Abstract
Background & Aims
Progression of alcohol-associated liver disease (ALD) is driven by genetic predisposition. The rs13702 variant in the lipoprotein lipase (LPL) gene is linked to non-alcoholic fatty liver disease. We aimed at clarifying its role in ALD.
Methods
Patients with alcohol-associated cirrhosis, with (n = 385) and without hepatocellular carcinoma (HCC) (n = 656), with HCC attributable to viral hepatitis C (n = 280), controls with alcohol abuse without liver damage (n = 366), and healthy controls (n = 277) were genotyped regarding the LPL rs13702 polymorphism. Furthermore, the UK Biobank cohort was analysed. LPL expression was investigated in human liver specimens and in liver cell lines.
Results
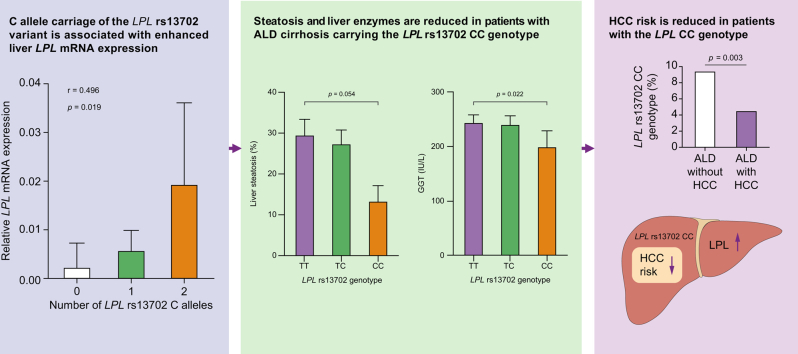
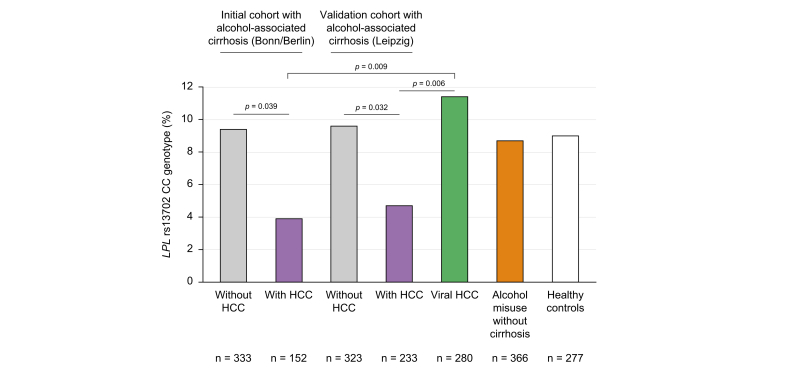
Frequency of the LPL rs13702 CC genotype was lower in ALD with HCC in comparison to ALD without HCC both in the initial (3.9% vs. 9.3%) and the validation cohort (4.7% vs. 9.5%; p <0.05 each) and compared with patients with viral HCC (11.4%), alcohol misuse without cirrhosis (8.7%), or healthy controls (9.0%). This protective effect (odds ratio [OR] = 0.5) was confirmed in multivariate analysis including age (OR = 1.1/year), male sex (OR = 3.0), diabetes (OR = 1.8), and carriage of the PNPLA3 I148M risk variant (OR = 2.0). In the UK Biobank cohort, the LPL rs13702 C allele was replicated as a risk factor for HCC. Liver expression of LPL mRNA was dependent on LPL rs13702 genotype and significantly higher in patients with ALD cirrhosis compared with controls and alcohol-associated HCC. Although hepatocyte cell lines showed negligible LPL protein expression, hepatic stellate cells and liver sinusoidal endothelial cells expressed LPL.
Conclusions
LPL is upregulated in the liver of patients with alcohol-associated cirrhosis. The LPL rs13702 high producer variant confers protection against HCC in ALD, which might help to stratify people for HCC risk.
Impact and implications
Hepatocellular carcinoma is a severe complication of liver cirrhosis influenced by genetic predisposition. We found that a genetic variant in the gene encoding lipoprotein lipase reduces the risk for hepatocellular carcinoma in alcohol-associated cirrhosis. This genetic variation may directly affect the liver, because, unlike in healthy adult liver, lipoprotein lipase is produced from liver cells in alcohol-associated cirrhosis.
Keywords: Cirrhosis, Alcohol-associated liver disease, HCC, LPL, rs13702, rs328
Abbreviations: ALD, alcohol-associated liver disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BCLC, Barcelona Clinic Liver Cancer; BSA, bovine serum albumin; FCS, foetal calf serum; FIB-4, fibrosis 4; HbA1c, glycated haemoglobin; HCC, hepatocellular carcinoma; HSCs, hepatic stellate cells; GADPH, glyceraldehyde 3-phosphate dehydrogenase; GGT, gamma-glutamyl transferase; LPL, lipoprotein lipase; LSECs, liver sinusoidal endothelial cells; MAF, minor allele frequency; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; OR, odds ratio; PNPLA3, patatin-like phospholipase domain-containing protein 3; T2DM, type 2 diabetes mellitus; UKB, UK Biobank
Graphical abstract
Highlights
-
•
The C allele of the LPL rs13702 variant was associated with reduced risk of ALD-HCC.
-
•
This association could be replicated on the population level in the UK Biobank cohort.
-
•
LPL is overexpressed in ALD cirrhosis, in contrast to HCV cirrhosis or non-cirrhotic liver.
-
•
LPL expression was not observed in hepatocytes but was in HSCs and LSECs.
Introduction
Heavy alcohol use, a common risk factor for liver disease, is reported by 6.6% of the adult population in the United States.1 Development, progression, and complications of alcohol-associated liver disease (ALD) and non-alcoholic liver disease are, next to lifestyle factors, strongly influenced by genetic predisposition.2 Thus, only 10–20% of patients with heavy alcohol abuse develop significant liver disease.2 Several common genetic variants that increase the risk for liver disease development have been identified. The most important is the I148M variant in the patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene, which is a risk factor both for cirrhosis and hepatocellular carcinoma (HCC), but its molecular background is still poorly understood.3 Some variants, such as MBOAT7 rs641738, mainly modulate the risk for cirrhosis,4 whereas others, such as the rs708113 polymorphism in the WNT3A-WNT9A locus are only associated with the risk for HCC.5 However, protective genetic variants, for example the rs429358 polymorphism in apolipoprotein E, the rs72613567 HSD17B13 locus, and rs2242652 in TERT have been unravelled.[6], [7], [8] Knowledge of these factors may both lead to a better understanding of the molecular pathways underlying liver disease development and the identification of potential drug targets, but also facilitate risk stratification, for example for HCC screening programmes.
In a genome-wide association study, the rs13702 locus in the lipoprotein lipase (LPL) gene has recently been linked to the risk of non-alcoholic fatty liver disease (NAFLD).9 In presence of the C allele at this locus, downregulation of LPL transcription by miR-410 is impaired,10 expression of LPL is increased in adipose tissue, and risk for NAFLD is decreased.9 LPL is a triglyceride lipase which is expressed in different metabolically active tissues, in particular on vascular endothelium, but only in foetal, not in adult healthy liver. It acts as an important modulator of lipid metabolism by hydrolysing triglycerides, which are transported in the blood in VLDL and chylomicrons, releasing free fatty acids, which can be taken up by parenchymal cells.[11], [12], [13] Hepatic expression of LPL has been found in patients with NAFLD,14,15 where it decreases after bariatric surgery.16 Interestingly, LPL expression in human NAFLD livers has been attributed to hepatic stellate cells, not to hepatocytes.17 Presence of a minor C allele of LPL rs13702 has been associated with lower triglyceride and higher HDL levels in the general population10 and presence of the CC genotype with favourable response to diet.18,19 Another minor variant of a LPL polymorphism, rs328, has reported to act similarly to rs13702 on serum lipid levels.20
We aimed at analysing if genetically modulated expression of LPL may be associated with development of cirrhosis or HCC in alcohol-associated liver disease.
