Abstract
GCN5 is a histone acetyltransferase (HAT) originally identified in Saccharomyces cerevisiae and required for transcription of specific genes within chromatin as part of the SAGA (SPT-ADA-GCN5 acetylase) coactivator complex. Mammalian cells have two distinct GCN5 homologs (PCAF and GCN5L) that have been found in three different SAGA-like complexes (PCAF complex, TFTC [TATA-binding-protein-free TAFII-containing complex], and STAGA [SPT3-TAFII31-GCN5L acetylase]). The composition and roles of these mammalian HAT complexes are still poorly characterized. Here, we present the purification and characterization of the human STAGA complex. We show that STAGA contains homologs of most yeast SAGA components, including two novel human proteins with histone-like folds and sequence relationships to yeast SPT7 and ADA1. Furthermore, we demonstrate that STAGA has acetyl coenzyme A-dependent transcriptional coactivator functions from a chromatin-assembled template in vitro and associates in HeLa cells with spliceosome-associated protein 130 (SAP130) and DDB1, two structurally related proteins. SAP130 is a component of the splicing factor SF3b that associates with U2 snRNP and is recruited to prespliceosomal complexes. DDB1 (p127) is a UV-damaged-DNA-binding protein that is involved, as part of a complex with DDB2 (p48), in nucleotide excision repair and the hereditary disease xeroderma pigmentosum. Our results thus suggest cellular roles of STAGA in chromatin modification, transcription, and transcription-coupled processes through direct physical interactions with sequence-specific transcription activators and with components of the splicing and DNA repair machineries.
In eukaryotes, genomic DNA is packaged by histones into nucleosomes that further fold to form higher-order chromatin structures. Eukaryotic cells have evolved two major enzymatic mechanisms to modify chromatin structure: (i) ATP-dependent nucleosome remodeling by multiprotein complexes that use the energy of ATP hydrolysis to alter the association of core histones with DNA and (ii) covalent modifications of core histones, including acetylation, that regulate core histone interactions with either DNA, adjacent nucleosomes, or other regulatory proteins (reviewed in references 7, 39, 64, and 74).
Reversible acetylation of specific lysine residues within the N-terminal tails of nucleosomal core histones has long been correlated with changes in chromatin that occur during transcription, replication, and DNA repair in vivo (reviewed in references 7, 61, and 65). Significant progress in understanding the role of nuclear histone acetylation came from the findings that the Saccharomyces cerevisiae transcription coactivator GCN5, and more recently other yeast and metazoan transcription cofactors, are histone acetyltransferases (HATs) and that several transcription corepressor complexes have histone deacetylases as integral subunits (reviewed in references 4 and 10). HATs differ in substrate specificity and may also modify nonhistone regulatory proteins, as originally demonstrated for p53 acetylation by p300 (27). Many nuclear HATs are also part of large multiprotein assemblies. These include yeast SAGA (SPT-ADA-GCN5 acetylase), ADA, NuA3, NuA4, and Elongator complexes, yeast and metazoan TFIID complexes, and human TFTC (TATA-binding protein [TBP]-free TBP-associated factor II [TAFII]-containing complex), PCAF, STAGA (SPT3-TAFII31-GCN5L acetylase), TIP60, and TFIIIC complexes (reviewed in references 10 and 24).
In yeast, GCN5 is an integral subunit of at least two distinct multiprotein HAT complexes, the ADA and SAGA complexes, that acetylate histones H3 and H2B within nucleosomes (25). The yeast SAGA complex is composed of (i) ADA adapter (coactivator) proteins (ADA1, ADA2, ADA3, ADA4 [GCN5], and ADA5 [SPT20]), (ii) SPT proteins (SPT3, SPT7, SPT8, and SPT20 [ADA5]), (iii) a subset of the yeast TAFIIs (yTAFIIs) (yTAFII17/20, yTAFII25, yTAFII60, yTAFII61/68, and yTAFII90), and (iv) a protein, Tra1, that is structurally related to members of the ATM/DNA-PK/phosphatidylinositol 3-kinase family (reviewed in references 10, 24, and 78). The ADA complex shares GCN5, ADA2, and ADA3 with SAGA but lacks all other SAGA subunits and has ADA-specific components (20). The SAGA complex, but not the ADA complex, interacts directly with various activators and potentiates activation domain-specific transcription in an acetyl coenzyme A (acetyl-CoA)-dependent manner on nucleosomal arrays in vitro (34, 72, 76).
Mammalian homologs of yeast GCN5 include PCAF and GCN5L (12, 62, 79, 82). PCAF and GCN5L proteins are encoded by distinct genes, and their expression is differential and complementary in various tissues (79, 82). However, GCN5L is essential for mouse development, whereas PCAF is dispensable (80, 81). Human GCN5L (hGCN5L) and PCAF form part of three distinct multiprotein HAT complexes: PCAF complex (54), TFTC (8), and STAGA (46). While still incompletely characterized, these human HAT complexes preferentially acetylate histone H3 and have related but not identical subunit compositions. All contain homologs of yeast SAGA subunits and a subset of TAFIIs that were originally found in TFIID but clearly lack TBP (reviewed in reference 24). Apart from a TFIID-like function for TFTC in transcription from “naked” DNA templates in vitro (77), the functions of these human TAFII-HAT complexes remain still largely unknown. More generally, the recent observations that yeast and metazoan transcriptional adapters and HATs are within large multiprotein complexes raises important questions as to the role(s) of the remaining protein subunits and whether HAT complexes have additional functions.
In the present study we report the identification of most of the protein subunits of the human STAGA complex. These include novel human proteins similar to yeast SAGA components. In addition, we show that human STAGA preferentially acetylates histone H3 within nucleosomes and mediates in vitro transcriptional activation by the chimeric Gal4-VP16 activator on a chromatin template through direct physical interactions with the VP16 activation domain. Furthermore, we demonstrate an association of STAGA in HeLa cells with spliceosome-associated protein 130 (SAP130) and with UV-damaged-DNA-binding factors, suggesting the possibility of additional functions for STAGA in transcription-coupled pre-mRNA splicing and DNA damage repair in vivo.
MATERIALS AND METHODS
Plasmids.
pFH-IRESneo was obtained by insertion of a Kozak consensus ATG and in-frame FLAG- and hemagglutinin (HA) epitope-coding sequences between the EcoRV and NotI sites of pIRES1neo (Clontech). hSPT3 and TAFII31 cDNAs were cloned into pFH-IRESneo to obtain, respectively, pFH:SPT3-IRESneo and pFH:TAF31-IRESneo. pGEX-5X-3-DDB1 (44) and pBJ5-FLAG-p125/DDB1 (33) have been described previously.
Stable cell lines, extract preparation, and protein complex purification.
HeLa S cells were transfected with pFH:SPT3-IRESneo or pFH:TAF31-IRESneo and selected with 500 μg of G418 (GIBCO) per ml. Single G418-resistent colonies expressing FLAG-HA-tagged proteins were expanded for nuclear extract preparation. Nuclear extracts were adjusted to 300 mM KCl (BC300) and 0.05% NP-40 and rotated with M2-agarose (Sigma) at 4°C for 3 to 6 h. After extensive washes with BC300–0.05% NP-40, proteins were eluted with 0.3 mg of FLAG peptide per ml in BC100–0.05% NP-40. M2 eluates were incubated with anti-HA antibody (12CA5; BAbCO) beads for 4 h at 4°C, the beads were washed extensively with BC400–0.1% NP-40, and proteins were eluted at 30°C with 2 mg of HA peptide per ml in BC100–0.1% NP-40. Alternatively, M2 eluates, adjusted to BC60–0.05% NP-40, were fractionated on S-Sepharose (Pharmacia). For purification of a STAGA complex lacking SAP130 (STAGA-s), nuclear extracts were loaded on a S-Sepharose column, the column was washed with BC100–0.05% NP-40, and bound proteins were step eluted with BC200–0.05% NP-40 and BC400–0.05% NP-40. The BC400 protein eluate was incubated with M2-agarose, the resin was washed extensively with BC320–0.05% NP-40, and bound proteins were eluted as described above.
Protein identification by MS.
