Abstract
Esophageal variceal (EV) bleeding is a severe medical emergency related to cirrhosis. Early identification of cirrhotic patients with at a high risk of EV bleeding is key to improving outcomes and optimizing medical resources. This study aimed to evaluate the feasibility of automated multimodal machine learning (MMML) for predicting EV bleeding by integrating endoscopic images and clinical structured data. This study mainly includes three steps: step 1, developing deep learning (DL) models using EV images by 12-month bleeding on TensorFlow (backbones include ResNet, Xception, EfficientNet, ViT and ConvMixer); step 2, training and internally validating MMML models integrating clinical structured data and DL model outputs to predict 12-month EV bleeding on an H2O-automated machine learning platform (algorithms include DL, XGBoost, GLM, GBM, RF, and stacking); and step 3, externally testing MMML models. Furthermore, existing clinical indices, e.g., the MELD score, Child‒Pugh score, APRI, and FIB-4, were also examined. Five DL models were transfer learning to the binary classification of EV endoscopic images at admission based on the occurrence or absence of bleeding events during the 12-month follow-up. An EfficientNet model achieved the highest accuracy of 0.868 in the validation set. Then, a series of MMML models, integrating clinical structured data and the output of the EfficientNet model, were automatedly trained to predict 12-month EV bleeding. A stacking model showed the highest accuracy (0.932), sensitivity (0.952), and F1-score (0.879) in the test dataset, which was also better than the existing indices. This study is the first to evaluate the feasibility of automated MMML in predicting 12-month EV bleeding based on endoscopic images and clinical variables.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-022-00724-6.
Keywords: Automated machine learning (AutoML), Multimodal machine learning (MMML), Computer vision (CV), Deep learning (DL), Esophageal variceal (EV) bleeding
Introduction
Portal hypertension is one of the most severe complications of cirrhosis [1]. It usually leads to several clinical symptoms, e.g., esophageal varices (EVs), spontaneous bacterial peritonitis, ascites, and hepatic encephalopathy. EVs are the most common type of gastroesophageal varices, with a prevalence of up to 85% in patients with decompensated cirrhosis [2]. EV bleeding is a severe medical emergency, with a 15% 6-week mortality rate in patients with poor liver conditions [3, 4]. In many cases, mortality does not occur due to bleeding but due to infections, hepatic encephalopathy, and liver failure [2]. Risk estimation is key to the clinical management of EV bleeding, which could largely lower the mortality rate.
In the past decade, there has been the remarkable progression in computer vision (CV)-based image analysis, owing to the significant development of deep learning (DL) algorithms. Medical images can be analyzed by CV models to help clinical practitioners make decisions more quickly and accurately [5]. Convolutional neural networks (CNNs) are a class of artificial neural networks based on the shared-weight architecture of sliding convolution kernels, most commonly applied to CV tasks [6]. Since 2020, transformer architecture has emerged as a competitive alternative to CNNs and has been increasingly applied in various CV tasks [7].
Gastroenterology is an early leader in bridging the gap between artificial intelligence and clinical practice [8, 9]. In endoscopy, a series of studies reported the application of CV models for endoscopy; the diagnosis of Helicobacter pylori [10] and gastric cancer [11] in upper endoscopy; and the automatic detection of lesions in capsule endoscopy [12].
In recent years, machine learning (ML) algorithms, as an alternative to conventional statistical methods, have shown promise in the field of clinical data analysis [13]. One of the outstanding advantages of the ML algorithm is to process complex relationships in big data [14]. Even though the ML algorithm has presented remarkable performance, the development of the models requires rich experience in the programming and knowledge of ML. Thus, it is challenging for clinical practitioners to adopt ML in their research. Currently, automated machine learning (AutoML) shows promise in closing the gap between ML and clinical researchers [15]. It could assist physicians in automating the procedure of model development [16]. Multimodal fusion is one of the vibrant fields of artificial intelligence [17]. It takes advantage of the complementarity of heterogeneous data and offers reliable classification. Multimodal machine learning (MMML) aims to develop models that can process and integrate features from multiple modalities [18]. It is now an emerging multidisciplinary research field. No previous study has reported the application of multimodal fusion in the prevention of EV bleeding.
Although a series of clinical studies concerning high-risk varices have been reported since the late 1980s [19, 20], the prediction of EV bleeding requires the long experience of endoscopists and is still less than stable [2, 3]. In this multicenter study, for the first time, we aimed to evaluate the feasibility of MMML models in 12-month EV bleeding prediction, integrating endoscopic images and clinical structured data.
Methods
Study Design
This was a retrospective cohort study. Hospitalized patients with cirrhosis were recruited from two hospitals, center #1: Jintan Affiliated Hospital of Jiangsu University, and center #2: The First Affiliated Hospital of Soochow University, between 2015 and 2021. This study was approved by the local Institutional Review Boards (approval number 2022098) and conducted in accordance with the Helsinki Declaration of 1975 as revised in 2013. All participants signed statements of informed consent before inclusion.
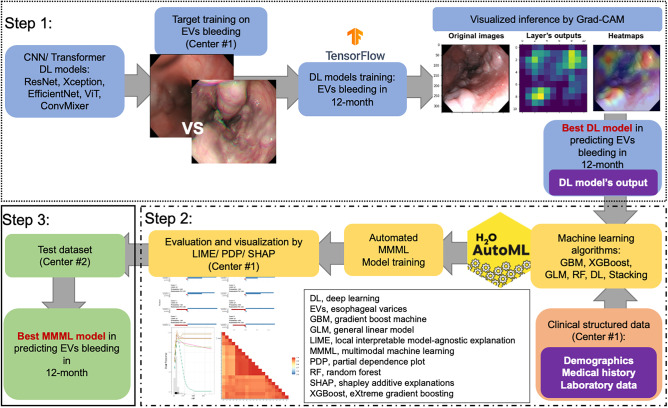
As shown in Fig. 1, our study mainly includes three steps: step 1, developing DL models on EV images (from center #1 Jintan Hospital) by 12-month EV bleeding; step 2, training and internally validating MMML models, integrating clinical structured data and the outputs of unweighted probabilities by deep learning models (from center #1 Jintan Hospital) to predict 12-month EV bleeding; step 3, externally testing the MMML models (at center #2 Soochow University).
Fig. 1.
