Abstract
Products containing BPA structural analog replacements have increased in response to growing public concern over adverse effects of BPA. Although humans are regularly exposed to a mixture of bisphenols, few studies have examined effects of prenatal exposure to BPA alternatives or bisphenol mixtures. In the present study, we investigate the effect of exposure to an environmentally-relevant, low-dose (150 ug/kg body weight per day) mixture of BPA, BPS, and BPF during gestation on the brain transcriptome in Long-Evans pups and dams using Tag RNA-sequencing. We also examined the association between dam licking and grooming, which also has enduring effects on pup neural development, and the transcriptomes. Associations between licking and grooming and the transcriptome were region-specific, with the hypothalamus having the greatest number of differentially expressed genes associated with licking and grooming in both dams and pups. Prenatal bisphenol exposure also had region-specific effects on gene expression and pup gene expression was affected more robustly than dam gene expression. In dams, the prelimbic cortex had the greatest number of differentially expressed genes associated with prenatal bisphenol exposure. Prenatal bisphenol exposure changed the expression of over 2000 genes in pups, with the majority being from the pup amygdala. We used Gene Set Enrichment Analysis (GSEA) to asses enrichment of gene ontology biological processes for each region. Top GSEA terms were diverse and varied by brain region and included processes known to have strong associations with steroid hormone regulation, cilium-related terms, metabolic/biosynthetic process terms, and immune terms. Finally, hypothesis-driven analysis of genes related to estrogen response, parental behavior, and epigenetic regulation of gene expression revealed region-specific expression associated with licking and grooming and bisphenol exposure that were distinct in dams and pups. These data highlight the effects of bisphenols on multiple physiological process that are highly dependent on timing of exposure (prenatal vs. adulthood) and brain region, and reiterate the contributions of multiple environmental and experiential factors in shaping the brain.
Keywords: Bisphenol, Maternal behavior, Prenatal, Amygdala, Gene expression
1. Introduction
Bisphenols are a group of chemicals widely used in plastics, thermal paper, dental sealants, personal care products, plastic food containers, epoxy resins, and other manufacturing products. Although there is overwhelming evidence of the harmful effects of bisphenol A well below the US FDA No Observed Adverse Effect Level (NOAEL) of 5 mg/kg body weight (bw) /day, bisphenols continue to be ubiquitous in our environment and food sources (Rochester, 2013). Bisphenols and bisphenol metabolites are detectable in human biosamples worldwide, including urine and plasma (Calafat et al., 2005; LaKind and Naiman, 2015; Liao et al., 2012; Liao and Kannan, 2013; Vandenberg et al., 2010). The primary routes of exposure for humans is oral, particularly through food processing procedures and leaching from food and beverage containers during storage and heating (Bhunia et al., 2013; Liao and Kannan, 2013), although bisphenols can also be absorbed through contact with thermal paper or use of personal care products (Björnsdotter et al., 2017; Gao and Kannan, 2020).
Bisphenol A (BPA) is the best-studied and most prevalent bisphenol, with over 6 billion pounds produced each year and a historical trend for increasing production as demand for products containing bisphenols increases (Flint et al., 2012; Huang et al., 2012; Owczarek et al., 2018). BPA is best known for its interactions with estrogen receptor (ER) alpha and beta, although it has a lower binding affinity for ERs than 17β-estradiol (Bolger et al., 1998; Kuiper et al., 1998). Despite lower affinity for ERs, low concentrations of BPA can stimulate similar cellular responses as estradiol, particularly for non-classic estrogenic activity, and many studies report nonmonotonic dose-response curves similar to endogenous hormone responses (Alonso-Magdalena et al., 2012; Viñas et al., 2013). Regardless of its estrogenic reputation, BPA can also interfere with the receptor signaling activity of other steroid hormone receptors, including androgen and thyroid hormone receptors (Richter et al., 2007; Sheng et al., 2012; Somogyi et al., 2016; Teng et al., 2013). BPA is capable of activating many transcription factors, which in turn have cascading effects on cellular processes (Krüger et al., 2008; Maertens et al., 2018; Quesada et al., 2002). Because BPA can interact with multiple molecular targets, exposure affects numerous physiological systems including immune function, reproduction, stress response, and tissue development, earning its reputation as a pervasive endocrine disruptor.
“BPA-free” products containing BPA structural analogs, often bisphenol F (BPF) or bisphenol S (BPF), in place of BPA have increased in the last decade in response to growing public concern over adverse effects of BPA. BPS was detected in 89 % and BPF was detected in 66 % of urine samples at concentrations comparable to BPA in a US sample (Lehmler et al., 2018; Liao et al., 2012; Liao and Kannan, 2013). While there is less research on these BPA alternatives, in vitro and in vivo evidence suggests that they have similar molecular properties as BPA and have comparable potency to BPA, if not higher potency, giving BPA alternatives the capacity to have detrimental effects on endocrine systems equal to that of BPA (Rochester and Bolden, 2015).
Developmental processes occurring during gestation, including brain maturation, are partly under direction of hormonal signaling of gonadal steroids (Berenbaum, 1998). Exposures to endocrine disruptors such as bisphenols during prenatal sensitive periods of brain development are especially consequential for neurodevelopment. BPA is capable of crossing the placental barrier and is detectable in the placenta, umbilical cord blood, amniotic fluid, breast milk, fetal serum, and fetal liver tissue (Cao et al., 2012; Chou et al., 2011; Ikezuki et al., 2002; Zhang et al., 2020; Zimmers et al., 2014). Human studies have found associations between prenatal maternal bisphenol levels and several neurodevelopmental disorders including autism spectrum disorder, schizophrenia, and attention-deficit/hyperactivity disorder (Henriksen et al., 2020; Mustieles et al., 2015; Naderi and Kwong, 2020; Rochester, 2013; Rochester et al., 2018; Stein et al., 2015). Higher levels of bisphenols were also found to be negatively associated with normative child neurodevelopment at two years of age in a study of 456 mother-infant dyads (Jiang et al., 2020).
Animal studies have identified changes in the brain resulting from prenatal BPA treatment including disruptions to neuronal migration and neocortical organization (Nakamura et al., 2007, 2006). Likewise, gestational exposure to BPA alternatives interferes with neuronal differentiation in human fetus-derived cell lines and disrupts prefrontal dopamine-serotonin systems in rat offspring (Castro et al., 2015; Fujiwara et al., 2018). Region-specific studies show prenatal BPA alters expression of genes associated with neuropsychiatric disorders such as Alzheimer’s disease, autism spectrum disorder, pathways affected by estrogen activity, and pathways related to synaptic organization and transmission (Arambula et al., 2018; Henriksen et al., 2020; Kanlayaprasit et al., 2021; Sukjamnong et al., 2020). The extensive, enduring developmental effects of bisphenol A on the brain are hypothesized to be at least partially a result of disruption of epigenetic regulation of gene expression (Kundakovic and Champagne, 2011; Wolstenholme et al., 2011). In accordance with this hypothesis, several studies have revealed differences in epigenetic regulators and DNA methylation following developmental BPA exposure in rodent offspring (Alavian-Ghavanini et al., 2018; Drobná et al., 2018; Kundakovic et al., 2013; Malloy et al., 2019).
Plasticity in brain development extends to sensitive periods in early postnatal life. Care-giving behaviors in particular provide essential somatosensory input that scaffolds brain development through specific systems including those involved in hormonal regulation, stress response, maternal care, social behavior, and cognition (Liu et al., 2000, 1997; Starr-Phillips and Beery, 2014; Weaver et al., 2004). For example, F1 pups receiving licking and grooming levels one standard deviation above the mean have higher adult levels of estrogen receptor (ER) alpha in brain regions governing maternal behavior in female rats and exhibit higher levels of licking and grooming toward their own offspring compared to pups receiving licking and grooming levels one standard deviation below the mean (Champagne et al., 2003a, 2003b; Francis et al., 1999). The same comparison of maternal licking and grooming has found recipients of the high licking and grooming phenotype have higher N-methyl-D-aspartate NMDA receptor subunit and brain-derived-neurotrophic factor mRNA expression (hippocampus), higher α1gamma-aminobutyric acid A receptor mRNA expression (hippocampus, medial prefrontal cortex, amygdala), higher glucocorticoid receptor mRNA expression (hippocampus), and reduced corticotropin-releasing hormone mRNA expression (hypothalamus; Caldji et al., 2003; Liu et al., 2000; Liu et al., 1997). Further studies have established that at least some of these changes can be attributed to alterations in epigenetic regulation associated with licking and grooming levels (e.g., Weaver et al., 2004; Champagne et al., 2006; Szyf et al., 2005). For example, simulated maternal grooming using a soft brush to supplement maternal care increased ER alpha promoter methylation and decreased ER alpha mRNA expression in the amygdala and preoptic area in females to levels equal to male pups, who typical receive a larger proportion of maternal licking and grooming (Edelmann and Auger, 2011; Kurian et al., 2010; Moore and Morelli, 1979). Thus, maternal contact with the litter affects gene expression in the pup brain during development with the influence of licking and grooming influencing systems that may also be affected by gestational bisphenol exposure.
In addition to being affected by licking and grooming, estrogen signaling also drives expression of maternal phenotype. Hormonal changes throughout pregnancy increase ER distribution in the MPOA and the reduction in progesterone and increase in estrogen at the end of pregnancy facilitate the onset of maternal behavior to coincide with parturition (Giordano et al., 1989; Siegel and Rosenblatt, 1978). Furthermore, the high licking and grooming phenotype described above is associated with increased ER alpha expression in the MPOA (Champagne et al., 2003a, 2003b). Importantly, animal studies have established that prenatal or perinatal bisphenol exposure below the NOAEL can affect dam care-giving behaviors. Dose-dependent effects of BPA on pup licking and grooming, arched-back nursing, and nest attendance have been reported in mice (Kundakovic et al., 2013; Palanza et al., 2002) and Sprague-Dawley rats (Della Seta et al., 2005). BPA increases ER alpha in the medial preoptic area in lactating rats (Aloisi et al., 2001), suggesting that the estrogenic-properties of bisphenols during the perinatal period are capable of disrupting neuronal and behavioral processes controlled by estrogen signaling in the maternal brain that may contribute to BPA-induced changes in maternal behavior.
Although bisphenols are pervasive in our environment, few studies have examined effects of exposure to BPA alternatives during the perinatal period. In the present study, we investigate the effect of bisphenol exposure during gestation on the transcriptome of pups and dams using Tag RNA-sequencing in the amygdala, hypothalamus, nucleus accumbens, hippocampus, and prelimbic cortex. We used an environmentally-relevant, low-dose mixture of BPA, BPS, and BPF well below the FDA NOAEL since the increasing use of BPS and BPF means humans are regularly exposed to a combination of bisphenols. We also examined the association between dam licking and grooming, which has neural underpinnings that overlap with those targeted by bisphenols, and the brain transcriptome. The amygdala, hypothalamus, nucleus accumbens, hippocampus, and prelimbic cortex were selected because of their sensitivity to actions of steroid hormones including estrogen, involvement in developmental programming by maternal licking and grooming, and role in maternal behavior. This design allows for comparison of effects postnatal maternal care and effects of prenatal exposure to an abundant endocrine disruptor on gene expression across multiple brain regions in both dams and pups.