In-gel tryptic digests of proteins from M2 affinity-purified STAGA resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels were analyzed by matrix-assisted laser desorption ionization–time-of-flight (mass spectrometry) (MALDI-TOF [MS]), liquid chromatography-ion trap-tandem MS, and MALDI- and nanospray-quadrupole-quadrupole TOF-MS. The mass spectral data were used to search the National Center for Biotechnology Information (NCBI) nonredundant and expressed sequence tag databases as previously described (reference 41 and references therein). The peptides identified are as follows: for STAF400 (TRRAP), GLSVDSAQEVK, NPADSISHVAYR, TATGAISAVFGR, LVEDNPSSLSLVEIYK, LAVDLSEVVIK, and YLQFVAALTDVNTPDETK; for STAF130 (SAP130), LPPNTNDEVDEDPTGNK, NFGDQPDIR, DYIVVGSDSGR, NVSEELDRTPPEVSK, MQGQEAVLAMSSR, AGNGQWASWIR, LTISSPLEAHK, SVAGGFVYTYK, SWLSYSYQSR, IVILEYQPSK, ILELLRPDPNTGK, TVLDPVTGDLSDTR, IVPGQFLAVDPK, TPVEEVPAAIAPFQGR, FLAVGLVDNTVR, AEVIMNYHVGETVLSLQK, NENQLIIFADDTYPR, WVTTASLLDYDTVAGADK, HGLEVSEMAVSELPGNPNAVWTVR, FSNTGEDWYVLVGVAK, and LGAVFNQVAFPLQYTPR; for STAF97 (hGCN5L), QIPVESVPGIR, TLILTHFPK, and TLPENLTLEDAK; for STAF65α (PAF65α), VHVSYLDGK, VALQDLQTNSK, KLTVEDFNR, AVLGDDPQLMK, KMPQLTASAIVSPHGDESPR, LFQTAFPAPYGPSPASR, ELYAFFGDSLATR, and GNLAPQGSVPSAVSSLTDDLLK; for STAF65β (PAF65β), GPVLSLAFSPNGK and LWDLASGTLYK; for STAF65γ (KIAA0764), YWGEIPISSSQTNR; for STAF54 (hADA3), VLEAETQILTDWQDK; for STAF42, KNLSEALGDNVK, YAFGSNVTPQPYLK, ISKEEFDLEAHR, EVIPTHTVYALNIER, and DILTSVVSR; for STAF31/32 (TAFII31), DMGITEYEPR, PSTPTLGTPTPQTMSVSTK, FTVQMPTSQSPAVK, DAQMMAQILK, DFLLDIAR, VINQMLEFAFR, and ASIPATSAVQNVLINPSLIGSK; for STAF28 (TAFII30), ASPAGTAGGPGAGAAAGGTGPLAAR and YTLTMEDLTPALSEYGINVK; and for STAF20/15 (TAFII20/15), LSPENNQVLTK and DVQLHLER.
Immunoprecipitations and GST pull-down assays.
Nuclear extracts from either FLAG-HA double-epitope-tagged hSPT3 (fh:SPT3) or control HeLa cells were mixed with M2-agarose in BC100, the suspension was rotated for 3 h at 4°C, and the beads were washed extensively with BC150–0.05% NP-40. Proteins were eluted with 0.3 mg of FLAG peptide per ml. Whole-cell extracts of HeLa cells transiently transfected with pBJ5-FLAG/DDB1 were adjusted to BC200 and rotated with M2-agarose. After extensive washes with BC200–0.05% NP-40 and BC100–0.05% NP-40, proteins were eluted with 0.5 mg of FLAG peptide per ml. Bacterially expressed glutathione S-transferase (GST), GST-DDB1, and GST-VP16 proteins immobilized on glutathione-agarose were incubated for 1 to 3 h at 4°C in BC100–0.05% NP-40 with nuclear extracts or affinity-purified STAGA; the resins were washed extensively with BC150–0.1% NP-40 (unless otherwise indicated), and bound proteins were eluted with Sarkosyl (0.2 to 0.5%).
HAT assay, chromatin assembly, and transcription.
HeLa core histones and native nucleosomes were purified as described previously (14, 46). HAT assays with 1 μg of core histones and nucleosomes were performed and analyzed on SDS-PAGE gels as described elsewhere (46). The S190 extract was prepared from Drosophila embryos 0 to 4 h after fertilization (37). The G5-MLP plasmid, which contains a 390-nucleotide G-less cassette, was assembled into chromatin with purified HeLa core histones in the S190 extract essentially as described previously (5). After chromatin assembly, Sarkosyl (0.05% final concentration) was added, and chromatin was immediately purified by gel filtration on a Sepharose CL-4B column (0.7 by 30 cm) equilibrated with EX-20 buffer (10 mM HEPES [pH 7.5], 20 mM KCl, 0.5 mM EDTA, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 10% glycerol, 0.01% NP-40). Micrococcal nuclease digestion was performed as previously described (11). In vitro transcription reactions were performed as schematized in Fig. 4C. Naked or chromatin-assembled G5-MLP plasmid DNA (25 ng) was preincubated with 40 ng of FLAG-tagged Gal4-VP16 in EX-20 buffer plus 0.5 mg of bovine serum albumin (BSA) per ml for 10 min at room temperature. The acetylation step was performed with 5 μl of M2-purified STAGA or the M2 mock-purified fraction for 30 min at 30°C in transcription buffer (10 mM Tris-HCl, 20 mM HEPES [pH 7.9], 60 mM KCl, 0.25 mg of BSA per ml, 6 mM MgCl2, 5 mM DTT, 30 mM ATP, 10 mM phosphocreatine, 0.5 μg of creatine phosphokinase, 12% glycerol, 10 U of RNasin) with or without 1 μM acetyl-CoA. Then HeLa nuclear proteins (35 μg) were added, and reaction mixtures were incubated for 30 min at 30°C. Transcription was initiated by addition of 0.5 mM UTP, 0.5 mM ATP, 12.5 μM CTP, 10 μCi [32P]CTP, and 0.1 mM 3′-O-methyl-GTP. After a 40-min incubation at 30°C, reactions were stopped by addition of 10 mM EDTA. Following a 20-min treatment at 37°C with 20 U of RNase T1 (Boehringer Mannheim), purified transcripts were analyzed on 7 M urea–8% polyacrylamide gels. Quantitation was performed on a PhosphorImager (Molecular Dynamics).
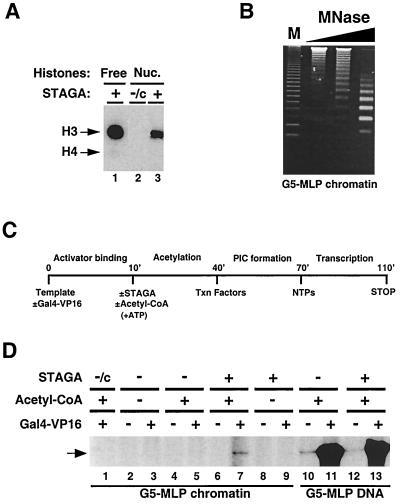
FIG. 4.
STAGA functions as a nucleosome-acetylating transcription coactivator on chromatin in vitro. (A) STAGA acetylates nucleosomes. Fluorography of an SDS-PAGE gel containing 1 μg of either purified HeLa core histones (Free) or native nucleosomes (Nuc.) acetylated with M2-purified STAGA (+) or the control M2-purifed fraction (-/c) in the presence of [3H]acetyl-CoA is shown. The presence of equal amounts of free and nucleosomal histones was confirmed by Coomassie staining (not shown). Arrows indicate the position of core histones H3 and H4. H4 acetylation is very weak and can be detected only with longer exposures with free histones. (B) Micrococcal nuclease digestion analysis of G5-MLP chromatin. Shown is an ethidium bromide-stained agarose gel containing a 123-bp DNA ladder (M) and the DNA products of a time course digestion of G5-MLP chromatin with micrococcal nuclease (MNase). (C) Diagram of the transcription reaction illustrating the order of addition of G5-MLP DNA or chromatin (Template), activator (Gal4-VP16), M2-purified STAGA, acetyl-CoA, ATP, nuclear extract (Txn Factors), and nucleoside triphosphates (NTPs). Times are in minutes. (D) Activator-dependent transcription stimulation by STAGA on chromatin in vitro. Autoradiogram of a urea gel containing specific transcripts (arrow) from transcription reactions performed with DNA or chromatin G5-MLP templates in the presence (+) or absence (−) of the indicated components as described for panel C.
EMSA.
For electrophoretic mobility shift assays (EMSA), the probe was a 239-bp HindIII DNA fragment from plasmid pG5T+I+ that had been labeled with Klenow enzyme and [32P]dCTP and either mock irradiated or UV irradiated at 254 nm using a UV-Stratalinker (Stratagene) at 5,000 or 15,000 J/m2. The binding reactions were performed at 25°C for 30 min with 0.1 ng of labeled probe and 4 to 5 μl of M2-purified STAGA or mock control fraction in a final volume of 8 to 10 μl containing 10 mM Tris-HCl (pH 7.9), 20 mM HEPES (pH 7.9), 10% glycerol, 50 mM KCl, 15 mM NaCl, 4 mM MgCl2, 5 mM DTT, 4 μg of BSA, 0.025% NP-40, 0.25 mM EDTA, 20 ng of poly[d(I-C)], and unlabeled UV-irradiated or mock-irradiated competitor DNA. For antibody supershift assays, STAGA was preincubated with 0.5 μl of diluted antisera (1:20 dilution in BC100 plus 0.5 mg of BSA per ml) for 15 min on ice before addition to binding reactions. Binding reactions were analyzed on 5% PAGE gels in TGE buffer (25 mM Tris-HCl, 190 mM glycine, 1 mM EDTA [pH 8.3]).