Flowchart of the study. Step 1: the development of DL-based CV models on esophageal variceal images (from center #1 Jintan Hospital) by 12-month EV bleeding; Step 2: training and internally validating MMML models, integrating clinical structured data and outputs by deep learning models (from center #1 Jintan Hospital), to predict 12-month EV bleeding; Step 3: externally test the MMML models (at Center #2 Soochow University). AutoML, automated machine learning; CV, computer vision; DL, deep learning; GBM, gradient boost machine; GLM, general linear model; MMML, multimodal machine learning; LIME, local interpretable model-agnostic explanation; PDP, partial dependence plot; RF, random forest; SHAP, SHapley additive explanations; XGBoost, eXtreme gradient boosting
Step 1: The Development of DL Models
Model Architectures
CNN-Based Architectures
Input layer: each image is normalized as 331 × 331 pixels, padded if necessary and then loaded into the pretrained CNN layers. Pretrained CNN layers include convolutional layers, average pooling layers, and fully connected layers (i.e., dense layer) with ReLU activation. Additional layers: subsequent to the pretrained CNN layers for feature extraction, four dense layers with ReLU activation and one dense layer with sigmoid activation replaced the original fully connected layers, which functioned as the classifier.
Transformer-Based Architectures
The transformer is characterized by synchronous input based on the self-attention mechanism. Vision transformer (ViT) architectures with the encoder and decoder parts of the transformer. The transformer encoder consists of three main components: input embedding, multihead attention, and feed-forward neural networks. Similar to the CNN-based architectures, following the transformer-based architectures, four dense layers with ReLU activation and one dense layer with sigmoid activation were added on the top, converting the extracted feature into the predictive probability.
Implementation
The CNN- or transformer-based models were transfer learning via Keras (TensorFlow framework as backbone). The Adam optimizer and the binary cross-entropy cost function, with a fixed learning rate of 0.0001 and a batch size of 32, were compiled in the model fitting. The code of computer vision models’ training is available to access: https://osf.io/ycxwr/?view_only=81b4f590605c472f9e979c854a573cce.
The Pretraining
The five CNN- or transformer-based models, including ResNet-50V2, Xception, EfficientNet-V2s, ViT-B16, and ConvMixer-768/32, were chosen. These DL models were previously trained on the ImageNet database (www.image-net.org). The pretrained models and parameters were obtained from Keras or TensorFlow Hub (https://hub.tensorflow.google.cn/). Given the limited number of esophageal variceal images (target training), a second pretraining on endoscopic images was performed. HyperKvasir is currently the largest image and video dataset of gastrointestinal endoscopy (https://datasets.simula.no/hyper-kvasir/) [21]. The data collect gastro- and colonoscopy images from Bærum Hospital in Norway and are partly labeled. We chose a total of 4000 cardia endoscopic images (esophagitis vs. normal cardia) for pretraining. In addition, one thousand cardia images from Jintan Hospital were also obtained for pretraining.
Target Training of DL Models
The esophageal variceal images were obtained from Jintan Hospital and saved in JPEG format. All images were rescaled to 331 × 331 pixels, and then the pixel values were normalized from 0–255 to 0–1. Based on the occurrence of bleeding 12 months after admission, the images were divided into two classes: control (no bleeding) vs. bleeding. The random split-sample method was used to divide the images into the training and validation datasets (7:3), comprising 571 images (308 control vs. 263 bleeding) and 239 images (112 control vs. 127 bleeding), individually. Via image augmentation, the number of images increased to 2000 images in the training dataset (1000 control vs. 1000 bleeding) and 400 images in the validation dataset (200 control vs. 200 bleeding). Detailed information on image augmentation is offered in Supplementary Fig. 1. Transfer learning was applied based on the aforementioned CNN- or transformer-based architectures by combining the existing feature extraction layers (frozen) with additional activation layers (training) for the learning of the target classification.
Visualization of DL Models
The visualization of the models was performed using Gradient-weighted Class Activation Mapping (Grad-CAM) [22]. Based on the outputs of the best binary-classification model, Grad-CAM technology was used to provide an inferential explanation by plotting heatmaps.
Step 2: The Development of the MMML Models
Criteria of Enrollment
In the two centers, hospitalized patients with cirrhosis were enrolled. The diagnosis of cirrhosis was made according to a combination of clinical, biochemical, and imaging signs or liver histology. Subjects were excluded if they (1) had prior EV bleeding, surveillance or treatment; (2) had noncirrhotic etiologies for portal hypertension; (3) had an episode of acute liver injury within 6 months due to drug-induced liver injury, acute alcoholic hepatitis, or infections; (4) were on dialysis; (5) had malignancy; or (6) were pregnant. Treatments for EVs included nonselective beta blockers, endoscopic ligation, sclerotherapy, banding, transjugular intrahepatic portosystemic shunt (TIPS), or laparoscopic splenectomy (LS) [23]. Alcohol consumption was defined according to the WHO criteria for moderate and heavy users: daily intake of 2–3 drinks and ≥ 4 drinks (12.5 g of ethanol/drink), respectively [24].
Clinical Data and Outcome
At admission, anthropometric, routine blood and biochemical tests were performed, as previously described, by researchers who were blinded to the study design [25]. The participants were followed for 12 months to determine EV rebleeding via phone calls and/or outpatient visits. During the follow-up, the patients did not receive the following interventions: endoscopic ligation, sclerotherapy, banding, TIPS, or LS.
H2O AutoML
H2O’s AutoML can be applied for automating the ML workflow, including automated training and tuning of a variety of models (https://www.h2o.ai). In addition, the platform offers various explainable methods for models and variables. H2O AutoML supports six common algorithms: DL, eXtreme gradient boosting (XGBoost), general linear regression (GLM), gradient boost machine (GBM), random forest (RF), and stacking.
Step 3: Evaluation of Models in the Test Dataset
The test dataset was collected from Soochow University. The criteria for enrollment were executed as Jintan Hospital.
Statistical Analysis
We fitted CV models using Python software (version: 3.9) and TensorFlow (2.8.0). Statistical analysis was performed using R version 4.0.4 (R Foundation for Statistical Computing). AutoML modeling was performed via H2O (cluster version: 3.36.0.2). Model performance was evaluated by accuracy, recall, precision, F1-score, sensitivity, specificity, and area under the receiver operating characteristic (ROC) curve (AUC) (pROC package 1.18.0). AutoML models were visualized with partial dependence plots (PDPs) and local interpretable model-agnostic explanation (LIME) (lime package 0.5.2). A two-sided p < 0.05 was considered statistically significant.
