anti-Tubulin, beta III isoform (Tuj1) (Millipore, Cat# MAB1637, RRID: AB_2210524), anti-phospho-Histone H2A.X(S139) (Cell Signaling Technology, Cat# 9718, RRID: AB_2118009), anti-phospho-Histone H2A.X(S139) (Millipore,Cat# 05-636, RRID: AB_309864), anti-GFP (Thermo Fisher Scientific, Cat# A-11122, RRID: AB_221569), anti-C9orf72 (GeneTex, Cat# GTX632041, RRID: AB_2784546), anti-Myc (Abmart, Cat# M20002, RRID: AB_2861172), ant-HA (Abmart, Cat# M20003, RRID: AB_2864345), anti-53BP1 (Cell Signaling Technology, Cat# 4937, RRID: AB_10694558), anti-Ku80 (Cell Signaling Technology, Cat# 2753, RRID: AB_2257526), anti-Neurofilament (Cell Signaling Technology, Cat# 2837 S, RRID: AB_823575), anti-Synapsin-1 (Cell Signaling Technology, Cat# 5297S, RRID: AB_ AB_2616578), anti-cleaved-caspase-3 (Cell Signaling Technology, Cat# 9661S, RRID: AB_2341188), anti-LC3B (Cell Signaling Technology, Cat# 3868S, RRID: AB_1151739), anti-P62 (Abcam, Cat# ab56416, RRID: AB_945626).
Fluorescent images were captured by Zeiss LSM 800 Confocal microscope and analyzed using ImageJ software.
Immunoblotting and immunoprecipitation
Whole cell protein was extracted with 1 × loading buffer (2.5 mM Tris-Cl (pH 6.8), 2% SDS, 0.1% Bromophenol blue, 10% glycerol, 8% 2-Hydroxy-1-ethanethiol in ddH2O). Immunoblot was detected with Omni-ECL™Femto Light Chemiluminescence Kit (ECL, Epizyme). Images were captured with Tanon 5200 and analyzed using ImageJ software.
For co-immunoprecipitation experiment, cells were lysed by using lysis buffer (15 mM Tris pH 7.4, 150 mM NaCl, 1% Triton) with protease inhibitor cocktail (APExBIO). Lysates were incubated with Flag M2 beads (Sigma) at 4 °C for 4 h. The immunoprecipitated samples were analyzed by immunoblot, or eluted with 3 × Flag peptide (GenScript, 0.2 mg/ml) for Mass spectrometry analysis.
The primary antibodies used in immunoblot experiments were following: anti-C9orf72 (GeneTex, Cat# GTX632041, RRID: AB_2784546), anti-C9orf72 (ProteinTech, Cat# 22637-1-AP, RRID: AB_10953528), anti-Flag (Sigma-Aldrich, Cat# F7425, RRID: AB_439687), anti-PRKDC (DNA-PKcs) (ABclonal, Cat# A1419, RRID: AB_2761049), anti-Histone H2A.X (ABclonal, Cat# A11540, RRID: AB_2758599), anti-GAPDH (Santa Cruz Biotechnology, Cat# sc-32233, RRID: AB_627679), anti-α Tubulin (Santa Cruz Biotechnology, Cat# sc-32293; RRID: AB_628412), anti-phospho-Histone H2A.X(S139) (Cell Signaling Technology, Cat# 9718, RRID: AB_2118009), anti-phospho-Histone H2A.X(S139) (Millipore, Cat# 05-636, RRID: AB_309864), anti-DNA-PK (phospho S2056) (Abcam, Cat# ab124918, RRID: AB_11001004), anti-Ku70 (Proteintech Cat# 10723-1-AP, RRID: AB_2218756), anti-Ku80 (Cell Signaling Technology, Cat# 2753, RRID: AB_2257526), anti-HA (Abmart, Cat# M20003, RRID: AB_2864345), anti-Myc (Abmart, Cat# M20002, RRID: AB_2861172), anti-Ligase 4 (Proteintech, Cat# 12695-1-AP, RRID: AB_2136253), anti-XRCC4 (Abcam, Cat# ab213729), anti-PAR (Cell Signaling Technology, Cat# 83732, RRID: AB_2749858), anti-GFP (Thermo Fisher Scientific, Cat# A-11122, RRID: AB_221569).
