Abstract
Discovering effective therapies is difficult for neurological and developmental disorders in that disease progression is often associated with a complex and interactive mechanism. Over the past few decades, few drugs have been identified for treating Alzheimer’s disease (AD), especially for impacting the causes of cell death in AD. Although drug repurposing is gaining more success in developing therapeutic efficacy for complex diseases such as common cancer, the complications behind AD require further study. Here, we developed a novel prediction framework based on deep learning to identify potential repurposed drug therapies for AD, and more importantly, our framework is broadly applicable and may generalize to identifying potential drug combinations in other diseases. Our prediction framework is as follows: we first built a drug-target pair (DTP) network based on multiple drug features and target features, as well as the associations between DTP nodes where drug-target pairs are the DTP nodes and the associations between DTP nodes are represented as the edges in the AD disease network; furthermore, we incorporated the drug-target feature from the DTP network and the relationship information between drug-drug, target-target, drug-target within and outside of drug-target pairs, representing each drug-combination as a quartet to generate corresponding integrated features; finally, we developed an AI-based Drug discovery Network (AI-DrugNet), which exhibits robust predictive performance. The implementation of our network model help identify potential repurposed and combination drug options that may serve to treat AD and other diseases.
Keywords: Network model, Deep learning, Drug repurposing, Drug combination therapy, Neurological and developmental disorders
Graphical Abstract
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and patients suffer from extensive neuropathological symptoms such as intracellular neurofibrillary tangles [1], [2]. Currently, AD is rising to be the sixth leading cause of death in the United States, and the number of AD dementia cases will grow to more than 100 million cases in 2050 if effective treatment is not found [3].
The biological mechanism of AD is complicated, and the development of AD is associated with a plethora of complex, progressive, interactive, and destructive processes that result in brain cell dysfunction and death [4], [5], [6], [7], [8]. Therefore, developing promising drugs for AD treatment is difficult to achieve [9], [10], [11], [12]. Over the past few decades, there have been only several drugs approved for the treatment of AD by the US Food and Drug Administration (FDA). For example, Tacrine was approved by the FDA in 1993, and Rivastigmine was identified in 1998 [13], [14], [15], [16], [17]. However, most of these potential drugs approved are intended for reducing AD-associated symptoms rather than leveraging the biological and functional networks in AD to cure the disease itself [18], [19]. In 2021, the first disease-modifying therapy (DMT) aducanumab was approved and became available for those with AD-associated mild cognitive impairment. As of January 2022, there were only 143 agents in clinical trials for AD therapeutics according to Clinicaltrials.gov [20].
There is an extremely high failure rate in AD drug discovery and effective therapy treatments are needed urgently given the fact that there are existing off-target side effects and suboptimal efficacy because of the complex disease pathobiology of AD development [9], [10], [11], [12]. Exerting effects on several pathogenic pathways associated with the disease is an effective strategy to combat drug resistance and disease heterogeneity, and more evidence proves that combination therapy, the use of multiple drugs to treat a disease, is a promising method to treat diseases, especially for complex disorders [21], [22], [23], [24]. Finding and identifying repurposed or combination therapies have made great progress in multiple complex diseases varying from cancer to infectious diseases [22], [23]. However, the discovery of effective therapies in AD is still limited in that there are few known therapies developed for AD treatment. As artificial intelligence advances, deep learning technologies have been successfully used to resolve various biomedicine questions, including regulatory genomics, epigenetics, cancer subtyping, therapeutic target identification, drug target interaction prediction and image analysis [25], [26], [27], [28], [29], [30], [31], [32]. Increasing evidence shows that deep learning, especially a graph neural network, is a suitable and promising modeling framework for drug repurpose and discovery [30], [31], [33].
To resolve the problem, we constructed an AD-specific repurposed drug network and target network and incorporated the association between drugs and targets, generating an AD-specific drug-target pair (DTP) network. Based on the reconstructed DTP network, we represented all drug-target pairs comprehensively by taking the advantage of the relationships between drug-drug, drug-target, and target-target. Furthermore, we characterized all potential drug combination therapies as quartets where each quartet consists of drug A, target A, drug B, and target B and meanwhile took account into the interactions between drug-drug, target-target, and drug-target between or outside of the drug-target pairs. Thereafter, we developed a deep learning-based model that could take quartet features as input and distinguish the true quartet from the false quartet. Here is our framework: the model first adopted a graph convolutional network to learn the features for each target-drug pair; using the feature representation of each quartet as an input, the architecture integrated with fully connected network worked on identifying promising quartets to treat AD [34], [35]. To validate our framework, despite the limited knowledge on existing AD drugs and combinations, we further extended and carried out drug repurposing predictions for additional biological contexts, specifically in three different cell lines (brain-related and brain-independent): DIPG25, U251 and A549. The results based on our data showed that the proposed framework achieved a robust performance when finding prospective repurposed drugs and potential combination drug pairs that may both serve as candidates for treating AD.
2. Materials and Methods
2.1. De novo repurposed drug discovery in AD
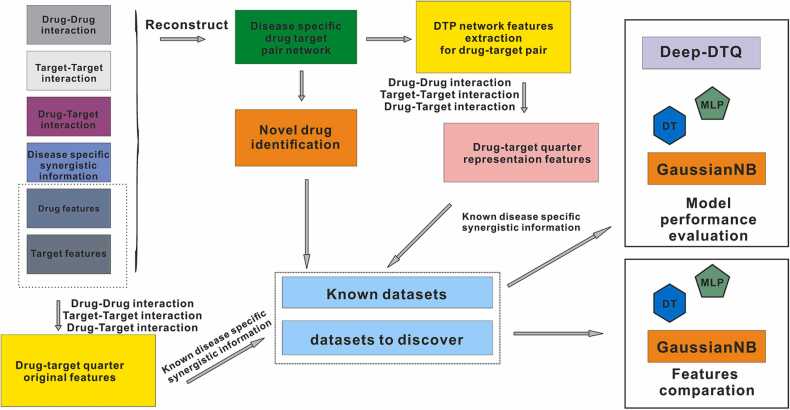
As shown in our workflow (Fig. 1), accessing known disease-specific synergistic information is the first step. Considering the absence of known synergistic gene pairs in AD development and progression, we first adopted OptiCon to identify synergistic regulators for combination therapy in AD [36]. OptiCon has been shown as an effective and robust tool for de novo identification of synergistic genes as candidates in a gene regulatory network based on network controllability theory. This algorithm first identifies a set of optimal control nodes (OCNs) that can maintain maximal control over deregulated pathways but minimal control over those disease-free pathways by using gene expression constraints in the network controllability framework and then identifies synergistic OCNs as key regulators and candidate targets for combination therapy in the disease-perturbed network based on synergy score taking into account disease-specific genetic mutation and gene functional interaction information.
Fig. 1.
The workflow of the Deep-DTQ prediction framework.
