Abstract
Why do some genomes stay small and simple, while others become huge, and why are some genomes more stable? In contrast to angiosperms and gymnosperms, liverworts are characterized by small genomes with low variation in size and conserved chromosome numbers. We quantified genome evolution among five Marchantiophyta (liverworts), measuring gene characteristics, transposable element (TE) landscape, collinearity, and sex chromosome evolution that might explain the small size and limited variability of liverwort genomes. No genome duplications were identified among examined liverworts and levels of duplicated genes are low. Among the liverwort species, Lunularia cruciata stands out with a genome size almost twice that of the other liverwort species investigated here, and most of this increased size is due to bursts of Ty3/Gypsy retrotransposons. Intrachromosomal rearrangements between examined liverworts are abundant but occur at a slower rate compared with angiosperms. Most genes on L. cruciata scaffolds have their orthologs on homologous Marchantia polymorpha chromosomes, indicating a low degree of rearrangements between chromosomes. Still, translocation of a fragment of the female U chromosome to an autosome was predicted from our data, which might explain the uniquely small U chromosome in L. cruciata. Low levels of gene duplication, TE activity, and chromosomal rearrangements might contribute to the apparent slow rate of morphological evolution in liverworts.
Keywords: genome evolution, duplication, transposable elements, sex chromosome evolution, collinearity
Significance.
The rate of genome evolution influenced by, for example, polyploidy, transposable element (TE) accumulation, and chromosomal rearrangements is generally high among plants and in particular within angiosperms. Bryophytes contain smaller and less variable genomes than other nonseed plants, suggesting that the pattern of genome size evolution differs between plant divisions in various aspects. However, the reasons for these observations are lacking as genomic information from bryophyte genomes is limited. Analyses of genome evolution among liverworts revealed a low rate of gene duplication and chromosomal rearrangements and rare bursts of TEs, which can explain the small size and limited variability of liverwort genomes.
Introduction
Some genomes stay small and simple, while others evolve to become huge (Pellicer and Leitch 2020). Likewise, some genomes show higher levels of stability than others, which might be related to the complexity and diversification of the organisms. High rates of genome size evolution, but not the actual size of the genome, correlate with high rates of speciation in angiosperms, consistent with previous predictions that genome size variability is linked to success in flowering plants (Puttick et al. 2015).
Angiosperm nuclear genomes vary 1,000-fold in DNA content, from around 60 Mb (Genlisea tuberosa) to 149,000 Mb (Paris japonica [Pellicer et al. 2018]). Whole-genome duplications (WGDs) can account for such variation but also accumulation of transposable elements (TEs; reviewed in Bennetzen and Wang [2014]). In addition, genome size dynamics is influenced by differences in rates of DNA removal, epigenetic silencing of TEs, natural selection, and drift (Tenaillon et al. 2010). Small angiosperm genomes appear to be reduced in size due to effective genome reduction, while genomes of species in the lycophyte genus Selaginella appear to be small due to a low rate of genome size increase (Baniaga et al. 2016). Gymnosperm genomes are on average large with low diversity between lineages (16-fold [Pellicer et al. 2018]). These large sizes are likely due to a slow constant accumulation of TEs together with slow removal of DNA (Bennetzen and Wang 2014). Genomes may also evolve through structural changes as a result of translocations, inversions, duplications, and deletions that shuffle the order and orientation of genomic sequences (Bennetzen and Wang 2014). These are often associated with ectopic recombination at repetitive elements and may act as a source of genetic novelty.
Bryophytes occupy a pivotal position in the phylogeny of land plants as the sister group to all other extant land plants. The diversification of bryophytes has been hypothesized to be slow due to an assumed slow rate of morphological change (Crum 1972). It has been speculated that this is accompanied by a slow rate of evolution at the molecular level (Stenøien 2008). Linde et al. (2021) showed that the molecular evolutionary rate, in terms of synonymous substitution rates, was slower compared with angiosperms but not as low as in gymnosperms.
A slow rate of diversification in bryophytes could also be due to a slow rate of structural changes in their genomes. In this study, we focus on liverworts (Marchantiophyta), one of the three major clades of the bryophytes, which are characterized by low variation in genome size and chromosome numbers (Berrie 1960; Bainard et al. 2013), by a lack of known ancient WGD events, and by smaller gene families (Bowman et al. 2017; One Thousand Plant Transcriptomes Initiative 2019). There are currently limited genomic data in bryophytes, with only a few genomes of phylogenetically divergent taxa available; thus, knowledge on the patterns and rates of structural changes is limited (Szövényi et al. 2021).
The aim of this study is to examine and quantify different aspects of structural genome evolution among liverworts. We study taxa representing contrasting divergence times to compare the changes of TE landscape complexity, sequence collinearity, and sex chromosome evolution to identify the processes underlying these structural changes.
Results
Low Gene Duplication Rate in Liverworts
It has been estimated that, on average, 65% of the annotated genes in plant genomes have a duplicate copy (Panchy et al. 2016). WGD and tandem duplication account for the majority of plant duplicates in angiosperms, but TE-based mechanisms and retrotransposition also generate a significant number of duplicates (Panchy et al. 2016). Using the same criteria as Panchy et al. (2016), we found that the estimates of duplicated genes among liverworts are similar and in lower range of those of the 45–84% observed in other land plants (table 1; supplementary fig. S1, Supplementary Material online [Panchy et al. 2016]).
Table 1.