Patients and methods
Patients
As the initial cohort, blood and clinical data were collected from 485 patients with alcohol-associated liver cirrhosis, 152 of them with HCC, from the University Hospital Bonn and the Berlin Department of Hepatology and Gastroenterology. The validation cohort, collected from the Division of Hepatology of the Leipzig University Medical Centre, comprised 323 patients with ALD and 233 patients with HCC attributable to ALD. In addition, 277 healthy individuals, 366 alcohol misusers without cirrhosis, and 280 patients with chronic hepatitis C and HCC were included in the study. Participants were considered as alcohol misusers without cirrhosis when a minimal amount of 60 g alcohol/day for women and 80 g alcohol/day for men was consumed for at least 10 years without resulting liver damage. Healthy controls included individuals from blood donation and cancer screening programmes who did not show clinical or laboratory signs of liver disease. Diagnosis of cirrhosis was based on liver biopsy or on a consistent clinical presentation supported by laboratory and imaging findings. We recorded demographic, standard laboratory and clinical data such as sex, age, and aetiology of cirrhosis. Participants were allocated to alcohol-associated cirrhosis if their history indicated average alcohol consumption to exceed 300 g ethanol/week after exclusion of other causes of cirrhosis such as HBV or HCV infection or haemochromatosis. HCC was diagnosed according to international guidelines.21
Apart from blood samples, liver specimens from liver explants from patients with cirrhosis with ALD and viral disease with and without HCC and tumour-free liver tissue gained by surgical resection of liver metastases were used for investigation of mRNA expression.
All controls and participants were of Caucasian ethnicity. Details of the study cohorts are shown in Table 1.
Table 1.
Demographic, clinical and laboratory details ofthe study cohorts.
| Alcohol-associated cirrhosis Bonn/Berlin |
Alcohol-associated cirrhosis Leipzig |
Viral HCC | Alcohol misuse without cirrhosis | Healthy controls | |||
|---|---|---|---|---|---|---|---|
| Without HCC | With HCC | Without HCC | With HCC | ||||
| Total number | 333 | 152 | 323 | 233 | 280 | 366 | 277 |
| Age, mean (range) | 58.1 (27–92) | 63.9 (36–87) | 54.8 (28–87) | 63.1 (47–79) | 59.6 (24–83) | 44.0 (28–81) | 65.0 (28–94) |
| Sex (% male/female) | 64.2/35.8 | 81.5/18.5 | 77.0/23.0 | 93.2/6.8 | 71.8/28.2 | 75.4/24.6 | 44.4/55.6 |
| Bilirubin [mg/dl], (Mean ± SD) | 3.91 ± 6.34 | 3.84 ± 5.56 | 4.17 ± 7.08 | 2.30 ± 3.25 | 2.74 ± 4.38 | 0.74 ± 0.63 | — |
| ALT [IU/L], (Mean ± SD) | 49.1 ± 151.5 | 52.4 ± 59.9 | 37.0 ± 37.0 | 48.0 ± 55.6 | 97.6 ± 148.3 | 45.2 ± 45.8 | — |
| AST [IU/L], (Mean ± SD) | 82.8 ± 237.7 | 90.5 ± 100.4 | 63.7 ± 48.8 | 88.0 ± 114.2 | 131.4 ± 172.8 | 55.6 ± 64.4 | — |
| GGT [IU/L], (Mean ± SD) | 199.9 ± 213.9 | 221.4 ± 185.5 | 194.0 ± 240.4 | 294.1 ± 379.3 | 138.2 ± 128.8 | 212.6 ± 396.1 | — |
| Platelet count [∗103/μl], (Mean ± SD) | 154.9 ± 143.9 | 140.2 ± 75.8 | 148.6 ± 82.3 | 153.2 ± 95.1 | 124.6 ± 91.9 | 228.8 ± 80.5 | — |
| MELD (Mean ± SD) | 16.3 ± 7.0 | 17.0 ± 8.1 | 17.9 ± 7.4 | 12.5 ± 5.9 | 15.8 ± 6.2 | — | — |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyl transferase; HCC, hepatocellular carcinoma; MELD, model for end-stage liver disease.
Ethical approval
The study protocol followed the ethical guidelines of the Helsinki Declaration and had been approved by the local ethics committees (Bonn: number 536/20; Leipzig: number 357/19-ek). Written informed consent was obtained from the participants before inclusion in this study.
Determination of LPL and PNPLA3 genotypes
Genomic DNA was isolated from 200 μl EDTA-blood using the QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) as suggested by the manufacturer’s protocol. Determination of the polymorphisms was performed by melting curve analysis using LightSNiP (SimpleProbe) assays from TIB-MolBiol (Berlin, Germany). A sample of 1 μl of DNA was mixed with 0.5 μl of the LightSNiP reagent mix, 5 μl of Blue Probe qPCR 2 × Mix (Biozym Scientific GmbH, Hessisch Oldendorf, Germany) and 3.5 μl PCR grade water (Invitrogen, Paisley, UK). The real-time PCR was performed by the LightCycler® 96 (Roche, Mannheim, Germany) according to the manufacturer’s protocol.
Cell culture
PLC/PRF/5 and LX-2 cells were cultured in RPMI 1640 medium containing l-glutamine (Thermo Fisher, Schwerte, Germany), 1% penicillin/streptomycin (PAN-Biotech GmbH, Aidenbach, Germany) and 10% foetal calf serum (FCS) (Biochrom, Berlin, Germany). Primary hepatocytes, liver sinusoidal endothelial cells, and primary hepatic stellate cells (HSCs; ScienCell™, Carlsbad, CA, USA) were cultivated in the appropriate media according to the manufacturer's instructions. All cell lines tested negative for Mycoplasma contamination.
Analysis of mRNA expression
RNA was isolated from liver explants with the GeneJET RNA Purification Kit (Thermo Scientific, Lithuania) according to manufacturer’s protocol. RNA was stored at -80 °C until cDNA synthesis was performed. Reverse transcription of the RNA was carried out with the QuantiTect® Reverse Transcription Kit (Qiagen, Hilden, Germany) resulting in cDNA without contamination of genomic DNA. A sample of 1 μl of cDNA was used as a template for the following real-time PCR on the LightCycler® 96 as recommended by the manufacturer. Samples of 5 μl of Blue S’Green qPCR 2 × Mix (Biozym Scientific GmbH, Hessisch Oldendorf, Germany), 1 μl of designated primers (5 μM) and 3 μl of PCR-grade water were added for the amplification of the desired gene products. LPL (forward: 5′-CTG CTG GCA TTG CAG GAA GTC T-3′, reverse: 5′-CAT CAG GAG AAA GAC TCG G-3′) and the housekeeping primers ribosomal protein L19 (forward: 5′-TGG GCA TAG GTA AGC GGA-3′, reverse: 5′-GCC TTG TCT GCC TTC AGC-3′) (IDT, Leuven, Belgium).
Normalisation was performed with the LightCycler® 96 SW 1.1 analysis software expressing the relative mRNA expression of LPL as 2(-ΔCt).