To this end, we accessed gene expression data in AD patients and non-AD patients as the control where there are a high-quality set of Illumina RNA sequencing data for patients with sporadic AD (n = 12; mean age=68) and cognitively healthy individuals (n = 10; mean age=68) from the temporal lobe of frozen postmortem brain tissues as previously described [37]. Besides, to evaluate the robustness of the AD-specific synergistic gene pairs discovered by OptiCon, we also accessed Illumina RNA sequencing data, derived from patients with sporadic AD (n = 8; mean age=91) and cognitively healthy individuals (n = 10; mean age=77) from the temporal lobe of frozen postmortem brain tissues as previously described [38]. For RNA expression datasets, we adopted stringtie to access fragments per kilobase of transcript per million mapped reads (FPKM) for each gene based on GRCh38, and used ballgown to perform differential expression based on adjusted p-value cutoff< = 0.05, and finally made gene expression file and gene deregulation score file ready for OptiCon inputs [39]. For a gene regulatory network for OptiCon, we adopted the comprehensive human gene regulatory network by integrating annotations from Reactome, KEGG and NCI-Nature pathway interaction database [40], [41], [42]. For AD-specific genes harboring somatic mutations, we accessed the AD-associated mutated gene sets where single nucleotide variations of these genes are found in AD brain and blood against non-AD counters as previously described [43]. Based on these data, we derived potential synergistic gene pairs for potential AD therapy.
3. DTDT quartet feature extraction
The features of each drug-target-drug-target (DTDT) quartet consist of combined features of two drug-target pairs and the interactions between and outside of these two drug-target pairs, including the interactions between drug A-drug B, drug A-target A, drug A-target B, drug B-target A, drug B-target B, and target A-target B. As for the features of each drug-target pair, it consists of drug feature, target feature, drug-target interaction and the direct relationships between the drug-target pair and other drug-target pairs in the AD-specific DTP network.
Drug features are defined by their chemical structures. Simplified molecular input line entry specification (SMILES) takes into account the chemical structures of drugs and could explicitly characterize the molecular structure of each drug and therefore classify these drugs into corresponding categories. We, here, took advantage of SMILES categorizing system to characterize the chemical properties of drugs, and the categories of each drug were chosen as features [44], [45]. If a drug was attributed to one group, then the group feature was set as 1, otherwise 0. To avoid the oversize of dimensionality in categories, we only chose the major categories to represent these drugs.
Protein features are defined by their compositional information, including both the chemical properties of amino acids and sequence information. We hypothesized that the interaction between a drug and its target protein is influenced by the hydrophobicity, polarity, and tertiary structure of the target protein [46]. Therefore, each amino acid in a protein sequence can be extracted as a chemical feature for this target protein. Based on previous research, common amino acids can be divided into six groups according to their chemical characteristics: strongly hydrophilic or polar acids (D, E, H, K, N, Q, R), strongly hydrophobic acids (A, F, I, L, M, V), weakly hydrophilic or hydrophobic acids (S, T, W, Y), cysteine (C), glycine (G), and proline (P) [47], [48]. We took into account the relative proportion of each amino acid as target protein sequence information and extracted the information as extra features. Finally, each target protein contains 26-dimensional features, including the relative proportion of each of the 20 amino acids in the protein and 6 classes of chemical characteristics.
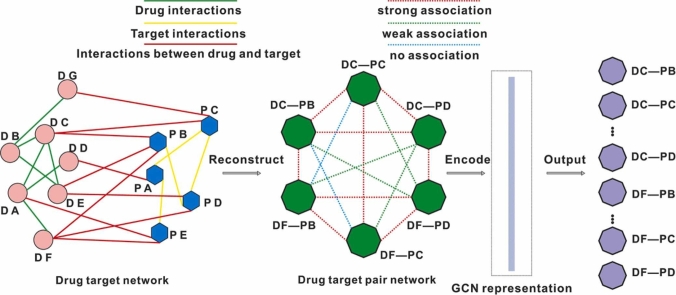
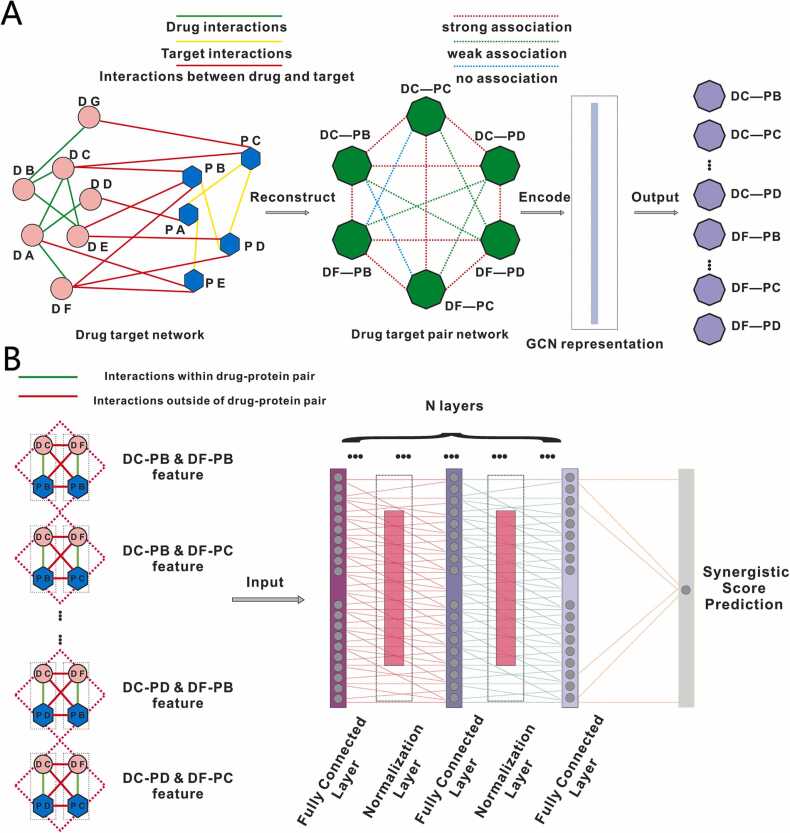
To integrate the interaction information between drugs and targets, we first constructed a DTP network based on a known drug-drug network, drug-target network, and target-target network. In this DTP network, each DTP node represents a drug and a target, and each edge represents one association between DTP nodes (Fig. 2A). These associations include strong association, weak association, and non-association. These relationships are defined as follows: strong association refers to two DTPs that share a common drug or target; weak association refers to two DTPs that don’t share any drug or target but there is an interaction between the drugs or the targets in these two DTPs; non-association refers to two DTPs that don’t share any drug or target and there isn’t direct interaction between the drugs or the targets in these two DTPs (Fig. 2A).
Fig. 2.
The deep learning model for candidate drug combination discovery in AD. (A) Disease-specific DTP network representation for drug-target pairs (DTP).(B) The architecture of the drug combinations prediction model.
The complete DTP network was formulated by adjacency matrix M:
whererepresents each DTP, represents the edges between DTPs and represents the associations between and depending on strong association, weak association, or non-association. Here, and represent two pair nodes: consists of drug a and target b, and consists of drug c and target d.
3.1. DTP feature representation based on graph convolutional neural networks
For a given constructed DTP network, it can be represented aswhere represents DTP nodes, represents the set of edges (associations between DTP nodes). Meanwhile, each node in the graph convolutional neural networks (GCN) contains its features [49], so the weighted adjacency matrix is:
where is the network’s adjacency matrix and is the identity matrix.
The inverse degree matrix can be calculated as follows:
Finally, the features of each DTP in the network can be calculated as follows:
where is the rectified linear unit function and is the feature vector of each DTP node, that is the combination of the features of a drug and a target for each DTP node.
4. Performance evaluation and cross validation
Here, if there were associations between the two drug-target pairs, we defined these two drug-target pairs as a positive quartet. Otherwise, we defined the two drug-target pairs as a negative quartet. More specifically, if one drug interacted with one synergistic target pair, and another drug interacted with another synergistic target pair, then we considered these drug target pairs as a potential drug-target quartet for AD.
