Abstract
The DNA microarray-based analysis of single nucleotide polymorphisms (SNPs) is important for the correlation of genetic variations and individual phenotypes, and for locating disease-causing genes. To facilitate the development of surfaces suitable for immobilization of oligonucleotides, we report here a novel method for the surface immobilization of DNA using pre-fabricated polyamidoamine (PAMAM) starburst dendrimers as mediator moieties. Dendrimers containing 64 primary amino groups in their outer sphere are covalently attached to silylated glass supports and, subsequently, the dendritic macromolecules are modified with glutaric anhydride and activated with N-hydroxysuccinimide. As a result of the dendritic PAMAM linker system the surfaces reveal both a very high immobilization efficiency for amino-modified DNA-oligomers, and also a remarkable high stability during repeated regeneration and re-using cycles. The performance of dendrimer-based DNA microarrays in the discrimination of SNPs is demonstrated.
INTRODUCTION
To elucidate the correlations between genetic variation and clinical phenotypic differences, the detection and analysis of single nucleotide polymorphisms (SNPs) is, for instance, of paramount interest for drug development. SNP-based pharmacogenomics will optimize the individualization of clinical therapy (1), such as autoimmune (2) and cardiovascular diseases (3), type II diabetes mellitus (4) and drug response in asthmatic therapies (5). To take advantage of the enormous potential of SNP databases, large numbers of SNP loci must be screened simultaneously and, thus, a reliable high-throughput method with the ability for automation is required. Besides the MALDI–TOF mass spectrometry (6–8), DNA microarrays have the potential to meet these demands (9). For example, the microarray-based analysis of several thousands of SNPs has recently been carried out (10–15). It is well established that the protocols employed for the immobilization of pre-fabricated nucleic acids largely affect the performance of the microarray. For instance, the binding capacity of the arrays surface can be increased significantly by the use of appropriate linker systems (16–18). Most commonly, the automated deposition of nucleic acids on amino-terminated surfaces, such as 3-aminopropyltriethoxysilane (APTS) or poly-l-lysine (PLL)-coated slides, is applied to generate microarrays. The use of such slides requires fixation steps, such as baking at elevated temperature or irradiation with UV light, leading to the formation of strong ionic interactions or a varying number of covalent bonds, respectively, between the surface and the DNA oligomers (19). A general problem is often associated with the limited stability of such arrays, leading to interference with stringent hybridization conditions and frequent regeneration steps. Furthermore, the spots of such surfaces often reveal inhomogeneous signal distributions (20). Thus, robust and homogenously chemically activated surfaces are required which allow for the covalent immobilization of oligomer probes. We have recently reported on dendritic linker systems attached to silica surfaces, enabling the highly efficient immobilization of amino-modified nucleic acids. These surfaces contain a thin layer of covalently immobilized and cross-linked polyamidoamine (PAMAM) starburst dendrimers (21). PAMAM dendrimers, initially developed by Tomalia et al. in the early 1980s (22), provide a high density of terminal amino groups at the outer sphere. We describe here the three-step preparation of a new type of dendrimer surface, containing non-cross-linked PAMAM moieties. The investigation of the surfaces for microarray production revealed a significantly increased binding capacity, highly homogeneous oligomer spots as well as a remarkable stability against regeneration procedures. Moreover, the dendrimer-based DNA microarrays were used to reliably discriminate SNPs.