Genomic Features of the Included Species and Subspecies
| Abbreviation | MPR | MPM | MPP | LC | MPA | AAN | AAG | KN | PP | SM | PA | AT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Assembly size (Mb) | 226 | 225 | 223 | 580 | 238 | 119 | 117 | 104 | 472 | 213a | 19,600 | 119 |
| Repeat content (%)b | 25 | 29 | 31 | 63 | 40 | nd | nd | nd | Nd | nd | nd | nd |
| No. of annotated genes | 19,138 | 18,806 | 17,374 | 16,768 | 14,478c | 14,629 | 25,846 | 17,055 | 32,458 | 22,285 | 28,354 | 27,655 |
| No. of duplicated genes | 7,312 | 6,636 | 6,379 | 6,901 | 5,761 | 6,578 | 13,602 | 7,497 | 16,659 | 13,718 | 14,598 | 18,302 |
| Percent duplicated genesd | 50 | 45 | 45 | 50 | 45 | 54 | 68 | 49 | 76 | 73 | 77 | 79 |
| Total gene length (Mb) | 71.9 | 62.3 | 56.1 | 57.1 | 44.4c | 28.8 | 52.6 | 81.8 | 107.7 | 37.9 | 89.3 | 65.6 |
| Mean gene length (bp) | 3,729 | 3,313 | 3227 | 3,404 | 3,070 | 1,972 | 2,034 | 4,768 | 3,271 | 1,701 | 3,148 | 2,375 |
| Mean CDS length (bp) | 1,215 | 1,239 | 1276 | 1,269 | 1,448 | 1,313 | 1,206 | 1,558 | 1,136 | 1,145 | 941 | 1,217 |
| Mean intron length (bp) | 347 | 373 | 301 | 363 | 322 | 172 | 133 | 499 | 264 | 111 | 1017 | 159 |
| Mean exon length (bp) | 256 | 287 | 302 | 349 | 241 | 272 | 339 | 294 | 373 | 213 | 312 | 306 |
| Exons per mRNA ratio | 5.2 | 5.6 | 5.8 | 5.3 | 6.0 | 4.8 | 4.5 | 6.6 | 5.4 | 5.6 | 3 | 5.3 |
Note.—MPP, Marchantia polymorpha ruderalis; MPM, Marchantia polymorpha montivagans; MPP, Marchantia polymorpha polymorpha; LC, Lunularia cruciata; MPA, Marchantia paleacea; AAN, Anthoceros angustus; AAG, Anthorceros agrestis; KN, Klebsormidium nitens; PP, Physcomitrella patens; SM, Selaginella moellendorffii; PA, Picea abies; AT, Arabidopsis thaliana. nd, not determined.
Gene statistics for P. abies were taken from Nystedt et al. (2013).
The assembled genome includes two haplotypes of ∼106 Mb (Banks et al. 2011).
Estimates include microsatellites and low complexity repeats.
Likely reflecting the lower quality of the annotation, not a reduced number of genes.
Number of duplicated genes divided by number of annotated genes longer than 150 aa.
In agreement with the low duplication rate, liverwort genomes contain a lower number of genes compared with most other clades of plants (table 1; supplementary fig. S1, Supplementary Material online). The liverwort estimates are within the range of those of the hornworts Anthoceros angustus and Anthorceros agrestis but lower than that of the lycophyte Selaginella moellendorffii. This is not surprising considering the lack of reported ancient WGDs in liverworts and hornworts (Bowman et al. 2017; Lang et al. 2018; Zhang et al. 2020). The average gene size of liverworts is similar to those of other plant clades (table 1). Despite the similar number of genes in Lunularia cruciata and Marchantia polymorpha (∼17,000 vs. ∼19,000; table 1), the estimated genome size of L. cruciata is more than twice that of M. polymorpha (table 1).
The Larger Size of the L. cruciata Genome Is Largely Explained by TE Content
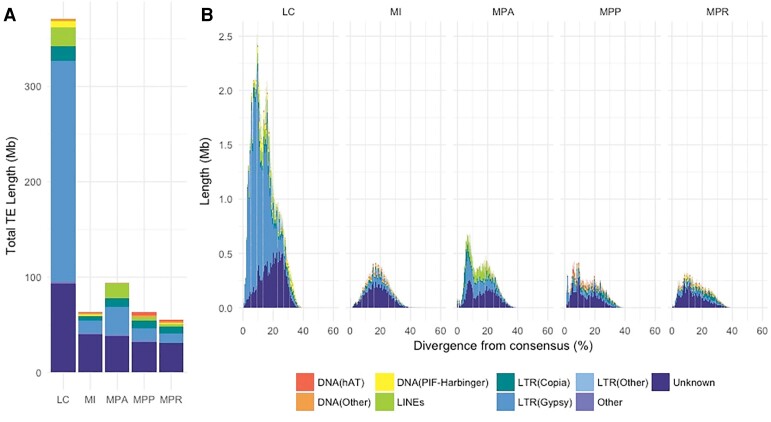
The estimated repeat contents of the liverwort assemblies range from 25% for M. polymorpha ruderalis to 63% for L. cruciata (table 1). The relatively high repeat content of L. cruciata is in agreement with its almost doubled genome size when compared with M. polymorpha. To examine the reasons behind this higher repeat content, the landscape of TEs in the liverwort genomes was characterized. Indeed, most of the genome inflation of L. cruciata is explained by expansion of TEs (fig. 2A, supplementary table S1, Supplementary Material online). The vast majority of this TE expansion emanates from a proliferation of one group of long terminal repeat (LTR) retrotransposons, the Ty3/Gypsy superfamily. The M. polymorpha ruderalis genome contains a similar amount of the two main retrotransposons superfamilies Ty1/Copia and Ty3/Gypsy (fig. 1A [Montgomery et al. 2020]), while L. cruciata contains a large excess of Ty3/Gypsy elements (40% of all repeats). The relatively large proportion of repeats in L. cruciata denoted “unknown” may contain liverwort-specific TEs or reflect problems in the assembly of repetitive sequences using short read data.
