Abstract
We review the emergence of the new field of organ-on-a-chip (OOAC) engineering, from the parent fields of tissue engineering and microfluidics. We place into perspective the tools and capabilities brought into the OOAC field by early tissue engineers and microfluidics experts. Liver-on-a-chip and heart-on-a-chip are used as two case studies of systems that heavily relied on the tissue engineering techniques and that were amongst the first OOAC models to be implemented, motivated by the need to better assess toxicity to human tissues in pre-clinical drug development. We review current challenges in OOAC that often stem from the same challenges in the parent fields, such as stable vascularization and drug absorption.
Keywords: Organ-on-a-Chip, Tissue Engineering, Microfabrication, Drug Testing, Disease Modeling, Microfluidics, High Throughput, Miniaturization
The need for high fidelity healthy and diseased human organ models
It has been a century since the first reported clinical toxicity incident [1]. During the labor-intensive and expensive process of early-stage drug development, each candidate will first be tested in a two-dimensional (2D) cell line-based model and selected ones will progress to animal studies. The 95% failure rate observed for drug candidates during the clinical trials attributes to the lack of preclinical models that faithfully recapitulate human physiology and disease phenotypes.
The field of organ-on-a-chip engineering expanded swiftly, fueled by the promise to provide such predictive human models, as a range of devices were developed to mimic vascular [2,3], liver [4,5], heart [6,7], gut [8], kidney [9], brain [10], and bone [11] tissues. Contrary to the conventional meaning of the word organ, organ-on-a-chip (OOAC) systems do not recapitulate the entire mini organ on a chip. Instead, they only provide specific tissue structures, such as a string of cardiac muscle, clusters of liver cells, or an epithelial-endothelial barrier, that allows scientists to capture the functional hallmarks of the organ that they wish to recapitulate such as contraction force, enzyme secretion or barrier function.
From tissue engineering, OOAC borrowed the concept of the biomimetic approach, mimicking in vitro the environmental parameters a cell encounters in the body, and from microfluidics, it borrowed the actual device where such mimicry occurs. Biomimicry occurs in tissue engineering through cell seeding in tailored scaffolds and addition of biomimetic physical cues and biochemical cues, followed by cultivation in macroscale bioreactors [12] (Figure 1A, Key Figure; Box 1). In organ-on-a-chip engineering, the macroscale bioreactor is now replaced by a microscale microfluidic device generated through various microfabrication methods, such as photolithography, 3D printing or injection molding (Figure 1A, Box 1), with clear advantages in the ability to reduce the number of required cells, precisely control topographical cues and integrate sensors. Often, a biomaterial is incorporated as a scaffold, or coated as a hydrogel into the microfluidic device and cells are seeded into open or closed microdevice wells (Figure 1A), to provide readouts that are amenable to high throughput testing requiring robotic manipulation as well as artificial intelligence powered modeling. Clearly, publications of landmark papers in both tissue engineering and microfluidics (Figure 1B), served as a catapult to initiate a wave of studies that moved the OOAC field forward[13–15].
Figure 1, Key Figure. Organs-on-a-chip arise from tissue engineering and microfluidics relying on microfabrication methods.
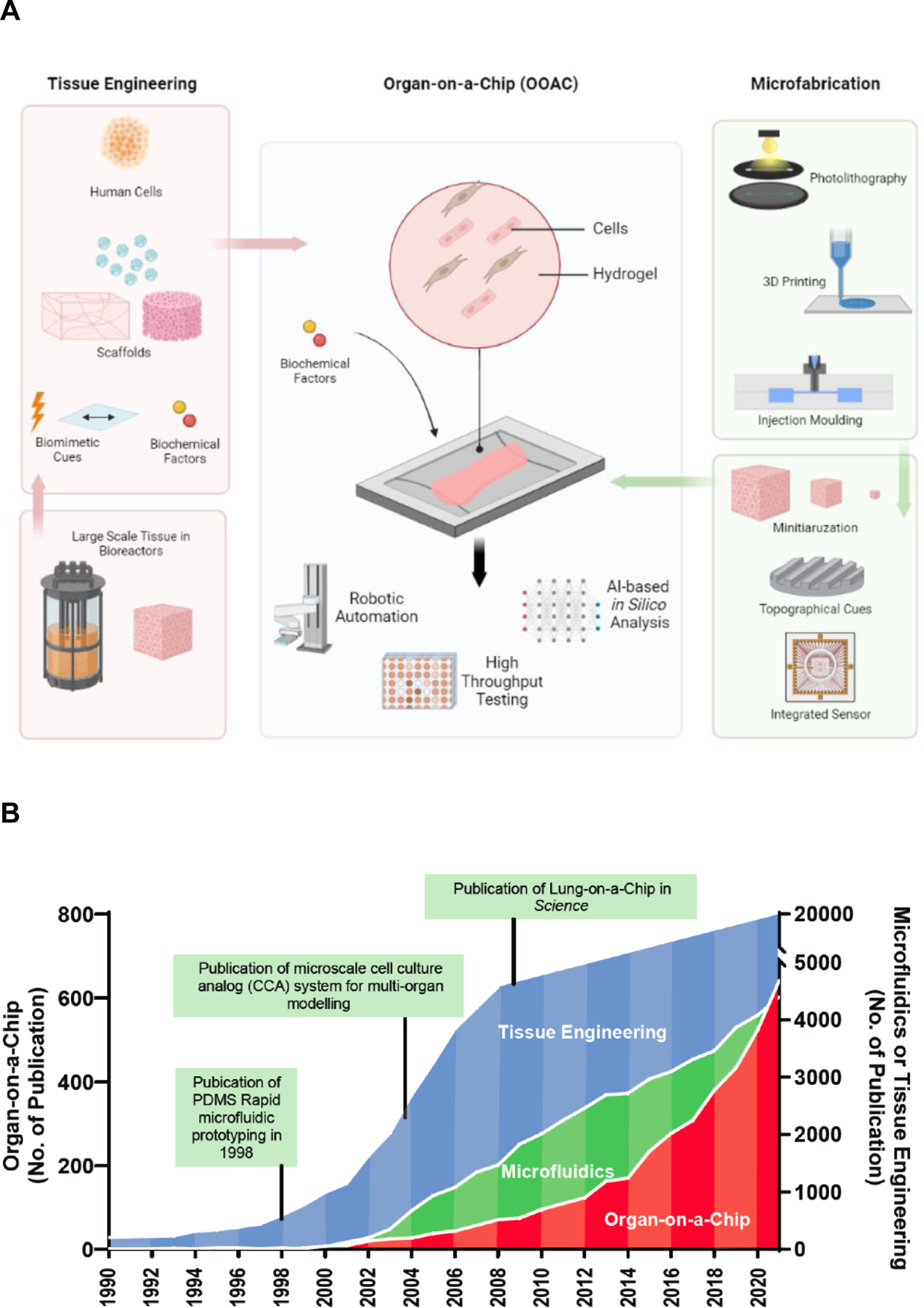
A) Tissue engineering and microfabrication synergistically converge into organs-on-a-chip. Tissue engineering is incorporating cells, biomaterials and biological factors in a bioreactor to create native-like large, engineered tissues for clinical applications. Microfabrication technologies, including 3D printing, hot embossing and injection molding, can create miniaturized bioreactors with built-in topographical features and biosensors. Organs-on-a-chip inherits these capacities from tissue engineering and microfabrication and resulting in platforms for drug screening and disease modeling. B) The exponential increase of organ-on-a-chip publications over the past decade. The number of publications annually with keywords “tissue engineering”, “microfluidics” and “organs-on-a-chip” from the National Library of Medicine database.
Box 1. Historical perspective on organ-on-a-chip engineering.
Organs-on-a-chip is an emerging technology that can bridge the gap between in vitro 2D multi-well cell culturing and in vivo animal models for drug discovery and human disease research. This field of science is a convergence of technological advances made in tissue engineering and microfluidics, that allows human organs to be miniaturized onto a hand-held platform, where biomechanical, biophysical and multicellular cues are integrated in a spatiotemporally controlled manner to facilitate the formation of 3D tissue and mimic essential organ-level functions. The term tissue engineering was coined in the late 1980s[119] and developed through the paradigm of the integrated use of biomaterials, cells and cm-scale bioreactors, as defined by Langer and Vacanti in the early 1990s[120], to build functional tissues for repair of injured or diseased human tissues and organs. Around the same time, the Whitesides Group detailed a fabrication technique that allows for precise control over substrate microstructure design and replica molding with the elastomer poly(dimethylsiloxane) (PDMS) [121]. These advances underpinned the emergence of the OOAC field.
