Abstract
We have developed a new class of probes for homogeneous nucleic acid detection based on the proposed displacement hybridization. Our probes consist of two complementary oligodeoxyribonucleotides of different length labeled with a fluorophore and a quencher in close proximity in the duplex. The probes on their own are quenched, but they become fluorescent upon displacement hybridization with the target. These probes display complete discrimination between a perfectly matched target and single nucleotide mismatch targets. A comparison of double-stranded probes with corresponding linear probes confirms that the presence of the complementary strand significantly enhances their specificity. Using four such probes labeled with different color fluorophores, each designed to recognize a different target, we have demonstrated that multiple targets can be distinguished in the same solution, even if they differ from one another by as little as a single nucleotide. Double-stranded probes were used in real-time nucleic acid amplifications as either probes or as primers. In addition to its extreme specificity and flexibility, the new class of probes is simple to design and synthesize, has low cost and high sensitivity and is accessible to a wide range of labels. This class of probes should find applications in a variety of areas wherever high specificity of nucleic acid hybridization is relevant.
INTRODUCTION
The measurement of nucleic acid hybridization under homogeneous, rather than heterogeneous, conditions provides a variety of advantages, such as convenience, real-time or in situ detection and accurate quantification for diverse applications (1). Since the earliest attempts nearly two decades ago to develop a homogeneous hybridization assay system based on fluorescence resonance energy transfer, efforts to explore more practical and general homogeneous systems have continued (2). This work reached fruition in recent years with the emergence of real-time PCR assays (3). A number of probe systems have so far been developed and new designs are still being proposed (4). Currently available probes include 5′-nuclease probes (5), molecular beacons (6) and their derivatives (7–9), adjacent probes (10) and fluorescent probes (11). Although more or less satisfactory for most common uses, these probes are difficult to design, synthesize and purify and are, therefore, expensive.
Current probes, either linear or conformationally constrained, when recognizing their targets undergo a direct hybridization reaction that involves hybridization between two single-stranded nucleic acids. Although nucleic acid hybridization is one of the most specific recognition events in nature, measurable physical changes that occur upon hybridization are rather small. Moreover, with direct hybridization it is difficult to achieve mismatch discrimination. Consequently, special structural modifications, such as hairpins (6), minor groove-binding groups (12) or peptide nucleic acids (11), must be used, which increases the difficulty and expense of probe preparation.
To overcome the shortcomings inherent in the direct hybridization mode, we report a new class of probes based on a mode of reaction that we call ‘displacement hybridization’, which differs from direct hybridization in that it has an additional competitor in the probe–target reaction system. An ideal competitor fulfills the following requirements: it must not be so competitive as to hinder the formation of perfectly matched probe–target hybrids but it must be sufficiently competitive to block non-specific hybridization. We hypothesized that a suitable competitor would be a single-stranded oligonucleotide that possesses the same sequence as the target in the binding region but is shorter than the probe. In this way, the competitor will form a stable duplex with the probe in the absence of the target and can be displaced in the presence of the target. We name this hybridization with enhanced specificity between a double-stranded and single-stranded oligonucleotide ‘displacement hybridization’. Regarding the duplex as a kind of probe, we labeled the original target-binding strand with a fluorophore at its 5′-end and labeled the competitor with a quencher at its 3′-end. We term this double-stranded molecule a ‘Yin-Yang’ probe, as the two strands complement each other and interact synergistically to enhance specificity.
MATERIALS AND METHODS
Reaction kinetics of displacement hybridization
An aliquot of 50 µl of double-stranded oligonucleotides (0.80 µM each strand), shown in each panel in Figure 1, was held at 25°C in a thermostat and its fluorescence monitored over time in a spectrofluorometer (Cary Eclipse, Varian). After confirming that the fluorescence did not change, 2 µl of 40 µM target oligonucleotide was added and the level of fluorescence was recorded at 15 s intervals. Double-stranded oligonucleotides were prepared by mixing the two oligonucleotides to the desired concentrations in 10 mM Tris–HCl, pH 8.0, containing 1.5 mM MgCl2 and heated at 94°C for 2 min, annealed at 50°C for 5 min and allowed to cool to room temperature. We named the strand complementary to the single-stranded oligonucleotide the ‘positive’ strand and the other strand the ‘negative’ strand. Custom oligodeoxyribonucleotides were synthesized and purified by Shanghai Shenyou Ltd.
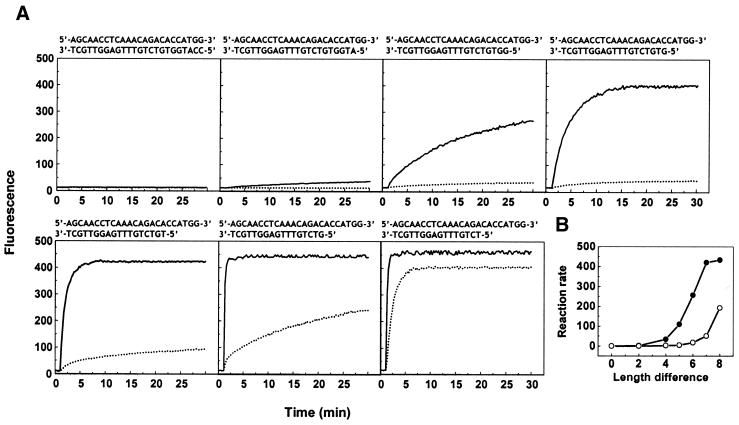
Figure 1.