RESULTS
Affinity purification of the human STAGA complex.
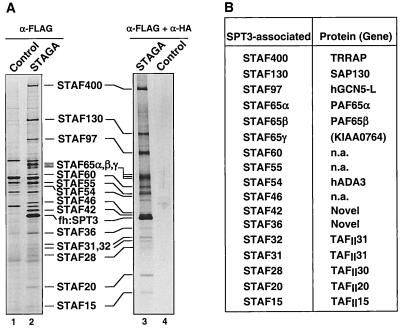
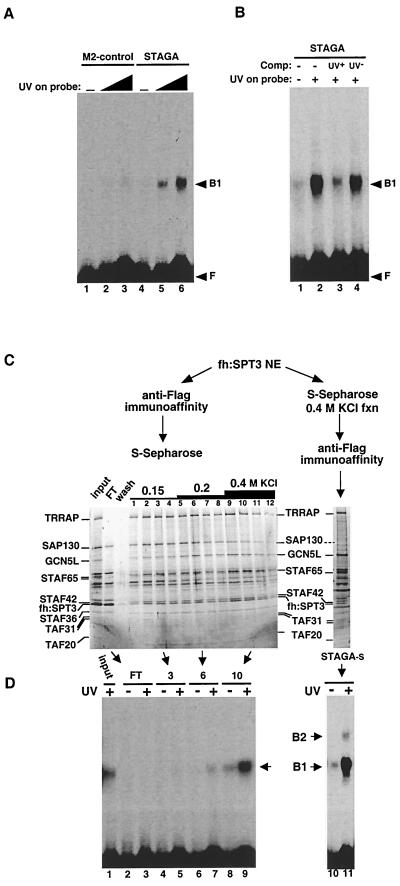
To further characterize the human STAGA complex, HeLa cell lines that stably express ectopic fh:SPT3 and fh:TAFII31 were established. Anti-FLAG immunoaffinity chromatography was used to purify epitope-tagged STAGA from nuclear extracts of fh:SPT3-expressing cells. Purified STAGA contained, besides fh:SPT3 (identified by immunoblotting and MS), at least 17 additional SPT3-associated factors (STAFs) (Fig. 1A, lane 2) that were absent in the mock-purified fraction derived from nuclear extracts of control (untransfected) HeLa cells (lane 1). Furthermore, STAFs remained specifically associated with fh:SPT3 after a subsequent purification on an anti-HA immunoaffinity resin (lane3) and coimmunopurified with untagged hSPT3 from nuclear extracts of fh:TAFII31-expressing cells (see below; see Fig. 3). Most STAFs also copurified during ion-exchange and protein-affinity chromatography and cosedimented in a glycerol gradient (see below; see Fig. 5 and 6; also data not shown). These data indicate that hSPT3 is in a large multiprotein complex with TAFII31 and other STAFs.
FIG. 1.
Affinity purification and composition of the human STAGA complex. (A) Immunopurified STAGA complex. Shown is a silver stain of gradient SDS-PAGE gels containing STAGA purified from nuclear extracts of the fh:SPT3-expressing cell line (lanes 2 and 3) and mock-purified fractions derived from control HeLa nuclear extracts (lanes 1 and 4) after affinity purification on M2-agarose (lanes 1 and 2) or after successive immunopurifications M2-agarose and anti-HA antibody resins (lanes 3 and 4). The positions of fh:SPT3 and STAFs with their approximate molecular masses are indicated. (B) Identity of STAFs determined by tandem MS. n.a., not analyzed.
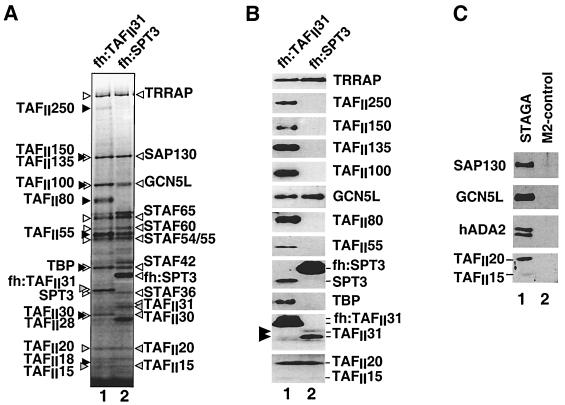
FIG. 3.
SDS-PAGE and Western blot analyses of complexes purified from cells expressing epitope-tagged fh:TAFII31 and fh:SPT3. (A) Silver-stained SDS-PAGE gel containing M2-agarose-purified complexes from nuclear extracts of cells expressing fh:TAFII31 (lane 1) and fh:SPT3 (lane 2). Black and white arrowheads indicate, respectively, TFIID-specific and STAGA-specific components; gray arrowheads point to TAFIIs shared by TFIID and STAGA. (B) Western blot analysis of the fh:TAFII31- and fh:SPT3-containing complexes in panel A. A double arrowhead indicates the expected position of native TAFII31 in the fh:TAFII31 complexes (lane 1). (C) Western blot analysis with specific SAP130, GCN5, hADA2, and TAFII20/15 antibodies of M2 affinity-purified STAGA from nuclear extracts of fh:SPT3 cells (lane 1) and of the mock-purified M2 fraction (lane 2) from control HeLa nuclear extracts (M2-control).
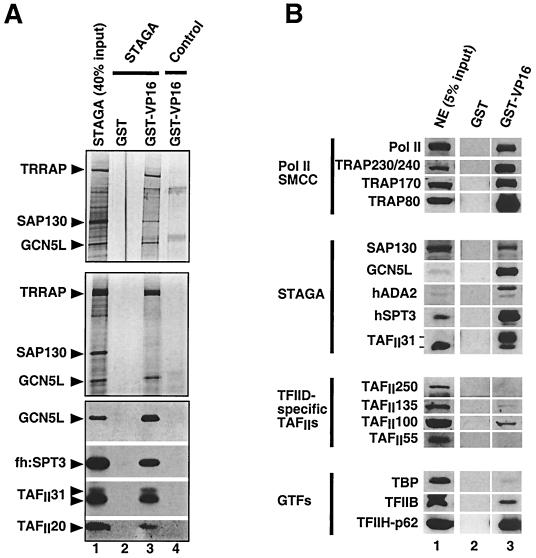
FIG. 5.
Specific interaction of STAGA with the VP16 transcription activation domain. (A) Direct physical interaction of STAGA with GST-VP16. M2-purified STAGA (lane 1) was incubated with GST or GST-VP16 proteins immobilized on glutathione-agarose, and the resins were washed with either 170 resin volumes of BC100–0.05% NP-40 (top panel) or 400 resin volumes of BC150–0.05% NP-40 (two bottom panels). STAGA components bound to GST (lane 2) and GST-VP16 (lane 3) resins were detected on silver-stained SDS-PAGE gels (two top panels) or by Western blotting (bottom panel). Lane 4 is similar to lane 3 but contains the M2 mock-purified fraction. (B) GST-VP16 selectively recruits STAGA and TRAP (SMCC) complexes from nuclear extracts. Western blot analysis of transcription factors from a HeLa nuclear extract (NE) that bind to GST and GST-VP16 resins.
FIG. 6.
UV-damaged-DNA-binding activity in STAGA. (A) UV-dependent DNA-binding activity in STAGA. EMSA analysis of DNA-binding activities in M2-purified STAGA (lanes 4 to 6) and in the control M2 mock-purified fraction (lanes 1 to 3) with a 32P-labeled DNA probe that was either untreated (lanes 1 and 4) or irradiated with increasing UV doses (5,000 J/m2 [lanes 2 and 5] and 15,000 J/m2 [lanes 3 and 6]). F, unbound free probe; B1, specific protein-DNA complex. (B) EMSA with M2-purified STAGA as in panel A with an untreated (-) and a UV-treated (+; 15,000 J/m2) probe and in the absence (-) and presence of unlabeled competitor (Comp) DNA probe (about a 10× molar excess) that was either untreated (UV−) or UV irradiated (UV+, 15,000 J/m2). (C) Chromatographic separation of distinct STAGA complexes. Two STAGA purification schemes are presented, starting from nuclear extracts (NE) of fh:SPT3-expressing cells as described in the text. Silver-stained SDS-PAGE gels containing the different STAGA fractions are shown. Input, M2-purifed STAGA complex (6 μl); FT, S-Sepharose unbound fraction (10 μl). The numbers 1 to 12 indicate S-Sepharose fractions (6 μl each). STAGA-s (8 μl) lacks SAP130. (D) SAP130 does not correlate with the UV-dependent DNA-binding activity in STAGA. EMSA analysis was done with STAGA fractions (5 μl each) in panel C with UV-irradiated (+, 5,000 J/m2) and nonirradiated (-) DNA probes. The left and right panels are EMSA results with the same probe run on the same gel but autoradiographed for 12 and 1 h, respectively. After normalization to TRRAP content, the UV-dependent DNA-binding activity in STAGA-s (lane 11) is about 10 times higher than that in the S-Sepharose fraction 10 (lane 9).