The term “organ-on-a-chip” was introduced in 2010 by Huh and Ingber’s “lung-on-a-chip” research[15]. Yet, the organ-on-a-chip research has been carried out much before this landmark paper was published. For example, Shuler and colleagues demonstrated the early concept of organ-level interaction with a cell culture analog system for drug toxicity assessment and introduced the idea of body-on-a-chip in the late 1990s[14].
Current organ-on-a-chip platforms can be commonly defined by some of these four characteristics: (i) presence and incorporation of multiple cell types to recapitulate heterogeneous cell population in a tissue (such as vascular, stromal, parenchymal, and immune cell types), (ii) microfabricated structures and microfluidic channel(s) to model native tissue under biophysical stimuli such as shear flow, (iii) an interface of membrane or pillar structures to mimic the transport of nutrients/drugs and oxygen or cellular trafficking that emulates the biochemical and cellular milieu of in vivo like setting and (iv) customized microstructured cellular compartment(s) that allow tissue constructs to recapitulate key organ functions.
Efforts in the OOAC are now directed towards cost-effective and streamlined manufacturing processes in a scalable manner. Fabrication with a fast turnaround time from device design to implementation and increasing throughput at all steps: fabrication, cultivation, sensing and data analysis are essential to the success of OOAC models.
Tissue construction in OOAC: learning from tissue engineering
The OOAC field directly applies the main tissue engineering paradigm: the integrated use of cells, biomaterials and a bioreactor (Box 2). In tissue engineering, cells are seeded into 3D scaffolds made of natural, synthetic, or mixed components, to form a specific tissue for the ultimate goal of implantation in the body to restore function in patients (Figure 2) [16,17]. Once the cells have established their phenotypes and started producing ECM, the supportive scaffold or hydrogel material is sometimes degraded and most often remodeled in both tissue engineering and OOAC [18,19] For example, in the Biowire II heart-on-a-chip device, induced pluripotent stem cell (iPSC) derived cardiomyocytes are initially seeded into a microfabricated device (a hot embossed polystyrene well with electrodes) using a collagen hydrogel[20]. As the cells remodel the initially provided hydrogel in the microbioreactor a contractile tissue is formed. In contrast to tissue engineering, OOAC microbioreactors assemble functional tissues from cells and biomaterials (most often hydrogel) to result in a functional hallmark that can be addressed under both healthy and disease states (Figure 2). The measurement of functional properties in response to drug and biologic libraries ideally can lead to readouts that can be used to understand mechanism and safety of the molecule required for regulatory approval, ultimately resulting in new therapies for patients.
Box 2. The role of tissue engineering and microfluidics in organ-on-a-chip engineering.
Essential to tissue engineering approaches is understanding the structure and function of the native organ. According to biomimetic principles, tissue engineers strive to recapitulate structure of the native organ, in order to achieve the tissue-specific function.[122] The microfabricated device enables that structural control, while cells provide the function.
The microfluidic and OOAC devices are still most often engineered from polydimethylsiloxane via soft lithography techniques, although recent reports provide elegant plastic-based solutions by engineering devices from polystyrene or acrylic[54]. Their one unifying feature is the dimensional scale, i.e. all processes are confined to the scale of ~100μm. This enables achieving a laminar fluid flow and often a creeping flow characteristic of the capillaries. Yet, this dimension overcomes one deadly limitation for tissue engineers- inability to achieve thick tissues due to oxygen diffusional limitations. In highly dense tissues such as the heart or the liver, when cultivation is attempted at cell densities similar to those in the body (~108cells/cm3), high metabolic demand will result in oxygen depletion within ~100μm in the absence of convection, i.e. for mass transport through the tissue that relies on diffusion alone [123]. Tissue engineers strive to overcome these issues by stable vascularization and interstitial flow or culture media perfusion through the defined conduits.[122] Confining the scale of the tissue to a microfluidic device eliminated the single most significant constraint that has plagued the tissue engineering field for decades and by doing so created an unlimited playing field for tissue engineers.
The cells are true tissue engineers and the one ingredient that effectively differentiates microfluidics and organs-on-a-chip. The OOAC devices are designed to mimic extracellular (ECM) cues, biochemical stimuli and physical forces the cells experience in the body to enable their establishment of functional tissues. The OOAC device itself, most often borrowed from microfluidics, is also used to effectively measure the key parameters, such as permeability or contractility, a feature that macroscale tissue engineering bioreactors most often lacked.
Achieving an authentic 3D culture that combines physiological relevance of multiple cell types, high throughput, reproducibility, easy translation from academic to industrial labs, and versatility remains challenging. Limitations of the parent fields, e.g. vascularization of tissue engineering, scalable manufacturing and drug absorption into the device of microfluidics, are still the main challenges of the OOAC field. Correlating the relevancy of OOAC models to the known clinical data and modeling human genetic disease remain exciting frontiers.
Figure 2. Convergence of organ-on-a-chip and tissue engineering principles.
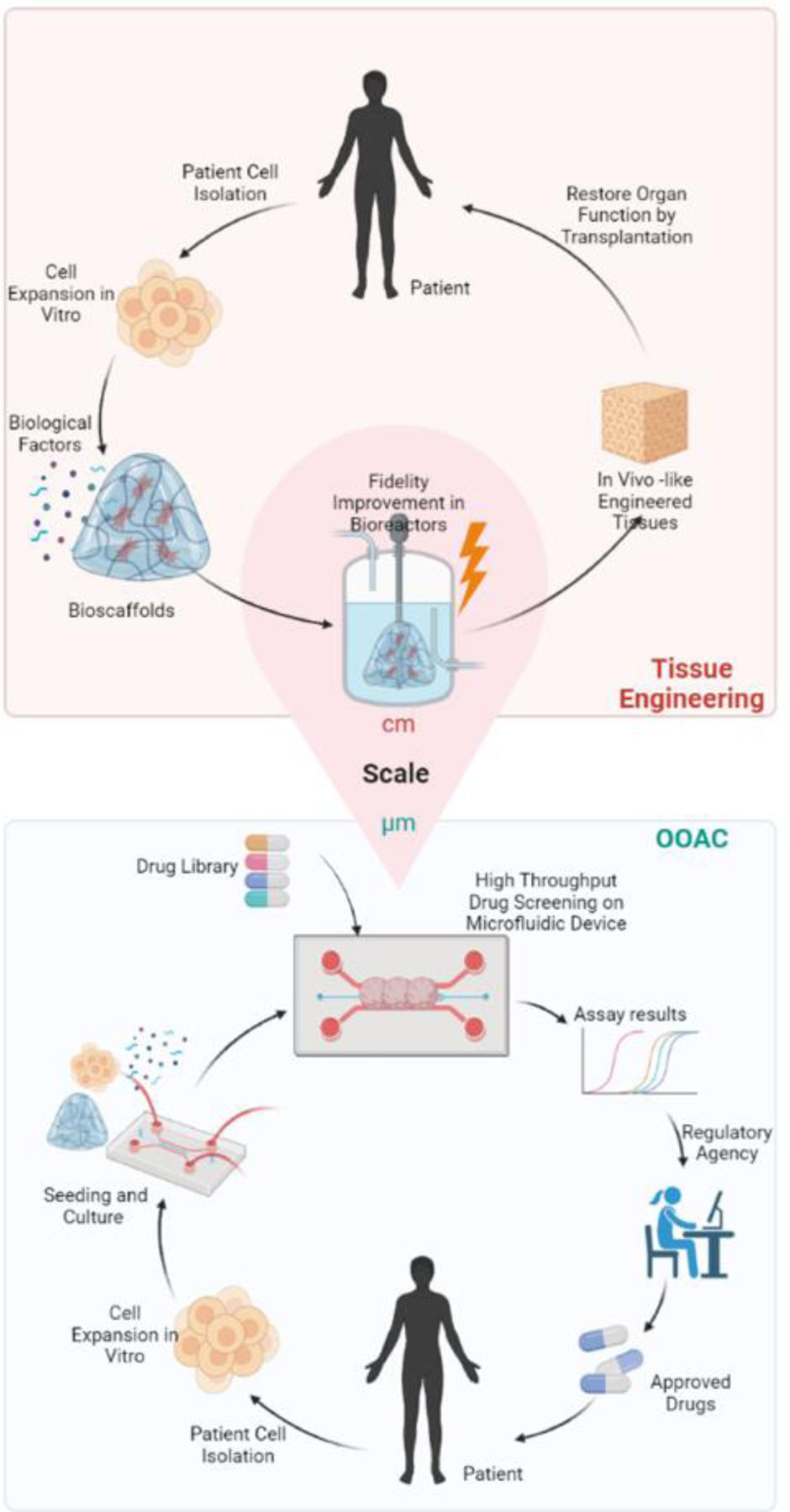
The tissue engineering pipeline incorporates cells (from patients), biomaterials, and biological stimuli to recapitulate the microenvironment of native tissue and to create large-sized functional tissue or cellular products for implantation. Tissue engineering products aim to replace the damaged tissue in the human body. As tissue replacement requires large tissue sizes, oxygen delivery and complete vascular integration remain an obstacle in the tissue engineering field. Organ-on-a-chip, on the other hand, can use the same tissue culture strategies as tissue engineering and create highly reproducible micrometer-sized tissues in microfabricated miniaturized bioreactors. Microfabrication can facilitate the integration of multiple elements to advance the organ-on-a-chip design: biological features, i.e. perfusable channels recapitulating blood flow; biosensors such as force sensors and multi-electrode arrays; and biological stimuli to promote tissue fidelity. The resulting device can provide functional evaluations of drug candidates and contributes to the validation and safety evaluations for regulatory approval before the drugs enter the market.