MATERIALS AND METHODS
Materials
DNA oligomers were purchased from Interactiva (Ulm, Germany). PAMAM starburst dendrimer was purchased as a 10% stock solution in methanol from Aldrich (prepared by Dendritec, MI). Unmodified microscope glass slides were obtained from Menzel Gläser (Braunschweig, Germany) and PLL-coated slides were obtained from Sigma (Deishofen, Germany). Chemicals and solvents were from Fluka (Neu-Ulm, Germany). Sequences used to discriminate SNPs: target T, 5′-Cy5-A CTC GCA AGC ACC ACC CTA TCA-3′; probe A, 5′-NH2-TGA TAG GGT GCT TGC GAG T-3′; probe B, 5′-NH2-TGA TAG GGT GCT TGG GAG T-3′; probe C, 5′- NH2-GA TAG GGT GCT GGC GAG T-3′; probe D, 5′- NH2-A TAG GGC GCT TGC GAG T-3′; probe E, 5′-NH2-TGA TAG GGT GCT GGG GAG T-3′; probe F, 5′- NH2-GA TAG GGC GCT TCG GAG T-3′; probe G, 5′-NH2-TGA TAG GGT GCT CGA GAG A-3′. Mismatched nucleotides are underlined. Sequences used to demonstrate the regeneration stability: probe A, 5′-NH2-TGA TAG GGT GCT TGC GAG T-3′; probe H, 5′-NH2-AGC GGA TAA CAA TTT CAC ACA GGA.
Technical equipment
Laser scanning system, GenePix4000 (Axon, CA); piezo-driven spotting device, Robodrop (BIAS, Bremen, Germany).
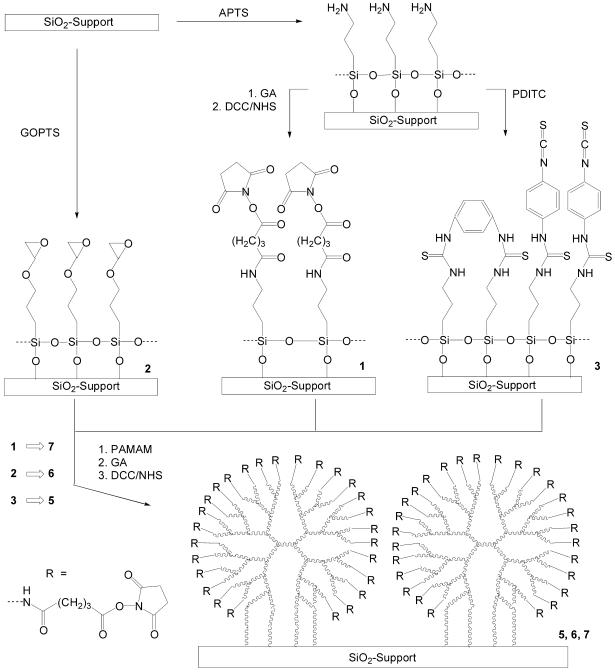
Preparation of activated surfaces
Amino-functionalized DNA oligomers were coupled to either conventional activated silica surfaces, or else to dendritic PAMAM surfaces. For this, PLL-coated slides 4 were used, as well as activated surfaces 1–3 and 5–7 prepared as depicted in Figure 1. Prior to activation, the substrates were cleaned by ultrasonic treatment in CH2Cl2, and subsequently immersed into a freshly prepared piranha solution (H2SO4:H2O2, 2:1) for 30 min. Silylation with APTS (Fluka) was performed as described previously (21). Briefly, the cleaned slides were treated with a mixture of ethanol:H2O:APTS (95:3:2 by volume) for 2 h under heavy stirring. To obtain surface 1, the conversion of surface amino groups into carboxyl groups was achieved by treatment with a saturated solution of glutaric anhydride (GA) in N,N-dimethylformamide (DMF) overnight. The slides were then carefully washed with DMF several times. Subsequently, carboxyl groups were activated with 1 mol l–1 N-hydroxysuccinimide (NHS)/1 mol l–1 N,N′-dicyclohexylcarbodiimide (DCC) in DMF for 1 h and washed with DMF. To prepare surface 2, the slides were incubated with glycidyloxypropyltrimethoxysilane (GOPTS; Fluka) as described elsewhere (23). To prepare surface 3, aminosilylated slides were submerged for 2 h in a solution containing 1,4-phenylenediisothiocyanate (PDITC, 10 mmol l–1; Fluka) in CH2Cl2, supplemented with pyridine (1%, v/v). Coating with dendrimers was carried out by treating modified surfaces 1–3 with a methanolic PAMAM solution (10% by weight; Aldrich). First, 100 µl were dropped upon the modified glass slide (76 × 26 mm). Thereafter, the slide was covered immediately with a second slide to form a sandwich-like arrangement. The slides were stored at room temperature for at least 12 h. Alternatively the slides can be stored for several weeks under nitrogen at –20°C. Excessive dendrimer was removed by washing twice with methanol and the surfaces were dried in a nitrogen stream. Next, the dendrimer-coated slides were activated with GA/NHS, similar to that described above, to obtain the dendritic linker systems 5, 6 or 7. The best results were observed when the activated surfaces were directly used for immobilization of nucleic acid probes.
