Abstract
We have taken a new approach to the identification of E2F-regulated promoters. After modification of a chromatin immunoprecipitation assay, we cloned nine chromatin fragments which represent both strong and weak in vivo E2F binding sites. Further characterization of three of the cloned fragments revealed that they are bound in vivo not only by E2Fs but also by members of the retinoblastoma tumor suppressor protein family and by RNA polymerase II, suggesting that these fragments represent promoters regulated by E2F transcription complexes. In fact, database analysis indicates that all three fragments correspond to genomic DNA located just upstream of start sites for previously identified mRNAs. One clone, ChET 4, corresponds to the promoter region for beclin 1, a candidate tumor suppressor protein. We demonstrate that another of the clones, ChET 8, is strongly bound by E2F family members in vivo but does not contain a consensus E2F binding site. However, this fragment functions as a promoter whose activity can be repressed by E2F1. Finally, we demonstrate that the ChET 9 promoter contains a consensus E2F binding site, can be activated by E2F1, and drives expression of an mRNA that is upregulated in colon and liver tumors. Interestingly, the characterized ChET promoters do not display regulation patterns typical of known E2F target genes in a U937 cell differentiation system. In summary, we have provided evidence that chromatin immunoprecipitation can be used to identify E2F-regulated promoters which contain both consensus and nonconsensus binding sites and have shown that not all E2F-regulated promoters show identical expression profiles.
The E2F family consists of six E2Fs which heterodimerize with one of two different DP proteins to create 12 different DNA binding transcriptional regulators (7, 9). The E2F factors can be divided into three subgroups: (i) E2F1, E2F2, and E2F3, which are highly related and display maximal expression in late G1 to early S phases; (ii) E2F4 and E2F5, which are less responsive to changes in proliferation and lack an N-terminal domain contained within E2Fs 1 to 3; and (iii) E2F6, a recently cloned E2F family member that lacks both the N-terminal region common to E2Fs 1 to 3 and the C-terminal transactivation domain common to E2Fs 1 to 5. Known E2F target genes include those for critical cell cycle regulators (e.g., cyclins, Cdks, and Cdk inhibitors), as well as important mediators of DNA synthesis (e.g., DNA polymerase alpha, DHFR, and thymidine kinase). Genes controlled by E2F show low promoter activity in quiescent and early G1 phase cells and high promoter activity in late G1 and S phase cells. Many studies have shown that the E2Fs can also bind to the pocket proteins retinoblastoma protein (Rb), p107, and p130, and it is believed that the interactions between the pocket proteins and the E2Fs are critical in E2F-mediated cell cycle regulation of transcription (7).
Several lines of evidence suggest that proper regulation of E2F target genes is critical to maintain normal cell proliferation. For example, many human tumors have suffered mutations in the regulators of E2F activity, suggesting that loss of E2F target gene regulation contributes to neoplastic transformation. Also, we and others have shown that overexpression of E2Fs has severe consequences in both normal and neoplastically transformed cells (19, 23, 24, 37, 40, 51). Studies such as these suggest that deregulation of certain E2F target genes is detrimental to proper cell growth control. E2F family members have been reported to bind to and regulate approximately 30 different target genes. However, the fact that E2F overexpression can have severe biological consequences without large changes in expression of these known target genes (reference 24 and unpublished data) suggests that E2F factors may regulate a set of target genes that have not been previously identified by the candidate gene approach. In fact, a recent microarray study suggested that hundreds of genes are affected by the overexpression of E2Fs (31), but the exact role E2F plays in the regulation of these genes needs to be examined in more detail.
Recently, a computer analysis of promoter databases was used to search for new E2F-regulated promoters (21). This study suggested that approximately 7% of mammalian promoters may be regulated by E2F factors. However, our previous approach had two main limitations. First, only previously characterized promoters are present in the current databases; therefore, promoters for as-yet-uncharacterized genes cannot be analyzed. Second, the computer-assisted approach was based on screening for sequences having high homology to the E2F consensus site. This consensus site was developed using a small subset of E2F binding sites identified in cell cycle-regulated promoters. If E2Fs bind to additional sites, either alone or in cooperation with other DNA binding proteins, these sites would be overlooked in the database search. Therefore, it seems clear that it is necessary to take an unbiased approach to identify additional E2F target genes. Accordingly, we have used a modification of a chromatin immunoprecipitation assay to clone novel E2F binding sites, most of which do not have strong homology to the E2F consensus site developed using cell cycle-regulated promoters. Interestingly, characterization of several of these novel E2F binding promoters revealed unique gene expression profiles.
MATERIALS AND METHODS
Cell culture and promoter and mRNA analyses.
HeLa cells were grown in 50% alpha minimal essential medium and 50% Joklik's medium with 5% supplemental calf serum (HyClone) and 1% penicillin-streptomycin (Gibco). The cells were grown to a density of 2 × 105 to 5 × 105 per ml before being harvested for cross-linking experiments. For all chromatin immunoprecipitation experiments presented in this study, asynchronously growing HeLa cells were used. For analysis of transcriptional properties of the ChET (for chromatin-precipitated E2F target) 8 promoter, a fragment was obtained by PCR using primers having sequences complementary to the −477 to + 94 region of the ChET 8 promoter plus restriction sites. The ChET 9 promoter studies were performed with a 293-bp segment of the ChET 9 clone. The PCR fragment was digested and inserted in either orientation into the HindIII site in pGL2 basic (Promega). For analysis of promoter activity and responsiveness to E2F1, NIH 3T3 cells (American Type Culture Collection) were maintained and transfected as described previously (14). pCMVE2F1 and the control vector pcDNA3 were described previously (25). U937 cells were maintained and RNA was prepared as described previously (8). Human tissue was procured at the University of Wisconsin Surgical Pathology Department; as required by our institutional review board protocol, the identities of the patients were unknown. The excess tissue was frozen after surgery, stored at −70°C, and prepared as described previously (11). Reverse transcription (RT)-PCR analyses were performed as described previously (11). Details of the primers used and the required hybridization temperatures can be found on our website at http://mcardle.oncology.wisc.edu/farnham. All primers were synthesized at the University of Wisconsin Biotechnology Center.
Chromatin immunoprecipitation.
Formaldehyde (Fisher Scientific) was added at a final concentration of 1% directly to cell culture media of nonadherent log-phase HeLa cells. Fixation proceeded at 22°C for 10 min and was stopped by the addition of glycine to a final concentration of 0.125 M. The HeLa cells were collected by centrifugation and rinsed in cold phosphate-buffered saline. The cell pellets were resuspended in swelling buffer (10 mM potassium acetate, 15 mM magnesium acetate, 0.1 M Tris [pH 7.6], 0.5 mM phenylmethylsulfonyl fluoride, and 100 ng of leupeptin and aprotinin/ml), incubated on ice for 20 min, and then Dounce homogenized. The nuclei were collected by microcentrifugation and then resuspended in sonication buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 0.5 mM phenylmethylsulfonyl fluoride, and 100 ng of leupeptin and aprotinin/ml) and incubated on ice for 10 min. Prior to sonication, 0.1 g of glass beads (212- to 300-μm diameter; Sigma) was added to each sample. The samples were sonicated on ice with an Ultrasonics sonicator at setting 10 for six 20-s pulses to an average length of approximately 1,000 bp and then microcentrifuged. The chromatin solution was precleared with the addition of Staphylococcus aureus protein A-positive cells for 15 min at 4°C. Prior to use, the Staph A cells were blocked with 1 μg of sheared herring sperm DNA/μl and 1 μg of bovine serum albumin/μl for at least 4 h at 4°C. Precleared chromatin from 107 cells was incubated with 1 μg of affinity-purified rabbit polyclonal antibody or no antibody and rotated at 4°C for approximately 12 to 16 h. Antibodies used included E2F1 no. 05-379 (UBI), E2F2 C-20 no. SC-633X (Santa Cruz), E2F3 C-18 no. SC-878X (Santa Cruz), E2F4 C-20 no. SC-866X (Santa Cruz), E2F5 E-19 no. SC999X (Santa Cruz), E2F6 E-20 no. SC-8366 (Santa Cruz), RNA polymerase II (a gift from David Bentley), p107 C-18 no. SC318X, p130 C-20 no. SC 317X, and Rb C-15 no. SC-50X. Immunoprecipitation, washing, and elution of immune complexes was carried out as previously described (3). Prior to the first wash, 20% of the supernatant from the reaction with no primary antibody for each time point was saved as total input chromatin and was processed with the eluted immunoprecipitates beginning at the cross-link reversal step. Cross-links were reversed by the addition of NaCl to a final concentration of 200 mM, and RNA was removed by the addition of 10 μg of RNase A per sample followed by incubation at 65°C for 4 to 5 h. The samples were then precipitated at −20°C overnight by the addition of 2.5 volumes of ethanol and then pelleted by microcentrifugation. The samples were resuspended in 100 μl of Tris-EDTA (pH 7.5), 25 μl of 5× proteinase K buffer (1.25% sodium dodecyl sulfate, 50 mM Tris [pH 7.5], and 25 mM EDTA), and 1.5 μl of proteinase K (Boehringer Mannheim) and incubated at 45°C for 2 h. Samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) followed by extraction with chloroform-isoamyl alcohol and then precipitated with 1/10 volume of 3 M NaOAc (pH 5.3), 5 μg of glycogen, and 2.5 volumes of ethanol. The pellets were collected by microcentrifugation, resuspended in 30 μl of H2O, and analyzed by PCR. A detailed protocol can be found at http://mcardle.oncology.wisc.edu/farnham.