Identification of STAGA components.
Identification of most STAFs was performed by MS and by searching the NCBI nonredundant and dBEST databases with the mass spectral data (see Materials and Methods). The results are summarized in Fig. 1B and described in more detail below. The complete list of peptide sequences can be found in Materials and Methods. The identity of most STAGA components was further verified by immunoblot analyses using specific antibodies (see below).
STAGA contains the transcription-transformation cofactor TRRAP, hGCN5L acetylase, novel human ADA-like and SPT-like cofactors, and a subset of TAFIIs.
The STAF400 protein band yielded six distinct peptide sequences corresponding to the TRRAP protein. TRRAP is an ATM-related protein that was originally identified through its interaction with the c-Myc and E2F1 transcription activation domains and that has been implicated in c-Myc-mediated oncogenic transformation in vitro (47). TRRAP is also present in human PCAF complex, TFTC, and TIP60 HAT complexes (8, 35, 73), and a yeast homolog, Tra1, is a subunit of the SAGA and NuA4 HAT complexes (3, 26, 59).
The STAF97 band yielded four peptide sequences belonging to hGCN5L. Two (TLPENLTLEDAK and SHPSAWPFMEPVK) are specific for hGCN5L. This is consistent with previous demonstrations that PCAF is either present in very small amounts or absent in HeLa cells (45, 62, 79).
The clustered STAF65 protein bands correspond to three distinct proteins (STAF65α, -β, and -γ). STAF65α (eight peptide sequences) and STAF65β (two peptide sequences) are, respectively, the PAF65α and PAF65β subunits originally found in the human PCAF complex. PAF65α and PAF65β share significant sequence similarities, respectively, with human TAFII80 and TAFII100, suggesting similar functions (53). Consistent with this, the yeast SAGA complex lacks PAF65 homologs but instead contains yeast TAFII60 and TAFII90, the bona fide homologs of human TAFII80 and TAFII100, respectively.
STAF65γ was identified by the peptide sequence YWGEIPISSSQTNR and is encoded by a gene (KIAA0764) of unknown function. BLAST searches of the NCBI protein databases revealed similarities with the yeast SAGA subunit SPT7. Alignment of both proteins using a MacVector program indicated that STAF65γ is similar (20% identity and 38% similarity) over its entire length to the yeast SPT7 C terminus, which includes a histone fold domain (Fig. 2A). This might suggest a function for STAF65γ within the STAGA complex comparable to the role of yeast SPT7 in maintaining the integrity of the SAGA complex (22, 25).
FIG. 2.
Amino acid sequence comparisons of human STAF65γ with S. cerevisiae (S.c.) SPT7 (A), STAF42 with S. cerevisiae ADA1 (B), and a domain of STAF42 with histone fold domains of human TAFII135 and histone H2A (C). (A and B) Alignment was performed with MacVector software. Identical and related amino acids are boxed and shaded, and the underlined sequence is the histone fold domain of yeast SPT7 (22). (C) The three α-helices (α1 to α3) of the histone H2A fold are schematized over their corresponding sequences. The underlined sequence indicates TAFII135 amino acids important for interactions with TAFII20 (23) that are conserved in STAF42.
The STAF54 protein band contained the hADA3 peptide sequence VLEAETQILTDWQDK. In addition, two forms of hADA2 with distinct electrophoretic mobilities on SDS-PAGE were also found in STAGA by Western blotting with specific antibodies (Fig. 3C). Whether this represents posttranslational modifications of hADA2 remains to be determined.
Analyses of the STAF42 protein band yielded five different peptide sequences for a novel human protein. A complete STAF42 amino acid sequence was assembled from several overlapping expressed sequence tags and human genomic sequences on chromosome 1q24–25 (GenBank accession numbers AL008639 and AL009182). STAF42 is a 335-amino-acid protein with a calculated molecular mass of 37.4 kDa and an estimated pI of 7.06. BLAST searches of the NCBI protein databases and a MacVector program alignment indicated an overall 37% similarity (19% identity) between human STAF42 and yeast ADA1 (Fig. 2B). Yeast ADA1 contributes to the structural integrity of the SAGA complex (63) and contains a histone H2A-like domain that is conserved in human TAFII135 and that interacts with the H2B-like domain of yTAFII61/68 (the homolog of human TAFII20) (23). This yADA1/TAFII135 H2A-like domain is conserved in human STAF42 (Fig. 2C). Since TAFII20 is also a subunit of the STAGA complex (see below), this suggests ADA1-like functions for STAF42, including possible direct interactions with TAFII20.
STAF31/32, STAF28, and STAF20/15 were identified as TAFII31 (eight peptides), TAFII30 (two peptides), and TAFII20/15 (two peptides), respectively. Interestingly, STAF32 corresponds to a modified form of TAFII31 that is specifically enriched in STAGA but absent in highly purified TFIID (data not shown). Significantly, in contrast to the TFTC complex, epitope-tagged STAGA lacks all the high-molecular-weight TAFIIs (Fig. 3B, lane 2), including TAFII250, -150, -135, -100, -80, and -55. This is in agreement with, and extends, previous observations for the untagged endogenous STAGA (46).
Immunopurification of TAFII31-containing complexes from HeLa cells expressing fh:TAFII31 confirmed its association with endogenous SPT3 and most STAFs, as well as with TFIID subunits (Fig. 3A and B and data not shown). Moreover, all the major proteins that coimmunopurified with fh:TAFII31 can be accounted for by subunits present in either TFIID or STAGA (Fig. 3A and B). In summary, the STAGA composition described above demonstrates that STAGA is highly related to the yeast SAGA complex and more or less similar to the human PCAF complex but is different from the TFTC and TFIID complexes.
Specific in vivo association of STAGA with a pre-mRNA splicing factor.
A total of 21 different tryptic peptide sequences identified STAF130 as the recently characterized SAP130. The specific association of SAP130 with highly purified STAGA was confirmed by Western blot analyses (Fig. 3C). SAP130 coimmunopurified with other STAFs under stringent conditions (300 mM KCl, 0.05% NP-40) on M2-agarose only from nuclear extracts of fh:SPT3-expressing cells (lane 1) and not from control HeLa nuclear extracts (lane 2). SAP130 is a component of the splicing factor complex SF3b, a submodule of the 17S U2 snRNP particle (17). Interestingly, significant amounts of STAF130 (SAP130) were associated with STAGA after two successive immunopurification steps (Fig. 1A, lane 3) at salt concentrations (300 to 400 mM KCl) that are known to disrupt SF3b association with U2 snRNP (6). Under these conditions, none of the STAFs that have been analyzed contained peptides belonging to other SF3b-U2 snRNP subunits. Moreover, significant amounts of SAP130 in the nuclear extract were also specifically coimmunoprecipitated with fh:SPT3 under more physiological salt concentrations (100 to 150 mM KCl) that are known to preserve the association of SF3b with the U2 snRNP particle (see Fig. 7C). SAP130 did not merely interact with overexpressed free fh:SPT3 but was preferentially associated with fh:SPT3-STAF complexes. Indeed, S-Sepharose fractionation of immunopurified STAGA separated free fh:SPT3 (found in the column flowthrough) from bound fractions that contained SPT3-STAF complexes and most of SAP130 (Fig. 6C). However, small amounts of SAP130 were also detected in the flowthrough fraction and STAGA-s could also be isolated (see below; see Fig. 5A and Fig. 6C). Furthermore, immunoblot analyses demonstrated that SAP130 also coimmunoprecipitated with fh:TAFII31 (data not shown). Altogether, these data demonstrate an interaction of SAP130 with STAGA in vivo and suggest that SAP130 might not be as tightly associated with STAGA as the other STAF subunits.
FIG. 7.