Cell sources and composition
Early studies in OOAC relied largely on cell lines[14,15], which were effective in recapitulating the tissue structure and validating the device operation. The field has since moved towards the use of primary cells for tissues that allow for such opportunity (e.g. skin[21]) or the use of adult stem cell derived cells (e.g. intestine[22]) and ultimately human-induced pluripotent stem cell (hiPSC) progeny that have the capacity to give rise to any tissue in the body [23,24](Figure 3A). The field of OOAC disease modeling in particular has been transformed from the conventional primary cells and cell lines to hiPSC differentiated cells.[25,26] Yet, it is important to place into context the limitations of each of these sources: primary cells cannot be obtained and propagated for some of the key tissues (e.g. neural or cardiac), due to the limited ability of the key tissue cells (neurons and cardiomyocytes) to proliferate[27], whereas stem cell derived progeny often suffer from limitations such as low level of maturation, resembling fetal rather than the adult cells.[17,23,24,28] Importantly, recent studies have focused on overcoming those maturation issues by either application of physical forces (e.g. electrical stimulation [17,29]), prolonged culture[30] (Figure 3B), the use of specialized culture media [31], or cultivation on polydimethylsiloxane (PDMS) substrates that are softer than tissue culture plastics [32].
Figure 3. Engineering strategies for tissue construction and platform fabrication.
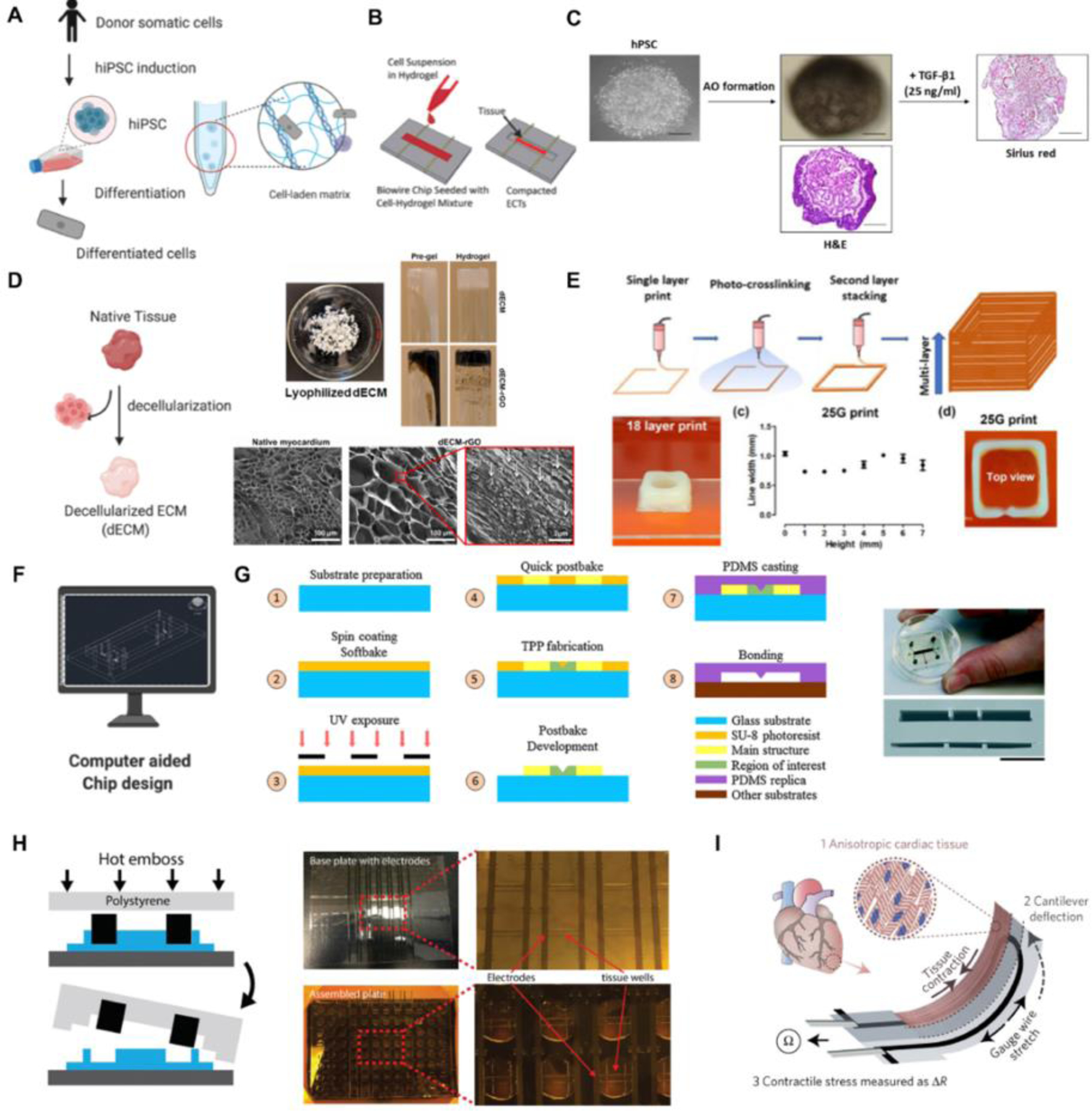
(A) Stem cell differentiation and cell encapsulation for the construction of engineered tissues. (B) Cell seeding for the construction of an engineered cardiac model on Biowire platform, reproduced with permission[17]. (C) Organoid formation through self-aggregation for fibrotic disease modeling, reproduced with permission [124]. (D) Construction of cardiac tissues with hydrogels comprised of ECM from porcine hearts, reproduced with permission [125]. (E) A multi-layer tissue model constructed with 3D bio-printing, reproduced with permission [125]. (F) Platform design using computer-aided tools. (G) Soft lithography and replica modeling procedures for the microfabrication of a PDMS device, reproduced with permission [126]. (H) Hot embossing process for the microfabrication of a scalable 96-well plate platform using tissue culture grade polystyrene, reproduced with permission[53]. (I) Microfabrication of an instrumented cardiac device via multi-material 3D printing, reproduced with permission [127].
