Abstract
R-loops are abundant and dynamic structures ubiquitously present in human cells both in the nuclear and mitochondrial genomes. They form in cis in the wake of transcription complexes and in trans apart from transcription complexes. In this review, we focus on the relationship between R-loops and topoisomerases, and cancer genomics and therapies. We summarize the topological parameters associated with the formation and resolution of R-loops, which absorb and release high levels of genomic negative supercoiling (Sc-). We review the deleterious consequences of excessive R-loops and rationalize how human type IA (TOP3B) and type IB (TOP1) topoisomerases regulate and resolve R-loops in coordination with helicase and RNase H enzymes. We also review the drugs (topoisomerase inhibitors, splicing inhibitors, G4 stabilizing ligands) and cancer predisposing genes (BRCA1/2, transcription, and splicing genes) known to induce R-loops, and whether stabilizing R-loops and thereby inducing genomic damage can be viewed as a strategy for cancer treatment.
Graphical Abstract
Graphical Abstract.
Topoisomerases (TOP1, TOP2 and TOP3B) can remove DNA supercoiling to prevent R-loop formation while TOP3B coordinates with helicase DDX5 and resolves R-loops by decatenation mechanism.
INTRODUCTION: OVERVIEW AND CLASSIFICATION OF THE 6 HUMAN TOPOISOMERASES
Different nucleic acids metabolic processes including replication, transcription, recombination, chromosome segregation and chromatin remodeling generate various topological constraints (torsional stress, DNA/RNA knots, DNA/RNA catenanes, hemicatenanes, etc.). The family of cellular enzymes that take care of these topological tensions are called topoisomerases. Topoisomerases function by breaking and rejoining nucleic acid backbones by forming transient enzyme-nucleic acid complexes between a topoisomerase catalytic tyrosine residue and one end of the broken nucleic acid. These covalent enzyme-nucleic acid catalytic intermediates are called ‘Topoisomerase cleavage complexes’ (TOPccs). Under normal conditions, TOPccs are transient and reverse readily to allow the rejoining of the nucleic acids backbones after the topological problems are resolved and topoisomerases are released (1–5).
Based on the number of strands they cleave, nature of their catalytic intermediates and changes in the DNA linking number after each single catalytic cycle, topoisomerases are broadly divided into two categories: type I and type II enzymes (Figure 1) (1,3,4,6). Type I topoisomerases cleave single-stranded DNA/RNA and change the linking number in steps of one. Type I topoisomerases are further classified into type IA and type IB enzymes based on distinct structural features and mechanisms of action (1–4). Humans have two different type IA enzymes: topoisomerase III-alpha (TOP3A) and topoisomerase III-beta (TOP3B) while yeast has only one isoform (Top3). Type IA topoisomerases form phosphotyrosyl linkage with the 5′-end of the broken nucleic acid (DNA/RNA) backbone; they require magnesium for catalytic activity and act by a ‘strand passage’ mechanism (the passage of a single-strand of nucleic acid through another) (1,2,7). TOP3A and TOP3B can relax hyper-negatively supercoiled/undertwisted DNA. TOP3A also resolves hemicatenanes structures formed during replication and recombination (7). TOP3B is unique as it can work on both DNA and RNA to resolve RNA catenanes and knots (8–11). Eukaryotic type IA enzymes work as multimeric complexes with their scaffolding proteins: RecQ- mediated genome instability protein 1 (RMI1) and RMI2 for TOP3A, and Tudor domain containing protein 3 (TDRD3) for TOP3B. TOP3A-RMI1-RIM2 (TRR) forms the dissolvasome complex with the Bloom syndrome protein (BLM) (BTRR complex). TOP3B-TDRD3 functions as a multimeric complex with fragile X mental retardation protein (FMRP) (3,9,11–13).
Figure 1.
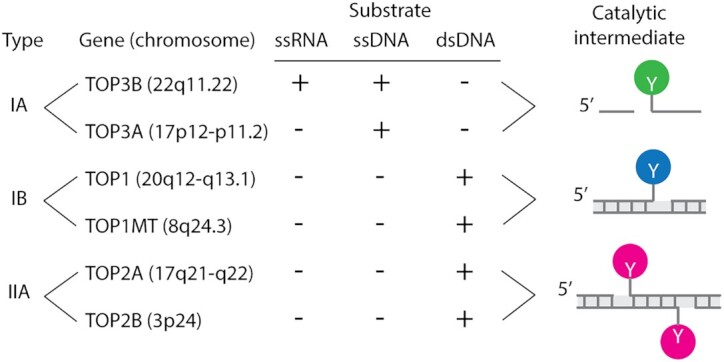
Classification and distinctive features of human topoisomerases. The catalytic intermediates (cleavage complexes) are schematically drawn at right (the Y’s refer to the catalytic tyrosyl residues that form the reversible cleavage complexes). Type IA topoisomerases cleave nucleic acids in single-stranded regions (ssRNA and ssDNA), whereas Type II topoisomerases cleave double-stranded DNA (dsDNA) only. Type IA topoisomerases are conserved in all domains of life and considered the most ancestral topoisomerase enzymes. TOP3B is the only RNA topoisomerase. Type IA enzymes can only relax hypernegative DNA. Type IB topoisomerases are highly efficient ‘swivelases’ (‘DNA untwisting enzymes’) removing both positive (Sc+) and negative DNA supercoiling (Sc−). TOP1 acts on the nuclear genome while TOP1MT acts on the mitochondrial genome (mtDNA). TOP2 enzymes relax both Sc+ and Sc− by forming DNA double-strand breaks (with a 4 bp stagger) in a DNA duplex region allowing the passage of another DNA segment through the cleavage complex. Upon completion of the DNA passage, TOP2 rejoins the DNA. TOP2-mediated strand passage is also essential for DNA decatenation during replication. Shaded rectangles represent canonical helical B-DNA with base pairs as short vertical bars.
Vertebrates have 2 type IB enzymes: a nuclear topoisomerase (TOP1) and a mitochondrial topoisomerase (TOP1MT). Both enzymes very efficiently and processively relax positively and negatively supercoiled DNA (Sc+ and Sc−, respectively). These type IB enzymes work by forming a phosphotyrosyl linkage with the 3'-end of the broken DNA and by a ‘controlled rotation’ mechanism in which the broken nucleic acids strand rotates around the intact strand. Their activity does not require divalent cations (Mg2+) or ATP and these type IB enzymes can function over a broad range of temperature and pH (1–4).
Type II topoisomerases act as homodimers. They cleave both strands of duplex DNA, forming 5′ phosphotyrosyl bonds with the ends of the broken strands with a four-base pair stagger (Figure 1). They require Mg2+ and ATP for changing the topology of nucleic acids by ‘duplex strand passage’ mechanism. By cleaving the DNA duplex, they allow a second DNA duplex to pass through their cleavage complex and change the DNA linking number in steps of two (1). Humans have two type II topoisomerases: topoisomerase II-alpha (TOP2A) and topoisomerase II-beta (TOP2B). Both relax Sc+ and Sc− DNA at sites where DNA duplexes cross each other. They resolve catenanes and DNA knots. TOP2A and TOP2B can also generate single-strand breaks when only one protein of the dimer breaks the DNA during the catalytic cycle (1–5, 14–16).
STRUCTURE, TOPOLOGY AND GENOMIC IMPACTS OF R-LOOPS
R-loops form by the hybridization of RNA with a complementary strand of double-stranded DNA. Hence, R-loops are triple-stranded structures consisting of DNA-RNA helical hybrids associated with the displaced single-stranded DNA (17). R-loops were first observed ≈47 years ago by R.L. White, D. Hogness, M. Thomas and R.W. Davis in Drosophila melanogaster rDNA and in biochemical experiments (18,19). R-loops form universally (in viruses, bacteria, plants and animals). Their length varies from 300 to 2500 bp (20–22) and they are generally highly dynamic with estimated half-lives of 10–15 minutes in vivo (17,23,24). They are distributed at accessible chromatin regions devoid of nucleosomes, at transcription start sites (TSS), gene promoters, transcription termination sites (TTS), in rDNAs, tRNA-coding genes, Ty elements, centromeres and telomeres, and in mitochondrial DNA, forming thousands of dynamic hotspots in the human genome (20,25–28).
R-loops form in cis (co-transcriptionally) when the newly transcribed RNA after exiting the RNA polymerase active site, threads back and anneals with the template DNA strand (Figure 2). Hence, highly transcribed genes, such as ribosomal genes and oncogenes are prone to R-loops. R-loops can also form in trans when RNAs (like long non-coding RNAs) produced in a distal locus, RNAs transcribed from homologous chromosomes or telomeric repeat RNA (TERRA) invade duplex DNA (20,28–35). In yeast, R-loops in trans are promoted by Rad51p and Rad52p, which also play important roles in D-loop formation and DNA strand exchange during homologous recombination-mediated DNA double-strand breaks (DSBs) repair (34). The mechanisms and fraction of R-loops formed in trans are still not very clear and the involvement of RAD51 in trans R-loop formation is also disputed; hence most R-loops are likely to form in cis in a cotranscriptional manner (32,36). Moreover, recent findings suggest that DNA double-strand breaks (DSBs) may have roles in inducing cellular R-loops (37,38). The local DNA sequence also contributes to R-loop formation. GC skews (C-rich template strand) favor the formation of 4-stranded guanosine quartet structures (G4s) on the displaced strand during transcription and produce more stable heteroduplex than AT sequences (25) (Figure 2). Additionally, the formation of G4s contribute to the stability of R-loops (39).
Figure 2.
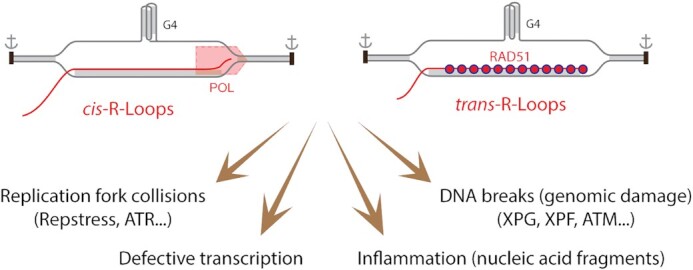
Deleterious effects of R-loops. Unresolved and excessive cis- and trans-R-loops have been reported to have multiple deleterious effects. Watson-Crick helical double-stranded nucleic acids are shaded in grey. DNA is shown in gray and RNA in red. A guanosine quartet structure is included as stabilizer of R-loops. RAD51 is implicated in the formation of trans-R-loops. Replication fork collisions give rise to replication stress (RepStress) and engagement of the ATR/CHK1 pathways. Stable R-loops also stall transcription complexes. DNA breaks are produced by the nucleotide excision repair nucleases (XPG and XPF), leading to DNA double-strand breaks and activation of ATM. Released nucleic acid fragments can initiate inflammation.