PCR mixtures contained 2 μl of immunoprecipitate or 2 μl of a 1:100 dilution of the total sample; 50 ng of each primer; 0.88 mM MgCl2; 0.2 mM (each) dATP, dCTP, dGTP, and dTTP; 1× thermophilic buffer (Promega); and 1.25 U of Taq DNA polymerase (Promega) in a total volume of 20 μl. Following 32 to 35 cycles of amplification, the PCR products were run on a 1.0% agarose gel and analyzed by ethidium bromide staining. The PCR primers used to analyze target genes can be found on our web site (http://mcardle.oncology.wisc.edu/farnham).
Cloning novel E2F targets.
Several modifications of the chromatin immunoprecipitation protocol were required for the cloning of novel target genes. The first modification occurred at the elution step. Instead of eluting twice using 150 μl of elution buffer each time, the immunoprecipitated chromatin was eluted from the Staph A cells once, using 30 μl of elution buffer. The eluate was then diluted with 270 μl of immunoprecipitation dilution buffer to a total volume of 300 μl, and a new aliquot of the same antibody as that used in the first immunoprecipitation was added for an overnight incubation at 4°C. The next morning, the samples were processed in the standard manner (i.e., they were washed and eluted, cross-links were reversed, and the samples were proteinase K digested, followed by phenol extraction and ethanol precipitation). At this point, an aliquot of the immunoprecipitated samples was used in a reaction with b-myb or dhfr primers to demonstrate that E2F targets had been selected. The remaining immunoprecipitated DNA was then treated with T4 DNA polymerase and cloned into either the zero-blunt vector (Invitrogen) or HincII-digested puc 19. Colonies having inserts were identified by restriction enzyme digestion using enzymes in the polylinker. Plasmids having inserts greater than 500 bp were chosen for further analysis. The sequence of each of the cloned fragments, details concerning the sequences of the primers used to analyze each clone, the required hybridization temperatures, and the product sizes can be found on our web site (http://mcardle.oncology.wisc.edu/farnham). All primers were obtained at the University of Wisconsin Biotechnology Center.
Electromobility shift assays.
In vitro E2F DNA binding activity was assayed by incubating about 6 μg of HeLa nuclear extract with 2.5 μg of sonicated salmon sperm DNA and 2 μl of 5X-500 buffer (100 mM HEPES [pH 7.4], 500 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 35% glycerol, and 5 mM NaF) in a total volume of 18 μl for 10 min at room temperature. Either a 34-bp double-stranded oligonucleotide containing the E2F site from the b-myb promoter or a 22-bp double-stranded oligonucleotide containing the E2F site from ChET 9 (both of which were end labeled using T4 DNA kinase and [γ-32P]ATP as described previously [28]) was then added in 2 μl of water, and the incubation continued for 20 min. The reactions were electrophoresed for 2 h on a 4% polyacrylamide gel that had been preelectrophoresed for 30 min. When competition assays were performed, the competitor DNA was included in the first incubation at a 50-fold molar excess to the labeled probe.
RESULTS
In vivo isolation of E2F binding sites.
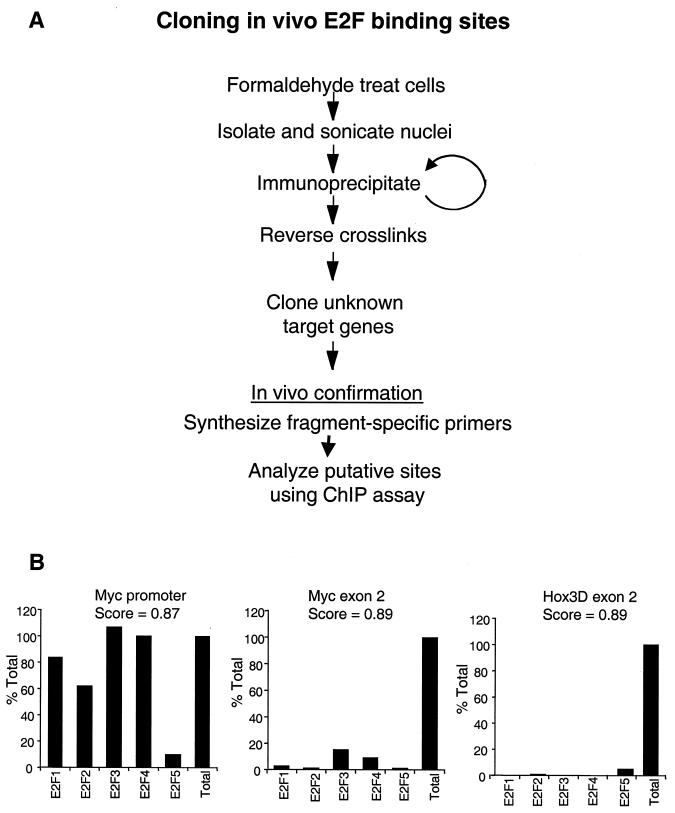
Conventional target gene identification methods, such as microarray analysis or subtractive hybridization, examine changes in gene expression profiles. Although valuable information can be obtained from such studies, the disadvantage of using these techniques to clone target genes of transcription factors is that the observed changes may in fact be the result of indirect pathways influencing gene expression patterns. For example, approximately 7% of the mRNAs on a human cDNA microarray responded to overexpression of an E2F (31). However, promoter analyses were not performed on these genes. Therefore it is not known if the genes in that study are regulated by promoters that contain consensus E2F binding sites or if the promoters are directly bound by E2F. In contrast, one major advantage of utilizing the chromatin immunoprecipitation method to identify novel target genes is that it selects for sites bound by the transcription factor of interest, thus eliminating the problem of indirect effects. Therefore, we have chosen to use chromatin immunoprecipitation as an unbiased approach to identify in vivo E2F binding sites (Fig. 1A).
FIG. 1.
(A) Schematic of the E2F chromatin immunoprecipitation cloning procedure. (B) Graphical representation of the results of a chromatin immunoprecipitation experiment measuring E2F binding at the Myc promoter, Myc exon 2, and Hox3D exon 2. Scores representing homology to the E2F consensus site as determined by computer analysis (21) are shown at the top of each graph. The y axis represents Imagequant quantitation of the amount of specific PCR products expressed as the percentage of antibody binding versus the amount of PCR product obtained using a standarized aliquot of input chromatin. The signal in the no-antibody lane was subtracted from each sample as a nonspecific binding background. The E2F family members used in the immunoprecipitation are shown on the x axis.