2. Methods
2.1. Animals
All animal protocols were approved by the University of Texas at Austin. Animals were housed in polycarbonate cages (19” × 10.5” × 8”) with aspen bedding (Nepco) and kept on a 12:12 h light cycle (lights off at 10 am EST) for the duration of the experiment. All animals were fed standard chow (Lab diet 5LL2; contains soy) ab libitum and water provided in glass bottles to limit bisphenol exposure through drinking water. Eight adult postnatal day (P) 60–70 Long-Evans females and six adult Long-Evans males were purchased from Charles River Labs and acclimated to the vivarium for at least two weeks before breeding. During breeding, P75–85 females were screened daily for receptive behavior and left with a breeder male overnight on the day lordosis was observed. All dams were weighed daily and socially housed throughout pregnancy until they were separated into individual cages a few days before giving birth. Day of birth was considered P0.
2.2. Experiment timeline
An overview of the experiment timeline is provided in Fig. 1A. Females were randomly assigned to mixed bisphenol group consisting of 150 μg/kg bodyweight total of equal parts bisphenol A (TCI #B0494, ≥99 %), bisphenol F (Alfa Aesar # A11471, ≥98 %), and bisphenol S (Alpha Aesar #A17342, ≥99 %) dissolved in corn oil (Acros Organics #405435000; n = 4) or vehicle (corn oil only; n = 4) group. Individual stock solutions made by dissolving 0.1 g of bisphenol A, S, and F each in 10 mL of corn oil and spun until the bisphenol was completely dissolved upon visual inspection. The working bisphenol mixture was made by adding 150 μl of each bisphenol stock solution to 9550 μl of corn oil for a final concentration of 0.45 μg/μl bisphenols in corn oil. The mixture was made on the first day of breeding and was stored at room temperature for 3.5 weeks until birth of all litters. The bisphenol mixture and corn oil vehicle were color-coded to blind the experimenter prior to group assignment and remained blind through tissue processing. Beginning on gestational day 8 (G8) through day of birth, dams were weighed and the volume of bisphenol mixture or vehicle was calculated based on body weight. The appropriate volume was then added to a quarter Nilla Wafer cookie and allowed to soak in to the cookie completely prior to being given to the dams. Cookies were placed in a clean holding cage with dams to ensure that every animal ate the entire cookie without interference from their cage mate until they were housed individually at the end of gestation. Dams were exposed only during gestation to examine the effects of bisphenol exposure specifically during pregnancy. On day of birth, litters were culled to 10 pups with equal numbers of males and females when possible. Dam and pups were left undisturbed except for weekly cage changes. Home cage recordings were taken daily through postnatal day 10. Brain tissue was collected from dams on P21, as this was the first day dams could be separated from the remainder of this litter. In order to evaluate the magnitude of effect relative to the last bisphenol exposure in pups compared to dams, we also collected brain tissue from pups on P21. The dam and litter were considered the unit of analysis for all measures; tissue was collected from a maximum of one male and one female from each litter. Pup sex was determined using anogenital distance. Littermates were weaned and used in another study.
Fig. 1.
Experimental Methods. A. Experiment Timeline. Pregnant dams were exposed to a mixture of bisphenols or vehicle from gestational day (G) 8 through day of birth. One-hour home cage recordings were taken one hour after lights out daily on postnatal day (P) 1–10 to score dam licking and grooming behavior. Whole brain tissue was collected from dams and pups on postnatal day 21. B. RNA-seq analysis pipeline. Flash frozen whole brain tissue was cut into 50 or 60 μM sections on a cryostat. Five brain regions were laser-dissected and RNA was extracted before samples underwent RNA sequencing. FASTQC was used on sequencing data before mapping using bowtie2. Genes were prefiltered to those with at least 10 count across all samples. Differential expression analysis was conducted using DESeq2 with licking and grooming and bisphenol exposure as factors. Gene Set Analysis was conducted on ranked differentially expressed genes (DEGs).
2.3. Dam licking and grooming behavior
Home cage behavior was recorded daily on P1–10 with Raspberry pi 3B+ minicomputers and NoIR infrared cameras positioned on each home cage in the colony. Infrared led lights above the cage allowed for recording during the dark phase. Pi cameras recorded simultaneously one hour after lights out and videos were automatically uploaded to the cloud, limiting effects from experimenter presence and time of day on maternal behavior. The amount of time dams spent licking and grooming any pup in each video was later scored by an observer blind to dam condition using Behavioral Observation Research Interactive Software (Friard and Gamba, 2016).
2.4. Tissue collection, staining and dissection
Dams and pups were sacrificed via rapid decapitation on P21. Whole brains were immediately extracted and placed in hexane on dry ice. Brain tissue was stored at − 80 degrees until cryosectioning. Brains were cut into sections on a cryostat set at − 19 degrees C and sections containing regions of interest were mounted on RNase-free PET membrane steel frames slides (Leica #11505190). Dam brains were sliced at 60 μM and pup brains were sliced at 50 μM to account for the smaller size of pup brains and ensure anatomical accuracy. Slides were immediately submerged into 75 % ethanol at − 20 °C for 2 min, stained with 1 % cresyl violet, then washed briefly in 75 % ethanol, 90 % ethanol, 100 % ethanol, and 100 % ethanol again for 1 min. Prepared slides were stored in falcon tubes containing silica bags at − 20 degrees until laser dissection.
The prelimbic cortex, nucleus accumbens (core and shell), hypothalamus, ventral hippocampus, and amygdala were laser dissected on Leica LMD7000. Total volume dissected was recorded for each brain region and did not differ across groups. Dissected tissue was immediately stored in 105 μl lysis buffer mixture (Thermo Fisher Scientific, Waltham, MA; MagMax Total RNA isolation kit, Cat. No. A27828), vortexed to homogenize, and stored at − 80 degrees C until RNA extraction.
2.5. RNA extraction
RNA was extracted from tissue samples using the MagMax Total RNA isolation kit on Kingfisher Flex system according to manufacturer instructions (Thermo Fisher Scientific, Waltham, MA; MagMax Total RNA isolation kit, Cat. No. A278280). RNA quality was determined using RNA 6000 Nano Assay (Agilent Technologies, Santa Clara, CA) and RNA concentration was determined with Quant-it RNA High Sensitivity assay kit (Thermo Fisher Scientific, Cat. No. Q33140). Total RNA extracted ranged from 140 ng to 470 ng per sample.
2.6. Tag-sequencing
25 μl RNA samples were Tag-sequenced at the Genome Sequence and Analysis Facility at the University of Texas at Austin. Tag-sequencing is designed to measure polyadenylated transcripts yielding data for differential gene expression analysis in well annotated genomes (Lohman et al., 2016; Meyer et al., 2011). Reads were sequenced on the NovaSeq 6000 SR100 with minimum reads of four million and target reads of five million.
2.7. Statistical analysis
Statistical analyses were conducted in R 4.0.3 (R Core Team, 2021). Gene expression data from each brain region for dams and pups was analyzed independently. Four samples across all brain regions were excluded from analysis: one from the dam hypothalamus due to technical error during tissue staining and three samples (dam nucleus accumbens, pup amygdala, and pup hippocampus) due to very low total raw counts likely resulting from tissue degradation prior to sequencing. Code for differential expression analysis, clustering, and GSEA is provided at https://github.com/lapphe/mixedbisphenols_rnaseq.
Linear mixed model analysis was used to analyze licking and grooming data with postnatal day and bisphenol exposure as fixed factors and dam/litter id as a random factor.
Licking and grooming time for each dam was averaged across all ten observations, scaled, centered prior to use in differential expression analysis as a continuous variable. Thus, differentially expressed genes related to licking and grooming are described as associations, where positive log2FoldChange indicates a positive relationship between licking and grooming and gene expression and negative log2FoldChange indicates a negative relationship between licking and grooming and gene expression.
2.7.1. RNA-seq analysis processing
The RNA-seq workflow overview is provided in Fig. 1B. Raw data were obtained from the Genome Sequence and Analysis Facility in fastq format. Files were preprocessed to remove adapter sequences, PCR duplicates, and low-quality reads using cutadapt v. 2.8 (Martin, 2011) and FastQC v. 0.11.9 was used to check quality control of the sample data (Andrews, 2010). Preprocessed reads were then aligned to the rat genome (rnor6) using bowtie2 (Langmead and Salzberg, 2012). 50–65 % of reads were mapped to the rat genome for all samples.
2.7.2. Differential expression analysis and clustering
Differential gene expression (DEG) analysis was performed using DESeq2 v 3.14 (Love et al., 2014) with a multifactor design containing bisphenol exposure and scaled and centered average licking and grooming scores as factors. Although pups of both sexes were included in the analysis, we did not use sex as a factor in the analysis due to the small group size of control males (n = 2), which also precluded us from examining the interaction between licking and grooming and bisphenol exposure. DEGs are reported for p values and false discovery rate (FDR)-corrected values (DESeq2 adjusted p values) using Benjamini-Hochberg correction with alpha set at .05 (Benjamini and Hochberg, 1995). DEGs were filtered using ∣log2(fold change)∣ criteria where indicated in results. Principal Component Analysis (PCA) visualization was conducted with normalized gene counts after variance stabilizing transformations using plotPCA tool. Hierarchical clustering analysis and heatmap visualization of the 100 most variable genes after DESeq2 normalization for each region were created using the pheatmap tool in R. Because bisphenol exposure and licking and grooming are known to affect estrogen systems, epigenetic regulators, and parental behavior, differential expression of genes in the “Response to estrogen” (GO:0043627), “Regulation of gene expression, epigenetic” (GO:0040029), and “Parental behavior” (GO:0060746) gene sets from the Broad Institute’s MSigDB database C5 Biological Pathways were examined in all transcriptome datasets.
2.7.3. Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed for enriched terms in the C5 Biological Processes gene set obtained from the Broad Institute’s MSigDB database using the msigdbr v. 7.4.1 R package (Subramanian et al., 2005). GSEA was run for each brain region for pups and dams using genes preranked by descending log2(fold change) from DESeq2 licking and grooming and bisphenol exposure analysis. GSEA was conducted with the fgsea v. 1.20.0 package using the fgseaMultilevel function, which uses an adaptive multilevel split Monte Carlo method capable of estimating low p values (Korotkevich et al., 2016). Parameters for fgsea were set to include terms with between 15 and 250 genes to avoid overly specific or general terms. Enriched pathways with a p value of less than. 01 were then collapsed using the fgsea collapsePathways function to maintain only independent terms and reduce redundancy in enriched terms.
3. Results
3.1. Licking and grooming was not affected by bisphenol exposure
The amount of time the dam spent licking and grooming pups was not different across postnatal days and was not affected by prenatal bisphenol exposure in this small cohort of dams (p > .05; Supplemental Fig. 1).
3.2. Bisphenol exposure and licking and grooming lead to differential gene expression
The total overall normalized counts of all genes for each sample were not different between Vehicle and Bisphenol groups for dams or pups in any brain region (Supplemental Fig. 2). However, differential expression analysis revealed many differentially expressed genes (DEGs) associated with licking and grooming and bisphenol exposure for dams and pups in the five brain regions. DEG counts are summarized in Table 1.