Figure 1.
Schematic illustration of the modification of glass surfaces to generate dendritic linker systems. Glass slides were silylated with APTS or GOPTS. APTS surfaces were reacted with GA/NHS to obtain surface 1. Treatment with PDITC led to surface 3. Subsequently, surfaces 1–3 were coated with PAMAM dendrimers, followed by activation with GA and NHS/DCC to yield surfaces 5–7.
Arraying and hybridization
Microarrays were spotted on the activated slides by using a piezo-driven spotting device (Robodrop). Concentrations of the arrayed 5′-amino-modified oligomer probes in water were 10 µmol l–1. Typically, volumes of 250 pl were deposited. Subsequent to spotting, the slides were incubated overnight in a chamber under saturated humidity. DNA microarrays spotted upon the dendritic linker systems described can be stored at –20°C for several months. Arrays spotted on PLL slides (Sigma) were additionally baked at 80°C for 2 h to dehydrate the spots. Hybridization experiments with DNA oligomers were carried out for 1 h in TETBS buffer (20 mmol l–1 Tris–HCl, 150 mmol l–1 NaCl, pH 7.35, 5 mmol l–1 EDTA, 0.05% v/v Tween-20), containing the 5′-Cy5-labeled complementary target oligonucleotide (1 nmol l–1). Subsequently, slides were washed thoroughly with 2× SSC (0.3 mmol l–1 NaCl, 30 mmol l–1 Tris–HCl)/0.1% sodium dodecylsulfate (SDS). Fluorescence intensity of hybridized features was detected using a fluorescence scanner (GenePix4000). When PCR products instead of DNA oligomers were employed as targets, single-stranded DNA was prepared with the aid of streptavidin-coated magnetic beads (Boehringer Mannheim, Mannheim, Germany) following the protocol supplied by the manufacturer. PCR products, 212 bp in length, were obtained from the 5′-non-coding region of hepatitis C virus (HCV) using a biotinylated forward primer and a Cy5-labeled reverse primer, similar to that described previously (24,25). The probe-loaded dendrimer glass slides were blocked overnight at 4°C, and the hybridization solution (10 mmol l–1 Tris–HCl pH 7.5, 1 mmol l–1 EDTA pH 7, 600 mmol l–1 NaCl, 1× Denhardt’s solution, 500 µg/ml DNA MB-grade, 50% v/v deionized formamide) containing 4 nmol l–1 of the target DNA was incubated on the slide for 1 h in a humidity-chamber at room temperature. Subsequently, the slide was washed for 15 min with 2× SSC containing 10 mmol l–1 EDTA.
RESULTS AND DISCUSSION
Surface activation
Although steric crowding on densely DNA oligomer-modified surfaces has been reported to limit hybridization efficiency (16), in our hands an insufficient loading on planar glass surfaces was often the reason for weak signal intensities and, in turn, for a limited sensitivity in nucleic acid hybridization analyses. Thus, to increase sensitivity, immobilization should occur through a flexible linker system providing a large number of reactive binding sites. In addition, the affinity of target molecules towards the linker matrix should be as low as possible to avoid high background intensities caused by non-specific binding during hybridization. The concept for the preparation of dendritic linker systems was based on the assumption that the highly functionalized surface of PAMAM starburst dendrimers attached to a solid support should help to maximize the immobilization capacity. Furthermore, we estimated that negatively charged carboxyl groups at the modified surfaces should lead to a reduced background. Thus, amino-modified PAMAM dendrimers were covalently attached in a first step either to an epoxy-modified surface 1 or, else, to the isothiocyanate- or carboxyl-modified surfaces 2 or 3, respectively (Fig. 1). Subsequently, the terminal amino groups of the dendrimers attached were converted into carboxyl groups by treatment with GA, followed by a final activation step with NHS/DCC allowing for the immobilization of, for instance, amino-modified biomolecules.
