Abstract
Duck circovirus genotype 2 (DuCV2) belongs to the genus Circovirus, family Circoviridae. It can generally cause lymphocyte atrophy and necrosis in ducks, which leads to immunosuppression. The function of the DuCV2 open reading frame 3 (ORF3) protein in viral pathogenesis in host cells remains unclear. Therefore, a series of studies based on ORF3 of the isolate DuCV GH01 strain (belonging to DuCV2) were carried out in duck embryo fibroblasts (DEFs) in this study. The results showed that the ORF3 protein could induce nuclear shrinkage and fragmentation in DEFs. Chromosomal DNA breakage was observed by TUNEL assay. The expression levels of caspase-related genes showed that ORF3 primarily promoted caspase 3 and caspase 9 expression. Furthermore, the protein expression levels of cleaved caspase 3 and cleaved caspase 9 in DEFs were enhanced by ORF3. Thus, ORF3 may activate the mitochondrial apoptosis pathway. When the 20 amino acid residues at the C-terminus of ORF3 (ORF3ΔC20) were deleted, the apoptosis rates were decreased. Moreover, compared to ORF3, ORF3ΔC20 downregulated the mRNA levels of cytochrome c (Cyt c), poly ADP-ribose polymerase (PARP) and apoptosis protease activating factor 1 (Apaf-1), which are the key molecules in the mitochondrial apoptotic pathway. Further study showed that ORF3ΔC20 could reduce the mitochondrial membrane potential (MMP). This study suggested that the DuCV2 ORF3 protein may primarily activate apoptosis through the mitochondrial pathway in DEFs, and this function is ORF3 C20 dependent.
Key words: duck circovirus genotype 2, ORF3 protein, DEF, apoptosis, mitochondrial apoptosis pathway
INTRODUCTION
Duck circovirus (DuCV) is a member of the genus Circovirus, family Circoviridae. It is an agent that causes duck immunosuppression by inducing lymphocyte death. This presents an opportunity for mixed infections (Todd, 2010). The DuCV genome is a single-stranded circular DNA with a length of approximately 1990 nt. According to the genomic sequence, DuCV has been divided into 2 genotypes, named DuCV1 and DuCV2 (Zhang et al., 2013). It mainly contains open reading frame 1 (ORF1) and ORF2, which encode Rep (the viral replication protein) and Cap (the viral structural protein), respectively. Another major protein, named ORF3, is encoded by the sequence located on the complementary strand of ORF1 and has been proven by sequence analysis (Xiang et al., 2012). The study also indicated that the DuCV2 ORF3 protein has apoptotic activity in sf9 cells (Xiang et al., 2012). Wu Zhuan-Chang et al. further found that the 20 amino acid residues at the C-terminus of DuCV2 ORF3 have a noncanonical nuclear location signal sequence (NLS) and are involved in the regulation of apoptosis by ORF3 in chicken fibroblast DF-1 cells (Wu et al., 2018). This finding indicates that the ORF3 protein is responsible for the pathogenesis of DuCV2. However, the function and basic molecular mechanism of the DuCV2 ORF3 protein in inducing apoptosis in duck embryo fibroblasts (DEFs), which are host cells of DuCV, have not yet been declared.
Apoptosis is a type of programmed cell death that is an active suicide process regulated by host cell genes in the process of cell growth, development, differentiation, and pathology (Nagata, 2018). The morphological characteristics of apoptosis include cell shrinkage and convolution, nuclear pyknosis, and fragmentation. Among these features, the most typical feature is pyknosis, which is the result of chromatin condensation (Zhang et al., 2018). According to different stimuli, apoptosis can be divided into 2 classical apoptosis signaling pathways: the mitochondrial pathway (intrinsic pathway) and the death receptor pathway (extrinsic pathway). The former is initiated with nonreceptor-mediated stimuli and generates intracellular signals that act directly on targets within the cell. Then, the mitochondrial permeability transition (MPT) pore is formed in the inner mitochondrial membrane, which results in a loss of the mitochondrial membrane potential (MMP) and release of cytochrome c (Cyt c) into the cytoplasm (Bock and Tait, 2020). Poly ADP-ribose polymerase (PARP) is a DNA nick-sensing enzyme involved in enhancing the translocation of Cyt c from mitochondria to the cytoplasm (Zhang et al., 2012). Cyt c can be further combined with caspase 9 and apoptosis protease activating factor 1 (Apaf-1) into a complex, namely, the apoptosome (Zhou et al., 2015; Wu et al., 2016). The death receptor apoptosis pathway is activated by members of the tumor necrosis factor (TNF) receptor superfamily. Subsequently, pro-caspase 8 is recruited and activated as caspase 8 (Tummers and Green, 2017). Both pathways ultimately result in cleavage of caspase 3, leading to DNA fragmentation, nucleoprotein degradation, apoptotic body formation, and final uptake by phagocytes (Jorgensen et al., 2017).
In this study, the activity of DuCV2 ORF3 protein-induced apoptosis in DEFs and its possible apoptosis signaling pathway were explored. ORF3 from the isolate DuCV GH01 strain (belonging to DuCV2) was employed. It was demonstrated that the ORF3 protein could induce apoptosis in DEFs. Moreover, the protein may primarily activate the mitochondrial apoptosis pathway. Specifically, the 20 amino acid residues at the C-terminus (C20) of ORF3 were involved in the induction of apoptosis in DEFs.