Results
The Development of the CV Models
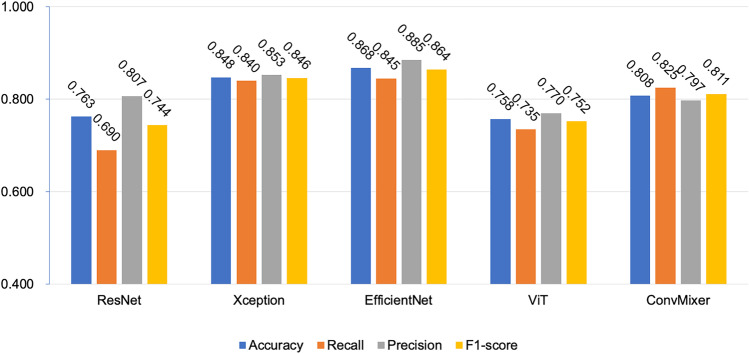
Eight hundred and ten endoscopic images of the EVs were obtained. After image augmentation, the number increased to 2400 (2000 in the training dataset and 400 in the validation dataset). After the two pretraining times, the five CNN- or transformer-based models were transfer learning on the EV endoscopic images at admission to the binary classification of 12-month bleeding. The performance of the five CV models in the validation dataset is shown in Fig. 2.
Fig. 2.
The performance of DL-based CV models in the validation dataset
EfficientNet achieved the highest accuracy of 0.868, followed by Xception (0.848) and ConvMixer (0.808) in the validation set. The recall, specificity, and F1-score of EfficientNet were 0.845, 0.885, and 0.864, respectively, which were significantly higher than the others.
The Heatmaps by Grad-CAM
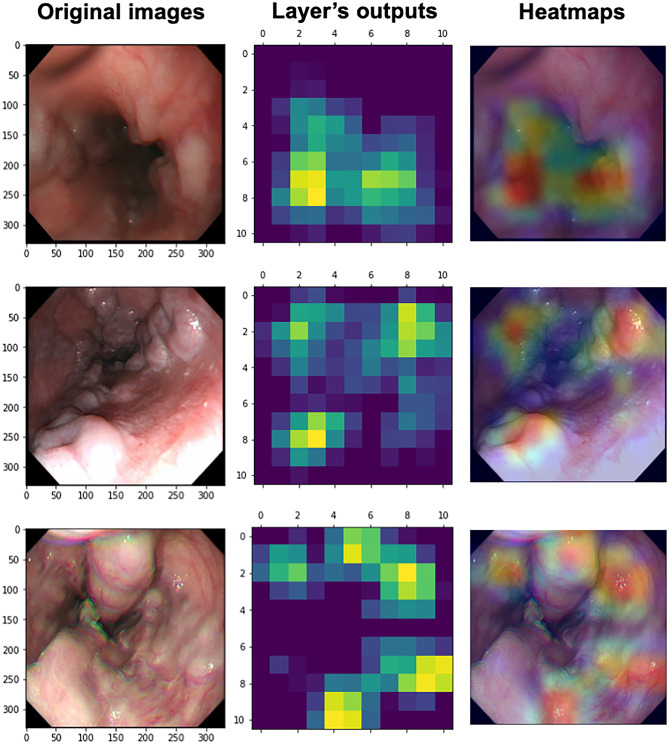
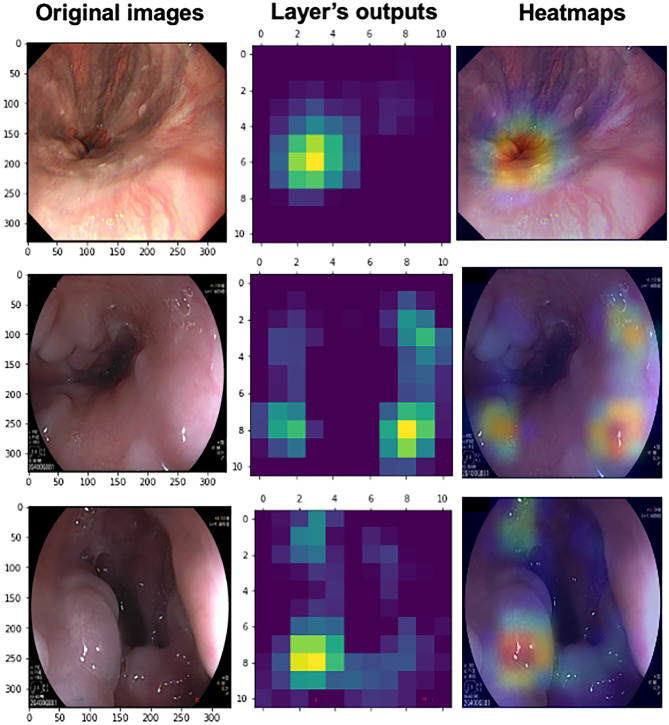
In Fig. 3, the Grad-CAM heatmaps were plotted and highlighted the potential regions of EV bleeding on the original images, inferred by the best binary-classification CV model, i.e., the EfficientNet model. Moreover, we also chose three typical incorrect cases in Fig. 4. The reasons for the misclassification included the confusion of varices and cardia, the underinflation, and the reflection of endoscopic light and bubbles.
Fig. 3.
Visualization of the EfficientNet model’s inference by Grad-CAM. The left column presents the original endoscopic images. The middle column shows the heatmaps based on the output of the feature extractor’s last layer of the EfficientNet model. The right column shows the Grad-CAM heatmaps covering the original images, which highlight the EV regions inferred by the EfficientNet model
Fig. 4.
Visualization of the misclassified images by Grad-CAM. The left column presents the original endoscopic images. The middle column shows the heatmaps based on the output of the feature extractor’s last layer of the EfficientNet model. The right column shows the Grad-CAM heatmaps covering the original images, which highlight the EV regions inferred by the EfficientNet model. The sample of the 1st row was misclassified due to the confusion of varices and cardia. The 2nd row shows the reflection of endoscopic light and bubbles. The 3rd row was not inflated enough
The Characteristics of Patients by Dataset
The study enrolled a total of 341 patients with cirrhosis from Jintan Hospital. The characteristics of the participants are listed in Table 1. The clinical indices, e.g., MELD score, Child‒Pugh score, APRI, and FIB-4, were also calculated.
Table 1.