(A) Reaction kinetics of displacement hybridization between double-stranded oligoucleotides and a perfectly complementary and a single nucleotide mismatch oligonucleotide. The sequences of the double-stranded oligonucleotides tested are listed at the top of each panel without showing modification groups. The perfectly complementary target was d(CCATGGTGTCTGTTTGAGGTTGCT) and the target containing a single nucleotide substitution was d(CCATGGTGTCTGTTTCAGGTTGCT), where underlining identifies the nucleotide substituent. Solid curves show the results obtained with the perfectly complementary oligonucleotide and dotted curves the results obtained with the single nucleotide mismatch oligonucleotide. (B) Dependence of the reaction rate on the difference in length between the positive and negative strands in the presence of the perfectly complementary oligonucleotide (solid circle) and the single mismatch oligonucleotide (open circle). The reaction rate is the fluorescence increase observed during the first minute of the reaction.
Preparation of double-stranded probes
To prepare double-stranded probes, 5 nmol of positive and negative strand were separately dissolved in 50 µl of 10 mM Tris–HCl, pH 8.0, containing 1.5 mM MgCl2. The two solutions were then mixed together and diluted to the desired concentration with the same buffer. The solution was denatured in a thermal cycler at 94°C for 2 min, annealed at 50°C for 5 min and allowed to cool to room temperature. Other oligonucleotides were dissolved in the same buffer as above unless indicated.
Specificity of double-stranded probes
The reaction kinetics of the double-stranded probe with four targets were measured as above. The probe concentration was 0.80 µM positive strand and 1.0 µM negative strand (0.80/1.0 µM). The final concentration of each target was 1.6 µM. To obtain the thermal denaturation curves, the same probe solutions with and without target were placed in a fluorometric thermal cycler (IQ iCycler; Bio-Rad). Denaturation was carried out at 94°C for 1 min and annealing was carried out at 70°C for 2 min. This was repeated for 41 cycles with a 1°C decrease in the annealing temperature each cycle. Fluorescence was measured during the annealing stage.
For photographs under UV light illumination, 2 µl of a 2-fold molar excess of target was added to 50 µl of probe. The probes and targets were as used above, except that the final probe concentration was 4.0/5.0 µM and the final target concentration was 8.0 µM. About 10 min later, photographs were taken at room temperature.
Comparison of double-stranded probe to linear probes
Four hybridization mixtures were prepared: double-stranded probe with perfectly complementary target; double-stranded probe with mismatched target; linear probe with perfectly complementary target; linear probe with mismatched target. Each mixture consisted of 50 µl of 1.0/1.2 µM double-stranded probe or 1.0 µM linear probe and was held at 25°C in a thermostat. After confirming that there was no change in fluorescence, 2 µl of 50 µM target oligonucleotide was added and the level of fluorescence was recorded at 15 s intervals.
Simultaneous discrimination between targets with a mixture of four probes
Four double-stranded probes were first prepared and then mixed together; the concentrations of each positive and negative strand were 1.0 and 2.0 µM, respectively. Each cuvette, containing 100 µl of the probe mixture, was held at 25°C in a thermostat and four pairs of excitation and emission wavelengths (λex/λem for FAM, 495/519 nm; λex/λem for HEX, 530/553 nm; λex/λem for TAMRA, 560/583 nm; λex/λem for Texas Red, 596/617 nm) were monitored concurrently over time in the spectrofluorometer. After confirming that there was no change in fluorescence, 4 µl of 50 µM target oligonucleotide was added and the fluorescence levels at different wavelengths were recorded at 15 s intervals. The fluorescence intensity of each fluorophore were normalized so that 0 represents fluorescence in the absence of target and 1 represents the perfectly complementary targets in response to the addition of an excess of perfectly complementary target.
Real-time PCR assays with double-stranded probes
Each 50 µl reaction contained 5 µl of DNA template (extracted from a volunteer’s blood cells), 0.2/0.24 µM double-stranded probe, 0.4 µM each primer and 2.0 mM MgCl2 in a PCR master mix (10 mM Tris–HCl, pH 8.3, 2.0 U Taq polymerase, 200 µM each dNTP and 50 mM KCl). After denaturation at 94°C for 5 min, 40 cycles of amplification (95°C for 30 s, 50°C for 30 s and 72°C for 1 min) were carried out in sealed tubes in a fluorometric thermal cycler (Rotor-Gene 2000; Corbett Research). Fluorescence was recorded at the annealing stage.
Real-time PCR assays with double-stranded primers
To test the utility of double-stranded primers in real-time PCR, the same human β-globin gene segment was amplified. Each 50 µl reaction contained 10 µl of DNA template and 0.4/0.6 µM double-stranded primers and other components were as above. PCR was conducted in a fluorometric thermal cycler (IQ iCycler; Bio-Rad) under the same conditions as above. Fluorescence was recorded at the annealing stage.
To compare the specificity of double-stranded primers with that of conventional primers, each 50 µl reaction contained SYBR Green I (Molecular Probes Inc.) at 20 000 times dilution. All other components and PCR conditions were as were used for testing double-stranded primers in real-time PCR. Fluorescence was recorded at the extension stage.
RESULTS
Reaction kinetics with double-stranded probes
We prepared a series of double-stranded oligonucleotides that share the same positive strand and possess negative strands of different length. Two single-stranded oligonucleotides were tested as the reaction target, one perfectly matched and another with a single nucleotide mismatch. Figure 1A shows the kinetic curves for the double-stranded oligonucleotides hybridized to each of the single-stranded oligonucleotides. When the positive and negative strands were of equal length, no reaction was observed. As their length difference increased, the rate of reaction became faster. When their length difference exceeded 7 nt, the reaction rate with the perfectly matched oligonucleotide changed little, indicating that the negative strand no longer exerted a significant effect on the reaction. However, the reaction rate with the mismatched oligonucleotide increased sharply at this point. Figure 1B shows the dependence of the reaction rate on the length difference of the two strands. Clearly, there is a marked reduction in the reaction rate with the mismatched oligonucleotide, suggesting a greater inhibitory effect of the negative strand on hybridization with the mismatched oligonucleotide.