Considering there could be many more potential negative quartets for AD compared to positive quartets, we adopted stratified 5-Fold cross-validation to evaluate the value of representation features that consist of DTP network features and interaction features between drugs and targets compared to original features that consist of drug features, target features, and interaction features between drugs and targets in multiple research objects, including AD and other contexts (such as cell lines DIPG25, U251 and A549). To guarantee a reliable performance evaluation, we devised multiple common machine learning models as benchmarks, including decision tree (DT), Gaussian Naive Bayes (GaussianNB), and Multi-layer Perceptron (MLP). We ran a grid search scheme and evaluated the efficacy of representation and original features based on parameter space in the aspect of F1 score, ROC AUC, and balanced accuracy.
After feature comparative analysis, the suitable configuration of DT, GaussianNB, and MLP models were found when identifying prospective drug-target quartets from false drug-target quartets in multiple biological objects. Furthermore, we compared our optimized Deep-DTQ to the optimized DT, GaussianNB, and MLP in the aspect of F1 score and ROC AUC in the same stratified 5-Fold cross-validation strategy.
5. Results
5.1. Construction of a deep learning model for AD drug repurposing
We first generated the feature vector of each DTDT node in the DTP network, and then developed a deep learning framework. As shown in Fig. 2B, the framework contained multiple layers. The input for this model is the features of DTDTs, which are represented by the combined features of two DTPs and the interactions of drugs and targets of these two DTPs.
Basically, the input was fed into fully connected network. Between the fully connected (FC) layers, we placed a group normalization layer, which divided the channels into multiple groups and then obtained the means and variance within each group for normalization. This strategy improved detection accuracy and made the model more transferrable to novel datasets, compared with other normalization methods like batch normalization and layer normalization [50]. After being transformed multiple times, the information was fed into the output layer consisting of one unit to make a final decision on whether the input DTDT is a qualified drug combination pair or not. It is worth noting the sigmoid activation function was applied in the output layer to achieve the classification task appropriately.
Here, multiple learning techniques were adopted to facilitate model training and to make the prediction model more accurate and robust. Dropout was the most common regularization method and a key component to training complicated deep neural network models, and it could set some neurons in the network to a ”dropped out” state during training in each iteration, which could average multiple networks during training and make the final trained model more generalized in essence [51]. We applied dropout to all the intermediates between FC layers in the model. As an effective regularization method, early stopping was also adopted to stop training with specific epoch patience once the model performance was no longer improved on the validation dataset to avoid overfitting. In addition, L1 norm and L2 norm were also combined for regularizing the parameter values in all FC layers. Lastly, for obtaining better local minima of parameters, adadelta learning rate method was adopted for gradient descent, which was proved to be robust to noisy gradient information, different model architecture, and diverse data modalities [52].
To access the configurations of Deep-DTQ managing to achieve robust predictive performance in multiple cell lines (disease), we constructed a huge hyperparameter space: the number of FC layers ranged from 2 to 8 with a step size was 1; a set of dropout rates between the dense layers were tested, ranging from 0 to 0.5, with a step size of 0.1; the batch size varied from 16 to 256, with a double fold way; L1 and L2 regularization penalty ranged from 1e‐5 to 1e‐1, with a ten-fold way; the initialized value of each parameter was sampled from a random uniform, random normal or ones distribution in the FC layers; The exponential linear unit (ELU), rectified linear activation (ReLU), hyperbolic tangent (Tanh) were applied to the FC layers randomly.
To get the best combination of hyperparameters in the high-dimensional space effectively, Bayesian optimization was adopted in the training process [53]. During the training process, as iteration grew, and the posterior distribution of the model’s cost function improved, the Bayesian optimization algorithm could further explore hyperparameter space which was worth exploring automatically, and seeking the best hyperparameter combination at a lower computation cost.
By taking advantage of Bayesian global optimization with gaussian processes, we set 200 init points in the hyperparameters space and allowed the training to continue at a maximum of 10 iterations. Based on our datasets, we finally got the best configurations of Deep-DTQ in the three cell lines or AD. The detailed hyperparameter configurations is shown in Supplementary Table 1.
6. Identification of AD-associated target gene pairs
We identified 171 synergistic gene pairs for AD (BH-adjusted empirical p-value<0.05), including MAML2-AGTR1 (p-value=0.000511), PSMC1-MAML2 (p-value=0.00155), MAML2-MTMR6 (p-value=0.001633), TOMM22-DOLPP1 (p-value=0.002193) and so on based on datasets as previously described [37]. Consistently, quite a few of these genes were previously found to be involved in AD development and progression. For example, the gene AGTR1 encodes type-1 angiotensin II receptor (AT1 receptor), which interacts with angiotensin II generated by angiotensin-converting enzyme, ACE. ACE-generated angiotensin II was reported to promote neurodegeneration and brain aging in AD patients [54], [55]. PSMC1 serves as a proteasome complex component and the overrepresentation of the component is associated with dysregulated proteostasis in AD pathophysiology [56]. Together, all these examples support the quality of our predictive framework. For a detailed list of the predicted synergistic gene pairs, please refer to Supplemental Table 2. To furthermore evaluate the robustness of the above identified synergistic gene pairs, we applied the OptiCon algorithm to the gene expression datasets as previously described [38], we found all those 171 synergistic gene pairs are shared out of 190 synergistic gene pairs identified by this dataset in Supplemental Table 3.
Considering there is little available information about AD treatment targets, not to mention synergistic gene pairs for AD, these identified synergistic gene pairs will be valuable candidates and resources for potential AD treatment.
6.1. Integration of drug-drug interactions and protein-protein interactions in AD networks
Based on the synergistic gene pairs we identified above, we collected drug-drug and drug-target interactions from DrugBank [57]. In addition, we obtained information on protein-protein interactions from HIPPIE database, and protein-protein interactions (PPIs) with scores greater than 0.5 were chosen to build a target-target network [58]. Based on available feature information of drugs and proteins, we filtered and finally got 38,288,600 drug-target interaction entries that cover 12,111 drugs and 4,975 proteins. Of 38,288,600 interaction entries, there are 29,382 real interactions between those drugs and proteins.
Limited to availability on the abovementioned database, we retrieved and accessed 10 available synergistic target pairs in AD. It was worth noting that five unique targets comprised these 10 synergistic target pairs, suggesting these five genes could be promising therapeutic targets for AD, and these genes included AGTR1 (P30556), FEN1 (P39748), PSMB2 (P49721), PDHB (P11177), and NADSYN1 (Q6IA69). We also took into account 125 targets that directly interacted with these 5 targets, resulting in a total of 130 AD‐associated targets in the AD-specific network. Next, we extracted 911 drugs that directly interacted with these targets. Furthermore, we considered another 461 drugs that interacted with more than 500 out of those 911 drugs in the drug-drug interaction network as potential drugs. These 461 potential drugs were those that did not have any known interactions with those AD-specific targets but they could be promising drugs for AD considering they frequently interacted with those drugs that were associated with AD-specific targets. Together, our analyses resulted in a total of 1,372 drugs in the AD network.
As for the features, target features were 26-dimension defined by their compositional information, including 6 categories of properties and the relative proportion of each of the 20 amino acids; Drug features were 737-dimension defined by their chemical structures based on shared drug categories by drugs in the AD network; Drug-target pair features were 763-dimension representation by GCN with target features, drug features and drug-drug network, drug-target network, target-target network; Drug-target quartet was 1538-dimension and its representation was as following: 2 drug-target pair features (1526-dimension), 2 drug-target interaction within drug-target pair (4-dimension), 1 target-target interaction (2-dimension), 1 drug-drug interaction (2-dimension), 2 drug-target interaction outside drug-target pair (4-dimension).