Western blot
Liver tissue and cell lines were homogenised in RIPA buffer (SERVA Electrophoresis GmbH, Heidelberg, Germany) supplemented with Halt™ Protease-Inhibitor-Cocktail 100 × (Life Technologies GmbH, Darmstadt, Germany) followed by a centrifugation step for 5 min at 300 × g. The protein concentration of the supernatant was determined using the DC Protein Assay Kit from Bio-Rad Laboratories GmbH (Feldkirchen, Germany) as per the manufacturer’s instructions. Samples of 30 μg of protein for cell lines and 20 μg of protein for liver tissue, respectively, were mixed with ROTI®Load 1 4 × concentrate (Carl Roth GmbH, Karlsruhe, Germany) and heated for 5 min at 95 °C. Equal sample volumes were size-fractionated using 10% SDS-PAGE and transferred onto a 0.2 μm Whatman™ Protran nitrocellulose membrane (Whatman GmbH, Dassel, Germany). Membranes were blocked for 30 min in 3% bovine serum albumin (BSA; PAN-Biotech GmbH, Aidenbach, Germany) in phosphate-buffered saline (Fisher Scientific GmbH, Schwerte, Germany) with 0.1% Tween 20 (Carl Roth, Karlsruhe, Germany) and incubated overnight with the appropriate antibody. Membranes were probed with a LPL mouse monoclonal antibody (clone ID: OTI2C12, OriGene Technologies, Rockville, US), a human/mouse LPL polyclonal antibody (AF7197, R&D Systems, Minneapolis, USA), and a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (sc-32233, Santa Cruz, Heidelberg, Germany) 1:500 in 3% BSA. Then, membranes were incubated with anti-mouse IgG (A9044, Merck, Darmstadt, Germany) (diluted 1:10,000 in 3% BSA for GAPDH and LPL monoclonal antibody) and with anti-goat IgG-HRP (sc-2020, Santa Cruz Biotechnology, Heidelberg, Germany) (diluted 1:5,000 in 3% BSA for LPL polyclonal antibody). The protein quantification was carried out with the Healthcare Amersham™ ECL Prime Western-Blot detection reagent (Fisher Scientific GmbH, Schwerte, Germany). For the detection of chemiluminescence and the following densiometric analysis the ChemDocTM MP Imaging System and the corresponding ImageLab Software (BioRad, Feldkirchen, Germany) was used.
UK Biobank cohort (population-based cohort)
In addition, we leveraged data from the UK Biobank (UKB) cohort – a prospective community cohort comprised of approximately half a million middle-aged individuals from the UK. Participants were interviewed between May 2006 to July 2010, where they completed a comprehensive health questionnaire, a physical examination, and donated biological specimens.22 To determine genotyping information, DNA was extracted from blood samples collected at enrolment and assayed using genotyping arrays.23 Information on health outcome events occurring after enrolment are supplied through linkage to UK mortality, hospital admission, and cancer registries.
Statistical analysis
The data from clinical cohorts were analysed with IBM SPSS Statistics software version 24 (IBM, New York, USA). Significant deviations from the Hardy-Weinberg-equilibrium were ruled out using an exact test (https://ihg.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl).
Comparison of genotypes was performed using the Pearson’s goodness-of-fit Χ2 test. Fisher’s exact test, Student’s t test, Wilcoxon–Mann–Whitney U test, and Spearman’s correlation were applied as appropriate and indicated. Univariate analysis followed by a multivariate forward binary regression was used to confirm the independency of the HCC risk factors with p <0.05 for inclusion and p >0.1 for exclusion of parameters.
In the UK Biobank cohort, HCC cases were identified by the presence of the International Classification of Diseases-10th edition code C22.0 recorded in an inpatient hospital admission, cause of death, or a cancer registration record, either before or after enrolment into the UKB. Patients without HCC were defined as controls. The association between rs13702 genotype and HCC case/control status was calculated via logistic regression, under an additive genetic model. Statistical adjustment was included for age, sex, and the first five principal components of genetic ancestry. We further extended this UKB analysis to capture: (a) 10 additional phenotypes of interest beyond HCC; and (2) two additional genetic variants: rs328 (LPL) and rs738409 (PNPLA3). The 10 additional phenotypes were: cirrhosis status; type 2 diabetes (T2DM), BMI, alanine aminotransferase (ALT), aspartate aminotransferase (AST), cholesterol, fibrosis 4 index (FIB-4), glycated haemoglobin (HbA1c), liver fat content, and triglycerides. The fields/methods used to derive these phenotypes are outlined in Tables S1 and S2. For binary phenotypes (e.g. HCC and diabetes), effect sizes are presented on the log odds ratio scale, which are symmetrical around the line of null effect. This was to ensure that our forest plots give equal weight (visually) to associations implying protection vs. associations implying harm. Genotype associations with continuous phenotypes (e.g. ALT and BMI) were determined using linear regression. Continuous phenotypes were first log-transformed to achieve approximate normality and then standardised into a z-score (i.e. with mean of zero and a standard deviation of one). This means the effect size represents the change in the phenotype (in terms of standard deviations) for each additional copy of the effect allele carried. In this way, the effect sizes for diverse continuous phenotypes can be compared ‘like-for-like’. All UKB associations were adjusted for age, sex, and the first five principal components of genetic ancestry. Analyses of UKB data were performed using Stata version 17 (StataCorp LLC, College Station, TX, USA).
Results
Study population
Details of the study cohorts, comprising the initial and the validation cohort of patients with alcohol-associated cirrhosis, with and without HCC, alcohol misusers without cirrhosis, and healthy controls as well as patients with HCC caused by viral hepatitis C, are shown in Table 1.
Genotype distribution
The distribution of LPL rs13702 genotypes was similar between healthy controls with a minor allele frequency (MAF) of 0.30, alcohol misuse without cirrhosis (MAF = 0.32) and the genotype distribution reported for the European population (MAF = 0.32).24 Similarly, irrespective of HCC, MAF was 0.29 in the initial cohort, 0.28 in the validation cohort, and 0.30 among patients with viral HCC. Genotype distributions of LPL rs13702 and rs238 are given in detail in Table 2.
Table 2.
Genotype distribution of the LPL and PNPLA3 polymorphisms.
| Genotype | Alcohol-associated cirrhosis Bonn/Berlin (n = 485) |
Alcohol-associated cirrhosis Leipzig (n = 556) |
Viral HCC (n = 280) | Alcohol misuse without cirrhosis (n = 366) | Healthy controls (n = 277) | ||
|---|---|---|---|---|---|---|---|
| Without HCC n = 333 | With HCC n = 152 | Without HCC n = 323 | With HCC n = 233 | ||||
| LPL rs13702 | |||||||
| TT | 160 (48.0%) | 80 (52.6%) | 171 (52.9%) | 119 (51.1%) | 142 (50.7%) | 164 (44.8%) | 138 (49.8%) |
| TC | 142 (42.6%) | 66 (43.4%) | 121 (37.5%) | 103 (44.2%)∗ | 106 (37.9%) | 170 (46.4%) | 114 (41.2%) |
| CC | 31 (9.3%) | 6 (3.9%)∗§ | 31 (9.6%) | 11 (4.7%)∗§ | 32 (11.4%) | 32 (8.7%) | 25 (9.0%) |
| LPL rs328 | |||||||
| CC | 281 (84.4%) | 127 (83.5%) | 275 (85.1%) | 190 (81.5%) | 224 (80.0%) | 293 (80.1%) | 230 (83.0%) |
| CG | 51 (15.3%) | 25 (16.5%) | 46 (14.3%) | 42 (18.1%) | 51 (18.2%) | 70 (19.1%) | 44 (15.9%) |
| GG | 1 (0.3%) | 0 (0.0%) | 2 (0.6%) | 1 (0.4%) | 5 (1.8%) | 3 (0.8%) | 3 (1.1%) |
| PNPLA3 rs738409 | |||||||
| CC | 130 (39.0%) | 38 (25.0%) | 140 (43.3%) | 59 (25.3%) | 152 (54.3%) | 212 (58.0%) | 165 (59.6%) |
| GC | 155 (46.5%)‡‡‡†††§ | 72 (47.4%)‡‡‡†††§ | 144 (44.6%)‡‡††§ | 126 (54.1%)∗∗∗‡‡‡†††§ | 100 (35.7%) | 129 (35.2%) | 97 (35.0%) |
| GG | 48 (14.4%)‡‡‡††† | 42 (27.6%)∗∗∗‡‡‡†††§ | 39 (12.1%)‡‡‡††† | 48 (20.6%)∗∗‡‡‡†††§ | 28 (10.0%) | 25 (6.8%) | 15 (5.4%) |
Statistical analysis with Pearson’s goodness-of-fit Χ2 test.