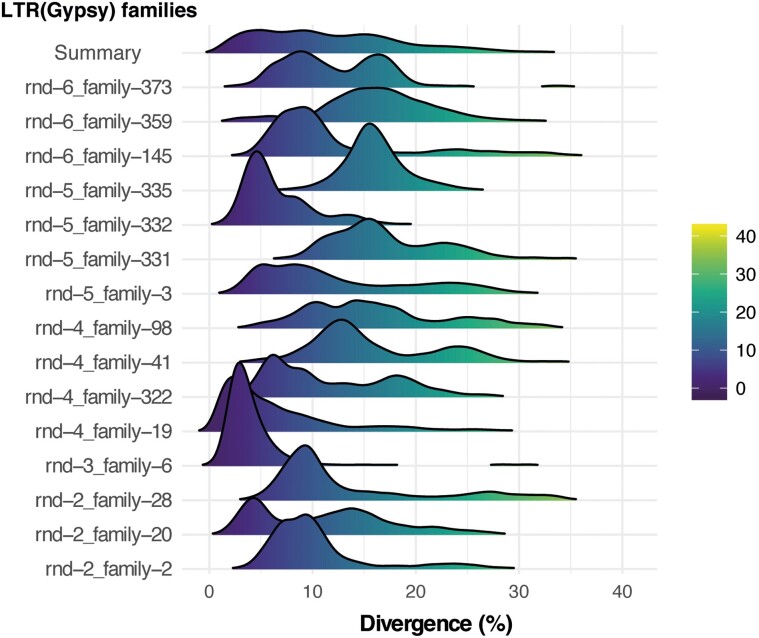
Fig. 2.
Individual Ty3/Gypsy families in L. cruciata were amplified at different time periods. Density plots of divergence from consensus for the 15 largest L. cruciata Ty3/Gypsy.
Fig. 1.
The Lunularia cruciata genome is larger due to a high abundance of Ty3/Gypsy retrotransposons. (A) Repeat content of the genome assemblies included in this study. (B) Abundance (in Mb) of repeats plotted against divergence from consensus (%) in 1% bins. LC, Lunularia cruciata; MI, Marchantia inflexa; MPA, Marchantia paleacea; MPP, Marchantia polymorpha subsp. polymorpha; MPR, Marchantia polymorpha subsp. ruderalis.
Bursts of TE transposition can be identified by examining the divergence of individual TEs from the TE family consensus sequence. In L. cruciata, more than one peak is obvious in the distribution of divergence time (fig. 1B), suggesting more than one burst of proliferation of Ty3/Gypsy elements. Similar but much less pronounced bursts were also evident in the other species that seem to have experienced a slower accumulation of TEs. A more detailed analysis of specific families of Ty3/Gypsy elements in L. cruciata reveals a complex pattern of proliferation of specific elements at several time points (fig. 2). This pattern supports the idea that specific elements may mutate to evade hosts’ silencing and proliferate for some time before host silencing reoccurs (Lisch and Slotkin 2011). A comparative analysis of Ty3/Gypsy and Ty1/Copia elements in L. cruciata and M. polymorpha ruderalis revealed that bursts seem to have occurred at similar time points in the two genomes, but the distributions of divergence are skewed toward lower divergence for L. cruciata, especially for Ty3/Gypsy elements (supplementary fig. S2, Supplementary Material online). This suggests either an increased transposon activity in the L. cruciata lineage or an increased efficiency of excision of transposed elements in the lineage leading to M. polymorpha. More data are needed to determine which of these alternatives are more important, but the latter hypothesis is supported by data on genome size evolution in liverworts (Bainard et al. 2013). The estimated number of full-length LTR retrotransposons, potentially functional, was 2,827 for the male genome of L. cruciata—considerably more than the 222 identified full-length LTRs in M. polymorpha ruderalis chromosome assembly. This difference supports more recent transposon insertions in L. cruciata.