Determination of hybridization capacities
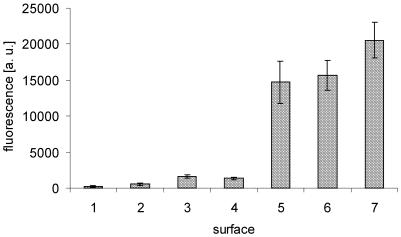
With respect to the signal intensities obtained from hybridization experiments, we initially compared a selection of surfaces described in Figure 1. For this, hybridization experiments were carried out with microarrays spotted on conventional surfaces 1–4, as well as on the dendrimer surfaces 5–7. Spotting and hybridization of microarrays was performed as described in Materials and Methods. With the exception of surface 4, which requires an additional baking step, all slides were treated under identical conditions. Subsequent to hybridization with a complementary Cy5-labeled oligomer probe, the signal intensity of the DNA spots was detected with a fluorescence scanner. As depicted in Figure 2, the conventional surfaces 1 and 2 were found to yield low hybridization signals. However, the isothiocyanate-functionalized surface 3 and the PLL slides 4 revealed slightly increased signal intensities. In contrast, a dramatic increase in signal intensity was obtained from the dendritic linker systems 5–7, and the PAMAM surface 7 revealed the highest hybridization capacity. Approximately 10-fold higher fluorescence signals were detected, as compared with surfaces 3 and 4. Most likely this is due to an increased amount of DNA probes tethered to the amino-functionalized dendrimers attached on the surface. The surfaces 5–7 offer additional advantages, since, due to the negatively charged carboxyl-terminated linker system, no further deactivation and passivation steps are required subsequent to oligomer attachment in order to obtain low background signal intensities. In contrast, when oligomers were arrayed on APTS, PDITC or PLL slides, the amino groups remaining had to be treated with, for instance, GA to reduce non-specific binding.
Figure 2.
Fluorescence intensity of hybridized spots on surfaces 1–7. Volumes of 250 pl of a 10 µmol l–1 solution of 5′-amino-modified DNA oligomers in water were arrayed on surfaces 1–7. Hybridization was performed with a 1 nmol l–1 solution of a Cy5-labeled complementary target oligomer. Hybridization signals were detected with a fluorescence scanner (GenePix4000, pmt500). The height of the histograms represents the average fluorescence pixel intensity of all spots.
Discrimination of SNPs
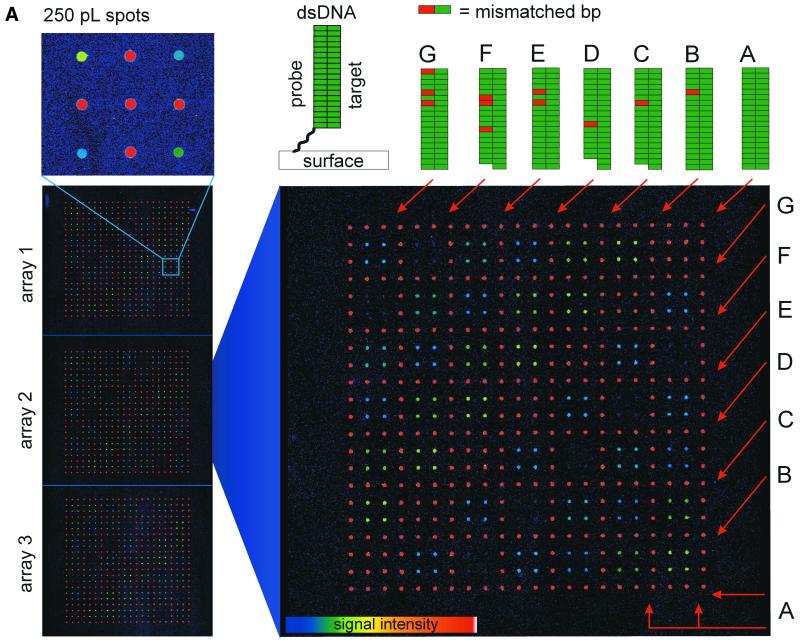
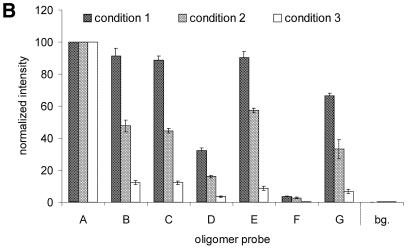
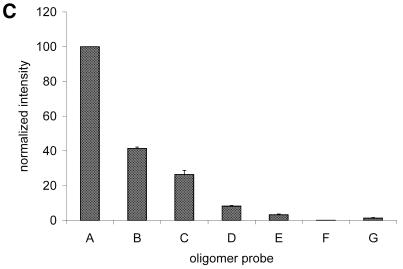
Further studies concerned the hybridization performance of an oligomer array based on dendrimer surface 7. Microarrays comprised of seven different single-stranded oligomer probes were spotted on surface 7, as depicted in Figure 3A. Oligomer A was fully complementary to the Cy5-modified target present in the hybridization buffer, while oligomers B to D contained a single mismatched nucleotide. Oligomer E contained two mismatches and oligomers F and G contained three mismatches at varying positions. The sequences of the oligomer probes are listed in Materials and Methods. Subsequent to hybridization, the microarrays were analyzed with a fluorescence imager and the average pixel