∗p <0.05; ∗∗p <0.01; ∗∗∗p <0.001 vs. cirrhosis without HCC. †p <0.05; ††p <0.01; †††p <0.001 vs. alcoholic controls. ‡p <0.05; ‡‡p <0.01; ‡‡‡p <0.001 vs. healthy controls. §p <0.01 vs. viral HCC.
HCC, hepatocellular carcinoma; LPL, lipoprotein lipase; PNPLA3, patatin-like phospholipase domain-containing protein 3.
LPL rs13702 CC genotype is protective against HCC in alcohol-associated cirrhosis
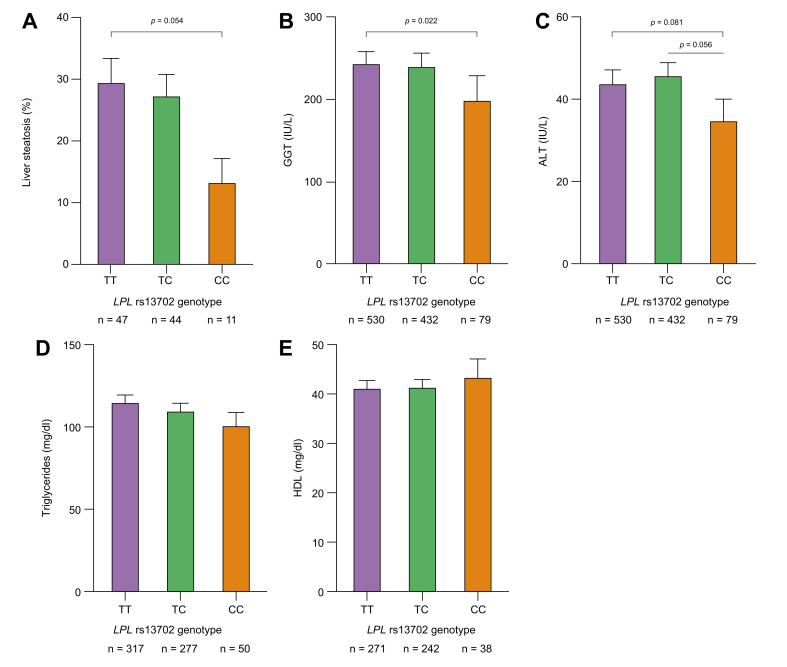
When we stratified patients with ALD concerning the presence of HCC, we found a significantly decreased frequency of the minor CC rs13702 LPL genotype compared with patients without HCC. This difference was confirmed in the validation cohort. Participants with viral HCC did not display a decreased frequency of the CC genotype (Fig. 1). When we compared histologically assessed levels of liver steatosis in participants with ALD between the genotypes, carriers of the CC genotype had lower liver fat content (Fig. 2A). Concerning liver enzymes, gamma-glutamyl transferase was significantly decreased in carriers of the CC genotype (Fig. 2B), with a numerically similar effect for ALT (Fig. 2C). Serum triglyceride levels decreased while HDL levels increased with the number of C alleles without meeting statistical significance (Fig. 2D and E). In the clinical cohorts, no association of the LPL rs13702 CC genotype to diabetes (p = 0.78) or obesity (p = 0.45), was found.
Fig. 1.
Frequency of the homozygous LPL rs13702 CC genotype among the study cohorts.
The homozygous LPL rs13702 CC genotype was less frequent among participants with HCC in alcohol-associated cirrhosis compared with participants without HCC and to participants with cirrhosis with HCC caused by viral hepatitis. Statistical analysis was with the Χ2 test. HCC, hepatocellular carcinoma; LPL, lipoprotein lipase.
Fig. 2.
Liver enzymes, hepatic steatosis, and lipid parameters in alcohol-associated cirrhosis according to the LPL rs13702 genotypes.
Carriers of the LPL rs13702 CC genotype display numerically lower levels of liver enzymes and triglyceride levels and decreased liver fat. (A) Liver steatosis as assessed by histology. (B) Gamma-glutamyl transferase (GGT). (C) Alanine aminotransferase (ALT). (C, D) Serum triglyceride levels. (E) Serum high-density lipoprotein levels according to the LPL rs13702 genotypes. Statistical analysis was with the Wilcoxon–Mann–Whitney U test. LPL, lipoprotein lipase.
The LPL rs328 polymorphism was not associated with alcoholic HCC in our cohort (Table 2).
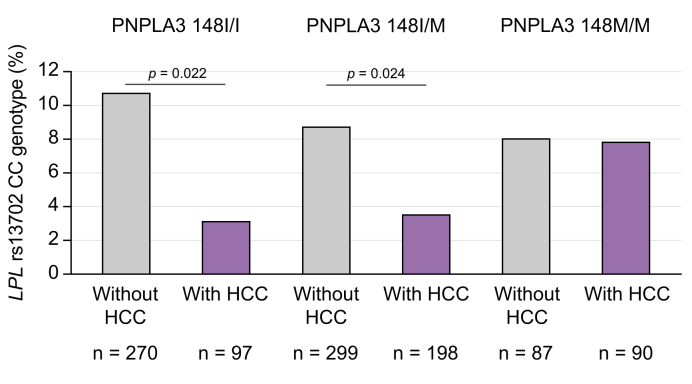
The protective effect of rs13702 LPL CC genotype is connected to PNPLA3 I148M genotype
As expected, carriage of at least one minor PNPLA3 rs738409 allele was replicated as a risk factor both for alcohol-associated cirrhosis and alcohol-associated HCC in our study cohorts (Table 2). Interestingly, the protective effect of the LPL rs13702 CC genotype was attenuated in individuals carrying the high-risk homozygous PNPLA3 GG genotype (Fig. 3). However, we found no statistical proof of interaction between LPL rs13702 CC and PNPLA3 I148M concerning the risk of developing HCC (p = 0.129; Table S3).
Fig. 3.
Frequency of the LPL rs13702 CC genotype in participants with alcohol-associated cirrhosis stratified for carriage of PNPLA3 I148M genotype.
The significant lower frequency of the LPL rs13702 CC genotype among participants with HCC was not found in carriers of the high risk PNPLA3 148 MM genotype. Statistical analysis was with the Χ2 test. HCC, hepatocellular carcinoma; LPL, lipoprotein lipase; PNPLA3: patatin-like phospholipase domain-containing protein 3.
The LPL rs13702 CC genotype is independently associated with HCC
To verify if carriage of the LPL rs13702 CC genotype is independently associated with occurrence of HCC in patients with alcohol-associated cirrhosis, we performed univariate and multivariate analysis of known risk factors for HCC. Univariate analyses in both cohorts of patients with alcohol-associated cirrhosis combined showed that in addition to the CC genotype, age, male sex, diabetes, and presence of the PNPLA3 148 M variant were associated with HCC (Table 3). Multivariate analysis confirmed age, male sex, diabetes, and PNPLA3 148 M as independent risk factors for HCC, whereas carriage of the LPL rs13702 CC genotype led to a decreased HCC risk (OR = 0.5; p = 0.03) (Table 3).