Briefly, following formaldehyde cross-linking of cells, chromatin is isolated and sheared to a desired length by sonication. Immunoprecipitation proceeds with an antibody to a factor of interest to selectively precipitate that protein and any DNA fragment cross-linked to it. Thus, DNA sequence elements associated with the desired protein in the context of the cellular environment are enriched in the immunoprecipitated sample. After reversal of the formaldehyde cross-links and purification of the DNA, the precipitated DNA fragments are cloned into a vector for isolation and further characterization of factor binding sites and functional relevance. Although the protocol for cloning E2F targets is similar to the standard chromatin immunoprecipitation assay that has been previously described (3, 33, 49), several modifications were made to the assay. Importantly, we performed two sequential chromatin immunoprecipitations using the same antibody for both the first and second steps. In preliminary studies, we found that a large number of nonspecific fragments were cloned if only one immunoprecipitation was performed. Therefore, the second immunoprecipitation was used to decrease the amount of nonspecific DNA present to enable more efficient cloning of the specific fragments. Also, a portion of the immunoprecipitated samples was retained prior to cloning to analyze known target genes as a positive control for the immunoprecipitation. Other modifications important for cloning the immunoprecipitated products can be found on our web site (http://mcardle.oncology.wisc.edu/farnham).
Previous studies have demonstrated the validity of using the chromatin immunoprecipitation protocol to identify site-specific interactions of transcription factors and promoter DNA in the context of a living cell. Importantly, we have shown that binding of E2F to target promoters is site specific. For example, the chromatin immunoprecipitation assay has been used to demonstrate that E2F4 binds with high affinity to the dhfr promoter (which is known to be regulated by E2F) but does not bind to the cad promoter (which is a Myc target gene) (3). Also, in a previous report (49) we showed that a site-specific mutation in the dhfr promoter eliminates binding of E2F4, as monitored by the chromatin immunoprecipitation assay. In addition, high-affinity binding of E2F family members to the myc promoter is abolished when a point mutation is introduced into the E2F site of that promoter (2). Finally, we have also shown that E2Fs do not bind to promoters which are regulated by liver-specific transcription factors (C. R. Graveel and P. J. Farnham, unpublished data). Clearly, the chromatin immunoprecipitation assay has been useful in demonstrating that not all cellular promoters bind E2Fs and that those that do require a specific site on the DNA for high-affinity in vivo binding. However, we felt that additional controls were needed prior to using this assay to clone novel targets. Namely, we wished to clone regulatory E2F binding sites and not clone random, nonfunctional E2F binding sites present in nonpromoter regions of the genome. Previous analysis of promoter and exon 2 databases suggested that E2F sites are found at a much higher frequency in promoters (21). However, sequences having high-score matches to the E2F consensus site (scores of 0.86 or better in the computer analysis) can be found in nonpromoter regions as well. To explore whether E2F is bound to these sites in living cells, we identified two such consensus sites located in the second exon of the myc and the hox3D genes. Because the myc promoter has previously been shown to be an E2F target (30, 34, 50), we examined binding of E2F family members to the exon sites in comparison to binding at the E2F site in the Myc promoter. As shown in Fig. 1B, E2F binding to the myc gene promoter is at much higher levels than binding to the myc and hox3D exon 2 regions in the same chromatin sample, even though the score match to the consensus E2F site is very high in the exon 2 regions.
Having assured ourselves that the chromatin immunoprecipitation assay can be used to detect promoter-specific and site-specific binding of E2F, we performed immunoprecipitations from HeLa cell chromatin using antibodies against either E2F1 or E2F4 and proceeded with the cloning procedure. We chose clones having inserts of 500 bp or greater and examined 11 clones obtained by immunoprecipitation with the E2F1 antibody and 7 clones obtained by immunoprecipitation with the E2F4 antibody for further analysis. The first step in the characterization of the ChET clones was to determine which ones contained bona fide E2F binding sites and which were false positives.
Confirmation of E2F binding.
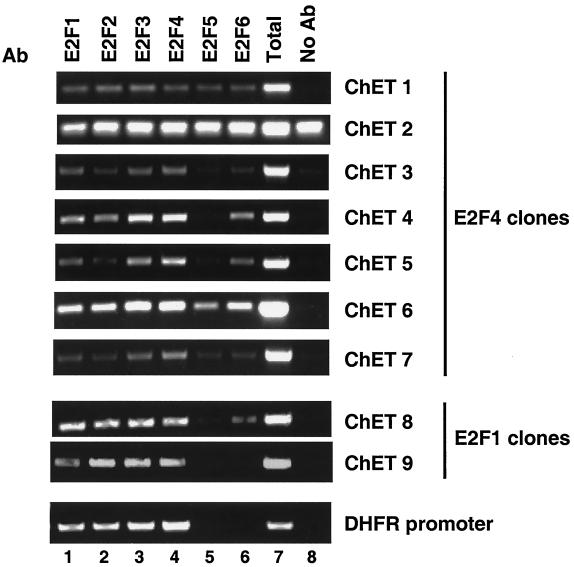
In vitro assays may bias results towards identifying sites which closely resemble the known consensus E2F binding site. In addition, any large genomic fragment may, by chance, contain a sequence resembling an E2F site and be scored as a positive in an in vitro assay. Therefore, we began confirmation of our cloned fragments using an in vivo assay. The ChET clone inserts were sequenced, and primers were made for use in chromatin immunoprecipitation experiments. Because the E2F binding site could be anywhere within the cloned fragment, primers were designed to analyze both ends of each clone. HeLa chromatin was immunoprecipitated using antibodies to E2Fs 1 to 6; a no-antibody negative control was also performed (Fig. 2). As a positive control, binding of E2F family members to the dhfr promoter was examined. For each primer set, a standardized aliquot of the input chromatin (total) was also analyzed. The ratio of the signals detected in the lanes containing samples immunoprecipitated by the various antibodies to the signal detected in the total lane allows a relative comparison representing the degrees of occupancy at the different sites. Figure 2 shows the results of a representative experiment using the primer sets that showed the highest-affinity E2F binding for each clone.
FIG. 2.
Confirmation of ChET clones by examining E2F binding in vivo. A representative chromatin immunoprecipitation experiment with HeLa cells is shown. Immunoprecipitation proceeded utilizing antibodies (Ab) against E2F1 (lane 1), E2F2 (lane 2), E2F3 (lane 3), E2F4 (lane 4), E2F5 (lane 5), and E2F6 (lane 6) or no antibody (lane 8). Following DNA purification, samples were subjected to PCR with primers designed for the individual E2F clones or the dhfr promoter as a control (labeled on the right). A portion of the total input was also examined by PCR (lane 7).
We began by testing the clones obtained using an E2F4 antibody in the immunoprecipitation step. Seven clones from the E2F4 immunoprecipitation were analyzed for E2F binding in vivo (Fig. 2). Of these, one clone (ChET 2) was found to be present in samples in which no antibody was added to the immunoprecipitation reaction mixture, indicating that binding was nonspecific. The remaining six clones were all bound specifically by E2F family members in numerous independent chromatin immunoprecipitation experiments. Although several of these clones (e.g., ChET 1) were of modest affinity, other clones (e.g., ChET 4) showed robust binding of E2Fs. Eleven clones obtained by immunoprecipitation with an E2F1 antibody were also examined to determine whether they were bound by E2Fs in vivo. We were unable to optimize primer sets to four of these clones for analysis in subsequent chromatin immunoprecipitation assays; these four clones will be included in future studies if optimal primer sets can be empirically determined. Therefore, we were left with seven clones for in vivo analysis. We could not confirm binding of E2Fs to four of these clones in subsequent chromatin immunoprecipitation assays; an example of one such false positive is shown below (see Fig. 4, ChET 10). However, three of the seven E2F1 clones contained bona fide E2F binding sites as determined by chromatin immunoprecipitation analysis. The ChET 8 and ChET 9 clones both contained high-affinity E2F binding sites in comparison to the binding detected at the well-characterized dhfr promoter (Fig. 2). Binding to the third E2F1 clone was of modest affinity (data not shown). In summary, of the 14 clones that we were able to examine using in vivo assays (7 obtained using an E2F1 antibody and 7 obtained using an E2F4 antibody), 9 were confirmed to contain bona fide in vivo E2F binding sites (Table 1). Although false positives are unavoidable, we believe that the two sequential immunoprecipitation reactions greatly enriched for clones containing bona fide E2F binding sites. It is important to note that the in vivo binding of E2Fs to the ChET clones has been confirmed in numerous experiments. For example, the experiments shown in Fig. 2 and below (see Fig. 4) are completely independent from each other and from the chromatin immunoprecipitation experiment used to clone the fragments. We have also confirmed binding of E2Fs to ChET 4, ChET 8, and ChET 9 in other human cell types (data not shown).