Design and preparation of double-stranded probes
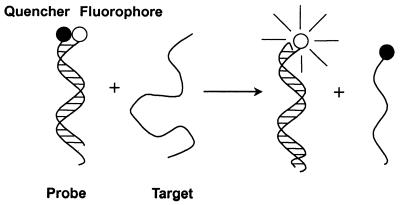
Figure 2 illustrates the design and working principles of the probe based on displacement hybridization. We labeled the 5′-end of the positive strand with a fluorophore and blocked its 3′-end with a phosphate group and the 3′-end of the negative strand was coupled to a quencher. When not bound to a target, the probe is non-fluorescent. After becoming bound to its target, the negative strand is displaced by the target and the fluorophore becomes fluorescent. New probes were made by titrating the positive strands with the negative strands to reach the lowest fluorescence. We noticed that, in most cases, the ratio of positive strand to negative strand was within 1.2 to 2.0. The heating procedures were used to prevent possible inter- or intramolecular secondary structure. When diluted to the desired concentration before use, no background fluorescence increase was observed, indicating that the double-stranded probes were very stable.
Figure 2.
Schematic drawings of a double-stranded probe and the working principle. The double-stranded probe is composed of two complementary oligonucleotides of different length. The longer positive strand is labeled with a fluorophore and the shorter negative strand is labeled with a quencher; the probe is non-fluorescent due to the close proximity of the fluorophore and the quencher. In the presence of target the negative strand is displaced by the target and the fluorophore becomes fluorescent.
Specificity of double-stranded probes
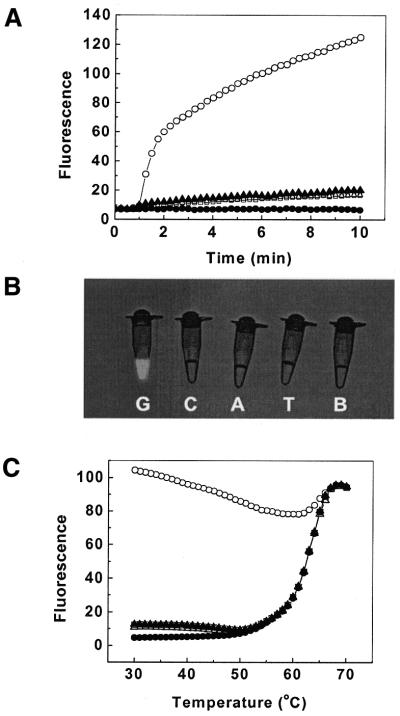
In order to show the specificity of the double-stranded probes, we prepared four targets that were identical except for one nucleotide at the same position. Figure 3A shows that hybridization of a double-stranded probe to the perfectly complementary target occurred between 10 and 20 times faster than hybridization to each of the three mismatched targets. To demonstrate this difference visually, similar reactions were carried out at a relatively high concentration (see Materials and Methods) and a photograph was taken by illuminating the reaction tubes with UV light to stimulate fluorescence. Figure 3B shows that fluorescence from the perfectly complementary probe–target hybridization can easily be discerned with the naked eye, while fluorescence from hybridization of the double-stranded probe to mismatched targets cannot be distinguished from the fluorescence seen in a tube without any target.
Figure 3.
(A) Reaction kinetics of the double-stranded probe with a perfectly complementary target possessing G (open circle) at the variable position and with targets possessing a single nucleotide C (open triangle), A (solid triangle) or T (open square) substitution at the same position. A solution containing no target (solid circle) was used as a control. The positive strand of the probe was FAM-d(AGCAACCTCAAACAGACACCAT) and the negative strand was d(TGTCTGTTTGAGGTTGCT)-dabcyl. The targets were d(CCATGGTGTCTGTTTNAGGTTGCT), where N is G, T, A or C. (B) Photographs of the perfectly complementary and single mismatch hybridization reactions. The perfectly complementary target (G) possessed a guanosine at the variable position, whereas the single mismatch targets (C, A and T) possessed either a cytidine, adenosine or thymidine at the same position. B identifies the control sample without target. (C) Denaturation curves of the perfectly complementary and single mismatch probe–target hybrids. All the nucleotide sequences used are as shown in (A).
We then measured the fluorescence of these tubes as a function of temperature. The fluorescence–temperature curves (Fig. 3C) indicated that at higher temperatures the double-stranded probes dissociated into single strands. When the temperature was decreased, the double-stranded probes renatured and only perfectly complementary targets took part in displacement hybridization. It is easy to see that the probe displayed complete discrimination between its perfectly matched target and the three single nucleotide mismatch targets in the temperature range 30–60°C. From these curves we can measure the melting temperature (Tm) of a probe and also predicate probe behavior if used in real-time nucleic acid amplification reactions. It is clear that target signals can be obtained during real-time PCR if fluorescence measurements are taken at temperatures that are lower than the probe melting point.
Comparing double-stranded with single-stranded probe
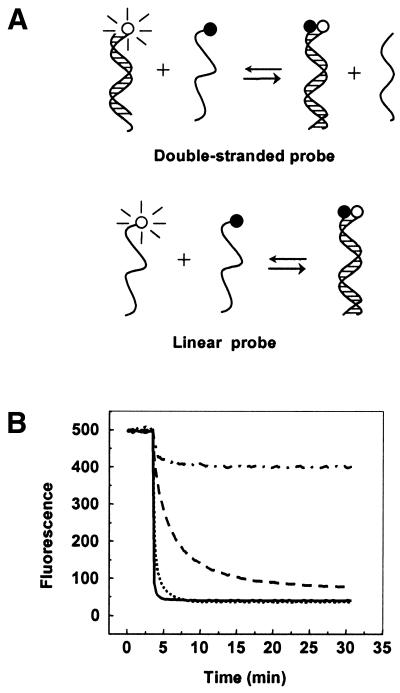
To further investigate the specificity-enhancing ability of the double-stranded probes, we compared the reaction kinetics of a double-stranded probe with an otherwise identical single-stranded linear probe in discriminating a single nucleotide mismatch. In order to observe hybridization of both of these probes to the target, we utilized the labeling scheme shown in Figure 4A. The single-stranded probe was the same as the positive strand of the double-stranded probe and was labeled at its 5′-end with FAM. The target oligonucleotides were labeled with dabcyl at their 3′-ends, so that binding to either probe would result in fluorescence reduction. The reaction between the single-stranded probe and its targets was completed quickly, with little difference between the perfectly matched and the single mismatch targets. However, the reactions of the double-stranded probe varied significantly with the target (Fig. 4B). A 5-fold fluorescence increase was observed with the perfectly matched target compared with the single mismatch target.