MATERIALS AND METHODS
Ethics Approval and Consent to Participate
The usage of duck embryos was approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (Permit Number: 2015-016), China. All methods used in animal experiments were performed according to the guidelines and regulations of the National Institutes of Health.
Cell Culture and Reagents
Duck embryos were purchased from a local duck farm in Sichuan Province, China. DEFs were prepared from 9- to 11-day-old duck embryos. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% newborn bovine serum (NBS, Gibco) at 37°C with 5% CO2.
Construction of ORF3-Related Plasmids
The DuCV2 ORF3 and DuCV2 ORF3ΔC20 fragments of the GH01 strain (GenBank ID: JX499186) containing the Flag gene were amplified by PCR using the ORF3 primers and ORF3ΔC20 primers (see Table 1), respectively. The two fragments were ligated with linearized pCAGGS and then transferred into competent E. coli DH5α cells. The plasmids were named pCAGGS-ORF3 and pCAGGS-ORF3ΔC20, respectively. To detect cellular sublocalization, DuCV2 ORF3 and DuCV2 ORF3ΔC20 were separately amplified by PCR using EGFP-ORF3 or EGFP-ORF3ΔC20 primers (see Table 1) to construct the plasmids pEGFP-ORF3 or pEGFP-ORF3ΔC20. After identification by PCR and restriction enzyme digestion, the positive clones were sequenced by Tsingke Biological Technology, China. Then, the corrected cells were stored at −80°C.
Table 1.
Primers used in this study.
| Target gene | Forward primer sequences | Reverse primer sequences | Accession number |
|---|---|---|---|
| ORF3 | CGGAATTCGCCACCATGGCTTCGCATCGGCGAGCTGGGAG | CCCTCGAGCTACTTATCGTCGTCATCCTTGTAATCCCTTTGAAGATTATGTTCATGTTTTAGAGT | JX499186 |
| ORF3ΔC20 | CGGAATTCGCCACCATGGCTTCGCTTCGGCCAG | CCGCTCGAGCTACTTATCGTCGTCATCCTTGTAATCTCGTCGGCGAGGA | |
| EGFP-ORF3 | CTAGCTAGCGCCACCATGGCTTCGCATCGGCGAG | CCCAAGCTTCCTTTGAAGATTATGTTCATGT | |
| EGFP-ORF3ΔC20 | CTAGCTAGCGCCACCATGGCTTCGCATCGGCGAG | CCCAAGCTTTCGTCGGAGAGGAAAAG | |
| q-caspase-3 | ACCGGACTGTCATCTCGTTC | CTTCACCATGGCTTAGCAAC | XM_005030494.2 |
| q-caspase-8 | ACAAAATGACAAGCCGACCC | TCTACGTCTGTCCCGTTCCG | XM_013094737.1 |
| q-caspase-9 | CTTTCCAGACTCCCTCGGGTA | CTTCCCCTTGGCAGATACGG | XM_013095294.1 |
| q-Apaf-1 | AGAGGGCACAAGGAAGCTATCA | AACTTACTACCATCAGGCGAAACA | XM_005024525.2 |
| q-Cyt c | TGAGGATACCCTGATGGAGTACTTG | CTTCGCAGTGGCATCTTTCAG | XM_005013559.2 |
| q-PARP | CTGGACTAAGTGCGTTGC | AGCCTCTGGAGGGAATA | XM_005014743 |
| q-β-actin | GATCACAGCCCTGGCACC | CGGATTCATACTCCTGCTT | NM_001310421.1 |
Sequences underlined are the protective bases. Restriction enzyme sites are in bold black (GAATTC: EcoR I; CTCGAG: Xho I; GCTAGC: Nhe I; AAGCTT: Hind III). Underlined bold black italics are Kozak sequences, and bold black italics are Flag sequences. q-: target genes for qRT-PCR measurement.
Western Blotting
The plasmids pCAGGS and pCAGGS-ORF3 were separately transfected into DEFs according to the manufacturer's instructions for Lipofectamine 3000 (Invitrogen, CA). The cells were collected and treated with 200 μL radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) containing 2 μL PMSF (Beyotime) at 48 h post transfection to detect ORF3 protein expression. To measure cleaved caspase 3 and cleaved caspase 9, cells were collected at 24 h, 48 h, and 72 h post transfection. Then, they were centrifuged at 12,000 rpm for 10 min at 4°C. The cell lysates were collected and separated by SDS‒PAGE, and then the gel was transferred to a PVDF membrane at 250 mA for 1 h on ice. The membrane was blocked with 5% nonfat milk for 1 h at room temperature. Then, anti-DYKDDDDK mouse monoclonal antibody (1:5,000, TransGen Biotech, Beijing, China), anti-cleaved caspase 3 rabbit monoclonal antibody (1:1,000, CST, Boston, MA), or anti-cleaved caspase 9 rabbit monoclonal antibody (1:1,000, CST) was used as the primary antibody, and HRP-conjugated goat anti-mouse IgG (1:5,000, Proteintech, Rosemont, IL) or HRP-conjugated goat anti-rabbit IgG (1:4,000, Proteintech) was used as the secondary antibody at room temperature for 1 h. Clarity Western ECL Substrate (Bio-Rad, CA) was used for membrane imaging.
