Abstract
We present a chromosome-length genome assembly and annotation of the Black Petaltail dragonfly (Tanypteryx hageni). This habitat specialist diverged from its sister species over 70 million years ago, and separated from the most closely related Odonata with a reference genome 150 million years ago. Using PacBio HiFi reads and Hi-C data for scaffolding we produce one of the most high-quality Odonata genomes to date. A scaffold N50 of 206.6 Mb and a single copy BUSCO score of 96.2% indicate high contiguity and completeness.
Significance.
We provide a chromosome-length assembly of the Black Petaltail dragonfly (Tanypteryx hageni), the first genome assembly for any non-libelluloid dragonfly. The Black Petaltail diverged from its sister species over 70 million years ago. T. hageni, like its confamilials, occupies fen habitats in its nymphal stage, a life history uncommon in the vast majority of dragonflies. We hope that the availability of this assembly will facilitate research on T. hageni and other petaltail species, to better understand their ecology and support conservation efforts.
Introduction
The Black Petaltail dragonfly (Tanypteryx hageni) (fig. 1A), found in montane habitats from California to British Columbia, is something of an evolutionary enigma. It is a member of the odonate family Petaluridae (known as “petaltails’ due to the broad, petal-like claspers at the end of the male abdomen), which is estimated to have originated approximately 150 million years ago (Ware et al. 2014).
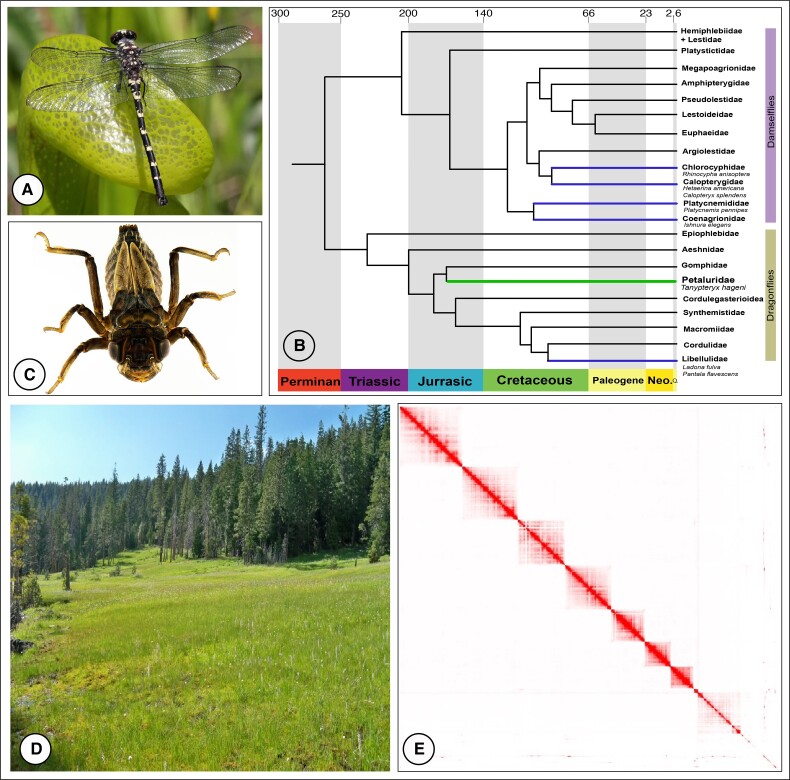
Fig. 1.
(A) T. hageni, adult. E. HiC contact map with the chromosome level assembly. (B) Phylogeny of Odonata (modified from Kohli et al. 2021). Families with a reference genome (The zygopterya Cholorcyphidae, Calopterygidae, Platycnemididae and Anisoptera Libellulidae) highlighted in purple, Petaluridae highlighted in green. (C) T. hageni, larvae, credit Marla Garrison. (D) A fen in Lassen National Forest (California, USA) where T. hageni was collected. (Upper right) E. Hi-C contact map visualizing the alignment to the reference genome assembly.