Figure 4.
(A) Labeling scheme for comparing double-stranded probe wuth linear probes. A fluorophore was linked to the end of the probe and a quencher was linked to the end of the target. (B) Comparison of reaction kinetics of double-stranded probe with the linear probe with matched and mismatched targets. While linear probes react equally with matched (solid line) and mismatched (dotted line) targets, double-stranded probes react well with matched (dashed line) but not with mismatched (dot–dash line) targets. The positive strand of the double-stranded probe and the sequence of the linear probe was FAM-d(AGCAACCTCAAACAGACACCAT) and the negative strand of the double-stranded probe was d(TGTCTGTTTGAGGTTGCT). The nucleotide sequence of the perfectly complementary target was d(ATGGTGTCTGTTTGAGGTTGCT)-dabcyl and the nucleotide sequence of the mismatched target was d(ATGGTGTCTGTTTCAGGTTGCT)-dabcyl, where the underlined nucleotide in each sequence identifies the difference between them.
Simultaneous discrimination between targets with a mixture of four probes
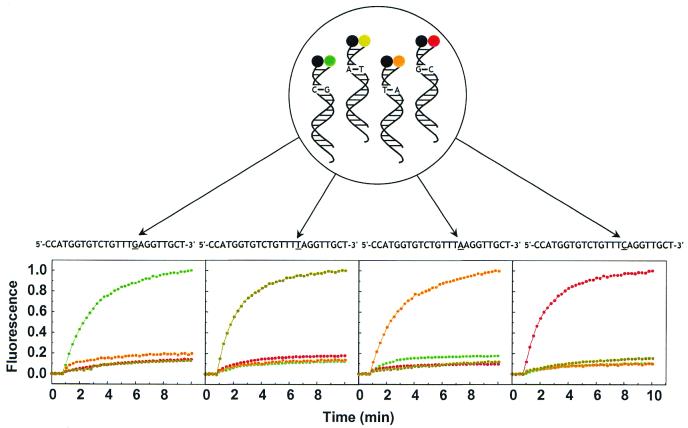
The labeling strategy in the double-stranded probe is similar to that in molecular beacons. Thus, as in molecular beacons, multicolor probes can be constructed. In order to demonstrate the ability of multiplex detection and allele discrimination, we carried out hybridization experiments with a mixture of four double-stranded probes and four oligonucleotide targets that differed from each other by only a single nucleotide. The double-stranded probes were complementary to these targets and were identical in all respects except that each was labeled with a different fluorophore and each possessed a different base pair in the probe sequence: FAM labeling of the double-stranded probe with a C-G pair; HEX labeling for an A-T pair, TAMRA labeling for a T-A pair; Texas Red labeling for a G-C pair. An equimolar mixture of these double-stranded probes was placed in four cuvettes. A different target was added to each cuvette and the fluorescence of each of the four double-stranded probes in each cuvette was monitored over time. Addition of a target resulted in a marked increase in the fluorescence of the double-stranded probe whose probe sequence was perfectly complementary to the targets, but did not result in any increase in the fluorescence of the other double-stranded probes that were present (Fig. 5). The color of the fluorescence in each cuvette identified which nucleotide was present in the target. These results indicate that only perfectly complementary targets can form a hybrid with a double-stranded probe and a single nucleotide mismatch between the target and the probe prevent hybridization. It can also be concluded that the double-stranded probes are extremely stable in the duplex and coexistence of different probes does not elicit any interference between them. A small amount of hybridization was observed in each cuvette, however, when the mismatch was a G-T base pair, as this is the least destabilizing of all the mismatches, a phenomenon that has also been observed for molecular beacons (13). When more than one target was added, the colors in the resulting fluorescence spectrum confirmed the identity of the target. Thus, multicolor double-stranded probes present in the same solution can distinguish alleles that differ form one another by as little as a single nucleotide.
Figure 5.
Detection of a single nucleotide difference using a mixture of four differently colored double-stranded probes. When a target oligonucleotide (sequence shown above each panel) was added to the mixture, only the double-stranded probe whose probe sequence was perfectly complementary to that target formed a hybrid and emitted its characteristic fluorescent color. Each kinetics curve is printed in the color of the double-stranded probe fluorescence (green, FAM; dark yellow, HEX, orange; TAMRA; red, Texas Red). The four double-stranded probes were FAM-d(AGCAACCTCAAACAGACACCAT)/d(TGTCTGTTTGAGGTTGCT)-dabcyl, HEX-d(AGCAACCTAAAACAGACACCAT)/d(TGTCTGTTTTAGGTTGCT)-dabcyl, TAMRA-d(AGCAACCTTAAACAGACACCAT)/d(TGTCTGTTTAAGGTTGCT)-dabcyl and Texas Red-d(AGCAACCTGAAACAGACACCAT)/d(TGTCTGTTTCAGGTTGCT)-dabcyl, where the underlined nucleotides identify the only differences among their sequences. The targets were d(CCATGGTGTCTGTTTNAGGTTGCT), where N was G, T, A or C.