Characteristics of the participants by dataset
| Train dataset | Validation dataset | Test dataset | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | Bleed | p | Control | Bleed | p | Control | Bleed | p | |
| Number | 205 | 70 | 57 | 9 | 119 | 42 | |||
| Male (%) | 125 (61.0) | 40 (57.1) | 0.672 | 32 (56.1) | 7 (77.8) | 0.389 | 78 (65.5) | 17 (40.5) | 0.008 |
| Age (years) | 59.99 (14.06) | 61.77 (13.29) | 0.354 | 59.68 (12.12) | 58.00 (13.38) | 0.704 | 59.90 (13.47) | 64.45 (13.44) | 0.061 |
| Etiology of Cirrhosis, n (%) | 0.230 | 0.999 | 0.986 | ||||||
| HBV | 177 (86.3) | 61 (87.1) | 52 (91.2) | 9 (100.0) | 97 (81.5) | 35 (83.3) | |||
| HCV | 5 (2.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (4.2) | 2 (4.8) | |||
| Others | 23 (11.2) | 9 (12.9) | 5 (8.8) | 0 (0.0) | 5 (4.2) | 2 (4.8) | |||
| Ascites (%) | 62 (30.2) | 22 (31.4) | 0.972 | 16 (28.1) | 4 (44.4) | 0.546 | 26 (21.8) | 12 (28.6) | 0.502 |
| Hepatic encephalopathy (%) | 62 (30.2) | 22 (31.4) | 0.280 | 2 (3.5) | 2 (22.2) | 0.151 | 19 (16.0) | 8 (19.0) | 0.826 |
| Alcohol consumption | 11 (5.4) | 25 (35.7) | < 0.001 | 5 (8.8) | 3 (33.3) | 0.121 | 8 (6.7) | 10 (23.8) | 0.006 |
| Body mass index (kg/m2) | 22.71 (3.45) | 23.27 (3.75) | 0.252 | 22.89 (3.27) | 24.12 (3.00) | 0.295 | 23.15 (3.12) | 22.92 (3.11) | 0.676 |
| Child–Pugh score | 7.00 [6.00, 9.00] | 8.00 [7.00, 10.00] | < 0.001 | 7.00 [6.00, 8.00] | 9.00 [8.00, 10.00] | 0.002 | 7.00 [6.00, 9.00] | 9.00 [8.00, 9.00] | < 0.001 |
| PLT (109/L) | 77.00 [51.00, 127.00] | 59.42 [28.93, 103.73] | 0.005 | 102.00 [48.00, 160.00] | 60.55 [43.68, 98.79] | 0.188 | 77.00 [54.50, 120.00] | 54.91 [26.28, 108.04] | 0.015 |
| TBIL (μmol/L) | 1.96 [1.12, 3.92] | 1.82 [1.15, 3.07] | 0.325 | 1.88 [1.16, 2.94] | 5.39 [2.37, 5.69] | 0.081 | 1.81 [1.23, 3.96] | 1.95 [1.40, 3.36] | 0.671 |
| Creatinine (mg/dl) | 0.75 [0.60, 0.96] | 0.77 [0.61, 1.01] | 0.593 | 0.69 [0.60, 0.89] | 0.79 [0.59, 0.99] | 0.472 | 0.74 [0.58, 0.87] | 0.66 [0.55, 0.85] | 0.405 |
| ALT (U/l) | 21.00 [13.58, 36.47] | 45.89 [41.80, 59.22] | < 0.001 | 23.17 [14.42, 31.71] | 47.92 [44.00, 87.19] | < 0.001 | 23.94 [15.15, 46.30] | 49.22 [41.20, 81.76] | < 0.001 |
| AST (U/l) | 30.87 [21.70, 51.03] | 54.85 [46.50, 71.09] | < 0.001 | 28.98 [20.79, 48.30] | 63.04 [47.57, 96.57] | 0.002 | 29.89 [23.45, 57.78] | 64.54 [52.92, 82.27] | < 0.001 |
| Albumin (g/l) | 29.80 [25.90, 33.80] | 27.35 [23.02, 31.70] | 0.005 | 29.70 [26.30, 34.60] | 30.10 [25.80, 38.50] | 0.709 | 29.80 [25.30, 33.70] | 27.00 [22.88, 29.35] | 0.001 |
| HDL-C (mg/dl) | 37.50 [27.09, 44.51] | 35.57 [27.41, 40.90] | 0.250 | 38.46 [33.28, 42.38] | 24.32 [21.22, 39.10] | 0.071 | 35.99 [24.92, 43.65] | 33.87 [25.23, 38.52] | 0.185 |
| PT (s) | 14.50 [13.00, 17.50] | 17.96 [15.50, 19.72] | < 0.001 | 14.30 [13.10, 15.80] | 17.82 [16.60, 20.38] | < 0.001 | 15.00 [13.50, 17.15] | 17.52 [16.28, 19.78] | < 0.001 |
| INR | 1.02 [0.87, 1.29] | 1.27 [1.10, 1.43] | < 0.001 | 0.96 [0.88, 1.10] | 1.25 [1.24, 1.50] | < 0.001 | 1.05 [0.91, 1.23] | 1.25 [1.10, 1.41] | < 0.001 |
| MELD score | 9.16 [5.16, 14.27] | 13.87 [10.19, 19.53] | < 0.001 | 7.09 [4.11, 10.17] | 20.09 [12.58, 23.01] | 0.001 | 9.76 [5.83, 13.13] | 13.81 [8.80, 17.52] | 0.001 |
| FIB-4 | 5.70 [3.18, 9.23] | 8.17 [4.82, 14.74] | < 0.001 | 4.76 [2.78, 7.79] | 11.57 [4.99, 13.32] | 0.051 | 5.64 [3.29, 9.11] | 9.44 [5.64, 22.67] | < 0.001 |
| APRI | 0.89 [0.46, 1.62] | 1.94 [1.10, 3.76] | < 0.001 | 0.88 [0.41, 1.32] | 2.18 [1.40, 5.53] | 0.005 | 1.00 [0.47, 1.93] | 2.51 [1.40, 5.49] | < 0.001 |
Classificational variables were presented as number (percentage). Normally distributed variables were presented as mean (standard deviation); variables with a skewed distribution were presented as median value (interquartile range). Etiology of cirrhosis (others) includes fatty liver diseases, autoimmune hepatic diseases, schistosomiasis, and etc.
APRI AST to Platelet Ratio Index, ALT alanine transaminase, AST aspartate aminotransferase, FIB-4 fibrosis 4 score, HBV hepatitis B virus, HCV hepatitis C virus, HDL-C high-density lipoprotein cholesterol, INR international normalized ratio, MELD model for end-stage liver disease, PLT platelet count, PT prothrombin time, TBIL total bilirubin
The Development of MMML Models
Based on the training and validation datasets, a series of MMML models were developed on the H2O AutoML platform. Supplementary Fig. 2 presents the heatmaps of all variables, while Supplementary Fig. 3 shows the model correlations. To further explain the relation between key variables (i.e., CV model, prothrombin time, alanine aminotransferase [ALT], aspartate aminotransferase [AST], and total bilirubin) and various models, partial dependence is plotted in Supplementary Fig. 4. Detailed information on the six models is listed in Supplementary Content 1.