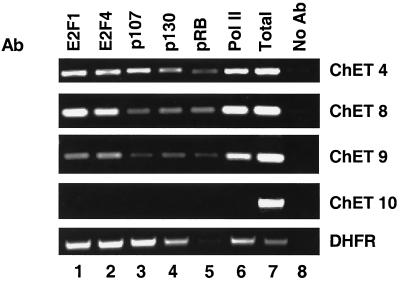
FIG. 4.
Characterization of the protein complexes bound in vivo to the ChET clones. A chromatin immunoprecipitation experiment was performed in HeLa cells utilizing antibodies (Ab) to E2F1 (lane 1), E2F4 (lane 2), p107 (lane 3), p130 (lane 4), Rb (lane 5), RNA polymerase II (Pol II; lane 6), or no antibody as a control (lane 8). An aliquot of the total input is also shown (lane 7). Primers to the ChET clones or the DHFR promoter were used in PCRs for analysis.
TABLE 1.
Summary of ChET cloning
| Parameter | Value
|
||
|---|---|---|---|
| E2F4a | E2F1a | Total (%) | |
| No. of fragments analyzed | 7 | 7 | 14 |
| No. of false positives | 1 | 4 | 5 (36) |
| No. of clones specifically bound by E2Fs | 6 | 3 | 9 (64) |
Antibody to the indicated E2F was used to clone the fragments.
Either an E2F1 or an E2F4 antibody was initially used for the cloning procedure; however, we found that the cloned sites did not reveal an E2F binding pattern to suggest family member binding specificity. Rather, binding of multiple E2F family members to each clone was detected in vivo (Fig. 2). These results, suggesting a lack of DNA binding specificity among the different E2Fs, are similar to those of our previous studies of known E2F target genes (49). E2F family members contain a highly conserved DNA binding domain; therefore, it is not surprising that these family members have the ability to bind to the same sequence in vivo. It is unlikely that our results indicate that multiple E2F family members are simultaneously binding to the same site, but the more likely explanation is that a dynamic exchange occurs at a given site and various E2Fs are trapped in different cells during the cross-linking procedure. There are precedents for this hypothesis, as dynamic exchange of the glucocorticoid receptor has been demonstrated in living cells (29). With this in mind, it is worth noting that the cloned sites can be separated into three basic categories. The first type (e.g., ChET 9) showed an in vivo binding pattern very similar to that of the dhfr promoter, i.e., strong binding of E2F1 to E2F4 and very little binding of E2F5 or E2F6. The second type (e.g., ChET 4 and ChET 8) showed strong binding of E2F1 to E2F4 and little binding of E2F5 but detectable binding of E2F6. Finally, the third type of site (e.g., ChET 1) showed equal low-affinity binding of E2F1 to E2F6. The significance of these distinct binding patterns is unknown, and further analysis will be required to elucidate the functional consequences, if any.
Genomic organization of ChET clones.
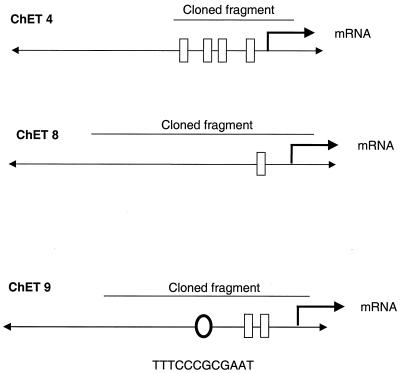
We next analyzed the sequences of the cloned fragments to determine if anything was known about their identities. Three of the high-affinity clones (ChET 4, ChET 8, and ChET 9) contained extremely GC-rich sequences, which is a hallmark of many E2F-regulated promoters. In addition, the PromoterInspector program (39) detected a promoter region in ChET 9 and the CpG Promoter program (15) detected a promoter-associated CpG island in ChET 4 and ChET 8. Most E2F-regulated promoters have E2F binding sites located in close proximity to the start site of transcription (21). If our cloned fragments do, in fact, represent promoters, then it is possible that the sequences just downstream of the E2F sites correspond to transcribed regions. To test this hypothesis, we first identified the region of the human genome corresponding to the cloned fragments and then compared several kilobases of sequence on either side of the cloned fragments to the GenBank database for a potential cDNA match (Fig. 3). Interestingly, we found that a previously identified but uncharacterized mRNA begins within the ChET 8 clone approximately 130 bp from one end. Also, a previously uncharacterized mRNA begins within ChET 9 (although the exact 5′ end of this mRNA has not yet been determined). Finally, the 5′ end of the mRNA encoding Beclin 1, a candidate tumor suppressor protein, is contained within the ChET 4 clone.
FIG. 3.
Genomic organization of the ChET clones. GenBank database searches were performed with the sequences corresponding to ChET 4, ChET 8, and ChET 9. After the locations of the clones were determined, further Blast searches were performed examining the sequences immediately adjacent to the cloned fragments. The locations of adjacent mRNAs are indicated by bent arrows. Positions of consensus Sp1 (rectangles) and E2F (oval) sites are also shown.
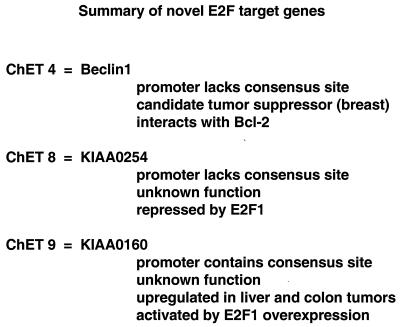
In summary, it appears that the three high-affinity E2F binding sites all correspond to promoter regions: ChET 8 is the promoter region for a 6,049-nt mRNA termed KIAA0254 (accession number D87443), which encodes a 1,009-amino-acid protein with no homology to other known proteins; ChET 9 is the promoter region for a 4,441-nt mRNA termed KIAA0160 (accession number D63881), which encodes an 803-amino-acid protein having extensive homology to the Drosophila protein Suppressor of zeste 12 [Su(z)12]; and ChET 4 is the promoter for a 2,098-nt mRNA encoding Beclin 1 (accession number AF139131), which is a candidate tumor suppressor gene. Of the five remaining E2F4 ChET clones, four can be identified in the human raw-sequence database. We have searched the GenBank database using several kilobases of surrounding sequence but have not identified mRNAs associated with these clones. Therefore, it is still unclear whether the remaining E2F4 ChET clones correspond to promoter regions.
Characterization of the novel E2F binding sites.
The next step in the characterization of the ChET clones was to determine the compositions of the protein complexes recruited to the clones in vivo. E2Fs bind directly to Rb, p107, and p130, and we have previously shown that these proteins are components of the transcription complexes formed on E2F target promoters in vivo (49). E2Fs also bind to basal transcription factors, such as TBP and TFIIH (35); therefore, it is likely that components of the RNA polymerase II transcription complex are also recruited to E2F target promoters. To examine these possibilities, chromatin immunoprecipitation experiments were performed using antibodies to E2Fs, pocket proteins, and RNA polymerase II (Fig. 4). For these and the remaining experiments, we chose to focus on the three clones which have the highest levels of in vivo E2F binding, ChET 4, ChET 8, and ChET 9. The data from the chromatin immunoprecipitation experiment shown in Fig. 4 suggest that, similar to known E2F binding sites, such as dhfr, the novel E2F binding sites also recruit pocket proteins. Interestingly, the binding profiles for the pocket proteins observed at the three novel clones vary in comparison to that observed at the dhfr promoter. The dhfr promoter is bound mainly by p107 and p130, with very little bound Rb, whereas the novel clones recruited almost equivalent levels of the three pocket proteins. Because the use of HeLa cells may have given altered pocket protein binding due to the expression of the viral E7 protein, we also examined pocket protein binding to the dhfr promoter and to the three ChET clones in U937 cells. We found very similar recruitment of pocket proteins to the E2F target promoters in the U937 cells and in HeLa cells (data not shown). In addition, the novel E2F binding sites recruited RNA polymerase II, suggesting that the three high-affinity clones indeed represent promoter sequences.