Real-time PCR detection with double-stranded probes
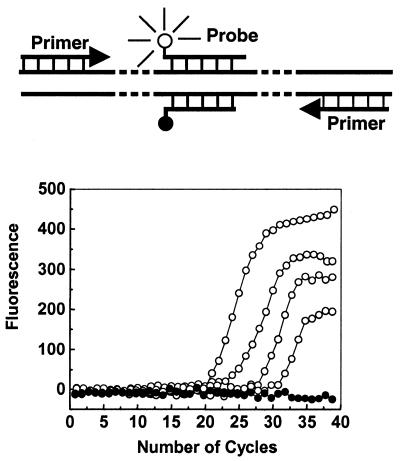
At concentrations similar to those of primers in PCR and at annealing temperatures, double-stranded probes rapidly interact with their targets and also undergo self-annealing, thus they can be used in real-time amplification assays. We demonstrated this in detection of the human β-globin gene. To construct the double-stranded probe, we chose a negative strand of 20 nt with a Tm close to that of the primers, and a positive strand of 24 nt in order to obtain a probe–target hybrid that melted ∼10°C higher. We call this probe the ‘24/20 probe’. We expected that at the annealing stage this probe would undergo self-annealing and become non-fluorescent in the absence of target. However, in the presence of the target, the positive strand of the probe would dissociate from the negative strand, bind to the target and become fluorescent. When the temperature was increased to allow extension of the primers (72°C) the two strands of the probe would dissociate from the target and would not interfere with chain extension. By measuring fluorescence intensity during the annealing stage of every cycle, PCR can be followed in a real-time format. Five reactions were preformed with serially diluted template. As expected, standard real-time detection curves were obtained (Fig. 6). Eleven double-stranded probes of different length (22/22–22/17 and 20/20–20/16) were investigated and they all worked well in real-time PCR assays, even those in which both strands were the same length. These observations demonstrate the great flexibility in the design of double-stranded probes.
Figure 6.
Real-time PCR with double-stranded probe. A 268 bp fragment of the human β-globin gene (GenBank accession no. HuMMB5E, –195 to +73) was amplified. The forward and reverse primers were d(GAAGAGCCAAGGACAGGTAC) (P1) and d(CAACTTCATCCACGTTCACC) (P2), respectively. The target sequence of the probe was located in the middle of the amplicon. The positive and negative strands of the probe were FAM-d(AGCAACCTCAAACAGACACCATGG)-PO4 and d(GGTGTCTGTTTGAGGTTGCT)-dabcyl, respectively. The templates (open circle) containing original extracted human DNA were serially diluted 10-fold starting from 50 to 0.05 ng DNA (from left to right). Water was used in place of the template for the negative control samples (solid circle). The upper part of the graph is an illustration of the double-stranded probe at the annealing stage when fluorescence was monitored.
Real-time PCR detection with double-stranded primers
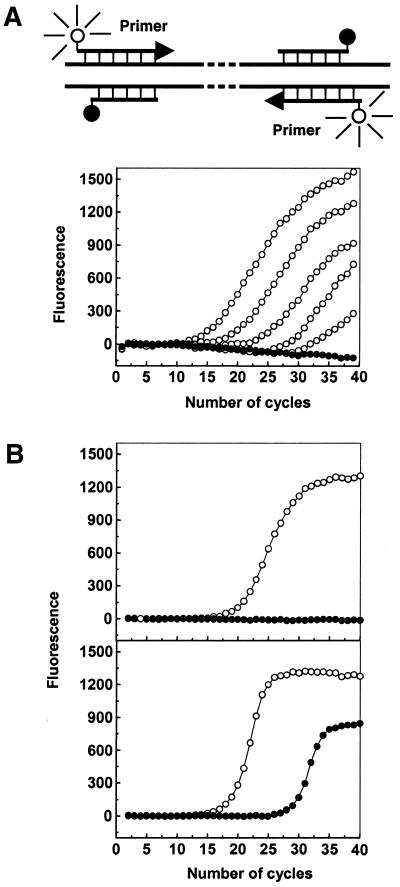
The distinct properties of double-stranded probes also prompted us to explore their use as specific primers for nucleic acid amplification. To turn double-stranded probes into double-stranded primers, modifications were introduced. First, their 3′-terminal phosphate groups were omitted so that they could serve as primers. Second, though not a must, the blunt ends of the probes were moved from the 5′- to 3′-end of the positive strands to let the extendable 3′-end assume a hybridized state. This caused a small separation between the fluorophore and the quencher; however, no significant background fluorescence increase was observed. In our experiments, double-stranded primers were used as both forward and reverse primer, though it could be only one of them. The results showed that all five pairs of primers (including primers 20/20–20/16) gave typical real-time PCR profiles, but slight background fluorescence increases appeared with primers 20/17 and 20/16, obviously due to their lower specificity. A typical real-time PCR profile is shown in Figure 7A.
Figure 7.
(A) Real-time PCR with double-stranded primers. The forward and reverse double-stranded primers were FAM-P1/d(GTACCTGTCCTTGGCTCTT)-dabcyl (positive strand/negative strand) and FAM-P2/d(GGTGAACGTGGATGAAGTT)-dabcyl, respectively. The positive samples (open circle) containing original extracted human DNA were serially diluted 10-fold starting from 100 to 0.01 ng; water was used in place of the template for the negative control (solid circle). The upper part of the graph is an illustration of double-stranded primers at the annealing stage when fluorescence was monitored. (B) Comparison of the specificity of PCR when double-stranded primers (upper) are used and when single-stranded primers (lower) are used. The forward and reverse double-stranded primers were P1/d(GTACCTGTCCTTGGCTCTT)-PO4 and P2/d(GGTGAACGTGGATGAAGTT)-PO4 and the conventional forward and reverse primers were P1 and P2, respectively. One hundred nanograms of extracted DNA was used as the target for positive samples (open circle) and water was used in place of the template for the negative samples (solid circle).