Table 3.
Regression analysis for possible HCC risk factors.
| Univariate analysis | ||||
|---|---|---|---|---|
| 95% CI |
||||
| Parameter | p | OR | Lower | Upper |
| Age | 0.000 | 1.084 | 1.067 | 1.101 |
| Sex (male) | 0.000 | 3.047 | 2.111 | 4.399 |
| Diabetes | 0.000 | 2.709 | 2.034 | 3.608 |
| PNPLA3 148 M | 0.000 | 2.077 | 1.573 | 2.742 |
|
LPL rs13702 CC |
0.003 |
0.442 |
0.254 |
0.769 |
|
Multivariate analysis∗ | ||||
|
95% CI |
||||
| Parameter | p | OR | Lower | Upper |
| Age | 0.000 | 1.075 | 1.056 | 1.095 |
| Sex (male) | 0.000 | 3.040 | 2.000 | 4.620 |
| Diabetes | 0.000 | 1.832 | 1.325 | 2.532 |
| PNPLA3 148 M | 0.000 | 2.002 | 1.433 | 2.796 |
| LPL rs13702 CC | 0.033 | 0.493 | 0.258 | 0.943 |
LPL, lipoprotein lipase; OR, odds ratio; PNPLA3: patatin-like phospholipase domain-containing protein 3.
Statistical analysis: univariate analysis with Pearson’s goodness-of-fit Χ2 test followed by multivariate forward binary regression analysis.
Carriage of the LPL rs13702 CC genotype is not related to stage or mortality in HCC
We analysed if the presence of the LPL rs13702 CC genotype might be associated with stage or outcome in patients with HCC. However, we did not find any association with Barcelona Clinic Liver Cancer (BCLC) stage at diagnosis, alpha-foetoprotein level at diagnosis, or survival (data not shown).
Liver LPL expression is increased in patients with alcohol-associated cirrhosis compared with controls
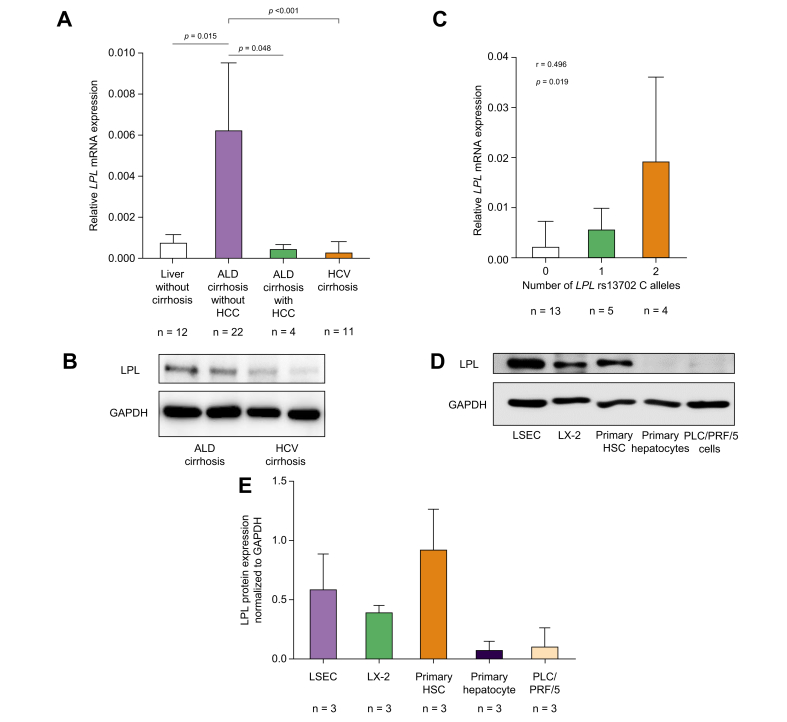
Because LPL is not expressed in healthy liver, we investigated if this held also true for alcohol-associated cirrhosis, given the association between HCC and the rs13702 variant influencing tissue expression.10 LPL mRNA expression was significantly increased in the livers from patients with alcohol-associated cirrhosis compared with viral cirrhotic livers, tumour-free, non-cirrhotic liver tissue gained by surgical resection of liver metastases, and ALD livers with HCC (Fig. 4A). A similar effect was seen at the protein level (Fig. 4B); however, only two specimens of liver from patients with ALD were available for protein analysis. Hepatic LPL mRNA expression in alcohol-associated cirrhosis was stepwise increased with the number of C alleles at the rs13702 locus (Fig. 4C).
Fig. 4.
LPL expression in human liver tissueandhepatic cells lines.
LPL mRNA expression is significantly increased in liver tissue from alcohol-associated cirrhosis compared with non-cirrhotic liver, HCV cirrhosis, and cirrhotic liver with HCC; it is associated with the number of C alleles at the LPL rs13702 locus and not found in hepatocytes, but HSCs and LSECs. (A) LPL mRNA expression in human liver tissue. (B) LPL protein expression in human tissue. (C) LPL mRNA expression correlated with the number of LPL rs13702 C alleles. (D) Representative Western blot for protein analysis in hepatic cell lines. (E) Quantified LPL protein expression in hepatic cell lines revealed negligible amounts in hepatocytes. Statistical analysis was with Student’s t test (A, C) and Spearman’s correlation (B). ALD, alcohol-associated liver disease; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCC: hepatocellular carcinoma; HSC, hepatic stellate cells; LPL, lipoprotein lipase; LSEC, liver sinusoidal endothelial cells.
LPL is predominantly expressed in HSCs and liver sinusoidal endothelial cells
In human NAFLD, it has been suggested by immunofluorescence staining that hepatic LPL expression does not originate from hepatocytes, but from HSCs.17 To verify this finding, we investigated LPL protein expression in various human liver cell lines. Although we found only minimal LPL expression in the hepatocyte cell line PLC/PRF/5 and in primary hepatocytes, LPL was expressed at much higher levels in liver sinusoidal endothelial cells (LSECs), primary HSCs, and the hepatic stellate cell line LX-2 at the protein level (Fig. 4D and E).
Validation at a population level in the UK Biobank cohort
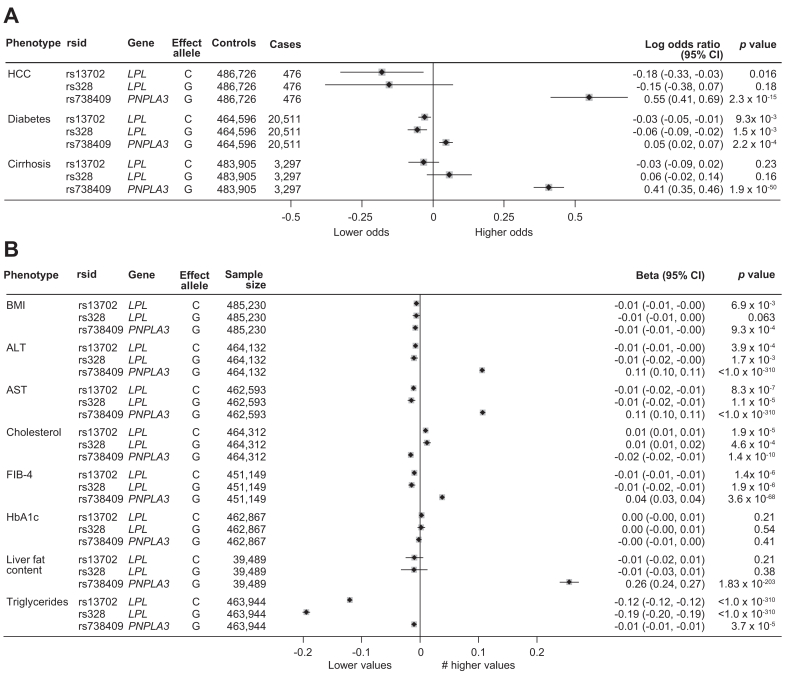
In the UKB cohort, rs13702:C was associated with a reduced risk of HCC (adjusted odds ratio: 0.83; 95% CI: 0.72–0.97; p = 0.016) (Fig. 5). Several metabolic traits and transaminase levels were also associated with rs13702:C, albeit the effect size was generally very small. An exception to this rule was the association between rs13702 and triglycerides where the effect size was much larger (beta: -0.12 decrease per C allele carried).