Nuclear Morphological Change Detection
The plasmids pCAGGS and pCAGGS-ORF3 were separately transfected into DEFs in 6-well plates. The cells were collected at 24 h, 48 h, and 72 h post transfection. Then, DAPI (1:100, Solarbio, Beijing, China) was used to stain the cells in the dark for 15 min at room temperature. Subsequently, the nuclei were observed under an inverted fluorescence microscope (Nikon).
Detection of DNA Fragmentation by TUNEL Assay
The plasmids pCAGGS and pCAGGS-ORF3 were transfected into DEFs as mentioned above. The cells were fixed with 4% paraformaldehyde at 24 h, 48 h, or 72 h post transfection for 1 h at room temperature. Then, they were permeabilized with 0.25% Triton for 10 min. The DNA fragmentation of the cells was detected by a TUNEL kit (Beyotime) according to the manufacturer's instructions. Fluorescence images were obtained by an inverted fluorescence microscope (Nikon).
Subcellular Localization Detection
The plasmids pEGFP-N1, pEGFP-ORF3, and pEGFP-ORF3ΔC20 were separately cotransfected with the pDsRed-mito plasmid. The cells were collected at 48 h post transfection to detect the subcellular localization of ORF3 or ORF3ΔC20. The cells were fixed with 4% paraformaldehyde at 4°C overnight. The cells were washed with PBS and then stained with DAPI (10 μg/mL, Solarbio) for 15 min at room temperature. After washing with PBS 3 times, the slides were imaged with a fluorescence microscope in inverted microscope configurations (Nikon).
Measurement of Apoptosis Rates by Flow Cytometry
The plasmid pCAGGS, pCAGGS-ORF3, or pCAGGS-ORF3ΔC20 was transfected into DEFs as previously described. The cells were collected at 48 h post transfection and detected by flow cytometry according to the manufacturer's instructions of the Annexin V-FITC/PI Apoptosis Kit (BD, NY). In brief, the DEFs were resuspended at 48 h after transfection. Annexin V-FITC (1:100 dilution) and PI (1:100 dilution) were added to the cells and mixed gently. The cells were incubated with the dyes in the dark for 15 min at room temperature. The apoptotic cells were then monitored by an LSRFortessa (BD).
Quantitative Real-Time PCR
DEFs were seeded into 6-well plates. The cells were transfected with different plasmids (pCAGGS, pCAGGS-ORF3 or pCAGGS-ORF3ΔC20) when they were grown to approximately 80%-90% confluence. Cells were collected for RNA extraction at 24 h, 48 h and 72 h post transfection. The RNAs were reverse transcribed into cDNA using a PrimeScript RT Reagent kit with gDNA Eraser (TaKaRa, Shiga, Japan). The primers for qRT‒PCR analysis are listed in Table 1. qRT‒PCR was performed according to the manufacturer's instructions for TB Green Premix Ex Taq (Tli RNaseH Plus) (TaKaRa) as follows: predenaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s and annealing/extension at 60°C for 30 s. The relative gene expression levels were calculated by 2−ΔΔCt.
Measurement of Mitochondrial Membrane Potential by JC-10 Assay
After DEFs were transfected with pCAGGS, pCAGGS-ORF3, or pCAGGS-ORF3ΔC20, the cells were collected at 24 h, 48 h or 72 h post transfection. The DEFs were treated according to the manufacturer's instructions for the JC-10 staining kit (Solarbio). Briefly, the cells were washed with PBS 3 times. Then, they were incubated with 200 μL JC-10 dye per well at 37°C for 20 min in the dark. Subsequently, JC-10 buffer was added to wash the cells 3 times. Fluorescence images were observed using an upright fluorescence microscope (Nikon). The OD value of each well was measured at 529 nm for monomers and at 590 nm for aggregates by a multifunctional microplate reader (Bio-Rad).
Statistical Analysis
The data are presented as the means ± SDs. Statistical significance was assessed using unpaired Student's t test by GraphPad Prism (version 8.0). The significant differences are annotated as *P < 0.05, ⁎⁎P < 0.01, and ⁎⁎⁎P < 0.001.
RESULTS
DuCV2 ORF3 Induces Apoptosis in DEFs
To investigate whether DuCV2 ORF3 induces apoptosis of DEFs, the recombinant plasmid pCAGGS-ORF3 was constructed. As shown in Figure 1A, a specific band with the expected size (approximately 12 kDa) was detected by western blotting, which indicated that ORF3 could be successfully expressed in DEFs after plasmid transfection. Subsequently, DNA fragmentation, one of the most specific findings in apoptosis, was detected by TUNEL assay (Chenlo et al., 2014). As shown in Figure 1B, positivity in the TUNEL assay was observed in DEFs after pCAGGS-ORF3 plasmid transfection. Moreover, DuCV2 ORF3 promoted DEF apoptosis in a time-dependent manner (Figure 1B). The above conclusion was also found by detecting cell and cell nuclear morphological changes. Compared with DEFs transfected with pCAGGS, cells transfected with pCAGGS-ORF3 exhibited significant cell fragmentation and nuclear fragmentation from 48 h to 72 h post transfection, as shown in Figure S1 and Figure 1C.
Figure 1.