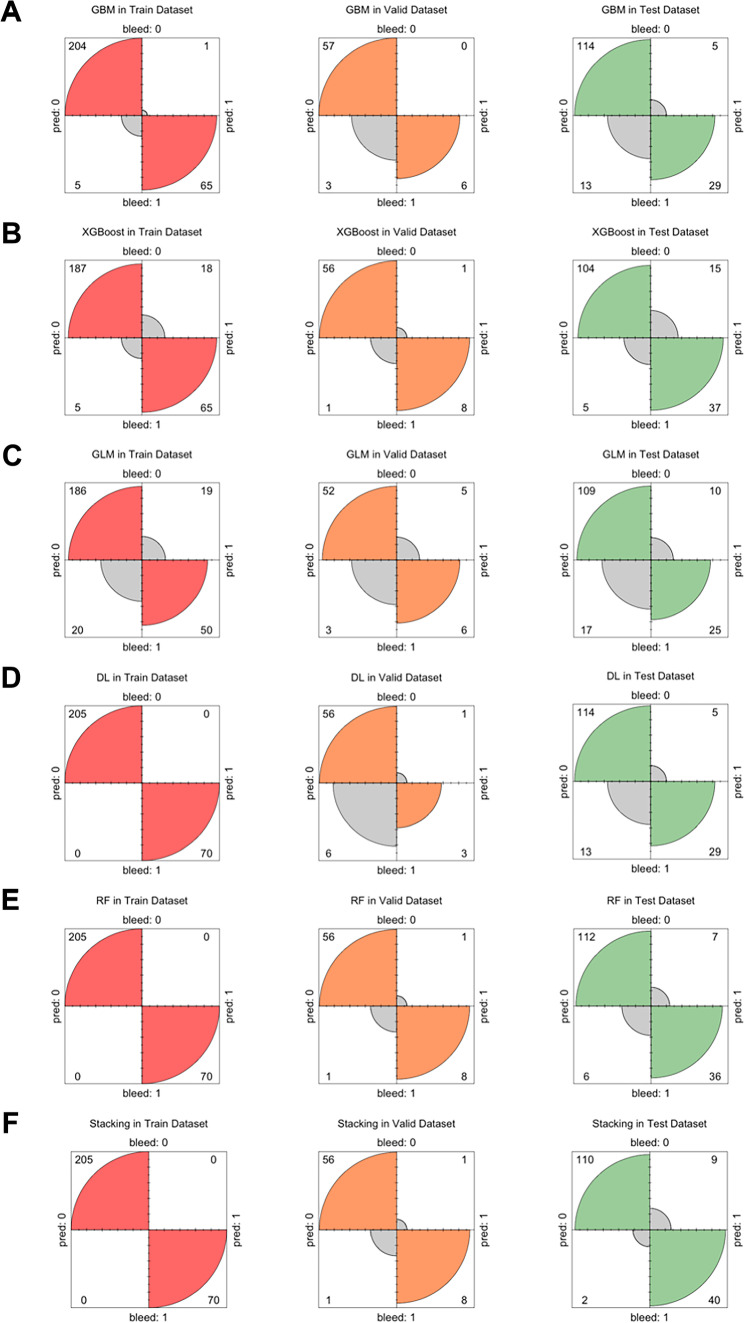
The confusion matrix of the six MMML models based on different algorithms is presented in Fig. 5 and Table 2. In the validation dataset, the stacking model achieved the highest AUC (0.998). Its model ID was StackedEnsemble_BestOfFamily_4_AutoML_1_20220313_114512, which consisted of six base models, i.e., one DL model, two RF models, one GBM, one XGBoost, and one GLM. In addition, all the MMML models were significantly better than the clinical indices, Child‒Pugh score (0.815), MELD score (0.856), APRI (0.791), and FIB-4 (0.704).
Fig. 5.
The confusion matrix of AutoML models in the datasets. DL, deep learning; RF, random forest; GBM, gradient boosting machine; GLM, general linear model; XGBoost, eXtreme gradient boosting
Table 2.
Performance of MMML models and clinical indexes by dataset
| Datasets | Models | Accuracy | Sensitivity | Specificity | Recall | Precision | F1-score | AUC |
|---|---|---|---|---|---|---|---|---|
| Training | ||||||||
| GBM | 0.978 | 0.929 | 0.995 | 0.929 | 0.985 | 0.956 | 0.998 | |
| GLM | 0.858 | 0.714 | 0.907 | 0.714 | 0.725 | 0.719 | 0.876 | |
| XGBoost | 0.916 | 0.929 | 0.912 | 0.929 | 0.783 | 0.850 | 0.970 | |
| RF | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| DL | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Stacking | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Child–Pugh | 0.495 | 0.900 | 0.356 | 0.900 | 0.323 | 0.475 | 0.662 | |
| MELD | 0.618 | 0.771 | 0.566 | 0.771 | 0.378 | 0.507 | 0.673 | |
| APRI | 0.720 | 0.600 | 0.761 | 0.600 | 0.462 | 0.522 | 0.725 | |
| FIB-4 | 0.644 | 0.586 | 0.663 | 0.586 | 0.373 | 0.456 | 0.645 | |
| Validation | ||||||||
| GBM | 0.952 | 0.667 | 1.000 | 0.667 | 1.000 | 0.800 | 0.990 | |
| GLM | 0.879 | 0.667 | 0.912 | 0.667 | 0.545 | 0.600 | 0.903 | |
| XGBoost | 0.970 | 0.889 | 0.982 | 0.889 | 0.889 | 0.889 | 0.994 | |
| RF | 0.970 | 0.889 | 0.982 | 0.889 | 0.889 | 0.889 | 0.983 | |
| DL | 0.894 | 0.333 | 0.982 | 0.333 | 0.750 | 0.462 | 0.984 | |
| Stacking | 0.970 | 0.889 | 0.982 | 0.889 | 0.889 | 0.889 | 0.998 | |
| Child–Pugh | 0.697 | 0.889 | 0.667 | 0.889 | 0.296 | 0.444 | 0.815 | |
| MELD | 0.909 | 0.667 | 0.947 | 0.667 | 0.667 | 0.667 | 0.856 | |
| APRI | 0.773 | 0.778 | 0.772 | 0.778 | 0.350 | 0.483 | 0.791 | |
| FIB-4 | 0.848 | 0.556 | 0.895 | 0.556 | 0.455 | 0.500 | 0.704 | |
| Test | ||||||||
| GBM | 0.888 | 0.690 | 0.958 | 0.690 | 0.853 | 0.763 | 0.968 | |
| GLM | 0.832 | 0.595 | 0.916 | 0.595 | 0.714 | 0.649 | 0.849 | |
| XGBoost | 0.876 | 0.881 | 0.874 | 0.881 | 0.712 | 0.787 | 0.935 | |
| RF | 0.919 | 0.857 | 0.941 | 0.857 | 0.837 | 0.847 | 0.976 | |
| DL | 0.888 | 0.690 | 0.958 | 0.690 | 0.853 | 0.763 | 0.937 | |
| Stacking | 0.932 | 0.952 | 0.924 | 0.952 | 0.816 | 0.879 | 0.975 | |
