Abstract
Multiple displacement amplification (MDA) based on isothermal random priming and high fidelity phi29 DNA polymerase-mediated processive extension has revolutionized the field of whole genome amplification by enabling the amplification of minute amounts of DNA, such as from a single cell, generating vast amounts of DNA with high genome coverage. Despite its advantages, MDA has its own challenges, one of the grandest being the formation of chimeric sequences (chimeras), which presents in all MDA products and seriously disturbs the downstream analysis. In this review, we provide a comprehensive overview of current research on MDA chimeras. We first reviewed the mechanisms of chimera formation and chimera detection methods. We then systematically summarized the characteristics of chimeras, including overlap, chimeric distance, chimeric density, and chimeric rate, as found in independently published sequencing data. Finally, we reviewed the methods used to process chimeric sequences and their impacts on the improvement of data utilization efficiency. The information presented in this review will be useful for those interested in understanding the challenges with MDA and in improving its performance.
Keywords: Chimeras, Chimeric sequences, Multiple displacement amplification, Whole genome amplification
1. Introduction
Cells are the basic building blocks of all living organisms [1] and contain genetic information in the form of DNA [2]. In biological and medical research, it is often necessary to understand the genetic information of a single cell rather than a group of cells [3], [4], [5], [6], [7], [8], [9], [10]. However, the amount of genomic DNA in one cell is often insufficient for sequencing [11]. Modern sequencing platforms require a minimum of 1 ng DNA for next-generation sequencing (NGS) and 1 μg for third-generation long-read sequencing [12], [13]. Long-read sequencing technologies, such as those provided by Pacific Biosciences and Oxford Nanopore Technology, also require high molecular weight DNA as input [12], [14], typically > 10 kb.
In some circumstances, such as in single-cell, metagenomics, or unculturable microorganism studies, it may be difficult to directly extract sufficient amounts of DNA of the required quality and length. To overcome this limitation, DNA can be amplified through whole genome amplification (WGA) technologies from as few as picograms to micrograms [11], [15]. Advances in DNA sequencing technologies have made it possible to sequence the entire genome of a single cell at an affordable cost through single-cell whole genome amplification (scWGA) techniques. All DNA molecules can be amplified parallelly, and single-cell genomics has emerged as an exciting field of its own [11], [16], [17], [18], [19], [20], [21], [22], [23], [24].
Multiple displacement amplification (MDA), a widely used WGA and scWGA approach, involves the binding of random hexamers to the denatured DNA followed by strand displacement synthesis using phi29 polymerase at a constant temperature [25]. This method is preferred due to its high processivity, strand displacement capacity, and a low error rate of 1 in 106–107 nucleotides [20], [26], [27]. Phi29 polymerase has higher fidelity compared to other strand-displacement DNA polymerases, such as Bst, Klenow, and T4. The primers used in MDA can be hybridized to the template at an optimum temperature (typically 30 °C) and are not as long as the primers used in other DNA polymerases that require higher reaction temperature and have lower randomness and reaction efficiency. Additionally, Phi29 polymerase has both high proofreading activity and strand displacement activity, whereas other DNA polymerases have only strong strand displacement activity (such as Bst and Klenow) or only strong proofreading activity (such as T4). The DNA amplified by MDA has a higher molecular weight (up to 100 kb) and better genome coverage than the output of other scWGA methods [3], [11], [23], [28], [29], [30], [31]. To further improve the performance of MDA, researchers have developed some customized strategies [32], including microwell MDA (MIDAS) [33], emulsion MDA (eMDA) [34], and microchannel MDA (μcMDA) [35], [36].
However, there are limitations to the use of MDA in DNA sequencing [11], [37], such as the formation of chimeras, which are amplification artifacts [38], [39], [40], [41] when two non-adjacent genome regions are joined together on one DNA molecule after MDA. Chimeras are considered a result of hyperbranched DNA generated by the strand displacement process [39], [40], [41] and cannot be used for genome assembling [42], [43], [44], [45], [46], [47]. This poses a significant challenge to single-cell sequencing studies of structural variants [23], [48], [49], [50] and can result in assembly errors in microbial genome [47] and macro-genomic studies [42], [43], [44].
Given that the amount of DNA involved in these studies is minimal, MDA is applied to amplify the DNA for subsequent sequencing, but the chimerism generated during the process can greatly impact the overall accuracy. Researches indicate that the chimerism in MDA sequencing data cannot be ignored and is attracting increasing attention, especially as single-cell studies have become a hot topic [13], [51], [52], [53], [54].
In this mini-review, we discuss the formation and systematic characteristics of MDA chimera [39] and present existing methodologies for detecting chimeras in MDA-treated short and long-read sequencing data from Sanger sequencing [38], NGS [39], [40], [41], [54], and third-generation sequencing platforms [51], [55]. We also provide information on the treatment of chimeras and their impact on the improvement of data utilization efficiency. Our review systematically compares and discusses different methods for chimera analysis, allowing researchers to choose appropriate ways to reduce the impacts of the MDA-induced chimeras.