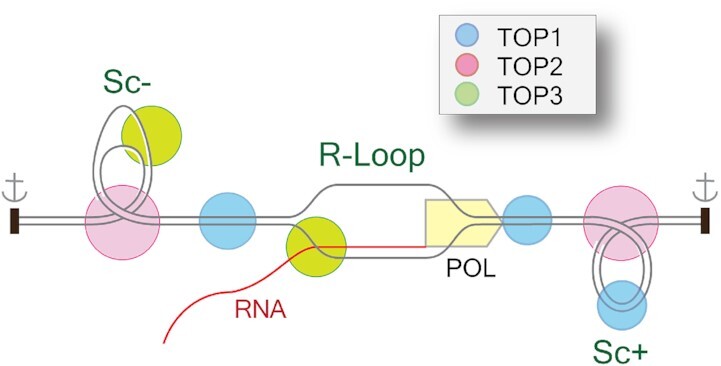
Formation of R-loops in a topologically confined chromatin region requires and absorbs large quantities of negative superhelicity (Sc-) (20,39,40). For the cis-R-loops, Sc- is generated by the translocating RNA polymerase complex, which generates Sc+ in front of the polymerase and equivalent Sc- behind the polymerase (Figure 3), as described in the ‘twin supercoiling model’ of Liu and Wang (41). For the trans-R-loops, Sc− is required for the formation of the RNA-DNA heteroduplex (Figure 4). The excellent review by Benham and Chedin (20) and Figure 4 explicit the topological considerations associated with R-loops, and how a 100 base pairs R-loop absorbs ≈11 negative superhelical turns (11 Sc−); with 10 generated by the helical twist on the RNA-DNA hybrid and 1 by the wrapping of the DNA single-strand around the heteroduplex (20).
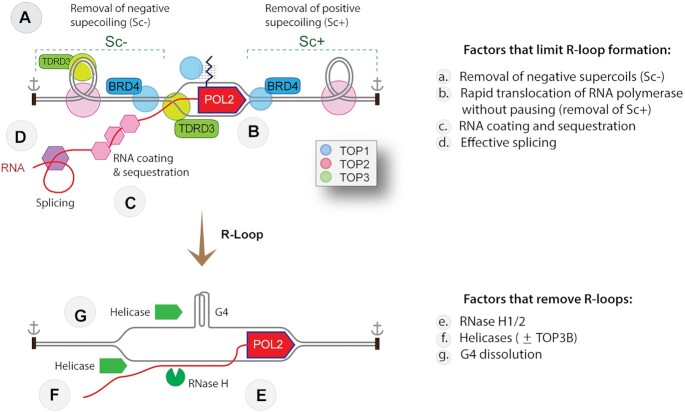
Figure 3.
Cellular mechanisms controlling R-loops associated with transcription (cis-R-loops). The top scheme represents an elongating RNA POL2 complex with its associated topological features within a chromatin topologically constrained domain whose boundaries are schematized as black rectangles with anchor signs. Duplex canonical DNA is shown as parallel lines (without showing the Watson-Crick helical turns for simplicity). Factors that limit R-loop formation and remove R-loops are listed at right, annotated in the figure, and discussed in the text. Symbols for the human topoisomerases are in the inset.
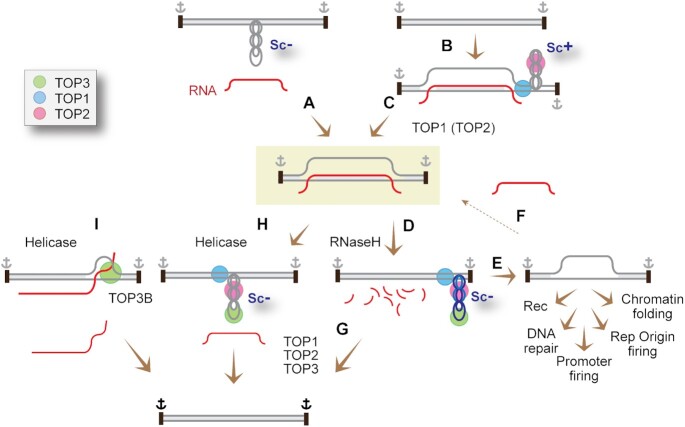
Figure 4.
Topological considerations for the formation (A−C) and removal (D−I) of R-loops. The formation of R-loops (A−C) is associated with (and requires) topological adjustments of the DNA within topologically constrained chromatin domains whose boundaries are schematized as black rectangles with anchor signs. (A) R-loops readily form in hypernegatively supercoiled (Sc−) domains (see Figure 2 for co-transcriptional R-loops formed in cis of transcription complexes). In doing so, R-loops constrain the Sc− (one negative superhelical turn (Sc−) per 10.5 bp of helical RNA-DNA hybrid). (B) to form R-loops by RNA strand invasion (R-loops formed in trans) (as in the case of CRISPR-Cas9 and TERRA), the DNA topology needs to be adjusted by removing the Sc+ (C) formed to compensate for hybridization of the RNA-DNA heteroduplex. (D) R-loops can be removed by degradation of the hybridized RNA by RNase H1 (and H2). (E) removal of the RNA releases the corresponding Sc−, promoting chromatin folding (one nucleosome absorbs one Sc−), replication origin and promoter firing, DNA repair and recombinations. (F) Sc- can help capture an RNA to reform R-loops. (G) TOP1 and TOP2 can relax the Sc− released by the digestion of RNA in double-stranded DNA, and TOP3 can relax the hypernegative Sc- in single-stranded segments generated by the hypernegative Sc−. (H) Helicases by dissociating the RNA from the heteroduplex also release the Sc− that was constrained in the R-loop (≈20 Sc− for a 200 bp R-loop). (I) By pushing the RNA−DNA hybrids to the end of the R-loops, the helicases can generate knotted DNA−RNA structures that are resolved (decatenated) by TOP3B in the single-stranded segment of the RNA and/or DNA.
An elegant model by Hanawalt et al. addresses another important topological problem faced by RNA during co-transcriptional R-loop formation (42). R-loop associated DNA-RNA hybrid generation requires the passage of the nascent RNA ‘tail’ through the negatively supercoiled DNA in the wake of RNA pol II and swiveling of the nascent RNA. This swiveling of RNA is problematic because the 3′-end of the nascent RNA in the transcription bubble cannot rotate and therefore for an R-loop to form behind the RNA pol II, the 5′-end of the nascent RNA must swivel. Swiveling of the 5′-end of the nascent RNA becomes increasingly difficult with increasing distance between the transcription start site and the site of R-loop initiation because the length of the tail increases. This causes reduced R-loop accumulation with greater distance from the transcription start site (42–46).
R-loops can be good (programmed or physiological) or detrimental (aberrant or pathological). Physiological R-loops contribute to diverse cellular metabolic processes including class switch recombination (22), mitochondrial replication (47,48), protection against promoter methylation (25,33,49), transcription termination (50), chromatin organization (helping recruitment of chromatin remodelers), chromosome segregation (51,52), DNA repair in transcribed regions, telomere repair and centromeric functions (33,35). R-loops have also been proposed to contribute to the repair of DSBs by promoting the recruitment of DNA repair proteins, by regulating DNA end-resection, and engaging alternating DNA repair pathways (37,38). The resolution of R-loops in confined chromatin regions can release large amount of Sc−, which favors chromatin contacts, the formation of non-B DNA structures (G4, cruciforms, Z-DNA), nucleosomes and protein-DNA interactions (20,39) (Figure 4, bottom right). R-loop-linked G-quadruplex structures in the non-template strand are also known to promote transcription by successive R-loop formation (53).
Unscheduled, pathological or aberrant R-loops originate primarily during transcription (32) and can cause genome instability (Figure 2). They can act as replication roadblocks, causing replication stress (RepStress), replication fork collapse and DNA breaks with activation of the ATR/CHK1 kinase pathway (54,55). Replication forks colliding with R-loops (associated with transcription) in a head-on orientation (HO) can result in more severe DNA damaging effects than co-directional (CD) collisions (52,56–61). R-loop-associated G4 structures can also affect replication fork progression in both HO and CD conditions by inducing fork stalling, CMG helicase-polymerase (polδ) uncoupling, and gaps in the nascent leading and lagging strand (62). Structure-specific endonucleases (XPG and XPF or FEN1 and XPF) can recognize and cleave at R-loop−ssDNA junctions in both replicating and non-replicating cells and cause DSBs and activation of ATM (55,63–65). R-loops may promote mutagenesis as the displaced single-stranded DNA is exposed to AID (activation-Induced cytidine deaminase), nucleases or endogenous genotoxic metabolites. R-loops can also prime unscheduled error-prone DNA synthesis (61). R-loops are linked with an increasing number of human diseases including neurological disorders [ataxia-ocular apraxia type 2 (AOA2), amyotrophic lateral sclerosis type 4 (ALS4), fragile X syndrome, Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD)], autoimmune diseases, and cancer (54,60,66–68). Their role in oncogenesis remains to be explored further.
GENERAL MECHANISMS LIMITING R-LOOPS WITH FOCUS ON DNA TOPOLOGY
The mechanisms protecting cells from excessive R-loops can be classified as prevention of R-loop formation and resolution of existing R-loops. The proposed molecular mechanisms by which topoisomerases control R-loops in human cells are outlined in Figures 3 and 4.
Factors limiting R-loop formation
In the case of cis-R-loops, failure to remove the Sc− generated by advancing transcription complexes promotes the formation of R-loops (20,69), and under normal conditions, human type IB topoisomerases (and type II topoisomerases at DNA crossovers; Figure 3a) can efficiently remove the Sc- generated by translocating RNA polymerases. Additionally type IB and II topoisomerases allow the translocation of polymerases by removing Sc + in duplex DNA. The activity of TOP1 is likely coupled with the C-terminal domain (CTD) of RNA POL2 and is stimulated by POL2 CTD phosphorylation by the bromodomain protein BRD4 (Figure 3a-b, top; see next section) (70). This is in contrast with TOP3B, which can only relax hypernegatively supercoiled DNA behind the RNA polymerases as TOP3B requires single-stranded DNA for binding, cleavage and strand-passage activity (Figure 3A) (12,26,65,71–78). The activity of TOP3B is coupled with its interaction with TDRD3, which also directly interacts with POL2 (Figure 3) (12).
The situation is different in bacteria, as they generally lack type IB enzymes. The bacterial type IA enzyme, Topo I (encoded by the topA gene) acts behind transcription complexes while gyrase acts in front of the transcription complexes to remove the Sc+. Of note, bacterial Topo I was the first topoisomerase reported to suppress cellular R-loops based on the observation that overexpression of RNase H suppressed the growth defect of E. coli topA mutants lacking Topo I (46,78–80). Recent findings suggest that both type IA topoisomerases (Topo I and Topo III) prevent the formation of R-loops and in turn unscheduled origin firing from RNase HI-sensitive origin of replication in the terminus (Ter) region of the bacterial chromosome (81,82). Another recent study showed that gyrase can also introduce excessive negative supercoils at replication-transcription head-on (HO) conflict regions and therefore promote the formation of pervasive R-loops in the Gram-positive bacterium Bacillus subtilis (83).