Slow Rate of Chromosomal Rearrangements in Liverworts
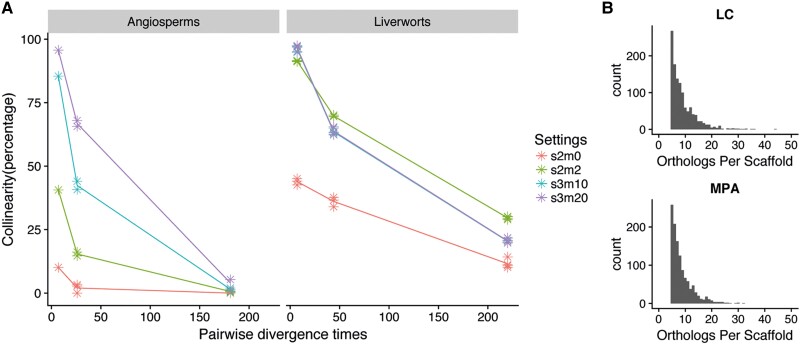
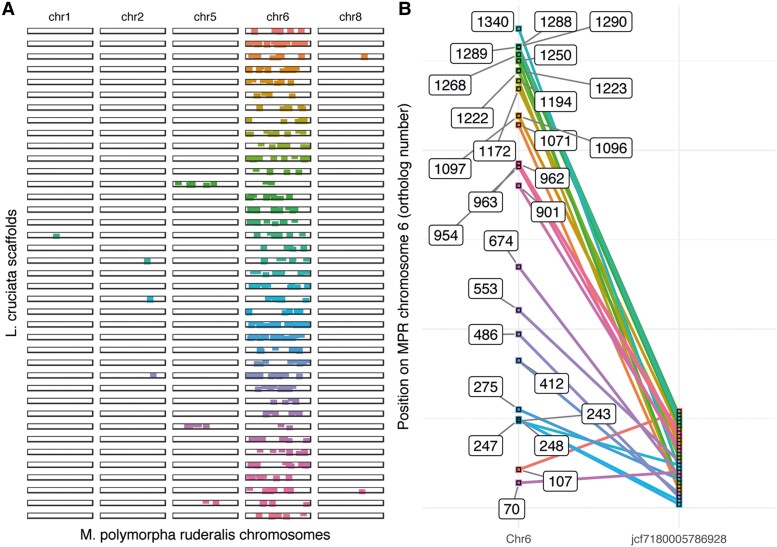
Marchantia polymorpha ruderalis and L. cruciata diverged some 220 Ma before the split of the last common ancestor of Arabidopsis thaliana and the basal angiosperm Amborella trichopoda (181 Ma [Kumar et al. 2017]). A comparison of the decrease of collinearity with divergence times between liverworts and angiosperms indicates a slower rate of chromosomal rearrangements in liverworts than in angiosperms (fig. 3). The assemblies of L. cruciata and Marchantia paleacea were comparatively more fragmented (fig. 3BandC). This could potentially bias the collinearity downward. Still, our data including these species suggest a slower decrease in collinearity for liverworts when compared with angiosperms. Within scaffold size limits, most L. cruciata single-copy genes have their orthologs on homologous M. polymorpha chromosomes, but they are commonly spread over the entire chromosome (fig. 4; supplementary fig. S3, Supplementary Material online).
Fig. 3.
Decrease in collinearity over time is slower in liverworts. (A) Percentage of collinear orthologous genes using MCscanX with different settings for s (number of genes required to call collinear blocks) and m (maximum gaps allowed). One dot represents one pairwise comparison and the lines are drawn through the averaged value for each estimated divergence time. (B) Due to the fragmented nature of the assemblies of L. cruciata (LC) and M. paleacea (MPA), as illustrated here as the distribution of orthologous single-copy genes per scaffold, the most commonly used default values of s = 5 and m = 25 are not useful for a valid comparison.
Fig. 4.
Chromosomal inversions can explain much of the decrease in collinearity between M. polymorpha and L. cruciata. (A) Illustration of rearrangements within and among chromosomes, exemplified by the 36 largest L. cruciata scaffolds with at least 10 orthologous hits on M. polymorpha subsp. ruderalis chromosome 6. Each squared dot represents one gene and all single-copy orthologous genes on the L. cruciata scaffolds are printed including the few genes having their orthologous hit on another chromosome than chromosome 6. The scaffolds are color-coded. (B) A close-up of the largest L. cruciata scaffold (in the number of orthologous single-copy genes) and its collinear relationship with M. polymorpha subsp. ruderalis chromosome 6. The genes are numbered after their order of appearance on chromosome 6—gaps in the numbering result from M. polymorpha genes that do not have an identified ortholog on this L. cruciata scaffold. The lines connecting orthologs have different colors for the sake of clarity.
Thus, our data suggest a multitude of chromosomal rearrangements, including inversions, since the separation of the lineages leading to L. cruciata and M. polymorpha. It should be noted that the frequency of translocations and other larger-scale-rearrangements (beyond the limited size of the L. cruciata scaffolds) is underestimated. Based on available data, it is not possible to estimate and compare rates within the two liverwort lineages. Thus, we cannot conclude if the rate of chromosomal rearrangements is associated with the higher TE activity in L. cruciata (Bennetzen and Wang 2014).
Sex Chromosome Evolution
Lunularia cruciata is dioicous, that is having separate male and female haploid individuals, with the presence of U or V sex chromosomes, respectively, and this is the presumed ancestral condition in liverworts (Berrie 1960; Bowman 2016; Iwasaki et al. 2021). In nonrecombining regions of the sex chromosomes, U and V chromosomes possess divergent gametolog pairs descended from a common ancestral autosomal gene. Previously, L. cruciata orthologs of M. polymorpha gametologs were identified from two transcriptomes representing female and male individuals (Iwasaki et al. 2021; supplementary table S3, Supplementary Material online). Many orthologs from the female transcriptome are descended from a presumed U-linked gene in the common ancestor of L. cruciata and M. polymorpha, with some others having an outgroup phylogenetic relationship with the two M. polymorpha gametologs (fig. 5; supplementary fig. S4, Supplementary Material online; supplementary table S3, Supplementary Material online). In contrast, whereas some male transcriptome orthologs descended from a presumed V-linked gene in the common ancestor or have an outgroup relationship, others are identical to the female transcriptome genes presumed descended from ancestral U-linked genes (fig. 5; supplementary fig. S4, Supplementary Material online; supplementary table S3, Supplementary Material online).