Table 1.
Counts of differentially expressed genes in pups and dams across five brain regions for licking and grooming and bisphenol exposure. The model was run using scaled and centered averages of time spent licking and grooming across all video recordings for each dam. Benjamini-Hochberg correction with alpha set at .05 was used to calculate FDR values.
| |
Licking/grooming |
Bisphenol vs. Vehicle |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
p < .05 |
FDR < 0.05 |
p < .05 |
FDR < 0.05 |
||||||||
| up | down | total | up | down | total | up | down | total | up | down | total | |
| Dams | ||||||||||||
| Hypothalamus | 344 | 216 | 560 | 23 | 2 | 25 | 53 | 29 | 82 | 0 | 0 | 0 |
| Prelimbic cortex | 139 | 169 | 308 | 0 | 0 | 0 | 282 | 301 | 583 | 1 | 7 | 8 |
| Hippocampus | 68 | 120 | 188 | 0 | 0 | 0 | 160 | 61 | 221 | 1 | 0 | 1 |
| Nucleus Accumbens | 109 | 76 | 185 | 5 | 0 | 5 | 53 | 38 | 91 | 0 | 0 | 0 |
| Amygdala | 44 | 39 | 83 | 0 | 0 | 0 | 122 | 134 | 256 | 0 | 0 | 0 |
| Total: | 704 | 620 | 1324 | 28 | 2 | 30 | 670 | 563 | 1233 | 2 | 7 | 9 |
| Pups | ||||||||||||
| Hypothalamus | 320 | 421 | 741 | 3 | 2 | 5 | 293 | 215 | 508 | 1 | 1 | 2 |
| Prelimbic cortex | 98 | 118 | 216 | 0 | 0 | 0 | 622 | 417 | 1039 | 26 | 6 | 32 |
| Hippocampus | 117 | 125 | 242 | 0 | 0 | 0 | 180 | 279 | 459 | 1 | 9 | 10 |
| Nucleus Accumbens | 80 | 116 | 196 | 0 | 0 | 0 | 430 | 308 | 738 | 80 | 116 | 196 |
| Amygdala | 168 | 94 | 262 | 1 | 0 | 1 | 1544 | 1618 | 3162 | 775 | 1003 | 1778 |
| Total: | 783 | 874 | 1657 | 4 | 2 | 6 | 3069 | 2837 | 5906 | 883 | 1135 | 2018 |
3.2.1. DEGs associated with licking and grooming in dams
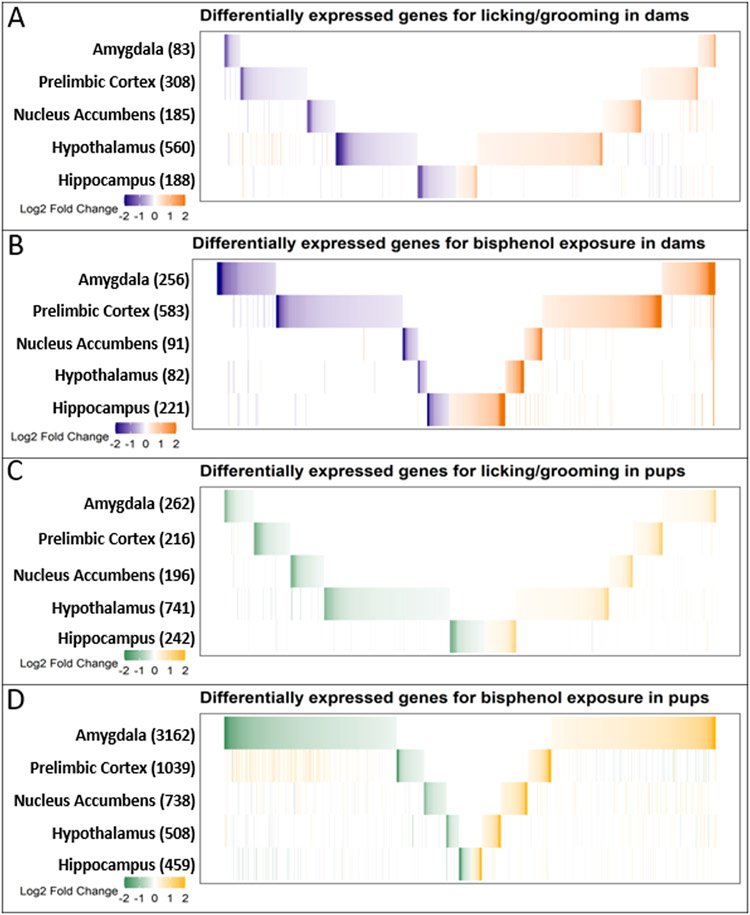
Exploratory analysis using unadjusted p values to determine DEGs found 1324 genes across all brain regions associated with licking and grooming. As shown in Fig. 2A, there was little overlap in DEGs across brain regions and when there was overlap, the direction of association was not consistent between regions. The majority of these DEGs were from the hypothalamus (560), followed by the prelimbic cortex (308), hippocampus (188), nucleus accumbens (185), and amygdala (83).
Fig. 2.
Exploratory analysis of differentially across brain regions. Genes with p < .05 in any of the five brain regions are displayed in the union heatmap. Total number of genes appear next to the region name. A. DEGs associated with licking and grooming in dams. B. DEGs as a results of bisphenol exposure in dams. C. DEGs associated with licking and grooming in pups. D. DEGs as a result of bisphenol exposure in pups.
Across all regions, 30 genes were significantly associated with dam licking and grooming (FDR<0.05). Unsurprisingly, the majority of those genes were from the hypothalamus (25 DEGs) and were positively associated with licking and grooming (23 DEGs; more licking and grooming corresponding to higher gene expression). Only two genes, Ctss and Glul, were negatively associated with licking and grooming. Five genes (Olfm1, C1ql3, Cck, Cnih2, and Slc17a7) from the nucleus accumbens were positively associated with licking and grooming. No other regions had DEGs that met the FDR < 0.05 criteria.
3.2.2. DEGs associated with licking and grooming in pups
Exploratory analysis using unadjusted p values found 1657 genes associated with licking and grooming across the five brain regions in pups (Table 1) and very little overlap of these DEGs across brain regions (Fig. 2C). Similar to dams, the majority of DEGs were in the hypothalamus (741), and roughly equal number of DEGS in the amygdala (262), hippocampus (242), prelimbic cortex (216), and nucleus accumbens (196). Only six genes were significantly differentially expressed (FDR <0.05). Five of these genes (Pcp4, Tac1, Penk, Wipf3, and Gda) were from the hypothalamus and one DEG (Hbb) was from the amygdala.
3.2.3. DEGs associated with bisphenol exposure in dams
Exploratory analysis using unadjusted p values found 1233 DEGs related to bisphenol exposure in dams and showed distinct effects by region resulting in little overlap in DEGs across the five regions measured (Fig. 2B). Where there was overlap, the direction of change was mostly in agreement across regions with a few exceptions. The transcriptome in prelimbic cortex was most affected, with 583 DEGs, followed by the amygdala (256), the hippocampus (221), hypothalamus (82), and hippocampus (221).
Considering only DEGs with FDR < 0.05, only 9 genes were differentially expressed with bisphenol exposure in dams across the five brains regions measured. Eight DEGs were in the prelimbic cortex, with seven genes downregulated and only Miox upregulated following bisphenol exposure. The remaining DEG, AABR07052744.1, was upregulated in the hippocampus in bisphenol-exposed dams.
3.2.4. DEGs associated with bisphenol exposure in pups
Across all brain regions, exploratory analysis (unadjusted p values) found 5906 genes associated with bisphenol exposure in pups (Table 1). Approximately half (3162) of these genes came from the pup amygdala, 1039 came from the prelimbic cortex, followed by 738 from the nucleus accumbens, 508 from the hypothalamus, and 459 from the hippocampus. Of genes that were differentially expressed in more than one region, the direction of change in expression was largely in agreement across the amygdala, nucleus accumbens, hypothalamus, and hippocampus. Interestingly, a subset of these genes in the prelimbic cortex showed opposite expression change in the prelimbic cortex compared to the amygdala (Fig. 2D).
In contrast to licking and grooming, bisphenol exposure significantly altered the expression of 2018 genes across all brain regions in pups (FDR <0.05). The overwhelming majority of the DEGs were in the pup amygdala (1778), followed by the nucleus accumbens (196), prelimbic cortex (32), hippocampus (10), and hypothalamus (2).
3.3. Clustering analysis
Hierarchical clustering with the top 100 most variable genes and principal component analysis using all genes for each region is shown in Fig. 3 (dams) and Fig. 4 (pups).
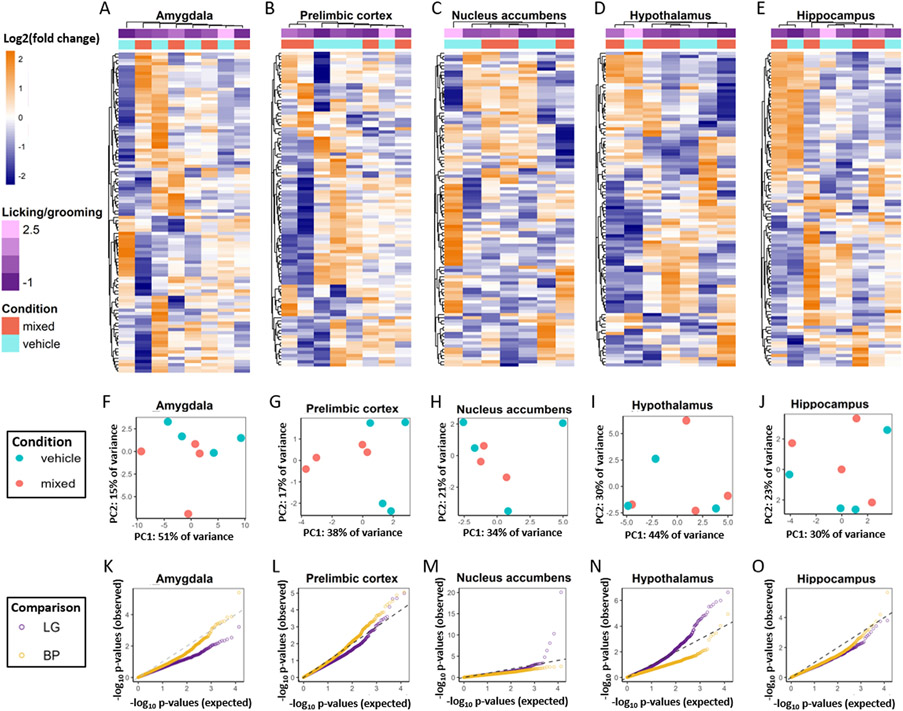
Fig. 3.
Results of differential expression analysis for dams. A-E. Heatmaps showing the log2FC for the top 100 most variable genes in each brain region. Genes are sorted along rows using hierarchical clustering of log2FC and samples are sorted across columns by expression profile. Licking and grooming levels and bisphenol condition for each sample are shown in top two rows. F-J. PCA plots for gene expression in each brain region. K-O. QQ plots showing the observed distribution of p values for licking and grooming and bisphenol exposure plotted against expected distribution of p values.