As mentioned above, human topoisomerases are also critical for removing the Sc+ in front of transcription complexes (Figure 3B). Dissipating Sc+ allows the efficient and rapid translocation of the transcription complexes and avoids RNA polymerase pausing with the potential of R-loop formation. Relaxation of Sc+ can be accomplished by both TOP1 and TOP2, with TOP1 acting in overwound duplex DNA and TOP2 at DNA crossovers (Figure 3B). Removal of Sc+ is also likely necessary for the formation of trans-R-loops in topological confined chromatin domains as R-loops act as sinks for Sc− (see above) (Figure 4B).
By coating nascent RNAs soon after they exit RNA pol II complexes, mRNA biogenesis and processing factors including RNA export proteins, splicing factors, cleavage and polyadenylation proteins prevent the invasion of the DNA double helix by the RNA and in turn prevent R-loop formation (Figure 3C) (27,61,84–91). This phenomenon was first observed in yeast THO mutants (the yeast THO complex is a multimer containing four proteins, Tho2, Hpr1, Mft1 and Thp2), which accumulate R-loops and display a hyper-recombination phenotype (92). In humans, the THO/TREX complex (formed with ALYREF and UAP56), THSC/TREX-2 (85,93–95), FIP1L (89) and splicing factors such as SRSF1 (86) are some of the RNA binding proteins (RBPs) that sequester nascent RNA and prevent RNA hybridization with the transcribed DNA. Mutations in the splicing factors U2AF1, SF3B1 and SRSF2 also cause increased levels of R-loops, as observed in myelodysplastic syndromes, acute myeloid leukemia and chronic myelomonocytic leukemia (96–99) (Figure 3D). The importance of efficient RNA splicing in preventing R-loops is supported by the induction of R-loops by the splicing inhibitor (100,101), pladienolide B (Plad-B) targeting SF3B1.
Removal and resolution of preformed R-loops
Ribonucleases H are the best-known factors for resolving R-loops (Figures 3C and 4d. These enzymes are conserved from bacteria to human and act by degrading the RNA moiety of RNA–DNA hybrids. Humans have two RNases H, RNase H1 (monomeric encoded by the RNH1 gene) and RNase H2 (trimeric consisting of RNH2A, RNH2B and RNH2C), which both can digest R-loops (60,66,78,86,91,102–105). Since RNase H enzymes fully remove the RNA, such a process releases the Sc- previously stored in R-loops, i.e. 11 Sc– for each 100-bp of a given processed R-loop (39). The released Sc– can promote the folding of chromatin, the formation of nucleosomes (each nucleosome requires 1 Sc–), the firing of replication origins and induce transcription, allow DNA repair or recombination (Figure 4e). Alternatively, the Sc- generated by RNases H has to be removed by topoisomerases: with TOP1 acting in underwound duplex DNA, TOP2 at DNA crossovers and TOP3B within DNA segments where the strands are separated by hypernegative superhelicity (Figure 4g).
RNA/DNA helicases also play an important role in R-loop removal (Figures 3F and 4I). Although the molecular mechanism is not established in vivo, based on their in vitro activities it is plausible that helicases act by unwinding the DNA–RNA hybrids of R-loops. The yeast protein Sen1 and its human homologue senataxin (SETX) were the first helicases identified and downregulation of human SETX causes accumulation of R-loops both at transcription pause sites, as well as at DSBs (50,106–108). A growing number of RNA helicases can also suppress R-loops including Aquarius (AQR) (from the same family as Sen1/SETX), members of DEAD box helicases (DDX1, DDX5, DDX19A, DDX21, DDX23) and DExH box (DHX9) helicase families, as well as BLM, FANCM helicase/translocase and WRN (55,63,88,109–125). How cells use these different helicases vs. RNase H remains to be established and is beyond the scope of this review. Of note, as in the case of RNase H described above, removal of R-loops by helicases within topologically confined DNA segments releases the Sc− previously sequestered in the R-loop, suggesting the coupling of R-loops with topoisomerases or the release of Sc- with chromatin opening after the unwinding of R-loops by helicases.
TOPOISOMERASES IB (TOP1 AND TOP1MT) AND R-LOOPS
TOP1’s connections with R-loop are summarized below and detailed with references in the prior and following section of this review:
TOP1 is abundant and extremely efficient at fully removing Sc− in duplex DNA, which enables TOP1 to dissipate the torsional tensions in the wake of RNA polymerase complexes (Figure 3), thereby preventing the persistent annealing of the nascent RNA with its template (Figure 3A).
TOP1’s efficiency at removing any amounts of Sc+ in front of translocating transcription complexes enables the rapid translocation of RNA polymerases along their DNA templates (Figure 3B).
Direct protein interactions couple TOP1 with RNA polymerase complexes (CTD of POL2; Figure 3, top).
TOP1 is concentrated in nucleoli where rDNA is transcribed at very high rates and R-loops tend to accumulate.
TOP1 has been proposed to stimulate RNA splicing, thereby avoiding the retention of long non-coding RNAs next to the DNA template and the stalling of transcription complexes [reviewed in (1,2)].
By removing Sc+ in topologically confined genomic segments, TOP1 can readily dissipate the Sc+ generated by R-loop formation in trans (Figure 4C).
TOP1 can remove the torsional stress released by the resolution of R-loops by RNase H and helicases (Figure 4G).
Historically, the connections of TOP1 with R-loops originated from the observations that human TOP1 could bind and phosphorylate splicing factors of the SR family (126–131) and that inhibiting TOP1 had direct impact in mRNA processing (132–134). As formation of mRNPs prevents R-loop formation, Tuduri et al. (76) postulated that TOP1 might prevent R-loop formation via phosphorylation of ASF/SF2 (as phosphorylation of SR proteins controls their recruitment to actively transcribing chromatin sites and subsequent mRNP formation). They found that TOP1-deficient human cells accumulate R-loop-induced DNA breaks and transcription-dependent replication stress (fork slowdown, stalling, and asymmetry) at actively transcribed genes. They also found that over-expression of RNase H1 suppressed the DNA damage and replication defect phenotypes and that yeast (S. cerevisiae) Top1 (lacking the kinase domain) could partially suppress these defects. Although the authors did not actually demonstrate the presence of higher levels of R-loops in TOP1-deficient cells and inferred the presence of R-loops from the fact that phenotypes were rescued by RNase H1, these results indicated that TOP1 could possibly suppress R-loop formation and subsequent DNA damage in actively transcribed genes in two separate ways: by relaxing the Sc− accumulated behind RNA polymerases and by promoting ASF/SF2 dependent mRNP formation and RNA sequestration (76).
Later it was found that yeast (S. cerevisiae) Top1 is also important for suppressing R-loops generated in actively transcribing ribosomal DNA region, and that Pol I transcription in the absence of Top1 was facilitated by RNase H activity (72). This study by El-Hage et al. (72) demonstrated that yeast strains lacking functional Top1 accumulate R-loops (as detected by chromatin immunoprecipitation analysis using the S9.6 antibody) over the 18S 5′-region causing defects in precursor ribosomal RNA (pre-rRNA) synthesis by RNA Pol I and accumulation of truncated pre-rRNA fragments. These defects in pre-rRNA synthesis were heightened in strains lacking both Top1 and Top2 activity [i.e. top1Δ top2-ts (temperature sensitive) strains at nonpermissive temperatures]. Higher levels of R-loops were also detected in strains lacking both Top1 and RNase H (72).
Recent publications suggest that, based on genome-wide R-loop analyses in TOP1-downregulated human cells, TOP1 plays much more complex roles in R-loop physiology (26,77). The study by Manzo et al. demonstrates that shRNA-mediated loss of TOP1 causes both loss and gain of R-loops for different group of genes in human embryonic kidney HEK293 cells. Long and highly transcribed genes, genes present in gene poor areas that cannot dissipate DNA supercoiling due to physical anchoring (for example: due to proximity to heterochromatin/ H3K9me3-mark and lamin B1 association) gained R-loops after TOP1 depletion. Conversely, TOP1 depletion caused loss of R-loops in genes that are of average length, reside in gene-rich neighborhoods, and are moderately expressed. These group of genes having decreased R-loop signals also co-localized with replication initiation regions, H3K27me3 Polycomb marks and components of the PRC complexes. A third group of genes displayed both loss and gain of R-loops after TOP1 depletion (26).
More recently, Promonet et al. highlighted TOP1’s role in removing persistent R-loops and torsional stress for a group of genes with replication and transcription machineries colliding in a head-on (HO) manner at their transcription termination sites (TTS). After shRNA-mediated depletion of TOP1 in HeLa cells, TTS of highly transcribed HO genes suffered replication stress (fork collapse due to persistent R-loops and accumulation of torsional stress) and DSBs as indicated by the accumulation of phosphorylated RPA32, γH2AX peaks and DSBs (detected by i-BLESS) (77).
One of the cofactors contributing to TOP1’s R-loop suppressing activity is the BET-BRD protein BRD2, which directly interacts with and stimulates TOP1 activity to prevent co-transcriptional R-loops (135). Another bromodomain protein BRD4 has also been shown to control TOP1 activity by regulating POL2 phosphorylation, which also contributes to TOP1-mediated R-loop suppression (70,135) (Figure 3, top). In addition, Li et al. reported that the helicase domain of RECQ5 interacts directly with TOP1 and induces TOP1 SUMOylation by the protein inhibitor of activated STAT protein 1 (PIAS1)–SRSF1 E3 ligase complex. The resulting K391/K436 SUMOylation of TOP1 was shown to promote TOP1−POL2 interactions and increase the efficient recruitment of RNA splicing factors that suppress R-loop formation (136). Confirming the role of TOP1 in suppressing R-loops and its connection with cancer, a recent study showed that the oncogene mutated HRAS (RAS) can increase TOP1 levels to minimize oncogene-induced R-loops and in turn increases replication-induced DNA damage (137).