While the family originated long before most recognized insect families, the geologic ages of its member species are even more extreme in relation with other animal species, leading members of Petaluridae to be considered as “living fossils.” Most extant petaltails are estimated to have appeared in the mid- to late-Cretaceous, with speciation being driven by major events like continental drift; T. hageni is estimated to have diverged from the sister species Tanypteryx pryeri (found in Japan) ∼73 million years ago, potentially diverging when the Beringian land bridge disappeared in the late Cretaceous (fig. 1B) (Fiorillo 2008; Ware et al. 2014). Often, species with long geological persistence have wide geographic ranges (Powell 2007; Hopkins 2011), and tend to be habitat generalists, or give rise to habitat generalists (Colles et al. 2009), but for T. hageni this is not the case—the persistence of this species (as well as other petaltails) is puzzling as the nymphs of T. hageni exclusively inhabit fens (fig. 1C) (Baird 2012), groundwater-driven habitats which host a number of specialist animals and plants. These habitats are characterized by soils saturated by groundwater, commonly found around springs and in riparian areas of headwater streams. Nymphs (fig. 1D) dig and maintain a burrow (a behavior displayed by a number of other petaltail species) that fills with water.
The research on fens in North America is sparse, but it is known that while fens make up a tiny fraction of the North American landscape, they contain a surprising proportion of the continent's biodiversity. The US Department of Agriculture observes that between 15% and 20% of the rare and uncommon plant species found in Deschutes National Forest (Oregon, USA) are found in fen ecosystems (US Department of Agriculture). It is estimated that fens are the most floristically diverse wetlands in the United States, and contain a high number of rare and endangered species (Bedford and Godwin 2003). Research on fens across the range of T. hageni is minimal but it is known that fens are degrading across the continental United States (Bedford and Godwin 2003). Thus, we have concerns not only for the survival of the Black Petaltail, but for the specialized habitats in which they live. There is little research regarding genomic adaptations to life in fens, so it is paramount to establish a baseline of understanding for this declining habitat.
Here, we present a chromosome-length assembly of the Black Petaltail. This genome will be a valuable tool for studying an organism that may be especially hard hit by climate change and habitat destruction. Additionally, this genome will shed light on an evolutionary enigma: the petaltail dragonflies have persisted for tens of millions of years, despite exclusively occupying fragile fen habitat as nymphs (Ware et al. 2014). Lastly, this genome will be an important resource in resolving the phylogeny of early divergences within Odonata, as no genome of any basal anisopteran is currently available.
Results and Discussion
Sequencing and Genome Size Estimation
We recovered >44.6 Gbp of sequence contained in HiFi reads, generated from 730 Gbp of raw sequence in subreads from two PacBio SMRT cells with a read N50 of 14.2 kbp. Smudgeplot showed strong evidence of a diploid genome (supplementary fig. S1, Supplementary Material online) and reported 1n = 14 monoploid coverage. The estimated genome size using kmers from HiFi reads with GenomeScope 2.0 (Ranallo-Benavidez et al. 2020) ranged from 1.47Gbp to 1.63 Gbp depending on the upper limit used, with an estimated 60.0–53.9% of unique sequence dependent upon the upper limit, (Ranallo-Benavidez et al. 2020), resulting in approximately 25 × coverage (supplementary fig. S2, Supplementary Material online).
In addition to the sequencing-based method, the T. hageni genome size was estimated by flow cytometry analysis using Drosophila melanogaster as the standard (supplementary fig. S3A, Supplementary Material online). The genome size of D. melanogaster has been estimated using next-gen sequencing methods and flow cytometry. According to the NCBI genome database, the median size of the D. melanogaster genome is 138.9 Mbp, whereas the lowest flow cytometry estimate is 156 Mbp (Nardon et al. 2005; Boulesteix et al. 2006). The T. hageni genome size estimates were calculated as the ratio of red mean fluorescence intensity (MFI) of the G1 and G2 peaks of the T. hageni samples (supplementary fig. S3B, Supplementary Material online) to the G1 and G2 peaks of the D. melanogaster standards (supplementary fig. S3C, Supplementary Material online) multiplied by the genome size estimates of D. melanogaster. Thus, the Seq control group estimates the T. hageni genome size using the D. melanogaster sequencing estimate, and the flow control group estimates the T. hageni genome size using the D. melanogaster flow cytometry genome size estimate. The first T. hageni genome size (2.26 Gbp) was calculated using Drosophila genome size 138.9 Mbp and the second T. hageni genome size (2.54 Gbp) was calculated using Drosophila genome size 156 Mbp.
While these estimates were quite a bit larger than our sequencing-based estimation, there is precedent for genome size estimates from flow cell cytometry being more than twice the size of sequencing based methods in insecta (Pflug et al. 2020). The largest genome size estimate for an Odonata is 2.36 pg (∼2.3 Gbp), with a mean size of 1.01pg (∼.99 Gbp) in Anisoptera (Ardila-Garcia and Gregory 2009), so it is likely that the flow cytometry estimates are overestimating the genome size of T. hageni.