Fig. 5.
Association of the LPL rs13702, LPL rs328 and PNPLA3 rs738409 minor alleles with liver and metabolic related phenotypes in the UK Biobank cohort.
Panels A and B include binary and continuous phenotypes, respectively. All associations are adjusted for age, sex, and the first five principal components of genetic ancestry. Details of the statistical analysis are provided in the Participants and methods section. ALT, alanine aminotransferase; AST, aspartate aminotransferase; FIB-4, fibrosis 4 index; HbA1c, glycated haemoglobin; HCC, hepatocellular carcinoma; T2DM, type 2 diabetes mellitus.
Carriage of rs328:G in LPL was also associated with a reduced risk of HCC and metabolic traits. However, although the effect sizes were generally comparable with rs13702, the statistical significance was weaker, likely reflecting the lower allele frequency of rs328:G. Neither rs13702:C (beta: -0.1; p = 0.21) nor rs328:G (beta: -0.01; p = 0.38) were associated with hepatic fat content assessed by MRI at the population level.
Discussion
Genetic case control studies are useful to investigate mechanisms for diseases where development spans several decades. This is particularly true for cirrhosis and HCC, in which many years of ongoing liver damage and genetic predisposition contribute to occurrence of disease.25 Several genetic risk loci have been identified and replicated for HCC in ALD, such as the PNPLA3 I148M variant and rs58542926 in the TM6SF2 gene.5 Knowledge of such risk factors may lead to better surveillance strategies in people at risk and elucidate the molecular basis of HCC development.
Analysing different cohorts of cases with HCC and controls with alcohol-associated cirrhosis, alcohol-abusers without liver disease, and healthy controls regarding the frequency of the LPL rs13702 genotypes, we found that the CC genotype was protective against development of HCC in two independent cohorts and after correction for age, sex, PNPLA3 I148M genotype, and diabetes. Interestingly, we noted that carriage of the CC genotype at the LPL rs13702 locus did not mitigate the risk for HCC in the high-risk group of homozygous PNPLA3 148 M carriers, underlining the complex interplay of different genetic risk factors and hinting at a shared molecular pathway. International guidelines recommend regular screening by ultrasound in all people affected by ALD and advanced fibrosis.21 As the number of patients with ALD is high – amounting to 60,000 in England and Wales in 2012, for example – and the incidence of alcohol abuse is rising,26 it will be essential to narrow down the number of people for screening to make efficient surveillance feasible. In NAFLD, a polygenic risk score comprising five different genetic risk loci showed a high negative predictive value of 94% for the development of HCC while limiting the number of individuals at risk to only 23% of the study population with cirrhosis.27 The discovery of new genetic variants such as at the LPL rs13702 locus may help to develop similar genetic risk scores for patients with ALD with an even better predictive capacity when interactions such as between PNPLA3 and LPL variants are considered. With an OR of 0.49, the LPL rs13702 CC genotype has a major impact on the individual risk of HCC. In addition, we investigated the LPL rs328 variant, which has a similar effect on lipid levels.20 Although we observed a numerically similar effect to the LPL rs13702 minor variant, it failed statistical significance for HCC risk. This fits in with reports from a previous study on lipid levels which indicated that rs328 is in linkage disequilibrium with rs13702 and exerts weaker effects.18
From a mechanistic point of view, the association of LPL genetic variation with the risk of HCC highlights the importance of lipid metabolism for HCC development in ALD. With PNPLA3,28 TM6SF2,29 and apolipoprotein E,6 other prominent genetic loci for HCC have been implicated in hepatic lipid accumulation. Although the underlying mechanisms leading to hepatic lipid accumulation have been partly unravelled, the molecular background behind the increased hepatocarcinogenesis is only incompletely understood. Among the factors supposed to facilitate HCC development and potentially linked to genetic variation are oxidative stress by overload with fatty acids and chronic inflammation.25 Concerning PNPLA3, we have reported that hepatocytes carrying PNPLA3 148 M secrete more pro-inflammatory chemokines when exposed to increased lipid levels, creating a tumorigenic environment.30 Although LPL is expressed in leukocytes, its effect on pro- or anti-inflammatory cell function has not been fully elucidated31 and may be dependent on the specific cell type and the composition of the lipid environment.
LPL has been known for many years as an important enzyme involved in lipid metabolism, regulating delivery of lipids to the tissues.31 Being functional as a dimer, it is expressed highly in endothelial cells, muscle, and adipose tissue.31 By abolishing the inhibitory effect of miR-410 on its gene expression, the minor variant of the rs13702 polymorphism leads to decreased circulating levels of triglycerides.10 Given the protective effect of the minor variant on hyperlipidaemia, it is not surprising that its presence is associated with a decreased risk for stroke, depending on the diet of the person,18 and a decreased risk for NAFLD.9 However, LPL expression has long been thought to occur only in foetal liver.12 Nevertheless, liver LPL mRNA levels were increased 3.6-fold in 16 patients with NAFLD compared with eight patients without steatosis undergoing bariatric surgery and correlated closely with liver fat content.14 Similarly, hepatic mRNA expression was reported as very low in 14 healthy controls, increased in 21 patients with NAFLD but highest in 54 patients with non-alcoholic steatohepatitis (NASH).17 Interestingly, immunofluorescence staining attributed the LPL expression in the liver not to hepatocytes, but to HSCs.17 Because carriage of a LPL rs13702 C allele has been shown to impact on triglyceride and HDL cholesterol levels in the general population10 and modulates the effect of diet,18 it is possible that the effect of the LPL rs13702 minor variant on HCC development might not have its mechanistical origin in liver tissue, but that the lipid content of lipoproteins taken up by the liver might be responsible for the differences in HCC risk. However, although impact of the rs13702 LPL genotype on serum lipid levels in our participants with alcohol-associated cirrhosis was only mild, we noted lower liver enzymes and lower hepatic fat content among individuals carrying the rs13702 CC genotype, supporting an impact on the liver metabolism. Because the effect on liver fat missed statistical significance at a population level, dietary habits such as alcohol abuse may be an important precondition.
Additionally, our study expands the knowledge on hepatic LPL expression by showing that LPL upregulation in the human liver is dependent on the number of C alleles at the rs13702 locus in alcohol-associated cirrhosis, highlighting the potential hepatic functional relevance of this genetic variant for metabolic liver disease. Finally, comparison of LPL expression in different liver derived cell lines supported the finding by Teratani et al.17 that HSCs are a major source of LPL. Although an important role for HSCs not only in hepatic fibrosis, but also in creating a tumorigenic microenvironment has been suspected based on their ability to produce growth factors and respective cytokines,32 evidence from clinical human studies has still been scarce. Our study now provides sound evidence that HSCs may be crucially involved in HCC development in humans. In line, the important role of cell metabolism for activation of HSCs has been reviewed recently.33 In addition, the most important genetic variation for HCC development, PNPLA3 I148M, not only modulates hepatocyte metabolism,34 but also promotes the profibrogenic activity of HSCs.35 Because high LPL expression was not limited to HSCs, but also found in LSECs, they might also contribute to the reduced risk for HCC.