Apart from genetic evidence that TOP1 suppresses R-loops, multiple studies have established that inhibition of TOP1 by anticancer drugs derived from camptothecin (CPT) (topotecan, irinotecan, belotecan) and the non-camptothecin indenoisoquinoline derivatives (138) can cause R-loops (63–65,101,111,139–141) (Table I). CPT-induced R-loops have been mapped at active divergent CpG-island (CGI) promoters after RNA polymerase stalling at actively transcribing genes including rDNA (140). CPT also causes R-loops in post-mitotic neuronal cells (65), as first demonstrated by Sordet et al. who showed that CPT-induced DNA damage response is attenuated by inhibiting transcription with DRB or by RNase H1 overexpression. CPT-induced R-loop formation can be explained by two molecular mechanisms. The trapping of TOP1 cleavage complexes (TOP1ccs) (see Figure 1) by CPT(138,142) can sequester TOP1, thereby decreasing the active TOP1 pool and in turn increasing Sc- density in cellular regions prone to R-loops (1,2,143). The trapping of TOP1cc can also induce covalent TOP1-DNA protein crosslinks (TOP-DPCs) (144,145), which stall RNA polymerases, induce their backtracking and generate R-loops. Additionally, CPT has been shown to inhibit transcription elongation while stimulating transcription initiation from promoter proximal sequences, which can also result in promoter associated R-loops (146). R-loop-derived cytoplasmic RNA–DNA hybrids are known to induce an innate immune response in cells and similarly the R-loops formed after TOP1 poisoning can lead to micronuclei formation and activation of the cGAS-STING pathway to mediate immune gene expression in HeLa cells (147,148). Topotecan-mediated inhibition of TOP1 action can also stabilize R-loops in neuron-specific noncoding RNA Snord116 leading to stalled transcription of antisense transcript to Angelman syndrome causing ubiquitin ligase encoding Ube3a (Ube3a-ATS) and activation of paternal UBE3A (149).
TOP3B AND R-LOOPS
TOP3B’s connections with R-loop suppression stems from two known molecular mechanisms summarized below and in Figure 5. They are detailed with references in the prior and following section of this review:
Figure 5.
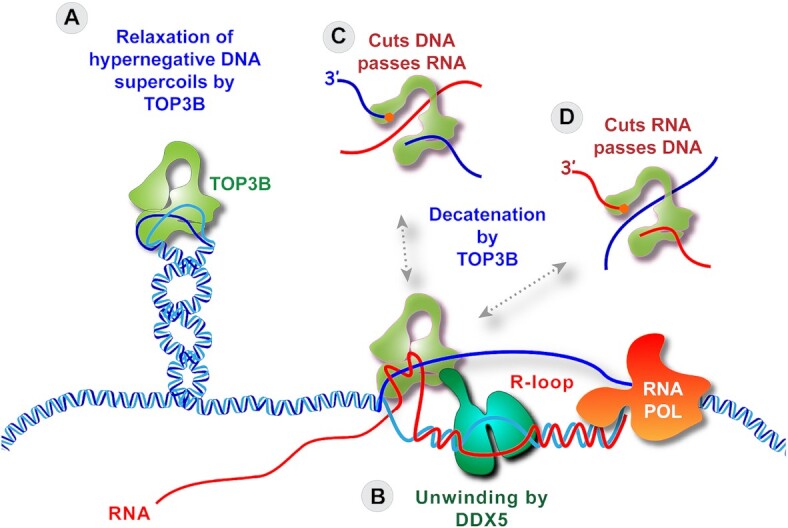
Proposed molecular mechanisms by which TOP3B can suppress R-loops (101). (A) By cleaving single-stranded DNA and passing the complementary strand through the gap, TOP3B can reduce hypernegative Sc and prevent the formation or destabilize R-loops. (B) Following the unwinding of the DNA–RNA heteroduplex of the R-loop by DDX5, TOP3B can decatenate the nucleic acids at the junction of the R-loop by cutting a DNA single-strand and passing the RNA through the gap (C), or by cutting the RNA and passing a DNA single-strand through the gap (D).
TOP3B is the only dual RNA-DNA topoisomerase (Figure 1) with potent decatenation activity, which enables TOP3B to bind and disassemble R-loops by decatenation (Figures 4I and 5B−D; and details below) (101).
TOP3B can reduce hypernegative supercoiling when the base pairs of the DNA double-helix are separated, allowing TOP3B to pass single-stranded DNA segments across each other (Figures 4H and 5A).
The role of TOP3B in suppressing R-loops is well established in cellular and biochemical systems (12,13,73–75,101,150). Recently a patient with bilateral renal cancer reportedly carried homozygous deletion for the TOP3B gene and had higher R-loop burden in lymphoblast cells (75). Another recent study in Drosophila (150) highlights the importance of top3b during aging as photoreceptor-specific depletion of top3b resulted in R-loops in photoreceptor neurons, which increased with age, indicating a critical role Top3b in suppressing these R-loops, promoting the expression of long genes with neuronal function and enhancing visual response during aging (150). Additionally, we recently reported that trapping of TOP3B cleavage complexes (TOP3Bccs) by genetic alteration of TOP3B (replacing the arginine 338 residue by tryptophan) causes R-loop accumulation in human HCT116 and HEK293 cells (10).
TOP3B’s scaffolding factor TDRD3, which reads asymmetric di-methylated arginine marks in core histones (H3R17me2a and H4R3me2a) and POL2 (R1810me2a) via its ‘Tudor domain’ has been proposed to target TOP3B to transcriptionally active CpG island (CGI) promoters to prevent R-loop accumulation (Figure 3, upper panel). TOP3B can relax hypernegative supercoil and this property of TOP3B is possibly important for prevention of R-loop formation (12). Consistent with this possibility, Huang et al. (73) highlighted the importance of arginine methylation of TOP3B in its C-terminal RGG motif (residues R833 and R835) for relaxing hypernegative supercoiling both in vitro and in vivo. Methylation-deficient TOP3B (R833/835K) was found defective in Sc- relaxation activity and shown to accumulate co-transcriptionally formed R-loops in in vitro assays and in TOP3B-TDRD3 targeting gene promoters in cells (73). TDRD3 knockout mice also show increased R-loops and DSBs at c-Myc and the Ig heavy chain (Igh) loci, which can promote oncogenic c-Myc/Igh translocations as frequently observed in Burkitt's lymphoma and multiple myeloma (12,68).
The first evidence that eukaryotic TOP3B resolves R-loops was reported in biochemical studies performed with Drosophila Top3b by Tao Hsieh and colleagues (74). They showed that Top3b resolves plasmid-based R-loops by cleaving the unpaired/ displaced strand of R-loop structures along their length (74). Recently, we have also established that, similar to Drosophila Top3b, human TOP3B can disassemble R-loop structures in vitro and cleave the single-stranded region of R-loops (101). In human colon cancer HCT116 and human embryonic kidney HEK293 cells, we showed (101) that TOP3B resolves both physiologically formed genotoxic/ aberrant R-loops and R-loops formed in response to two independent R-loop inducing agents; CPT, which traps TOP1 (see previous section), and pladienolide B (Plad-B), which inhibits the spliceosome (96,97,110,111,139,140,147) by stabilizing a transition state of the SF3B complex (151,152). We also showed that TOP3B can resolve cellular R-loops induced by downregulation of senataxin (SETX) (101).
Direct evidence for the physical association of TOP3B with R-loops has recently been provided in human cells treated with CPT or Plad-B, or deficient for Senataxin (by siSETX transfection) (101). TOP3B was detected in the R-loops by DNA-RNA hybrid immunoprecipitation and Western blotting. Notably, the kinetics of TOP3B cleavage complexes (TOP3Bccs) was the same as the kinetics of R-loop formation (within minutes and transient in the case of CPT, and progressive over several hours in the case of Plad-B), and the TOP3Bccs were found associated both RNA and DNA, consistent with TOP3B acting as R-loop decatenase (101). We proposed a model (Figure 5) where TOP3B interacts with the helicase DDX5 (independently of TDRD3) in an epistatic manner to resolve cellular R-loops. DDX5, an ATP-dependent RNA helicase is widely known to interact with and resolve R-loops both in vitro and in vivo in promoters, near the transcription start sites, transcription termination sites and at DSBs (31,32,115,119–123). We also identified other TOP3B interacting helicases (interacting with TOP3B independently of TDRD3 and nucleic acid) with known roles in R-loop metabolism including SNRNP200, HELZ2, DHX9, DDX3X, DDX17, DDX21, DDX6, DHX15, DHX19A, RECQL and Aquarius (AQR). R-loop unwinding by DDX5 helicase and possibly other R-loop unwinding TOP3B-coupled helicases possibly generate intertwined DNA-RNA molecules that are resolved by TOP3B-mediated decatenation (cleavage and strand passage activity on single-stranded DNA and RNA regions of R-loops) (101) (Figure 5).
CPT treatment induces R-loops mostly in promoters (CpG-islands) at active TSS (139,140,147). The distribution of Plad-B-induced R-loops is not well-established. A recent study (100) demonstrated that these R-loops map to intergenic regions, extending downstream of POL2 transcribing genes. These newly formed R-loops after Plad-B treatment cover ∼50 kb of total genome and caused by read-through transcription after the poly-A signals of a few hundred genes (100). On the contrary, after senataxin (SETX) depletion, R-loops tend to accumulate at transcription pause sites, as well as at DSB sites (50,106–108). As TOP3B can resolve R-loops formed by different mechanisms (CPT, Plad-B treatments and siSETX transfection) it can be speculated that TOP3B can work on and resolve R-loops formed at diverse genomic location i.e. in transcription start site of active CGI promoters (R-loops associated with CPT treatment), transcription pause sites, DSBs (formed after senataxin depletion) and in intergenic regions, extending downstream of RNAPII transcribing genes (linked with Plad-B treatment) (50,64,96,97,100,106–108,110,111,139,140,147,153,154).
Another recent study demonstrated that in MCF7 breast cancer cell DHX9-TOP3B interaction is TDRD3 dependent and TDRD3-DHX9 forms separate complexes and works independently of TOP3B to resolve promoter-associated R-loops (13). Further detailed studies are warranted to explore the mechanism by which DHX9 and other TOP3B interacting helicases are coupled with TOP3B to suppress excessive R-loops formed in cancer cells and tumors.
R-LOOPS AND CANCER
Because of their association with high transcription and their potential to induce DNA breaks and a mutator phenotype, R-loops have been associated with oncogenes and tumor suppressor genes. Yet, the identification and quantification of R-loops remains challenging. The 9.6 antibody is not routinely usable in the clinic and appropriate controls are necessary even in tissue culture settings due to the binding of the antibody to non-R-loops structures such as double-stranded RNA (20,155).
Table 1 lists some of the known tumor suppressor genes associated with R-loops including BRCA1, BRCA2, ATRX, BLM, WRN, Fanconi anemia genes (FANCA, FANCD2, FANACI, FANCJ, and FANCM), TOP3B or splicing factors (SF3B1, U2AF1, SRSF2). It also lists known oncogenes associated with increased R-loops including HRAS, EWS-FLI1, SS18–SSX1. Mutations/inactivation of TOP3B and R-loops have also been proposed to be oncogenic based on the observation that a patient with renal carcinoma presented with TOP3B deletion in his cancer cells (75).
Table 1.