We constructed our training data set, which included 228 positive drug-target quartets, and 8,298,982 negative drug-target quartets (10 * 911 * 911–228). In addition, we constructed a potential drug combination discovery pool that consisted of 461 potential drugs and 2,125,210 (10 * 461 * 461) drug-target quartets. Afterward, as a cross-validation, we retrieved synergistic drug information in independent biological contexts (using a diverse set of cell lines: DIPG25, U251 and A549) in DrugCombDB, and created three cell line-specific DTP networks and further constructed corresponding drug-target quartets datasets based on the same method as AD [59].
Taking similar approaches, we constructed the following three cell line-specific networks. In the DIPG25 specific network, there were 1,630 drugs, 45 targets, and 946 potential drugs; target features were 26-dimension, drug features were 1,011-dimension, and final drug-target quartet was 2,086-dimension. In the U251 specific network, there were 1,968 drugs, 74 targets, and 878 potential drugs; target features were 26-dimension, drug features were 1084-dimension, and final drug-target quartet was 2,232-dimension. In the A549 specific network, there were 1,989 drugs, 80 targets, and 886 potential drugs; target features are 26-dimension, drug features are 1093-dimension, and final drug-target quartet is 2250-dimension.
6.2. Representation features based on the DTP network outperformed original features
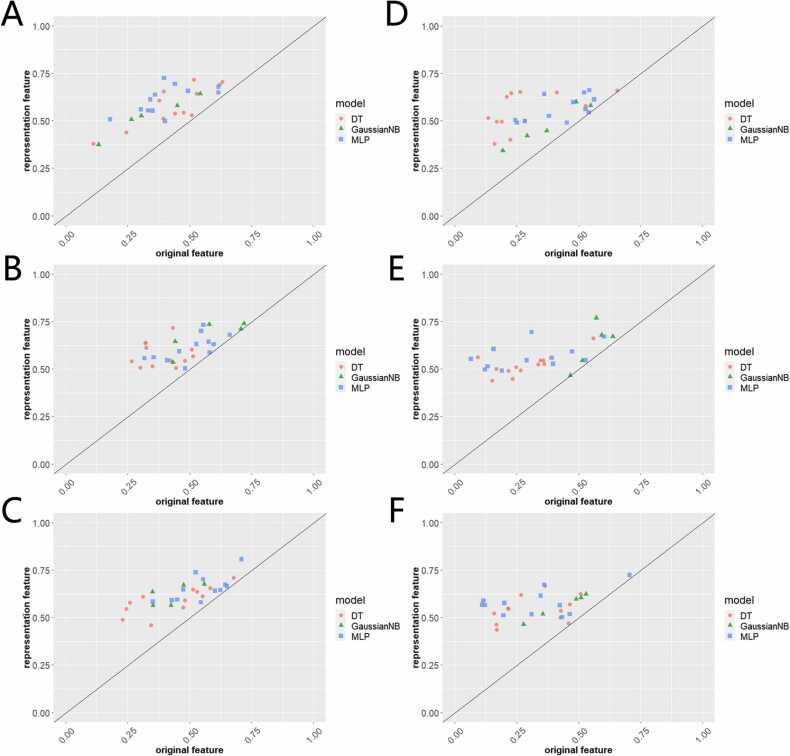
To evaluate the predictive efficacy of the representation features extracted from the DTP network, we constructed three sets of DT, GaussianNB, MLP models with variable hyperparameter space, and compared the predictive performance for each model with the same configurations based on representation features and original features. In the A549 cell line, the representation features showed better predictive performance in the aspect of F1 score, ROC AUC, and balanced accuracy while keeping the same model and the same configuration (Fig. 3A-C), suggesting the DTP network is able to encode the drug-target pair efficiently and comprehensively and generate more characteristic features for each drug-target in cell line-specific context. For example, for MLP with logistic activation and 200 hidden layer size, representation features achieved F1 score at 0.725 while original features got 0.400; for DT with entropy criterion and 3 min sample splits, representation features achieved ROC AUC at 0.567 while original features got 0.514. As for the DIPG25 cell line, representation features outperformed the original again (Fig. 3D-E). For example, for GaussianNB with 3e-09 var smoothing, representation features achieved balanced accuracy at 0.623 while original features only got 0.530.
Fig. 3.
Evaluation of predictive efficacy for representation.(A) Predictive F1 score comparison between representation features and original features in A549.(B) Predictive ROC AUC comparison between representation features and original features in A549.(C) Predictive balanced accuracy comparison between representation features and original features in A549.(D) Predictive F1 score comparison between representation features and original features in DIPG25.(E) Predictive ROC AUC comparison between representation features and original features in DIPG25.(F) Predictive balanced accuracy comparison between representation features and original features in DIPG25.
Besides, similar trends were observed in AD and U251 (Supplement Fig. 1A-F), suggesting the represented features are capable of conserving the intrinsic features in a disease-specific way. The detailed configurations and predictive performance are shown in Supplementary Table 4.
7. Our network model outperforms existing methods
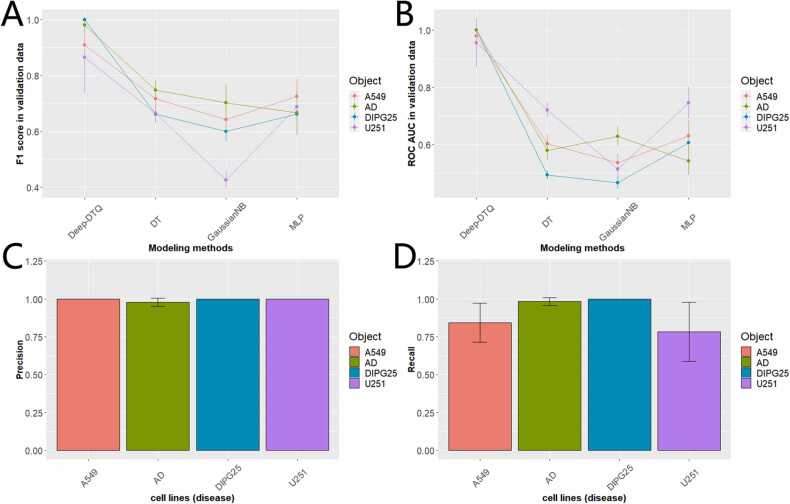
After Bayesian optimization, we got customized optimized Deep-DTQ for multiple cell lines (disease). To explore the predictive performance of Deep-DTQ, we compared the model to the abovementioned optimized DT, GaussianNB, and MLP after grid search optimization in the aspect of F1 score and ROC AUC based on stratified 5-Fold cross-validation. As shown in Fig. 4A, Deep-DTQ achieved better F1 scores compared with the other three models in all cell lines (disease). For example, Deep-DTQ achieved F1 at 1.0 ± 0 (SD), 0.86 ± 0.129 (SD), 0.91 ± 0.077 (SD), and 0.98 ± 0.025 (SD) respectively in DIPG25, U251, A549, and AD; however, DT only got 0.66 ± 0.027 (SD), 0.67 ± 0.037 (SD), 0.72 ± 0.037 (SD), and 0.75 ± 0.035 (SD), GaussianNB only got 0.60 ± 0.037 (SD), 0.43 ± 0.033 (SD), 0.64 ± 0.048 (SD), and 0.70 ± 0.065 (SD), and MLP only got 0.66 ± 0.048 (SD), 0.69 ± 0.058 (SD), 0.73 ± 0.061 (SD), and 0.67 ± 0.080 (SD) in corresponding cell lines (disease). Similarly, Deep-DTQ achieved better ROC AUC compared to the other models as shown in Fig. 4B.