We wished to further characterize a subset of the ChET promoter clones. We chose the ChET 8 and ChET 9 clones for an initial promoter analysis. If we are correct in assuming that the ChET 8 clone represents the promoter for the KIAA0254 mRNA, then it should have promoter activity when inserted in the correct orientation with the transcription start site upstream of the luciferase cDNA. The ChET 8 fragment was cloned in both orientations (forward and reverse) upstream of the luciferase cDNA and transfected into NIH 3T3 cells, and promoter activity was measured. The cdc2 promoter-luciferase reporter construct was used in these experiments as a positive control. As shown in Fig. 5A, only the forward orientation of the ChET 8 fragment showed high promoter activity, whereas the reverse orientation did not. In fact, the ChET 8 promoter was considerably more active than the cdc2 promoter, which is a strong E2F-regulated promoter. Importantly, the orientation of the ChET 8 fragment that showed promoter activity was the correct orientation to drive KIAA0254 mRNA transcription.
FIG. 5.
Transient-transfection analysis of ChET promoter-luciferase reporters. (A) A transient-transfection experiment in NIH 3T3 cells was performed with a segment of ChET 8 cloned in either the forward or reverse orientation upstream of luciferase. The y axis of the graph represents the relative luciferase units, with the transfected material shown on the x axis. pGL2 represents the luciferase vector lacking a promoter. (B) A transient-transfection analysis was performed with the ChET 8 promoter-luciferase reporter transfected into NIH 3T3 cells in the presence of 2 μg of an E2F1 expression vector (cytomegalovirus [CMV] E2F1) or the pCDNA3 vector as a control. The cdc2 promoter-luciferase reporter construct was used as a control in both panels A and B. (C) Transient-transfection analysis of the ChET 9-luciferase reporter construct was performed in NIH 3T3 cells. A graphical representation of the results is shown with the dhfr promoter used as a positive control. (D) Overexpression of E2F1 upregulates ChET 9 promoter activity. Cotransfection experiments were performed containing 2 μg of a CMV E2F1 expression construct with the ChET 9-luciferase construct or the dhfr-luciferase reporter vector as a control. Results of the luciferase assay are shown in the graph as indicated for panel A.
We have previously shown that E2F site-containing promoters can be activated by cotransfection with E2F1 (25). To determine if binding of E2F influences the transcriptional activity of the ChET 8 promoter, we cotransfected the ChET 8 promoter-luciferase reporter construct with an E2F1 expression construct, again using cdc2-luciferase as a positive control (Fig. 5B). We found that, as expected, E2F1 overexpression activated the cdc2 promoter. In contrast, E2F1 repressed transcription from the ChET 8 promoter. Although the exact mechanism by which E2F1 represses the ChET 8 promoter is still unknown, preliminary analyses indicate that squelching (i.e., the sequestration of coactivators and/or general transcription factors by a transactivation domain) is most likely not the mechanism. For example, an E2F1 construct with the entire transactivation domain deleted is still a potent repressor of ChET 8 promoter activity (data not shown). The potential relevance of these findings will be discussed further below (see Discussion).
The ChET 9 fragment was also cloned into the luciferase reporter vector for further analysis to determine if indeed this cloned sequence contains promoter activity. A transient transfection analysis of the ChET 9 luciferase reporter construct demonstrated that the ChET 9 clone has promoter activity (Fig. 5C) which is approximately equal to the activity of the dhfr promoter (Fig. 5D). In addition, the overexpression of E2F1 stimulates ChET 9 promoter activity (Fig. 5D). An extensive characterization of the ChET 9 promoter is in progress; however, it is worth noting that deletion of the region containing the consensus E2F site eliminates the E2F1-mediated transactivation (data not shown). These results indicate that the ChET 9 clone has promoter activity and indeed contains an E2F site which is functional in a transient overexpression system.
Localization of potential E2F sites.
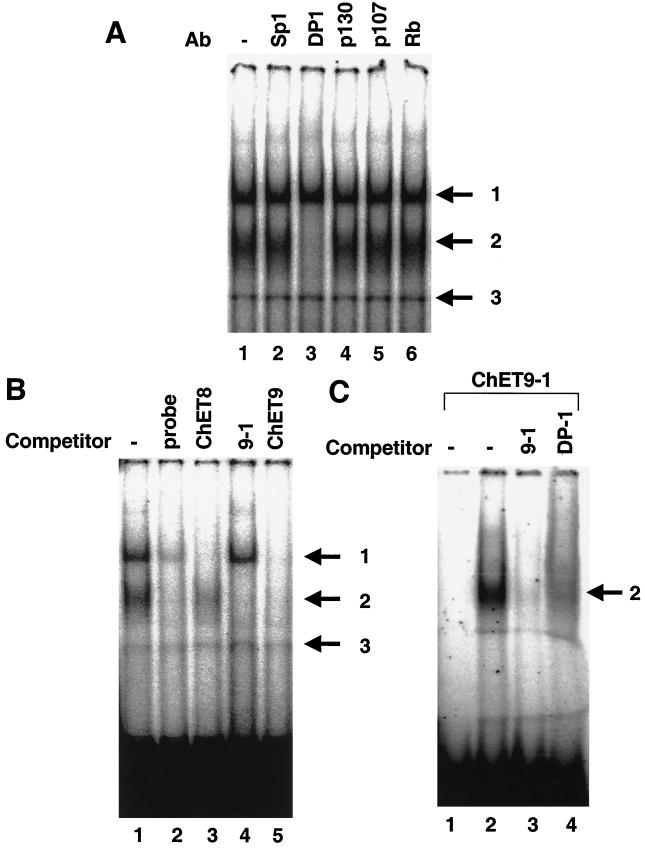
Visual inspection of the sequences corresponding to the three cloned promoter fragments bound by E2F family members in vivo revealed that only one clone, ChET 9, contains a consensus E2F site. Using a computer program which has previously been shown to effectively predict consensus E2F binding sites (21), we next examined the clones for matches to the E2F consensus sequence. The cutoff previously used for identifying E2F sites was a 0.86 similarity to the consensus (21). Using this criterion, once again the only clone containing a consensus E2F site was ChET 9. We thought that it might be possible to identify the E2F binding sites within the ChET clones by using an in vitro assay. Therefore, electromobility shift assays (EMSA) were performed using the well-characterized E2F binding site in the b-myb promoter as a probe. Two bands (complexes 1 and 2) which represent specific protein-DNA complexes were observed, as determined by antibody disruption (Fig. 6A) and competition with the unlabeled probe (Fig. 6B); an additional nonspecific band (complex 3) was also observed in most reactions. Because each of the six E2Fs heterodimerizes with DP-1, we used the DP-1 antibody to show that complex 2 contains an E2F-DP heterodimer (Fig. 6A). Although complex 1 is specifically competed by the probe (Fig. 6B), the DP-1 antibody did not disrupt it. We (reference 24 and data not shown) and others (41) have previously shown that a low-mobility complex which binds specifically to E2F probes cannot be identified by using antibodies to the different E2F family members. It is possible that this complex represents an uncharacterized E2F family member, or the epitope may be obscured by another protein. To distinguish these possibilities, we employed antibodies to the pocket proteins. However, antibodies to the pocket proteins did not disrupt complex 1 (Fig. 6A). Therefore, the identities of the proteins composing complex 1 are still unknown.
FIG. 6.