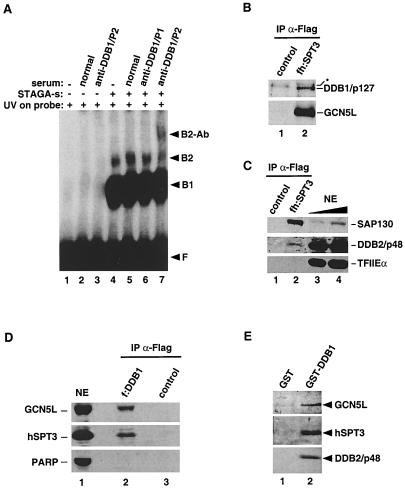
STAGA associates with UV-DDB factors in vivo and in vitro. (A) DDB1 contributes to the UV-dependent DNA-binding activity in STAGA. EMSA analysis was done with a UV-irradiated DNA probe and either buffer (lanes 1 to 3) or purified STAGA-s (lanes 4 to 7) that was untreated (lanes 1 and 4) or preincubated with either normal rabbit serum (normal), rabbit anti-DDB1 peptide 1 antibodies (anti-DDB1/P1), or rabbit anti-DDB1 peptide 2 antibodies (anti-DDB1/P2). F, unbound probe; B1 and B2, specific protein-DNA complexes; B2-Ab, specific B2-antibody complex. (B and C) Endogenous DDB1 and DDB2 associate with epitope-tagged STAGA in HeLa nuclei. Western blot analysis of anti-FLAG/M2 immune precipitates from nuclear extracts of either normal HeLa cells (lanes 1) or HeLa cells stably expressing fh:SPT3 (lanes 2) is shown. (B) An asterisk indicates a nonspecific product (lanes 1 and 2). (C) SAP130, DDB2 (p48), and TFIIEα in crude nuclear extracts (NE, lanes 3 to 4) and in immune precipitates (lanes 1 to 2). (D) Endogenous STAGA associates with epitope-tagged DDB1 in HeLa nuclei. A Western blot of anti-FLAG/M2 immune precipitates from nuclear extracts of HeLa cells left untransfected (control) or transiently transfected with an f:DDB1 expression vector is shown. PARP, poly-ADP-ribose polymerase; NE, crude HeLa nuclear extract. (E) GST-DDB1 recruits DDB2 and STAGA components (GCN5L and hSPT3) from nuclear extracts in vitro. Western blot analysis of pull-down assays using a HeLa nuclear extract and either GST or GST-DDB1 proteins immobilized on glutathione-agarose is shown.
STAGA is an acetyl-CoA-dependent transcription coactivator on a chromatin-assembled template in vitro.
The existence of at least three different human SAGA-like complexes (i.e., PCAF complex, TFTC, and STAGA), and the fact that splicing factors have so far not been described in association with any other coactivator and/or HAT complex, raised the question of whether STAGA is involved in transcription regulation in a manner similar to that of the yeast SAGA complex or whether it is a specialized human complex dedicated to other RNA, DNA, or chromatin transactions.
We originally addressed a possible transcription function of STAGA by using various nonchromatinized (naked) DNA templates and either crude nuclear extracts immunodepleted of both TFIID and STAGA or systems reconstituted with purified general transcription factors and RNA polymerase II. In all cases we were unable to observe STAGA-dependent transcription in the absence of TFIID or TBP (data not shown). This further differentiates STAGA from TFTC, which was shown previously to have TFIID-like functions (77). Furthermore, STAGA neither influenced basal transcription nor potentiated activator (Gal4-VP16)-dependent transcription in crude nuclear extracts and in PC4-dependent purified systems containing either TFIID or TBP; moreover, addition of acetyl-CoA had no effect in these systems (data not shown; also see below).
We then investigated whether STAGA might function primarily at the chromatin level. In agreement with previous observations (46) purified STAGA preferentially acetylated histone H3 in equimolar mixtures of all four free core histones (Fig. 4A, lane 1). Moreover, immunopurified STAGA (Fig. 4, lane 3), but not the control mock-purified fraction (lane 2), also acetylated predominantly H3 within nucleosomes, albeit with a lower efficiency (compare lanes 1 and 3). To test for a possible transcription function of STAGA on chromatin in vitro, a plasmid DNA containing five Gal4-binding sites upstream of the adenovirus major late core promoter and a G-less cassette (G5-MLP) was assembled into nucleosome arrays by using a Drosophila embryo S190 assembly extract complemented with purified HeLa core histones. The nucleosomal G5-MLP template (“crude chromatin”) was either used directly in transcription reactions or further purified by gel filtration. This step removes S190 factors (including HATs, acetyl-CoA, and ATP) that are present in the assembly reaction but does not completely remove ATP-dependent chromatin remodeling activities (19) (see below). Agarose gel analysis of the purified G5-MLP chromatin template digested with micrococcal nuclease indicated a circa-180-bp regular DNA ladder, which confirmed the assembly of 19 physiologically spaced nucleosomes on the G5-MLP plasmid (Fig. 4B).
Similar to previous observations with a different activator (retinoic acid receptor/retinoid X receptor) and coactivator (p300+TIF2) (19), and also with unpurified chromatin templates and HeLa nuclear extract as a source of general and basal transcription factors, we observed robust activation by the potent chimeric activator Gal4-VP16 that was completely dependent on the VP16 activation domain but was not further stimulated by the addition of purified STAGA (data not shown). Purified chromatin was then used to specifically address a potential role of STAGA in transcription activation by Gal4-VP16. Purified G5-MLP (chromatin or plasmid) DNA templates were preincubated in the presence or absence of Gal4-VP16, purified STAGA, and acetyl-CoA (note that ATP was added during the acetylation step in all reactions), and transcription was initiated by the addition of nuclear extract and nucleoside triphosphates (Fig. 4C). As expected, efficient basal transcription and strong (117-fold) activation by Gal4-VP16 were observed from the naked G5-MLP DNA template in the absence of added STAGA (Fig. 4D, lanes 10 and 11). Addition of immunopurified STAGA, in the presence of acetyl-CoA, had no significant effect on either basal or activated (93-fold) transcription from this naked DNA template (Fig. 4D, lanes 12 and 13). In stark contrast, the purified G5-MLP chromatin template was transcriptionally silent in reactions lacking immunopurified STAGA, in both the absence and presence of Gal4-VP16 and acetyl-CoA (Fig. 4D, lanes 2 to 5). Addition of immunopurified STAGA to reactions lacking acetyl-CoA also failed to activate transcription from the chromatin template (Fig. 4D, lanes 8 and 9). However, in the presence of acetyl-CoA, immunopurified STAGA stimulated transcription from the G5-MLP chromatin template in a Gal4-VP16 activator-dependent manner (Fig. 4D, lanes 6 and 7). Under the same conditions, addition of a control mock-purified fraction had no effect (lane 1). Interestingly, the coactivator activity of STAGA appears to require the function of ATP-dependent chromatin remodeling factors endogenous to the system since no transcription was observed when ATP was omitted during the acetylation and preinitiation complex formation steps (data not shown).
Since STAGA-mediated transcription activation was dependent on the presence of the Gal4-VP16 activator, we investigated the possibility that STAGA could be recruited to the promoter via direct interactions with the VP16 activation domain. As shown in Fig. 5A, purified STAGA bound efficiently to a GST-VP16 fusion protein immobilized on glutathione-agarose (lane 3) but not to the control GST resin (lane 2). Interestingly, SAP130 appeared to bind less tightly than the other STAGA subunits to the GST-VP16 resin (Fig. 5A) and, unlike most other subunits, did not remain associated with GST-VP16 after more extensive washes of the resin (Fig. 5A, middle and bottom panels). To further assess the relative strength and specificity of the STAGA-VP16 interaction, we compared the recruitment by immobilized GST-VP16 of endogenous (untagged) STAGA and other previously reported VP16-interacting factors in a crude HeLa cell nuclear extract. As shown in Western blot analysis (Fig. 5B), all STAGA subunits tested (including SAP130) were specifically recruited from nuclear extracts by GST-VP16 (lane 3) but not by the GST control resin (lane 2). Moreover, STAGA subunits (with the exception of SAP130) were highly enriched in the GST-VP16-bound fraction (lane 3) compared with the input nuclear extract (lane 1). As expected, RNA polymerase II and components of the TRAP (SMCC) coactivator complex (36) were also specifically recruited by the GST-VP16 resin (Fig. 5B, compare lanes 2 and 3) but were only moderately enriched in the VP16-bound fraction (compare lanes 1 and 3). TFIIB and TFIIH (Fig. 5B) were also specifically recruited (compare lanes 2 and 3) but were not enriched compared with the input nuclear extract (lanes 1 and 3). Strikingly, under the same conditions, TBP and TFIID-specific TAFIIs did not interact significantly (or interacted only very weakly) with GST-VP16 (Fig. 5B, lanes 3). Altogether, these results demonstrate that STAGA is a bona fide transcription coactivator on chromatin-assembled promoters in vitro.
STAGA interacts in vivo and in vitro with UV-damaged-DNA-binding proteins.
The STAGA-associated protein SAP130 belongs to a family of structurally related proteins that include the 160-kDa cleavage and polyadenylation specificity factor (CPSF160) and the UV-damaged-DNA-binding protein DDB1 (13, 17, 48). In fact, SAP130 is more closely related in amino acid sequence to DDB1 (22% identity) than to CPSF160 (47) (data not shown). The 127-kDa DDB1 and the 48-kDa DDB2 proteins interact to constitute the active UV-damaged-DNA-binding factor UV-DDB (reviewed in reference 66). The striking structural similarity between SAP130 and DDB1 suggested the possibility of common interacting target factors and/or a function of SAP130 in recognition of UV-damaged DNA. Thus, we tested for UV-damaged-DNA-binding activity in purified STAGA. By EMSA we detected a UV dose-dependent DNA-binding activity that was specifically associated with immunopurified STAGA, compared with the M2 mock-purified fraction (Fig. 6A, lanes 1 to 3 versus 4 to 6). This activity was competed specifically with DNA that had been UV irradiated (Fig. 6B, lane 3) but not with nonirradiated DNA (lane 4). The same results were obtained by using unrelated DNA probes, indicating that the binding is independent of DNA sequence and length (data not shown).