Genome Assembly and QC
Our contig assembly was generated with hifiasm v.0.16.1 (Cheng et al. 2021) and submitted to NCBI to identify possible contaminants. Following the removal of two possible contaminants (identified as Illumina adapters by NCBI), the assembly was 1.69 Gbp in length, contained 2,133 contigs, and had a contig N50 > 4 Mbp (supplementary Table S1, Supplementary Material online). After scaffolding with Hi-C data, we generated a highly contiguous assembly that was 1.70 Gbp in length with a scaffold N50 > 206.6 Mbp, with 90.5% of base pairs assigned to nine chromosome, matching previous estimates of the haplotype of T. hageni (Kuznetsova and Golub 2020) (supplementary Table S1 and fig. S7, Supplementary Material online, fig. 1E for contact maps). After filtering out contigs that were assigned to proteobacteria, mollusca, cnidaria, and bacteroidetes by megablastv.2.9.0 (Camacho et al. 2009) and had a GC content >42% or <34%, and replaced mitochondrial contigs with the mitochondrial genome assembly, resulting in a final assembly length of 1.68 Gbp with a scaffold N50 > 206.6 Mbp (supplementary Table S1, Supplementary Material online) and an overall GC content of 37.98% (supplementary Table S2, Supplementary Material online). We note that this final assembly size is slightly larger than the sequencing based estimates (supplementary fig. S2, Supplementary Material online), and smaller than the flow cytometry estimates (supplementary fig. S3, Supplementary Material online). As far as we are aware, Calopteryx splendens and Pantala flavescens, are the only other Odonata genome assembly for which there are genome size estimates. In the case of C. splendens, the authors estimated the genome assembly size to be 1.7 Gbp using flow cytometry, while the (illumina based) assembly length was 1.6 Gbp (Ioannidis et al. 2017). The genome assembly size of Pantala flavescens was slightly larger than a k-mer-based genome size estimation, generated using Feulgen image analysis.
We recovered 96.8% of universal single-copy orthologs (including 96.2% single and complete, 0.6% duplicated, and an additional 1.06% fragmented) from the BUSCO (Manni et al. 2021) Insecta database indicating a high level of completeness, especially when compared with most publicly available Odonata genomes (Table 1). The low level of duplicated single copy orthologs also indicates that the built-in haplotype purging step in hifiasm successfully removed duplicate contigs.
Table 1.
Comparison of Publicly Available Odonata Genomes
| Family | Assembly level | Assembly size (Mb) | N50 (Mb) | Scaffolds | Number of Annotated Genes | Single copy complete BUSCO Score of Assembly | Source | |
|---|---|---|---|---|---|---|---|---|
| Tanypteryx hageni | Petaluridae | Chromosome | 1.68 | 206.6 | 1,033 | 22,261 | C:96.8%[S:96.2%,D:0.6%],F:1.0%,M:2.2%,n:1367 | Authors |
| Ladona fulva | Libellulidae | Contig | 1.16 | 0.06 | 9,411 | 18,817 | C:95.5%[S:93.6%,D:1.9%],F:2.1%,M:2.4%,n:1367** | NCBI |
| Pantala flavescens | Libellulidae | Chromosome | 0.66 | 553 | 43 | 15,354 | C:96.9%[S:93.6%,D:3.3%],n:1367 | (Liu et al. 2022) |
| Ischnura elegans | Coenagrionidae | Chromosome | 1.66 | 123.6 | 110 | 27,076 | C:97.2%[S:96.4%,D:0.8%], F:1.3%,M:1.5%,n:1367 | (Price et al. 2022) |
| Hetaerina americana | Calopterygidae | Scaffold | 1.63 | 86.1* | 1583* | N/A | C:97.8%[S:96.3%,D:1.5%],F:0.5%,M:1.7%,n:1367 | NCBI |
| Platycnemis pennipes | Platycnemididae | Chromosome | 1.65 | 144.8 | 88 | N/A | C:96.9%[S:96.3%,D:0.6%],F:1.4%,M:1.7%,n:1367** | NCBI |
| Rhinocypha anisoptera | Chlorocyphidae | Contig | 1.55 | 0.41* | 754,445* | N/A | C:87.7%[S:71.0%,D:16.7%],F:4.2%,M:8.1%,n:1367** | NCBI |
| Calopteryx splendens | Calopterygidae. | Contig | 1.63 | 0.42* | 8,896* | 22,523 | C:95.0%[S:94.6%,D:0.4%],F:3.0%,M:2.0%,n:1367** | (Ioannidis et al. 2017) |
After blobtools analysis, 19.22% of our genome was assigned to chordata. (supplementary fig. S4, Supplementary Material online). This included two of the chromosome-length scaffolds, which had nearly identical GC proportions and coverage to the other chromosome-length scaffolds (supplementary fig. S4, Supplementary Material online). It is unlikely that this was due to contamination, as our Hi-C experiment assigned these to chromosomes. We hypothesized that this could be due to a lack of coverage of the lineage Petaluridae in the BLAST database. To test this, we characterized the top BLAST hits of publicly available Petaluridae transcriptomes. In the transcriptomes of Phenes raptor (Suvorov et al. 2021), T. pryeri (Suvorov et al. 2021), and T. hageni (Misof et al. 2014) 23%, 10%, and 10% of contigs had a top blast hit of chordata (supplementary fig. S5, Supplementary Material online). This suggests that a lack of database coverage for Petaluridae could be a likely explanation for this phenomenon. It appears that the genomes of Petaluridae have greatly diverged from what is covered in databases, leading to erroneous BLAST characterizations. We also observed this phenomenon when blasting the genome of the Atlantic Horseshoe Crab (Limulus polyphemus), where 9.7% of BLAST hits mapped to Chordata (supplementary fig. S6, Supplementary Material online). The Atlantic Horseshoe Crab also has a highly repetitive genome, and lack of database coverage may be resulting in BLAST results outside of the phylum.