In vitro EMSA of cloned fragments. In each panel (arrows), complex 3 represents a nonspecific band, complex 2 indicates an E2F-DP complex, and complex 1 indicates a complex containing proteins that have not yet been identified. (A) Supershift EMSA was performed to determine the components of the gel-shifted complexes. Reaction mixtures containing HeLa nuclear extract were incubated with an antibody (Ab) to Sp1 (lane 2), DP-1 (lane 3), p130 (lane 4), p107 (lane 5), or Rb (lane 6) or no antibody (lane 1) followed by incubation with the b-myb E2F site labeled as a probe. (B) EMSA competition experiments using the E2F site from the b-myb promoter as a probe. Oligonucleotides corresponding to the unlabeled probe (lane 2), a fragment spanning the ChET 8 transcription start site (lane 3), an oligonucleotide corresponding to the consensus E2F site from ChET 9 (lane 4), or the ChET 9 fragment (lane 5) were used as competitors. (C) EMSA using the ChET 9 E2F site as a probe. A double-stranded oligonucleotide containing the consensus E2F site within the ChET 9 fragment (the sequence is shown in Fig. 3) was radiolabeled and used for EMSA. The probe was incubated with HeLa nuclear extract and the unlabeled probe (lane 3), a DP-1 antibody (lane 4), or extract alone (lane 2). Lane 1 represents an aliquot of the probe without extract incubation. An arrow to the right of the gel image indicates the specific E2F-DP complex.
There are no consensus E2F binding sites located in the ChET 8 clone; therefore, it was not apparent which region was responsible for the recruitment of E2F to the promoter region. Because E2F sites are often found within 50 bp of the transcription start site (21), we prepared a fragment which surrounds the transcription start site for use as a competitor in an EMSA competition experiment (Fig. 6B). Competition was observed by using this promoter-proximal fragment; other regions of the ChET 8 clone did not compete the probe (data not shown). Further analyses will have to be performed to determine the exact location of the E2F site within the ChET 8 clone. Because competition was not complete in the in vitro EMSA experiments, an alternative in vivo method for assaying binding activity may need to be developed to localize the E2F site within the cloned fragment. Although a complete mutational analysis of the promoter region is beyond the scope of this initial study, we have recently shown that the region required for responsiveness to E2F in the transient-expression assay (Fig. 5B) resides in the promoter-proximal fragment shown as a competitor in Fig. 6B (unpublished data).
EMSA competition experiments utilizing the oligonucleotide corresponding to the consensus E2F site in the ChET 9 promoter revealed that this site is an efficient competitor of the DP-E2F-DNA complex formed on the b-myb E2F site (Fig. 6B, lane 4). Interestingly, additional sequences located in other regions of the ChET 9 fragment are required to compete binding of complex 1 (Fig. 6B, lane 5). To confirm that E2F has the ability to bind directly to the E2F site identified in ChET 9, we performed EMSA experiments utilizing a radiolabeled probe containing the E2F site located in ChET 9. As shown in Fig. 6C, a single lower-mobility complex is detected when nuclear extract is incubated with the ChET 9 probe. The addition of a DP-1 antibody to this reaction ablated the complex, indicating that this complex contains a DP-1–E2F heterodimer. Thus, the consensus site in ChET 9 competes binding to the E2F site in the b-myb promoter and binds to a protein complex which can be disrupted by a DP-1 antibody. Taken together with the promoter-reporter assays, our results suggest that the consensus E2F site in ChET 9 contributes to in vivo binding and E2F responsiveness.
Characterization of expression patterns.
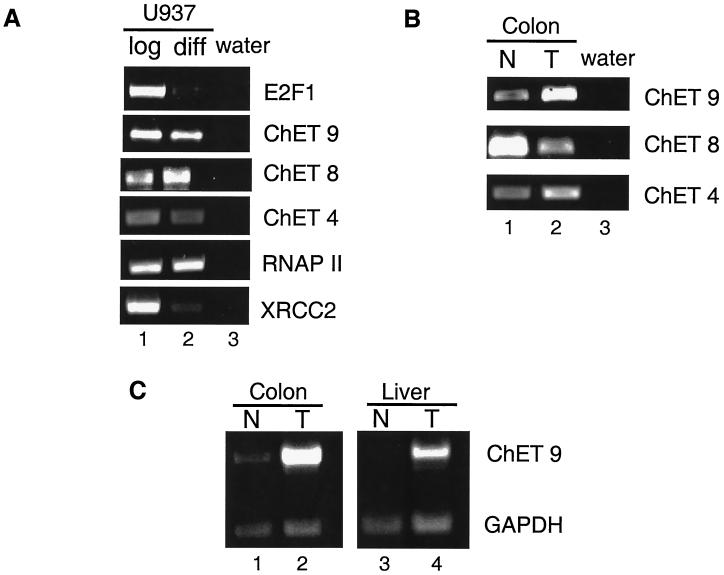
Many E2F-regulated promoters display cell cycle stage-specific transcription patterns. However, almost without exception, the previously identified E2F-regulated promoters were identified from a pool of promoters already known to be cell cycle regulated (42). It seemed possible that promoters identified by using an unbiased approach might show different transcriptional regulation. Therefore, we monitored the expression levels of the mRNAs driven by ChET 4, ChET 8, and ChET 9 in a cell differentiation system. U937 cells were forced to differentiate by treating them with retinoic acid for 5 days, and then RNA was prepared from log-phase (growing) and arrested (differentiated) cells. As a control, we monitored the expression of E2F1, an E2F-regulated gene that has been shown to be downregulated when growing cells exit the cell cycle (14, 18, 32, 43). As expected, levels of E2F1 mRNA decreased upon differentiation and cell cycle arrest (Fig. 7A). In contrast, the levels of mRNA for the three novel clones did not decrease significantly in the U937 cell population upon differentiation, which suggests that these promoters are not cell cycle regulated. These data indicate that E2F family members may regulate both cell cycle- and non cell cycle-responsive promoters. To determine whether another unbiased approach would also yield constitutively active E2F-regulated promoters, we examined the mRNAs for two promoters that were predicted to be regulated by E2F from a computer-assisted identification of consensus E2F binding sites in the promoter regions. The large subunit of RNAPII and XRCC2 were both identified in a previous study which scanned the eukaryotic promoter databases for E2F target promoters (21). Chromatin immunoprecipitation analysis indicated that indeed the promoters for RNAPII large subunit and XRCC2 are bound by E2F (data not shown). Analysis of mRNA levels in the U937 cells indicated that one of the two computer-identified genes was constitutively expressed whereas the other was downregulated upon differentiation. Therefore, four of the five E2F-bound promoters identified by using unbiased approaches are not regulated upon differentiation of U937 cells. It is also important to note that although the amount of E2F1 declines during differentiation of U937 cells, the overall amount of E2F activity remains high due to the constitutive expression of E2F4. In fact, we have shown that the amount of E2F4 bound to the ChET promoters is unchanged after differentiation of U937 cells (data not shown). Perhaps only those E2F target genes which are uniquely responsive to E2F1 versus E2F4 will show a decline in activity upon differentiation.
FIG. 7.
mRNA expression profiles of the three high-affinity ChET clones. (A) RT-PCR analysis of mRNA expression levels in RNA obtained from either U937 log-phase (lane 1) or differentiated (diff; lane 2) cells. RT-PCR primers complementary to E2F1, ChET 9, ChET 8, ChET 4, RNA polymerase (RNAP) II, or XRCC2 were used as indicated on the right. A water control is shown in lane 3. (B) RT-PCR analysis of RNA from either normal (N) colon (lane 1) or colon tumor (T; lane 2). Primer sets to the specific mRNAs are indicated. (C) RT-PCR analysis of ChET 9 mRNA expression in the RNA obtained from either human normal colon (lane 1), colon tumor (lane 2), normal liver (lane 3), or liver tumor (lane 4). The normal colon and colon tumor samples are the same as those shown in panel B with GAPDH primers added as a loading control.
E2F target genes have been suggested to be critical regulators of cell growth control. Therefore, we also examined whether the expression of ChET 4, ChET 8, and ChET 9 is altered during neoplastic transformation. We first compared the levels of mRNAs in samples of human normal colon and colon tumor taken from the same patient (Fig. 7B). In RT-PCR analysis, ChET 4 had slightly higher levels of mRNA expression in tumor tissue, ChET 8 showed a slight decrease in mRNA levels in the tumor sample, and ChET 9 was significantly upregulated in the colon tumor tissue. To determine if upregulation of ChET 9 mRNA was specific to colon tumors, we also compared normal liver and liver tumor mRNA levels. To control for equivalent RNA sample concentrations, primers for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were included in the reaction mixtures. The results show that ChET 9 mRNA was upregulated in both tumor types (Fig. 7C), which suggests that ChET 9 expression may be generally deregulated in human tumors. We have also confirmed that ChET 9 mRNA is upregulated in eight of nine additional human colon tumor samples (unpublished data).