The specificity-enhancing effect of the double-stranded primers was demonstrated by comparison with single-stranded primers. To make the two experimental conditions as close as possible, we used unlabeled double-stranded primers and in both cases a non-specific fluorogenic dye, SYBR Green I, was used for real-time amplification detection. Figure 7B shows that there was significant background amplification in the conventional PCR negative control, but no such background amplification when double-stranded primers were used.
DISCUSSION
The displacement hybridization reaction between a double-stranded oligonucleotide and a single-stranded oligonucleotide is spontaneous. A recent study (14) indicated that the overall displacement rate is a combination of two kintetic pathways: dissociative and sequential displacement. The contribution from the dissociative pathway is predominant at temperatures close to the melting point of the duplex, whereas the contribution from the sequential displacement pathway prevails at lower temperatures and when the concentration of the displacing target is high. Single-nucleotide heterogeneities encountered during displacement stop the process (15), resulting in a high degree of discrimination between perfectly complementary and single mismatch targets. As a result, superior allele discrimination is achieved than is possible by hybridization alone. Our labeling scheme will facilitate further studies on the mechanism of displacement hybridization.
One characteristic of the double-stranded probes is that after probe–target hybridization, the distance between the fluorophore and the quencher changes from close proximity to totally free separation. This property not only endows them with much higher sensitivity than other probes by using common fluorophores and quenchers, but also enables the use of special labels to further improve their performance. Gold nanoparticles have been shown to be extremely efficient fluorescence quenchers and have been applied in molecular beacons (16). Quantum dots are a new type of extremely bright nanoparticle fluorophore (17). Both of these nanoparticles could be used with double-stranded probes to improve detection sensitivity; however, they will encounter difficulties in dual labeled probes.
Although the double-stranded DNA probes demonstrated here are made up of two complementary oligodeoxyribonucleotides of different length, this need not always be the case. For RNA detection the two strands can be the same length, since RNA can form more stable duplexes than DNA. The two strands also need not be blunt at the labeling end. For labeling with fluorescence energy transfer dyes, such as long lifetime chelates (18), the ends of the two strands can be several bases apart to permit optimal energy transfer. Another important characteristic of double-stranded probes is that the ratio of positive strands to negative strands can be altered. To ensure high specificity, the number of negative strands is usually somewhat greater than the number of positive strands.
It has already been reported that conformationally constrained probes, like molecular beacons, possess higher specificity than corresponding linear probes (19). Double-stranded probes can also be regarded as conformationally constrained probes, due to the existence of a stable double-stranded state. Since the specificity of a molecular beacon is determined by both the loop and stem sequence and length, whereas the specificity of a double-stranded probe is determined by the length difference between the two strands, it is difficult to conduct a reasonable comparison of specificity between them. However, two obvious differences can still be noticed from a comparison of the denaturation curves of double-stranded probes with those of molecular beacons. One is the sharper transition in the curves of double-stranded probes compared with those observed with a variety of molecular beacons (6,13). This indicates that the bimolecular duplex denatures faster than an intramolecular duplex, due to the greater entropy change of the former. An even sharper transition has been observed with gold nanoparticle-labeled oligonucleotide probes (20). A sharper transition means lower background and higher sensitivity when detection is carried out at below the Tm of the probe in real-time PCR. Another difference is the much wider window between the curves of the perfectly complementary and single mismatch targets with double-stranded probes than with molecular beacons. In fact, we have not observed any significant fluorescence increase in denaturation curves of mismatched targets with double-stranded probes designed for single-nucleotide discrimination, whereas it is common for molecular beacons designed for this purpose to fluoresce at low temperature (21). As already noted, the window is wider with molecular beacons than with linear probes (13,19), which explains the fact that molecular beacons are superior to TaqMan probes in single mutation detection in real-time PCR (22). However, since the windows with molecular beacons are still not wide enough to cover low temperatures, these probes failed to discriminate single mutations in isothermal amplification systems without further modification (23). These observations suggest that double-stranded probes exhibit potentially greater specificity and sensitivity than molecular beacons and may extend their use in single mutation detection to both PCR and isothermal amplification systems.
When combined with nucleic acid amplification detection, an ideal probe should be, in addition to its high sensitivity and specificity, flexible enough to satisfy a variety of stringent requirements in practical use. The probe should be simple to design, easy to synthesize and robust in use. Single-dye modification makes the double-stranded probe much easier to synthesize and purify, and thus cheaper, than dual-labeled probes. The robustness of the double-stranded probes is reflected in the observation that 11 different double-stranded probes designed for the same PCR worked well under the same conditions. We concluded that in PCR, any double-stranded probe with a Tm between the annealing temperature and 72°C can detect the amplicon efficiently. This is because the double-stranded probes have a sharp transition in the denaturation curve and detection conducted below the melting point can indicate the amplicon concentration without background interference. The color multiplexing ability represents another merit of double-stranded probes, which can find applications in simultaneous detection of clinically related genes, random mutations, infectious agents and single nucleotide polymorphisms. Melting curve analysis using adjacent probes has proved to be a good method for Tm multiplexing (24). However, Tm multiplexing has technical challenges in addition to the optimizations often necessary for multiplexing primer sets and is restricted to current instrumentation and available fluorescent energy donor–acceptor pairs (25). No more than four alleles have been simultaneously detected by Tm multiplexing, whereas five or more mutation detection has been achieved by color multiplexing (9,26). Unlike Tm multiplexing, color multiplexing can be finished in real-time without subsequent manipulations; if subsequent manipulation is carried out, multiplexing capacity can be further improved (27).