Annotation
Here 55.12% of the genome was classified as repetitive using RepeatModeler2v2.0.1 (Flynn et al. 2020) and RepeatMasker v4.1.2 (Smit et al. 2013). 26.14% of the genome was classified as “unclassified repetitive elements’‘, 15.73% as DNA transposons, 11.38% as retroelements, and 1.53% as rolling circles. We identified 22,261 protein coding genes. The annotated protein set contained 89.4% of BUSCO insecta genes, with 6.7% of the BUSCO genes duplicated, and another 4.7% fragmented.
Mitochondrial Genome
Conclusion
Here we present one of the most complete Odonata genomes to date. As the first non-libelluloid Anisopteran genome, and the first genome of an odonate habitat specialist, this assembly will be a valuable tool for understanding the biology of the Black Petaltail, resolving the phylogeny of Anisoptera, and will provide general insights into long-persisting species.
Materials and Methods
Specimen Collection, DNA Extraction, and Sequencing
We collected an immature nymph live from a burrow near Cherry Hill Campground (Lassen National Forest, California, USA) in fall of 2020. The specimen was flash frozen in liquid nitrogen and stored in a −80 °C freezer prior to extraction. High molecular weight DNA was extracted from a single individual using the Qiagen Genomic-tip kit.
Another specimen was collected from the same location in spring 2022 for Hi-C library generation. It was also flash frozen in liquid nitrogen, and sent for Hi-C analysis by DNA Zoo, who used the hemolymph for Hi-C library preparation.
High molecular weight DNA was sheared to 18 kbp using a Diagenode Megaruptor and prepared into a sequencing library using the PacBio HiFi SMRTbell Express Template Kit 2.0. The library was sequenced on two PacBio Sequel II 30 h SMRT cells in CCS mode at the BYU sequencing center.
D. melanogaster heads and T. hageni head and thorax tissue were placed into ice cold Galbraith's buffer containing RNase A and crushed using a Dounce homogenizer. Lysate was then transferred to FACS tubes, through a filter. Samples were stained with propidium iodide (PI) diluted in Galbraith's buffer (Galbraith et al. 2001) (1 mg/mL) for 3 h. Two experiments were performed, one on the BD Accuri C6 cytometer and another on the Beckman Cytoflex cytometer, with two data points being retrieved from the Accuri experiment and one from the Cytoflex experiment. During the Accuri experiment, two data points were obtained from the ratios of red mean fluorescence intensities of the G1 and G2 peaks of one Drosophila sample and one T. hageni sample. During the cytoflex experiment, only the G1 peak of the Drosophila and T. hageni samples were compared. Thus, two biological replicates are presented with an additional technical replicate. The total quantity of DNA content within the T. hageni samples was calculated as the ratio of red MFI of the G1 and G2 peaks of the T. hageni samples to the G1 and G2 peaks of the D. melanogaster standards multiplied by the genome size estimates of D. melanogaster. As demonstrated by other insect genome estimates obtained by next-generation sequencing technology and flow cytometry, including D. melanogaster, flow cytometry analysis may result in larger genome size estimates compared to sequencing methods (Pflug et al. 2020).