The role of LPL in advanced liver disease and the mechanism by which LPL upregulation decreases hepatic tumorigenesis are not known. Similar to PNPLA3, which differs functionally in mice and humans, findings on LPL function in mice and humans are contradictory. Human LPL high producers are protected from NAFLD.9 Similarly, our data suggest that overexpression of LPL is beneficial regarding HCC development in human ALD. However, knockout of LPL in HSCs in a murine NASH model reduced hepatic fibrosis.17 The major differences in the experimental design concerning disease – NASH vs. ALD, species – murine vs. human, and stage of liver disease – fibrosis vs. HCC might account for these apparently contradictory results. Most importantly, experimental knockout of a protein which results in complete absence might lead to different effects compared with differences in the level of expression. LPL knockout mice survive birth for only hours.36 In support of the concept that LPL expression in hepatocytes is irrelevant to liver disease, knockout of LPL specifically in hepatocytes did not change hepatic fat content.37 Genetic LPL deficiency in humans is a very rare and often severe condition, that leads to high blood triglyceride levels and recurrent pancreatitis, with fatty liver being clinically not relevant in these individuals.38,39 Thus, knockout of LPL only in HSCs may not reflect the complex interplay involving several tissues in humans. It seems plausible that LPL upregulation in human NAFLD and ALD occurs in compensation to pathological changes in lipid metabolism, as shown for chronic alcohol abuse,40 which would explain why genetic high producers are to some extent protected from hepatic liver overload, NAFLD and HCC. Intrahepatic fat content is recognised as a risk factor for HCC in humans,41 which is reflected by the results from murine HCC models.42 However, the hypothesis that LPL is upregulated in compensation to hepatic fat overload, which results in reduction of hepatic fat, lipotoxicity, and subsequently HCC development, has still to be proven.
Our findings are based on a case control study, with the well-known limitations of this approach. Most importantly, we cannot exclude that some of the controls will develop HCC in the future or would have developed HCC if they had not died before from another complication of liver cirrhosis, such as bacterial sepsis. Such issues could be only resolved in a prospective study including only people at diagnosis of cirrhosis. However, although such studies are highly welcome to assess the so far known risk factors for HCC, this approach would likely need decades, because HCC formation in cirrhosis is a process spanning many years. Next, we cannot prove an additive effect, meaning that people carrying only one LPL rs13702 C allele have an HCC risk in between wildtype and homozygous CC allele carriers in our cohort. This might be because of a threshold effect: LPL mRNA expression in the liver was much more increased in homozygous compared with heterozygous CC allele carriers (Fig. 4C) and the effect on liver enzyme levels and hepatic fat was much more pronounced in homozygous CC allele carriers (Fig. 2A–C). LPL activity is tightly controlled also on a post-transcriptional level,43 for example by the inhibitor ANGPTL3, which is mainly expressed in the liver,44 so that the effect of a single C allele might be diminished considerably. In addition, our study may be underpowered to show an additive effect: studies showing an additive effect on lipid levels in the blood included more than 27,00010 or more than 7,10018 individuals. Nevertheless, as illustrated in Fig. 5, analysis of the UK Biobank cohort showed an additive effect.
The correlation of hepatic LPL mRNA expression with the number of rs13702 C alleles in patients with alcohol-associated cirrhosis is limited by the rather small number of specimens which we analysed. However, higher LPL expression in carriers of a C allele has been described before in non-liver tissue.9 Given that patients with alcoholic-associated cirrhosis carry two C alleles in only 10%, analysis of more liver specimens from those individuals is challenging.
Next, we cannot exclude that genetic variation at LPL rs13702 is not causally related to HCC, but only in linkage with the real associated factor. Mechanistic studies in humans are nearly impossible, and LPL mice models do not fully reflect the human phenotype. Because LPL was highly upregulated in liver tissue of patients with ALD and expression levels correlated to the number of rs13702 C alleles, it is highly likely that LPL is crucially involved in advanced ALD. Measurement of intrahepatic LPL activity might link LPL rs13702 genotypes mechanistically more closely to HCC development, but we did not have enough specimens to analyse LPL activity.
In summary, we report a novel genetic variant in LPL associated with decreased occurrence of HCC in ALD.
Financial support
This study was funded by the Deutsche Krebshilfe (70114349) to HDN. FSt was funded by grant SNF 310030_169196 from the Swiss National Fund. JN received funding by the German Research Foundation (DFG SFB/TRR 57 and SPP1937) and the Hector foundation (M88). The funding organisations had no role in the design of the study, and collection, analysis, and interpretation of the data, or writing of the manuscript.
Authors contributions
Collected specimen and data: PL, AK, JF, SB, WS, HI, CB, FSt, MS, MMS. Performed the experiments: FSc, CM, HDN, BK, BL. Analysed the data PL, HDN, FSc, HI, SB. Wrote the manuscript PL, HDN, FSc. Critically revised the manuscript JN, FSt, CPS.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of interest
None of the authors has any conflict of interest in relation to the contents of this manuscript.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
The authors thank all study participants, clinicians, and administrative staff who contributed to this study. We also acknowledge the UK Biobank resource: application number 8764.
Footnotes
Author names in bold designate shared co-first authorship.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100684.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Kranzler H.R., Soyka M. Diagnosis and pharmacotherapy of alcohol use disorder: a review. JAMA. 2018;320:815–824. doi: 10.1001/jama.2018.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seitz H.K., Bataller R., Cortez-Pinto H., Gao B., Gual A., Lackner C., et al. Alcoholic liver disease. Nat Rev Dis Primer. 2018;4 doi: 10.1038/s41572-018-0014-7. http://www.nature.com/articles/s41572-018-0014-7 [DOI] [PubMed] [Google Scholar]
- 3.Pingitore P., Romeo S. The role of PNPLA3 in health and disease. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:900–906. doi: 10.1016/j.bbalip.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Buch S., Stickel F., Trépo E., Way M., Herrmann A., Nischalke H.D., et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 5.Trépo E., Caruso S., Yang J., Imbeaud S., Couchy G., Bayard Q., et al. Common genetic variation in alcohol-related hepatocellular carcinoma: a case-control genome-wide association study. Lancet Oncol. 2022;23:161–171. doi: 10.1016/S1470-2045(21)00603-3. [DOI] [PubMed] [Google Scholar]
- 6.Innes H., Nischalke H.D., Guha I.N., Weiss K.H., Irving W., Gotthardt D., et al. The rs429358 locus in apolipoprotein E is associated with hepatocellular carcinoma in patients with cirrhosis. Hepatol Commun. 2022;6:1213–1226. doi: 10.1002/hep4.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stickel F., Lutz P., Buch S., Nischalke H.D., Silva I., Rausch V., et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatology. 2020;72:88–102. doi: 10.1002/hep.30996. [DOI] [PubMed] [Google Scholar]