Double-stranded probes, when used as primers, could improve nucleic acid amplification in many ways. First, double-stranded primers are natural ‘hot-start’ primers, since they are in duplex form and cannot hybridize with each other at temperatures lower than their Tm values. This also minimizes non-specific annealing in the course of amplification. Recently, a hairpin-like structure also showed this specificity (28). In contrast, common ‘hot-start’ methods, such as wax barriers (29) and the use of polymerase antibodies (30) or engineered polymerases (31), can function only at the beginning of the amplification. For non-probe real-time detection, double-stranded primers are superior to dual-dye-labeled Amplifluore primers (32) and the more recent self-reporting PNA–DNA primers (33), since neither of the latter can distinguish specific from non-specific amplification. Like double-stranded probes, double-stranded primers also have color multiplexing ability, which can be of great use in allele-specific amplification. Recently, Donohoe et al. (34) achieved Tm multiplexing allele-specific PCR with a combination of three techniques. Despite its complexity, the obvious limitation of this approach, as stated by the author, is that optimization and discrimination of GC-rich PCR products is difficult as they used a long GC tail in one primer to obtain Tm discrimination. We reason that using double-stranded primers labeled with different colors, allele-specific PCR can be easily accomplished regardless of the composition of the amplicons.
Other researchers have recognized the usefulness of the signal-generating ability of ‘strand displacement’ (35–37). However, these methods were based on the ‘branch migration’ mechanism, which requires the ‘probe strand’ to be entirely complementary to the target nucleic acid and the displaced ‘signal strand’ to be separated before measurement, hindering their use for homogeneous detection. Later, Morrison et al. (38) designed a kind of double-stranded probe consisting of two complementary oligonucleotides of equal length for use in homogeneous detection of PCR products. However, the slow kinetics and poor signal obtained prevented further efforts. Two other approaches using labeled oligonucleotide duplexes in real-time PCR have recently been reported. One is the AmpliSensor probe (39) and the other is an improved 5′-nuclease probe (40). The former was used as a ‘signal primer’ in asymmetrical semi-nested PCR assays and the latter was used to improve the detection sensitivity of the 5′-nuclease assay. In both cases the potential specificity-enhancing ability of the probe was ignored. This was also seen in a later work screening the binding affinity of peptide–oligonucleotide conjugates (41).
The significance of the double-stranded probes we propose here lies in their extreme specificity, their spontaneity of reaction and their sensitivity and simple preparation. Their abilitiy to discriminate single-nucleotide substitutions enables them to be used in a wide range of applications in molecular diagnostics, especially in mutation detection and genotyping. This high specificity can also be extended to improve antisense therapy. The spontaneity of their interaction with targets can be used to trace mRNAs in living cells and to construct biosensors and new biochip detection devices. Double-stranded probes could be suitable for gene quantification when combined with various nucleic acid amplification systems for real-time detection. The simplicity and low cost of double-stranded probes should make them good alternatives to currently used probes. Finally, it should be realized that double-stranded probes and all their applications demonstrated herein actually embody the specificity-enhancing ability of displacement hybridization. This new reaction mode should be applicable in any circumstances when high specificity is needed.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Fred Russell Kramer and Sanjay Tyagi for insightful reviews of the manuscript and enlightening discussions, and Shuang Chen and Yangjian Cheng for technical assistance. This work was partially supported by grant C97013 from the Fujian Natural Science Foundation and a grant from the Key Laboratory of Marine Biology Engineering of the Third Institute of Oceanography, Xiamen, China.
REFERENCES
- 1.Foy C.A. and Parkes,H.C. (2001) Emerging homogeneous DNA-based technologies in the clinical laboratory. Clin. Chem., 47, 990–1000. [PubMed] [Google Scholar]
- 2.Cantor C.R. (1996) Lighting up hybridization. Nat. Biotechnol., 14, 264. [DOI] [PubMed] [Google Scholar]
- 3.Schweitzer B. and Kingsmore,S. (2001) Combining nucleic acid amplification and detection. Curr. Opin. Biotechnol., 12, 21–27. [DOI] [PubMed] [Google Scholar]
- 4.Lakowicz J.R. (1999) Principles of Fluorescence Spectroscopy, 2nd Edn. Kluwer Academic/Plenum Publishers, New York, NY.