Sequencing QC and Genome Assembly
We generated HiFi reads from raw subreads with PacBio SMRTlink. We then used the HiFi reads to estimate genome size with Genomescope 2.0 (Ranallo-Benavidez et al. 2020). We generated read histograms with KMC3 (Kokot et al. 2017), and tested three different higher upper bounds of 10,000, 100,000, and 1,000,00 with a k of 21. We also assessed the reads with smudgeplot (Ranallo-Benavidez et al. 2020) by using the script “smudgeplot.py” to estimate the upper and lower bounds of the plot, which returned estimates of 970 and 10 respectively, and used these estimates, and a k of 21, when generating the final smudgeplot. We generated an initial contig assembly with hifiasm v.0.16.0 (Cheng et al. 2021), and submitted the assembly to NCBI, through the genome submission tool, to check for contamination.
To generate chromosome length scaffolds, we used High-throughput chromosome conformation capture (Hi-C). The DNA Zoo consortium (dnazoo.org) generated an in situ Hi-C library using the protocol described in Rao et al. (2014). Hi-C data were then aligned to the draft assembly using Juicer (Durand et al. 2016), and the candidate chromosome length genome assembly was built using 3D-DNA (Dudchenko et al. 2017). The resulting contact maps (fig. 1E, supplementary fig. S7, Supplementary Material online) were manually reviewed using Juicebox Assembly Tools (Durand et al. 2016; Dudchenko et al. 2018). Manual curation involves identify visually anomalous patterns in the heatmap (see e.g., Rao et al. 2014; Durand et al. 2016; Dudchenko et al. 2017, 2018; Harewood et al. 2017; Lapp et al. 2018)). The full process has been described in detail (Dudchenko et al. 2018). Interactive contact maps were generated using juicebox.js (Robinson et al. 2018), for both the draft and reference assembly and are publicly available at www.dnazoo.org/assemblies/Tanypteryx_hageni.
We screened for contamination with taxon-annotated GC-coverage plots using BlobTools v1.1.1 (Laetsch and Blaxter 2017). We mapped all Hi-Fi reads against the final assembly using minimap2 v2.1 (Li 2018), sorted the bam file with samtools v1.13 (Danecek et al. 2021) using the command samtools sort, and assigned taxonomy with megablast (Shiryev et al. 2007) using the parameters: task megablast and -e-value 1e-25. We calculated coverage using the blobtools function map2cov, created the blobtools database using the command blobdb, and generated the blobplot with the command blobtools plot. After examining the blobplot we removed contigs blasting to proteobacteria, bacteroides, cnidaria and mollusca, and had a GC content >42% or < 34%.
To investigate whether excessive megablast assignments to chordata were due to a lack of database coverage for Petaluridae, we used BLAST to classify the transcriptomes of T. hageni (Misof et al. 2014), and other members of Petaluridae, T. pryeri and P. raptor (Suvorov et al. 2021), against the Genbank nucleotide database, using the same parameters as above: task megablast and -e-value 1e-25.
Quality Control
We generated all contiguity stats with assembly-stats (Trizna 2020). We also ran BUSCO (Manni et al. 2021) on the other publicly available Odonata genomes for comparison (Table 1) using the Insecta ODB10 database, in genome mode with the flag –long to retrain BUSCO for more accurate identification of genes.
Annotation
We first modeled and masked the repetitive elements of the scaffold and chromosome-level assemblies using RepeatModeler2 (Flynn et al. 2020). We then annotated the masked, scaffold-level assembly using MAKER v3.01.03 (Campbell et al. 2014). We ran a homology-only MAKER run using the 1kite T. hageni transcriptome (Misof et al. 2014), the transcriptomes of the Petaluridae T. pryeryi and P. raptor (Suvorov et al. 2021), and the complete annotated protein sets of Ladona fulva, Pantala flavescens, and Ischnura elegans. We trained Augustus (Stanke et al. 2008, 2006) on the identified protein sets using BUSCO and the insecta dataset (Manni et al. 2021), and ran MAKER a second time to generate ab-initio gene predictions. We then mapped these proteins to the chromosome level assembly using miniprot v0.4 (Li 2022) and re-trained Augustus (Stanke et al. 2008, 2006) using the scaffold-level coding sequences, with 1000 base pairs surrounding each sequence as the training set. As this annotation resulted in a high number of genes, and a less-than ideal BUSCO score we also mapped the protein set of Pantala flavescens (Liu et al. 2022) to the masked chromosome level assembly using miniprot v0.4(Li 2022), and extracted the mapped Pantala proteins, the protein set from the augustus annotation, and the protein set of the mapped proteins from their respective gff files with gffread (Pertea 2022). We combined all three protein sets and clustered the proteins at 80* similarity with CD-HIT v4.8.1 (Fu et al. 2012). We then mapped this clustered protein set back to the chromosome-level assembly with miniprot (Li 2022), and used BLAST (Shiryev et al. 2007) to align the candidate proteins to all Arthropoda proteins available on NCBI using the parameters -outfmt “6 sseqid pident evalue qseqid -max_target_seqs 1 -max_hsps 1 -num_threads 16 -evalue 1e-15”. We only retained proteins with a significant BLAST hit for our final annotation, and used BUSCO, using the Insecta database, to assess annotation completeness.