Cancer relevant genes linked with R-loops
| Genes | Details/remarks | References |
|---|---|---|
| SRSF2, U2AF1, SF3B1, U2AF2, and ZRSR2 |
Mutations in these splicing factors cause pausing of POL2, increased R-loops, replication stress and genomic instability Linked with myelodysplastic syndromes (MDSs), acute myeloid leukemia (AML), uveal melanoma, bladder, pancreatic and lung cancer |
(96–98,100,162–170) |
| BRCA1 | BRCA1 regulates transcription elongation and its inactivation induces R-loops at the 5′ end of genes with promoter-proximal POL2 pausing (by counteracting its binding partner COBRA1), transcription and R-loops at 3′ transcription termination pause sites (with senataxin), R-loops at centromeric α-satellite repeats, telomeric TERRA R-loops Luminal progenitor cells of BRCA1-associated breast tumors accumulate R-loops Ewing sarcoma phenocopies BRCA1-deficient tumors. BRCA1 sequestration by transcription machinery and R-loop structures causes HR defect In MYCN-amplified human neuroblastoma cells, BRCA1 limits MYCN driven R-loop formation in promoter-proximal regions |
(54,171–177) |
| BRCA2 | Interacts with POL2 and regulates R-loops formed at promoter-proximal pause sites at the 5′ end of genes Associates with TREX-2 to prevent R-loop accumulation Regulates DNA-RNA hybrid level at DSBs by recruiting RNase H2 Associates with and promotes DDX5 activity to control R-loops at DSBs |
(93,121,178–180) |
| FANCA, FANCD2, FANACI, FANCJ, and FANCM | Reduce R-loops at transcription–replication conflict regions In FANCD2-/- patient-derived lymphoblast R-loops cause replication fork arrest at common fragile site (CFS) |
(181–184) |
| EWS–FLI1 | Fusion of N-terminal of EWSR1 and C-terminal of FLI1 protein. Regulates R-loops by controlling POL2 elongation step and via interaction with splicing machinery |
(175) |
| TOP3B | TOP3B deletion is linked increased accumulation of R-loops in renal cancer cells from patient | (75) |
| ATRX (alpha thalassemia/mental retardation X-linked) | Mutated in ALT-positive human tumors (brain cancers, pancreatic neuroendocrine tumors, chondroblastomas, and osteosarcomas) and implicated in alternative lengthening of telomeres activation Chromatin remodeling factor; Controls R-loops in telomeres and rDNA repeats. Interacts with RNAs and prevent DNA-RNA hybrid formation in R-loops |
(185–188) |
| RecQ helicases |
BLM possibly unwinds DNA-RNA hybrid of R-loops and protects cells from genome instability WRN plays role in activation of ATR/CHK1, and ATM signaling pathways and protect cells from R-loop induced genome instability |
(124,125,189–191) |
| SS18–SSX1 oncoprotein |
Fusion of the SS18 subunit of BAF chromatin remodeling complex with SSX Promotes R-loops and replication stress in synovial sarcoma |
(192) |
| HRAS | Increases global transcription and R-loop formation | (193) |
Based on the prevalence of R-loops in cancers and on the genomic instability resulting from excessive R-loops, a plausible hypothesis is that cancer cells with alterations in genes that increase R-loops should be vulnerable to drugs that further enhance R-loops (Table 2) or to drugs that preferentially target cancer cells with high R-loop burden (Table 3). As discussed in the prior sections of this review, both TOP1 and splicing inhibitors can be included in these categories. These drugs can be viewed as mostly enhancing the DNA damaging effects of R-loops (see Figure 2) and inducing replication blocks that need ATR/CHK1 activity. Yet, the oncolytic effects of TOP1 and of splicing inhibitors are not solely due to an increase in R-loop burden. Thus, searching for specific R-loop inducers, such as TOP3B, RNase H and helicase inhibitors can be viewed as potential avenues for anticancer drug development.
Table 2.
Pharmacological agents that induce or stabilize R-loops
| Drugs/inhibitors/agents | Target/mechanism of action/remarks | References |
|---|---|---|
| Camptothecin (CPT) and analogues | Traps TOP1 cleavage complexes (TOP1ccs) | (101,139,140,147) |
| JQ1 | Bromodomain and extraterminal domain (BET) protein family inhibitor | (135,194,195) |
| Pladienolide B (Plad-B) | Inhibits the splicing factor SF3B1and in turn inhibits splicing |
(96–98,100,110,162,163) |
| Crosslinking agents like Reactive natural aldehydes and mitomycin c | Degrade BRCA2 causing accumulation of R-loops Can increase R-loop levels, especially in cells without a functioning FA pathway by physical stalling of the transcription machinery which promote R-loop formation |
(181,182,196,197) |
| Pyridostatin/ PDS, Braco-19, and FG | Bind and stabilize G4 quadruplex structures in the displaced strand of R loops which in turn stabilizes DNA: RNA hybrids and helps extension of cotranscriptional R loops Can target BRCA1/2 deficient cancers |
(198) |
| CX-5461 | Inhibits RNA polymerase I transcription and G4 quadruplex ligand Selectively kills tumors with BRCA1/2 deficiency |
(199,200) |
| Romidepsin | HDAC Inhibitor | (201) |
| VX970 and other ATR inhibitors | ATR inhibition and increase replication stress | (30,192,202) |
| SMG1 inhibitor | Inhibits serine/threonine-protein kinase SMG1 and non-sense mediated decay | (99) |
| Anticancer sulphonamides | Target U2AF-related splicing factor coactivator of activating protein-1 for proteasomal degradation | (98,203) |
Table 3.
Drugs that kill cells with elevated R-loop levels
| Drug | Details/remarks | References |
|---|---|---|
| Trabectedin and lurbinectedin | Form DNA adducts and enhance R-loop associated DNA damage and replication stress in actively transcribing region Target cancers with mutations BRCA1/2 and Fanconi anemia protein |
(204–206) |
| ATR inhibitors (VE-821) | ATR inhibition in cells with high R-loop burden (after depletion of splicing factor SRSF1, helicase senataxin) leads to increase in MUS81–EME1-generated DNA damage Kill myelodysplastic syndrome (MDS) and acute myeloid leukemia cancer cells with mutations in splicing factors U2AF1 and high R-loop burden Kills Ewing sarcoma cells with high R-loop burden |
(54,96,175,207) |
| Olaparib (PARP1 inhibitor) | Ewing sarcoma phenocopies BRCA1-deficient tumors. BRCA1 sequestration by transcription machinery and R-loop structures cause HR defect and sensitize to PARP inhibitors | (54,175) |
CONCLUSIONS AND FUTURE CHALLENGES
In this review we summarized the roles of type IA and type IB topoisomerases in R-loop metabolism and the importance of DNA topology for the suppression and induction of R-loops. Yet, several areas remain unexplored. Although, the rDNA repeats are known to contain many R-loops due to their high transcription, the biological impact, and the resolution of excessive R-loops in rDNA remains understudied.
Another part of the human genome where R-loops deserve further understanding is the mitochondrial DNA (mtDNA). Replication of mtDNA has been proposed to be associated with R-loops but this possibility remains controversial (156). Like TOP1, TOP1MT efficiently relaxes both Sc– and Sc+ (157,158). Furthermore, mitochondria do not contain detectable TOP3B and limited TOP2 enzymes (159,160) and no RNase H2 (161). Genetic deletion of TOP1MT increases the Sc- of mtDNA with D-loop alterations (159). Yet, mitochondrial R-loops have not been mapped under these conditions.
The TOP3A-interacting helicase BLM has been reported to resolve in R-loops (125). Yet, it is unknown whether TOP3A, which like TOP3B only works on single-stranded regions in hypernegatively supercoiled DNA plays any role in R-loop homeostasis.
Finally, one of the challenges to study and take advantage of the increasing biological knowledge on R-loops is the development of simple and quantitative assays to measure R-loops in tissues and cancer samples.
DATA AVAILABILITY
No new data were generated or analysed in support of this research.
ACKNOWLEDGEMENTS
We wish to thank Dr Xi Yang for contributing to the drawing of Figure 5, and all members of the Laboratory of Molecular Pharmacology, Developmental Therapeutics Branch for their input.
Contributor Information
Sourav Saha, Developmental Therapeutics Branch & Laboratory of Molecular Pharmacology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Yves Pommier, Developmental Therapeutics Branch & Laboratory of Molecular Pharmacology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
FUNDING
Center for Cancer Research, the Intramural Program of the National Cancer Institute, NIH [Z01-BC-006161].
Conflict of interest statement. None declared.