To address whether SAP130 is required for the UV-dependent DNA-binding activity associated with STAGA, we fractionated immunopurified STAGA by ion-exchange chromatography on S-Sepharose (Fig. 6C). Whereas most STAGA bound to S-Sepharose, excess free fh:SPT3 and small amounts of SAP130 did not (Fig. 6C, lane FT). Elution of STAGA subunits with increasing salt concentrations allowed the isolation of three major STAGA populations that differed in the relative stoichiometry of their SAP130 and GCN5L subunits. The 150 mM KCl fractions (Fig. 6C, lanes 1 to 4) had substoichiometric amounts of GCN5L compared to SAP130 and other STAFs. STAGA in the 200 mM KCl fractions (lanes 5 to 8) had apparently stoichiometric amounts of both SAP130 and GCN5L, whereas the 400 mM KCl fractions (lanes 9 to 12) had substoichiometric amounts of SAP130 compared to GCN5L and other STAFs. Representative fractions of each step elution (i.e., fractions 3, 6, and 10) having similar concentrations of TRRAP, fh:SPT3, STAF42, STAF36, and TAFII20 were tested for UV-dependent DNA-binding activity. As shown in Fig. 6D (left panel), the high-salt STAGA fraction 10, which contained substoichiometric amounts of SAP130, was enriched in UV-dependent DNA-binding activity (lanes 8 and 9), but other fractions containing SAP130, including the Sepharose unbound fraction, were inactive (lanes 2 to 7). This was not due to the presence of inhibitory activities in fractions defective in DNA-binding, because their addition to fraction 10 had no effect (data not shown). Similar results were obtained after peak fractions of each step elution were pooled (data not shown). These results demonstrate that SAP130, in the absence of most STAGA components (i.e., in the S-Sepharose unbound fraction), has no detectable UV-dependent DNA-binding activity. Moreover, the concentration of SAP130 in STAGA-containing fractions inversely correlated with the STAGA-associated UV-dependent DNA-binding activity. To further confirm this, we purified STAGA-s by first fractionating nuclear extracts of fh:SPT3-expressing cells on S-Sepharose. The column was washed extensively, and bound proteins were eluted with buffer containing 200 and 400 mM KCl. STAGA in the 400 mM KCl eluate was directly affinity purified on M2-agarose (Fig. 6C, right panel). This SAP130-free STAGA-s complex had the highest UV-dependent DNA-binding activity, which resulted in the appearance of a secondary (B2) complex by EMSA (Fig. 6D, lanes 10 and 11).
These data suggested that a protein(s) different from SAP130 was responsible for the UV-dependent DNA-binding activity in STAGA. We tested for the presence of the bona fide DDB1 protein in the protein-DNA complexes observed with STAGA-s by using specific anti-DDB1 antibodies in electrophoretic mobility supershift assays. The UV-dependent B2 complex formed by STAGA-s was supershifted specifically by an anti-DDB1 (peptide 2) antiserum (Fig. 7A, lane 7) that efficiently recognizes native DDB1 but not by either normal rabbit serum (lane 5) or an anti-DDB1 (peptide 1) antiserum (lane 6) that is inefficient in recognizing native DDB1 (42). Consistent with the association of DDB1 with STAGA-s purified under high-salt conditions (see above), Western blot experiments confirmed that DDB1 in nuclear extracts of fh:SPT3-expressing cells also coimmunoprecipitated with fh:SPT3 and hGCN5L under more physiological salt concentrations (Fig. 7B, lane 2).
It has been reported previously that, at high enough concentrations, UV-DDB forms two protein-DNA complexes (B1 and B2) with UV-irradiated DNA that are both dependent on DDB1 and DDB2 subunits (21, 33, 51). However, antibodies to tagged DDB1 can supershift only the B2 complex (33). This also was observed with the UV-DDB activity in STAGA when probed with the anti-DDB1 (peptide 2) antibody (Fig. 7A, lane 7) and further suggested that DDB2 was also associated with STAGA. In agreement with this, immunoprecipitation of fh:SPT3 containing STAGA also coimmunoprecipitated DDB2 from nuclear extracts (Fig. 7C, lane 2). Under the same conditions, other nuclear factors, including TFIIE (Fig. 7C) and CPSF160 (data not shown), were not associated with fh:SPT3. To further confirm the interaction in vivo of UV-DDB with STAGA, HeLa cells were transiently transfected with an expression vector encoding FLAG-tagged DDB1 (f:DDB1). Immunoprecipitations with M2-agarose (Fig. 7D) demonstrated a specific association of endogenous hGCN5L and hSPT3 with f:DDB1 in nuclear extracts of transfected (lane 2) but not untransfected (lane 3) cells. As a control, another very abundant chromatin-modifying enzyme, poly-ADP-ribose polymerase, was not immunoprecipitated with f:DDB1 (Fig. 7D, lane 2). These results show that UV-DDB components interact in vivo with STAGA. Under the conditions used, however, only substoichiometric amounts of endogenous UV-DDB components were purified in association with STAGA, which contrasts with the efficient copurification of SAP130.
Finally, we asked whether immobilized GST-DDB1 fusion protein could recruit endogenous STAGA from HeLa cell nuclear extracts. As shown by Western blot analysis (Fig. 7E), GST-DDB1 (lane 2), but not a control GST resin (lane 1), bound DDB2 (bottom panel), hGCN5L (top panel), hSPT3 (middle panel), and TAFII31 (data not shown). A number of other transcription factors, including components of the TRAP (SMCC) complex (TRAPs 230, 240, and 80), TFIIA, TFII-I, and TFIIIB90, were not detected in the bound fraction of the GST-DDB1 resin. Notably, GST-DDB1 also failed to recruit SAP130 (data not shown). This, however, correlates with the above observation that purified STAGA complexes lacking SAP130 are enriched in UV-DDB activity (Fig. 6C and D). In conclusion, these results demonstrate a specific interaction, in vitro and in HeLa cells, of the DNA damage recognition and repair factor UV-DDB with STAGA and suggest that UV-DDB might preferentially interact with a form of STAGA that is not associated with SAP130.
DISCUSSION
This report presents the purification, structural characterization, and functional analysis of the human STAGA complex. We demonstrate that STAGA is a coactivator complex that potentiates, in an acetyl-CoA-dependent manner, activator-dependent transcription in vitro on chromatin templates. Furthermore, we show specific interactions of STAGA with pre-mRNA splicing and UV-damaged-DNA recognition and repair factors in vivo. These results provide the first evidence for a direct association of a component of the general splicing machinery with a transcription coactivator complex and suggest additional roles of STAGA in nucleosomal histone acetylation during DNA damage recognition and/or repair of chromatin in vivo.
Structural relationships of human STAGA to yeast SAGA, other human SAGA-like complexes, and TFIID.
The subunit composition of STAGA reveals a striking complexity and structural resemblance to the yeast SAGA and the human PCAF and TFTC complexes. STAGA appears to be more closely related to the PCAF complex than to the TFTC complex. Indeed, besides their different HATs (hGCN5L versus PCAF), STAGA and PCAF complexes share many subunits that include hSPT3, hADA2, hADA3, PAF65α, PAF65β, TAFII31, TAFII30, TAFII20/15, and TRRAP. Only a subset of these (hGCN5L, hSPT3, hADA3, PAF65β, TAFII31, TAFII30, TAFII20/15, and TRRAP) were also found in the human TFTC complex. Moreover, both STAGA and PCAF complexes lack high-molecular-weight TAFIIs (i.e., TAFII55, TAFII80, TAFII100, TAFII135, and TAFII150) that are uniquely shared by TFTC and TFIID (8). Consistent with this, and in contrast to the TFIID-like function of the TFTC complex (77), STAGA could not substitute for TFIID in our in vitro transcription assays.
We also report the identification within STAGA of two novel histone fold-containing protein subunits: STAF65γ, which is encoded by the KIAA0764 gene of previously unknown function, and STAF42, a novel histone H2A-like protein. STAF65γ and STAF42 have significant similarity, respectively, to SPT7 and ADA1 components of the yeast SAGA complex. Thus, our results show that all yeast SAGA subunits described so far, with the notable exception of SPT8 and SPT20 (ADA5), have human counterparts within STAGA, suggesting that human STAGA is a bona fide functional homolog of yeast SAGA (see also below). Note that it remains possible that homologs of yeast SPT8 and SPT20 are among the few STAF proteins that were not analyzed by MS.
The 130-kDa STAGA-associated factor was identified as SAP130 (see below). The corresponding yeast homolog (Rse1p) has not yet been reported to interact with yeast SAGA, which could point to a potentially significant difference between human and yeast cells. Alternatively, by analogy to the relatively loose association of SAP130 with human STAGA, Rse1p might also dissociate during the multiple chromatographic steps required to purify the native SAGA complex.