DuCV2 ORF3 promoted apoptosis in DEFs. (A) The expression of recombinant ORF3 protein was detected by western blotting. (B) Apoptosis (green) was measured by TUNEL assay at 24 h, 48 h, and 72 h after plasmid pCAGGS-ORF3 or pCAGGS transfection. Scale bars, 100 μm. (C) Morphological changes in the nucleus were measured by DAPI from 0 h to 72 h post-plasmid transfection. Dotted arrows indicate nuclear fragmentation.
The Mitochondrial Pathway of Apoptosis Was Stimulated by DuCV2 ORF3
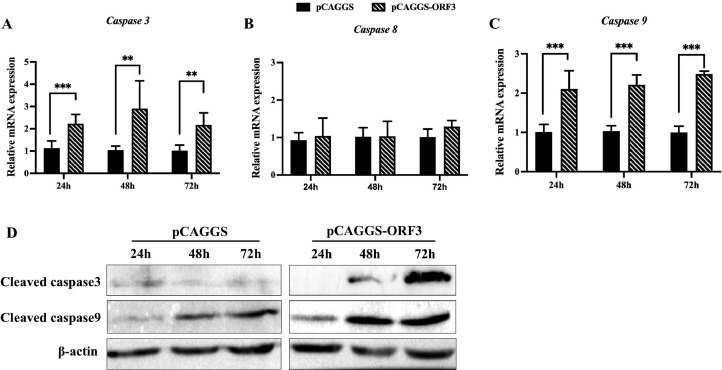
Caspase 3 is activated by both the mitochondrial apoptosis pathway and the death receptor apoptosis pathway (Asadi et al., 2022). Here, the expression levels of caspase 3 were measured at different times after transfection of DEFs with plasmid pCAGGS-ORF3 or pCAGGS. The results showed that ORF3 might stimulate canonical apoptosis signaling throughout the experimental period (Figure 2A). Caspase 8 and caspase 9, essential initiating caspases separately required for apoptotic signaling via the death receptor pathway and mitochondrial pathway (Chen et al., 2007; Mandal et al., 2020), were employed to define the DuCV2 ORF3-induced apoptotic pathway. The ORF3 protein was unable to stimulate the expression of caspase 8 (Figure 2B), and it could significantly upregulate the expression of caspase 9 (P < 0.001, Figure 2C) at 24 h, 48 h and 72 h after plasmid pCAGGS-ORF3 transfection. Subsequently, cleaved caspase 3 and cleaved caspase 9 were detected by western blotting. The results showed that under ORF3 stimulation, the protein expression levels of cleaved caspase 3 and cleaved caspase 9 were enhanced in a time-dependent manner (Figure 2D). Thus, we suspected that DuCV2 ORF3 induces apoptosis in DEFs mainly through the mitochondrial pathway.
Figure 2.
ORF3 activated mitochondrial apoptosis. (A–C) The relative mRNA expression levels of caspase 3 (A), caspase 8 (B), and caspase 9 (C). DEFs were transfected with the pCAGGS-ORF3 or pCAGGS plasmid. Total cellular RNA was extracted at 24 h, 48 h, and 72 h post transfection, and the target genes were analyzed by qRT‒PCR. Mean values ± SDs (n = 3–6) are shown. ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001. (D) The measurement of cleaved caspase 3 and cleaved caspase 9 by western blotting at different time points post pCAGGS-ORF3 or pCAGGS plasmid transfection.
DuCV2 ORF3 C20 Was Involved in the Regulation of DEF Apoptosis
According to the genome, DuCV has been divided into 2 genotypes, named DuCV1 and DuCV2 (Fu et al., 2011). Compared with the DuCV1 ORF3 protein, the C-terminus of the DuCV2 ORF3 protein has 20 more amino acid residues, which is a noncanonical NLS of DuCV2 ORF3 in DF-1 cells (Wu et al., 2018). However, DuCV2 ORF3 was distributed in the cytoplasm of DEFs with or without C20, indicating that C20 did not affect the localization of DuCV2 ORF3 in DEFs (Figure 3A). Nonetheless, C20 affected the proapoptotic role of ORF3 in DEFs, as indicated in Figure 3B. The cells were stained with an Annexin V-FITC/PI apoptosis kit (BD) at 48 h post plasmid (pCAGGS, pCAGGS-ORF3 or pCAGGS-ORF3ΔC20) transfection, and apoptotic rates were measured by flow cytometry. The proportion of apoptotic cells stimulated by both ORF3 and ORF3ΔC20 was significantly higher than the proportion of cells stimulated by the pCAGGS vehicle (9.66% ± 1.00%, P < 0.01, Figure 3B). Moreover, the percentages of apoptotic cells (including early apoptotic cells and late apoptotic cells) in the pCAGGS-ORF3 and pCAGGS-ORF3ΔC20 groups were 20.11% ± 4.58% and 13.44% ± 1.19%, respectively (Figure 3B). The apoptosis rate of the pCAGGS-ORF3 group was higher than that of the pCAGGS-ORF3ΔC20 group (P = 0.0504, Figure 3B). The above results indicated that DuCV2 ORF3 and ORF3ΔC20 could induce apoptosis in DEFs and that ORF3 C20 was involved in the regulation of apoptosis in DEFs.
Figure 3.