REFERENCES
- 1. Pommier Y., Nussenzweig A., Takeda S., Austin C. Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 2022; 23:407–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pommier Y., Sun Y., Huang S.N., Nitiss J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001; 70:369–413. [DOI] [PubMed] [Google Scholar]
- 4. Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. [DOI] [PubMed] [Google Scholar]
- 5. Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011; 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel P.S., Krishnan R., Hakem R. Emerging roles of DNA topoisomerases in the regulation of R-loops. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2022; 876-877:503450. [DOI] [PubMed] [Google Scholar]
- 7. Bizard A.H., Hickson I.D. The many lives of type IA topoisomerases. J. Biol. Chem. 2020; 295:7138–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmad M., Xu D., Wang W. Type IA topoisomerases can be “magicians” for both DNA and RNA in all domains of life. RNA Biol. 2017; 14:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J. et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013; 16:1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saha S., Sun Y., Huang S.N., Baechler S.A., Pongor L.S., Agama K., Jo U., Zhang H., Tse-Dinh Y.C., Pommier Y. DNA and RNA cleavage complexes and repair pathway for TOP3B RNA- and DNA-protein crosslinks. Cell Rep. 2020; 33:108569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stoll G., Pietilainen O.P.H., Linder B., Suvisaari J., Brosi C., Hennah W., Leppa V., Torniainen M., Ripatti S., Ala-Mello S. et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013; 16:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y., McBride K.M., Hensley S., Lu Y., Chedin F., Bedford M.T. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell. 2014; 53:484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan W., Al-Hadid Q., Wang Z., Shen L., Cho H., Wu X., Yang Y. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res. 2021; 49:8573–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berger J.M., Gamblin S.J., Harrison S.C., Wang J.C. Structure and mechanism of DNA topoisomerase II. Nature. 1996; 379:225–232. [DOI] [PubMed] [Google Scholar]
- 15. Nitiss J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009; 9:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uuskula-Reimand L., Wilson M.D. Untangling the roles of TOP2A and TOP2B in transcription and cancer. Sci. Adv. 2022; 8:eadd4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanz L.A., Hartono S.R., Lim Y.W., Steyaert S., Rajpurkar A., Ginno P.A., Xu X., Chedin F. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016; 63:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas M., White R.L., Davis R.W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White R.L., Hogness D.S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977; 10:177–192. [DOI] [PubMed] [Google Scholar]
- 20. Chedin F., Benham C.J. Emerging roles for R-loop structures in the management of topological stress. J. Biol. Chem. 2020; 295:4684–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malig M., Hartono S.R., Giafaglione J.M., Sanz L.A., Chedin F. Ultra-deep coverage single-molecule R-loop footprinting reveals principles of R-loop formation. J. Mol. Biol. 2020; 432:2271–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003; 4:442–451. [DOI] [PubMed] [Google Scholar]
- 23. Crossley M.P., Bocek M.J., Hamperl S., Swigut T., Cimprich K.A. qDRIP: a method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res. 2020; 48:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castillo-Guzman D., Chedin F. Defining R-loop classes and their contributions to genome instability. DNA Repair (Amst.). 2021; 106:103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manzo S.G., Hartono S.R., Sanz L.A., Marinello J., De Biasi S., Cossarizza A., Capranico G., Chedin F. DNA topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 2018; 19:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan Y.A., Aristizabal M.J., Lu P.Y., Luo Z., Hamza A., Kobor M.S., Stirling P.C., Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014; 10:e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wahba L., Costantino L., Tan F.J., Zimmer A., Koshland D S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016; 30:1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ariel F., Lucero L., Christ A., Mammarella M.F., Jegu T., Veluchamy A., Mariappan K., Latrasse D., Blein T., Liu C. et al. R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell. 2020; 77:1055–1065. [DOI] [PubMed] [Google Scholar]
- 30. Barroso S., Herrera-Moyano E., Munoz S., Garcia-Rubio M., Gomez-Gonzalez B., Aguilera A. The DNA damage response acts as a safeguard against harmful DNA-RNA hybrids of different origins. EMBO Rep. 2019; 20:e47250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cloutier S.C., Wang S., Ma W.K., Al Husini N., Dhoondia Z., Ansari A., Pascuzzi P.E., Tran E.J Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol. Cell. 2016; 62:148. [DOI] [PubMed] [Google Scholar]
- 32. Gomez-Gonzalez B., Aguilera A. Origin matters: spontaneous DNA-RNA hybrids do not form in trans as a source of genome instability. Curr. Genet. 2021; 67:93–97. [DOI] [PubMed] [Google Scholar]
- 33. Niehrs C., Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020; 21:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wahba L., Gore S.K., Koshland D The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013; 2:e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fernandes R.V., Feretzaki M., Lingner J. The makings of TERRA R-loops at chromosome ends. Cell Cycle. 2021; 20:1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lafuente-Barquero J., Garcia-Rubio M.L., Martin-Alonso M.S., Gomez-Gonzalez B., Aguilera A. Harmful DNA:RNA hybrids are formed in cis and in a Rad51-independent manner. Elife. 2020; 9:e56674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marnef A., Legube G. R-loops as Janus-faced modulators of DNA repair. Nat. Cell Biol. 2021; 23:305–313. [DOI] [PubMed] [Google Scholar]
- 38. Petermann E., Lan L., Zou L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat. Rev. Mol. Cell Biol. 2022; 23:521–540. [DOI] [PubMed] [Google Scholar]
- 39. Stolz R., Sulthana S., Hartono S.R., Malig M., Benham C.J., Chedin F. Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:6260–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Drolet M., Broccoli S., Rallu F., Hraiky C., Fortin C., Masse E., Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 2003; 8:d210–d221. [DOI] [PubMed] [Google Scholar]
- 41. Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Belotserkovskii B.P., Hanawalt P.C. Mechanism for R-loop formation remote from the transcription start site: topological issues and possible facilitation by dissociation of RNA polymerase. DNA Repair (Amst.). 2022; 110:103275. [DOI] [PubMed] [Google Scholar]
- 43. Belotserkovskii B.P., Hanawalt P.C. Topology and kinetics of R-loop formation. Biophys. J. 2022; 121:3345–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gowrishankar J., Leela J.K., Anupama K. R-loops in bacterial transcription: their causes and consequences. Transcription. 2013; 4:153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Drolet M., Brochu J. R-loop-dependent replication and genomic instability in bacteria. DNA Repair (Amst.). 2019; 84:102693. [DOI] [PubMed] [Google Scholar]
- 46. Masse E., Drolet M. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J. Mol. Biol. 1999; 294:321–332. [DOI] [PubMed] [Google Scholar]
- 47. Pohjoismaki J.L., Holmes J.B., Wood S.R., Yang M.Y., Yasukawa T., Reyes A., Bailey L.J., Cluett T.J., Goffart S., Willcox S. et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010; 397:1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu B., Clayton D.A. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 1996; 15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 49. Grunseich C., Wang I.X., Watts J.A., Burdick J.T., Guber R.D., Zhu Z., Bruzel A., Lanman T., Chen K., Schindler A.B. et al. Senataxin mutation reveals how R-loops promote transcription by blocking DNA methylation at gene promoters. Mol. Cell. 2018; 69:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skourti-Stathaki K., Proudfoot N.J., Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kabeche L., Nguyen H.D., Buisson R., Zou L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 2018; 359:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crossley M.P., Bocek M., Cimprich K.A. R-loops as cellular regulators and genomic threats. Mol. Cell. 2019; 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee C.Y., McNerney C., Ma K., Zhao W., Wang A., Myong S. R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat. Commun. 2020; 11:3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wells J.P., White J., Stirling P.C. R loops and their composite cancer connections. Trends Cancer. 2019; 5:619–631. [DOI] [PubMed] [Google Scholar]
- 55. Brickner J.R., Garzon J.L., Cimprich K.A. Walking a tightrope: the complex balancing act of R-loops in genome stability. Mol. Cell. 2022; 82:2267–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gan W., Guan Z., Liu J., Gui T., Shen K., Manley J.L., Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011; 25:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamperl S., Bocek M.J., Saldivar J.C., Swigut T., Cimprich K.A. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell. 2017; 170:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lang K.S., Hall A.N., Merrikh C.N., Ragheb M., Tabakh H., Pollock A.J., Woodward J.J., Dreifus J.E., Merrikh H. Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell. 2017; 170:787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wellinger R.E., Prado F., Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006; 26:3327–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garcia-Muse T., Aguilera A. Loops: from physiological to pathological roles. Cell. 2019; 179:604–618. [DOI] [PubMed] [Google Scholar]
- 61. Aguilera A., Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 62. Kumar C., Batra S., Griffith J.D., Remus D The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions. Elife. 2021; 10:e72286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sollier J., Stork C.T., Garcia-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014; 56:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cristini A., Ricci G., Britton S., Salimbeni S., Huang S.N., Marinello J., Calsou P., Pommier Y., Favre G., Capranico G. et al. Dual processing of R-loops and topoisomerase I induces transcription-dependent DNA double-strand breaks. Cell Rep. 2019; 28:3167–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sordet O., Redon C.E., Guirouilh-Barbat J., Smith S., Solier S., Douarre C., Conti C., Nakamura A.J., Das B.B., Nicolas E. et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009; 10:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Santos-Pereira J.M., Aguilera A. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 67. Perego M.G.L., Taiana M., Bresolin N., Comi G.P., Corti S. R-loops in motor neuron diseases. Mol. Neurobiol. 2019; 56:2579–2589. [DOI] [PubMed] [Google Scholar]
- 68. Richard P., Manley J.L. Loops and links to human disease. J. Mol. Biol. 2017; 429:3168–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006; 59:723–730. [DOI] [PubMed] [Google Scholar]
- 70. Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H. et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016; 165:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Drolet M., Bi X., Liu L.F. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 1994; 269:2068–2074. [PubMed] [Google Scholar]
- 72. El Hage A., French S.L., Beyer A.L., Tollervey D Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010; 24:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang L., Wang Z., Narayanan N., Yang Y. Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 2018; 46:3061–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilson-Sali T., Hsieh T.S. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase iiibeta. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7974–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang T., Wallis M., Petrovic V., Challis J., Kalitsis P., Hudson D.F. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019; 9:190222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tuduri S., Crabbe L., Conti C., Tourriere H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009; 11:1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Promonet A., Padioleau I., Liu Y., Sanz L., Biernacka A., Schmitz A.L., Skrzypczak M., Sarrazin A., Mettling C., Rowicka M. et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 2020; 11:3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Drolet M., Phoenix P., Menzel R., Masse E., Liu L.F., Crouch R.J. Overexpression of rnase H partially complements the growth defect of an Escherichia coli delta topA mutant: r-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Phoenix P., Raymond M.A., Masse E., Drolet M. Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J. Biol. Chem. 1997; 272:1473–1479. [DOI] [PubMed] [Google Scholar]