Fig. 4.
Results of differential expression analysis for pups. A-E. Heatmaps showing the log2FC for the top 100 most variable genes in each brain region. Genes are sorted along rows using hierarchical clustering of log2FC and samples are sorted across columns by expression profile. Licking and grooming levels and bisphenol condition for each sample are shown in top two rows. F-J. PCA plots for gene expression in each brain region where bisphenol treatment group is shown by color and pup sex indicated by symbol. K-O. QQ plots showing the observed distribution of p values for licking and grooming and bisphenol exposure plotted against expected distribution of p values.
3.3.1. Clustering analysis for dam samples
Bisphenol-exposed samples did not clearly cluster together in any brain region. However, in the hypothalamus, sample clustering appears to be arranged by high, medium, and low levels of licking and grooming. Visualization of the first two PCA factors for normalized gene counts for all regions (Fig. 3F-J) reiterates the lack of clustering by bisphenol exposure, with the exception of the prelimbic cortex where PC1 (38 % of variance) appears to separate Vehicle and Bisphenol samples (Fig. 3G). QQ plots showing the observed versus expected distribution of p values for licking and grooming and bisphenol exposure (Fig. 3K-O) indicate that licking and grooming was more strongly associated with gene expression in the dam hypothalamic (Fig. 3N) and nucleus accumbens (Fig. 3M) transcriptome and bisphenol exposure had a marginally larger effect on the prelimbic cortex transcriptome compared to licking and grooming (Fig. 3L). Other brain regions did not show substantial deviations in observed p values in the transcriptome profile for either licking and grooming or bisphenol exposure in dams.
3.3.2. Clustering analysis for pup samples
In the pup amygdala and prelimbic cortex, the top 100 most variable genes clustered by bisphenol exposure (Fig. 4A & B). This was confirmed with visualization of the top two factors from PCA analysis on normalized gene counts of all genes in these two regions (Fig. 4F & G) and the hippocampus (Fig. 4J). Separation by bisphenol group on PC1 was particularly prominent in the amygdala, where samples also clustered by pup sex along PC2. Hierarchical clustering in the nucleus accumbens and hypothalamus did not show clear patterns of clustering by bisphenol exposure or licking and grooming (Fig. 4C-E). There was also not separation by bisphenol exposure in those regions, although PC1 separated hypothalamic samples by sex (Fig. 4 H-J). QQ plots underscore the strong association of gene expression in the pup amygdala with bisphenol exposure (Fig. 4k). The QQ plot for the prelimbic cortex, nucleus accumbens, and hippocampus also indicate that the transcriptome is more affected by bisphenol exposure than licking and grooming in those regions, although less dramatically than the amygdala (Fig. 4L, M, & O). The QQ plot for the pup hypothalamus shows deviation from the expected distribution for p values for both licking and grooming and bisphenol exposure with a slightly stronger effect of licking and grooming (Fig. 4N).
3.4. GSEA analysis
3.4.1. GSEA for dam transcriptomes
Supplemental Table 3 presents the significant GO biological processes terms for each brain region for bisphenol exposure and licking and grooming for dams from GSEA analysis. Cilium-related terms appeared among the top five terms with a positive normalized enrichment score in the amygdala, hippocampus, and nucleus accumbens for bisphenol exposure and thippocampus and hypothalamus for licking and grooming. Similarly, “axonemal dynein complex assembly” and “axoneme assembly” were among the top five terms in the amygdala and were driven by similar core genes as the cilium-related terms. Several top terms were associated with processes known to have strong associations with steroid hormone regulation for both bisphenol exposure and licking and grooming, including “development of primary female sexual characteristics” (bisphenol, amygdala), “cellular process involved in reproduction in multicellular organism” (bisphenol, hippocampus), “response to dexamethasone” (bisphenol, nucleus accumbens), and “mating” (licking and grooming, prelimbic cortex). Notably, the term “response to dexamethasone” in the nucleus accumbens had a particularly small FDR value and relatively large normalized enrichment score of − 2.09 for bisphenol exposure.
3.4.2. GSEA for pup transcriptomes
The significant terms for GSEA analysis for bisphenol and licking and grooming for pups are shown in Supplemental Table 4. Similar to the dam GSEA results, “cilium movement” and “axoneme assembly” appeared among the top five terms in the hippocampus and hypothalamus for bisphenol exposure. Each brain region also had at least one metabolic and biosynthetic process-related term in the top five for bisphenol exposure.
Top enriched terms for licking and grooming for pups included several terms related to mitochondrial processes: “response to mitochondrial polarization” (amygdala), “ATP synthesis coupled electron transport” (hippocampus), “mitochondrial respiratory chain complex assembly” (hippocampus), “mitochondrial translation termination” (hippocampus), and “respiratory electron transport chain” (hypothalamus). In the nucleus accumbens, top enriched terms contained several terms with negative normalized enrichment scores related to cardiovascular development. Also noteworthy are the “oxidative phosphorylation” and “ribose phosphate biosynthetic process” terms enriched with licking and grooming in the nucleus accumbens with FDR < 0.05.
3.5. Hypothesis-driven differential gene expression
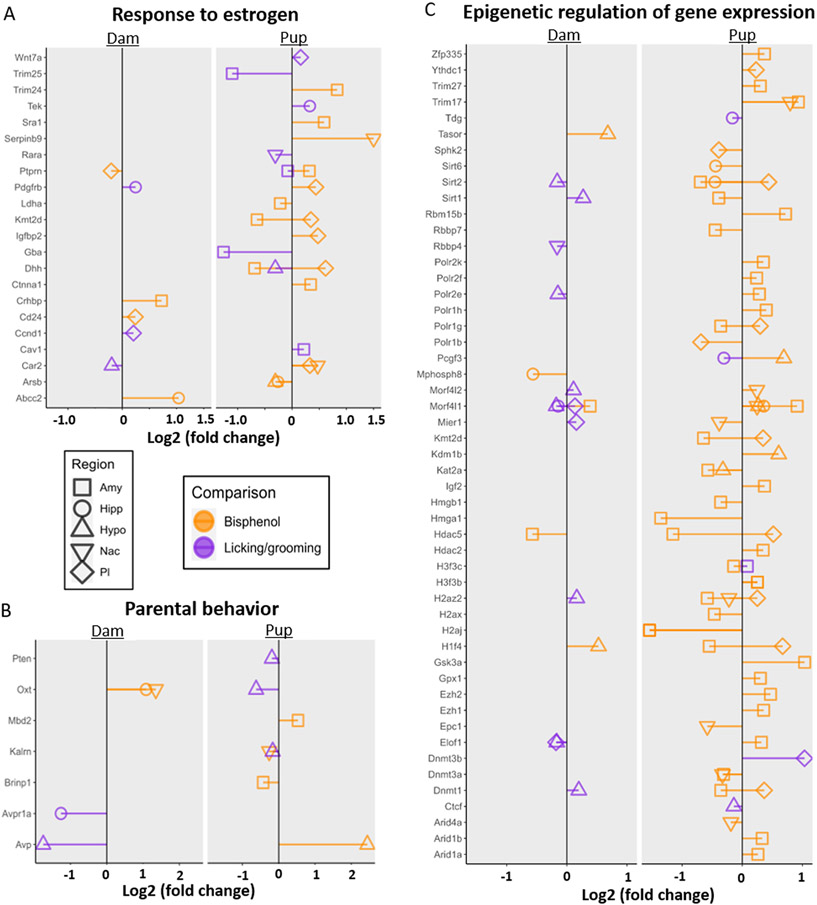
Both bisphenol exposure and postnatal licking and grooming are known to affect estrogen pathways, maternal behavior, and molecular epigenetic regulators, so we explored expression of genes in the “Response to Estrogen”, “Parental Behavior”, and “Regulation of gene expression, epigenetic” biological processes terms. Of the 71 genes in the “Response to estrogen” gene ontology biological process term, expression of 11 genes (15.49%) were significantly affected by licking and grooming and expression of 15 genes (21.12 %) were significantly affect by bisphenol exposure in dams and pups (p < .05; Fig. 5A). Bisphenol exposure lead to differential expression of more estrogen response genes across all brain regions in pups affecting twelve genes, with the majority of the genes differentially expressed in the amygdala and prelimbic cortex (p < .05). Similar to dam gene expression, licking and grooming affected fewer estrogen response DEGs across all brain regions compared to bisphenol exposure, downregulating five genes and upregulating three genes (p < .05).
Fig. 5.
Hypothesis-driven differential gene expression. A. Log2 fold changes for genes in the “Response to Estrogen” (GO:0043627) Biological Processes gene list for dams and pups. B. Log2 fold changes for genes in the “Parental Behavior” (GO:0060746) Biological Processes gene list for dams and pups. C. Log2 fold changes for genes in the “Regulation of gene expression, epigenetic” (GO:0040029) Biological Processes gene list for dams and pups. Only data for genes with p value < .05 are shown. Fold change associated with Bisphenol exposure are shown in orange and Licking and grooming are shown in purple. Symbol shape denotes the brain region.
Licking and grooming was associated with five (45.45 %) of the 11 genes in the “Parental behavior” term and differences in expression were limited to the hypothalamus in pups and hypothalamus and hippocampus in dams (Fig. 5B). The effects of bisphenol exposure on parental behavior genes was more variable across brain regions in dams and pup, also affecting expression of five genes (45.45 %).
Finally, differential expression of 205 genes belonging to the “Regulation of gene expression, epigenetic” were examined for bisphenol exposure and licking and grooming (Fig. 5C). Bisphenol exposure affected expression of 41 (20.00 %) of these genes. The majority of the bisphenol-induced changes in gene expression were in pups, with only five genes (2.48 %) differentially expressed in dams. Only 15 genes (7.46 %) related to epigenetic control of gene expression were significantly associated with licking and grooming. In contrast to bisphenol exposure, more (4.98 %) of those significant genes were differentially expressed in dams, with only five genes (2.49 %) differentially expressed by licking and grooming in pups.
4. Discussion
We compared the relative effects of two factors known to affect brain and behavior, postnatal maternal care and prenatal bisphenol exposure, by examining the associations between these factors and the transcriptome in multiple brain regions in pups and dams at weaning. Our data demonstrate region-specific effects of a prenatal bisphenol A, S, and F mixture on the brain transcriptome in pups and, to a lesser extent, dams. Dam postnatal licking and grooming during P1–10 was also associated with gene expression in pups and dams, particularly in the hypothalamus. Hypothesis-driven and GSEA showed that genes associated with bisphenol exposure and licking and grooming belong to diverse biological processes including cell signaling, epigenetic regulation of gene expression, mitochondrial processes, parenting behavior, and other processes under hormonal regulation. BPA has been the focus of most previous work examining effects of bisphenols on the brain and our data expand that work by showing that exposure to an environmentally-relevant mixture of bisphenols affects the brain transcriptome at a dose well below the FDA NOAEL. These data may help inform risk for bisphenol exposure of pregnant women and infants.