Fig. 4.
Evaluation of predictive performance for Deep-DTQ. (A) F1 score predictive performance comparison for Deep-DTQ, DT, GaussianNB, and MLP.(B) ROC AUC predictive performance comparison for Deep-DTQ, DT, GaussianNB, and MLP.(C) Precision predictive performance comparison for Deep-DTQ in AD, A549, DIPG25, and U251.(D) Recall predictive performance comparison for Deep-DTQ in AD, A549, DIPG25, and U251.
Furthermore, we explored the precision and recall for Deep-DTQ in identifying prospective drug-target quartets from false drug-target quartets. As shown in Fig. 4C, Deep-DTQ achieved almost perfect precision where A549 got 1.0 ± 0 (SD), AD got 0.98 ± 0.028 (SD), DIPG25 got 1.0 ± 0 (SD) and U251 got 1.0 ± 0 (SD). More interestingly, Deep-DTQ even achieved an excellent recall score where A549 got 0.84 ± 0 (SD), AD got 0.98 ± 0.026 (SD), DIPG25 got 1.0 ± 0 (SD) and U251 got 0.78 ± 0.195 (SD). Considering Depp-DTQ achieved intriguing recall and precision, it’s promising to apply the model to discover novel drug combinations for cell lines and AD. The detailed predictive performance in this part is shown in Supplementary Table 5.
7.1. Our deep learning model further predicts high-confidence candidate drug combinations for AD
Applying our model to the drug combination pool, we identified 233,269 potential AD drug combinations out of 2,125,210 combinations (Supplemental Table 6). Within 233,269 potential combinations, there were 5 target genes, and these included AGTR1, FEN1, NADSYN1, PDHB, and PSMB2. Interestingly, these 5 target genes had previously been shown to be associated with brain impairment and AD progression via multiple forms, including mutations, RNA expression, and proteomics, suggesting these genes could be promising therapeutic targets for AD treatment [54], [55], [60], [61], [62], [63], [64]. Furthermore, we found AGTR1, FEN1, and PSMB2 were druggable genes based on genome-wide association analysis as previously reported [65]. There were 461 unique drugs in our potential drug combinations, which covered most of the potential drugs we identified before. More interestingly, 61 drugs out of these 461 drugs had been tested in AD clinical trial studies at various stages according to ClinicalTrials.gov [18], including completed, active, and recruiting. After filtering targets and drugs, we finally got 6067 high-confidence drug combinations and 227,202 potential drug combinations for AD (Supplemental Table 6). Clinical trials on these 61 drugs from ClinicalTrails.gov as of 2022 are available in Supplemental Table 6. In addition, we further applied Deep-DTQ to identify novel drug combinations for A549, DIPG25, and U251, which are available in GitHub (https://github.com/Pan-bio/drug-combination), to facilitate relevant research.
8. Discussion
In this paper, we first built a DTP network where the nodes are drug-target pairs and the edges are the associations between drug-target pairs. By integrating drug features, target features, and the relationship information between drugs and targets, we generated representative low-dimensional features for each drug-target pair. Further, we constructed a quartet network where a quartet consists of two drug-target pairs and the quartet features are characterized by combining the features of corresponding drugs and targets, and the relationships within and outside of drug-target pairs. After obtaining the features for each quartet node in the DTDT network, we developed a deep learning-based model to identify true quartets from false quartets. Based on performance evaluation, we showed the representation features learned from disease specific DTP network behaved better than original features; in addition, our model Deep-DTQ achieved a robust predictive performance compared to traditional machine learning methods. Finally, we identified multiple novel drug-target quartets that may serve as drug combination therapy for treating AD. Although our research scheme was applied to AD, it is broadly applicable to other disease or cell line contexts, such as A549, DIPG25 and U251.
According to the abovementioned published databases, we identified five unique genes that comprised synergistic target pairs discovered in AD, including AGTR1 (P30556), FEN1 (P39748), PSMB2 (P49721), PDHB (P11177), and NADSYN1 (Q6IA69). In fact, previous research reports support that these five genes are linked to cognitive impairment in human and may serve as promising therapeutic targets for AD. The abnormal expression level of AGTR1 is associated with increasing protein kinase C that can promote the accumulation of amyloid Aβ, leading to AD progression [66], [67]. In addition, AGTR1 participates in the biological process of Aβ-degrading enzyme, and bioinformatics analysis such as protein-protein interaction network analysis and functional module analysis indicate that AGTR1 plays a role in the progression of AD [68], [69], [70], [71]. As base excision repair gene, FEN1 often harbor variants associated with DNA damage caused by oxidative stress in nerve cells that contribute to AD [60], [72], [73]. In addition, abnormal methylation of FEN1, and neutrophil infiltration resulting from FEN1 are also significantly associated with AD progression [74], [75]. As a key component of proteasome encoded by PSMB2, several studies have suggested that the aberrational expression of PSMB2 is associated with AD [64], [76], [77]. Deregulated co-regulation of PSMB2 with has-miR-423–5p and RPL30 in brain regions is found to be involved in cognitive impairment and PSMB2 is used as potential preclinical biomarkers for AD identification [76], [77], [78], [79]. As a part of the PDH complex responsible for mitochondrial function, the aberrational expression of PDHB and tau is found in AD [62]. In addition, multiple independent bioinformatics analyses such as network analysis have indicated PDHB as a potential key gene related to AD pathology [62], [80], [81], [82]. As for NADSYN1, there are several pieces of evidence showing that genetic polymorphisms of NADSYN1 are associated with cognitive impairment [83], [84], [85], [86]. Aberrant protein expression of NADSYN1 is also observed in cognitive impairment patients [87]. Taken together, these above studies support the validity of the synergistic gene pairs we identified.
In order to represent the DTP information comprehensively, a large, complicated matrix must be constructed. In our case, we took into account 125 targets that directly interact with 5 core targets and 461 drugs that interact with more than 500 out of core 911 drugs, and these features amounted to tens of billions. To identify more potential drug target quartets for AD treatment, a larger adjacent matrix should be constructed after adding more potential targets and drugs, which places a high burden on computational needs.
Another limitation is that few drugs have been identified for treating AD over the past few years, and there is little known information regarding combination drugs for AD. Therefore, despite our cross-validations using independent methods, quite a few of the potential combination drugs we identified from our model cannot receive support from the literature or clinical studies. As technology advances and more data is accumulated, the situation may be improved in the future.
Funding
This work was supported by the National Institutes of Health grants R35GM133658 (to S.Y.), R01AG066749 (to X.J.) and R35GM137836 (to N.S.). S.Y. was also supported by Scialog Award# 28706 sponsored jointly by Chan Zuckerberg Initiative, Research Corporation for Science Advancement (RCSA) and the Cottrell Foundation.
CRediT authorship contribution statement
Xingxin Pan: Conceptualization, Methodology, Formal analysis, Data curation, Writing – original draft. Jun Yun: Validation, Investigation, Visualization. Zeynep H. Coban Akdemir: Data curation, Validation. Xiaoqian Jiang: Resources, Writing – review & editing, Funding acquisition. Erxi Wu: Resources, Writing – review & editing. Jason H. Huang: Resources, Project administration, Writing – review & editing. Nidhi Sahni: Conceptualization, Resources, Writing – review & editing, Supervision, Funding acquisition. S. Stephen Yi: Conceptualization, Methodology, Investigation, Resources, Writing – original draft, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
No conflicts of interest are disclosed by the authors.