| Child–Pugh | 0.596 | 0.762 | 0.538 | 0.762 | 0.368 | 0.496 | 0.686 | |
| MELD | 0.534 | 0.929 | 0.395 | 0.929 | 0.351 | 0.510 | 0.680 | |
| APRI | 0.665 | 0.786 | 0.622 | 0.786 | 0.423 | 0.550 | 0.739 | |
| FIB-4 | 0.770 | 0.452 | 0.882 | 0.452 | 0.576 | 0.507 | 0.703 |
APRI AST to Platelet Ratio Index, DL deep learning, FIB-4 fibrosis 4 score, GBM gradient boost machine, GLM general linear model, MELD model for end-stage liver disease, MMML multimodal machine learning, RF random forest, XGBoost eXtreme gradient boosting
Visualization of the Stacking Model
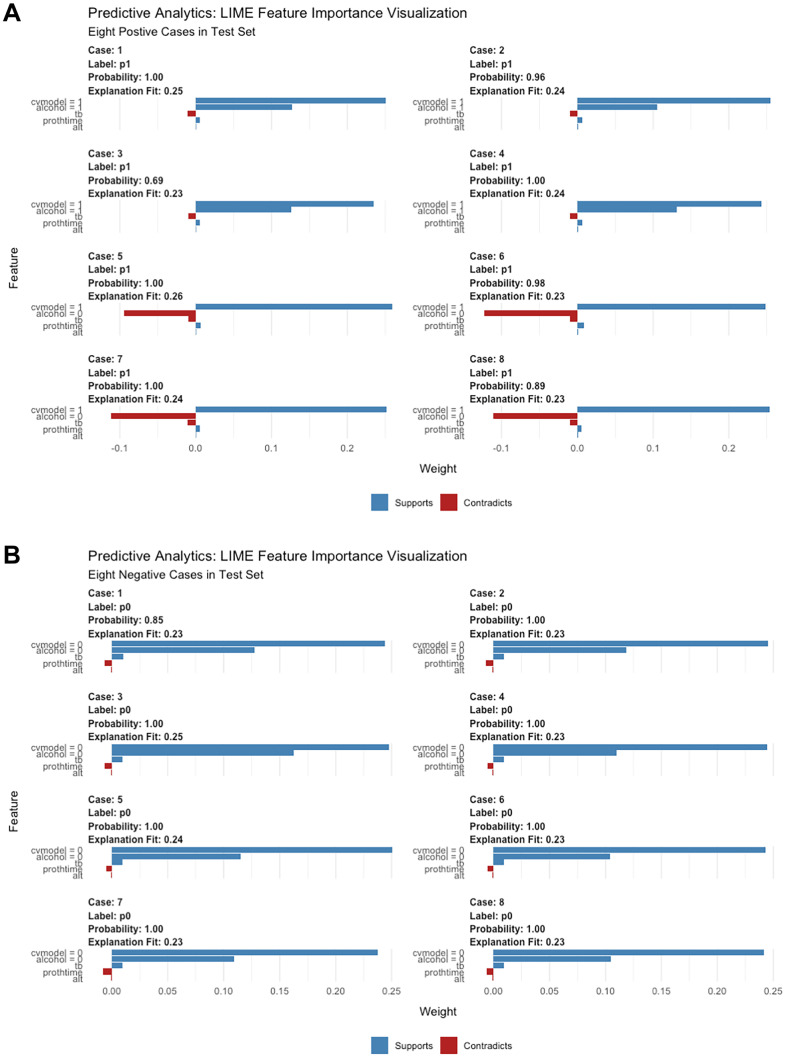
To better understand the stacking model, we used LIME to visualize how the key variables contribute to the model’s output. The CV model output, alcohol consumption, total bilirubin, prothrombin time, and ALT were regarded as important variables in the prediction. Their contributions are semiquantitatively presented in Fig. 6.
Fig. 6.
Local Interpretable Model-Agnostic Explanations (LIME) plots of the stacking model. Interpretation of sample prediction requires random drawing of samples to make model predictions and observe the model through the LIME algorithm. It shows the key variables’ contribution to the positive (A) and negative (B) outcomes
Evaluation of the Models in the Test Dataset
Finally, we evaluated the six MMML models in the test dataset (Fig. 5 and Table 2). According to the AUC, the RF (0.976) and stacking (0.975) models reached the highest values. However, in consideration of the nature of the screening tools, the stacking model was the best model, owing to the highest sensitivity (0.952). Additionally, it also showed the best accuracy (0.932) and F1-score (0.879).
Discussion
In this study, we first developed five DL-based CV models to classify EV images by 12-month EV bleeding; then, we fitted a series of MMML models integrating the clinical structured data and the outputs of the best CV model to predict 12-month EV bleeding on the H2O AutoML platform. Finally, we externally tested the models and found the stacking model to be the best model. This study, for the first time, evaluated the feasibility of multimodal fusion in predicting EV bleeding.
The clinical management of variceal bleeding depends on the stages in the natural history of portal hypertension [1]. Nonselective β-blockers combined with endoscopic ligation are now the first-line treatment of primary prophylaxis. The present evidence shows that the risk of variceal bleeding increases with the impairment of liver function, variceal sizes and risk features (i.e., red wales, red spots, diffuse redness) [2]. Early identification of high-risk patients with EV bleeding is essential to improve the outcomes of patients with cirrhosis.