4.1. Postnatal licking and grooming is associated with differential gene expression in the hypothalamus of dams and pups
Postnatal licking and grooming had the strongest associations with the dam and pup hypothalamus in terms of number of differentially expressed genes (FDR <0.05; Table 1). This is expected given the importance of the medial preoptic area (MPOA) of the hypothalamus as a key site for integrating multisensory pup cues and hormonal signals to facilitate maternal behavior (Kohl et al., 2017). In pups, licking and grooming had smaller effects on pup gene expression overall compared to bisphenol exposure. Licking and grooming is a discrete mechano-tactile stimulus thought to elicit a specific constrained physiological response in pups, distinct even from other types of maternal contact, so it is unsurprising that the correlation with gene expression in the brain was less extensive in pups in comparison to effects of a systemic exposure like bisphenol administration (Hellstrom et al., 2012; Kojima et al., 2012).
4.1.1. Licking and grooming and dam gene expression
Exploration of the neurobiology of the licking and grooming phenotype has mainly focused on the roles of ovarian hormones in the dam brain as facilitators of this behavior. The actions of estrogens and oxytocin in the dam hypothalamus have received particular attention for their associations with maternal licking, but these were not among the list of 25 significant genes in this study. Notably, DEG analysis also identified five genes that were positively associated with licking and grooming in the dam nucleus accumbens, a region thought to mediate parental responsiveness and shown to have differential dopamine response in high and low licking dams during pup licking bouts (Table 1; Champagne et al., 2004; Kohl et al., 2017). Further exploration these maternal hypothalamic and nucleus accumbens DEGs in the context of licking and grooming and maternal behavior more generally are warranted.
4.1.2. Licking and grooming affected hypothalamic gene expression but not Esr1 in pups
Postnatal licking and grooming received affects later-life maternal phenotype, consistent with changes in gene expression in the hypothalamus in our data. Female offspring of low licking and grooming dams (one standard deviation below the mean) have reduced oxytocin receptor binding and mRNA, reduced ER alpha and ER beta mRNA, and reduced estrogen-induced cFos expression in the MPOA compared to offspring of high licking and grooming dams (Champagne et al., 2001, 2003b, 2006; Peña et al., 2013). In contrast to those findings, Esr1 expression was not related to licking and grooming in this study and hypothalamic Oxt expression was reduced with higher licking and grooming levels. Prior work has also identified enduring changes in epigenetic markers, including elevated Esr1 promoter DNA methylation, reduced H3K9me3 at Esr1 promoter, and reduced Stat5 binding to the Esr1 promoter in offspring of low versus high licking and grooming dams in the hypothalamus, which may contribute to the sustained effects of licking and grooming in the hypothalamus across the lifespan (Champagne et al., 2006; Peña et al., 2013). Ctcf a transcriptional repressor, had an inverse relationship with licking and grooming in the pup hypothalamus. Although our data show the hypothalamic transcriptome is closely associated with licking and grooming relative to other brain regions, the small sample size, use of both male and female pups, and inability to contrast the low vs. high extremes of licking and grooming behavior may have occluded some of the gene-specific changes observed in previous studies.
4.2. Gestational bisphenol exposure affects brain gene expression in pups more than in dams
Prenatal bisphenol exposure had a highly robust effect on the pup brain transcriptome compared to gestationally exposed dams. Over 2000 genes were differentially expressed in bisphenol exposed pups, while only nine genes were differentially expressed in bisphenol exposed dams across all five brain regions (FDR <0.05). Pups were exposed during a developmental sensitive period for neuronal differentiation, migration, and synaptic development so disruption to those processes by bisphenols are likely to have larger long-term effects than exposure during adulthood. In addition, human and animal studies demonstrate that bisphenols cross the placental barrier and there may be accumulation of bisphenols in amniotic fluid with reduced capacity of the fetus to metabolize and clear bisphenols from the body. Overall, these processes would result in higher exposure levels in developing offspring compared to the mother during these sensitive windows of development (Ikezuki et al., 2002; Mielke and Gundert-Remy, 2009).
4.2.1. Bisphenols and dam gene expression
The limited significant exposure-associated changes in gene expression in dams were predominantly in the prelimbic cortex (Table 1; Supplemental Table 1). There is little work examining the effect of adult exposure of bisphenols in dams, but at least one study shows altered gene expression in the prefrontal cortex of adult male and female rats following an injection of 50 ug/kg bw /day for four days (Castro et al., 2013). Furthermore, low dose BPA during adolescence is shown to alter spine density on pyramidal cells in the medial prefrontal cortex of adult Sprague Dawley rats (Bowman et al., 2014). Together with our findings, these two studies suggest that adult exposure to bisphenols is capable of inducing changes in prefrontal gene expression that may have functional consequences.
4.2.2. Prenatal bisphenols and pup gene expression
The effect of prenatal bisphenol exposure on the pup transcriptome was more robust than licking and grooming in all brain regions except the hypothalamus, where licking and grooming and bisphenol exposure had similar effects on the number of differentially expressed genes. While bisphenol exposure DEGs were found in all five brain regions in pups, the overwhelming majority of DEGs were in the amygdala (Table 1; Supplemental Table 2). The second-most affected pup brain region was the nucleus accumbens, where the expression of 196 genes were changed in pups. Prenatal BPA enhances the effects of methamphetamine and changes in limbic dopamine systems, but this is the first study to examine transcriptome-wide changes specifically in the nucleus accumbens (Narita et al., 2006; Suzuki et al., 2003).
Several candidate gene approach studies have focused on differences in Esr1, Esr2, Oxtr, and Oxt in some of the brain regions included in our analyses (Arambula et al., 2018; Cao et al., 2013; Kundakovic et al., 2013; Witchey et al., 2019). Unfortunately, Esr2 expression was very low in all samples and was excluded during the prefiltering step of the analysis pipeline. We did not find differential expression of Esr1 in pups and dams in our dataset, although females had higher hypothalamic expression than males as expected (Supplementary fig. 3). At least one other study reported no effect of prenatal BPA exposure on Esr1 or Esr2 in the MPOA or anteroventral periventricular nucleus of Sprague Dawley rats on day of birth using in situ hybridization (Cao et al., 2013). This contrasts several other findings collectively showing the bisphenol effect on ERs in the amygdala and hypothalamus are age-dependent such that bisphenol effects are generally associated with reduced expression before weaning and increased expression after weaning (Patisaul, 2020; Rebuli et al., 2014; Arambula et al., 2016; Kundakovic et al., 2013). Oxt and Oxtr expression were also unchanged by bisphenols in our data, conflicting with other reports of sex and dose-specific increases (Arambula et al., 2016, 2018; Witchey et al., 2019; Adewale et al., 2011).
Transcriptome-wide approaches provide a larger picture of gene profile changes following gestational bisphenol exposure. A Consortium Linking Academic and Regulatory Insights of BPA Toxicity (CLARITY) study showed prenatal BPA affects the neonatal amygdala transcriptome on P1, predominantly for genes involved in synaptic organization and transmission (Arambula et al., 2018). These effects were especially evident in females relative to males, and PCA analysis showed samples clustered by sex but not bisphenol treatment group. However, our data show the amygdala PC1 (64 % of the variance) primarily separated samples by bisphenol exposure and PC2, which accounts for only 11 % of variance, separated samples by sex (Fig. 5F). A similar RNA-seq CLARITY study found dose-dependent hypothalamic and hippocampal transcriptional changes after gestational BPA in P1 pups, with the hypothalamus exhibiting greater transcriptional changes relative to the hippocampus (Arambula et al., 2016). Notably, their hypothalamic effects of BPA were largely male-specific, and the relatively small number of hypothalamic DEGs identified in the present study may be attributed to excluding pup sex as a factor in the analysis, especially given the separation of hypothalamic male samples along PC1 (Fig. 5J). Similarly, two studies found sex-specific effects of 5 mg/kg bw/day prenatal BPA exposure on hippocampal gene expression in P1–2 rat pups for genes related to neuronal viability, neuritogenesis, learning/memory, and genes implicated in autism spectrum disorder (ASD) etiology (Thongkorn et al., 2021, 2019). Differential expression of ASD-related genes following BPA exposure are reported in the prefrontal cortex of neonatal rats (5 mg/kg bw/day; Kanlayaprasit et al., 2021) and P28 male mice (5 mg BPA/kg in diet; Henriksen et al., 2020) while lower doses (5 ug/kg bw/day) of prenatal BPA, BPF, or BPS can have unique effects on juvenile and adult expression of genes related to dopamine and serotonin systems (Castro et al., 2015, 2013). Our data complements previous work by showing that bisphenol-induced expression profiles are mostly unique to each brain region in pups and the direction of expression for particular genes may be in opposing directions (see amygdala vs prelimbic cortex; Fig. 2).
The region-specificity and variety of biological systems affected by bisphenols is reflected in the diversity of the top biological process terms, including familiar processes known to be disrupted by BPA, such as immune-related and metabolic terms. Surprisingly, three cilium and axoneme-related terms reached the FDR level of significance for the hypothalamus, perhaps suggesting a role for bisphenols in the development of ciliopathies. Cilium and axoneme genes also play important roles in several signaling pathways in the brain, organization of the forebrain, neuronal migration, and learning and memory (Park et al., 2019). Understanding the functional consequences for behavior and cognition connected to these expression profiles alone and in concert will help bridge the gap with human work linking prenatal bisphenol levels, child neurodevelopment, and heightened susceptibility to some neurodevelopmental disorders (Henriksen et al., 2020; Mustieles et al., 2015; Naderi and Kwong, 2020; Rochester, 2013; Rochester et al., 2018; Stein et al., 2015).
4.3. Postnatal licking and grooming and prenatal bisphenols affect genes related to estrogen response, prenatal behavior, and epigenetic regulation of gene expression
In a hypothesis-driven analysis, we examined the expression of genes involved in estrogen response, parental behavior, and epigenetic regulation of gene expression. The overall pattern of greater effects of bisphenol exposure on the pup brain compared to dams extend to these analyses, whereas licking and grooming associations are more equally distributed between dams and pups. The majority of bisphenol-induced changes in gene expression for this analysis are in the amygdala, prelimbic cortex, and nucleus accumbens for pups. Notably, licking and grooming and bisphenol exposure sometimes has opposing effects on expression of a particular gene. This underscores the complexity in disentangling effects of prenatal bisphenol exposure on pup outcomes and underscores the need to evaluate maternal care in these developmental studies.
4.3.1. Estrogen response genes are more affected by bisphenols in pups than dams
Mechanistically, estrogen activity is at the intersection of parental behavior and bisphenol exposure, so we predicted that these factors related to estrogen signaling could alter expression of genes known to be responsive to estrogen in the maternal and pup brain. Contrary to our hypothesis, dams had very few estrogen response genes affected by either factor compared to pups (Fig. 5A). However, there were several significant estrogen response genes in pups associated with bisphenol exposure in the amygdala and prelimbic cortex, although with nearly no concordance of significant genes between regions. Genes associated with licking and grooming were distributed across the brain regions for pups. These findings indicate prenatal bisphenol-induced effects in the brain are partially mediated through disruption to estrogen pathways, particularly in pups, though without altering expression of Esr1.