Just as early tissue engineers quickly realized that a tissue such as myocardium or liver cannot be made effectively with only one cell type, OOAC engineers immediately implemented co-culture approaches in their devices. This was achieved by e.g. establishing an endothelial/epithelial co-culture system on a permeable membrane within an organ-on-a-chip device to achieve barrier function in systems such as lung[15], kidney[33] or gut-on-a-chip[34]. The supporting populations such as fibroblasts are one of the critical components of constructing healthy models for functional liver and heart OOAC as well as development of the fibrotic tissue models[35,36]. It is important for tissue chips to incorporate fibroblasts of the same genetic background in a physiologically relevant ratio. Additional supporting cells such as macrophages are also being explored to augment tissue assembly[37]. They can also be introduced in the fluids perfusing vascularized tissues to recapitulate the cell’s pathological environment during the inflammatory cascade[38].
Biomaterials
The biomaterials that fueled the growth of tissue engineering are most often used to support the cells in OOAC devices. Hydrogels have played a central role in OOAC where they are used as scaffolds to guide the growth of tissues. [39–43] Hydrogels can be made either from natural (e.g., collagen, fibrin, gelatin, alginate, chitosan, HA, etc.) or synthetic polymers (e.g., PLA, PEG- derivatives, PVA, etc.), and can be crosslinked through covalent or noncovalent bonds. Scaffold-free constructs such as tissue spheroids and organoids can be formed through self-aggregation by suspending cells in low-attachment plates or using magnetic levitation[44] followed by embedding them into the microstructured OOAC devices. Decellularized ECM (dECM) within microphysiological systems have advantages as they preserve the intrinsic biochemical and topographical micro-environments[39]; e.g. decellularized porcine cardiac ECM in a heart-on-a-chip device has been used to support functional cardiac tissue assembly from iPSC-derived cardiomyocytes[40] (Figure 3C and D).
Synthetic polymers (e.g. elastomers) are becoming increasingly popular in in vitro modeling field since their properties can be engineered for specific applications[43]. They can be produced in large uniform quantities and have reproducible properties such as tensile strength, elastic modulus, and degradation rate. Photo-crosslinkable hydrogels, including poly(ethylene glycol) diacrylate (PEGDA), gelatin methacryloyl (GelMA) and methacrylated hyaluronic acid (MeHA), are among the most commonly used [41,42]
OOAC device seeding and cultivation
Seeding cell-laden hydrogel into the densely packed and precisely organized structure to exhibit organ-specific functions is an important step of tissue construction (Figure 3E). Manual loading of the cell-gel mixture by pipetting into a device is still most common [17,38,45–47], but it is not suitable for high-through studies, and often results in unsatisfying reproducibility due to inconsistent seeding. Using the techniques of 3D printing, electrospinning, automated liquid handlers and the combination of two or more of these processes some of these advantages can be overcome[48](Figure 3E).
OOAC device fabrication: learning from microfluidics
A variety of microfabrication techniques including soft lithography, replica modeling, hot embossing along with rapidly evolving 3D printing have been leveraged to engineer OOAC platforms and devices (Box 2, Figure 1). The surface chemistry of the materials for chip fabrication can significantly affect cell behavior and drug effects[46,47], so platform material and corresponding manufacturing techniques should be carefully selected.
Soft lithography and replica molding for PDMS device fabrication
Polydimethylsiloxane (PDMS), a silicon-based organic polymer, has been the most popular material for the fabrication of OOAC platforms[49]. Its desirable features include easy handling, cost-effectiveness, biocompatibility, optical clearance for observation, permeability to gases and mechanical flexibility. PDMS-based devices can be easily fabricated using standard soft lithography techniques followed by replica molding [50] (Figure 3F and 3G). However, there are considerable limitations associated with these devices. First, soft lithography and replica molding remain a multi-step process that requires access to the clean room, dedicated equipment, and substantial manual handling. Inherently, these steps constrain the throughput of device fabrication and reproducibility. Second, the high absorption of hydrophobic molecules into PDMS limits its application in drug screening[47]. Moreover, PDMS tends to swell under various conditions, especially in hydrocarbon-based solvents[46].
Fabrication of plastic devices
Plastic devices will only adsorb but not absorb small molecules such as drug candidates, a limitation that can be easily overcome via device coating steps with e.g. Pluronic or bovine serum albumin[17]. Thus, thermoplastic-based materials such as polymethylmethacrylate, polycarbonate, and polystyrene are rapidly becoming the second most popular class of materials used in OOAC microfabrication[51]. These materials have low electrical conductivity and high chemical stability, and they are well suited for mass production at a low cost[52]. Hot embossing is a widespread thermoforming method for manufacturing thermoplastic devices. It is suitable for producing devices in well-plate formats at significantly increased throughput compared to soft lithography and PDMS replica molding[53]. Briefly, this process involves heating a thermoplastic sheet to its glass transition temperature and pressing it against a thermally durable template. Silicon, metal, or PDMS templates have been used as the hot embossing masters, given their ability to resist deformation under heat and pressure. Micromilling of acrylic and polymethylmethacrylate is also effectively used for rapid prototyping of complex plastic OOAC devices[54] (Figure 3H). To further scale-up the manufacturing throughput requires injection molding and laser welding technologies that easily reduce the production costs after the initial stages of research and development.[55]
Incorporation of sensing and stimulating capabilities
A meaningful organ-on-chip model cannot be achieved without retrieving data at characteristic points in the system and incorporating additional components of the device such as stimulus loading components and sensors (Figure 3I). For example, embedding field stimulation or point stimulation electrodes that mimic the action of pacemaker cells allows the conditioning of cardiac tissue chips, thus allowing the tissue to reach a higher level of maturity, as well as control of beating rate during drug testing [17]. Embedding electrodes at scale requires complex multi-material fabrication techniques[53]. A variety of in vitro models have adopted external mechanical stimulation such as mechanical stretch and compression to evaluate fibrotic remodeling[56]. Additionally, thin and elastic microporous membranes allow for establishment of a selective barrier function and mimicking the breathing motion in a lung-on-a-chip device, via lateral membrane extension[15]. Micropillar arrays for force readouts were fabricated using a multi-layer microlithography technique and replica molding[56]. Microelectrode array (MEA) was incorporated to measure electrophysiological activities on device, including the electrical signal propagation across tissues and electrophysiology of adjacent cell clusters.[57,58] A high-speed mechano-active multielectrode array can realize the strain amplitude modulation and electrophysiological recordings at the same time[59].
Heart- and liver-on-a-chip: case studies for the implementation of tissue engineering in OOAC
Contributions of tissue engineering to delineating liver cell culture conditions
Highly simplified in vitro assays of culturing primary hepatocytes or hepatic cell lines on plastic culture dishes have been attempted for predicting a particular aspect of drug metabolism and toxicity. However, these models generally failed due to the rapid de-differentiation of the primary hepatocytes and, consequently, the loss of metabolic function over time [60,61].
Early tissue engineers spearheaded liver OOAC efforts by delineating critical contributions of appropriate matrix and heterotypic cell-cell interactions. As demonstrated by the Yarmush group and others, culture of primary hepatocytes sandwiched between two collagen layers [62] has shown promise in retaining drug-metabolizing capabilities and transporter expression on the sinusoidal membranes [63,64]. This technique remains a gold standard in liver cell culture. The Bhatia group pioneered the concept of heterotypic interactions with hepatocyte/fibroblast co-culture in islands of defined scale, to recapitulate short-range signaling between the two cell types [65]. Most early models often focused on cellular survival and maintaining hepatocyte-like activity where albumin synthesis and urea secretion are used as system readouts [65,66]. Similarly, Cho and co-workers demonstrated that by seeding a layer of fibroblast as a feeder layer the liver-specific functions of the hepatocyte, including intracellular albumin generation, urea synthesis, and glycogen storage were significantly enhanced due to the increased heterotypic contact [66]. In most cases, these early static models did not recapitulate the sinusoidal flow that would influence uptake and efflux of compounds.