The relatedness of STAF42 to histone H2A and TAFII135 also suggests the presence of a histone octamer-like structure within STAGA similar to that proposed for TFIID and yeast SAGA (reviewed in references 22 and 23). Moreover, the structural resemblance between STAGA and TFIID further extends to their association with the structurally related pre-mRNA processing factors SAP130 and CPSF160 and might also indicate a conservation at the functional level (see below).
Transcription coactivator functions of STAGA on chromatin.
In accord with the structural similarities of human STAGA to the yeast SAGA coactivator complex, we also have shown that the STAGA complex can be recruited by an acidic activation domain (VP16), acetylates predominantly histone H3 within nucleosomes, and exhibits acetyl-CoA-dependent coactivator functions on chromatin templates in vitro (Fig. 4 and 5). Thus, in a manner similar to that of yeast SAGA (34, 72), STAGA recruitment by activators might target its nucleosomal acetylation functions to the promoter. The fact that under our conditions STAGA and acetyl-CoA do not influence promoter activity on naked DNA templates, either in nuclear extracts or in purified transcription systems (Fig. 4D and data not shown), is consistent with a selective requirement of the STAGA HAT activity in nucleosome acetylation. Our in vitro results, however, do not exclude additional roles of STAGA in facilitating or stabilizing activator-chromatin interactions or the possibility that STAGA might also acetylate other nonhistone proteins and/or function as an adapter to help recruit the basal transcription machinery. Consistent with this possibility, several yeast SAGA components have been shown to interact with TBP (TAFIIs, SPT3, and SPT8) and acidic activators (ADAs). Furthermore, the largest STAGA subunit, TRRAP, is a protein that interacts with c-Myc and E2F1 transcription activation domains (47).
Interestingly, the VP16 activation domain recruits preferentially STAGA and components of the TRAP (SMCC) coactivator complex from nuclear extracts relative to components of the general transcription machinery (TBP, TFIIB, and TFIIH) and TFIID- and TFTC-specific TAFIIs (Fig. 5B). This contrasts (but is not incompatible) with earlier observations and models identifying direct interactions of activators with TFIID or TBP, TFIIB, and/or TFIIH as critical steps in activation (reviewed in reference 56). Our observation, however, correlates with (i) the fact that TFIID-specific TAFIIs are not generally required for activation in yeast (50, 75), (ii) the recent demonstration that transcription activation by Gal4-VP16 in crude HeLa nuclear extracts does not require TFIID-specific TAFIIs but depends on human SRB7 (53), a component of the TRAP (SMCC) complex (reviewed in reference 45), and (iii) the dependence on yeast SRB4 (a component of the yeast SRB/Mediator complex) for TBP recruitment to promoters that do not require TFIID-specific TAFIIs in vivo (43). The preferential recruitment of STAGA from nuclear extracts by the VP16 activation domain might also suggest a function of STAGA early during the activation process (e.g., in chromatin modification). This would be consistent with the observed ordered recruitment of hGCN5 and general transcription factors to the beta interferon promoter (2) and the requirement in vitro and in certain cases in yeast cells for activator-dependent nucleosome acetylation by GCN5 or SAGA prior to ATP-dependent chromatin remodeling at the promoter by the SWI-SNF complex (28, 58). However, a possible additional role of STAGA or SAGA at later stages cannot be excluded (see above).
STAGA links coactivators to a component of the splicing machinery.
A surprising but significant finding is the identification of the spliceosome-associated protein SAP130 as a STAGA-associated factor in vivo. Human SAP130 is a component of the splicing factor SF3b, a U2 snRNP-associated protein complex that is essential for spliceosome assembly (17) and is enriched in nuclear interchromatin granule clusters, or “speckles” (49). Both SF3b and SF3a complexes associate with the 12S U2 snRNP to form the functional 17S U2 snRNP particle that ultimately stably interacts, in an ATP-dependent manner, with the intronic pre-mRNA branch point site. The precise function of SAP130 is unknown. The fact that SAP130 is the only subunit within SF3b that cannot be UV cross-linked to pre-mRNA in the prespliceosome complex suggests a non-RNA-binding function (reviewed in references 13, 17, and 40). The yeast homolog of SAP130, Rse1p, is essential both for spliceosome assembly in vitro and efficient pre-mRNA splicing in vivo (13). Intriguingly, while significant amounts of SAP130 were found to be associated with highly purified STAGA (Fig. 1A, lane 3), other components of SF3b or U2 snRNP were not detected. However, SAP130 is not as stably associated with STAGA as the other bona fide STAF subunits.
What could be the role of SAP130 interaction with STAGA? One possibility is that SAP130, in association with STAGA, has a novel function unrelated to splicing. Alternatively, SAP130 might function as an adapter to link STAGA and transcription regulators to the general splicing machinery. Indeed, pre-mRNA processing events can occur cotranscriptionally through physical interactions of capping, splicing, and 3′-end processing factors with the hyperphosphorylated C-terminal domain of the large subunit of RNA polymerase II (reviewed in reference 31). Moreover, a promoter-directed “C-terminal-domain-loading” mechanism might exist to recruit pre-mRNA processing factors during the early transcription initiation and elongation phase, an idea supported by the TFIID-mediated recruitment of CPSF to the elongating RNA polymerase II (16). The structural similarities between TFIID and STAGA complexes and between their respective associated factors, CPSF160 and SAP130, thus suggest a possible function for STAGA-SAP130 interactions in the recruitment of the general splicing machinery to actively transcribed genes. This is in accord with the recruitment of SAP130 and STAGA components in nuclear extracts by the VP16 transcription activation domain (Fig. 5) and is consistent with the recent finding that U2 snRNP is incorporated early into the prespliceosomal E complex in an ATP-independent manner (18). A role of transcription factors and associated cofactors in the regulation of pre-mRNA splicing is also supported by evidence of promoter-dependent alternative splicing in vivo (15). Moreover, immunofluorescence studies showing a preferential localization of highly acetylated histone H3-containing chromatin at the periphery of interchromatin granule clusters also suggest a possible functional colocalization of STAGA and/or other H3-acetylating HATs with sites of active cotranscriptional splicing in the nucleus (30).
Interaction of STAGA with UV-DDB and XPE factors involved in DNA damage repair and the hereditary disease xeroderma pigmentosum.
We have shown that STAGA associates in HeLa cells with DDB1 and DDB2 components of the UV-damaged-DNA-binding factor UV-DDB. UV-DDB recognizes various DNA-distorting lesions, including those induced by UV irradiation and certain anticancer drugs and has been implicated in global genomic nucleotide excision repair (NER) in vivo, a pathway that includes the repair of the nontranscribed strand of expressed genes (reviewed in references 38 and 66). Missense mutations in the DDB2 gene and the absence of UV-DDB activity are associated with deficient NER in a subset of individuals with the autosomal recessive disease xeroderma pigmentosum of group E (XPE), which is characterized by UV sensitivity and a high incidence of skin cancer (reference 51 and references therein). However, UV-DDB is not required for transcription-coupled NER of transcribed strands in vivo and is not required in vitro for NER in systems reconstituted with purified factors (1). This has led to the suggestion that UV-DDB might be specifically required in vivo for global genomic NER within chromatin (32, 55, 66), a notion that is consistent with the inhibition of NER by nucleosomes (reviewed in references 48 and 68). Significantly, additional roles for UV-DDB have also been proposed, including a function as a transcription partner for E2F1 (29).
The in vivo and in vitro interactions of STAGA with UV-DDB presented here support a role of UV-DDB in DNA repair within chromatin and further suggest possible mechanisms. In a manner analogous to SAGA and STAGA recruitment by promoter-bound activators, the interaction of UV-DDB with STAGA might target the nucleosome acetylase activity of GCN5L to damaged chromatin sites in order to facilitate the assembly and/or function of the NER machinery on nucleosomes. As for transcription activation, a highly stable interaction of STAGA with DDB1 might not be necessary; indeed, only a transient acetylation by SAGA is sufficient to mark nucleosomes for subsequent remodeling by the SWI-SNF complex at the yeast PHO8 promoter in vivo (58). This also would be consistent with the observed enhanced DNA repair synthesis of hyperacetylated nucleosomes in vivo (57) and the facilitation of NER on synthetic dinucleosomes by the ATP-dependent chromatin remodeling factor ACF (71).
In an alternative (but not mutually exclusive) model, STAGA might preferentially recruit UV-DDB and the basal repair machinery to active genes through acetylation-mediated chromatin unfolding and/or through direct physical recruitment of UV-DDB to the damaged sites. This possibility is consistent with a role of STAGA as a transcription coactivator on chromatin and with previously observed interactions of UV-DDB with viral and cellular transcription activators (29, 42, 44). In addition, this model might also provide a rationale for the observed efficient repair of nontranscribed promoter and coding sequences in certain genes (references 67, 69, and 70 and references therein) and the surprisingly efficient repair of the nontranscribed strands of active genes in differentiated cells that have an otherwise very inefficient global genomic repair (reviewed in reference 52). In any case, our results suggest a possible role of STAGA in NER within chromatin that parallels recent reports linking components of the human TIP60 HAT complex and the yeast INO80 ATP-dependent chromatin remodeling complex to the repair of DNA breaks in vivo (35, 60).