Animals
Mice were housed in a room at 24 °C in a 24 h light/dark cycle with access to food and water in the Animal Facility at Shanghai Model Organisms Center in Shanghai. Male mice, P30-P180 of age, were used in the study. The C9orf72−/− mice (#027068, Jackson Laboratory) [36] were purchased from the Jackson Laboratory and genotyped with forward primer (AATGG CTGAA CCTGA TCACT G), and reverse primer (AAAGG TAGCC GCCAA CAAG). All mouse experiments were conducted in accordance with guidelines of Shanghai Institutional Animal Care and Use Committee (IACUC) and under an approved IACUC protocol of ShanghaiTech University.
Intracerebroventricular AAV Injection
The AAV viruses were generated and injected as previously described [71]. Briefly, the DNA encoding poly-GR was synthesized and inserted into an AAV expression vector containing inverted repeats of serotype 2. The expression of GFP and poly-GR was under the control of CAG promoter. AAV particles were packaged into serotype 9 type capsid and purified by Applied Biological Materials (abm). Pups at postnatal day 0 were cryoanesthetized on ice. The AAV was injected into the cerebral ventricle by the glass micropipette (Drummond Scientific Company, #5-000-1001-X10). Each cerebral ventricle was injected with 1 μl (5 × 109 genomes/μl) AAV.
Primary neuron culture
Primary cortical neurons were cultured as previously described [69]. Briefly, the cortex was isolated from P0 mice and kept in ice-cold Hank’s balanced salt solution (Cellgro, 21-022-CMR). The tissues were incubated with 0.25% Trypsin at 37 °C for 20 min. Dissociated cells were plated onto poly-D-lysine (Sigma Aldrich) coated dishes or coverslips and cultured in the neuronal culture medium (Neurobasal (Thermo Fisher) +2% B27 (Thermo Fisher) +1% Glutamax (Thermo Fisher)). Neurons were cultured in the incubator with 37 °C in 5% CO2. The neuronal culture medium was changed by half in every two days.
NPCs and neurons differentiation
The human neural progenitor stem cells (NPCs) were maintained in DMEM/F12 medium with 0.5% N2, 1% B27, and 20 μg/ml bFGF. NPCs were differentiated into neurons in the absence of bFGF for at least 3 weeks, with 10 μM ROCK inhibitor (Y-27632; Calbiochem) during the first 24 h.
Generation of knock-out cells and knock-down cells
Gene-modified cells by CRISPR-Cas9 were generated as previously described [70]. Briefly, the guidance RNAs: GCA ACA GCT GGA GAT GGC GG, and GGA TTT CTC CAT TTA GAG TG for C9orf72 were inserted into BbsI site of PX330-GFP construct and verified by sequencing. For C9orf72 gene knockout, U2OS, HEK293T, and iPSCs were transfected with PX330-GFP construct. The GFP-positive cells were sorted by FACS and placed into the 96-well plate, with each well seeded with a single cell. The cloned cells originated from an individual cell were verified by genomic DNA sequencing and Immunoblotting.
The siRNAs were purchased from GenePharma, C9orf72 siRNA 5′-GGUCA GAGUA UUAUU CCAAT T-3′ and 5′-UUGGA AUAAU ACUCU GACCT T-3′. Ku70 siRNA 5′-GGAGU CGUCA GAUUA UACUT T-3′ and 5′-AGUAU AAUCU GACGA CUCCT T-3′. DNA-PKcs siRNA 5′-GGCAC UAAAU UCCUU AUAUT T-3′ and 5′-AUAUA AGGAA UUUAG UGCCT T-3′.
Laser microirradiation and live imaging
Laser microirradiation experiment was performed as described previously [49]. Briefly, the U2OS cells were transfected with GFP, GFP-Lig4, GFP-C9orf72, respectively. The transfected cells were pre-sensitized with Hoechst 33342 (Meilunbio) in the final concentration of 10 μg/ml at 37 °C for 10 min. The living cell images were captured by Nikon Yokogawa CSU-W1 SoRa spinning disk confocal microscopy. The region of interest of 1 μm width was selected for cells and the 405 nm laser was set to 100% transmission to scan the selected ROI for 200 ms to induce DNA double-strand break. Images were captured 5 times per second for 1 min.