Acknowledgements
The authors acknowledge the Biomedical Research Computing Facility at UT Austin, and Texas Advanced Computing Center (TACC) for assistance.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.02.004.
Contributor Information
Zeynep H. Coban Akdemir, Email: Zeynep.H.CobanAkdemir@uth.tmc.edu.
Xiaoqian Jiang, Email: Xiaoqian.Jiang@uth.tmc.edu.
Erxi Wu, Email: erxi.wu@bswhealth.org.
Jason H. Huang, Email: jason.huang@bswhealth.org.
Nidhi Sahni, Email: nsahni@mdanderson.org.
S. Stephen Yi, Email: stephen.yi@austin.utexas.edu.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary Figure 1. Evaluation of predictive efficacy for representation. (A) Predictive F1 score comparison between representation features and original features in AD. (B) Predictive ROC AUC comparison between representation features and original features in AD. (C) Predictive balanced accuracy comparison between representation features and original features in AD. (D) Predictive F1 score comparison between representation features and original features in U251. (E) Predictive ROC AUC comparison between representation features and original features in U251. (F) Predictive balanced accuracy comparison between representation features and original features in U251.
.
References
- 1.Wenk G.L. Neuropathologic changes in Alzheimer's disease. J Clin Psychiatry. 2003;64:7–10. [PubMed] [Google Scholar]
- 2.Goedert M., Spillantini M.G. A century of Alzheimer's disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 3.Association As. Alzheimer's disease facts and figures. Alzheimer's Dement. 2019;2019(15):321–387. [Google Scholar]
- 4.Karch C.M., Goate A.M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amemori T., Jendelova P., Ruzicka J., Urdzikova L.M., Sykova E. Alzheimer’s disease: mechanism and approach to cell therapy. Int J Mol Sci. 2015;16:26417–26451. doi: 10.3390/ijms161125961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinney J.W., Bemiller S.M., Murtishaw A.S., Leisgang A.M., Salazar A.M., Lamb B.T. Inflammation as a central mechanism in Alzheimer's disease. Alzheimer's Dement: Transl Res Clin Interv. 2018;4:575–590. doi: 10.1016/j.trci.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crews L., Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer's disease. Hum Mol Genet. 2010;19:R12–R20. doi: 10.1093/hmg/ddq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mucke L. Alzheimer's disease. Nature. 2009;461:895–897. doi: 10.1038/461895a. [DOI] [PubMed] [Google Scholar]
- 9.Cummings J. What can be inferred from the interruption of the semagacestat trial for treatment of Alzheimer's disease? Biol Psychiatry. 2010;68:876–878. doi: 10.1016/j.biopsych.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Becker R.E., Greig N.H. Increasing the success rate for Alzheimer's disease drug discovery and development. Expert Opin Drug Discov. 2012;7:367–370. doi: 10.1517/17460441.2012.672409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullane K., Williams M. Alzheimer's therapeutics: continued clinical failures question the validity of the amyloid hypothesis—but what lies beyond? Biochem Pharmacol. 2013;85:289–305. doi: 10.1016/j.bcp.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Castellani R.J., Perry G. Pathogenesis and disease-modifying therapy in Alzheimer's disease: the flat line of progress. Arch Med Res. 2012;43:694–698. doi: 10.1016/j.arcmed.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Eagger S.A., Levy R., Sahakian B.J. Tacrine in Alzheimer's disease. Lancet. 1991;337:989–992. doi: 10.1016/0140-6736(91)92656-m. [DOI] [PubMed] [Google Scholar]
- 14.Rogers S.L., Friedhoff L.T. The efficacy and safety of donepezil in patients with Alzheimer's disease: results of a US multicentre, randomized, double-blind, placebo-controlled trial. Dement Geriatr Cogn Disord. 1996;7:293–303. doi: 10.1159/000106895. [DOI] [PubMed] [Google Scholar]
- 15.Rösler M., Bayer T., Anand R., Cicin-Sain A., Gauthier S., Agid Y., Dal-Bianco P., Stähelin H.B., Hartman R., Gharabawi M. Efficacy and safety of rivastigmine in patients with Alzheimer's disease: international randomised controlled trialCommentary: another piece of the Alzheimer's jigsaw. Bmj. 1999;318:633–640. doi: 10.1136/bmj.318.7184.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson D., Murray Ji, Group G.R. Galantamine: a randomized, double‐blind, dose comparison in patients with Alzheimer's disease. Int J Geriatr Psychiatry. 2001;16:852–857. doi: 10.1002/gps.409. [DOI] [PubMed] [Google Scholar]
- 17.Farlow M.R., Graham S.M., Alva G. Memantine for the treatment of Alzheimer’s disease. Drug Saf. 2008;31:577–585. doi: 10.2165/00002018-200831070-00003. [DOI] [PubMed] [Google Scholar]
- 18.Zarin D.A., Tse T., Williams R.J., Califf R.M., Ide N.C. The ClinicalTrials.gov results database-update and key issues. N Engl J Med. 2011;364:852–860. doi: 10.1056/NEJMsa1012065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liggins C., Snyder H.M., Silverberg N., Petanceska S., Refolo L.M., Ryan L., Carrillo M.C. International Alzheimer's disease research portfolio (IADRP) aims to capture global Alzheimer's disease research funding. Alzheimer's Dement. 2014;10:405–408. doi: 10.1016/j.jalz.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Cummings J., Lee G., Nahed P., Kambar M.E.N., Zhong K., Fonseca J., Taghva K. Alzheimer's disease drug development pipeline: 2022. Alzheimer's Dement: Transl Res Clin Interv. 2022;8 doi: 10.1002/trc2.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabner B.A., Roberts T.G. Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 22.Al-Lazikani B., Banerji U., Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 23.Hammer S.M., Saag M.S., Schechter M., Montaner J.S., Schooley R.T., Jacobsen D.M., Thompson M.A., Carpenter C.C., Fischl M.A., Gazzard B.G. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society–USA panel. Jama. 2006;296:827–843. doi: 10.1001/jama.296.7.827. [DOI] [PubMed] [Google Scholar]
- 24.Stephenson D., Perry D., Bens C., Bain L.J., Berry D., Krams M., Sperling R., Dilts D., Luthman J., Hanna D. Charting a path toward combination therapy for Alzheimer’s disease. Expert Rev Neurother. 2015;15:107–113. doi: 10.1586/14737175.2015.995168. [DOI] [PubMed] [Google Scholar]
- 25.Pan X., Huang L.F. Multi-omics to characterize the functional relationships of R-loops with epigenetic modifications, RNAPII transcription and gene expression. Brief Bioinforma. 2022:23. doi: 10.1093/bib/bbac238. bbac238. [DOI] [PubMed] [Google Scholar]