- 5.Holland P.M., Abramson,R.D., Watson,R. and Gelfand,D.H. (1991) Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl Acad. Sci. USA, 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 7.Whitcombe D., Theaker,J., Guy,S.P., Brown,T. and Little,S. (1999) Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol., 17, 804–807. [DOI] [PubMed] [Google Scholar]
- 8.Kandimalla E.R. and Agrawal,S. (2000) Cyclicons as hybridization-based fluorescent primer-probes: synthesis, properties and application in real-time PCR. Bioorg. Med. Chem., 8, 1911–1916. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi S., Marras,S.A.E. and Kramer,F.R. (2000) Wavelength-shifting molecular beacons. Nat. Biotechnol., 18, 1191–1196. [DOI] [PubMed] [Google Scholar]
- 10.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–131, 134–138. [DOI] [PubMed] [Google Scholar]
- 11.Isacsson J., Cao,H., Ohlsson,L., Nordgren,S., Svanvik,N., Westman,G., Kubista,M., Sjoback,R. and Sehlstedt,U. (2000) Rapid and specific detection of PCR products using light-up probes. Mol. Cell Probes, 14, 321–328. [DOI] [PubMed] [Google Scholar]
- 12.Kutyavin I.V., Afonina,I.A., Mills,A., Gorn,V.V., Lukhtanov,E.A., Belousov,E.S., Singer,M.J., Walburger,D.K., Lokhov,S.G., Gall,A.A. et al. (2000) 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res., 15, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 14.Reynaldo L.P., Vologodskii,A.V., Neri,B.P. and Lyamichev,V.I. (2000) The kinetics of oligonucleotide replacements. J. Mol. Biol., 297, 511–520. [DOI] [PubMed] [Google Scholar]
- 15.Biswas I., Yamamoto,A. and Hsieh,P. (1998) Branch migration through DNA sequence heterology. J. Mol. Biol., 279, 795–806. [DOI] [PubMed] [Google Scholar]
- 16.Dubertret B., Calame,M. and Libchaber,A.J. (2001) Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol., 19, 365–370. [DOI] [PubMed] [Google Scholar]
- 17.Chan W.C. and Nie,S. (1998) Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science, 281, 2016–2018. [DOI] [PubMed] [Google Scholar]
- 18.Selvin P.R. (2000) The renaissance of fluorescence resonance energy transfer. Nature Struct. Biol., 7, 730–734. [DOI] [PubMed] [Google Scholar]
- 19.Bonnet G., Tyagi,S., Libchaber,A. and Kramer,F.R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA, 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taton T.A., Mirkin,C.A. and Letsinger,R.L. (2000) Scanometric DNA array detection with nanoparticle probes. Science, 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- 21.Marras S.A., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 22.Täpp I., Malmberg,L., Rennel,E., Wik,M. and Syvänen,A.-C. (2000) Homogeneous scoring of single-nucleotide polymorphisms: comparison of the 5′-nuclease TaqMan assay and molecular beacon probes. Biotechniques, 28, 732–738. [DOI] [PubMed] [Google Scholar]
- 23.de Baar M.P., Timmermans,E.C., Bakker,M., de Rooij,E., van Gemen,B. and Goudsmit,J. (2001) One-tube real-time isothermal amplification assay to identify and distinguish human immunodeficiency virus type 1 subtypes A, B and C and circulating recombinant forms AE and AG. J. Clin. Microbiol., 39, 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernard P.S. and Wittwer,C.T. (2000) Homogeneous amplification and variant detection by fluorescent hybridization probes. Clin. Chem., 46, 147–148. [PubMed] [Google Scholar]
- 25.Bernard P.S., Ajioka,R.S., Kushner,J.P. and Wittwer,C.T. (1998) Homogeneous multiplex genotyping of hemochromatosis mutations with fluorescent hybridization probes. Am. J. Pathol., 153, 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol., 16, 359–363. [DOI] [PubMed] [Google Scholar]
- 27.Lee L.G., Livak,K.J., Mullah,B., Graham,R.J., Vinayak,R.S. and Woudenberg,T.M. (1999) Seven-color, homogeneous detection of six PCR products. Biotechniques, 27, 342–349. [DOI] [PubMed] [Google Scholar]
- 28.Kaboev O.K., Luchkina,L.A., Tret’iakov,A.N. and Bahrmand,A.R. (2000) PCR hot start using primers with the structure of molecular beacons (hairpin-like structure). Nucleic Acids Res., 28, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hebert B., Bergeron,J., Potworowski,E.F. and Tijssen,P. (1993) Increased PCR sensitivity by using paraffin wax as a reaction mix overlay. Mol. Cell Probes, 7, 249–252. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg D.E., Rybalkin,I., Chen,S., Mukhamedova,N., Vlasik,T., Siebert,P.D. and Chenchik,A. (1994) TaqStart antibody: “hot start” PCR facilitated by a neutralizing monoclonal antibody directed against Taq DNA polymerase. Biotechniques, 16, 1134–1137. [PubMed] [Google Scholar]
- 31.Kebelmann-Betzing C., Seeger,K., Dragon,S., Schmitt,G., Moricke,A., Schild,T.A., Henze,G. and Beyermann,B. (1998) Advantages of a new Taq DNA polymerase in multiplex PCR and time-release PCR. Biotechniques, 24, 154–158. [DOI] [PubMed] [Google Scholar]
- 32.Nazarenko I.A., Bhatnagar,S.K. and Hohman,R.J. (1997) A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res., 25, 2516–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiandaca M.J., Hyldig-Nielsen,J.J., Gildea,B.D. and Coull,J.M. (2001) Self-reporting PNA/DNA primers for PCR analysis. Genome Res., 11, 609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donohoe G.G., Laaksonen,M., Pulkki,K., Ronnemaa,T. and Kairisto,V. (2000) Rapid single-tube screening of the C282Y hemochromatosis mutation by real-time multiplex allele-specific PCR without fluorescent probes. Clin. Chem., 46, 1540–1547. [PubMed] [Google Scholar]
- 35.Vary C.P., McMahon,F.J., Barbone,F.P. and Diamond,S.E. (1986) Nonisotopic detection methods for strand displacement assays of nucleic acids. Clin. Chem., 32, 1696–1701. [PubMed] [Google Scholar]
- 36.Ellwood M.S., Collins,M., Fritsch,E.F., Williams,J.I., Diamond,S.E. and Brewen,J.G. (1986) Strand displacement applied to assays with nucleic acid probes. Clin. Chem., 32, 1631–1636. [PubMed] [Google Scholar]
- 37.Vary C.P. (1987) A homogeneous nucleic acid hybridization assay based on strand displacement. Nucleic Acids Res., 15, 6883–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison L.E., Halder,T.C. and Stols,L.M. (1989) Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal. Biochem., 183, 231–244. [DOI] [PubMed] [Google Scholar]
- 39.Wang C.N., Wu,K.Y. and Wang,H.T. (1995) Quantitative PCR using the AmpliSensor assay. In Dieffennach,C.W. and Dveksler,G.S. (eds), PCR Primer: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 193–202.
- 40.Di C. and Joseph,L. (1998) Fluorescence detection assay for homogeneous PCR hybridization systems, US patent 5,716,784.
- 41.Harrison J.G., Liu,X. and Balasubramanian,S. (1999) Screening for oligonucleotide binding affinity by a convenient fluorescence competition assay. Nucleic Acids Res., 27, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]