intensities of the hybridization spots were determined (Fig. 3B). To optimize stringency, three hybridization conditions were experimentally tested in succession. First, the microarrays were hybridized under low stringent conditions at room temperature in TETBS (condition 1 in Fig. 3B). The stringency was increased by an additional washing step at an elevated temperature (2× SSC/0.1% SDS, 51°C, 5 min; condition 2 in Fig. 3B). In a third protocol, the TETBS buffer was supplemented with 50% formamide (condition 3 in Fig. 3B). The signal intensities detected indicated that under high stringency condition 3, the single mismatched base pairs in oligomers B and C are discriminated with a loss of signal intensity of ∼90%. The hybridization efficiency of D is decreased by ∼95%, possibly due to the additional absence of two terminal nucleotides at its 5′-end. As expected, oligonucleotide probe E, containing two mismatched nucleotides, yielded hybridization signals smaller than oligonucleotides B and C. Oligomer G showed a slightly decreased signal intensity compared with E, probably due to the terminal position of the third mismatch. Oligomer F showed almost no affinity for the target DNA, probably due to the centralized position of three mismatches.
Figure 3.
(A) Discrimination of SNPs with a dendrimer-based DNA microarray. Seven different single-stranded oligomer probes were spotted at a microscope glass slide with surface 7 to form three identical microarrays. First, a square lattice was spotted with the fully complementary oligomer A to verify the long-range homogeneity of the glass slide. Subsequently, quadrupols of the complementary A and mismatched B to G oligomers were arrayed diagonally at the unoccupied squares. The 5′-Cy5-labeled target oligomer T (fully complementary to probe A) was used for hybridization. Double-stranded DNA oligomers formed by hybridization are schematically depicted at the top. The enlargement of nine 250 pl spots with an average diameter of 170 µm demonstrates the highly homogenous signal distribution within the individual spots. (B) Quantitative analysis of the microrray hybridization of oligonucleotide target T, shown in (A). The height of the histograms corresponds to the normalized fluorescence intensities of the spots and thus are indicative for the hybridization efficiencies of oligomer probes A to G. In addition, the fluorescence intensity of the background (bg) is shown. SNP discrimination was performed under varying conditions: condition 1, hybridization at room temperature in TETBS; condition 2, hybridization at room temperature in TETBS, followed by an additional washing step at 51°C (Tm = 59°C) for 5 min; and condition 3, hybridization in TETBS/formamide (1:1 v/v) at room temperature. (C) Hybridization with PCR products. The identical microarray shown in (A) was regenerated and incubated with a Cy5-labeled 212mer target DNA (see text for details). The height of the histograms corresponds to the normalized fluorescence intensities of the spots. The number and relative positions of the mismatches are identical as shown for oligonucleotide target T.