1.1. The multiple displacement amplification
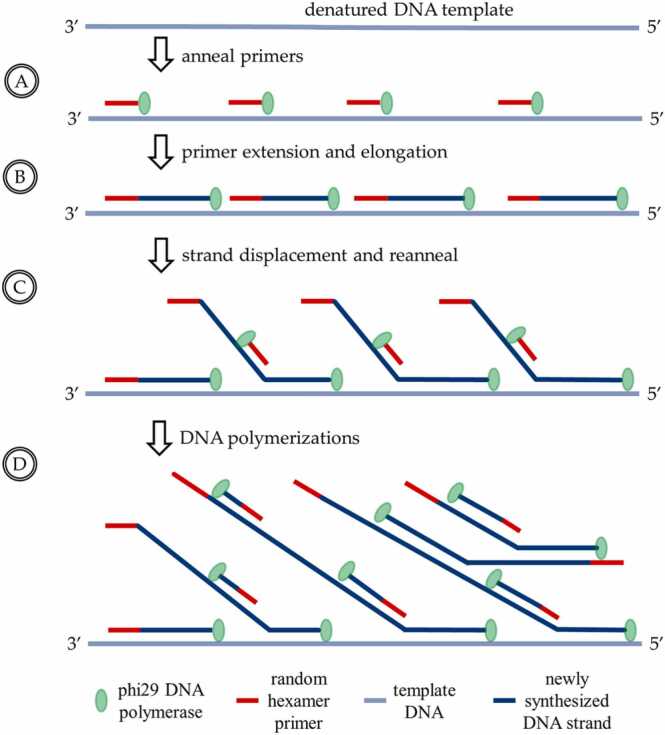
As mentioned in the introduction, MDA [11], [25], [32], [37] is a 30 °C isothermal DNA amplification process mediated by phi29 DNA polymerase. The iterative process begins with the annealing of random hexamer primers to denatured ssDNA templates. DNA synthesis then starts at multiple sites on the template (Fig. 1A) and continues uninterruptedly along the template until encountering a downstream primer (Fig. 1B). The phi29 polymerase has strong displacement activity, causing the downstream strand to be displaced gradually from its 5′-end as the upstream elongation of DNA strand continues (Fig. 1C and Fig. 2A). This results in the formation of complex branch structures composed of a large amount of partly displaced strands (Fig. 1D). The branched molecules can serve as new templates upon the binding of new random primers and polymerase, leading to exponential growth in a total amount of DNA. Although the branch growth generates clusters of DNA molecules instead of linear dsDNA, the rule of only two ssDNA molecules being able to hybridize at the same site applies, allowing the products to be used for sequencing library construction. The amplification ends with heat inactivation of phi29 polymerase, after which a sufficient amount of DNA is collected.
Fig. 1.
Experimental process of MDA with phi29 DNA polymerase: (A) The random hexamer primers (represented by the short red lines) anneal to the denatured single-strand DNA (ssDNA) template (represented by the light blue lines). (B) The phi29 DNA polymerase (represented by the green spheres) synthesizes DNA along the ssDNA template from the positions where the primers have annealed, until it reaches the adjacent newly synthesized double-strand DNA (dsDNA, represented by the dark blue lines). (C) The phi29 DNA polymerase continues the DNA replication process, replacing the newly synthesized DNA strand and continuing the DNA polymerization, while primers anneal to the newly synthesized DNA. (D) DNA polymerization initiates along the new DNA strands, resulting in the formation of a network of hyperbranched DNA structure and generating very long DNA fragments (ranging from 2 kb to 70 kb) with low amplification error rates.
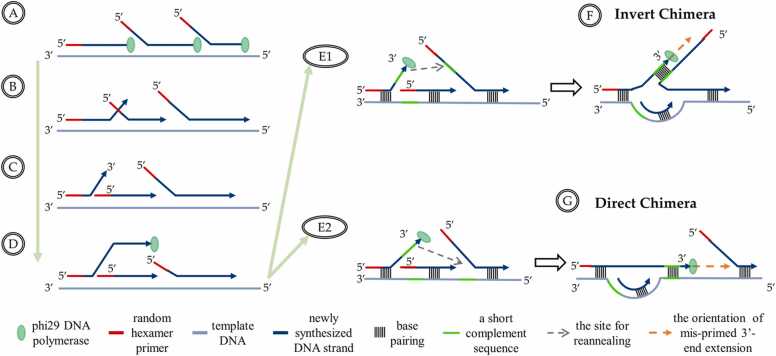
Fig. 2.
The mechanisms underlying the 5′-end displacement by phi29 DNA polymerase, 3′-end displacement by branch migration, and chimera formation with direct sequences and inverted sequences during the MDA process. Mispriming events occur during MDA due to the present of similar sequences (green lines in right Figure) on adjacent templates. (A) The MDA reaction proceeds through the strand displacement mechanism, with phi29 DNA polymerase extending 3′-termini while concurrently displacing any downstream copies starting from their 5′-ends. (B, C) Branch migration reaction displaces 3′-terminis with an equilibrium between competing secondary structures. (D) Phi29 DNA polymerase binds to the displaced 3′-ends, allowing it to prime on the nearby regions with different coordinates. (E1) The displaced 3′-ends re-anneal to a nearby new DNA template strand (represented by the light gray dotted arrow), which can be a synthesized and displaced ssDNA with a free 5′-end or an unrelated strand if there exists a complementary pair (green line) between the displaced 3′-end and the free 5′-end. (F) Elongation continues (represented by the orange dotted arrow) by phi29 DNA polymerase along the new template strand, generating inverted chimeras. (E2) The displaced 3′-ends re-anneal to the same template that has a similar base sequence (represented by green line in E2) but not at the same region. (G) Elongation continues (represented by orange dotted arrow) by phi29 DNA polymerase along template strand, generating direct chimeras.
1.2. The mechanism of MDA chimera formation
The MDA process involves the simultaneous synthesis of multiple complementary strands from different sites on a single template strand, leading to the formation of hyperbranched structures [38], [39], [41], [54], [56], [57], [58]. The branched DNA structures have numerous single-stranded 5′-ends, many of which are eventually converted into dsDNA by random hexamer primers. This results in vast amounts of 5′-ends of ssDNA being freely dissociated from the template and available for priming. The branch-migration mechanism, which is an entropically advantageous mode of DNA strand displacement with alternative forms, is predicted to appear in equilibrium [59], [60]. The displaced ssDNA also competes for re-annealing with the template [39], which can cause the 3′-end of the extended DNA to fall off (Fig. 2B and Fig. 2C). The single-stranded intermediate states can anneal to alternative secondary structures with other ssDNA (Fig. 2D), including the displaced strand (Fig. 2E1) or the original template (Fig. 2E2) with complementary base pairs. This random mispriming event of displaced strands on unexpected genomic regions will lead to the formation of MDA chimeras (Fig. 2F and Fig. 2G).
The chimeras can be classified into two types: inverted chimeras and direct chimeras [39], [41], [56]. The formation mechanisms of these two types of chimeras are different [39], [54]. In direct chimeras, the displaced 3′-termini re-anneal on the same template strand in a region that is similar but not identical to the original primer site. The genomic distance between the displaced 3′-termini and the priming 5′-end is ignored in the amplicon. In inverted chimeras, the displaced 3′-termini anneal on a new template strand, which can be a displaced ssDNA with a free 5′-end or an irrelevant strand. The mis-primed 3′-end is extended according to the new template strand from the priming site by phi29 DNA polymerase, leading to the joining of two regions that are not expected to appear on the same strand. During the re-annealing, DNA strands tend to choose a site that is spatially close to themselves and contains a short complementary sequence in order to reduce the hybridization barrier potential. Sometimes, the mispriming events can occur with only 1 or 0 complementary bases, as the temporary annealing of the 3′-ends can be rapidly stabilized by the extension of the phi29 DNA polymerase.