Liver-on-a-chip: a platform for recapitulating liver drug metabolism and disease states
As the field evolved, researchers began to model specific features of the liver, for example, sinusoidal flow, and demonstrate their effect on hepatocyte adsorption, distribution, metabolism, and excretion (ADME) relevant activities. These liver-on-a-chip systems usually consist of hydrogel encapsulated hepatocytes or 3D liver spheroids behind a thin, porous, membrane [5,67] or micropillars [68] that permits the transport of metabolites and small proteins, while protecting the seeded hepatocytes from the effects of shear stress ultimately achieving improved hepatic functions over days in culture compared to the static conditions [68]. Similarly, utilizing a dual microchannel configuration system, both Kang and colleagues and Prodanov and colleagues separately demonstrated a long-term mimicry of the liver sinusoid on a chip platform for up to four weeks, when co-culturing hepatocytes with endothelial cells and stellate cells under continuous perfusion [4,69]. Recently, Tan and co-workers developed a high-throughput microfluidic system that can accommodate 96 perfusable miniature liver-on-a-chip systems in 384-well format [70]. To simplify the complex metabolic conversion of the drug into the active components and study their effect on downstream organs, Sung and co-workers. developed a micro-physiological system featuring liver, tumor, and bone marrow interconnected by a single perfusable channel, capturing metabolism of Tegafur to 5-FU and downstream death of colon cancer cells (Figure 4A) [71]. Using a similar configuration, Chen and colleagues showed a 3 to 10-fold higher expression of common cytochrome P450 (CYP) enzymes (CYP1A1 and CYP3A4) activity, compared to traditional culture [72]. By linking three tissues – liver, heart, and lung – in a closed circulatory perfusion system, Skardal and co-workers demonstrated unexpected secondary effects on cardiac organoids (e.g. loss of contractile machinery) upon bleomycin treatment, an anti-cancer chemotherapy drug that can cause pulmonary inflammation and fibrosis (Figure 4B) [73], which was the result of interleukin-1β released from lung epithelium. In a recent study by Yin and colleagues, the use of human-induced pluripotent stem-cell (hiPSC) derived organoids to model liver and heart revealed that although clomipramine can be actively metabolized by liver organoids, the active metabolite of anti-depressant causes significant impairment to cardiac contractile activity and calcium flux (Figure 4C) [74].
Figure 4: Representative application of organ-on-chip system in drug screening and toxicity testing.
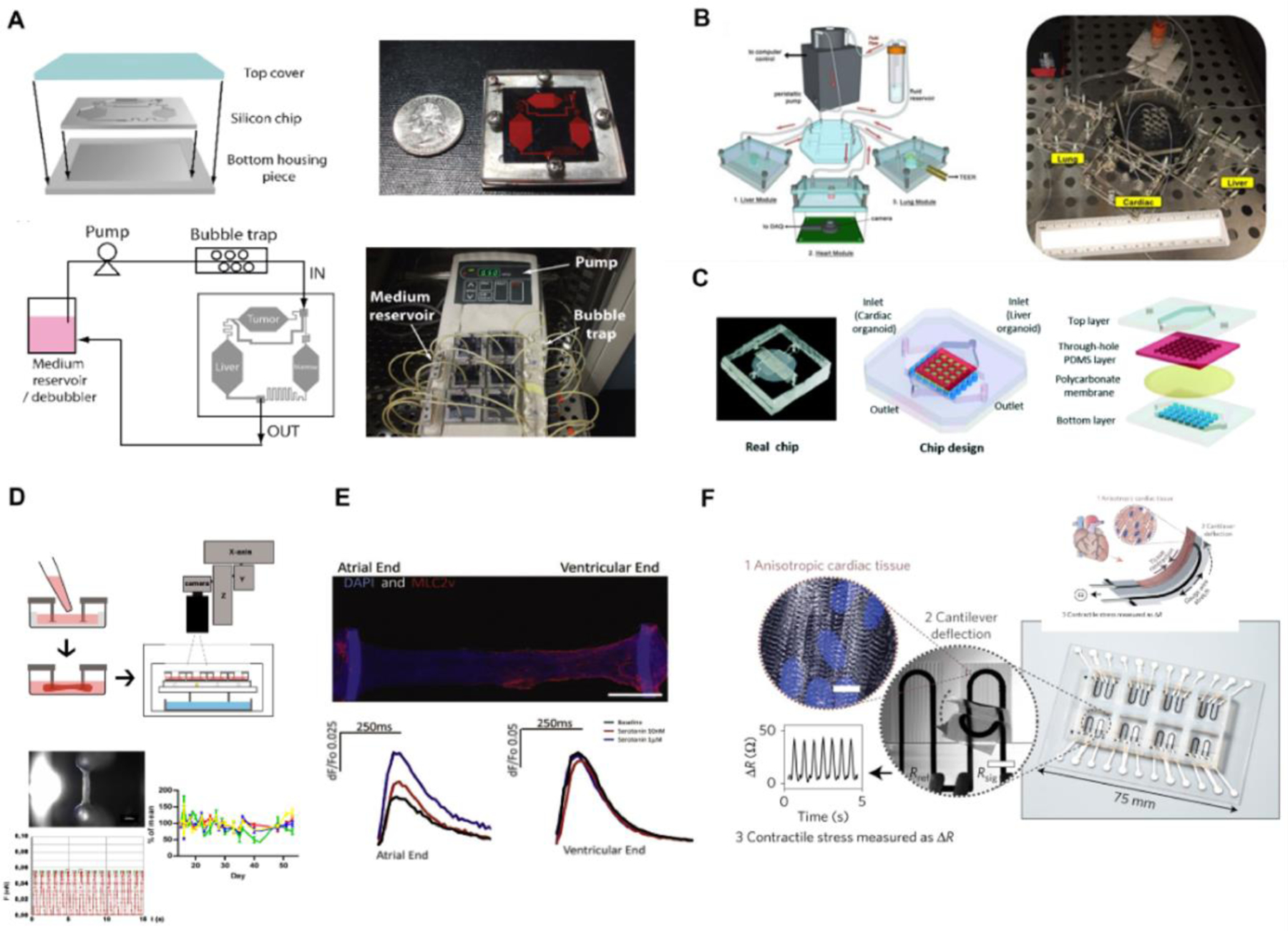
Liver: Panel A-C; Heart: Panel D-F.
(A) Schematics and photographs of a silicon-based perfusable liver-on-a-chip system connected to tumor tissue and bone marrow tissue downstream to study the effect of active metabolite 5-fluorouracil. Reproduced with permission[71]. (B) Schematic and photograph demonstrating a “plug and play” design of liver-on-a-chip hardware system connected to adjacent heart-on-a-chip and lung-on-a-chip systems, demonstrating an altered drug response in heart contraction rate. Reproduced with permission[73]. (C) A design of integrated organ-on-chip system with liver organoid in the top layer and heart tissue organoid at the bottom layer. Metabolism-dependent cardiac toxicity of clomipramine was assessed through a change in heart contraction rate. Reproduced with permission[74]. (D) Schematic overviews of a post-based heart-on-a-chip system for studying heart output through post deflection. Optical video recording of post deflection can generate contraction force, frequency, and relaxation velocity through post geometry and elastic properties. Reproduced with permission[97]. (E) An immunostaining image of heteropolar atrial and ventricular cardiac engineered tissue on Biowire II system. Serotonin only affects the calcium transient to the atrial region of the cardiac construct in a concentration-dependent manner. Reproduced with permission[17]. (F) A schematic and immunostaining image of cardiac muscular thin-film technology with embedded multilayer cantilevers to measure the contraction of an anisotropic cardiac tissue. A soft strain gauge sensor embedded in the PDMS film measures the changes of resistance corresponding to cardiac contractile outputs. Reproduced with permission[7].
In addition to studying drug metabolism and toxicity effect, researchers have begun utilizing the liver-on-a-chip system in disease modeling, such as non-alcoholic fatty liver disease (NAFLD) [75]. The liver-on-a-chip model has also been used to emulate the cancer metastasis cascade [76,77].