These results demonstrate that identical arrays can be employed for repeated hybridization experiments under varying conditions and that the regeneration stability of the dendrimer surface is extremely useful for optimizing the hybridization procedure. Furthermore, despite the simplicity of the spotting solutions, remarkable high and homogeneous signal distributions within the individual spots were obtained (Fig. 3A). No additional spotting additives were required, which are usually applied with PLL, APTS or aldehyde surfaces to obtain homogenous spot features (20). This allows for minimized signal deviations of the data, and thus minimizes experimental errors.
The identical microarrays shown in Figure 3A were employed to discriminate mismatches in longer target DNA. Subsequent to a storage period of almost 6 months at –20°C, the microarrays were regenerated as described below and hybridized with Cy5-labeled 212mer single-stranded DNA, obtained by PCR from the 5′-non-coding region of HCV, containing the identical sequence as target oligonucleotide T (24,25). As shown in Figure 3C, SNPs were also clearly detectable in this hybridization assay. One mismatch led to a decrease in signal intensity of ∼60–92%, depending on the position of the mismatch. Two mismatches showed only 3% of the fluorescence intensity of the perfect match, and three mismatches decreased the signal to only 2%. Thus, no significant difference was observed in the hybridization of PCR products, as compared with the hybridization of oligonucleotides. Moreover, the latter experiments once more demonstrate the extraordinary high physicochemical stability of microarrays produced with dendrimer-coated glass slides.
Regeneration stability
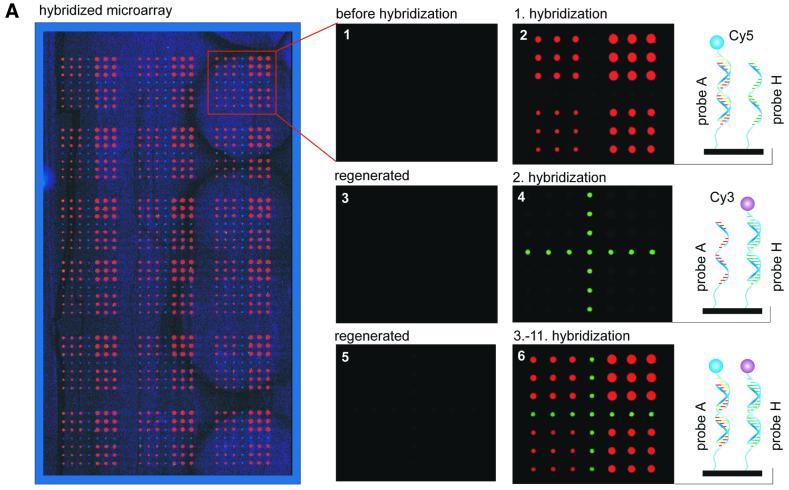
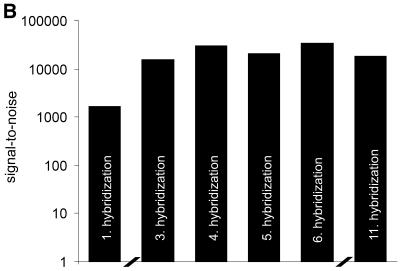
To further investigate the stability of dendrimer surface 7 against repeated regeneration procedures, microarrays containing two different oligomer probes A and H (sequences in Materials and Methods) were spotted as shown in Figure 4A. The first hybridization was performed with the 5′-Cy5-labeled target oligomer complementary to probe A. Hybridized oligomer microarrays were fully regenerated by a 5 min treatment with NaOH (50 mmol l–1)/SDS (0.1%) at 60°C. Fluorescence imaging confirmed that this procedure was sufficient to remove >99% of the target attached. Slides were then hybridized with the 5′-Cy3-labeled target oligomer complementary to probe H. The slides were regenerated again and, subsequently, further hybridizations were performed with both target oligomers simultaneously (Fig. 4A). Up to 10 regeneration and rehybridization procedures were carried out without any loss of signal intensity. Moreover, after the first regeneration cycle a significant increase in signal-to-noise ratio was observed due to the decreased background intensity (Fig. 4B).