1.3. The detection and identification of MDA chimeras
An overview of detection methods mainly used in studies on MDA chimeras is presented in Table 1, with each method described as follows.
Table 1.
Overview of detection methods of major MDA chimeras’ studies.
| Studies | Sample | Platform | Read length (bp) | Total read (M) | Total bases (G) | Involved software | Detection Tools | Time (h) | Features and limitations |
|---|---|---|---|---|---|---|---|---|---|
| Zhang et al. 2006 Nature Biotechnology[38] | Prochlorococcus | Sanger | ∼ 800 | 0.01 | 0.01 | Phrap and BLAST | / | / | The earliest study, high chimeric rate, but lower throughput, smaller genome, and no tool. |
| Lasken et al. 2007 BMC Biotechnology[39] | single Escherichia coli | 454 Life Sciences | ∼ 100 | 0.11 | 0.01 | BLAST | / | / | Revealed the mechanisms of chimera formation, but lower throughput, smaller genome, low chimeric rate, no tool. |
| Tu et al. 2015 Plos One[40] | Human | Illumina | 101 | 772.59 | 78.03 | SOAP2 | / | ∼ 126 | Explored chimeras in bigger genome and large-scale sequencing data, but took lots of time, missed many chimeras, and no tool. |
| Lu et al. 2019 Int. J. Mol. Sci.[41] | single Human cell | Illumina | 150 | 603.31 | 90.50 | BWA | ChimeraMiner | ∼ 55 | Improved chimeras detection tool and analyzed chimeras in human single cell MDA sequencing data for the first time. |
| Yuya et al. 2021 DNA Research[51] | Phageomes | PacBio Sequel | 5551 | 0.33 | 1.83 | LAST | SACRA | ∼ 630 | Novel tool for pre-processing chimeras in PacBio reads and reducing chimeric rate, but small genome, no overlap sequence analysis, and no comprehensively exploration. |
| Lu et al. 2022 bioRxiv[55] | Human | PacBio Sequel | 3821 | 3.55 | 13.57 | minimap2 | 3rd-ChimeraMiner | ∼ 168 | Tool for analyzing chimeras in human long-read sequencing data and transferring the chimeras to normal reads, but DNA not from single cell just single cell level. |
1.3.1. Chimera detection in Sanger sequencing data of single cells
Chimera detection in Sanger sequencing data of single cells is a critical issue that has been studied in recent years. The use of cloned libraries derived from MDA products of single cells is a routine practice in whole-genome shotgun sequencing. In 2006 [38], Zhang et al. tested different post-amplification enzymatic treatments of the same MDA-amplified DNA to decrease chimeras and improve genome assembly in the presence of chimeric sequences. They found that using S1 nuclease resulted in partial removal of chimera, but the chimeric rate was not significantly reduced. However, the combination of three treatments: phi29 polymerase debranching, S1 nuclease digestion, and DNA polymerase I nick translation, resulted in a low chimeric rate of 6.25 %. Skipping any of these treatments increased the chimeric rate.
The authors also developed IterativeAssembler, a tool for multiple rounds of genome assembly and chimeric sequence detection. The tool is mainly based on an iterative assembly procedure that involves assembling total reads into contigs, aligning total reads with the contigs, breaking chimeric sequences at the chimeric point, and feeding the resultant sequences for the next round of assembly. The iterative assembly procedure is repeated until the chimeric rate stops increasing.
The above research highlights the challenges posed by chimeric sequences derived from MDA reactions in genome assembly procedures and indicates that MDA can introduce an unusually high percentage of chimeras into sequencing data, leading to difficulties in accurate genome assembly. The detection method used in the study demonstrates that these chimeric sequences can be split into two parts and mapped to two distinct regions of the reference genome.
This study uses Sanger sequencing platforms, which provide long reads (about 800 bp) amplified by MDA from the DNA of a single bacterial cell but have limited throughput [61], [62], [63]. These platforms are not suitable for large-scale studies and are being placed by more advanced sequencing technologies. In addition, the amplified DNA in this study was from bacteria with relatively small genome size [64], and the formation and structural features of chimeras still need to be explored through further in-depth analysis.
1.3.2. Chimeras detection in 454 sequencing data of a single E. coli cell by MDA
In 2007 [39], Lasken et al. made a significant contribution to the field of single-cell whole genome sequencing by discovering 475 MDA chimeras in a sample of 108,944 totally uniquely mapped reads from a single E. coli cell. The MDA products were sequenced using the 454 sequencing platform [65], which is a low-throughput technology that has been discontinued since 2016 [66].
The authors established a standard for detecting MDA chimeras by identifying reads that had two > 20 bp non-continuous sequences that were joined together and aligned to non-contiguous regions of the reference genome. The authors, for the first time, categorized the chimeras based on the orientation of the pairs of aligned read parts (they were either mapped to the same strand of the genome or the opposite strand of the genome), the number of overlapping bases in the chimeric junction, and the size of the intervening genome region spanned by the chimeric junction. They also provided new insights into the reaction mechanisms that generate chimeras during MDA and expanded our understanding of the MDA reaction.
However, there are limitations to this study. The sequencing data were generated using a low-throughput platform, and only 475 chimeras were discovered, which is fairly low number and far away from a significant level, resulting in a lack of representativeness. Furthermore, the genome of E. coli is simple [67] and cannot efficiently reflect the chimerism formation. In addition, no tool or pipeline was developed for detecting MDA chimeras from the whole genome sequencing data. With the advancement of sequencing technologies and comprehensive usage of MDA in NGS, it is hoped that this study can be expanded in the future.