During the past 40 years, Baveno conferences aimed to define key events in portal hypertension, to review the progress on diagnosis and therapy and to issue evidence-based recommendations for management [3]. Since the Baveno VI, the criteria (platelet count > 150*109/L and liver stiffness measurement [LSM] < 20 kPa) have emphasized the application of transient elastography in the evaluation of portal hypertension [3, 26]. The Baveno VI criteria were introduced in 2015 to rule out varices needing treatment. Then, the expanded Baveno VI (platelet count > 110*109/L and LSM < 25 kPa) and stepwise platelet-MELD criteria (platelet count > 150*109/L and MELD = 6) were proposed. Nawalerspanya et al. [27] compared the three criteria in compensated cirrhosis. They found that the expanded Baveno VI and the platelet-MELD criteria showed higher specificities than the original criteria. Despite the increasingly common use of transient elastography, it is still not widely available in developing countries. Thus, a more practical tool to reliably estimate the risk of EV bleeding would be clinically useful.
Colli et al. [28] performed a Cochrane meta-analysis. They found that the platelet count/spleen diameter ratio could be a tool to rule out adults without varices before endoscopy. However, its ability to predict variceal bleeding was limited. Previous studies developed a variety of prediction models to estimate EV bleeding risk. Dong et al. [29] developed an ML-based scoring system, named the EVendo score, to screen patients with EVs and varices needing treatment based on the international normalized ratio, AST, platelet counts, urea nitrogen, hemoglobin, and presence of ascites. It identified patients with EVs (AUROC 0.84) and patients with varices needing treatment (AUROC 0.74). Agarwal et al. [30] reported the feasibility of ML-based models to predict the first episode of EV bleeding in patients with compensated advanced chronic liver disease. The accuracy of the XGBoost model was 93.7% and 85.7% in the internal and external validation cohorts, respectively. They found that endoscopic classification and LSM were the key variables of the model.
DL has been increasingly adopted in a series of clinical CV tasks, especially in medical image classification and segmentation [31]. The application of DL-based CV models to endoscopic examination could help in analyzing lesions in real time. Recent studies report that automated-trained MMML models achieve good performance in the field of complex analysis and disease risk prediction [15, 18].
In this multicenter study, first, five CNN- or transformer-based CV models were transfer learning to the binary classification of EV endoscopic images at admission, according to EV bleeding events during the 12-month follow-up. The EfficientNet model achieved the highest accuracy. EfficientNet is a CNN architecture and scaling method that uniformly scales all dimensions with a set of fixed scaling coefficients [32]. EfficientNet achieved state-of-the-art accuracy on CIFAR-100 (91.7%) and other transfer learning datasets, with significantly fewer parameters.
Subsequently, we collected the clinical structured data at admission, combined with the outputs of the EfficientNet model, which were then loaded into the H2O AutoML platform. H2O’s AutoML is an automated ML workflow with various explainable and visualized methods. The AutoML workflow processed the multimodalities mentioned above and exported six MMML models to predict 12-month EV bleeding. In the internal validation dataset, the stacking model’s performance was better than the five others (i.e., XGBoost, GBM, DL, GLM, and RF) and the existing scoring systems (i.e., Child‒Pugh, MELD, APRI, and FIB-4) [33]. The CV model output, alcohol consumption, total bilirubin, prothrombin time, and ALT were found to be the key variables in the prediction of the stacking model.
Furthermore, we tested the models and found that the stacking model was still the best model in the external dataset. Stacking is a way of combining multiple classification or regression models [34]. It is usually composed of two layers: the first layer consists of the base models that predict the outputs, and the second layer is a meta-classifier or regressor that takes the first-layer output as an input and generates new predictions. In our study, the stacking model consisted of six base models (two RF models, one DL, one GBM, one XGBoost, and one GLM).
Our study featured automated MMML models that integrated clinical structured data and CV model outputs. To the best of our knowledge, there have been no previous reports. However, our study has some limitations. First, we did not collect transient elastography data; thus, we failed to compare the performance with the Baveno criteria. In addition, we focused on only one outcome, i.e., 12-month bleeding. Therefore, further studies are required to evaluate the models in the prediction of rebleeding, readmission, and mortality. Last, the number of enrolled patients was limited, which required more data for validation.
Conclusion
In this study, for the first time, we evaluated the feasibility of automated MMML, integrating DL-based CV features and clinical structured data, in predicting 12-month EV bleeding. A stacking model was developed and showed practicable performance. Our study may offer insights into further clinical research processes and multimodal data, e.g., medical images and structured variables.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
Study concept and design (JZ, YH), acquisition of data (XZ, XG, CY), analysis and interpretation of data (JX, LL, JG, MY), drafting of the manuscript (YW), administrative, technical, or material support (XL, GX, JZ), and study supervision (YH). All authors have made a significant contribution to this study and have approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China [82000540]; Medical Education Collaborative Innovation Fund of Jiangsu University [JDY2022018]; Science and Technology Plan (Apply Basic Research) of Changzhou City [CJ20210006]; the Youth Program of Suzhou Health Committee [KJXW2021051 and KJXW2019001]. No funding body had any role in the design of the study and collection, analysis, interpretation of data, or in writing the manuscript.
Data Availability
The datasets and R codes in the study are available from the corresponding author on reasonable request. The code of computer vision models’ training is available to access: https://osf.io/ycxwr/?view_only=81b4f590605c472f9e979c854a573cce.
Declarations
Research Involving Human Participants and/or Animals
This study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (the IRB approval number 2022098). All procedures performed in studies involving human participants were in accordance with the Helsinki Declaration of 1975 as revised in 2013.
Informed Consent
Informed consent was obtained from all individual participants included in the study. The authors affirm that human research participants provided informed consent for publication of the images in Fig. 3.
Conflict of Interest
The authors declare no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Wang, Yu Hong, Yue Wang, and Xin Zhou contribute equally.