Figure 4.
(A) Regeneration stability of DNA microarrays based on surface 7. Microarrays comprised of two different oligomer probes, A and H, which were spotted on dendritic surface 7. Volumes ranged from 250 pl up to 1 nl. The microarrays were first hybridized with a 5′-Cy5-labeled target oligomer A′, complementary to probe A (enlargement 2). Slides were regenerated (enlargement 3), and subsequently hybridized with the Cy3-labeled target oligomer H′, complementary to probe H (enlargement 4). Further hybridizations were performed simultaneously with both target oligomers (enlargement 6). The double-stranded formed nucleic acids are schematically depicted at the right side. (B) Signal-to-noise ratio of re-hybridized microarrays. The height of the histograms represents the average signal-to-noise ratio of spots hybridized with the Cy5-labeled target A′. The signal-to-noise ratio was obtained from the signal intensities of the hybridized spots measured at pmt500, divided by the background signal intensity caused by non-specific binding of the target.
CONCLUSIONS
We have here reported on the generation of a novel linker system comprising PAMAM dendrimers covalently attached to planar glass surfaces. Compared with conventional APTS or PLL slides usually employed, DNA microarrays prepared by means of the PAMAM surfaces show significantly increased signal intensity and, with that, a higher sensitivity in hybridization analyses can be obtained. As demonstrated by the discrimination of SNPs, oligomer probes attached to the dendritic linker system reveal highly specific hybridization properties. Furthermore, the dendrimer-based microarrays can be recovered several times and no additional spotting additive is necessary to obtain highly homogeneous spots.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Deutsche Forschungsgemeinschaft (DFG), the Land Bremen and the Federal Ministry for Education and Research (BMBF) under contract 11833A.
REFERENCES
- 1.McCarthy J.J. and Hilfiker,R. (2000) The use of single-nucleotide polymorphisms maps in pharmacogenomics. Nat. Biotechnol., 18, 505–508. [DOI] [PubMed] [Google Scholar]
- 2.Goldfeld A.E., Leung,J.Y., Sawyer,S.A. and Hartl,D.L. (2000) Post-genomics and the neutral theory: variation and conservation in the tumor necrosis factor-α promotor. Gene, 261, 19–25. [DOI] [PubMed] [Google Scholar]
- 3.Wu A.H.B. and Tsongalis,G.J. (2001) Correlation of polymorphisms to coagulation and biochemical risk factors for cardiovascular diseases. Am. J. Cardiol., 87, 1361–1366. [DOI] [PubMed] [Google Scholar]
- 4.Thameem F., Wolford,J.K., Bogardus,C. and Prochazka,M. (2001) Analysis of PBX1 as a candidate gene for type 2 diabetes mellitus in pima indians. Biochim. Biophys. Acta, 1518, 215–220. [DOI] [PubMed] [Google Scholar]
- 5.Drysdale C.M. (2000) Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl Acad. Sci. USA, 97, 10483–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin T.J. and Smith,L.M. (2000) Single-nucleotide polymorphism analysis by MALDI–TOF mass spectrometry. Trends Biotechnol., 18, 77–84. [DOI] [PubMed] [Google Scholar]
- 7.Guo B. (1999) Mass spectrometry in DNA analysis. Anal. Chem., 71, 333R–337R. [DOI] [PubMed] [Google Scholar]
- 8.Isola N.R., Allman,S.L., Golovlev,V.V. and Chen,C.H. (2001) MALDI–TOF mass spectrometric method for detection of hybridized DNA oligomers. Anal. Chem., 73, 2126–2131. [DOI] [PubMed] [Google Scholar]
- 9.Helmberg A. (2001) DNA-microarrays: novel techniques to study aging and guide gerotologic medicine. Exp. Gerontol., 36, 1189–1198. [DOI] [PubMed] [Google Scholar]