1.3.3. Chimeras detection in human whole genome sequencing data
In 2015 [40], based on the characteristics of MDA chimeras identified in previous studies, Tu et al. divided MDA chimeras into two categories: insertion and single-end chimeras. They established a bioinformatics pipeline using the Short Oligonucleotide Analysis Package 2 (SOAP2) alignment software [68] for recognizing MDA chimeras in paired-end (PE) sequencing data [69] from the Illumina HiSeq sequencing platform [70], [71], [72]. The pipeline first distinguished the insertion chimeras by observing the mapped genome strand orientations of PE reads after they were aligned to the hg19 human reference genome. The authors then collected candidate single-end chimeric reads, which were mapped and unmapped reads with the initial 30 nucleotides (regarded as a seed) perfectly mapped to the hg19 and the remaining nucleotides unmapped to the same location and used a self-designed subsection alignment strategy to search for all single-end chimeras. The strategy involved seed extension and local alignment and was applied repeatedly to the candidate single-end chimeric reads.
The author further comprehensively characterized MDA chimeras in large-scale high-throughput human whole genome sequencing data. The pipeline accurately distinguished the insertion chimeras and the single-end chimeras based on their own characteristics using the subsection-alignment strategy and provided detailed structural visualization of the single-end chimeras through the use of two structurally statistical indicators: the length of overlap sequences and the distance between two subsections of the single-end chimeras. The author reported that the pipeline improved data utilization efficiency but noted that it was tedious, time-consuming, complicated, and omitted some single-end chimeras due to its limitations, such as the lack of complete analysis tool and the exclusion of split-reads from SOAP2 alignment results.
1.3.4. Chimeras detection in human single-cell whole genome sequencing data
Along with the wide application of single-cell sequencing, the presence of chimeras in single-cell MDA products has become an obstacle hindering the comprehensive analysis of sequencing data [7], [73], [74], [75]. To address this issue, a faster and more accurate detection tool is desired. In 2019 [41], Lu et al. introduced ChimeraMiner, an improved chimeric read detection pipeline designed to detect chimeras from human whole genome sequencing data in MDA. The pipeline utilizes the Burrows-Wheeler Aligner [76], [77] Maximal Exact Matches MEM (BWA-MEM) [78], a novel and widely used DNA alignment tool in genome studies, for aligning PE reads to the hg19 reference genome.
ChimeraMiner identifies insertion chimeras from the read pairs aligned to the same genome strand and considers soft-clipped alignment reads as candidate single-end chimeras. It breaks the soft-clipped alignment reads into two or more segments, constructs the adjacent segments as new read-pairs, maps the new read-pairs to the hg19 reference genome, and searches for overlapped sequences to identify valid single-end chimeras using a solo cyclical alignment. The process takes less than half of the time of the previous pipeline [56] based on SOAP2 and could accurately identify all types of MDA chimeras in every single-cell dataset.
The use of BWA-MEM makes ChimeraMiner more suitable for analyzing MDA-related whole genome sequencing data and single-cell whole genome sequencing data. Most importantly, the tool’s ability to rapidly integrate single-end chimeras into subsequent bioinformatics analysis enhances the utilization efficiency of MDA sequencing data, especially single-cell MDA sequencing data. However, ChimeraMiner cannot be applied to detect chimeras in the third-generation sequencing (TGS) MDA data due to differences in the read lengths between NGS and TGS data.
1.3.5. Chimeras detection in long-read sequencing data
The advancement in sequencing platforms has enabled the sequencing of DNA molecules with lengths > = 20 kb. Long-read sequencing [79] using DNA products amplified by phi29 DNA polymerase-based MDA has become a popular choice. This technology has been utilized to perform more detailed analyses of single-cell genomes [80], [81] and has led to the discovery of novel somatic variations, structural variations [82], [83], and repeat regions of genomes [12], [13], which were hard to study in single cells before [84], [85]. As a result, there is a better understanding [86] of somatic variation, mutation rates, and the functional effects of these genomic elements.
Long-read sequencing has potential applications beyond human cells and could result in improved genome assemblies for single cellular organisms that are difficult to culture in the laboratory [51]. However, the amplified DNA produced by phi29 DNA polymerase-based MDA contains numerous chimeric reads that are derived from artificial sequences connecting discontinuous DNA regions. These chimeric reads can hamper proper downstream analysis and become more pronounced with increasing read lengths.
1.3.5.1. Identification of MDA chimeras in PacBio reads from low-biomass phageomes
Kiguchi et al. developed a novel bioinformatics tool called the Split Amplified Chimeric Read Algorithm (SACRA) to address the challenge of long-read sequencing of low-biomass samples. SACRA is designed to correct chimeric reads in PacBio data obtained from MDA products of low-biomass human gut phageomes [51]. The algorithm identifies MDA chimeras in PacBio reads by aligning error-corrected PacBio reads to phageomes genome using LAST [87] and then removing alignments with < 95 % identity or < 50-bp aligned length. PacBio reads with ≥ 50-bp unaligned sequences are considered chimeras originating from MDA.
SACRA proves to be highly effective and accurate in pre-processing MDA chimeras, markedly reducing the average chimera ratio from 72 % to 1.5 %. However, Kiyuchie et al. did not consider the overlap sequence of the detected chimeras, which is the most important feature of chimera. Additionally, the study focused on human gut phageomes, which have a relatively small genome and may not be representative of species with more complex genomes. Therefore, further research is needed to more comprehensively explore MDA chimeras in long-read sequencing data from more representative species.
1.3.5.2. Identification of MDA chimeras in PacBio reads from human single-cell
Recently, Lu et al. detected and characterized chimeric reads in PacBio long-read sequencing reads by coupling MDA and PacBio single-molecule sequencing platform. They developed 3rd-ChimeraMiner, a novel bioinformatics tool for recognizing and classifying MDA chimeras in PacBio long reads [55]. The tool first aligned PacBio reads to the hg19 reference genome using minimap2, and considered the mapped reads with an “SA” tag as segmented mapped reads (SMRs). SMRs with the soft-clipped alignments were considered candidate chimeric reads, which were then segmented and sorted based on their alignment position. The local alignment strategy was employed to search for the overlap sequences between adjacent segments and identify valid MDA single-end chimeras.
3rd-ChimeraMiner is the first bioinformatic tool for analyzing chimeras in long-read sequencing data of human single-cell MDA products. It explored the distribution and proportion of MDA chimeras in human PacBio sequencing data and transformed MDA chimeras into normal reads based on the chimeric points and the strand orientation. By applying the 3rd-ChimeraMiner for MDA long-read sequencing data at the single cell level, Lu et al. found that MDA chimeras were ubiquitous, and the use of 3rd-ChimeraMiner improved the full-length mapping ratio and utilization efficiency of PacBio sequencing data.