- 80. Usongo V., Nolent F., Sanscartier P., Tanguay C., Broccoli S., Baaklini I., Drlica K., Drolet M. Depletion of rnase HI activity in Escherichia coli lacking DNA topoisomerase I leads to defects in DNA supercoiling and segregation. Mol. Microbiol. 2008; 69:968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brochu J., Vlachos-Breton E., Sutherland S., Martel M., Drolet M. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet. 2018; 14:e1007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Broccoli S., Phoenix P., Drolet M. Isolation of the topB gene encoding DNA topoisomerase III as a multicopy suppressor of topA null mutations in Escherichia coli. Mol. Microbiol. 2000; 35:58–68. [DOI] [PubMed] [Google Scholar]
- 83. Lang K.S., Merrikh H. Topological stress is responsible for the detrimental outcomes of head-on replication-transcription conflicts. Cell Rep. 2021; 34:108797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Aguilera A. mRNA processing and genomic instability. Nat. Struct. Mol. Biol. 2005; 12:737–738. [DOI] [PubMed] [Google Scholar]
- 85. Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003; 12:711–721. [DOI] [PubMed] [Google Scholar]
- 86. Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 87. Li X., Manley J.L. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006; 20:1838–1847. [DOI] [PubMed] [Google Scholar]
- 88. Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009; 35:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012; 26:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tresini M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers J.A., van I.W.F., Grosveld F.G., Medema R.H., Hoeijmakers J.H. et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015; 523:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wahba L., Amon J.D., Koshland D., Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011; 44:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jimeno S., Rondon A.G., Luna R., Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002; 21:3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bhatia V., Barroso S.I., Garcia-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 94. Castellano-Pozo M., Garcia-Muse T., Aguilera A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep. 2012; 13:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dominguez-Sanchez M.S., Barroso S., Gomez-Gonzalez B., Luna R., Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011; 7:e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nguyen H.D., Leong W.Y., Li W., Reddy P.N.G., Sullivan J.D., Walter M.J., Zou L., Graubert T.A. Spliceosome mutations induce R loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 2018; 78:5363–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nguyen H.D., Yadav T., Giri S., Saez B., Graubert T.A., Zou L. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell. 2017; 65:832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen L., Chen J.Y., Huang Y.J., Gu Y., Qiu J., Qian H., Shao C., Zhang X., Hu J., Li H. et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol. Cell. 2018; 69:412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cheruiyot A., Li S., Nonavinkere Srivatsan S., Ahmed T., Chen Y., Lemacon D.S., Li Y., Yang Z., Wadugu B.A., Warner W.A. et al. Nonsense-mediated RNA decay is a unique vulnerability of cancer cells harboring SF3B1 or U2AF1 mutations. Cancer Res. 2021; 81:4499–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Castillo-Guzman D., Hartono S.R., Sanz L.A., Chédin F. SF3B1-targeted splicing inhibition triggers global alterations in transcriptional dynamics and R-loop metabolism. 2020; bioRxiv doi:08 June 2020, preprint: not peer reviewed 10.1101/2020.06.08.130583. [DOI]
- 101. Saha S., Yang X., Huang S.N., Agama K., Baechler S.A., Sun Y., Zhang H., Saha L.K., Su S., Jenkins L.M. et al. Resolution of R-loops by topoisomerase III-beta (TOP3B) in coordination with the DEAD-box helicase DDX5. Cell Rep. 2022; 40:111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Amon J.D., Koshland D RNase H enables efficient repair of R-loop induced DNA damage. Elife. 2016; 5:e20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lee D.Y., Clayton D.A. RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Genes Dev. 1997; 11:582–592. [DOI] [PubMed] [Google Scholar]
- 104. Maul R.W., Chon H., Sakhuja K., Cerritelli S.M., Gugliotti L.A., Gearhart P.J., Crouch R.J. R-loop depletion by over-expressed rnase H1 in mouse B cells increases activation-induced deaminase access to the transcribed strand without altering frequency of isotype switching. J. Mol. Biol. 2017; 429:3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lockhart A., Pires V.B., Bento F., Kellner V., Luke-Glaser S., Yakoub G., Ulrich H.D., Luke B. RNase H1 and H2 are differentially regulated to process RNA-DNA hybrids. Cell Rep. 2019; 29:2890–2900. [DOI] [PubMed] [Google Scholar]
- 106. Alzu A., Bermejo R., Begnis M., Lucca C., Piccini D., Carotenuto W., Saponaro M., Brambati A., Cocito A., Foiani M. et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012; 151:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cohen S., Puget N., Lin Y.L., Clouaire T., Aguirrebengoa M., Rocher V., Pasero P., Canitrot Y., Legube G. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 2018; 9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mischo H.E., Gomez-Gonzalez B., Grzechnik P., Rondon A.G., Wei W., Steinmetz L., Aguilera A., Proudfoot N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011; 41:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chakraborty P., Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.). 2011; 10:654–665. [DOI] [PubMed] [Google Scholar]
- 110. Chakraborty P., Huang J.T.J., Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018; 9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cristini A., Groh M., Kristiansen M.S., Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018; 23:1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hodroj D., Recolin B., Serhal K., Martinez S., Tsanov N., Abou Merhi R., Maiorano D An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017; 36:1182–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Song C., Hotz-Wagenblatt A., Voit R., Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017; 31:1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Li L., Germain D.R., Poon H.Y., Hildebrandt M.R., Monckton E.A., McDonald D., Hendzel M.J., Godbout R. DEAD Box 1 facilitates removal of RNA and homologous recombination at DNA double-strand breaks. Mol. Cell. Biol. 2016; 36:2794–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mersaoui S.Y., Yu Z., Coulombe Y., Karam M., Busatto F.F., Masson J.Y., Richard S. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019; 38:e100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lafuente-Barquero J., Luke-Glaser S., Graf M., Silva S., Gomez-Gonzalez B., Lockhart A., Lisby M., Aguilera A., Luke B. The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage. PLos Genet. 2017; 13:e1007136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pan X., Chen Y., Biju B., Ahmed N., Kong J., Goldenberg M., Huang J., Mohan N., Klosek S., Parsa K. et al. FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci. Rep. 2019; 9:19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Silva B., Pentz R., Figueira A.M., Arora R., Lee Y.W., Hodson C., Wischnewski H., Deans A.J., Azzalin C.M. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 2019; 10:2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kang H.J., Eom H.J., Kim H., Myung K., Kwon H.M., Choi J.H. Thrap3 promotes R-loop resolution via interaction with methylated DDX5. Exp. Mol. Med. 2021; 53:1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kim S., Kang N., Park S.H., Wells J., Hwang T., Ryu E., Kim B.G., Hwang S., Kim S.J., Kang S. et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020; 48:7218–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sessa G., Gomez-Gonzalez B., Silva S., Perez-Calero C., Beaurepere R., Barroso S., Martineau S., Martin C., Ehlen A., Martinez J.S. et al. BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repairdouble dagger. EMBO J. 2021; 40:e106018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Villarreal O.D., Mersaoui S.Y., Yu Z., Masson J.Y., Richard S. Genome-wide R-loop analysis defines unique roles for DDX5, XRN2, and PRMT5 in DNA/RNA hybrid resolution. Life Sci. Alliance. 2020; 3:e202000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yu Z., Mersaoui S.Y., Guitton-Sert L., Coulombe Y., Song J., Masson J.Y., Richard S. DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer. 2020; 2:zcaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tan J., Wang X., Phoon L., Yang H., Lan L. Resolution of ROS-induced G-quadruplexes and R-loops at transcriptionally active sites is dependent on BLM helicase. FEBS Lett. 2020; 594:1359–1367. [DOI] [PubMed] [Google Scholar]
- 125. Chang E.Y., Novoa C.A., Aristizabal M.J., Coulombe Y., Segovia R., Chaturvedi R., Shen Y., Keong C., Tam A.S., Jones S.J.M. et al. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol. 2017; 216:3991–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rossi F., Labourier E., Forne T., Divita G., Derancourt J., Riou J.F., Antoine E., Cathala G., Brunel C., Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996; 381:80–82. [DOI] [PubMed] [Google Scholar]
- 127. Labourier E., Rossi F., Gallouzi I.E., Allemand E., Divita G., Tazi J. Interaction between the N-terminal domain of human DNA topoisomerase I and the arginine-serine domain of its substrate determines phosphorylation of SF2/ASF splicing factor. Nucleic Acids Res. 1998; 26:2955–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rossi F., Labourier E., Gallouzi I.E., Derancourt J., Allemand E., Divita G., Tazi J. The C-terminal domain but not the tyrosine 723 of human DNA topoisomerase I active site contributes to kinase activity. Nucleic Acids Res. 1998; 26:2963–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Soret J., Gabut M., Dupon C., Kohlhagen G., Stevenin J., Pommier Y., Tazi J. Altered serine/arginine-rich protein phosphorylation and exonic enhancer-dependent splicing in mammalian cells lacking topoisomerase I. Cancer Res. 2003; 63:8203–8211. [PubMed] [Google Scholar]
- 130. Girstun A., Ishikawa T., Staron K. Effects of SRSF1 on subnuclear localization of topoisomerase I. J. Cell. Biochem. 2019; 120:11794–11808. [DOI] [PubMed] [Google Scholar]
- 131. Straub T., Grue P., Uhse A., Lisby M., Knudsen B.R., Tange T.O., Westergaard O., Boege F. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J. Biol. Chem. 1998; 273:26261–26264. [DOI] [PubMed] [Google Scholar]
- 132. Solier S., Barb J., Zeeberg B.R., Varma S., Ryan M.C., Kohn K.W., Weinstein J.N., Munson P.J., Pommier Y. Genome-wide analysis of novel splice variants induced by topoisomerase I poisoning shows preferential occurrence in genes encoding splicing factors. Cancer Res. 2010; 70:8055–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Solier S., Ryan M.C., Martin S.E., Varma S., Kohn K.W., Liu H., Zeeberg B.R., Pommier Y. Transcription poisoning by Topoisomerase I is controlled by gene length, splice sites, and miR-142-3p. Cancer Res. 2013; 73:4830–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Miao Z.H., Player A., Shankavaram U., Wang Y.H., Zimonjic D.B., Lorenzi P.L., Liao Z.Y., Liu H., Shimura T., Zhang H.L. et al. Nonclassic functions of Human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 2007; 67:8752–8761. [DOI] [PubMed] [Google Scholar]
- 135. Kim J.J., Lee S.Y., Gong F., Battenhouse A.M., Boutz D.R., Bashyal A., Refvik S.T., Chiang C.M., Xhemalce B., Paull T.T. et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019; 33:1751–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Li M., Pokharel S., Wang J.T., Xu X., Liu Y. RECQ5-dependent sumoylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat. Commun. 2015; 6:6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sarni D., Barroso S., Shtrikman A., Irony-Tur Sinai M., Oren Y.S., Aguilera A., Kerem B Topoisomerase 1-dependent R-loop deficiency drives accelerated replication and genomic instability. Cell Rep. 2022; 40:111397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Thomas A., Pommier Y. Targeting topoisomerase I in the era of precision medicine. Clin. Cancer Res. 2019; 25:6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Marinello J., Bertoncini S., Aloisi I., Cristini A., Malagoli Tagliazucchi G., Forcato M., Sordet O., Capranico G. Dynamic effects of topoisomerase I inhibition on R-loops and short transcripts at active promoters. PLoS One. 2016; 11:e0147053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Marinello J., Chillemi G., Bueno S., Manzo S.G., Capranico G. Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res. 2013; 41:10110–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Groh M., Lufino M.M., Wade-Martins R., Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLos Genet. 2014; 10:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009; 109:2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Koster D.A., Palle K., Bot E.S., Bjornsti M.A., Dekker N.H. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007; 448:213–217. [DOI] [PubMed] [Google Scholar]
- 144. Sun Y., Saha L.K., Saha S., Jo U., Pommier Y. Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways. DNA Repair (Amst.). 2020; 94:102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Sun Y., Saha S., Wang W., Saha L.K., Huang S.N., Pommier Y. Excision repair of topoisomerase DNA-protein crosslinks (TOP-DPC). DNA Repair (Amst.). 2020; 89:102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ljungman M., Hanawalt P.C. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis. 1996; 17:31–35. [DOI] [PubMed] [Google Scholar]