- 26.Pan X., Liu B., Wen X., Liu Y., Zhang X., Li S., Li S. Vol. 10. 2019. D-GPM: a deep learning method for gene promoter methylation inference; p. 807. (Genes). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B., Liu Y., Pan X., Li M., Yang S., Li S.C. DNA methylation markers for pan-cancer prediction by deep learning. Genes. 2019;10:778. doi: 10.3390/genes10100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao F., Wang W., Tan M., Zhu L., Zhang Y., Fessler E., Vermeulen L., Wang X. DeepCC: a novel deep learning-based framework for cancer molecular subtype classification. Oncogenesis. 2019;8:1–12. doi: 10.1038/s41389-019-0157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X., Burgman B., Wu E., Huang J.H., Sahni N., Yi S.S. i-Modern: Integrated multi-omics network model identifies potential therapeutic targets in glioma by deep learning with interpretability. Comput Struct Biotechnol J. 2022;20:3511–3521. doi: 10.1016/j.csbj.2022.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao T., Hu Y., Valsdottir L.R., Zang T., Peng J. Identifying drug–target interactions based on graph convolutional network and deep neural network. Brief Bioinforma. 2021;22:2141–2150. doi: 10.1093/bib/bbaa044. [DOI] [PubMed] [Google Scholar]
- 31.Torng W., Altman R.B. Graph convolutional neural networks for predicting drug-target interactions. J Chem Inf Model. 2019;59:4131–4149. doi: 10.1021/acs.jcim.9b00628. [DOI] [PubMed] [Google Scholar]
- 32.Jiang X., Missel R., Toloubidokhti M., Li Z., Gharbia O., Sapp J.L., Wang L. Springer; 2021. Label-free physics-informed image sequence reconstruction with disentangled spatial-temporal modeling; pp. 361–371. (International Conference on Medical Image Computing and Computer-Assisted Intervention). [Google Scholar]
- 33.Lin X., Quan Z., Wang Z.-J., Ma T., Zeng X. KGNN: knowledge graph neural network for drug-drug interaction prediction. IJCAI. 2020:2739–2745. [Google Scholar]
- 34.Defferrard M., Bresson X., Vandergheynst P. Convolutional neural networks on graphs with fast localized spectral filtering. Adv Neural Inf Process Syst. 2016;29:3844–3852. [Google Scholar]
- 35.Hinton G., Deng L., Yu D., Dahl G.E., Mohamed A.-R., Jaitly N., Senior A., Vanhoucke V., Nguyen P. Sainath TN: deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Process Mag. 2012;29:82–97. [Google Scholar]
- 36.Hu Y., Chen C.-h, Ding Y.-y, Wen X., Wang B., Gao L., Tan K. Optimal control nodes in disease-perturbed networks as targets for combination therapy. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-10215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nativio R., Lan Y., Donahue G., Sidoli S., Berson A., Srinivasan A.R., Shcherbakova O., Amlie-Wolf A., Nie J., Cui X. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat Genet. 2020;52:1024–1035. doi: 10.1038/s41588-020-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuno Y., Abolhassani N., Mazzei G., Sakumi K., Saito T., Saido T.C., Ninomiya T., Iwaki T., Yamasaki R., Kira J.-i. MUTYH actively contributes to microglial activation and impaired neurogenesis in the pathogenesis of Alzheimer’s disease. Oxid Med Cell Longev. 2021:2021. doi: 10.1155/2021/8635088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J.S., Lee J., Jung E.S., Kim M.-H., Kim I.B., Son H., Kim S., Kim S., Park Y.M., Mook-Jung I. Brain somatic mutations observed in Alzheimer’s disease associated with aging and dysregulation of tau phosphorylation. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M Veselinovic A., B Veselinovic J., V Zivkovic J., M Nikolic G. Application of SMILES notation based optimal descriptors in drug discovery and design. Curr Top Med Chem. 2015;15:1768–1779. doi: 10.2174/1568026615666150506151533. [DOI] [PubMed] [Google Scholar]
- 45.Öztürk H., Ozkirimli E., Özgür A. A comparative study of SMILES-based compound similarity functions for drug-target interaction prediction. BMC Bioinforma. 2016;17:1–11. doi: 10.1186/s12859-016-0977-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z.-C., Huang M.-H., Zhong W.-Q., Liu Z.-Q., Xie Y., Dai Z., Zou X.-Y. Identification of drug–target interaction from interactome network with ‘guilt-by-association’principle and topology features. Bioinformatics. 2016;32:1057–1064. doi: 10.1093/bioinformatics/btv695. [DOI] [PubMed] [Google Scholar]
- 47.Li F.-M., Wang X.-Q. Identifying anticancer peptides by using improved hybrid compositions. Sci Rep. 2016;6:1–6. doi: 10.1038/srep33910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pánek J., Eidhammer I., Aasland R. A new method for identification of protein (sub) families in a set of proteins based on hydropathy distribution in proteins. Protein: Struct Funct Bioinforma. 2005;58:923–934. doi: 10.1002/prot.20356. [DOI] [PubMed] [Google Scholar]
- 49.Kipf T.N., Welling M.: Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:160902907 2016.
- 50.Wu Y., He K.: Group normalization. In Proceedings of the European conference on computer vision (ECCV). 2018: 3–19.
- 51.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 52.Zeiler M.D. Adadelta: an adaptive learning rate method. arXiv Prepr arXiv. 2012;12125701 [Google Scholar]
- 53.Snoek J., Larochelle H., Adams R.P. Practical bayesian optimization of machine learning algorithms. Adv Neural Inf Process Syst. 2012:25. [Google Scholar]
- 54.Forrester S.J., Booz G.W., Sigmund C.D., Coffman T.M., Kawai T., Rizzo V., Scalia R., Eguchi S. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol Rev. 2018;98:1627–1738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labandeira-Garcia J.L., Rodríguez-Perez A.I., Garrido-Gil P., Rodriguez-Pallares J., Lanciego J.L., Guerra M.J. Brain renin-angiotensin system and microglial polarization: implications for aging and neurodegeneration. Front Aging Neurosci. 2017;9:129. doi: 10.3389/fnagi.2017.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q., Ma C., Gearing M., Wang P.G., Chin L.-S., Li L. Integrated proteomics and network analysis identifies protein hubs and network alterations in Alzheimer’s disease. Acta Neuropathol Commun. 2018;6:1–19. doi: 10.1186/s40478-018-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alanis-Lobato G., Andrade-Navarro M.A., Schaefer M.H. HIPPIE v2. 0: enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw985. gkw985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H., Zhang W., Zou B., Wang J., Deng Y., Deng L. DrugCombDB: a comprehensive database of drug combinations toward the discovery of combinatorial therapy. Nucleic Acids Res. 2020;48:D871–D881. doi: 10.1093/nar/gkz1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwiatkowski D., Czarny P., Toma M., Korycinska A., Sowinska K., Galecki P., Bachurska A., Bielecka-Kowalska A., Szemraj J., Maes M. Association between single-nucleotide polymorphisms of the hOGG1, NEIL1, APEX1, FEN1, LIG1, and LIG3 genes and Alzheimer's disease risk. Neuropsychobiology. 2016;73:98–107. doi: 10.1159/000444643. [DOI] [PubMed] [Google Scholar]
- 61.Alaylıoğlu M., Dursun E., Genç G., Şengül B., Bilgiç B., Gündüz A., Apaydın H., Kızıltan G., Gürvit H., Hanağası H. Genetic variants of vitamin D metabolism-related DHCR7/NADSYN1 locus and CYP2R1 gene are associated with clinical features of Parkinson’s disease. Int J Neurosci. 2020:1–11. doi: 10.1080/00207454.2020.1820502. [DOI] [PubMed] [Google Scholar]