Mitochondrial Genome Assembly
We assembled and annotated the mitochondrial genome using Mitohifi (Allio et al. 2020; Uliano-Silva et al. 2021) on the scaffolded assembly, using the default parameters and the mitochondrial genome of Anax parthenope (Ma et al.) as a reference. We aligned the mitochondrial genome with the mitochondrial genomes of the Corduliidae Epothalmia elegans (Wang et al. 2019), the Macromiidae Macromia daimoji (Kim et al.), the Libellulidae Pantala flavescens (David et al.), the Gomphidae Gomphus vulgatissimus, the Corduligastridae Cordulegaster boltonii (Hovmöller 2006), the Aeshnidae Anax parthenope (Ma et al.), and the zygopteran Pseudolestes mirabilis (Yu et al. 2021), representing each of the families of Anisoptera for which there is a mitochondrial genome assembly, in AliView (Larsson 2014) using MUSCLE v3.8.31(Edgar 2004).
Supplementary Material
Acknowledgments
Hi-C data for the Black Petaltail were created by the DNA Zoo Consortium (dnazoo.org). DNA Zoo sequencing effort is supported by Illumina, Inc.; IBM; and the Pawsey Supercomputing Center. E.L.A. was supported by the Welch Foundation (Q-1866), a McNair Medical Institute Scholar Award, an NIH Encyclopedia of DNA Elements Mapping Center Award (UM1HG009375), a US-Israel Binational Science Foundation Award (2019276), the Behavioral Plasticity Research Institute (NSF DBI-2021795), NSF Physics Frontiers Center Award (NSF PHY-2019745), and an NIH CEGS (RM1HG011016-01A1).
This work was also supported by a college undergraduate research award from the Department of Life Sciences at Brigham Young University.
Contributor Information
Ethan R Tolman, American Museum of Natural History, Department of Invertebrate Zoology, New York, New York; The City University of New York Graduate Center, New York, New York.
Christopher D Beatty, Program for Conservation Genomics, Department of Biology, Stanford University.
Jonas Bush, Department of Plant and Wildlife Sciences, Brigham Young University, Provo, Utah.
Manpreet Kohli, American Museum of Natural History, Department of Invertebrate Zoology, New York, New York; Department of Natural Sciences, Baruch College, New York, New York.
Carlos M Moreno, Department of Microbiology and Molecular Biology, Brigham Young University, Provo, Utah.
Jessica L Ware, American Museum of Natural History, Department of Invertebrate Zoology, New York, New York.
Ruqayya Khan, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas.
Chirag Maheshwari, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas.
David Weisz, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas.
Olga Dudchenko, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas; The University of Western Australia, UWA School of Agriculture and Environment, Crawley, Washington, Australia.
Erez Lieberman Aiden, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas; Center for Theoretical Biological Physics and Department of Computer Science, Rice University, Houston, Texas; The University of Western Australia, UWA School of Agriculture and Environment, Crawley, Washington, Australia; Broad Institute of MIT and Harvard, Cambridge, Massachusetts; Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech, Pudong, China; LOEWE Centre for Translational Biodiversity Genomics (LOEWE-TBG), Frankfurt, Germany.
Paul B Frandsen, Department of Plant and Wildlife Sciences, Brigham Young University, Provo, Utah; LOEWE Centre for Translational Biodiversity Genomics (LOEWE-TBG), Frankfurt, Germany; Data Science Lab, Office of the Chief Information Officer, Smithsonian Institution, Washington, DC.