1.4. The characteristics of MDA chimeras
Chimeras, both inverted and direct (Fig. 3A and Fig. 3B), are recognized as a phenomenon where adjacent segments of the chimeras can be located consecutively but adjacently on a chromosome, either on the same strand or reverse strand of the reference genome [41]. These adjacent segments are connected by an overlap. Previous studies [38], [41], [51] have demonstrated that the number of phi29 DNA polymerase-mediated MDA chimeras is substantial and statistically significant. Here, we summarize the representative characteristics of MDA chimeras.
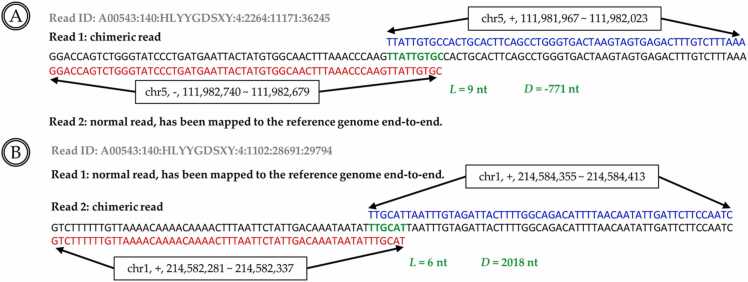
Fig. 3.
(A) Two adjacent segments of a read mapped to the reverse strands of the reference genome (+/-). This read was defined as an inverted chimera. (B) Two adjacent segments of a read are mapped to the same strands of the reference genome (-/-) at different regions. This read is defined as a direct chimera. Moreover, the green sequences in A and B represent the overlapping sequences of the chimeras, between the adjacent segments of the chimeras. D is the chimeric distance between the end coordinate of the former segment and the start coordinates of the following segment. L is the length of the overlapping sequences between two adjacent segments of the chimeras.
1.4.1. The overlap sequence and the chimeric distance
MDA chimeras are often characterized by the presence of overlapping sequences between the former segments, which initiate priming on the new template, and the lagging segments of the DNA. The overlapping sequence is found to be highly similar to the tail of the former segment and the reverse extended sequence of the lagging segment, and its length distribution is one of the statistical indicators of chimeras.
Based on the mechanisms of MDA chimera formation, scientists have identified many potential templates, especially with a short length, in the human genome for the formation of MDA chimeras. These potential templates are referred to as chimeric hotspots [88]. These hotspots can be composed of a pair of the same sequences on the same DNA strand in direct chimeras or a pair of reverse complementary sequences on the same DNA strand in inverted chimeras.
The average length of MDA products is theorized to be 12 kb based on the kinetics of phi29 DNA polymerase, but segmentation far greater than 10 kb apart is less likely to occur in the same amplicon. Studies have shown that the formation of chimeras is an intra-molecular process [11], [13], [39], [54], [58], and the distance between the adjacent segments of chimeras, or the chimeric distance, is usually < 10 kb. The distribution of chimeric distance is another statistical indicator of chimeras.
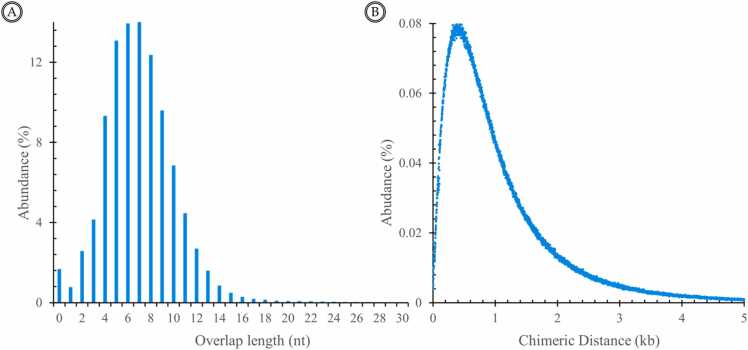
Inverted chimeras are the most abundant type in all visible chimeras and have the simplest structure, making them a popular subject for analyzing chimeric distance and overlap sequence length. The abundance distribution of chimeric distance in inverted chimeras has been found to have an approximate bimodal distribution with a peak of 250–300 nucleotides in the range from 0 to 5000 nucleotides (Fig. 4A), while the distribution of the overlap sequence length is similar to a Poisson distribution with a peak of 7 nucleotides, ranging from 5 to 8 nucleotides in most chimeras (Fig. 4B).
Fig. 4.
Features of the inverted chimeras. (A) The distribution of chimeric distance of two adjacent segments of the reads is an approximate bimodal, with the absolute value of the chimeric distance shown in the X-axis. (B) The abundance distribution of the length of overlapping sequences between two adjacent segments of the inverted chimeras in the NGS data. The abundance of each length is shown as the percentage of the number.
1.4.2. The randomness of chimeras
Previous studies explored the relationship between the formation of chimeras and various factors in DNA sequencing. The ratio of insertion chimeras over single-end chimeras has been found to be positively correlated with the insertion fragment length of the sequencing library [40]. Additionally, the number of inverted chimeras in a chromosome is positively correlated with the length of the chromosome [40], [88]. In other words, more inverted chimeras are observed in a longer chromosome [88].
In a study of single-neuron sequencing analysis of L1 retro-transposition, the authors used PCR to amplify a chimera candidate in six different samples. They found that the PCR products were only detected in one sample [54], suggesting that each chimera event is unique to a given sample. In a sense, the generation of chimeras during MDA is random, but the large genome size and magnification fold of MDA can lead to a unique preference for specific regions of the genome in each individual experiment.
The concept of chimeric hotspots was discussed in the subsection titled “The overlap sequence and the chimeric distance” in the study “The detection and identification of MDA chimeras”. However, it remains unclear whether MDA-generated chimeras have any preferred hotspots and what factors contribute to their formation. Tu et al. conducted research on inverted chimeras and the criteria of chimeric hotspot selection [88]. They systematically screened the chimeric hotspots in the human reference genome and elaborately analyzed factors affecting chimeric hotspot selection, such as chromosome distribution, overlap length, overlap GC content, and genomic distance between two segments. Such analysis of chimera formation could assist in improving the MDA reaction conditions and reducing the occurrence of chimeras.