DISCUSSION
To our knowledge, this is the first demonstration that the chromatin immunoprecipitation assay can be used to clone promoters which are direct in vivo targets of a mammalian site-specific DNA binding protein. This provides a powerful new approach to examine direct transcription factor targets in an unbiased manner which does not rely on previous characterization of a consensus sequence or a prior knowledge of gene expression patterns. Although others have used similar approaches to isolate genomic fragments (6, 36, 38), those studies did not use subsequent experiments to confirm in vivo binding of the factor of interest to the isolated DNA. Due to this lack of in vivo confirmation, it is difficult to assess the validity of the previous protocols. Also, sequence analysis of the clones isolated in the previous studies indicated that the cloned fragments corresponded to nonpromoter regions, such as introns (5, 10, 22, 45, 46). One study did find that 3 out of 43 clones isolated after in vitro incubation of genomic DNA with purified Ets-1 protein were promoters; however, the authors did not confirm in vivo binding of ETS1 to these 3 clones or to the other 40 isolated clones. Therefore, it is difficult to be sure if any of the clones in that study were bona fide in vivo targets of ETS1.
Utilizing the chromatin immunoprecipitation assay to clone fragments bound by E2F family members, we found that 64% (9 of 14) of the clones characterized were bona fide in vivo E2F binding sites. Characterization of the three highest-affinity clones revealed that they correspond to promoter regions (Fig. 8), providing validation that novel E2F-regulated promoters can be isolated by using this protocol. Future studies will be performed to determine whether any of the remaining clones are promoters. A recent study using high-density microarray analysis (31) found that about 7% of the mRNAs represented on the microarray were responsive to overexpression of E2Fs. These results suggest that a high percentage of mammalian genes might be regulated by direct binding of E2F to the promoter region. If this estimate of the number of E2F target genes is correct, then it is not surprising that we did not isolate one of the several dozen well-characterized E2F target promoters in the set of nine positive clones that we analyzed. However, one of our ChET clones (ChET 9, which corresponds to the promoter region of the KIAA0160 gene) was shown to be upregulated by E2F overexpression in the microarray analysis (31). The fact that this gene was isolated by two independent screening methods for E2F target genes provides strong evidence that this promoter is indeed a direct target of E2F family members and that the chromatin immunoprecipitation cloning technique can identify E2F-regulated promoters.
FIG. 8.
Summary of information obtained relating to the ChET promoter clones.
One of the E2F4 clones, ChET 4, displayed high-affinity E2F binding in vivo and corresponded to the promoter region for the beclin 1 gene. Beclin 1 was isolated through its ability to interact with bcl-2 and has been postulated to possess tumor suppressor activity in breast cancer (1). It is interesting that the gene for a potential tumor suppressor protein was isolated as an E2F target gene because E2F regulation is thought to play a significant role in tumorigenisis. Further experiments examining the nature of the role E2F plays in Beclin 1 regulation may provide further insight into the role of E2F in tumor development.
We found that two of the high-affinity E2F binding clones did not contain E2F sites which closely matched the consensus sequence. It is important to note that others have previously shown that site-specific DNA binding proteins can regulate transcription through sequence elements that diverge from the consensus. For example, CREB, Ets-1, and AML1 can regulate expression of the human T-cell receptor beta chain promoter through nonconsensus binding sites (12) and a nonconsensus site mediates regulation of the atrial natriuretic factor by serum response factor (13). Computer inspection suggests that the ChET clones may contain multiple low-affinity E2F sites, each of which diverges from the known consensus. Perhaps a combination of weak binding sites allows for cooperative recruitment of the E2F complex in vivo. It is also possible that the promoter context may greatly influence E2F binding efficiency within the cellular environment. We have previously shown that some (e.g., CCAAT and YY1) but not all (e.g., Oct1, Ap2, and NF1) transcription factor sites can synergize with E2F sites to activate transcription (47). It is possible that this synergy was mediated by cooperative DNA binding. Also, others have shown that Sp1 can physically interact with E2F family members and that binding of Sp1 can influence the occupancy of a nearby E2F site (20, 27). Each of the three characterized ChET clones contains at least one consensus Sp1 binding site (Fig. 3). Finally, others have shown that E2Fs can interact with other sequence-specific DNA binding proteins, such as C/EBPα (17, 44). Interestingly, we have recently shown that E2F1 can be recruited to promoters which contain C/EBPα binding sites but lack E2F consensus sites (Graveel and Farnham, unpublished). It remains to be determined if C/EBPα and/or other protein-protein interactions are mediating the recruitment of E2F to the promoters we have cloned. However, recruitment of E2F through the recognition sequence of another DNA binding protein could explain why some of the cloned fragments failed to show robust competition of a consensus E2F site in vitro.
To date, the majority of well-characterized E2F target promoters have been shown to be cell cycle regulated and activated by E2F overexpression. In contrast, our three novel E2F target promoters are constitutively expressed in growing versus differentiated U937 cells. It is perhaps not surprising that E2F target promoters isolated using an unbiased approach show expression profiles different from those of the well-characterized E2F target promoters. According to microarray analyses, hundreds of genes are regulated by E2F family members (16, 31). It is highly unlikely that this large number of mRNAs, which encode proteins having highly diverse biological functions, will all show exactly the same expression pattern in all cell types.
It is interesting that the mRNA produced by each of the three novel promoters displayed unique expression profiles when normal versus tumor human primary samples were examined; one mRNA was constitutively expressed, one mRNA was downregulated in the tumor sample, and one mRNA was highly upregulated in tumor RNA. Interestingly, one of the promoters that we cloned which displayed high-affinity binding in vivo was shown to be repressed, not activated, by E2F1. Although most E2F target genes studied to date are activated in response to overexpression of E2F1, it has been shown that the cyclin D1 promoter is also repressed by E2F1 (48). In addition, the recent microarray analysis by Muller et al. provided evidence that E2Fs can both activate and repress cellular genes, although their data did suggest that most E2F-mediated repression was indirect (31). Additional evidence supporting E2F-mediated repression of the ChET 8 promoter can be extrapolated from a recent study examining the cell cycle fluctuations of thousands of human mRNAs (4). We have extracted the expression profiles of E2F1 and KIAA0254, the mRNA driven by the ChET 8 promoter, from the published microarray data. Interestingly, ChET 8 mRNA levels are inversely related to E2F1 mRNA levels (data not shown). Collectively, these findings support a role for E2F1 in repression of the ChET 8 promoter. Further experiments need to be performed to characterize similarities and differences between the promoters which are directly activated and those which are directly repressed upon overexpression of E2F1. However, these observations suggest that the nature and context of the E2F binding site may influence the role that E2F plays in regulation of a promoter.
In summary, the data presented in this paper establish the basis for cloning novel promoters regulated by specific transcription factors through chromatin immunoprecipitation techniques. Our initial data suggest that the E2F consensus binding sequence may not account for all potential in vivo E2F targets, possibly due to the roles of interacting proteins within the cellular environment. Importantly, the possibility that E2F family members can regulate promoters that lack consensus binding sites may aid in the understanding of microarray studies which show that hundreds of mRNAs can respond to overexpression of E2Fs (16, 31). Also, we find it most interesting that the expression profiles of the genes identified by using this unbiased approach are quite different from the expression profiles of the previously characterized E2F target genes. Finally, of particular interest are ChET 9 and ChET 4. ChET 9 contains a consensus E2F binding site and shows high-affinity binding in vivo and in vitro. Interestingly, the KIAA0160 mRNA which is transcribed by ChET 9 is upregulated in two different tumor types. The protein encoded by the KIAA0160 mRNA has high homology to a Drosophila protein called Su(z)12. This protein was isolated as a suppressor of a mutation of the gene for zeste, a site-specific DNA binding transcription factor. Although no characterizations of Su(z)12 have been performed; another suppressor of zeste, Su(z)2, is known to be a locus-specific chromosome binding protein. Therefore, it is possible that KIAA0160 will be involved in transcriptional regulation. ChET 4, which shows high-affinity E2F in vivo binding but does not contain a consensus E2F site, is the promoter region for the beclin 1 gene, a putative tumor suppressor gene. The Beclin 1 protein is thought to effect the degradation of cellular proteins and has been shown to be significantly downregulated in human breast carcinomas (26). Our future studies will be focused on understanding the role of Beclin 1 and KIAA1060 in neoplastic transformation.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grant CA45250 (to P.J.F.), CA07175 (an NCI Cancer Center Core grant), HG01696 and GM61503 (to M.Q.Z.), and training grant CA09681 (A.S.W.) from the National Institutes of Health.