The effect of C20 on ORF3-induced apoptosis in DEFs. (A) The subcellular localization of ORF3-related proteins was detected by immunofluorescence assay. The DuCV2 ORF3 and ORF3ΔC20 genes were ligated into plasmid pEGFP-N1. The subcellular localization of ORF3 or ORF3ΔC20 protein was stained with EGFP. The cytoplasm and nucleus were stained with pDsRed-mito proteins and DAPI, respectively. (B) The apoptosis rates of DEFs were measured by flow cytometry. DEFs were transfected with pCAGGS, pCAGGS-ORF3 or pCAGGS-ORF3ΔC20. The cells were collected 48 h after transfection. Annexin V-FITC (1:100 dilution) and PI (1:100 dilution) were used to stain the cells, which were monitored by flow cytometry. Mean values ± SDs (n = 4) are shown. ⁎⁎P < 0.01.
Mitochondrial Apoptotic Capacity of DuCV2 ORF3 Was Dependent on its C20
To further assess the role of the ORF3 C20 protein in the ORF3-induced mitochondrial apoptosis pathway, the mRNA levels of Cyt c, PARP, and Apaf-1 were detected. The expression levels of Cyt c induced by ORF3 were upregulated significantly compared with those induced by ORF3ΔC20 at 24 h post transfection (P < 0.01, Figure 4A). The expression levels of PARP and Apaf-1 gradually increased in the pCAGGS-ORF3 group during the experimental period and peaked at 72 h post transfection (Figure 4A). PARP and Apaf-1 were only weakly expressed in the pCAGGS-ORF3ΔC20 group (Figure 4A). This result suggested that ORF3 C20 plays a role in promoting the expression of the PARP and Apaf-1 genes.
Figure 4.
C20 was involved in regulating the mitochondrial apoptosis induced by DuCV2 ORF3 in DEFs. DEFs were transfected with pCAGGS, pCAGGS-ORF3 or pCAGGS-ORF3ΔC20. The cells were collected at 24 h, 48 h, and 72 h post transfection to (A) measure the expression levels of Cyt c, PARP, and Apaf-1 by qRT‒PCR; (B) detect MMP by JC-10; and (C) analyze the ratio of monomers/aggregates. Scale bars, 50 μm. Mean values ± SDs (n = 3–6) are shown. *P < 0.05, ⁎⁎P < 0.01, ⁎⁎⁎P < 0.001.
Reduced mitochondrial membrane potential (MMP) is one of the hallmarks of mitochondrial apoptosis pathway activation (Lee et al., 2020). To further determine whether ORF3 C20 affected the ORF3-induced mitochondrial apoptosis pathway, MMP in DEFs was detected by JC-10 assay (Yang et al., 2021). JC-10 dye is concentrated in the mitochondrial matrix, with aggregates forming in normal cells, and fluoresces red. In apoptotic cells, JC-10 enters the cytoplasm as a monomer and emits green fluorescence. The green fluorescence of DEF cells in the pCAGGS-ORF3 group was stronger than that in the pCAGGS-ORF3ΔC20 group (Figure 4B). The ratio of JC-10 monomers to aggregates in DEFs transfected with pCAGGS-ORF3 was significantly higher than that in DEFs transfected with pCAGGS-ORF3ΔC20 at 48 and 72 h after transfection (P < 0.01, Figure 4C). This result indicated that DuCV2 ORF3 was dependent on its C20 in the induction of mitochondrial apoptosis in DEFs.
DISCUSSION
Epidemiological investigations have found that DuCV coinfects waterfowl with other viruses and has shown an increasing trend in recent years. DuCV infection leads to atrophy and necrosis of lymphocytes, which provides conditions for the invasion of other viruses. Since DuCV cannot be cultured in vitro, studies on the pathogenic mechanism of DuCV are very limited. Previous studies have mainly focused on DuCV Cap (structural protein) and Rep (replication-related protein), while studies on the ORF3 protein are relatively limited. The present study found that DuCV2 ORF3 was directly related to the apoptotic capacity of DEFs, which is consistent with the function of chicken anemia virus (CAV) ORF3 (named apoptin) and porcine circovirus (PCV) (Hough et al., 2015; Kucharski et al., 2016). Therefore, ORF3 is an important protein in the pathogenesis of DuCV2.
The ORF3 proteins of different circoviruses can induce apoptosis through different apoptotic pathways. CAV apoptin phosphorylates Nurr77 protein, thereby regulating the release of Apaf-1 protein, reducing mitochondrial membrane potential, and further activating caspase 9 and caspase 3 to induce the mitochondrial apoptosis pathway (Maddika et al., 2005; Chaabane et al., 2014; Zhang et al., 2021). PCV1 ORF3 protein induced caspase-independent apoptosis and cleavage of PARP in 293T cells (Chaiyakul et al., 2010). In addition, PCV2 ORF3 protein could activate caspase 3 and caspase 8 through the death receptor apoptosis pathway in PK15 cells (Liu et al., 2005; Xu et al., 2020). This finding indicates that the apoptosis pathway induced by ORF3 is related not only to circovirus species but also to circovirus genotype. Moreover, PCV2 ORF3 induces apoptosis through a p53-dependent pathway, not a caspase 8-dependent pathway, in melanoma cells and mouse primary splenocytes (Teras et al., 2018). Here, it was found that the DuCV2 ORF3 protein could 1) reduce the MMP of DEFs; 2) upregulate the expression of Cyt c, PARP, and Apaf-1; and 3) increase the expression of caspase 9, caspase 3, cleaved caspase 9, and cleaved caspase 3. Therefore, we speculated that the DuCV2 ORF3 protein induced apoptosis in DEFs through the mitochondrial pathway, which was consistent with the apoptosis pathway induced by CAV apoptin in human tumor cells (Zhang et al., 2021).