4.3.2. Bisphenol exposure and licking and grooming affect expression of parental genes equally in dams and pups
In coordination with other hormonal changes, increases in estrogen during late pregnancy facilitate neuroendocrine changes foundational to the onset of maternal behavior in rodents (Rosenblatt, 1994; Russell et al., 2001). Disrupting this process with bisphenols may modify the expression of genes known to be involved in parental care. Furthermore, maternal phenotype is known to be partially developmentally programed, so expression of parental genes associated with bisphenol exposure and licking and grooming received may also be observed in pups. Despite the absence of an effect of prenatal bisphenols on dam licking and grooming in our study, we found that bisphenols increased the expression of Oxt in the hippocampus and nucleus accumbens in dams (Fig. 5B). Higher licking and grooming corresponded to downregulation of dam hippocampal Avpr1a and hypothalamic Avp, which has been shown to promote maternal behaviors including nursing and pup retrieval (Fig. 5B; Bosch and Neumann, 2008). In contrast, bisphenol exposure increased Avp by more than 2-fold in the pup hypothalamus, which is consistent with a previous report of increased hippocampal Avp following pre and postnatal 2500 μg/kg/day BPA exposure until weaning in adult female Sprague-Dawley rats (Cheong et al., 2018). Changes in Avp may affect maternal behavior in female F1 offspring toward their own pups.
4.3.3. Expression of epigenetic genes are primarily associated with licking and grooming in dams and with bisphenol exposure in pups
Both licking and grooming and bisphenol exposure have enduring epigenetic effects on pups (Cariati et al., 2020; Champagne et al., 2006; Chang et al., 2016; Rahmani et al., 2018; Strakovsky and Schantz, 2018; Weaver et al., 2004). While our data are consistent with these prior findings, licking and grooming affected fewer epigenetic genes and fold changes in significant genes were generally small compared to bisphenol exposure, with the exception of Dnmt3b in prelimbic cortex which had increased expression with higher licking and grooming levels. Prenatal BPA exposure has been shown to have dose and sex-specific effects on Dmnt1 and Dmnt3a in the prefrontal cortex and hypothalamus in mice, both of which were also affected by bisphenols in our data although in different brain regions (Kundakovic et al., 2013). Disruption to epigenetic modulators by bisphenols may account for the long-lasting effects of bisphenols, which in the current study impact the transcriptome three weeks after the final exposure.
In dams, the majority of differentially expressed epigenetic genes were related to licking and grooming and found largely in the hypothalamus and prelimbic cortex. The comparatively smaller effect of bisphenols on epigenetic regulators of gene expression matches the relatively smaller effect of bisphenols on the dam transcriptome in general, which may reflect the timing of analyses of the dam transcriptome.
4.4. Gestational exposure to BPA, BPF and BPS mixture did not affect maternal behavior
Despite evidence of dose-dependent changes in maternal behavior in adult females exposed to bisphenols during pregnancy from other studies, we did not observe any difference in the total time spent licking and grooming between vehicle and bisphenol exposed rat dams. Palanza et al. (2002) showed that adult exposure to 10 ug/kg bw/day BPA on gestational day 14–18 reduced nest attendance and nursing while CD1 mice (Palanza et al., 2002). Sprague-Dawley rat dams exposed to 40 ug/kg bw/day throughout gestation and lactation had reductions in pup licking and grooming duration and frequency, anogenital licking and grooming frequency, and duration of arched back nursing (Della Seta et al., 2005). Likewise, Kundakovic et al. (2013) examined the effects of 2, 20, and 200 ug/kg bw /day BPA during gestational day 0–19 in BALB/c mice and found quadratic changes in licking and grooming and arched back nursing with increasing dose such that the largest dose had greater frequency of those behaviors. Similarly, BPS exposure at 200 ug/kg body weight/day, but not 2 ug/kg body weight/day, in CD-1 mice increased the percentage of time dams spent on the nest on P14, but did not change the percent of time spent licking and grooming or repairing the nest (Catanese and Vandenberg, 2017). Collectively, these studies show a pattern of changes in specific maternal behaviors following bisphenol exposure with reductions at lower exposures and increases in pup-directed on-nest behaviors with higher exposures. Given our small sample size, we were likely not sufficiently powered to detect these effects.
Furthermore, other work has shown interactions between gestation bisphenol exposure and cross fostering on anxiety-like behavior and social interaction with a conspecific in mice, which could be taken into consideration with a larger sample size (Cox et al., 2010).
4.5. Study limitations
Comparing the transcriptome across multiple brain regions in both dam and pups from the same sample is a strength of the study. However, this produces a large number of samples per dam/litter unit, precluding the use of a large sample size. Thus, despite evidence that the effects of gestational BPA on gene expression is sex-specific, we were not able to separate our data by pup sex. The limited number of dam/litter units also means the full distribution of low to high licking and grooming phenotypes was not present, limiting our ability to test effects across the full phenotype range and our power to detect the previously documented relationship between licking and grooming and gene expression (Champagne et al., 2003a, 2003b). In addition, effects of BPA are often nonmonotonic and our study included only one mixed bisphenol dose. Whether the effects on the transcriptome depend on bisphenol dose or specific type of bisphenol (BPA vs “safe” alternatives BPS or BPF) rather than the combination of bisphenols cannot be determined with this data and is a topic of ongoing investigation in our lab. Furthermore, brain tissue was collected on P21, three weeks after the final exposure and 11 days after the final behavioral observation. Because of this timing, we may have missed transient changes in gene expression present earlier in development that may have cascading effects on neurodevelopment and the transcriptomic effects we observed may be those that are only chronically different or are secondary effects of licking and grooming and prenatal bisphenol exposure.
5. Conclusions
We demonstrate that a low dose of a mixture of BPA, BPS, BPF during gestation affects the transcriptome of dam and pups three weeks after the final exposure. Licking and grooming was not changed by bisphenol exposure, but nevertheless was associated with gene expression in the dam and pup hypothalamus. Bisphenols had a greater effect on pups than on dams in terms of number of differentially expressed gene and effects were brain region-specific. Strikingly, the pup amygdala was most affected by bisphenols. These data provide a starting point to evaluate the relative contributions of licking and grooming and exposure to multiple bisphenols on gene expression in the brain and compare effects on dam and pups. Future work should more closely evaluate the functional consequences of these changes in gene expression, especially in the pup amygdala, as well as consider the interaction between prenatal bisphenols at different doses, a more complete repertoire of maternal behavior, and neurobiological and behavioral outcomes in pup and dam.
Supplementary Material
Funding
Funding for this study was provided by R01ES030950–01A1 from NIEHS.
Footnotes
CRediT authorship contribution statement
Hannah Lapp: Methodology, Investigation, Formal analysis, Visualization, Writing- original draft, Software, Amy Margolis: Conceptualization, Methodology, Writing- reviewing and editing, Funding acquisition, Resources, Frances Champagne: Conceptualization, Methodology, Supervision, Investigation, Writing- reviewing and editing, Funding acquisition, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.neuro.2022.08.014.
References
- Adewale HB, Todd KL, Mickens JA, Patisaul HB, 2011. The impact of neonatal bisphenol – a exposure on sexually dimorphic hypothalamic nuclei in the female rat. NeuroToxicology 32, 38–49. 10.1016/j.neuro.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavian-Ghavanini A, Lin P-I, Lind PM, Risén Rimfors S, Halin Lejonklou M, Dunder L, Tang M, Lindh C, Bornehag C-G, Rüegg J, 2018. Prenatal bisphenol A exposure is linked to epigenetic changes in glutamate receptor subunit gene Grin2b in female rats and humans. Sci. Rep 8, 11315 10.1038/S41598-018-29732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisi AM, Della Seta D, Ceccarelli I, Farabollini F, 2001. Bisphenol-A differently affects estrogen receptors-alpha in estrous-cycling and lactating female rats. Neurosci. Lett 310, 49–52. 10.1016/s0304-3940(01)02092-4. [DOI] [PubMed] [Google Scholar]
- Alonso-Magdalena P, Ropero AB, Soriano S, García-Arévalo M, Ripoll C, Fuentes E, Quesada I, Nadal Á, 2012. Bisphenol-A acts as a potent estrogen via non-classical estrogen triggered pathways. Mol. Cell. Endocrinol., Health Impacts Endocr. Disrupters 355, 201–207. 10.1016/j.mce.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Andrews S, 2010. FastQC: A quality control tool for high throughput sequence data. [Google Scholar]
- Arambula SE, Belcher SM, Planchart A, Turner SD, Patisaul HB, 2016. Impact of low dose oral exposure to bisphenol A (BPA) on the neonatal rat hypothalamic and hippocampal transcriptome: a CLARITY-BPA consortium study. Endocrinology 157, 3856–3872. 10.1210/en.2016-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arambula SE, Jima D, Patisaul HB, 2018. Prenatal bisphenol A (BPA) exposure alters the transcriptome of the neonate rat amygdala in a sex-specific manner: a CLARITY-BPA consortium study. NeuroToxicology 65, 207–220. 10.1016/j.neuro.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Berenbaum SA, 1998. How hormones affect behavioral and neural development: introduction to the special issue on “gonadal hormones and sex differences in behavior. Dev. Neuropsychol 14, 175–196. 10.1080/87565649809540708. [DOI] [Google Scholar]
- Bhunia K, Sablani SS, Tang J, Rasco B, 2013. Migration of chemical compounds from packaging polymers during icrowave, conventional heat treatment, and storage. Compr. Rev. Food Sci. Food Saf 12, 523–545. 10.1111/1541-4337.12028. [DOI] [PubMed] [Google Scholar]
- Björnsdotter MK, de Boer J, Ballesteros-Gómez A, 2017. Bisphenol A and replacements in thermal paper: a review. Chemosphere 182, 691–706. 10.1016/j.chemosphere.2017.05.070. [DOI] [PubMed] [Google Scholar]
- Bolger R, Wiese TE, Ervin K, Nestich S, Checovich W, 1998. Rapid screening of environmental chemicals for estrogen receptor binding capacity. Environ. Health Perspect 106, 551–557. 10.1289/ehp.98106551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Neumann ID, 2008. Brain vasopressin is an important regulator of maternal behavior independent of dams’ trait anxiety. Proc. Natl. Acad. Sci. USA 105, 17139–17144. 10.1073/pnas.0807412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RE, Luine V, Khandaker H, Villafane JJ, Frankfurt M, 2014. Adolescent bisphenol-A exposure decreases dendritic spine density: role of sex and age. Synapse 68, 498–507. 10.1002/syn.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL, 2005. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ. Health Perspect 113, 391–395. 10.1289/ehp.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldji C, Diorio J, Meaney MJ, 2003. Variations in maternal care alter GABAA receptor subunit expression in brain regions associated with fear. Neuropsychopharmacology 28, 1950–1959. 10.1038/sj.npp.1300237. [DOI] [PubMed] [Google Scholar]