The screening of 196 billion chimeric hotspots yielded 36.7 million inverted chimeras from MDA-amplified sequencing data. Two datasets were analyzed to evaluate the selective preference in chimeras for hotspots. No clear preference was observed in the distribution of chimeras and hotspots across chromosomes. However, hotspots with an overlap of 12–13 nucleotides were most found to be more susceptible to mispriming as templates in chimera formation. In addition, a periodic selective preference was noticed in the GC content of the overlapping sequence, which was found to be related to the sequence denaturation temperature. The distance between two chimeric segments showed a preference for 80 and 280 nucleotides.
In summary, the formation of chimeras is a random event that occurs when the free 3′-end of a molecule randomly anneals to a nearby template with corresponding chimeric hotspots that can be primed to generate chimeras.
1.4.3. Chimeric rate
In 2006 [38], phi29 DNA polymerase-mediated MDA chimeras were first discovered when sequencing genomes from single cells by cloning. At that time, the chimeric rate ranged from 17.00 % to 19.28 % (Table 2 and Fig. 5). However, after sequential treatment with phi29 polymerase debranching, S1 nuclease digestion, and DNA polymerase I nick translation, the chimeric rate decreased to 6.25–8.33 %, which was still statistically significant. An unusually high percentage of chimeras limited the quality of genome assemblies, and an iterative assembling procedure was developed to computationally remove almost half of the chimeras to generate an assembly of higher quality. Despite these efforts, a comprehensive analysis of the composition and features of chimeras is still lacking.
Table 2.
The chimeric rate and density in previous studies.
| Studies | Read length (bp) | Chimeric rate | Density (events/1 Mb) |
|---|---|---|---|
| Zhang et al. 2006 Nature Biotechnology[38] | ∼800 | 19.30% | 251 |
| Lasken et al. 2007 BMC Biotechnology[39] | ∼100 | 0.45% | 45 |
| Tu et al. 2015 Plos One[40] | 202 | 1.80% | 89 |
| Lu et al. 2019 Int. J. Mol. Sci.[41] | 300 | 4.56% | 152 |
| Yuya et al. 2021 DNA Research[51] | 5551 | 72% | 356 |
| Lu et al. 2022 bioRxiv[55] | 3821 | 76.16% | 469 |
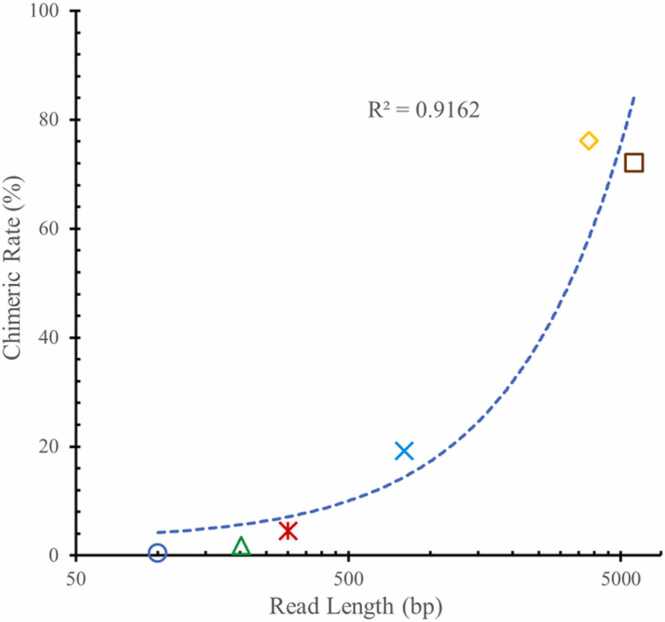
Fig. 5.
Distribution of chimeric rate versus the mean length of the sequence reads, including both inverted and direct chimeras. The chimera rate is calculated as the ratio of the number of chimeras to the number of total sequenced reads. The distribution in the figure shows that the chimeric rate is positively correlated with the mean read length.
In 2007 [39], phi29 DNA polymerase-mediated MDA chimeras were characterized for the first time in single E. coli whole-genome sequencing data. In 2015 [40] and 2019 [41], MDA chimeras were systematically analyzed in human MDA whole-genome sequencing data and human single-cell MDA whole-genome sequencing data, respectively. The chimeric rates were 0.4 % for E. coli, around 6 % for human MDA sequencing data and 0.93–4.68 % for human single-cell MDA sequencing data. In these studies, the inverted chimera was the dominant type of single-end chimeras, whether in E. coli low-throughput sequencing data (85 %) or in human high-throughput sequencing data (∼91 %).
Coupling MDA and PacBio single-molecule sequencing platforms has enabled the analysis of chimeras in MDA products sequenced by long-read sequencing. In PacBio metagenomic-sequencing data of MDA-amplified phageomes DNA [51], an average of 72 % of PacBio reads were chimeras, which is unexpectedly high. In PacBio human-sequencing data [55], the chimeric rates increased with the increasing amplification fold, ranging from 42 % (MDA products initiated from multi-cells, amplification fold of 102) to over 76 % (MDA products initiated from DNA amounts of the single-cell level, amplification fold of 106). The inverted chimera remained the dominant type of chimera (average 89%), consistent with the results of previous NGS analyses. After analysis, 99.92 % of recognized chimeras were found absent in the original genome.
1.4.4. Chimeric density
About one chimera event was detected every 22 kb [39] of MDA-magnified DNA from a single E. coli cell in the 454 sequencers (Table 2), while the rate decreased to about one chimera per 100 kb [54] in human MDA products sequenced by the Illumina platform. However, the frequency of the chimeric incident is relatively high in the third-generation sequencing data.