Supplementary material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
The draft and unfiltered reference assembly are publicly available on DNA Zoo's website (www.dnazoo.org/assemblies/Tanypteryx_hageni). The final assembly file, annotation files, and the mitochondrial genome alignment are available on figshare (project link: https://figshare.com/projects/A_Chromosome-length_Assembly_of_the_Black_Petaltail_Tanypteryx_hageni_Dragonfly/151584. DOI: genome assembly 10.6084/m9.figshare.21390975, genome annotation file 10.6084/m9.figshare.21390966, annotated proteins 10.6084/m9.figshare.21390972, repeatmasker output 10.6084/m9.figshare.21391029, and mitochodnrial genome alignment 10.6084/m9.figshare.21824424). In addition to the figshare repository, the final assembly file is also being processed by NCBI and will be available under the accession number JAJPWW000000000. Illumina reads used for HiC scaffolding are available at https://www.ncbi.nlm.nih.gov/sra/SRX18440492%5Baccn%5D. The Pacbio Hifi reads are available under the NCBI BioProject accession PRJNA743558.
Literature Cited
- Allio R, et al. 2020. Mitofinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour 20:892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila-Garcia AM, Gregory TR. 2009. An exploration of genome size diversity in dragonflies and damselflies (Insecta: Odonata). J Zool 278:163–173. [Google Scholar]
- Baird I. 2012. The wetland habitats, biogeography and population dynamics of Petalura gigantea (Odonata: Petaluridae) in the Blue Mountains of New South Wales. PhD, The University of Western Sydney.
- Bedford BL, Godwin KS. 2003. Fens of the United States: distribution, characteristics, and scientific connection versus legal isolation. Wetlands 23:608–629. [Google Scholar]
- Boulesteix M, Weiss M, Biémont C. 2006. Differences in Genome Size Between Closely Related Species: The Drosophila melanogaster Species Subgroup. Mol Biol Evol 23:162–167. doi: 10.1093/molbev/msj012 [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics 48:4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods 18:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles A, Liow LH, Prinzing A. 2009. Are specialists at risk under environmental change? Neoecological, paleoecological and phylogenetic approaches. Ecol Lett 12:849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David FJ, et al. 2021. The first complete mitochondrial genome of the migratory dragonfly Pantala flavescens Fabricius, 1798 (Libellulidae: Odonata). Mitochondrial DNA B Resour 6:808–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DNA Zoo . DNA Zoo.www.dnazoo.org(Accessed July 27, 2022a).
- Dudchenko O, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, et al. 2018. The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. doi:10.1101/254797.
- Durand NC, et al. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo AR. 2008. Dinosaurs of Alaska: implications for the cretaceous origin of beringia. In: The terrane puzzle: new perspectives on paleontology and stratigraphy from the North American Cordillera. Blodgett, RB, Stanley, GD. Jr, editors. Vol. 442Geological Society of America. doi: 10.1130/2008.442(15). [DOI] [Google Scholar]
- Flynn JM, et al. 2020. Repeatmodeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA 117:9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D W, Lambert G M, Macas J, Dolezel J. 2001. Analysis of nuclear DNA content and ploidy in higher plants. Curr Protoc Cytom. Chapter 7:Unit 7.6. doi: 10.1002/0471142956.cy0706s02 [DOI] [PubMed] [Google Scholar]
- Harewood L, et al. 2017. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol 18:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins MJ. 2011. How species longevity, intraspecific morphological variation, and geographic range size are related: a comparison using late Cambrian trilobites. Evolution 65:3253–3273. [DOI] [PubMed] [Google Scholar]
- Hovmöller R. 2006. Molecular phylogenetics and taxonomic issues in dragonfly systematics (Insecta: Odonata). [Google Scholar]
- Ioannidis P, et al. 2017. Genomic features of the damselfly Calopteryx splendens representing a sister clade to most insect orders. Genome Biol Evol 9:415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Jeong SY, Wang AR, An J, Kim I. 2021. Complete mitochondrial genome sequence of Macromia daimoji Okumura, 1949 (Odonata: Macromiidae). Mitochondrial DNA B Resour 3:365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli M, et al. 2021. Evolutionary history and divergence times of Odonata (dragonflies and damselflies) revealed through transcriptomics. iScience 24:103324. doi: 10.1016/j.isci.2021.103324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokot M, Długosz M, Deorowicz S. 2017. KMC 3: counting and manipulating k-mer statistics. Bioinformatics 33:2759–2761. [DOI] [PubMed] [Google Scholar]
- Kuznetsova VG, Golub NV. 2020. A checklist of chromosome numbers and a review of karyotype variation in Odonata of the world. Comp Cytogenet 14:501–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML. 2017. BlobTools: Interrogation of genome assemblies. F1000Res. 6:1287. doi: 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- Lapp SA, et al. 2018. Pacbio assembly of a plasmodium knowlesi genome sequence with hi-C correction and manual annotation of the SICAvar gene family. Parasitology 145:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. Aliview: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2022. lh3/miniprot.https://github.com/lh3/miniprot. (Accessed October 13, 2022).