- 10.Fan J.B., Chen,X., Halushka,M.K., Berno,A., Huang,X., Ryder,T., Lipshutz,R.J., Lockhart,D.J. and Chakravarti,A. (2000) Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res., 10, 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J., Kruglyak,L., Stein,L., Hsie,L., Topaloglou,T., Hubbell,E., Robinson,E., Mittmann,M., Morris,M.S., Shen,N., Kilburn,D., Rioux,J., Nusbaum,C., Rozen,S., Hudson,T.J., Lipshutz,R., Chee,M. and Lander,E.S. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 12.Lindblad-Toh K., Tanenbaum,D.M., Daly,M.J., Winchester,E., Lui,W.O., Villapakkam,A., Stanton,S.E., Larsson,C., Hudson,T.J., Johnson,B.E., Lander,E.S. and Meyerson,M. (2000) Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol., 18, 1001–1005. [DOI] [PubMed] [Google Scholar]
- 13.Fan J.B., Chen,X., Halushka,M.K., Berno,A., Huang,X., Ryder,T., Lipshutz,R., Lockhart,D.J. and Chakravarti,A. (2000) Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res., 10, 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raitio M., Lindroos,K., Laukkanen,M., Pastinen,T., Sistonen,P., Sajantila,A. and Syvänen,A.C. (2001) Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays. Genome Res., 11, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erdogan F., Kirchner,R., Mann,W., Ropers,H.-H. and Nuber,U.A. (2001) Detection of mitochondrial single nucleotide polymorphisms using a primer elongation reaction on oligonucleotide microarrays. Nucleic Acids Res., 29, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21 (Suppl.), 5–9. [DOI] [PubMed] [Google Scholar]
- 17.Maskos U. and Southern,E. (1992) Oligonucleotide hybridisations on glass supports: a novel linker for oligonucleotide synthesis and hybridisation properties of oligonucleotides synthesised in situ. Nucleic Acids Res., 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shchepinov M.S., Case-Green,S.C. and Southern,E. (1997) Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res., 25, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung V.G., Morley,M., Aguilar,F., Massimi,A., Kucherlapati,R. and Childs,G. (1998) Making and reading microarrays. Nature Genet., 21 (Suppl.), 15–19. [DOI] [PubMed] [Google Scholar]
- 20.Diehl F., Grahlmann,S., Beier,M. and Hoheisel,J.D. (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res., 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benters R., Niemeyer,C.M. and Wöhrle,D. (2001) Dendrimer-activated solid supports for nucleic acid- and protein-microarrays. Chem. Biol. Chem., 2, 686–694. [DOI] [PubMed] [Google Scholar]
- 22.Tomalia D.A., Naylor,A.M. and Goddard,W.A. (1990) Starburst dendrimers: molecular-level control of size, shape, surface chemistry, topology and flexibility from atoms to macroscopic matter. Angew. Chem. Int. Ed. Engl., 29, 138–175. [Google Scholar]
- 23.Piehler J., Brecht,A., Valiokas,R., Liedberg,B. and Gauglitz,G. (2000) A high-density poly(ethylene glycol) polymer brush for immobilization on glass-type surfaces. Biosens. Bioelectron., 15, 473–481. [DOI] [PubMed] [Google Scholar]
- 24.Boldt L., Gersdorf,H., Niemeyer,C., Holtkamp,F., Bischoff,R., Sälter,W., Adler,M., Kayser,O., WolfS,M., Jüptner,W. and Blohm,D. (1998) Biosensors ’98; 5th World Congress on Biosensors, Poster presentation, Berlin.
- 25.Niemeyer C., Boldt,L., Ceyhan,B. and Blohm,D. (1999) Evaluation of single-stranded nucleic acids as carriers in the DNA-directed assembly of macromolecules. J. Biomol. Struct. Dyn., 17, 527–538. [DOI] [PubMed] [Google Scholar]