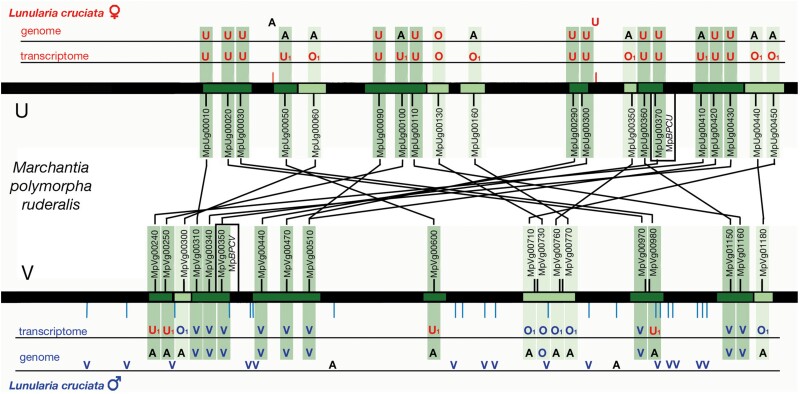
Fig. 5.
Many L. cruciata orthologs of M. polymorpha sex chromosome genes are also sex-specific. The M. polymorpha U and V chromosome schematics are central (Bowman et al. 2017; Montgomery et al. 2020; Iwasaki et al. 2021). Gametologs are listed between the U and V chromosomes, with phylogenetically conserved U- and V-specific genes represented by short outward pointing lines (supplementary table S4, Supplementary Material online). Gametolog pairs predicted to have been in nonrecombining regions, presumably on sex chromosomes of the common ancestors of Lunularia and Marchantia, are in dark green, whereas those predicted to have been in recombining regions (either pseudoautosomal or autosomal) in the common ancestor are in light green (Iwasaki et al. 2021). The results of phylogenetic analyses of L. cruciata orthologs of M. polymorpha gametologs (supplementary fig. S4 and table S4, Supplementary Material online) are presented for the male and female transcriptomes and genomes. Orthologs are classified as U or V if the detected ortholog is more closely related to the U or V M. polymorpha gametolog, respectively, than the L. cruciata sequences to each other. Orthologs are defined as O (outgroup) if the two M. polymorpha gametologs are more closely related to each other than the L. cruciata ortholog. If the male and female transcriptomes possess the same gametolog, it is demarcated with a “1.” For the genome, if the gene sequence is specific to the female or male genome, it is demarcated in red (U or O) or blue (V or O), respectively. If the sequence is found in both female and male genomes, it is demarcated in black (A, autosomal or pseudoautosomal). In each case where a single sequence is found in the transcriptomes, the gene is predicted to be (pseudo)autosomal. Note that although many of the L. cruciata genes represented are predicted to reside on the sex chromosomes, the chromosomal order and orientation of the genes in L. cruciata is unknown.
Although the structure of L. cruciata sex chromosomes is presently unresolved, genes residing on the U or V chromosomes should be present only in the female or male genomes, respectively. We first confirmed that in each case the gametolog orthologs of the female and male genomes matched those of the female and male transcriptomes, respectively (fig. 5; supplementary fig. S4, Supplementary Material online; supplementary table S3, Supplementary Material online). We next examined whether the gametolog orthologs were unique to the female or male genomes, suggesting that the sequences reside on the U or V chromosome, or whether they were shared, which would suggest that sequences are autosomal (or pseudoautosomal). In each case where gametolog ortholog pairs were identified with distinct sex-specific sequences, they are located on genomic scaffolds that are unique to either the female or male. In contrast, in every case where the male and female gametolog orthologs have similar or identical sequences, the genomic scaffolds on which these genes reside are found in both the female and male genomes. Lastly, at least some orthologs of phylogenetically conserved M. polymorpha U- and V-specific genes may also be U- or V-linked in L. cruciata as they reside on scaffolds containing sequences found in genomic assemblies derived from only one sex (fig. 5; supplementary table S4, Supplementary Material online). This is in contrast to scaffolds presumed to be autosomal where corresponding scaffolds in the opposite sex are typically >99% identical.
Discussion
It has been hypothesized that bryophytes, being structurally simple organisms with a predominantly haploid life cycle, have experienced slow molecular evolution, similar to their seemingly slow morphological diversification. Linde et al. (2021) showed that the silent site substitution rates of bryophytes are not lower when compared with vascular plants as a group. Bryophyte rates were lower than for angiosperms but higher than gymnosperms; thus, low substitution rates cannot alone explain a difference in morphologic evolution.
Bryophytes are characterized by small genomes. The mean (median in parentheses) of bryophyte genome sizes has been estimated to be 244 Mb (205 Mb), 1,844 Mb (751 Mb), and 504 Mb (433 Mb) for hornworts, liverworts, and mosses, respectively (Pellicer et al. 2018). Liverworts are subdivided into three classes: the species-rich classes Marchantiopsida, Jungermanniopsida, and the smaller class Haplomitriopsida. Species belonging to Marchantiopsida show limited genome sizes (∼5-fold variation [Pellicer and Leitch 2020]) compared with Jungermanniopsida (∼100-fold variation [Pellicer and Leitch 2020]). This variation is very limited compared with the angiosperm lineage, where genome sizes vary 2,440-fold with a mean (median in parentheses) of 5,020 Mb (1,663 Mb), and although some of the largest genomes are observed within the angiosperms, their genome sizes are skewed toward smaller genomes (Pellicer et al. 2018).