Pull-down assay
The pull-down assay was performed as described previously [72]. Briefly, HEK293T cells were transfected with pEGFP-C1-Ku70, pEGFP-C1-Ku80, pEGFP-C1-hDNAPKcs FAT, HA-C9orf72, pcDNA3.4-C9orf72-Flag and p3xFLAG-CMV-7.1-hDNAPKcs-KIP domain constructs respectively. After 48 h, the cells were harvested with RIPA buffer (50 mM Tris, 300 mM NaCl, 1% Triton X-100, 10% Glycerine). The protein was purified with the corresponding beads (GFP-Trap agarose (chromotek, #gta-20), anti-HA IP Resin (Genscript, #L00777) or ANTI-FLAG(R) M2 Affinity Gel (Merck/Millipore, # A2220-5ML)) at 4 °C for 2 h. Beads were collected by centrifugation at 5000 g for 2 min and washed with RIPA buffer for 3 times. Proteins were eluted with 100 μl elution buffer (50 mM Citric, 150 mM NaCl) in 1 M Tris (pH 9.0) for neutralization.
The recombinant HA-C9orf72 or Flag-C9orf72 (0.2 μg each) was mixed anti-HA or anti-Flag beads in pull-down buffer (50 mM Tris, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 10 mM MgCl2, 1% NP-40, pH 8.0) respectively, and incubated at 4 °C for 2 h. The beads were collected after washing with PBS for 2 times, and incubated with 0.2 μg recombinant proteins or in 500 μl pull-down buffer (0.1% SDS added) at 4 °C for 6 h. For DNA-PKcs pull-down experiment, cell lysate was supernatant from HEK293T cells lysed in RIPA buffer.
Chromatin fraction
Chromatin fraction was performed as previously described [45]. HEK293T cells were treated with 5 μM ETP overnight and collected with 10% FBS DMEM medium after dissociation with 0.25% Typsin-EDTA. After washing with PBS for 3 times, cells (4 × 106) were resuspended in 200 μl buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.34 M Sucrose, 0.1% Triton X-100,10% glycerol, 1 mM DTT, 10 mM NaF, 1 mM Na2VO3, and protease inhibitor cocktail), and incubation on ice for 5 min. After centrifugation at 3000 g for 5 min, the nuclei were washed with buffer A for 3 times and resuspended with 200 μl buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 10 mM NaF, 1 mM Na2VO3, 1 mM PPi and protease inhibitor), followed by incubation on ice for 10 min. After centrifugation at 1700 g for 5 min, the pellet was washed 3 times with buffer B. The chromatin fraction was collected by centrifugation at 13,000 g for 5 min, and resuspended in loading buffer.
Long amplicon PCR
The long amplicon PCR was performed as previously described [73]. Cortical neurons from AAV mice with indicated genotypes were resuspended in 500 μl lysis buffer (4 M Urea, 10 mM EDTA, 0.5% Sarkosyl, 0.1 M Tris-Cl, pH 8.0) supplemented with PK (1 μg/μl) and incubated in water bath at 52 °C for 12 h. Lysates were subjected to purification process by using phenol-chloroform extraction.
LA PCR were performed as previous describe. Briefly, a ~6.3 kb DNA region of Dntt was amplified by KOD-Plus-Neo (TOYOBO), and accumulated DNA lesions in genomic DNA was quantified by Image J. The primers are as follows: forward: 5′-TCAAA GCGAT TCAGA AAGTC CCAGT AAA-3′; reverse: 5′-TCAGA CCCAC ATAGC AATGA GGAGA TAG-3′. A 117-bp amplicon was amplified from mitochondria DNA by using the following primers: forward: 5′-CCCAG CTACT ACCAT CATTC AAGT-3′; reverse: 5′-GATGG TTTGG GAGAT TGGTT GATGT-3′.