Researchers have found that there is a nonclassical NLS region within the C-terminal 20 amino acids of the DuCV2 ORF3 protein, which can regulate the localization of the DuCV2 ORF3 protein in the nucleus of DF-1 and inhibit the apoptosis-inducing activity of the protein (Wu et al., 2018). To this end, C20 of the DuCV2 ORF3 was truncated to study the apoptotic effect of ORF3 C20 in DEFs. Both proteins (DuCV2 ORF3 and DuCV2 ORF3ΔC20) were mainly localized in the cytoplasm. Furthermore, ORF3ΔC20 reduced the apoptosis of DEFs compared to ORF3, which indicated that C20 is an important functional part of DuCV2 ORF3 in the induction of apoptosis in DEFs. The reason for the different subcellular localization of the DuCV2 ORF3ΔC20 protein in DF-1 and DEF cells is presumed to be affected by cell type. Studies have shown that the subcellular localization of CAV apoptin varies in different cell lines. In most human tumor cell lines, CAV apoptin is mainly localized in the nucleus (Danen-Van Oorschot et al., 2003; Noteborn, 2009). In human nontumor cells, it is mainly localized in the cytoplasm (Poon et al., 2005; Heckl et al., 2008). These features could also be found with the PCV ORF3 protein (Hough et al., 2015). Therefore, we speculated that the DuCV2 ORF3 protein may also be similar to CAV apoptin and PCV ORF3, that is, the cellular sublocalization varies with different cell types. In addition, subcellular localization may be one of the main factors affecting the apoptosis-inducing activity of the protein (Schat, 2009). The apoptotic activity of CAV apoptin is stronger in human tumor cell lines than in nontumor cells (Heilman et al., 2006). Thus, DuCV2 ORF3 had weaker apoptotic activity in DEFs (Figure 3B) than in DF-1 cells (Wu et al., 2018), which ultimately led to differences in the regulation of the apoptotic capacity of the ORF3 C20 proteins. Additionally, we hypothesize that when ORF3 is localized in the cytoplasm, the length of the ORF3 protein positively correlates with the strength of the protein to induce apoptosis.
In addition, the phosphorylation site of the protein may be another major factor affecting the apoptosis-inducing activity of the protein (Liu et al., 2007; Schat, 2009). CAV apoptin is phosphorylated in tumor cells but not in nontumor cells when studying its apoptosis mechanism (Rohn et al., 2002; Malla et al., 2020). It has been reported that the Thr108 residue of CAV apoptin is phosphorylated in tumor cells, allowing it to localize to the nucleus (Lai et al., 2017). If Thr108 is dephosphorylated, Thr107 phosphorylation compensates for this effect (Lanz et al., 2012). However, CAV apoptin was not phosphorylated at Thr108 in nontumor cells and could not inhibit the activity of the chromosomal region maintenance 1 (CRM1) protein in the nucleus, allowing the protein to enter the cytoplasm (Poon et al., 2005). Further studies showed that phosphorylation at the Thr56 and Thr61 residues of apoptin during CAV replication mediates checkpoint kinase 1/2 (Chk1/2) protein to regulate its nuclear localization and apoptotic activity (Kucharski et al., 2016; Feng et al., 2020). Therefore, phosphorylation of the ORF3 protein may be one of the main factors affecting its localization and biological function. We predicted the phosphorylation sites of the DuCV2 ORF3 protein using NetPhos3.1 online analysis software. The DuCV2 ORF3 (GH01 strain) protein has a total of 10 potential phosphorylation sites (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1), which are Ser2, Tyr12, Ser19, Thr23, Ser26, Ser55, Ser61, Tyr69, Ser71, and Thr89. There is a potential phosphorylation site, Thr89, at C20 of the DuCV2 ORF3 protein. We speculate that the phosphorylation site of Thr89 of the DuCV2 ORF3 C20 protein may be the main phosphorylation site of the ORF3 protein to exert a proapoptotic function. However, the exact mechanism remains to be proven.
In conclusion, the DuCV2 ORF3 protein can induce apoptosis in DEFs through the mitochondrial pathway. We found that the ORF3 C20 protein did not affect the distribution of the DuCV2 ORF3 protein in DEFs but could be involved in the apoptotic activity of the protein in DEFs. This study provides theoretical support for further exploration of the pathogenic mechanism of DuCV2.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (32172833/31872475), Natural Science Foundation of Sichuan Province (2022NSFSC0078/2022NSFSC0079), earmarked fund for China Agriculture Research System of MOF and MARA, and Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2021-18).
DISCLOSURES
The authors declare no competing interests.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102533.