We thank David Bentley for the RNA polymerase II antibody, Alexander Kel for exon 2 computer sequence analysis, Scott Eberhardy, Carrie Graveel, and Tadge Kanjo for RNA samples, Julie Wells for technical assistance, and members of the Farnham laboratory for helpful discussions.
REFERENCES
- 1.Aita V M, Liang X H, Murty V V, Pincus D L, Yu W, Cayanis E, Kalachikov S, Gilliam T C, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 2.Albert T, Wells J, Funk J-O, Pullner A, Raschke E-E, Stelzer G, Meisterernst M, Farnham P J, Eick D. The chromatin structure of the dual c-Myc promoter P1/P2 is regulated by separate elements. J Biol Chem. 2001;276:20482–20490. doi: 10.1074/jbc.M100265200. [DOI] [PubMed] [Google Scholar]
- 3.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bussemaker H J, Li H, Siggia E D. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–174. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Kaminsky S, Maouche-Chretien L, Vitelli L, Vinit M-A, Blanchard I, Yamamoto M, Peschle C, Romeo P-H. Chromatin immunoselection defines a TAL-1 target. EMBO J. 1998;17:5151–5160. doi: 10.1093/emboj/17.17.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Belle I, Mercola D, Adamson E D. Method for cloning in vivo targets of the Egr-1 transcription factor. BioTechniques. 2000;29:162–169. doi: 10.2144/00291rr03. [DOI] [PubMed] [Google Scholar]
- 7.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 8.Eberhardy S E, D'Cunha C A, Farnham P J. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J Biol Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- 9.Farnham P J, Slansky J E, Kollmar R. The role of E2F in the mammalian cell cycle. Biochim Biophys Acta. 1993;1155:125–131. doi: 10.1016/0304-419x(93)90001-s. [DOI] [PubMed] [Google Scholar]
- 10.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 11.Graveel C R, Jatkoe T, Madore S J, Holt A L, Farnham P J. Expression profiling and identification of novel genes in hapatocellular carcinomas. Oncogene. 2001;20:2704–2712. doi: 10.1038/sj.onc.1204391. [DOI] [PubMed] [Google Scholar]
- 12.Halle J P, Haus-Seuffert P, Woltering C, Stelzer G, Meisterernst M. A conserved tissue-specific structure at a human T-cell receptor beta-chain core promoter. Mol Cell Biol. 1997;17:4220–4229. doi: 10.1128/mcb.17.8.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hines W A, Thorburn J, Thorburn A. A low-affinity serum response element allows other transcription factors to activate inducible gene expression in cardiac myocytes. Mol Cell Biol. 1999;19:1841–1852. doi: 10.1128/mcb.19.3.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 15.Ioshikhes I P, Zhang M Q. Large-scale human promoter mapping using CpG islands. Nat Genet. 2000;26:61–63. doi: 10.1038/79189. [DOI] [PubMed] [Google Scholar]
- 16.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins J R. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen L M, Iwama A, Lodie T A, Sasaki K, Felsher D W, Golub T R, Tenen D G. c-Myc is a critical target for C/EBPα in granulopoiesis. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 19.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 20.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kel A E, Kel-Margoulis O V, Farnham P J, Bartley S M, Wingender E, Zhang M Q. Computer-assisted identification of cell cycle-related genes—new targets for E2F transcription factors. J Mol Biol. 2001;309:99–120. doi: 10.1006/jmbi.2001.4650. [DOI] [PubMed] [Google Scholar]
- 22.Kim J H, Hui P, Yue D, Aycock J, Leclerc C, Bjoring A R, Perkins A S. Identification of candidate target genes for EVI-1, a zinc finger oncoprotein, using a novel selection strategy. Ocogene. 1998;17:1527–1538. doi: 10.1038/sj.onc.1202331. [DOI] [PubMed] [Google Scholar]
- 23.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee T A, Farnham P J. Exogenous E2F1 is growth inhibitory before, during, and after neoplastic transformation. Oncogene. 2000;19:2257–2268. doi: 10.1038/sj.onc.1203556. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Slansky J E, Myers D J, Drinkwater N R, Kaelin W G, Farnham P J. Cloning, chromosomal location, and characterization of mouse E2F1. Mol Cell Biol. 1994;14:1861–1869. doi: 10.1128/mcb.14.3.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang X H, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 27.Lin S Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 29.McNally J G, Muller W G, Walker D, Wolford R, Hager G L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 30.Moberg K H, Logan T J, Tyndall W A, Hall D J. Three distinct elements within the murine c-myc promoter are required for transcription. Oncogene. 1992;7:411–421. [PubMed] [Google Scholar]
- 31.Muller H, Bracken A P, Vernell R, Moroni M C, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner J D, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. . (Erratum, 15:4660, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 34.Oswald F, Lovec H, Möröy T, Lipp M. E2F-dependent regulation of human MYC: trans-activation by cyclins D1 and A overrides tumour suppressor protein functions. Oncogene. 1994;9:2029–2036. [PubMed] [Google Scholar]
- 35.Pearson A, Greenblatt J. Modular organization of the E2F1 activation domain and its interaction with general transcription factors TBP and TFIIH. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- 36.Phelps D E, Dressler G R. Identification of novel Pax-2 binding sites by chromatin precipitation. J Biol Chem. 1996;271:7978–7985. doi: 10.1074/jbc.271.14.7978. [DOI] [PubMed] [Google Scholar]
- 37.Qin X-Q, Livingston D M, Kaelin W G, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson L, Panayiotakis A, Papas T S, Kola I, Seth A. ETS target genes: identification of egr1 as a target by RNA differential display and whole genome PCR techniques. Proc Natl Acad Sci USA. 1997;94:7170–7175. doi: 10.1073/pnas.94.14.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherf M, Klingenhoff A, Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoteInspector: a novel context analysis approach. J Mol Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- 40.Shan B, Lee W-H. Deregulated expression of E2F1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh P, Wong S H, Hong W. Overexpression of E2F-1 in rat embryo fibroblasts leads to neoplastic transformation. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slansky J E, Farnham P J. The role of the transcription factor E2F in the growth regulation of DHFR. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 43.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. . (Erratum, 13:7201.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slomiany B A, D'Arigo K L, Kelly M M, Kurtz D T. C/EBPα inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol Cell Biol. 2000;20:5986–5987. doi: 10.1128/mcb.20.16.5986-5997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strutt D I, White R A H. Characterisation of T48: a target of homeotic gene regulation in Drosophila embryogenesis. Mech Dev. 1994;46:27–39. doi: 10.1016/0925-4773(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 46.Tomotsune D, Shoji H, Wakamatsu Y, Kondoh H, Takahashi N. A mouse homologue of the Drosophila tumor suppressor gene 1(2)gl controlled by HoxC8 in vivo. Nature. 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- 47.van Ginkel P R, Hsiao K-M, Schjerven H, Farnham P J. E2F-mediated growth regulation requires transcription factor cooperation. J Biol Chem. 1997;272:18367–18374. doi: 10.1074/jbc.272.29.18367. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe G, Albanese C, Lee R J, Reutens A, Vairo G, Henglein B, Pestell R G. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells J, Boyd K E, Bartley S M, Farnham P J. Binding specificity of E2F and pocket protein family members in living cells. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong K K, Zou X, Merrell K T, Patel A J, Marcu K B, Chellappan S, Calame K. v-Abl activates c-myc transcription through the E2F site. Mol Cell Biol. 1995;15:6535–6544. doi: 10.1128/mcb.15.12.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:1–5. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]