Comet assay
Comet assay was performed as described previously [74]. A small tank was built with cover glasses on a clean glass slide. 0.5% low melting point agarose (Yeasen) (37 °C) was prepared to mix with cells (about 20,000 cells in 100 μl low melting point agarose) and was put in the small tank. Glass slides containing cell samples were placed at 4 °C for 15 min. Cover glasses were taken off and samples were lysed in the lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris-base, 1% Triton X-100, 1% DMSO, pH 10.0) at 4 °C for 1 h followed by washing for 3 times. Samples were placed into electrophoresis buffer (300 mM NaOH, 1 mM EDTA) for 20 min and then electrophoresis was performed in an ice bath for 20 min (approx. 0.8–1.5 V/cm). After electrophoresis, the samples were neutralized with neuralization buffer (0.4 M Tris-Cl, pH 7.5) 3 times. Slides were drained and stained with syber green I (Meilunbio) for 20 min. The images were captured by Zeiss LSM 800 Confocal microscope and analyzed using ImageJ software [75].
DNA damage repair assay
DNA damage repair assay was performed as described previously [50, 76]. NHEJ reporter or HR reporter plasmids were digested with HindIII (NEB) in vitro to induce DNA double-strand breaks. Linear plasmids were purified and transfected together with mCherry as a transfection control into HEK293T cells. Alt-NHEJ reporter plasmids (synthesized by GENEWIZ) were transfected to HEK293T cells with I-sceI and mCherry to induce DSB in vivo. Two days after transfection, the cells were fixed and the fluorescent images were captured and analyzed by ImageJ software.
Behavior test
Behavior test was performed as described previously [77]. For the grip test, the strength of front limbs and hind limbs were measured with a grip strength meter, 5 times for each mouse. For the rotarod test, each mouse performed 3 trails each day for 3 d, with an accelerating rotarod, starting from 4 rpm and accelerating to 40 rpm in 5 minutes. The latency time in the third day was recorded and analyzed. In all the behavioral test, ≥6 mice per genotype were tested.
Statistical analysis
Quantification and statistical analyses were performed using GraphPad Prism 8 software (GraphPad). Student’s t test, One-way ANOVA, and Two-way ANOVA were used to analyze data as indicated. Statistical analysis between two groups were performed by paired or unpaired two-tailed t-test. ANOVA was used for comparisons among multiple groups. The p-value of < 0.05 was considered significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Data are mean. All representative images were obtained from experiments that were repeated at least three times.
Supplementary information
Acknowledgements
We thank XL, CF, and ZY from the Molecular Imaging Core Facility (MICF); PH, and CZ from the Multi-Omics Core Facility (MOCF); and YX, XR, and JL from the Molecular and Cell Biology Core Facility (MCBCF) at the School of Life Science and Technology, ShanghaiTech University for providing technical support. We also thank W. Wang (Interdisciplinary Research Center on Biology and Chemistry, China) for providing C9ALS iPSCs, V. Gorbunova (University of Rochester, USA) for providing NHEJ reporter constructs, and D. Spector (Cold Spring Harbor Laboratory, USA) for providing U2OS-LacO cells. This study was supported by grants from the National Natural Science Foundation of China (91949117, 31871044 to LL).
Author contributions
LL and LYH designed the experiments and wrote the manuscript. LYH and JQL performed and analyzed most of the experiments; JJC generated C9orf72−/− NPCs and performed neuron differentiation. CNC, SSS, and YHS contributed to data analyses.
Data availability
The data generated to support the finding of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Our study did not require ethical approval.
Footnotes
Edited by M Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Liying He, Jiaqi Liang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01074-0.