Low Levels of Gene Duplication in Marchantiophyta
A low degree of gene duplication has been reported for M. polymorpha ruderalis (Bowman et al. 2017). Our data now suggest that the percentage of duplicated genes and tandem duplicated is similar between all the studied Marchantiopsida liverworts despite a large phylogenetic distance in the case of L. cruciata and M. polymorpha ruderalis. The low number of duplicated genes in Marchantiophyta liverworts can to a large extent be explained by a lack of ancient WGD events (Bowman et al. 2017). A considerable number of paralogs were all the same detected in liverworts, probably to a large extent resulting from tandem or segmental duplications (fig. 1). Still, tandem duplications in Marchantiophyta are at the lower range compared with angiosperms (Bowman et al. 2017). Gene number estimates within Marchantiophyta are between those of the hornwort A. angustus and A. agrestis and similar to those of the charophyte alga Klebsormidium nitens. The lower gene number estimate in A. angustus is consistent with a reported high rate of gene family loss in this species (Zhang et al. 2020). As gene duplication may increase evolutionary potential (Ohno 2013), the low gene numbers of Marchantiophyta species are consistent with a reduced evolvability of the group.
Bursts of Transposable Element Accumulation in L. cruciata
Despite a limited variation in genome size among representatives of Marchantiopsida, the genome of L. cruciata is approximately twice the size when compared with that of M. polymorpha. This difference is mainly due to a higher copy number of a few families of TEs in L. cruciata that are the result of several bursts of amplification. These results show that such bursts, that are common in angiosperms, also occur in liverworts. However, extrapolating from our results this kind of burst seems rare, at least among Marchantiopsida. Even though the TE content is generally lower in the studied liverworts when compared with other most plant species, a similar collection of elements was observed. The most common TE elements were Ty3/Gypsy and Ty1/Copia, as has been reported in other plants (Voytas et al. 1992; Suoniemi et al. 1998). The ratio of Ty3/Gypsy to Ty1/Copia elements varies in the studied liverwort species, with Ty3/Gypsy being the most abundant in L. cruciata, whereas the two are present in similar proportions in M. paleacea. Most of this variation can be explained by multiple bursts of Gypsy families in L. cruciata.
The larger transposon accumulation over time in Lunularia when compared with the Marchantia species is not associated with any known radiation, and the order Lunulariales is considered monotypic with a single species, L. cruciata, in contrast to the much more speciose Marchantiales (Söderström et al. 2016).
Low Rate of Chromosomal Rearrangements in Marchantiopsida
Overall, there is a much higher average microsynteny across placental mammals compared with angiosperms (Zhao and Schranz 2019). Wang et al. (2012) reported 0.6–6.7% collinear ortholog pairs in pairwise comparisons between monocot and eudicot species that diverged approximately 160 Ma (Kumar et al. 2017). Because different methods and thresholds have been used, those values are not directly comparable with the values obtained in the present study. Still, regardless of the settings used in our analysis, the percentage collinear gene pairs between Marchantiopsida liverwort species pairs is always higher than those obtained from angiosperm pairs. Thus, a relatively low rate of chromosomal rearrangements is suggested for species within Marchantiopsida. Although the fragmented assembly of L. cruciata precludes a direct comparison of rates of inversions versus translocations, our results are consistent with the conclusion that structural changes within plant chromosomes are far more abundant than those between chromosomes (Dvorak et al. 2018). Linde et al. (2020) found that chromosome 2 of M. polymorpha subsp. montivagans was distinctly more divergent than any other parts of the genome in pairwise comparisons with the other subspecies of M. polymorpha. A higher degree of structural rearrangements on chromosome 2 of subsp. montivagans suggested that it might have been more strongly protected against introgression than other parts of the genome. This chromosome also had a higher proportion of transposons.
Sex Chromosome Evolution in L. cruciata
Among liverworts, the karyotype of L. cruciata with the V chromosome larger than the U is unusual and in contrast to the vast majority of liverworts with dimorphic sex chromosomes in which the U is larger than the V (Lorbeer 1934; Sergio and Viana 1973). A translocation of a fragment of the U chromosome to an autosomal location could account for this unique karyotype. The pattern of L. cruciata gametolog evolution is consistent with the hypothesis that a segment composed of both previously nonrecombining regions (including three gametologs and one U-specific gene) and potentially ancestrally pseudoautosomal regions was translocated from the U to an autosome. Consistent with this hypothesis and as predicted by Muller's ratchet (Muller 1932), the corresponding V-gametologs appear to have been lost.
Conclusions
Our analyses reveal clear differences in the patterns of genome evolution between liverworts and angiosperms resulting in smaller and less variable liverwort genomes. First, although angiosperms have gone through several rounds of ancient WGDs, no ancient WGD has been identified for liverworts. Even though tandem duplications are not uncommon in liverworts, their genomes generally contain fewer genes. This difference could restrict the evolutionary potential for liverworts, as less new genetic material is available for mutation, drift, and selection to act upon. Second, the rate of structural changes in terms of both genome size and rearrangements seems low when compared with angiosperms. These differences might be the result of lower transposon activity. We did observe bursts of primarily LTR TEs in liverworts, similar to what is prevalent in angiosperms. However, such events seem to be much more rare in liverworts. As transposon activity is thought to provide additional mutations that might speed up evolution in terms of both rates of adaptation and speciation (Chénais et al. 2012; Rebollo et al. 2012), the association between a low TE activity and a low rate of evolutionary change in liverworts is not unexpected.
Materials and Methods
Genome Assembly and Annotation
Genomes of five Marchantiopsida liverwort species/subspecies were included: M. polymorpha subsp. ruderalis, M. polymorpha subsp. polymorpha, M. polymorpha subsp. montivagans, Marchantia inflexa, and L. cruciata. Assembly and annotation of M. polymorpha subsp. ruderalis are publicly available (Bowman et al. 2017; Montgomery et al. 2020; Iwasaki et al. 2021) and so is the assembly of M. inflexa (Marks et al. 2019), which was used for only repeat characterization. Sequencing, assembly, and annotation of M. polymorpha subsp. polymorpha and subsp. montivagans are described in Linde et al. (2020). The genome of M. paleacea subsp. diptera was sequenced and assembled as described in Radhakrishnan (2017) and summarized in Linde et al. (2020).