- 8.Buch S., Innes H., Lutz P.L., Nischalke H.D., Ju Marquardt, Fischer J., et al. Genetic variation in TERT modifies the risk of hepatocellular carcinoma in alcohol-related cirrhosis: results from a genome-wide case-control study. Gut. 2023;72:381–391. doi: 10.1136/gutjnl-2022-327196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghodsian N., Abner E., Emdin C.A., Gobeil É., Taba N., Haas M.E., et al. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson K., Nettleton J.A., Rotllan N., Tanaka T., Smith C.E., Lai C.-Q., et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am J Hum Genet. 2013;92:5–14. doi: 10.1016/j.ajhg.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., He P.-P., Zhang D.-W., Zheng X.-L., Cayabyab F.S., Yin W.-D., et al. Lipoprotein lipase: from gene to atherosclerosis. Atherosclerosis. 2014;237:597–608. doi: 10.1016/j.atherosclerosis.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Merkel M., Weinstock P.H., Chajek-Shaul T., Radner H., Yin B., Breslow J.L., et al. Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J Clin Invest. 1998;102:893–901. doi: 10.1172/JCI2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon J.H., Kim K., Choi S.H. Lipoprotein lipase: is it a magic target for the treatment of hypertriglyceridemia. Endocrinol Metab. 2022;37:575–586. doi: 10.3803/EnM.2022.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westerbacka J., Kolak M., Kiviluoto T., Arkkila P., Sirén J., Hamsten A., et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 15.Yoshimoto S., Loo T.M., Atarashi K., Kanda H., Sato S., Oyadomari S., et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 16.Pardina E., Baena-Fustegueras J.A., Llamas R., Catalán R., Galard R., Lecube A., et al. Lipoprotein lipase expression in livers of morbidly obese patients could be responsible for liver steatosis. Obes Surg. 2009;19:608–616. doi: 10.1007/s11695-009-9827-5. [DOI] [PubMed] [Google Scholar]
- 17.Teratani T., Tomita K., Furuhashi H., Sugihara N., Higashiyama M., Nishikawa M., et al. Lipoprotein lipase up-regulation in hepatic stellate cells exacerbates liver fibrosis in nonalcoholic steatohepatitis in mice. Hepatol Commun. 2019;3:1098–1112. doi: 10.1002/hep4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corella D., Sorlí J.V., Estruch R., Coltell O., Ortega-Azorín C., Portolés O., et al. MicroRNA-410 regulated lipoprotein lipase variant rs13702 is associated with stroke incidence and modulated by diet in the randomized controlled PREDIMED trial. Am J Clin Nutr. 2014;100:719–731. doi: 10.3945/ajcn.113.076992. [DOI] [PubMed] [Google Scholar]
- 19.Hammad S.S., Eck P., Sihag J., Chen X., Connelly P.W., Lamarche B., et al. Common variants in lipid metabolism-related genes associate with fat mass changes in response to dietary monounsaturated fatty acids in adults with abdominal obesity. J Nutr. 2019;149:1749–1756. doi: 10.1093/jn/nxz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W., Apostol G., Schreiner P.J., Jacobs D.R., Boerwinkle E., Fornage M. Associations of lipoprotein lipase gene polymorphisms with longitudinal plasma lipid trends in young adults: the Coronary Artery Risk Development in Young Adults (CARDIA) study. Circ Cardiovasc Genet. 2010;3:179–186. doi: 10.1161/CIRCGENETICS.109.913426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Association for The Study of the Liver, European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. Plos Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., et al. Hepatocellular carcinoma. Nat Rev Dis Primer. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 26.Williams R., Aspinall R., Bellis M., Camps-Walsh G., Cramp M., Dhawan A., et al. Addressing liver disease in the UK: a blueprint for attaining excellence in health care and reducing premature mortality from lifestyle issues of excess consumption of alcohol, obesity, and viral hepatitis. Lancet. 2014;384:1953–1997. doi: 10.1016/S0140-6736(14)61838-9. [DOI] [PubMed] [Google Scholar]
- 27.Bianco C., Jamialahmadi O., Pelusi S., Baselli G., Dongiovanni P., Zanoni I., et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J Hepatol. 2021;74:775–782. doi: 10.1016/j.jhep.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.BasuRay S., Wang Y., Smagris E., Cohen J.C., Hobbs H.H. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc Natl Acad Sci U S A. 2019;116:9521–9526. doi: 10.1073/pnas.1901974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longo M., Meroni M., Paolini E., Erconi V., Carli F., Fortunato F., et al. TM6SF2/PNPLA3/MBOAT7 loss-of-function genetic variants impact on NAFLD development and progression both in patients and in in vitro models. Cell Mol Gastroenterol Hepatol. 2022;13:759–788. doi: 10.1016/j.jcmgh.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nischalke H.D., Lutz P., Bartok E., Krämer B., Langhans B., Frizler R., et al. The PNPLA3 I148M variant promotes lipid-induced hepatocyte secretion of CXC chemokines establishing a tumorigenic milieu. J Mol Med. 2019;97:1589–1600. doi: 10.1007/s00109-019-01836-3. [DOI] [PubMed] [Google Scholar]
- 31.Chang C.L. Lipoprotein lipase: new roles for an “old” enzyme. Curr Opin Clin Nutr Metab Care. 2019;22:1115. doi: 10.1097/MCO.0000000000000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coulouarn C., Clément B. Stellate cells and the development of liver cancer: therapeutic potential of targeting the stroma. J Hepatol. 2014;60:1306–1309. doi: 10.1016/j.jhep.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Trivedi P., Wang S., Friedman S.L. The power of plasticity-metabolic regulation of hepatic stellate cells. Cell Metab. 2021;33:242–257. doi: 10.1016/j.cmet.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.BasuRay S., Smagris E., Cohen J.C., Hobbs H.H. The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology. 2017;66:1111–1124. doi: 10.1002/hep.29273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruschi F.V., Claudel T., Tardelli M., Caligiuri A., Stulnig T.M., Marra F., et al. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatololgy. 2017;65:1875–1890. doi: 10.1002/hep.29041. [DOI] [PubMed] [Google Scholar]
- 36.Weinstock P.H., Bisgaier C.L., Aalto-Setälä K., Radner H., Ramakrishnan R., Levak-Frank S., et al. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest. 1995;96:2555–2568. doi: 10.1172/JCI118319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G., Xu J.-N., Liu D., Ding Q., Liu M.-N., Chen R., et al. Regulation of plasma lipid homeostasis by hepatic lipoprotein lipase in adult mice. J Lipid Res. 2016;57:1155–1161. doi: 10.1194/jlr.M065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nierman M.C., Rip J., Twisk J., Jjm Meulenberg, Kastelein J.J.P., Stroes E.S.G., et al. Gene therapy for genetic lipoprotein lipase deficiency: from promise to practice. Neth J Med. 2005;63:14–19. [PubMed] [Google Scholar]
- 39.Maltais M., Brisson D., Gaudet D. Non-alcoholic fatty liver in patients with chylomicronemia. J Clin Med. 2021;10:669. doi: 10.3390/jcm10040669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider J., Liesenfeld A., Mordasini R., Schubotz R., Zöfel P., Kubel F., et al. Lipoprotein fractions, lipoprotein lipase and hepatic triglyceride lipase during short-term and long-term uptake of ethanol in healthy subjects. Atherosclerosis. 1985;57:281–291. doi: 10.1016/0021-9150(85)90040-1. [DOI] [PubMed] [Google Scholar]
- 41.Paul B., Lewinska M., Andersen J.B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berardi D.E., Bock-Hughes A., Terry A.R., Drake L.E., Bozek G., Macleod K.F. Lipid droplet turnover at the lysosome inhibits growth of hepatocellular carcinoma in a BNIP3-dependent manner. Sci Adv. 2022;8 doi: 10.1126/sciadv.abo2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S.A., Kersten S., Qi L. Lipoprotein lipase and its regulators: an unfolding story. Trends Endocrinol Metab. 2021;32:48–61. doi: 10.1016/j.tem.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kosmas C.E., Bousvarou M.D., Sourlas A., Papakonstantinou E.J., Peña Genao E., Echavarria Uceta R., et al. Angiopoietin-like protein 3 (ANGPTL3) inhibitors in the management of refractory hypercholesterolemia. Clin Pharmacol Adv Appl. 2022;14:49–59. doi: 10.2147/CPAA.S345072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.