In conclusion, we have identified novel histone fold-containing proteins as subunits of the human STAGA complex and shown that STAGA is a transcription coactivator-HAT complex and a bona fide human homolog of the yeast SAGA complex. The intriguing previous observation that DDB1 belongs to a family of structurally related proteins that include CPSF160 and the splicing factor SAP130 raised the possibility of common and/or related roles and interacting factors for these apparently functionally distinct proteins (13, 17). Our results support this notion by the identification of STAGA as a common interacting partner for DDB1 and SAP130, while CPSF160 might preferentially associate with the related TFIID complex (16). These observations further suggest possible additional functions of STAGA in splicing and DNA repair of chromatin, two processes that are coupled to transcription in vivo.
ACKNOWLEDGMENTS
We thank G. Chu for pBJ5-FLAG-p125/DDB1, S. Berger for hADA2 antiserum, J. Manley for CPSF160 antibodies, R. Lamb for pGEX-5X-3-DDB1, S. Linn for p48 (DDB2) antibodies, S. McMahon and M. Cole for TRRAP antiserum, B. Slagle for XAP1 (DDB1) peptide antibodies, A. Krutchinsky for valuable contributions, and J. Fu and C. Bhattacharyya for technical assistance. We are also grateful to Cedric S. Wesley for the help in establishment of the large population of Drosophila flies.
This work was supported by grants from the NIH to R.G.R. (CA42567) and B.T.C. (RR00862).
ADDENDUM
Similar to the results reported here for STAGA, studies published while this article was under review have shown that TFTC is recruited to UV-damaged DNA (9) and that the PCAF complex facilitates transcription of chromatin in vitro (49a).
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 3.Allard S, Utley R T, Savard J, Clarke A, Grant P A, Brandl C J, Pillus L, Workman J L, Côté J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 5.Becker P B, Tsukiyama T, Wu C. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 6.Behrens S-E, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradbury E M. Reversible histone modifications and the chromosome cell cycle. BioEssays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 8.Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- 9.Brand M, Moggs J G, Mustapha O-A, Fabrice L, Dilworth F J, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biol Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 11.Bulger M, Kadonaga J T. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262. [Google Scholar]
- 12.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspary F, Shevchenko A, Wilm M, Séraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Côté J, Utley T, Workman J L. Basic analysis of transcription factor binding to nucleosomes. Methods Mol Genet. 1995;6:108–152. [Google Scholar]
- 15.Cramer P, Cáceres J F, Cazalla D, Kadener S, Muro A F, Baralle F E, Kornblihtt A R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 16.Dantonel J-C, Murthy K G K, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 17.Das B K, Xia L, Paladjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 19.Dilworth F J, Fromental-Ramain C, Yamamoto K, Chambon P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol Cell. 2000;6:1049–1058. doi: 10.1016/s1097-2765(00)00103-9. [DOI] [PubMed] [Google Scholar]
- 20.Eberharter A, Sterner D E, Schieltz D, Hassan A, Yates III J R, Berger S L, Workman J L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J Biol Chem. 1999;274:20027–20033. doi: 10.1074/jbc.274.28.20027. [DOI] [PubMed] [Google Scholar]
- 22.Gangloff Y-G, Sanders S L, Romier C, Kirschner D, Weil P A, Tora L, Davidson I. Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol Cell Biol. 2001;21:1841–1853. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangloff Y-G, Werten S, Romier C, Carré L, Poch O, Moras D, Davidson I. The human TFIID components TAFII135 and TAFII20 and yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol Cell Biol. 2000;20:340–351. doi: 10.1128/mcb.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant P A, Berger S L. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- 25.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 26.Grant P A, Schieltz D, Pray-Grant M G, Yates III J R, Workman J L. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 27.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 28.Hassan A H, Neely K E, Workman J L. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 29.Hayes S, Shiyanov P, Chen X, Raychadhuri P. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol Cell Biol. 1998;18:240–249. doi: 10.1128/mcb.18.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendzel M J, Kruhlak M J, Bazett-Jones D P. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol Biol Cell. 1998;9:2491–2507. doi: 10.1091/mbc.9.9.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 32.Hwang B J, Ford J M, Hanawalt P C, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang B J, Toering S, Francke U, Chu G. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol Cell Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda K, Steger D J, Eberharter A, Workman J L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikura T, Ogryzko V V, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 36.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 37.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 38.Keeney S, Eker A P M, Brody T, Vermeulen W, Bootsma D, Hoeijmakers J H J, Linn S. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc Natl Acad Sci USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 40.Krämer A, Grüter P, Gröning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krutchinsky A N, Zhang W, Chait B T. Rapidly switchable matrix-assisted laser desorption/ionization and electrospray quadrupole-time-of-flight mass spectrometry for protein identification. J Am Soc Mass Spectrom. 2000;11:493–504. doi: 10.1016/S1044-0305(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 42.Lee T-H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virology. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X-Y, Bhaumik S R, Green M R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 44.Lin G Y, Paterson R G, Richardson C D, Lamb R A. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology. 1998;249:189–200. doi: 10.1006/viro.1998.9317. [DOI] [PubMed] [Google Scholar]
- 45.Malik S, Roeder R G. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biol Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 46.Martinez E, Kundu T K, Fu J, Roeder R G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 47.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 48.Meijer M, Smerdon M J. Accessing DNA damage in chromatin: insights from transcription. BioEssays. 1999;21:596–603. doi: 10.1002/(SICI)1521-1878(199907)21:7<596::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 49.Mintz P J, Patterson S D, Neuwald A F, Spahr C S, Spector D L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Mizuguchi G, Vassilev A, Tsukiyama T, Nakatani Y, Wu C. ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J Biol Chem. 2001;276:14773–14783. doi: 10.1074/jbc.M100125200. [DOI] [PubMed] [Google Scholar]
- 50.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcription activation in yeast. Nature. 1996;383:188–190. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 51.Nichols A F, Itoh T, Graham J A, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 52.Nouspikel T, Hanawalt P C. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oelgeschläger T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 54.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X-J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 55.Otrin V R, Kuraoka I, Nardo T, McLenigan M, Eker A P M, Stefanini M, Levine A S, Wood R D. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol Cell Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 57.Ramanathan B, Smerdon M J. Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J Biol Chem. 1989;264:11026–11034. [PubMed] [Google Scholar]
- 58.Reinke H, Gregory P D, Hörz W. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol Cell. 2001;7:529–538. doi: 10.1016/s1097-2765(01)00200-3. [DOI] [PubMed] [Google Scholar]
- 59.Saleh A, Schieltz D, Ting N, McMahon S B, Litchfield D W, Yates III J R, Lees-Miller S P, Cole M D, Brandl C J. Tra1p is a component of the yeast Ada-Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- 60.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 61.Smerdon M J. DNA repair and the role of chromatin structure. Curr Opin Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 62.Smith E R, Belote J M, Schiltz R L, Yang X-J, Moore P A, Berger S L, Nakatani Y, Allis C D. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26:2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 65.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 66.Tang J Y, Hwang B J, Ford J M, Hanawalt P C, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teng Y, Li S, Waters R, Reed S H. Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial Rad16 requisite for repairing upstream control sequences. J Mol Biol. 1997;267:324–337. doi: 10.1006/jmbi.1996.0908. [DOI] [PubMed] [Google Scholar]
- 68.Thoma F. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyases and nucleotide excision repair. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tijsterman M, Tasseron-de Jong J G, van de Putte P, Brouwer J. Transcription-coupled and global genome repair in the Saccharomyces cerevisiae RPB2 gene at nucleotide resolution. Nucleic Acids Res. 1996;24:3499–3506. doi: 10.1093/nar/24.18.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tu Y, Tornaletti S, Pfeifer G P. DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. EMBO J. 1996;15:675–683. [PMC free article] [PubMed] [Google Scholar]
- 71.Ura K, Araki M, Saeki H, Masutani C, Ito T, Iwai S, Mizukoshi T, Kanaeda Y, Hanaoka F. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 2001;20:2004–2014. doi: 10.1093/emboj/20.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 73.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati M L, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 74.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 76.Wallberg A E, Neely K E, Gustafsson J-A, Workman J L, Wright A P H, Grant P A. Histone acetyltransferase complexes can mediate transcriptional activation by the major glucocorticoid receptor activation domain. Mol Cell Biol. 1999;19:5952–5959. doi: 10.1128/mcb.19.9.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 78.Winston F, Sudarsanam P. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- 79.Xu W, Edmondson D G, Roth S Y. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18:5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu W, Edmondson D G, Evrard Y A, Wakamiya M, Behringer R R, Roth S Y. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 81.Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]