References
- 1.Gines P, Krag A, Abraldes JG, Sola E, Fabrellas N, Kamath PS. Liver cirrhosis. Lancet. 2021;398:1359–1376. doi: 10.1016/S0140-6736(21)01374-X. [DOI] [PubMed] [Google Scholar]
- 2.de Franchis R, Bosch J, Garcia-Tsao G, Reiberger T, Ripoll C, Baveno VIIF: Baveno VII - Renewing consensus in portal hypertension. J Hepatol, 2021 [DOI] [PMC free article] [PubMed]
- 3.de Franchis R, Baveno VIF. Expanding consensus in portal hypertension: Report of the Baveno VI Consensus Workshop: Stratifying risk and individualizing care for portal hypertension. J Hepatol. 2015;63:743–752. doi: 10.1016/j.jhep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Collaborators GBDC. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:245–266. doi: 10.1016/S2468-1253(19)30349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J, et al. Convolutional Neural Network Technology in Endoscopic Imaging: Artificial Intelligence for Endoscopy. Clin Endosc. 2020;53:117–126. doi: 10.5946/ce.2020.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mondal AK, Bhattacharjee A, Singla P, Prathosh AP. xViTCOS: Explainable Vision Transformer Based COVID-19 Screening Using Radiography. IEEE J Transl Eng Health Med. 2022;10:1100110. doi: 10.1109/JTEHM.2021.3134096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang TL, Carneiro G. Artificial intelligence in gastrointestinal endoscopy. J Gastroenterol Hepatol. 2021;36:5–6. doi: 10.1111/jgh.15344. [DOI] [PubMed] [Google Scholar]
- 9.Visaggi P, et al. Artificial Intelligence in the Diagnosis of Upper Gastrointestinal Diseases. J Clin Gastroenterol. 2022;56:23–35. doi: 10.1097/MCG.0000000000001629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bang CS, Lee JJ, Baik GH. Artificial Intelligence for the Prediction of Helicobacter Pylori Infection in Endoscopic Images: Systematic Review and Meta-Analysis Of Diagnostic Test Accuracy. J Med Internet Res. 2020;22:e21983. doi: 10.2196/21983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho BJ, et al. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy. 2019;51:1121–1129. doi: 10.1055/a-0981-6133. [DOI] [PubMed] [Google Scholar]
- 12.Mori Y, et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]
- 13.Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284:603–619. doi: 10.1111/joim.12822. [DOI] [PubMed] [Google Scholar]
- 14.Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faes L, et al. Automated deep learning design for medical image classification by health-care professionals with no coding experience: a feasibility study. Lancet Digit Health. 2019;1:e232–e242. doi: 10.1016/S2589-7500(19)30108-6. [DOI] [PubMed] [Google Scholar]
- 16.Hung AJ, Chen J, Gill IS. Automated Performance Metrics and Machine Learning Algorithms to Measure Surgeon Performance and Anticipate Clinical Outcomes in Robotic Surgery. JAMA Surg. 2018;153:770–771. doi: 10.1001/jamasurg.2018.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi S, et al. Multimodal Fusion With Reference: Searching for Joint Neuromarkers of Working Memory Deficits in Schizophrenia. IEEE Trans Med Imaging. 2018;37:93–105. doi: 10.1109/TMI.2017.2725306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al'Aref SJ, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J. 2019;40:1975–1986. doi: 10.1093/eurheartj/ehy404. [DOI] [PubMed] [Google Scholar]
- 19.North Italian Endoscopic Club for the S, Treatment of Esophageal V: Prediction of the first variceal hemorrhage in patients with cirrhosis of the liver and esophageal varices. A prospective multicenter study. N Engl J Med 319:983–989, 1988 [DOI] [PubMed]
- 20.The general rules for recording endoscopic findings on esophageal varices. Jpn J Surg 10:84–87, 1980 [DOI] [PubMed]
- 21.Borgli H, et al. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data. 2020;7:283. doi: 10.1038/s41597-020-00622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, et al. A Multi-Label Deep Learning Model with Interpretable Grad-CAM for Diabetic Retinopathy Classification. Annu Int Conf IEEE Eng Med Biol Soc. 2020;1560–1563:2020. doi: 10.1109/EMBC44109.2020.9175884. [DOI] [PubMed] [Google Scholar]
- 23.Roberts D, et al.: Treatment for bleeding oesophageal varices in people with decompensated liver cirrhosis: a network meta-analysis. Cochrane Database Syst Rev 4:CD013155, 2021 [DOI] [PMC free article] [PubMed]
- 24.Zhou Y, Zheng J, Li S, Zhou T, Zhang P, Li HB: Alcoholic Beverage Consumption and Chronic Diseases. Int J Environ Res Public Health 13, 2016 [DOI] [PMC free article] [PubMed]
- 25.Wang Y, Yu W, He M, Huang Y, Wang M, Zhu J. Serum cytoskeleton-associated protein 4 as a biomarker for the diagnosis of hepatocellular carcinoma. Onco Targets Ther. 2019;12:359–364. doi: 10.2147/OTT.S189425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa M, et al. The Baveno VI criteria for predicting esophageal varices: validation in real life practice. Rev Esp Enferm Dig. 2017;109:704–707. doi: 10.17235/reed.2017.5052/2017. [DOI] [PubMed] [Google Scholar]
- 27.Nawalerspanya S, Sripongpun P, Chamroonkul N, Kongkamol C, Piratvisuth T. Validation of original, expanded Baveno VI, and stepwise & platelet-MELD criteria to rule out varices needing treatment in compensated cirrhosis from various etiologies. Ann Hepatol. 2020;19:209–213. doi: 10.1016/j.aohep.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Colli A, et al.: Platelet count, spleen length, and platelet count-to-spleen length ratio for the diagnosis of oesophageal varices in people with chronic liver disease or portal vein thrombosis. Cochrane Database Syst Rev 4:CD008759, 2017 [DOI] [PMC free article] [PubMed]
- 29.Dong TS, et al.: Machine Learning-based Development and Validation of a Scoring System for Screening High-Risk Esophageal Varices. Clin Gastroenterol Hepatol 17:1894–1901 e1891, 2019 [DOI] [PubMed]
- 30.Agarwal S, et al. Development of a machine learning model to predict bleed in esophageal varices in compensated advanced chronic liver disease: A proof of concept. J Gastroenterol Hepatol. 2021;36:2935–2942. doi: 10.1111/jgh.15560. [DOI] [PubMed] [Google Scholar]
- 31.Dana J, et al.: Conventional and artificial intelligence-based imaging for biomarker discovery in chronic liver disease. Hepatol Int, 2022 [DOI] [PMC free article] [PubMed]
- 32.Abedalla A, Abdullah M, Al-Ayyoub M, Benkhelifa E. Chest X-ray pneumothorax segmentation using U-Net with EfficientNet and ResNet architectures. PeerJ Comput Sci. 2021;7:e607. doi: 10.7717/peerj-cs.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng H, Qi X, Guo X: Diagnostic Accuracy of APRI, AAR, FIB-4, FI, King, Lok, Forns, and FibroIndex Scores in Predicting the Presence of Esophageal Varices in Liver Cirrhosis: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 94:e1795, 2015 [DOI] [PMC free article] [PubMed]
- 34.Ghasemian A, Hosseinmardi H, Galstyan A, Airoldi EM, Clauset A. Stacking models for nearly optimal link prediction in complex networks. Proc Natl Acad Sci U S A. 2020;117:23393–23400. doi: 10.1073/pnas.1914950117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and R codes in the study are available from the corresponding author on reasonable request. The code of computer vision models’ training is available to access: https://osf.io/ycxwr/?view_only=81b4f590605c472f9e979c854a573cce.