References
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14:544–58. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci. 2017;20:1225–35. doi: 10.1038/nn.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–23. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 8.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 9.Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao Z, Liu L, Tao Z, Wang R, Ren H, Sun H, et al. Motor dysfunction and neurodegeneration in a C9orf72 mouse line expressing poly-PR. Nat Commun. 2019;10:2906. doi: 10.1038/s41467-019-10956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SY, Lopez-Gonzalez R, Krishnan G, Phillips HL, Li AN, Seeley WW, et al. C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat Neurosci. 2019;22:851–62. doi: 10.1038/s41593-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–88.e17. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med. 2018;24:1136–42. doi: 10.1038/s41591-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–55.e5. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–91. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farg MA, Konopka A, Soo KY, Ito D, Atkin JD. The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum Mol Genet. 2017;26:2882–96. doi: 10.1093/hmg/ddx170. [DOI] [PubMed] [Google Scholar]
- 18.Nihei Y, Mori K, Werner G, Arzberger T, Zhou Q, Khosravi B, et al. Poly-glycine-alanine exacerbates C9orf72 repeat expansion-mediated DNA damage via sequestration of phosphorylated ATM and loss of nuclear hnRNPA3. Acta Neuropathol. 2020;139:99–118. doi: 10.1007/s00401-019-02082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrade NS, Ramic M, Esanov R, Liu W, Rybin MJ, Gaidosh G, et al. Dipeptide repeat proteins inhibit homology-directed DNA double strand break repair in C9ORF72 ALS/FTD. Mol Neurodegener. 2020;15:13. doi: 10.1186/s13024-020-00365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R, Xu X, Hao Z, Zhang S, Wu D, Sun H, et al. Poly-PR in C9ORF72-related amyotrophic lateral sclerosis/frontotemporal dementia causes neurotoxicity by clathrin-dependent endocytosis. Neurosci Bull. 2019;35:889–900. doi: 10.1007/s12264-019-00395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maor-Nof M, Shipony Z, Lopez-Gonzalez R, Nakayama L, Zhang YJ, Couthouis J, et al. p53 is a central regulator driving neurodegeneration caused by C9orf72 poly(PR) Cell. 2021;184:689–708.e20. doi: 10.1016/j.cell.2020.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Gonzalez R, Yang D, Pribadi M, Kim TS, Krishnan G, Choi SY, et al. Partial inhibition of the overactivated Ku80-dependent DNA repair pathway rescues neurodegeneration in C9ORF72-ALS/FTD. Proc Natl Acad Sci USA. 2019;116:9628–33. doi: 10.1073/pnas.1901313116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35:1779.e5–e13. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao S, MacNair L, McGoldrick P, McKeever PM, McLean JR, Zhang M, et al. Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann Neurol. 2015;78:568–83. doi: 10.1002/ana.24469. [DOI] [PubMed] [Google Scholar]
- 25.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24:313–25. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q, Jiang J, Gendron TF, McAlonis-Downes M, Jiang L, Taylor A, et al. Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat Neurosci. 2020;23:615–24. doi: 10.1038/s41593-020-0619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao Q, Liang C, Chang Q, Zhang W, Yang M, Chen JF. C9orf72 deficiency promotes motor deficits of a C9ALS/FTD mouse model in a dose-dependent manner. Acta Neuropathol Commun. 2019;7:32. doi: 10.1186/s40478-019-0685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su MY, Fromm SA, Zoncu R, Hurley JH. Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD. Nature. 2020;585:251–5. [DOI] [PMC free article] [PubMed]
- 30.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, et al. Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 2016;12:e1006443. doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35:1656–76. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016;2:e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Burberry A, Wang JY, Sandoe J, Ghosh S, Udeshi ND, et al. The C9orf72-interacting protein Smcr8 is a negative regulator of autoimmunity and lysosomal exocytosis. Genes Dev. 2018;32:929–43. doi: 10.1101/gad.313932.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee HC, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med. 2016;8:347ra93. doi: 10.1126/scitranslmed.aaf6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–9. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCauley ME, O’Rourke JG, Yanez A, Markman JL, Ho R, Wang X, et al. C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature. 2020;585:96–101. [DOI] [PMC free article] [PubMed]
- 38.Wang T, Liu H, Itoh K, Oh S, Zhao L, Murata D, et al. C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metab. 2021;33:531–46.e9. doi: 10.1016/j.cmet.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laflamme C, McKeever PM, Kumar R, Schwartz J, Kolahdouzan M, Chen CX, et al. Implementation of an antibody characterization procedure and application to the major ALS/FTD disease gene C9ORF72. Elife. 2019;8. [DOI] [PMC free article] [PubMed]