The female genome of L. cruciata is described in Linde et al. (2021). BUSCO (Simão et al. 2015), the eukaryote_odb9 lineage data set, and CEGMA were used to assess the completeness of the assembly, which was 87.4% (89.1%) according to BUSCO and 90.32% (95.56%) according to CEGMA, with the rates in parenthesis also including fragmented/partial hits. GenomeScope (http://qb.cshl.edu/genomescope/) was used to verify the estimates for genome size and repeat content using a kmer-based statistical approach on the raw reads giving an estimate of the genome size (581 Mb) similar to the assembly size (580 Mb) but a lower estimate of the repeat content (35%) compared with the value estimated as described below in the repeat annotation section, likely due to the failure to detect older and more divergent repeats. Annotation of L. cruciata was performed as described in Linde et al. (2020). BUSCO was used to assess the completeness of the annotations. The M. polymorpha ruderalis annotation version 3.1, which is considered to be more or less complete, had a completeness of 97.4 (97.7%). The completeness of the annotations of M. polymorpha subsp. montivagans, subsp. polymorpha, L. cruciata, and M. paleacea was 92.8% (97.8%), 86.8% (96.7%), 85.8 (95.4%), and 90.8% (96.7%), respectively (fragmented hits included in numbers within parenthesis).
The genome of a male L. cruciata was assembled using the data set SRR8246012 (Dong et al. 2019). The assembly was performed using MaSuRCA version 3.4.2 (Zimin et al. 2013), with pair end, using default settings. The genome size of male L. cruciata was estimated on the basis of size of the female genome (581 Mb) and a 519 Mb genome was assembled. BUSCO (Simão et al. 2015), together with the eukaryota data set, was used to assess the completeness of the assembly, which was 93.4% (97.7%), with the rate in parenthesis including not only the complete but also the fragmented/partial hits. Assembly statistics are given in supplementary table S2, Supplementary Material online. Both L. cruciata assemblies with annotations are available at genomevolution.org (IDs 64629, 64630, and 64631).
Sex Chromosome Phylogenetics
Predicted L. cruciata gametolog orthologs were identified from available genome and transcriptome sequence data (Dong et al. 2019; One Thousand Plant Transcriptomes Initiative 2019; Iwasaki et al. 2021). Nucleotide sequences were manually aligned as amino acid translations using Se-Al v2.0a11 (http://tree.bio.ed.ac.uk/software/seal/). Ambiguously aligned sequences were removed and alignments of nucleotides were employed in subsequent Bayesian phylogenetic analysis using MrBayes 3.2.1 (Huelsenbeck and Ronquist 2001; Huelsenbeck et al. 2001). The Bayesian analysis for the nucleotide data set was run for 500,000 generations, which was sufficient for convergence of the two simultaneous runs (split frequencies < 0.05). To allow for the burn-in phase, the initial 50% of the total number of saved trees was discarded. The graphic representation of the trees was generated using the FigTree (version 1.4.0) software (http://tree.bio.ed.ac.uk/software/figtree/). Nucleotide alignments are provided in supplementary file S1, Supplementary Material online.
Supplementary Material
Acknowledgments
We thank Jacques Dainat and the Swedish Bioinformatics Advisory Program (SciLifeLab, Uppsala) for bioinformatics support and mentorship. We thank the Swedish Research Council VR (2011-5609 and 2014-522 to U.L.) and the Australian Research Council (DP200100225 to J.L.B.) for economic support. We also thank the National Genomics Infrastructure (NGI)/Uppsala Genome Center, SciLifeLab, and Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) for their assistance in massive parallel sequencing and computational infrastructure.
Contributor Information
Anna-Malin Linde, Department of Plant Ecology and Evolution, Evolutionary Biology Centre, Uppsala University, Sweden.
Shilpi Singh, School of Biological Sciences, Monash University, Melbourne, Victoria, Australia.
John L Bowman, School of Biological Sciences, Monash University, Melbourne, Victoria, Australia.
Magnus Eklund, Department of Plant Ecology and Evolution, Evolutionary Biology Centre, Uppsala University, Sweden.
Nils Cronberg, Biodiversity, Department of Biology, Lund University, Sweden.
Ulf Lagercrantz, Department of Plant Ecology and Evolution, Evolutionary Biology Centre, Uppsala University, Sweden.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
All sequence data analyzed in this research were previously published as described above. Genome assemblies and annotations for L. cruciata were produced based on published data and are available at genomevolution.org (IDs 64629, 64630, and 64631).