Appendix. Supplementary materials
REFERENCES
- Asadi M., Taghizadeh S., Kaviani E., Vakili O., Taheri-Anganeh M., Tahamtan M., Savardashtaki A. Caspase-3: structure, function, and biotechnological aspects. Biotechnol. Appl. Biochem. 2022;69:1633–1645. doi: 10.1002/bab.2233. [DOI] [PubMed] [Google Scholar]
- Bock F.J., Tait S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020;21:85–100. doi: 10.1038/s41580-019-0173-8. [DOI] [PubMed] [Google Scholar]
- Chaabane W., Cieslar-Pobuda A., El-Gazzah M., Jain M.V., Rzeszowska-Wolny J., Rafat M., Stetefeld J., Ghavami S., Los M.J. Human-gyrovirus-apoptin triggers mitochondrial death pathway-nur77 is required for apoptosis triggering. Neoplasia. 2014;16:679–693. doi: 10.1016/j.neo.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyakul M., Hsu K., Dardari R., Marshall F., Czub M. Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus. J. Virol. 2010;84:11440–11447. doi: 10.1128/JVI.01030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Guerrero A.D., Huang L., Shabier Z., Pan M., Tan T.H., Wang J. Caspase-9-induced mitochondrial disruption through cleavage of anti-apoptotic BCL-2 family members. J. Biol. Chem. 2007;282:33888–33895. doi: 10.1074/jbc.M702969200. [DOI] [PubMed] [Google Scholar]
- Chenlo P.H., Curi S.M., Pugliese M.N., Ariagno J.I., Sardi-Segovia M., Furlan M.J., Repetto H.E., Zeitler E., Cohen M., Mendeluk G.R. Fragmentation of sperm DNA using the TUNEL method. Actas Urol. Esp. 2014;38:608–612. doi: 10.1016/j.acuro.2014.02.022. [DOI] [PubMed] [Google Scholar]
- Danen-Van Oorschot A.A., Zhang Y.H., Leliveld S.R., Rohn J.L., Seelen M.C., Bolk M.W., Van Z.A., Erkeland S.J., Abrahams J.P., Mumberg D., Noteborn M.H. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J. Biol. Chem. 2003;278:27729–27736. doi: 10.1074/jbc.M303114200. [DOI] [PubMed] [Google Scholar]
- Feng C., Liang Y.K., Teodoro J.G. The role of apoptin in chicken anemia virus replication. Pathogens. 2020;9:294. doi: 10.3390/pathogens9040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G.H., Shi S.H., Huang Y., Cheng L.F., Peng C.X., Wan C.H., Chen H.M., Lin F., Lin J.S. Genetic diversity and genotype analysis of duck circovirus. Avian Dis. 2011;55:311–318. doi: 10.1637/9584-102010-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Heckl S., Regenbogen M., Sturzu A., Gharabaghi A., Feil G., Beck A., Echner H., Nagele T. Value of apoptin's 40-amino-acid C-terminal fragment for the differentiation between human tumor and non-tumor cells. Apoptosis. 2008;13:495–508. doi: 10.1007/s10495-007-0174-5. [DOI] [PubMed] [Google Scholar]
- Heilman D.W., Teodoro J.G., Green M.R. Apoptin nucleocytoplasmic shuttling is required for cell type-specific localization, apoptosis, and recruitment of the anaphase-promoting complex/cyclosome to PML bodies. J. Virol. 2006;80:7535–7545. doi: 10.1128/JVI.02741-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough K.P., Rogers A.M., Zelic M., Paris M., Heilman D.W. Transformed cell-specific induction of apoptosis by porcine circovirus type 1 viral protein 3. J. Gen. Virol. 2015;96:351–359. doi: 10.1099/vir.0.070284-0. [DOI] [PubMed] [Google Scholar]
- Jorgensen I., Rayamajhi M., Miao E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017;17:151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski T.J., Ng T.F., Sharon D.M., Navid-Azarbaijani P., Tavassoli M., Teodoro J.G. Activation of the chicken anemia virus apoptin protein by Chk1/2 phosphorylation is required for apoptotic activity and efficient viral replication. J. Virol. 2016;90:9433–9445. doi: 10.1128/JVI.00936-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai G.H., Lien Y.Y., Lin M.K., Cheng J.H., Tzen J.T.C., Sun F.C., Lee M.S., Chen H.J., Lee M.S. VP2 of chicken anaemia virus interacts with apoptin for downregulation of apoptosis through de-phosphorylated threonine 108 on. Apoptin. Sci. Rep. 2017;7:14799. doi: 10.1038/s41598-017-14558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz H.L., Florea B.I., Noteborn M.H.M., Backendorf C. Development and application of an in vitro apoptin kinase assay. Anal. Biochem. 2012;421:68–74. doi: 10.1016/j.ab.2011.10.030. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Kalimuthu K., Park Y.S., Luo X., Choudry M.H.A., Bartlett D.L., Lee Y.J. BAX-dependent mitochondrial pathway mediates the crosstalk between ferroptosis and apoptosis. Apoptosis. 2020;25:625–631. doi: 10.1007/s10495-020-01627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chen I., Kwang J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 2005;79:8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhu Y., Chen I., Lau J., He F., Lau A., Wang Z.L., Karuppannan A.K., Kwang J. The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection. J. Virol. 2007;81:9560–9567. doi: 10.1128/JVI.00681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddika S., Booy E.P., Johar D., Gibson S.B., Ghavami S., Los M. Cancer-specific toxicity of apoptin is independent of death receptors but involves the loss of mitochondrial membrane potential and the release of mitochondrial cell-death mediators by a Nur77-dependent pathway. J. Cell Sci. 2005;118(Pt 19):4485–4493. doi: 10.1242/jcs.02580. [DOI] [PubMed] [Google Scholar]