- Liu H, et al. 2022. Chromosome-level genome of the globe skimmer dragonfly (Pantala flavescens). GigaScience 11:giac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Hu Y, Wu J. 2021. The complete mitochondrial genome of Anax parthenope (Odonata: Anisoptera) assembled from next-generation sequencing data. Mitochondrial DNA B Resour 5:2817–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Nardon C. 2005. Is genome size influenced by colonization of new environments in dipteran species?. Mol Ecol 14:869–878. doi: 10.1111/j.1365-294X.2005.02457.x. [DOI] [PubMed] [Google Scholar]
- Pertea G. 2022. gpertea/gffread.https://github.com/gpertea/gffread(Accessed October 13, 2022).
- Pflug JM, Holmes VR, Burrus C, Johnston JS, Maddison DR. 2020. Measuring genome sizes using read-depth, k-mers, and flow cytometry: methodological comparisons in beetles (Coleoptera). G3 Genes|Genomes|Genetics 10:3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MG. 2007. Geographic range and genus longevity of late paleozoic brachiopods. Paleobiology 33:530–546. [Google Scholar]
- Price BW, et al. 2022. The genome sequence of the blue-tailed damselfly, Ischnura elegans (Vander Linden, 1820). doi: 10.12688/wellcomeopenres.17691.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. 2020. Genomescope 2.0 and smudgeplot for reference-free profiling of polyploid genomes. Nat Commun 11:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2018. Juicebox.js provides a cloud-based visualization system for hi-C data. Cell Syst 6:256–258.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryev SA, Papadopoulos JS, Schäffer AA, Agarwala R. 2007. Improved BLAST searches using longer words for protein seeding. Bioinformatics 23:2949–2951. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013. RepeatMasker Open-4.0.http://www.repeatmasker.org.
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644. [DOI] [PubMed] [Google Scholar]
- Stanke M, Tzvetkova A, Morgenstern B. 2006. AUGUSTUS At EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol 7:S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A, et al. 2021. Deep ancestral introgression shapes evolutionary history of dragonflies and damselflies. Syst Biol:syab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizna M. 2020. assembly_stats 0.1.4. doi: 10.5281/zenodo.3968775. [DOI]
- Uliano-Silva M, Nunes JGF, Krasheninnikova K, McCarthy SA. 2021. marcelauliano/MitoHiFi: mitohifi_v2.0. doi: 10.5281/zenodo.5205678. [DOI]
- US Department of Agriculture . The Fen Phenomenon An Elusive Ecosystem.https://www.fs.usda.gov/Internet/FSE_DOCUMENTS/fseprd504763.pdf. (Accessed April 14, 2022).
- Wang J-D, Wang C-Y, Zhao M, He Z, Feng Y. 2019. The complete mitochondrial genome of an edible aquatic insect Epophthalmia elegans (Odonata: Corduliidae) and phylogenetic analysis. Mitochondrial DNA Part B 4:1381–1382. [Google Scholar]
- Ware JL, et al. 2014. The petaltail dragonflies (Odonata: Petaluridae): Mesozoic habitat specialists that survive to the modern day. J Biogeogr 41:1291–1300. [Google Scholar]
- Yu D-N, et al. 2021. Increasing 28 mitogenomes of Ephemeroptera, Odonata and Plecoptera support the Chiastomyaria hypothesis with three different outgroup combinations. PeerJ 9:e11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The draft and unfiltered reference assembly are publicly available on DNA Zoo's website (www.dnazoo.org/assemblies/Tanypteryx_hageni). The final assembly file, annotation files, and the mitochondrial genome alignment are available on figshare (project link: https://figshare.com/projects/A_Chromosome-length_Assembly_of_the_Black_Petaltail_Tanypteryx_hageni_Dragonfly/151584. DOI: genome assembly 10.6084/m9.figshare.21390975, genome annotation file 10.6084/m9.figshare.21390966, annotated proteins 10.6084/m9.figshare.21390972, repeatmasker output 10.6084/m9.figshare.21391029, and mitochodnrial genome alignment 10.6084/m9.figshare.21824424). In addition to the figshare repository, the final assembly file is also being processed by NCBI and will be available under the accession number JAJPWW000000000. Illumina reads used for HiC scaffolding are available at https://www.ncbi.nlm.nih.gov/sra/SRX18440492%5Baccn%5D. The Pacbio Hifi reads are available under the NCBI BioProject accession PRJNA743558.