- Cao J, Rebuli ME, Rogers J, Todd KL, Leyrer SM, Ferguson SA, Patisaul HB, 2013. Prenatal bisphenol A exposure alters sex-specific estrogen receptor expression in the neonatal rat hypothalamus and amygdala. Toxicol. Sci 133, 157–173. 10.1093/toxsci/kft035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X-L, Zhang J, Goodyer CG, Hayward S, Cooke GM, Curran IHA, 2012. Bisphenol A in human placental and fetal liver tissues collected from Greater Montreal area (Quebec) during 1998–2008. Chemosphere 89, 505–511. 10.1016/j.chemosphere.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Cariati F, Carbone L, Conforti A, Bagnulo F, Peluso SR, Carotenuto C, Buonfantino C, Alviggi E, Alviggi C, Strina I, 2020. Bisphenol A-induced epigenetic changes and its effects on the male reproductive system. Front. Endocrinol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro B, Sánchez P, Torres JM, Ortega E, 2013. Effects of adult exposure to bisphenol A on genes involved in the physiopathology of rat prefrontal cortex. PLOS One 8, e73584. 10.1371/journal.pone.0073584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro B, Sánchez P, Torres JM, Ortega E, 2015. Bisphenol A, bisphenol F and bisphenol S affect differently 5α-reductase expression and dopamine–serotonin systems in the prefrontal cortex of juvenile female rats. Environ. Res 142, 281–287. 10.1016/j.envres.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Catanese MC, Vandenberg LN, 2017. Bisphenol S (BPS) alters maternal behavior and brain in mice exposed during pregnancy/lactation and their daughters. Endocrinology 158, 516–530. 10.1210/en.2016-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Diorio J, Sharma S, Meaney MJ, 2001. Naturally occurring variations in maternal behavior in the rat are associated with differences in estrogen-inducible central oxytocin receptors. Proc. Natl. Acad. Sci. USA 98, 12736–12741. 10.1073/pnas.221224598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Weaver I, Diorio J, Sharma S, Meaney M, 2003a. Natural variations in maternal care are associated with estrogen receptor alpha expression and estrogen sensitivity in the medial preoptic area. Endocrinology 144, 4720–4724. 10.1210/en.2003-0564. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Francis DD, Mar A, Meaney MJ, 2003b. Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiol. Behav 79, 359–371. 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Chretien P, Stevenson CW, Zhang TY, Gratton A, Meaney MJ, 2004. Variations in nucleus accumbens dopamine associated with individual differences in maternal behavior in the rat. J. Neurosci 24, 4113–4123. 10.1523/JNEUROSCI.5322-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Weaver ICG, Diorio J, Dymov S, Szyf M, Meaney MJ, 2006. Maternal care associated with methylation of the estrogen receptor-α1b promoter and estrogen receptor-α expression in the medial preoptic area of female offspring. Endocrinology 147, 2909–2915. 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Chang H, Wang D, Xia W, Pan X, Huo W, Xu S, Li Y, 2016. Epigenetic disruption and glucose homeostasis changes following low-dose maternal bisphenol A exposure. Toxicol. Res 5, 1400–1409. 10.1039/c6tx00047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong A, Johnson SA, Howald EC, Ellersieck MR, Camacho L, Lewis SM, Vanlandingham MM, Ying J, Ho S-M, Rosenfeld CS, 2018. Gene expression and DNA methylation changes in the hypothalamus and hippocampus of adult rats developmentally exposed to bisphenol A or ethinyl estradiol: a CLARITY-BPA consortium study. Epigenetics 13, 704–720. 10.1080/15592294.2018.1497388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W-C, Chen J-L, Lin C-F, Chen Y-C, Shih F-C, Chuang C-Y, 2011. Biomonitoring of bisphenol A concentrations in maternal and umbilical cord blood in regard to birth outcomes and adipokine expression: a birth cohort study in Taiwan. Environ. Health 10, 94. 10.1186/1476-069X-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Gatewood JD, Howeth C, Rissman EF, 2010. Gestational exposure to bisphenol A and cross-fostering affect behaviors in juvenile mice. Horm. Behav 58, 754–761. 10.1016/j.yhbeh.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Seta D, Minder I, Dessì-Fulgheri F, Farabollini F, 2005. Bisphenol-A exposure during pregnancy and lactation affects maternal behavior in rats. Brain Res. Bull 65, 255–260. 10.1016/j.brainresbull.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Drobná Z, Henriksen AD, Wolstenholme JT, Montiel C, Lambeth PS, Shang S, Harris EP, Zhou C, Flaws JA, Adli M, Rissman EF, 2018. Transgenerational effects of bisphenol A on gene expression and DNA methylation of imprinted genes in brain. Endocrinology 159, 132–144. 10.1210/en.2017-00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann MN, Auger AP, 2011. Epigenetic impact of simulated maternal grooming on estrogen receptor alpha within the developing amygdala. Brain Behav. Immun 25, 1299–1304. 10.1016/j.bbi.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint S, Markle T, Thompson S, Wallace E, 2012. Bisphenol A exposure, effects, and policy: a wildlife perspective. J. Environ. Manag 104, 19–34. 10.1016/j.jenvman.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Francis D, Diorio J, Liu D, Meaney MJ, 1999. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science 286, 1155–1158. 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- Friard O, Gamba M, 2016. BORIS: a free, versatile open-source event-logging software for video/audio coding and live observations. Methods Ecol. Evol 7, 1325–1330. 10.1111/2041-210X.12584. [DOI] [Google Scholar]
- Fujiwara Y, Miyazaki W, Koibuchi N, Katoh T, 2018. The Effects of low-dose Bisphenol A and Bisphenol F on neural differentiation of a fetal brain-derived neural progenitor cell line. Front. Endocrinol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C-J, Kannan K, 2020. Phthalates, bisphenols, parabens, and triclocarban in feminine hygiene products from the United States and their implications for human exposure. Environ. Int 136, 105465 10.1016/j.envint.2020.105465. [DOI] [PubMed] [Google Scholar]
- Giordano AL, Siegel HI, Rosenblatt JS, 1989. Nuclear estrogen receptor binding in the preoptic area and hypothalamus of pregnancy-terminated rats: correlation with the onset of maternal behavior. Neuroendocrinology 50, 248–258. 10.1159/000125230. [DOI] [PubMed] [Google Scholar]
- Hellstrom IC, Dhir SK, Diorio JC, Meaney MJ, 2012. Maternal licking regulates hippocampal glucocorticoid receptor transcription through a thyroid hormone–serotonin–NGFI-A signalling cascade. Philos. Trans. R. Soc. Lond. B Biol. Sci 367, 2495–2510. 10.1098/rstb.2012.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen AD, Andrade A, Harris EP, Rissman EF, Wolstenholme JT, 2020. Bisphenol A exposure in utero disrupts hypothalamic gene expression particularly genes suspected in autism spectrum disorders and neuron and hormone signaling. Int. J. Mol. Sci 21, 3129. 10.3390/ijms21093129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Wong CKC, Zheng JS, Bouwman H, Barra R, Wahlstróm B, Neretin L, Wong MH, 2012. Bisphenol A (BPA) in China: a review of sources, environmental levels, and potential human health impacts. Environ. Int., Emerg. Environ. Health Issues Mod. China 42, 91–99. 10.1016/j.envint.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Ikezuki Y, Tsutsumi O, Takai Y, Kamei Y, Taketani Y, 2002. Determination of bisphenol A concentrations in human biological fluids reveals significant early prenatal exposure. Hum. Reprod 2839–2841. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Li J, Xu S, Zhou Y, Zhao H, Li Y, Xiong C, Sun X, Liu H, Liu W, Peng Y, Hu C, Cai Z, Xia W, 2020. Prenatal exposure to bisphenol A and its alternatives and child neurodevelopment at 2 years. J. Hazard. Mater 388, 121774 10.1016/j.jhazmat.2019.121774. [DOI] [PubMed] [Google Scholar]
- Kanlayaprasit S, Thongkorn S, Panjabud P, Jindatip D, Hu VW, Kikkawa T, Osumi N, Sarachana T, 2021. Autism-related transcription factors underlying the sex-specific effects of prenatal bisphenol A exposure on transcriptome-interactome profiles in the offspring prefrontal cortex. Int. J. Mol. Sci 22, 13201 10.3390/ijms222413201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Autry AE, Dulac C, 2017. The neurobiology of parenting: a neural circuit perspective. BioEssays 39, e201600159. 10.1002/bies.201600159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Stewart RA, Demas GE, Alberts JR, 2012. Maternal contact differentially modulates central and peripheral oxytocin in rat pups during a brief regime of mother–pup interaction that induces a filial huddling preference. J. Neuroendocr 24, 831–840. 10.1111/j.1365-2826.2012.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A, 2016. Fast gene set enrichment analysis. Bioinformatics, 10.1101/060012. [DOI] [Google Scholar]
- Krüger T, Long M, Bonefeld-Jørgensen EC, 2008. Plastic components affect the activation of the aryl hydrocarbon and the androgen receptor. Toxicology 246, 112–123. 10.1016/j.tox.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Kuiper GGJM, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, Gustafsson J-Å, 1998. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology 139, 4252–4263. 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Champagne FA, 2011. Epigenetic perspective on the developmental effects of bisphenol A. Brain Behav. Immun 25, 1084–1093. 10.1016/j.bbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Gudsnuk K, Franks B, Madrid J, Miller RL, Perera FP, Champagne FA, 2013. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc. Natl. Acad. Sci. USA 110, 9956–9961. 10.1073/pnas.1214056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Olesen KM, Auger AP, 2010. Sex differences in epigenetic regulation of the estrogen receptor-α promoter within the developing preoptic area. Endocrinology 151, 2297–2305. 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaKind JS, Naiman DQ, 2015. Temporal trends in bisphenol A exposure in the United States from 2003–2012 and factors associated with BPA exposure: spot samples and urine dilution complicate data interpretation. Environ. Res 142, 84–95. 10.1016/j.envres.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmler H-J, Liu B, Gadogbe M, Bao W, 2018. Exposure to bisphenol A, bisphenol F, and bisphenol S in U.S. adults and Children: The National Health and Nutrition Examination Survey 2013–2014. ACS Omega 3, 6523–6532. 10.1021/acsomega.8b00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Kannan K, 2013. Concentrations and profiles of bisphenol A and other bisphenol analogues in foodstuffs from the United States and their implications for human exposure. J. Agric. Food Chem 61, 4655–4662. 10.1021/jf400445n. [DOI] [PubMed] [Google Scholar]