- Malla W.A., Arora R., Khan R.I.N., Mahajan S., Tiwari A.K. Apoptin as a tumor-specific therapeutic agent: current perspective on mechanism of action and delivery systems. Front. Cell Dev. Biol. 2020;8:524. doi: 10.3389/fcell.2020.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R., Barron J.C., Kostova I., Becker S., Strebhardt K. Caspase-8: the double-edged sword. Biochim. Biophys. Acta Rev. Cancer. 2020;1873 doi: 10.1016/j.bbcan.2020.188357. [DOI] [PubMed] [Google Scholar]
- Nagata S. Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 2018;36:489–517. doi: 10.1146/annurev-immunol-042617-053010. [DOI] [PubMed] [Google Scholar]
- Noteborn M.H.M. Proteins selectively killing tumor cells. Eur. J. Pharmacol. 2009;625:165–173. doi: 10.1016/j.ejphar.2009.06.068. [DOI] [PubMed] [Google Scholar]
- Poon I.K., Oro C., Dias M.M., Zhang J.P., Jans D.A. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 2005;65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- Rohn J.L., Zhang Y.H., Aalbers R.I., Otto N., Den Hertog J., Henriquez N.V., Van De Velde C.J., Kuppen P.J., Mumberg D., Donner P., Noteborn M.H. A tumor-specific kinase activity regulates the viral death protein Apoptin. J. Biol. Chem. 2002;277:50820–50827. doi: 10.1074/jbc.M208557200. [DOI] [PubMed] [Google Scholar]
- Schat K.A. Chicken anemia virus. Curr. Top. Microbiol. Immunol. 2009;331:151–183. doi: 10.1007/978-3-540-70972-5_10. [DOI] [PubMed] [Google Scholar]
- Teras M., Viisileht E., Pahtma-Hall M., Rump A., Paalme V., Pata P., Pata I., Langevin C., Boudinot S.R. Porcine circovirus type 2 ORF3 protein induces apoptosis in melanoma cells. BMC Cancer. 2018;18:1237. doi: 10.1186/s12885-018-5090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D. Circoviruses: immunosuppressive threats to avian species: a review. Avian. Pathol. 2010;29:373–394. doi: 10.1080/030794500750047126. [DOI] [PubMed] [Google Scholar]
- Tummers B., Green D.R. Caspase-8: regulating life and death. Immunol. Rev. 2017;277:76–89. doi: 10.1111/imr.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.C., Lee S., Malladi S., Chen M.D., Mastrandrea N.J., Zhang Z.W., Bratton S.B. The Apaf-1 apoptosome induces formation of caspase-9 homo- and heterodimers with distinct activities. Nat. Commun. 2016;7:13565. doi: 10.1038/ncomms13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.C., Zhang R.H., Li Y.M., Shao D.H., Chen H., Jiang S.J., Ma Z.Y., Wang X. C-terminal 20 residues of ORF3 protein of duck circovirus genotype 2 regulates the nuclear localization and inhibits apoptotic activity of ORF3 protein. Vet. Microbiol. 2018;214:21–27. doi: 10.1016/j.vetmic.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Xiang Q.W., Wang X., Xie Z.J., Sun Y.N., Zhu Y.L., Wang S.J., Liu H.J., Jiang S.J. ORF3 of duck circovirus: a novel protein with apoptotic activity. Vet. Microbiol. 2012;159:251–256. doi: 10.1016/j.vetmic.2012.03.045. [DOI] [PubMed] [Google Scholar]
- Xu Y.L., Zheng J.G., Sun P.P., Guo J.H., Zheng X.Z., Sun Y.G., Fan K.H., Yin W., Li H.Q., Sun N. Cepharanthine and Curcumin inhibited mitochondrial apoptosis induced by PCV2. BMC Vet. Res. 2020;16:345. doi: 10.1186/s12917-020-02568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M.Y., Wang L., Ni M., Neuber B., Wang S.M., Gong W.J., Sauer T., Schubert M.L., Huckelhoven-Krauss A., Xia R.X., Ge J., Kleist C., Eckstein V., Sellner L., Muller-Tidow C., Dreger P., Schmitt M., Schmitt A. Dual effects of cyclooxygenase inhibitors in combination with CD19.CAR-T cell immunotherapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.670088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.J., Lau S.S., Monks T.J. A dual role for poly (ADP-Ribose) Polymerase-1 during caspase-dependent apoptosis. Toxicol. Sci. 2012;128:103–114. doi: 10.1093/toxsci/kfs142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.T., Zhang X.C., Cheng A.C., Wang M.S., Yin Z.Q., Huang J., Jia R.Y. Apoptosis triggered by ORF3 proteins of the circoviridae family. Front. Cell Infect. Microbiol. 2021;10 doi: 10.3389/fcimb.2020.609071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Y., Chen X., Gueydan C., Han J.H. Plasma membrane changes during programmed cell deaths. Cell Res. 2018;28:9–21. doi: 10.1038/cr.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Jia R.Y., Lu Y.Y., Wang M.S., Zhu D.K., Chen S., Yin Z.Q., Chen X.Y., Cheng A.C. Identification, genotyping, and molecular evolution analysis of duck circovirus. Gene. 2013;529:288–295. doi: 10.1016/j.gene.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Zhou M.Y., Li Y.N., Hu Q., Bai X.C., Huang W.Y., Yan C.Y., Scheres S.H.W., Shi Y.G. Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev. 2015;29:2349–2361. doi: 10.1101/gad.272278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.