- 41.Amick J, Roczniak-Ferguson A, Ferguson SM. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell. 2016;27:3040–51. doi: 10.1091/mbc.e16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, et al. Activity-Induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–98. doi: 10.1016/S0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore S, Berger ND, Luijsterburg MS, Piett CG, Stanley FKT, Schrader CU, et al. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat Commun. 2019;10:241. doi: 10.1038/s41467-018-08111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Yu M, Bai Y, Yu J, Jin F, Li C, et al. Elevated CXorf67 Expression in PFA Ependymomas suppresses DNA repair and sensitizes to PARP inhibitors. Cancer Cell. 2020;38:844–56.e7. doi: 10.1016/j.ccell.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–82. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 48.Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 2013;16:1008–15. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–91. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci USA. 2004;101:7624–9. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–24. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 52.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–84. doi: 10.1128/MCB.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lempiainen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009;28:3067–73. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–48. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double-strand breaks. J Biol Chem. 2005;280:14709–15. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 56.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–40. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–23. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 59.Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, et al. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci USA. 2014;111:10209–14. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8:1819–31. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, An R, Umanah GK, Park H, Nambiar K, Eacker SM, et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016;354. [DOI] [PMC free article] [PubMed]
- 62.Dawson TM, Dawson VL. Mitochondrial mechanisms of neuronal cell death: potential therapeutics. Annu Rev Pharm Toxicol. 2017;57:437–54. doi: 10.1146/annurev-pharmtox-010716-105001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park H, Kam TI, Dawson TM, Dawson VL. Poly (ADP-ribose) (PAR)-dependent cell death in neurodegenerative diseases. Int Rev Cell Mol Biol. 2020;353:1–29. doi: 10.1016/bs.ircmb.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 64.McGurk L, Mojsilovic-Petrovic J, Van Deerlin VM, Shorter J, Kalb RG, Lee VM, et al. Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol Commun. 2018;6:84. doi: 10.1186/s40478-018-0586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murata MM, Kong X, Moncada E, Chen Y, Imamura H, Wang P, et al. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol Biol Cell. 2019;30:2584–97. doi: 10.1091/mbc.E18-10-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan J, Dawson TM, Dawson VL. Cell death mechanisms of neurodegeneration. Adv Neurobiol. 2017;15:403–25. doi: 10.1007/978-3-319-57193-5_16. [DOI] [PubMed] [Google Scholar]
- 67.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9:1175–83. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 68.Fischbach A, Kruger A, Hampp S, Assmann G, Rank L, Hufnagel M, et al. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2018;46:804–22. doi: 10.1093/nar/gkx1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong Z, Chen W, Chen C, Wang H, Cui W, Tan Z, et al. CUL3 deficiency causes social deficits and anxiety-like behaviors by impairing excitation-inhibition balance through the promotion of cap-dependent translation. Neuron. 2020;105:475–90.e6. doi: 10.1016/j.neuron.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Cao Y, Wu H, Ye X, Zhu Z, Xing G, et al. Enzymatic activity of the scaffold Protein Rapsyn for Synapse formation. Neuron. 2016;92:1007–19. doi: 10.1016/j.neuron.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tung YT, Peng KC, Chen YC, Yen YP, Chang M, Thams S, et al. Mir-17 approximately 92 confers motor neuron subtype differential resistance to ALS-associated degeneration. Cell Stem Cell. 2019;25:193–209.e7. doi: 10.1016/j.stem.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 72.Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–97. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitra J, Guerrero EN, Hegde PM, Liachko NF, Wang H, Vasquez V, et al. Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc Natl Acad Sci USA. 2019;116:4696–705. [DOI] [PMC free article] [PubMed]
- 74.Kruman II, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, et al. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004;41:549–61. doi: 10.1016/S0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- 75.Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2014;2:457–65. doi: 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen C, Lu Y, Zhang B, Figueiredo D, Bean J, Jung J, et al. Antibodies against low-density lipoprotein receptor-related protein 4 induce myasthenia gravis. J Clin Invest. 2013;123:5190–202. doi: 10.1172/JCI66039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated to support the finding of this study are available from the corresponding author upon reasonable request.