Contributions of tissue engineering to delineating conditions for functional myocardial tissue formation
Early tissue engineers were consistently focused on engineering a patch for the repair of infarcted or otherwise impaired myocardial function (e.g. as a result of congestive heart disease)[78–80]. In the process, they made important advances that subsequently fueled the progress of heart-on-a-chip engineering. These advances include recognizing the importance of non-myocytes in creation of functional cardiac tissue in vitro, incorporating mechanical and electrical stimulation to drive differentiated cell phenotype, and defining conditions that lead to controllable hydrogel matrix compaction during tissue remodeling. Importantly, human adult cardiomyocytes are terminally differentiated and cannot be expanded from biopsies.[27] Early tissue engineers developed important 3D culture methods relying on neonatal rat cardiomyocytes as a model cell population, a full decade before culture of cardiomyocytes from human pluripotent stem cells became a reality[81–83]. Early validation of OOAC approaches, also relied on neonatal rat cardiomyocytes [84].
Cardiac fibroblasts make up to 50% of the total myocardial digestate[85] and overgrow in 2D culture. The importance of fibroblasts in engineering functional cardiac patches[86] has been confirmed by multiple independent laboratories leading to the consensus use of FBs in cardiac tissue engineering in vitro and in vivo[87–89].
Electromechanical coupling is critical for the function of the native cardiac muscle. Zimmermann and Eschenhagen pioneered the use of mechanical stimulation, first static stretch, then unidirectional dynamic stimulation and finally auxotonic stimulation [83,90,91]. We first used chronic electrical field stimulation to enable assembly of individual neonatal rat cardiomyocytes into a functional and differentiated cardiac tissue [82]. Current cardiac tissue engineering as well as OOAC systems include at least one of these complex electromechanical stimuli[92,93].
Ability to control cell-gel compaction was also elegantly demonstrated by Zimmermann and Eschenhagen, by creating a tissue using collagen or fibrin hydrogels first anchored to two pieces of Velcro® then to a pair of posts [83,90,91]. Subsequently, the Bursac Lab demonstrated the creation of complex and highly conductive tissues using cell-gel compaction and ultimately, the creation of very large contractile cardiac patches[79,94].
Heart-on-chip: a synergistic approach to provide relevant clinical assessment
Most heart-on-a-chip studies describe the development of cylindrical structures of human ventricular myocardium from iPSC-derived cardiomyocytes, with diameters of several hundred micrometers, anchored at two ends of the OOAC platform[40,95,96] [97] (Figure 4D), although cultivation of muscle strips in microfluidic channels is also present[98,99]. Their contraction force can be measured either through deflection of polydimethylsiloxane (PDMS) posts[25] or wires[17]. Recent studies report more sophisticated systems that enable independent control of loading and shortening[100].
Nunes and colleagues, and Zhao and colleagues, modified an electrical stimulation protocol originally developed for cardiac tissue engineering to mature these strings of tissue by ramping electrical stimulation frequency (up to 6Hz) [101], eliminated the use of drug-absorbing PDMS[20], and enabled non-invasive continuous measurement of contraction force by imaging (Figure 4E) created defined atrial, ventricular and atrioventricular tissues from human iPSCs. The initial experimental design ramped the frequency up to 3Hz expecting cell maturation, as heart rate peaks at 3Hz at 7 weeks gestation[101]. Ramping the frequency to 6Hz was originally designed with the expectation that heart failure conditions would arise with rapid pacing. Surprisingly, the data demonstrated superior cell maturation in the 6Hz vs 3Hz protocol [101]. Importantly, Ronaldson-Bouchard and colleagues designed cardiac microtissues of unprecedented maturity level using similar intensity training of increasing contractile pacing rate between 1–6Hz and starting from cardiomyocytes at an earlier stage of differentiation (day 12 vs day 20)[29].
Cardiac tissue contractile performance can be evaluated from deflection of PDMS cantilevers seeded with cardiomyocytes[102,103]. By incorporating the system in a fluidic channel, Agarwal and co-workers provided an automated fluidic control that allowed for washout of drug dosage after assessment[103].
The generally low throughput of optical sensing methods of contractility motivates the development of new approaches. Lind and colleagues embedded flexible strain gauge sensors through multi-material 3D printing techniques into PDMS cantilevers (Figure 4F) [7]. The embedded sensors provided an electrical approach to continuously measure contractile properties [7,104].
Whereas models of healthy human myocardium are needed to effectively screen for drug toxicity, drug discovery critically depends on the availability of the appropriate models of diseased myocardium. To this end, progress has been made on the models of genetically inherited heart disease, caused by mutations in a single protein such as cardiomyopathy as a result of sarcomeric protein titin truncations [25] or mitochondrial protein taffazin mutations [26] and hypertrophic cardiomyopathy resulting from LIM protein mutations to name a few [16]. Williams and co-workers modeled a polygenic cardiac disease, left ventricular hypertrophy, using cells from patients with high blood pressure who are enrolled in the NIH HyperGen study[105]. Wang and Mastikhina induced cardiac fibrosis through a direct mixture of fibroblasts and cardiomyocytes in the same tissue or chemically applying a known pro-fibrotic cytokine, TGF-β.[106] Wang and colleagues developed a model of drug-induced cardiomyopathy, by chronically treating Biowire II cardiac tissues with Angiotensin II for 3 weeks[107]. These models of interstitial cardiac fibrosis were found to have a proteomic signature closer to the human fibrotic myocardium than the animal model of aortic stenosis[108]. Ma and co-workers developed filamentous matrices through a photocurable hybrid polymer and demonstrated that mechanical cues from different stiffness fibers could affect the contractile function of their microtissues [109].
Concluding remarks and future perspectives
Organ-on-a-chip platform technology is promising as it offers a controllable microenvironment to facilitate the understanding of tissue biology and enable drug screening. Current platforms do have their shortcomings (see Outstanding Questions) that interestingly stem from the parent fields of tissue engineering and microfluidics (Box 2).
Outstanding Questions.
How much tissue maturation is enough to capture key physiological responses in OOAC?
How many cell types should we add to the OOAC to faithfully recapitulate the organ function?
Where in the drug development pipeline should OOAC devices be placed: before animal studies, in parallel to animal studies, or completely replace animal studies?
What is needed to be able to replace human clinical trials, i.e. run the trial on a chip instead of with human volunteers?
What are the appropriate scaling approaches when connecting multiple OOACs on a single platform?
As production throughput increases, cultivation and analysis become bottlenecks, thus what is the highest possible throughput of organ-on-a-chip systems?
Since OOAC systems are high-content (although not high-throughput at this time), how can we effectively handle the extensive amount of physiological data as the throughput increases?
What is the minimal cultivation time required to achieve physiologically relevant functional readouts?
How can we design more simplified, user-friendly, and reproducible systems?
Which physiological factors are we currently missing in the OOAC devices, e.g. appropriate topographical cues, more refined metabolism, etc?
Which factors should we further refine, e.g. ECM, mechanical, or electrical stimulation?
Can we create universal culture media?
How can we establish cell-cell circuits in OOAC devices, so cells can support themselves without extensive exogenous supplements, as they do in the body?
What is needed to achieve full organ morphogenesis on a chip starting from human iPSC?
Can we better model patient-specific OOACs by using isogenic cell sources?
How does the multi-parametric functional evaluation translate to patient-specific clinical outcomes?
Most OOAC devices still only incorporate 2–3 main cell types (e.g. cardiomyocytes, endothelial cells and fibroblasts), often falling short of the physiological complexity of cell composition of the native organ, the same limitation the original tissue engineers encountered. Achieving stably perfusable vasculature over weeks is still not possible. In liver-on-a-chip, incorporation of bile ducts and liver cholangiocytes remains a challenge, whereas in the cardiac field, incorporation of resident macrophages, autonomic neurons, epicardial cells and endocardial cells is yet to be routinely performed. This limitation could potentially be overcome through the integrated use of organoids, which provide superior cellular fidelity, into OOAC devices as discussed elsewhere and development of new directed differentiation protocols [110]. Importantly, as the cell heterogeneity increases, the control of spatial cell positioning has to increase as well. For instance, hepatocytes in the body are exposed to an oxygen gradient that influences their gene expression and results in different metabolic activities.[111] However, only a few microfluidic platforms have focused on reproducing the liver “zonation” through the generation of biochemical, oxygen, and hormonal gradients, and subsequent zonal drug toxicity responses in hepatocytes [112,113]. OOAC devices with perfusable channels are of growing interest due to their ability to include hemodynamic and inflammatory components for modeling more complex diseases. It is still challenging to model both innate and adaptive immune responses, involving synergetic response of multiple cell types (e.g., macrophages, neutrophils, and mast cells) along with their associated cytokines and growth factors. For appropriate introduction of adaptive immune components (T and B cells) the tissues have to be matched to the patient. This only works well if the primary tissue cells and blood can be obtained from the patient readily. However, the protocols to derive T and B cells from iPSC are still emerging [114], limiting the wide applicability at this time.