- Liao C, Liu F, Alomirah H, Loi VD, Mohd MA, Moon H-B, Nakata H, Kannan K, 2012. Bisphenol S in urine from the United States and seven asian countries: occurrence and human exposures. Environ. Sci. Technol 46, 6860–6866. 10.1021/es301334j. [DOI] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, Sharma S, Pearson D, Plotsky PM, Meaney MJ, 1997. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 277, 1659–1662. 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- Liu D, Diorio J, Day JC, Francis DD, Meaney MJ, 2000. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat. Neurosci 3, 799–806. 10.1038/77702. [DOI] [PubMed] [Google Scholar]
- Lohman BK, Weber JN, Bolnick DI, 2016. Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol. Ecol. Resour 16, 1315–1321. 10.1111/1755-0998.12529. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/S13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens A, Tran V, Kleensang A, Hartung T, 2018. Weighted gene correlation network analysis (WGCNA) reveals novel transcription factors associated with bisphenol A dose-response. Front. Genet 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloy MA, Kochmanski JJ, Jones TR, Colacino JA, Goodrich JM, Dolinoy DC, Svoboda LK, 2019. Perinatal bisphenol A exposure and reprogramming of imprinted gene expression in the adult mouse brain. Front. Genet 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17, 10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV, 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol 20, 3599–3616. 10.1111/j.1365-294X.2011.05205.x. [DOI] [PubMed] [Google Scholar]
- Mielke H, Gundert-Remy U, 2009. Bisphenol A levels in blood depend on age and exposure. Toxicol. Lett 190, 32–40. 10.1016/j.toxlet.2009.06.861. [DOI] [PubMed] [Google Scholar]
- Moore CL, Morelli GA, 1979. Mother rats interact differently with male amd female offspring. J. Comp. Physiol. Psychol 93, 677–684. 10.1037/h0077599. [DOI] [PubMed] [Google Scholar]
- Mustieles V, Pérez-Lobato R, Olea N, Fernández MF, 2015. Bisphenol A: human exposure and neurobehavior. NeuroToxicology 49, 174–184. 10.1016/j.neuro.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Naderi M, Kwong RWM, 2020. A comprehensive review of the neurobehavioral effects of bisphenol S and the mechanisms of action: new insights from in vitro and in vivo models. Environ. Int 145, 106078 10.1016/j.envint.2020.106078. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Itoh K, Yaoi T, Fujiwara Y, Sugimoto T, Fushiki S, 2006. Murine neocortical histogenesis is perturbed by prenatal exposure to low doses of bisphenol A. J. Neurosci. Res 84, 1197–1205. 10.1002/jnr.21020. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Itoh K, Sugimoto T, Fushiki S, 2007. Prenatal exposure to bisphenol A affects adult murine neocortical structure. Neurosci. Lett 420, 100–105. 10.1016/j.neulet.2007.02.093. [DOI] [PubMed] [Google Scholar]
- Narita M, Miyagawa K, Mizuo K, Yoshida T, Suzuki T, 2006. Prenatal and neonatal exposure to low-dose of bisphenol-A enhance the morphine-induced hyperlocomotion and rewarding effect. Neurosci. Lett 402, 249–252. 10.1016/j.neulet.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Owczarek K, Kudłak B, Simeonov V, Mazerska Z, Namieśnik J, 2018. Binary mixtures of selected bisphenols in the environment: their toxicity in relationship to individual constituents. Molecules 23, 3226. 10.3390/molecules23123226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanza PL, Howdeshell KL, Parmigiani S, vom Saal FS, 2002. Exposure to a low dose of bisphenol A during fetal life or in adulthood alters maternal behavior in mice. Environ. Health Perspect 110, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Jang HJ, Lee JH, 2019. Roles of primary cilia in the developing brain. Front. Cell. Neurosci 13, 218. 10.3389/fncel.2019.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patisaul HB, 2020. Achieving CLARITY on bisphenol A, brain and behaviour. J. Neuroendocrinol 32, e12730 10.1111/jne.12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña CJ, Neugut YD, Champagne FA, 2013. Developmental timing of the effects of maternal care on gene expression and epigenetic regulation of hormone receptor levels in female rats. Endocrinology 154, 4340–4351. 10.1210/en.2013-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada I, Fuentes E, Viso-León MC, Soria B, Ripoll C, Nadal A, 2002. Low doses of the endocrine disruptor Bisphenol-A and the native hormone 17β-estradiol rapidly activate the transcription factor CREB. FASEB J. 16, 1671–1673. 10.1096/fj.02-0313fje. [DOI] [PubMed] [Google Scholar]
- Rahmani S, Pour Khalili N, Khan F, Hassani S, Ghafour-Boroujerdi E, Abdollahi M, 2018. Bisphenol A: what lies beneath its induced diabetes and the epigenetic modulation? Life Sci. 214, 136–144. 10.1016/j.lfs.2018.10.044. [DOI] [PubMed] [Google Scholar]
- Rebuli ME, Cao J, Sluzas E, Delclos KB, Camacho L, Lewis SM, Vanlandingham MM, Patisaul HB, 2014. Investigation of the effects of subchronic low dose oral exposure to bisphenol A (BPA) and ethinyl estradiol (EE) on estrogen receptor expression in the juvenile and adult female rat hypothalamus. Toxicol. Sci 140, 190–203. 10.1093/toxsci/kfti074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CA, Taylor JA, Ruhlen RL, Welshons WV, vom SFS, 2007. Estradiol and bisphenol A stimulate androgen receptor and estrogen receptor gene expression in fetal mouse prostate mesenchyme cells. Environ. Health Perspect 115, 902–908. 10.1289/ehp.9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester JR, 2013. Bisphenol A and human health: a review of the literature. Reprod. Toxicol 42, 132–155. 10.1016/j.reprotox.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Rochester JR, Bolden AL, 2015. Bisphenol S and F: a systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ. Health Perspect 123, 643–650. 10.1289/ehp.1408989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester JR, Bolden AL, Kwiatkowski CF, 2018. Prenatal exposure to bisphenol A and hyperactivity in children: a systematic review and meta-analysis. Environ. Int 114, 343–356. 10.1016/j.envint.2017.12.028. [DOI] [PubMed] [Google Scholar]
- Rosenblatt JS, 1994. Psychobiology of maternal behavior: contribution to the clinical understanding of maternal behavior among humans. Acta Paediatr. Suppl 397, 3–8. 10.1111/j.1651-2227.1994.tb13259.x. [DOI] [PubMed] [Google Scholar]
- Russell JA, Douglas AJ, Ingram CD, 2001. Chapter 1 Brain preparations for maternity – adaptive changes in behavioral and neuroendocrine systems during pregnancy and lactation. An overview. In: Progress in Brain Research, The Maternal Brain. Elsevier, pp. 1–38. 10.1016/S0079-6123(01)33002-9. [DOI] [PubMed] [Google Scholar]
- Sheng Z-G, Tang Y, Liu Y-X, Yuan Y, Zhao B-Q, Chao X-J, Zhu B-Z, 2012. Low concentrations of bisphenol a suppress thyroid hormone receptor transcription through a nongenomic mechanism. Toxicol. Appl. Pharmacol 259, 133–142. 10.1016/j.taap.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Siegel HI, Rosenblatt JS, 1978. Duration of estrogen stimulation and progesterone inhibition of maternal behavior in pregnancy-terminated rats. Horm. Behav 11, 12–19. 10.1016/0018-506X(78)90054-5. [DOI] [PubMed] [Google Scholar]
- Somogyi V, Horváth TL, Tóth I, Bartha T, Frenyó LV, Kiss DS, Jócsák G, Kerti A, Naftolin F, Zsarnovszky A, 2016. Bisphenol A influences oestrogen- and thyroid hormone-regulated thyroid hormone receptor expression in rat cerebellar cell culture. Acta Vet. Hung 64, 497–513. 10.1556/004.2016.046. [DOI] [PubMed] [Google Scholar]
- Starr-Phillips EJ, Beery AK, 2014. Natural variation in maternal care shapes adult social behavior in rats. Dev. Psychobiol 56, 1017–1026. 10.1002/dev.21182. [DOI] [PubMed] [Google Scholar]
- Stein TP, Schluter MD, Steer RA, Guo L, Ming X, 2015. Bisphenol A exposure in children with autism spectrum disorders. Autism Res. 8, 272–283. 10.1002/aur.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakovsky RS, Schantz SL, 2018. Impacts of bisphenol A (BPA) and phthalate exposures on epigenetic outcomes in the human placenta. Environ. Epigenet 4 10.1093/eep/dvy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukjamnong S, Thongkorn S, Kanlayaprasit S, Saeliw T, Hussem K, Warayanon W, Hu VW, Tencomnao T, Sarachana T, 2020. Prenatal exposure to bisphenol A alters the transcriptome-interactome profiles of genes associated with Alzheimer’s disease in the offspring hippocampus. Sci. Rep 10, 9487. 10.1038/S41598-020-65229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Mizuo K, Nakazawa H, Funae Y, Fushiki S, Fukushima S, Shirai T, Narita M, 2003. Prenatal and neonatal exposure to bisphenol-a enhances the central dopamine d1 receptor-mediated action in mice: enhancement of the methamphetamine-induced abuse state. Neuroscience 117, 639–644. 10.1016/S0306-4522(02)00935-1. [DOI] [PubMed] [Google Scholar]
- Szyf M, Weaver ICG, Champagne FA, Diorio J, Meaney MJ, 2005. Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Front. Neuroendocrinol 26, 139–162. 10.1016/j.yfrne.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Teng C, Goodwin B, Shockley K, Xia M, Huang R, Norris J, Merrick BA, Jetten AM, Austin CP, Tice RR, 2013. Bisphenol A affects androgen receptor function via multiple mechanisms. Chem. -Biol. Interact 203, 556–564. 10.1016/j.cbi.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongkorn S, Kanlayaprasit S, Jindatip D, Tencomnao T, Hu VW, Sarachana T, 2019. Sex differences in the effects of prenatal bisphenol A exposure on genes associated with Autism Spectrum Disorder in the hippocampus. Sci. Rep 9, 3038. 10.1038/s41598-019-39386-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongkorn S, Kanlayaprasit S, Panjabud P, Saeliw T, Jantheang T, Kasitipradit K, Sarobol S, Jindatip D, Hu VW, Tencomnao T, Kikkawa T, Sato T, Osumi N, Sarachana T, 2021. Sex differences in the effects of prenatal bisphenol A exposure on autism-related genes and their relationships with the hippocampus functions. Sci. Rep 11, 1241. 10.1038/s41598-020-80390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJR, Schoenfelder G, 2010. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect 118, 1055–1070. 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñas R, Goldblum RM, Watson CS, 2013. Rapid estrogenic signaling activities of the modified (chlorinated, sulfonated, and glucuronidated) endocrine disruptor bisphenol A. Endocr. Disruptors 1, e25411. 10.4161/endo.25411. [DOI] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ, 2004. Epigenetic programming by maternal behavior. Nat. Neurosci 7, 847–854. 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Witchey SK, Fuchs J, Patisaul HB, 2019. Perinatal bisphenol A (BPA) exposure alters brain oxytocin receptor (OTR) expression in a sex- and region-specific manner: a CLARITY-BPA consortium follow-up study. Neurotoxicology 74, 139–148. 10.1016/j.neuro.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme JT, Rissman EF, Connelly JJ, 2011. The role of Bisphenol A in shaping the brain, epigenome and behavior. Horm. Behav., Spec. Issue.: Behav. Epigenetics 59, 296–305. 10.1016/j.yhbeh.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, He Y, Zhu H, Huang X, Bai X, Kannan K, Zhang T, 2020. Concentrations of bisphenol A and its alternatives in paired maternal–fetal urine, serum and amniotic fluid from an e-waste dismantling area in China. Environ. Int 136, 105407 10.1016/j.envint.2019.105407. [DOI] [PubMed] [Google Scholar]
- Zimmers SM, Browne EP, O’Keefe PW, Anderton DL, Kramer L, Reckhow DA, Arcaro KF, 2014. Determination of free Bisphenol A (BPA) concentrations in breast milk of U.S. women using a sensitive LC/MS/MS method. Chemosphere 104, 237–243. 10.1016/j.chemosphere.2013.12.085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.