In human-sequencing data obtained using the PacBio platform, the average distance of adjacent chimeras generated increased with the amplification fold, ranging from 2 kb for MDA products amplified from single cells with an amplification fold of 106 to 7 kb for MDA products from multiple cells with an amplification fold of 102. The chimeric density, which is defined as the chimerism events that occur in 1 Mb region of the genome, is calculated using the following formula:
1.5. The processing of MDA chimeras
Under normal circumstances, the presence of the insertion chimeras in the sequencing data is usually considered useful information as these reads can be fully mapped to the reference genome. However, single-end chimeras (both inverted and direct chimeras) are often seen as useless data and are discarded due to their incompatibility with the reference genome. In most bioinformatics studies, chimeras can be filtered after mapping, but sometimes they can cause problems. For example, in the identification of structural variations (SVs) in MDA sequencing data, chimeras can lead to false positive SVs and increase the validation effort by a factor of 200-fold [89]. In constructing contigs and scaffolds in genome assembly of metagenomics of low-biomass samples, chimeras can connect non-contiguous genome regions and make the de novo genome assembly process more challenging [42], [44], [45]. This is especially true in MDA sequencing data with long reads, which have high chimeric rates. Thus, dealing with chimeric reads after detection is a crucial issue for related studies.
To overcome these problems, chimeras can be split into two or more shorter sequences based on the chimeric points and then be transformed to the correct orientation based on their detected strand orientation. After that, these shorter reads can be treated as normal reads and re-mapped to the reference genome for further bioinformatics analysis. This process can significantly improve the utilization of sequencing data, especially for PacBio long-read sequencing data.
The use of IterativeAssembler to process MDA chimeras in Sanger sequencing data of cloned libraries derived from MDA reactions showed that the longest contig was increased from 35.4 kb to 58.3 kb, and the percentage of misassembled contigs dropped from 20 % to 13 % [38]. ChimeraMiner was able to remove 83.82 % of false positive SVs introduced by single-cell MDA and improved the recognition of SVs in single-cell NGS data [41]. By converting chimeras detected by 3rd-ChimeraMiner to normal reads in PacBio MDA data and re-aligning to the reference genome hg19, the full-length mapping ratio was increased from 14.47 % to 79.92 % in single cell-level PacBio sequencing data, with an average 97.77 % inversion of PacBio sequencing data being removed [55].
2. Conclusion
This mini-review offers an overview of chimeras generated during phi29 DNA polymerase-mediated MDA. We first summarize the chimera formation mechanisms and current bioinformatics methods of detecting chimeras in MDA sequencing data. We then systematically explore the characteristics of chimeras, including chimeric distance, overlapping sequence, chimeric rate, and chimeric density in different sequencing platforms and biological samples. Finally, we review the methods for processing MDA chimeras and discuss their impact effect. Despite many challenges, this review provides insights into optimizing MDA conditions, reducing the influence of chimeras in genome sequencing, and improving sequencing data utilization efficiency through chimeras integration in bioinformatics analysis.
Funding
This work was supported by the project BK20211513 of the Natural Science Foundation of Jiangsu Province, the project 61571121 of National Natural Science Foundation of China and the Key Research and Development Project of Jiangsu Province (BE2022804).
CRediT authorship contribution statement
J.T., and Z.L., conceived and designed the manuscript; N.L., performed the bioinformatics analysis, the data statistic and figure plot; N.L., contributed analysis tools; J.T., N.L., and Y.Q, wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of interests
There are no conflicts to declare.
Acknowledgments
We thank the Big Data Center of Southeast University for providing the facility support on the numerical calculations in this paper. We also thank Ms. Qiongdan Zhang from Southeast University for language editing.
Contributor Information
Zuhong Lu, Email: zhlu@seu.edu.cn.
Jing Tu, Email: jtu@seu.edu.cn.
References
- 1.Turner W. The cell theory, past and present. J Anat Physiol. 1890;24:253–287. [PMC free article] [PubMed] [Google Scholar]
- 2.Avery O.T., Macleod C.M., McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type Iii. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang L., Ma F., Chapman A., Lu S., Xie X.S. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genom Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 4.Li G.W., Xie X.S. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 6.Navin N.E. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConnell M.J., et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcy Y., et al. Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navin N., et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gawad C., Koh W., Quake S.R. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 12.Fan X., et al. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22:195. doi: 10.1186/s13059-021-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hård J., et al. Long-read whole genome analysis of human single cells. bioRxiv. 2021;2021 doi: 10.1101/2021.04.13.439527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenger A.M., et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan J.H.J., et al. Experimental and bioinformatics considerations in cancer application of single cell genomics. Comput Struct Biotechnol J. 2021;19:343–354. doi: 10.1016/j.csbj.2020.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telenius H., et al. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 17.Troutt A.B., McHeyzer-Williams M.G., Pulendran B., Nossal G.J. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci USA. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., et al. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., et al. Whole genome amplification from a single cell – implications for genetic-analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blainey P.C. The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 23.Hou Y., et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience. 2015;4 doi: 10.1186/s13742-015-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L., Ma F., Chapman A., Lu S.J., Xie X.S. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genom Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 25.Dean F.B., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garmendia C., Bernad A., Esteban J.A., Blanco L., Salas M. The bacteriophage-Phi-29 DNA-polymerase, a proofreading enzyme. J Biol Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 27.Zhang D.Y., Brandwein M., Hsuih T., Li H.B. Ramification amplification: a novel isothermal DNA amplification method. Mol Diagn. 2001;6:141–150. doi: 10.1054/modi.2001.25323. [DOI] [PubMed] [Google Scholar]
- 28.Lasken R.S. Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol. 2007;10:510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Lasken R.S. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. [DOI] [PubMed] [Google Scholar]
- 30.Xu X., et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao Y., et al. Rapid droplet multiple displacement amplification based on the droplet regeneration strategy. Anal Chim Acta. 2021;1141:173–179. doi: 10.1016/j.aca.2020.10.031. [DOI] [PubMed] [Google Scholar]
- 32.Long N., Qiao Y., Xu Z., Tu J., Lu Z. Recent advances and application in whole-genome multiple displacement amplification. Quant Biol. 2020;8:279–294. doi: 10.1007/s40484-020-0217-2. [DOI] [Google Scholar]
- 33.Gole J., et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Y.S., et al. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proc Natl Acad Sci USA. 2015;112:11923–11928. doi: 10.1073/pnas.1513988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., et al. 1D-reactor decentralized MDA for uniform and accurate whole genome amplification. Anal Chem. 2017;89:10147–10152. doi: 10.1021/acs.analchem.7b02183. [DOI] [PubMed] [Google Scholar]