While multiple endothelial and epithelial cell types can be cultured in different channel compartments to mimic a tissue microenvironment on the microfluidic platform[15], the engineered tissue microenvironment is not easily scalable and in most cases not easily accessible for tissue withdrawal. These challenges come from the very nature of the microfluidic and microfabricated devices themselves. As most platforms operate in a closed format[77], it is challenging to retrieve intact tissue to study the change in genetic profiling in target organ due to environmental cues or to detect secreted biomolecule factors without the factors being over-diluted or absorbed into the microfluidic lining. Secondly, quantification analysis with current platforms is complicated. Also, most of these systems do not provide real-time functional readouts of a 3D vascularized tissue. Thirdly, most organ-on-a-chip platforms do not connect multiple organs with a single vascular system, although recent studies move towards this requirement[115]. As multiple compartments are connected, fluid handling becomes an issue, due to excess dead volume. The effectiveness in modeling inter-organ trafficking of live cells in the circulation system across multiple organs also decreases as multiple organs are connected. One of the obstacles in biomedical applications is reproducibility, specifically as related to translation from academic to industrial laboratories, as operation of OOAC devices requires retraining of scientific and technical staff. This limitation can be overcome by designing devices in a footprint of a well plate with open access for liquid handling[43].
Some of the throughput challenges on both the fabrication and tissue establishment side could be addressed by utilization of 3D printing and additive manufacturing technologies, especially if sensors are 3D printed into plastic platforms created by scalable techniques such as injection molding and hot embossing. Additionally, advances in 3D bioprinting facilitate the packing of cells into organ-like structures to accurately and simultaneously capture multiple aspects of human physiology, and use microtissues as the ink themselves [116,117]. We envision future organ-on-a-chip systems requiring synergistic incorporation of both heterogeneity of relevant cells and better controlled spatiotemporal compartments to achieve appropriate biochemical gradients and thus, enabling a more comprehensive drug screening program. As the political and regulatory landscape changes with the recent bill that ended the outdated FDA mandate requiring experimental drugs to be tested on animals[118] before they could be used on humans in clinical trials, this new and improved generation of human OOAC models will deliver on the promise of precision medicine.
Highlights.
Conceived to address challenges in modeling human biology, organ-on-a-chip (OOAC) engineering has grown enormously from tissue engineering to capture specific hallmarks of human tissues.
The evolution of OOAC models is intertwined with progress in tissue engineering by applying cellular components, biomaterials, and tissue culture technologies.
The manufacture of OOAC devices incorporates microfabrication techniques, material chemistry, and biosensing applications.
Current OOAC platforms focus on constructing higher-order structures using tissue engineering principles. For example, liver- and heart-on-chips are used to study drug metabolism, toxicity, and pathology.
Limitations in the OOAC field include achieving systematic biomimicry and stability. Synergistic incorporation of advanced cell culture and manufacturing techniques can expand the impact of OOAC platforms.
Acknowledgements
This work is funded by the Canadian Institutes of Health Research (CIHR) Foundation Grant FDN-167274, Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN 326982-10), NSERC-CIHR Collaborative Health Research Grant (CHRP 493737-16), National Institutes of Health Grant 2R01 HL076485 and Heart and Stroke Foundation Grants-in-Aid (G-16-00012711 and G-18-0022356). MR was supported by Killam Fellowship and Canada Research Chair. The authors acknowledge the Canada Foundation for Innovation and the Ontario Research Fund, Project #36442 for funding of the Centre for Organ-on-a-Chip Engineering. Some components of schematics were created with BioRender.com.
Glossary
- Tissue engineering
A biomedical engineering discipline that uses cells and biomaterials to produce human tissues, primarily for the repair or improvement of organ functions
- Organ-on-a-chip
An engineered biosystem containing miniature tissues grown inside a microfabricated chip to mimic organ-specific functions
- Biomaterials
Natural or synthetic substances that are used to interact with biological systems in biomedical applications
- Microfabrication
A set of techniques that are used to convert substrate materials into complex systems with miniature structures of micrometer scale or smaller
- Microfluidics
A discipline that involves study, precise control, and manipulation of fluids that are constrained to chambers or channels of microscale, typically below 100 μm
- Soft lithography
A family of techniques for patterning and fabricating micro-scale structures using elastomeric materials including stamps, molds, and photomasks
- Hot embossing
A fabrication technology with high replication accuracy involves the use of glass transition temperature and high pressure to transfer microscale structures from a master mold into a polymer substrate
- Disease modeling
The development of application of tools (animals, cell-based assays, or computational systems) that can recapitulate the pathological processes in the actual disease conditions, and aid the study of disease mechanisms and prediction of potential treatment approaches
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
M.R and Y.Z. are inventors on an issued patent describing Biowire II technology that has been licensed to Valo Health. M.R. and Y.Z. receive licensing revenue from this invention.
References:
- 1.Thomson GW (1956) Quinidine as a cause of sudden death. Circulation 14, 757–765 [DOI] [PubMed] [Google Scholar]
- 2.Jeon JS et al. (2014) Generation of 3D functional microvascular networks with human mesenchymal stem cells in microfluidic systems. Integr Biol (Camb) 6, 555–563. 10.1039/c3ib40267c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan DTT et al. (2017) A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 17, 511–520. 10.1039/c6lc01422d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prodanov L et al. (2016) Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng 113, 241–246. 10.1002/bit.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma LD et al. (2018) Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip 18, 2547–2562. 10.1039/c8lc00333e [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y et al. (2019) Rapid Wire Casting: A Multimaterial Microphysiological Platform Enabled by Rapid Casting of Elastic Microwires (Adv. Healthcare Mater. 5/2019). Advanced Healthcare Materials 8, 1970019. 10.1002/adhm.201970019 [DOI] [PubMed] [Google Scholar]
- 7.Lind JU et al. (2017) Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nature materials 16, 303–308. 10.1038/nmat4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jalili-Firoozinezhad S et al. (2019) A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 3, 520–531. 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musah S et al. (2017) Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng 1 10.1038/s41551-017-0069 [DOI] [PMC free article] [PubMed]
- 10.Booth R and Kim H (2012) Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB). Lab Chip 12, 1784–1792. 10.1039/c2lc40094d [DOI] [PubMed] [Google Scholar]
- 11.Lee JH et al. (2012) Microfluidic 3D bone tissue model for high-throughput evaluation of wound-healing and infection-preventing biomaterials. Biomaterials 33, 999–1006. 10.1016/j.biomaterials.2011.10.036 [DOI] [PubMed] [Google Scholar]
- 12.Rafatian N et al. (2021) Drawing Inspiration from Developmental Biology for Cardiac Tissue Engineers. Adv Biol (Weinh) 5, e2000190. 10.1002/adbi.202000190 [DOI] [PubMed] [Google Scholar]
- 13.Duffy DC et al. (1998) Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Analytical Chemistry 70, 4974–4984. 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 14.Sin A et al. (2004) The Design and Fabrication of Three-Chamber Microscale Cell Culture Analog Devices with Integrated Dissolved Oxygen Sensors. Biotechnology Progress 20, 338–345. 10.1021/bp034077d [DOI] [PubMed] [Google Scholar]