- 62.Petyuk V.A., Chang R., Ramirez-Restrepo M., Beckmann N.D., Henrion M.Y., Piehowski P.D., Zhu K., Wang S., Clarke J., Huentelman M.J. The human brainome: network analysis identifies HSPA2 as a novel Alzheimer’s disease target. Brain. 2018;141:2721–2739. doi: 10.1093/brain/awy215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng Y.-S., Tang C.-W., Peng Y.-Y., Chang H., Chen C.-L., Guo S.-L., Wu L.-C., Huang M.-C., Lee H.-C. Comparative functional genomic analysis of Alzheimer’s affected and naturally aging brains. PeerJ. 2020;8 doi: 10.7717/peerj.8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puthiyedth N., Riveros C., Berretta R., Moscato P. Identification of differentially expressed genes through integrated study of Alzheimer’s disease affected brain regions. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finan C., Gaulton A., Kruger F.A., Lumbers R.T., Shah T., Engmann J., Galver L., Kelley R., Karlsson A., Santos R. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag1166. eaag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pang C., Yang H., Hu B., Wang S., Chen M., Cohen D.S., Chen H.S., Jarrell J.T., Carpenter K.A., Rosin E.R. Identification and analysis of Alzheimer’s candidate genes by an amplitude deviation algorithm. J Alzheimer'S Dis Park. 2019:9. doi: 10.4172/2161-0460.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saavedra J.M. Evidence to consider Angiotensin II receptor blockers for the treatment of early Alzheimer’s disease. Cell Mol Neurobiol. 2016;36:259–279. doi: 10.1007/s10571-015-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quitterer U., AbdAlla S. Improvements of symptoms of Alzheimers disease by inhibition of the angiotensin system. Pharmacol Res. 2020;154 doi: 10.1016/j.phrs.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 69.Hemming M.L., Selkoe D.J. Amyloid β-protein is degraded by cellular angiotensin-converting enzyme (ACE) and elevated by an ACE inhibitor. J Biol Chem. 2005;280:37644–37650. doi: 10.1074/jbc.M508460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vijh D., Imam M.A., Haque M.M.U., Das S., Islam A., Malik M.Z.: Network pharmacology and bioinformatics approach reveals the therapeutic activity and action mechanism of curcumin in Alzheimer disease. 2022. [DOI] [PubMed]
- 71.Zhang B., Gaiteri C., Bodea L.-G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwiatkowski D., Czarny P., Toma M., Jurkowska N., Sliwinska A., Drzewoski J., Bachurska A., Szemraj J., Maes M., Berk M. Associations between DNA damage, DNA base excision repair gene variability and Alzheimer's disease risk. Dement Geriatr Cogn Disord. 2016;41:152–171. doi: 10.1159/000443953. [DOI] [PubMed] [Google Scholar]
- 73.Wang H., Lautrup S., Caponio D., Zhang J., Fang E.F. DNA damage-induced neurodegeneration in accelerated ageing and Alzheimer’s disease. Int J Mol Sci. 2021;22:6748. doi: 10.3390/ijms22136748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coppedè F., Tannorella P., Stoccoro A., Chico L., Siciliano G., Bonuccelli U., Migliore L. Methylation analysis of DNA repair genes in Alzheimer’s disease. Mech Ageing Dev. 2017;161:105–111. doi: 10.1016/j.mad.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 75.Yin P., Xue Y., Wang T., Zhong D., Li G. The therapeutic targets of fingolimod (FTY720) are involved in pathological processes in the frontal cortex of Alzheimer's disease patients: a network pharmacology study. Front Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.609679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J., Gutala R., Hwang Y., Kim J.M., Konu O., Ma J., Li M. Strain‐and region‐specific gene expression profiles in mouse brain in response to chronic nicotine treatment. Genes Brain Behav. 2008;7:78–87. doi: 10.1111/j.1601-183X.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- 77.Orre M., Kamphuis W., Dooves S., Kooijman L., Chan E.T., Kirk C.J., Dimayuga Smith V., Koot S., Mamber C., Jansen A.H. Reactive glia show increased immunoproteasome activity in Alzheimer’s disease. Brain. 2013;136:1415–1431. doi: 10.1093/brain/awt083. [DOI] [PubMed] [Google Scholar]
- 78.Nair V.D., Ge Y. Alterations of miRNAs reveal a dysregulated molecular regulatory network in Parkinson’s disease striatum. Neurosci Lett. 2016;629:99–104. doi: 10.1016/j.neulet.2016.06.061. [DOI] [PubMed] [Google Scholar]
- 79.Kaur G., Poljak A., Sachdev P.: Deep proteome analysis of plasma reveals novel biomarkers of mild cognitive impairment and Alzheimer’s disease: A longitudinal study. bioRxiv 2022.
- 80.Drummond E., Pires G., MacMurray C., Askenazi M., Nayak S., Bourdon M., Safar J., Ueberheide B., Wisniewski T. Phosphorylated tau interactome in the human Alzheimer’s disease brain. Brain. 2020;143:2803–2817. doi: 10.1093/brain/awaa223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monaco A., Amoroso N., Bellantuono L., Lella E., Lombardi A., Monda A., Tateo A., Bellotti R., Tangaro S. Shannon entropy approach reveals relevant genes in Alzheimer’s disease. PloS One. 2019;14 doi: 10.1371/journal.pone.0226190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu W., Yang Y., Jin Y., Song C., Ye X., Zhu W.: Identification of Key Genes and Pathways Asscociated with Alzheimer's Disease Based on Bioinformatics Databases. Available at SSRN 3996123.
- 83.Xie S., Jiang M., Liu H., Xue F., Chen X., Zhu X. Association of Vitamin D anabolism-related gene polymorphisms and susceptibility to Uterine Leiomyomas. Front Genet. 2022;13 doi: 10.3389/fgene.2022.844684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu X., Wu P., Shen L., Jiao B., Liao X., Wang H., Peng J., Lin Z. DHCR7 rs12785878 T>C polymorphism is associated with an increased risk of early onset of Alzheimer's disease in chinese population. Front Genet. 2021;12 doi: 10.3389/fgene.2021.583695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jorde R., Mathiesen E.B., Rogne S., Wilsgaard T., Kjærgaard M., Grimnes G., Schirmer H. Vitamin D and cognitive function: the Tromsø study. J Neurol Sci. 2015;355:155–161. doi: 10.1016/j.jns.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 86.Alaylıoğlu M., Dursun E., Genc G., Şengül B., Bilgiç B., Gündüz A., Apaydın H., Kızıltan G., Gürvit H., Hanağası H. Genetic variants of vitamin D metabolism-related DHCR7/NADSYN1 locus and CYP2R1 gene are associated with clinical features of Parkinson’s disease. Int J Neurosci. 2022;132:439–449. doi: 10.1080/00207454.2020.1820502. [DOI] [PubMed] [Google Scholar]
- 87.Quiroz-Baez R., Hernández-Ortega K., Martínez-Martínez E. Insights into the proteomic profiling of extracellular vesicles for the identification of early biomarkers of neurodegeneration. Front Neurol. 2020;11 doi: 10.3389/fneur.2020.580030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary Figure 1. Evaluation of predictive efficacy for representation. (A) Predictive F1 score comparison between representation features and original features in AD. (B) Predictive ROC AUC comparison between representation features and original features in AD. (C) Predictive balanced accuracy comparison between representation features and original features in AD. (D) Predictive F1 score comparison between representation features and original features in U251. (E) Predictive ROC AUC comparison between representation features and original features in U251. (F) Predictive balanced accuracy comparison between representation features and original features in U251.