- 36.Li J., et al. Accurate and sensitive single-cell-level detection of copy number variations by micro-channel multiple displacement amplification (mucMDA) Nanoscale. 2018;10:17933–17941. doi: 10.1039/c8nr04917c. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Navin N.E. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K., et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 39.Lasken R.S., Stockwell T.B. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 2007;7:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu J., et al. Systematic characteristic exploration of the chimeras generated in multiple displacement amplification through next generation sequencing data reanalysis. PLOS One. 2015;10 doi: 10.1371/journal.pone.0139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu N., et al. ChimeraMiner: an improved chimeric read detection pipeline and its application in single cell sequencing. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20081953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigue S., et al. Whole genome amplification and De novo assembly of single bacterial cells. PLOS One. 2009;4 doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chitsaz H., et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol. 2011;29:915–U214. doi: 10.1038/nbt.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nurk S., et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kogawa M., Hosokawa M., Nishikawa Y., Mori K., Takeyama H. Obtaining high-quality draft genomes from uncultured microbes by cleaning and co-assembly of single-cell amplified genomes. Sci Rep. 2018;8 doi: 10.1038/s41598-018-20384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arroyo Mühr L.S., et al. De novo sequence assembly requires bioinformatic checking of chimeric sequences. PLOS One. 2020;15 doi: 10.1371/journal.pone.0237455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lariviere D., Mei H., Freeberg M., Taylor J., Nekrutenko A. Understanding trivial challenges of microbial genomics: an assembly example. bioRxiv. 2018;347625 doi: 10.1101/347625. [DOI] [Google Scholar]
- 48.Voet T., et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013;41:6119–6138. doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Loo P., Voet T. Single cell analysis of cancer genomes. Curr Opin Genet Dev. 2014;24:82–91. doi: 10.1016/j.gde.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Guan P., Sung W.K. Structural variation detection using next-generation sequencing data A comparative technical review. Methods. 2016;102:36–49. doi: 10.1016/j.ymeth.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 51.Kiguchi Y., Nishijima S., Kumar N., Hattori M., Suda W. Long-read metagenomics of multiple displacement amplified DNA of low-biomass human gut phageomes by SACRA pre-processing chimeric reads. DNA Res: Int J rapid Publ Rep Genes Genomes. 2021;28 doi: 10.1093/dnares/dsab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linheiro R., Archer J. CStone: a de novo transcriptome assembler for short-read data that identifies non-chimeric contigs based on underlying graph structure. PLOS Comput Biol. 2021;17 doi: 10.1371/journal.pcbi.1009631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong H., et al. Cas9-based local enrichment and genomics sequence revision of megabase-sized shark IgNAR loci. J Immunol. 2022;208:181–189. doi: 10.4049/jimmunol.2100844. [DOI] [PubMed] [Google Scholar]
- 54.Evrony G.D., et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu N., et al. 3rd-ChimeraMiner: a pipeline for integrated analysis of whole genome amplification generated chimeric sequences using long-read sequencing. bioRxiv. 2022;2013 doi: 10.1101/2022.08.13.503872. [DOI] [Google Scholar]
- 56.Tu J., et al. Systematic characteristic exploration of the chimeras generated in multiple displacement amplification through next generation sequencing data reanalysis. PLOS One. 2015;10 doi: 10.1371/journal.pone.0139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evrony G.D., Lee E., Park P.J., Walsh C.A. Resolving rates of mutation in the brain using single-neuron genomics. eLife. 2016;5 doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evrony G.D., et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kornberg A., Baker T.A. DNA Replication. Wh Freeman; New York: 1992. [Google Scholar]
- 60.Lee C.S., Davis R.W., Davidson N. A physical study by electron microscopy of the terminally repetitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. J Mol Biol. 1970;48:1–22. doi: 10.1016/0022-2836(70)90215-9. [DOI] [PubMed] [Google Scholar]
- 61.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith L.M., Fung S., Hunkapiller M.W., Hunkapiller T.J., Hood L.E. The synthesis of oligonucleotides containing an aliphatic amino group at the 5′ terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis. Nucleic Acids Res. 1985;13:2399–2412. doi: 10.1093/nar/13.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith L.M., et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 64.Fournier, P.-E. & Raoult, D. in Infectious Diseases (Third Edition) (eds Jonathan Cohen, Steven M. Opal, & William G. Powderly) 86–91 (Mosby, 2010).
- 65.Margulies M., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Archer C.T., et al. The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli. BMC Genom. 2011;12:9. doi: 10.1186/1471-2164-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li R.Q., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 69.Kaper F., et al. Whole-genome haplotyping by dilution, amplification, and sequencing. Proc Natl Acad Sci USA. 2013;110:5552–5557. doi: 10.1073/pnas.1218696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ossowski S., et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venu R.C., et al. In: Fungal Genomics: Methods and Protocols. Xu Jin-Rong, H. Bluhm Burton H., editors. Humana Press; 2011. pp. 167–178. [Google Scholar]
- 72.Mardis E.R. DNA sequencing technologies: 2006-2016. Nat Protoc. 2017;12:213–218. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- 73.Marcy Y., et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLOS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baslan T., et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong X., et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat Methods. 2017;14:491. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr arXiv. 2013;1303:3997. [Google Scholar]
- 79.Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kraft F., Kurth I. Long-read sequencing to understand genome biology and cell function. Int J Biochem Cell Biol. 2020;126 doi: 10.1016/j.biocel.2020.105799. [DOI] [PubMed] [Google Scholar]
- 81.Amarasinghe S.L., et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenko Bizjan B., et al. Challenges in identifying large germline structural variants for clinical use by long read sequencing. Comput Struct Biotechnol J. 2020;18:83–92. doi: 10.1016/j.csbj.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quan C., Lu H., Lu Y., Zhou G. Population-scale genotyping of structural variation in the era of long-read sequencing. Comput Struct Biotechnol J. 2022;20:2639–2647. doi: 10.1016/j.csbj.2022.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sedlazeck F.J., et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wenger A.M., et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mantere T., Kersten S., Hoischen A. Long-read sequencing emerging in medical genetics. Front Genet. 2019;10 doi: 10.3389/fgene.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tu J., et al. Hotspot selective preference of the chimeric sequences formed in multiple displacement amplification. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18030492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiao X., et al. Structural alterations from multiple displacement amplification of a human genome revealed by mate-pair sequencing. PLOS One. 2011;6 doi: 10.1371/journal.pone.0022250. [DOI] [PMC free article] [PubMed] [Google Scholar]