Literature Cited
- Bainard JD, Forrest LL, Goffinet B, Newmaster SG. 2013. Nuclear DNA content variation and evolution in liverworts. Mol Phylogenet Evol. 68:619–627. [DOI] [PubMed] [Google Scholar]
- Baniaga AE, Arrigo N, Barker MS. 2016. The small nuclear genomes of Selaginella are associated with a low rate of genome size evolution. Genome Biol Evol. 8:1516–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332:960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Wang H. 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol. 65:505–530. [DOI] [PubMed] [Google Scholar]
- Berrie GK. 1960. The chromosome numbers of liverworts (Hepadcae and Anthocerotae). Trans Br Bryol Soc. 3:688–705. [Google Scholar]
- Bowman JL. 2016. A brief history of Marchantia from Greece to genomics. Plant Cell Physiol. 57:210–229. [DOI] [PubMed] [Google Scholar]
- Bowman JL, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171:287–304.e15. [DOI] [PubMed] [Google Scholar]
- Chénais B, Caruso A, Hiard S, Casse N. 2012. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509:7–15. [DOI] [PubMed] [Google Scholar]
- Crum H. 1972. Geographic origins of the mosses of North America's Eastern deciduous forest. Hattori Bot Lab J. 35:269–298. [Google Scholar]
- Dong S, et al. 2019. The amount of RNA editing sites in liverwort organellar genes is correlated with GC content and nuclear PPR protein diversity. Genome Biol Evol. 11:3233–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, et al. 2018. Structural variation and rates of genome evolution in the grass family seen through comparison of sequences of genomes greatly differing in size. Plant J. 95:487–503. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314. [DOI] [PubMed] [Google Scholar]
- Iwasaki M, et al. 2021. Identification of the sex-determining factor in the liverwort Marchantia polymorpha reveals unique evolution of sex chromosomes in a haploid system. Curr Biol. 31:5522–5532.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- Lang D, et al. 2018. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93:515–533. [DOI] [PubMed] [Google Scholar]
- Linde A-M, Eklund DM, Cronberg N, Bowman JL, Lagercrantz U. 2021. Rates and patterns of molecular evolution in bryophyte genomes, with focus on complex thalloid liverworts, Marchantiopsida. Mol Phylogenet Evol. 165:107295. [DOI] [PubMed] [Google Scholar]
- Linde A-M, Sawangproh W, Cronberg N, Szövényi P, Lagercrantz U. 2020. Evolutionary history of the Marchantia polymorpha complex. Front Plant Sci. 11:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D, Slotkin RK. 2011. Strategies for silencing and escape: the ancient struggle between transposable elements and their hosts. Int Rev Cell Mol Biol. 292:119–152. [DOI] [PubMed] [Google Scholar]
- Lorbeer G. 1934. Die Zytologie der Lebermoose mit besonderer Berücksichtigung allgemeiner Chromosomenfragen Tl 1 [thesis]. Leipzig: Borntraeger. [Google Scholar]
- Marks RA, Smith JJ, Cronk Q, Grassa CJ, McLetchie DN. 2019. Genome of the tropical plant Marchantia inflexa: implications for sex chromosome evolution and dehydration tolerance. Sci Rep. 9:8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SA, et al. 2020. Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Curr Biol. 30:573–588.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. 1932. Some genetic aspects of sex. Am Nat. 66:118–138. [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- Ohno S. 2013. Evolution by gene duplication. New York: Springer Science & Business Media. [Google Scholar]
- One Thousand Plant Transcriptomes Initiative . 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H. 2016. Evolution of gene duplication in plants. Plant Physiol. 171:2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Hidalgo O, Dodsworth S, Leitch IJ. 2018. Genome size diversity and its impact on the evolution of land plants. Genes (Basel) 9:E88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Leitch IJ. 2020. The plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytol. 226:301–305. [DOI] [PubMed] [Google Scholar]
- Puttick MN, Clark J, Donoghue PCJ. 2015. Size is not everything: rates of genome size evolution, not C-value, correlate with speciation in angiosperms. Proc R Soc B: Biol Sci. 282:20152289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan G. 2017. Tracing the evolution of the arbuscular mycorrhizal symbiosis in the plant lineage [doctoral thesis]University of East Anglia. [cited 2022 Jan 3]. Available from: https://ueaeprints.uea.ac.uk/id/eprint/66949/. [Google Scholar]
- Ramírez-Sánchez O, Pérez-Rodríguez P, Delaye L, Tiessen A. 2016. Plant proteins are smaller because they are encoded by fewer exons than animal proteins. Genom, Proteom Bioinform. 14:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 46:21–42. [DOI] [PubMed] [Google Scholar]
- Sergio C, Viana MJ. 1973. Sur la fertilité de Lunularia cruciata (L.) Dum. au Portugal. Rev Fac Cienc Ser C Cienc Nat. 17:703–719. [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Söderström L, et al. 2016. World checklist of hornworts and liverworts. PhytoKeys 59:1–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenøien HK. 2008. Slow molecular evolution in 18S rDNA, rbcL and nad5 genes of mosses compared with higher plants. J Evol Biol. 21:566–571. [DOI] [PubMed] [Google Scholar]
- Suoniemi Annu, Tanskanen Jaakko, Schulman Alan H. 1998. Gypsy‐like retrotransposons are widespread in the plant kingdom. The Plant Journal 13(5):699–705. 10.1046/j.1365-313X.1998.00071.x [DOI] [PubMed] [Google Scholar]
- Szövényi P, Gunadi A, Li F-W. 2021. Charting the genomic landscape of seed-free plants. Nat Plants. 7:554–565. [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS. 2010. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15:471–478. [DOI] [PubMed] [Google Scholar]
- Voytas D F, Cummings M P, Koniczny A, Ausubel F M, Rodermel S R. 1992. copia-like retrotransposons are ubiquitous among plants. Proceedings of the National Academy of Sciences 89(15):7124–7128. 10.1073/pnas.89.15.7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. 2012. MCScanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. 2020. The hornwort genome and early land plant evolution. Nat Plants. 6:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Schranz ME. 2019. Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc Natl Acad Sci U S A. 116:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, et al. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data analyzed in this research were previously published as described above. Genome assemblies and annotations for L. cruciata were produced based on published data and are available at genomevolution.org (IDs 64629, 64630, and 64631).