- 147. Marinello J., Arleo A., Russo M., Delcuratolo M., Ciccarelli F., Pommier Y., Capranico G. Topoisomerase I poison-triggered immune gene activation is markedly reduced in human small-cell lung cancers by impairment of the cGAS/STING pathway. Br. J. Cancer. 2022; 127:1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Crossley M.P., Song C., Bocek M.J., Choi J.H., Kousorous J., Sathirachinda A., Lin C., Brickner J.R., Bai G., Lans H. et al. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature. 2023; 613:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Powell W.T., Coulson R.L., Gonzales M.L., Crary F.K., Wong S.S., Adams S., Ach R.A., Tsang P., Yamada N.A., Yasui D.H. et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Jauregui-Lozano J., Escobedo S., Easton A., Lanman N.A., Weake V.M., Hall H. Proper control of R-loop homeostasis is required for maintenance of gene expression and neuronal function during aging. Aging Cell. 2022; 21:e13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Cretu C., Agrawal A.A., Cook A., Will C.L., Fekkes P., Smith P.G., Luhrmann R., Larsen N., Buonamici S., Pena V. Structural basis of splicing modulation by antitumor macrolide compounds. Mol. Cell. 2018; 70:265–273. [DOI] [PubMed] [Google Scholar]
- 152. Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007; 3:570–575. [DOI] [PubMed] [Google Scholar]
- 153. Yeo A.J., Becherel O.J., Luff J.E., Cullen J.K., Wongsurawat T., Jenjaroenpun P., Kuznetsov V.A., McKinnon P.J., Lavin M.F. R-loops in proliferating cells but not in the brain: implications for AOA2 and other autosomal recessive ataxias. PLoS One. 2014; 9:e90219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Yuce O., West S.C. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol. 2013; 33:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Chedin F., Hartono S.R., Sanz L.A., Vanoosthuyse V. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J. 2021; 40:e106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Shokolenko I., Alexeyev M. Mitochondrial DNA: consensuses and controversies. DNA (Basel). 2022; 2:131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zhang H., Barcelo J.M., Lee B., Kohlhagen G., Zimonjic D.B., Popescu N.C., Pommier Y. Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhang H., Meng L.H., Zimonjic D.B., Popescu N.C., Pommier Y. Thirteen-exon-motif signature for vertebrate nuclear and mitochondrial type IB topoisomerases. Nucleic Acids Res. 2004; 32:2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Zhang H., Zhang Y.W., Yasukawa T., Dalla Rosa I., Khiati S., Pommier Y. Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases iialpha and iibeta in vertebrate mitochondria. Nucleic Acids Res. 2014; 42:7259–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Menger K.E., Rodríguez-Luis A., Chapman J., Nicholls T.J. Controlling the topology of mammalian mitochondrial DNA. Open Biology. 2021; 11:210168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009; 276:1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sorrells S., Nik S., Casey M.J., Cameron R.C., Truong H., Toruno C., Gulfo M., Lowe A., Jette C., Stewart R.A. et al. Spliceosomal components protect embryonic neurons from R-loop-mediated DNA damage and apoptosis. Dis. Model Mech. 2018; 11:dmm031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wan Y., Zheng X., Chen H., Guo Y., Jiang H., He X., Zhu X., Zheng Y. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J. Cell Biol. 2015; 209:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Furney S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de la Grange P., Roman-Roman S. et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013; 3:1122–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Harbour J.W., Roberson E.D., Anbunathan H., Onken M.D., Worley L.A., Bowcock A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013; 45:133–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Martin M., Masshofer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B. et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013; 45:933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., Pellagatti A., Wainscoat J.S., Hellstrom-Lindberg E., Gambacorti-Passerini C. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011; 365:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Biankin A.V., Waddell N., Kassahn K.S., Gingras M.C., Muthuswamy L.B., Johns A.L., Miller D.K., Wilson P.J., Patch A.M., Wu J. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012; 491:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E., Cho J., Suh J., Capelletti M., Sivachenko A. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012; 150:1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer. 2016; 16:413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zhang X., Chiang H.C., Wang Y., Zhang C., Smith S., Zhao X., Nair S.J., Michalek J., Jatoi I., Lautner M. et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 2017; 8:15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L. et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 2015; 57:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Racca C., Britton S., Hedouin S., Francastel C., Calsou P., Larminat F. BRCA1 prevents R-loop-associated centromeric instability. Cell Death. Dis. 2021; 12:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Vohhodina J., Goehring L.J., Liu B., Kong Q., Botchkarev V.V. Jr, Huynh M., Liu Z., Abderazzaq F.O., Clark A.P., Ficarro S.B et al. BRCA1 binds TERRA RNA and suppresses R-loop-based telomeric DNA damage. Nat. Commun. 2021; 12:3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Gorthi A., Romero J.C., Loranc E., Cao L., Lawrence L.A., Goodale E., Iniguez A.B., Bernard X., Masamsetti V.P., Roston S. et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018; 555:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Herold S., Kalb J., Buchel G., Ade C.P., Baluapuri A., Xu J., Koster J., Solvie D., Carstensen A., Klotz C. et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature. 2019; 567:545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Chiang H.C., Zhang X., Li J., Zhao X., Chen J., Wang H.T., Jatoi I., Brenner A., Hu Y., Li R. BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic Acids Res. 2019; 47:5086–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Shivji M.K.K., Renaudin X., Williams C.H., Venkitaraman A.R. BRCA2 Regulates transcription elongation by RNA polymerase II to prevent R-loop accumulation. Cell Rep. 2018; 22:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. D’Alessandro G., Whelan D.R., Howard S.M., Vitelli V., Renaudin X., Adamowicz M., Iannelli F., Jones-Weinert C.W., Lee M., Matti V. et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 2018; 9:5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Gomez-Gonzalez B., Sessa G., Carreira A., Aguilera A. A new interaction between BRCA2 and DDX5 promotes the repair of DNA breaks at transcribed chromatin. Mol Cell Oncol. 2021; 8:1910474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Garcia-Rubio M.L., Perez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., Aguilera A. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015; 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Schwab R.A., Nieminuszczy J., Shah F., Langton J., Lopez Martinez D., Liang C.C., Cohn M.A., Gibbons R.J., Deans A.J., Niedzwiedz W. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell. 2015; 60:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Madireddy A., Kosiyatrakul S.T., Boisvert R.A., Herrera-Moyano E., Garcia-Rubio M.L., Gerhardt J., Vuono E.A., Owen N., Yan Z., Olson S. et al. FANCD2 Facilitates replication through common fragile sites. Mol. Cell. 2016; 64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Bhatia V., Herrera-Moyano E., Aguilera A., Gomez-Gonzalez B. The role of replication-associated repair factors on R-loops. Genes (Basel). 2017; 8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yan Q., Wulfridge P., Doherty J., Fernandez-Luna J.L., Real P.J., Tang H.Y., Sarma K. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat. Commun. 2022; 13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Nguyen D.T., Voon H.P.J., Xella B., Scott C., Clynes D., Babbs C., Ayyub H., Kerry J., Sharpe J.A., Sloane-Stanley J.A. et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017; 18:914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011; 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Udugama M., Sanij E., Voon H.P.J., Son J., Hii L., Henson J.D., Chan F.L., Chang F.T.M., Liu Y., Pearson R.B. et al. Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4737–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Urban V., Dobrovolna J., Huhn D., Fryzelkova J., Bartek J., Janscak P. RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J. Cell Biol. 2016; 214:401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Marabitti V., Lillo G., Malacaria E., Palermo V., Sanchez M., Pichierri P., Franchitto A. ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. 2019; 47:3485–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Marabitti V., Valenzisi P., Lillo G., Malacaria E., Palermo V., Pichierri P., Franchitto A. R-loop-associated genomic instability and implication of WRN and WRNIP1. Int. J. Mol. Sci. 2022; 23:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Jones S.E., Fleuren E.D.G., Frankum J., Konde A., Williamson C.T., Krastev D.B., Pemberton H.N., Campbell J., Gulati A., Elliott R. et al. ATR is a therapeutic target in Synovial Sarcoma. Cancer Res. 2017; 77:7014–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Kotsantis P., Silva L.M., Irmscher S., Jones R.M., Folkes L., Gromak N., Petermann E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016; 7:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Edwards D.S., Maganti R., Tanksley J.P., Luo J., Park J.J.H., Balkanska-Sinclair E., Ling J., Floyd S.R. BRD4 Prevents R-loop formation and transcription-replication conflicts by ensuring efficient transcription elongation. Cell Rep. 2020; 32:108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lam F.C., Kong Y.W., Huang Q., Vu Han T.L., Maffa A.D., Kasper E.M., Yaffe M.B. BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nat. Commun. 2020; 11:4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Tan S.L.W., Chadha S., Liu Y., Gabasova E., Perera D., Ahmed K., Constantinou S., Renaudin X., Lee M., Aebersold R. et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017; 169:1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Liang Z., Liang F., Teng Y., Chen X., Liu J., Longerich S., Rao T., Green A.M., Collins N.B., Xiong Y. et al. Binding of FANCI-FANCD2 complex to RNA and R-loops stimulates robust FANCD2 monoubiquitination. Cell Rep. 2019; 26:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. De Magis A., Manzo S.G., Russo M., Marinello J., Morigi R., Sordet O., Capranico G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Sanij E., Hannan K.M., Xuan J., Yan S., Ahern J.E., Trigos A.S., Brajanovski N., Son J., Chan K.T., Kondrashova O. et al. CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 2020; 11:2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J. et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017; 8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Safari M., Litman T., Robey R.W., Aguilera A., Chakraborty A.R., Reinhold W.C., Basseville A., Petrukhin L., Scotto L., O’Connor O.A. et al. R-loop-mediated ssDNA breaks accumulate following short-term exposure to the HDAC inhibitor romidepsin. Mol. Cancer Res. 2021; 19:1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Gomez-Gonzalez B., Barroso S., Herrera-Moyano E., Aguilera A. Spontaneous DNA-RNA hybrids: differential impacts throughout the cell cycle. Cell Cycle. 2020; 19:525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Uehara T., Minoshima Y., Sagane K., Sugi N.H., Mitsuhashi K.O., Yamamoto N., Kamiyama H., Takahashi K., Kotake Y., Uesugi M. et al. Selective degradation of splicing factor caperalpha by anticancer sulfonamides. Nat. Chem. Biol. 2017; 13:675–680. [DOI] [PubMed] [Google Scholar]
- 204. Casado J.A., Rio P., Marco E., Garcia-Hernandez V., Domingo A., Perez L., Tercero J.C., Vaquero J.J., Albella B., Gago F. et al. Relevance of the Fanconi anemia pathway in the response of human cells to trabectedin. Mol. Cancer Ther. 2008; 7:1309–1318. [DOI] [PubMed] [Google Scholar]
- 205. Monk B.J., Lorusso D., Italiano A., Kaye S.B., Aracil M., Tanovic A., D’Incalci M Trabectedin as a chemotherapy option for patients with BRCA deficiency. Cancer Treat. Rev. 2016; 50:175–182. [DOI] [PubMed] [Google Scholar]
- 206. Tumini E., Herrera-Moyano E., San Martin-Alonso M., Barroso S., Galmarini C.M., Aguilera A. The antitumor drugs trabectedin and lurbinectedin induce transcription-dependent replication stress and genome instability. Mol. Cancer Res. 2019; 17:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Matos D.A., Zhang J.M., Ouyang J., Nguyen H.D., Genois M.M., Zou L. ATR protects the genome against R loops through a MUS81-triggered feedback loop. Mol. Cell. 2020; 77:514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.