- 15.Huh D et al. (2010) Reconstituting Organ-Level Lung Functions on a Chip. Science 328, 1662–1668. 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riaz M et al. (2022) Muscle LIM Protein Force-Sensing Mediates Sarcomeric Biomechanical Signaling in Human Familial Hypertrophic Cardiomyopathy. Circulation 145, 1238–1253. doi: 10.1161/CIRCULATIONAHA.121.056265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y et al. (2019) A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 176, 913–927 e918. 10.1016/j.cell.2018.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown BN and Badylak SF (2014) Extracellular matrix as an inductive scaffold for functional tissue reconstruction. Transl Res 163, 268–285. 10.1016/j.trsl.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swinehart IT and Badylak SF (2016) Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev Dyn 245, 351–360. 10.1002/dvdy.24379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y et al. (2020) Engineering microenvironment for human cardiac tissue assembly in heart-on-a-chip platform. Matrix Biology 85–86, 189–204. 10.1016/j.matbio.2019.04.001 [DOI] [PMC free article] [PubMed]
- 21.Wufuer M et al. (2016) Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Scientific Reports 6, 37471. 10.1038/srep37471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasendra M et al. (2018) Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Scientific Reports 8, 2871. 10.1038/s41598-018-21201-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi K and Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 25.Hinson JT et al. (2015) HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986. 10.1126/science.aaa5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G et al. (2014) Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20, 616–623. 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmann O et al. (2009) Evidence for cardiomyocyte renewal in humans. Science 324, 98–102. 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y et al. (2020) Towards chamber specific heart-on-a-chip for drug testing applications. Adv Drug Deliv Rev 165–166, 60–76. 10.1016/j.addr.2019.12.002 [DOI] [PMC free article] [PubMed]
- 29.Ronaldson-Bouchard K et al. (2018) Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. 10.1038/s41586-018-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.<j/>(2013)Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem cells and development 22, 1991–2002. 10.1089/scd.2012.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callaghan NI et al. (2022) Advanced physiological maturation of iPSC-derived human cardiomyocytes using an algorithm-directed optimization of defined media components. bioRxiv, 2022.2010.2010.507929. 10.1101/2022.10.10.507929 [DOI]
- 32.Dhahri W et al. (2020) Abstract 291: In Vitro Matured Human Embryonic Stem Cell-derived Cardiomyocytes Form Grafts With Enhanced Structure and Improved Electromechanical Integration in Injured Hearts. Circulation Research 127, A291–A291. doi: 10.1161/res.127.suppl_1.291 [DOI] [PubMed] [Google Scholar]
- 33.Zhou M et al. (2016) Development of a Functional Glomerulus at the Organ Level on a Chip to Mimic Hypertensive Nephropathy. Sci Rep 6, 31771. 10.1038/srep31771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HJ et al. (2012) Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174. 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 35.Wang EY et al. (2019) Biowire Model of Interstitial and Focal Cardiac Fibrosis. ACS Central Science 5, 1146–1158. 10.1021/acscentsci.9b00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang EY et al. (2022) Intersection of stem cell biology and engineering towards next generation in vitro models of human fibrosis. Front Bioeng Biotechnol 10, 1005051. 10.3389/fbioe.2022.1005051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graney PL et al. (2020) Macrophages of diverse phenotypes drive vascularization of engineered tissues. Sci Adv 6. ARTN eaay6391 10.1126/sciadv.aay6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu RXZ et al. (2022) Vasculature-on-a-chip platform with innate immunity enables identification of angiopoietin-1 derived peptide as a therapeutic for SARS-CoV-2 induced inflammation. Lab Chip 22, 1171–1186. 10.1039/d1lc00817j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsui JH et al. (2021) Tunable electroconductive decellularized extracellular matrix hydrogels for engineering human cardiac microphysiological systems. Biomaterials 272, 120764. 10.1016/j.biomaterials.2021.120764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwan J et al. (2016) Anisotropic engineered heart tissue made from laser-cut decellularized myocardium. Scientific Reports 6, 32068. 10.1038/srep32068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuvellier M et al. (2021) 3D culture of HepaRG cells in GelMa and its application to bioprinting of a multicellular hepatic model. Biomaterials 269, 120611. 10.1016/j.biomaterials.2020.120611 [DOI] [PubMed] [Google Scholar]
- 42.Christoffersson J et al. (2019) Fabrication of modular hyaluronan-PEG hydrogels to support 3D cultures of hepatocytes in a perfused liver-on-a-chip device. Biofabrication 11, 015013. 10.1088/1758-5090/aaf657 [DOI] [PubMed] [Google Scholar]
- 43.Lai BFL et al. (2020) Recapitulating Pancreatic Tumor Microenvironment through Synergistic Use of Patient Organoids and Organ-on-a-Chip Vasculature. Advanced Functional Materials n/a, 2000545. 10.1002/adfm.202000545 [DOI] [PMC free article] [PubMed]
- 44.Langhans SA (2018) Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front Pharmacol 9, 6–6. 10.3389/fphar.2018.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai BFL et al. (2021) A well plate-based multiplexed platform for incorporation of organoids into an organ-on-a-chip system with a perfusable vasculature. Nat Protoc 16, 2158–2189. 10.1038/s41596-020-00490-1 [DOI] [PubMed] [Google Scholar]
- 46.Raj M K and Chakraborty S (2020) PDMS microfluidics: A mini review. Journal of Applied Polymer Science 137, 48958. 10.1002/app.48958 [DOI] [Google Scholar]
- 47.Wang JD et al. (2012) Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann Biomed Eng 40, 1862–1873. 10.1007/s10439-012-0562-z [DOI] [PubMed] [Google Scholar]
- 48.Feinberg AW and Miller JS (2017) Progress in three-dimensional bioprinting. MRS Bulletin 42, 557–562. 10.1557/mrs.2017.166 [DOI] [Google Scholar]
- 49.Quirós-Solano WF et al. (2018) Microfabricated tuneable and transferable porous PDMS membranes for Organs-on-Chips. Scientific Reports 8, 13524. 10.1038/s41598-018-31912-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia Y and Whitesides GM (1998) SOFT LITHOGRAPHY. Annual Review of Materials Science 28, 153–184. 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- 51.Tsao C-W (2016) Polymer Microfluidics: Simple, Low-Cost Fabrication Process Bridging Academic Lab Research to Commercialized Production. Micromachines 7. 10.3390/mi7120225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gencturk E et al. (2017) Advances in microfluidic devices made from thermoplastics used in cell biology and analyses. Biomicrofluidics 11, 051502–051502. 10.1063/1.4998604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y et al. (2019) A Multimaterial Microphysiological Platform Enabled by Rapid Casting of Elastic Microwires. Adv Healthc Mater 8, e1801187. 10.1002/adhm.201801187 [DOI] [PubMed] [Google Scholar]
- 54.Park S and Young EWK (2021) E-FLOAT: Extractable Floating Liquid Gel-Based Organ-on-a-Chip for Airway Tissue Modeling under Airflow. Advanced Materials Technologies 6, 2100828. 10.1002/admt.202100828 [DOI] [Google Scholar]
- 55.Juang YJ and Chiu YJ (2022) Fabrication of Polymer Microfluidics: An Overview. Polymers (Basel) 14. 10.3390/polym14102028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asmani M et al. (2018) Fibrotic microtissue array to predict anti-fibrosis drug efficacy. Nature Communications 9, 2066. 10.1038/s41467-018-04336-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim M et al. (2022) Multimodal Characterization of Cardiac Organoids Using Integrations of Pressure-Sensitive Transistor Arrays with Three-Dimensional Liquid Metal Electrodes. Nano letters 22, 7892–7901. 10.1021/acs.nanolett.2c02790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melle G et al. (2020) Intracellular Recording of Human Cardiac Action Potentials on Market-Available Multielectrode Array Platforms. Front Bioeng Biotechnol 8, 66. 10.3389/fbioe.2020.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imboden M et al. (2019) High-speed mechano-active multielectrode array for investigating rapid stretch effects on cardiac tissue. Nat Commun 10, 834. 10.1038/s41467-019-08757-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauschke VM et al. (2016) Massive rearrangements of cellular MicroRNA signatures are key drivers of hepatocyte dedifferentiation. Hepatology 64, 1743–1756. 10.1002/hep.28780 [DOI] [PubMed] [Google Scholar]
- 61.Gerets HHJ et al. (2012) Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol Toxicol 28, 69–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novik EI et al. (2008) Augmentation of EB-directed hepatocyte-specific function via collagen sandwich and SNAP. Biotechnology Progress 24, 1132–1141. 10.1002/btpr.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamilton GA et al. (2001) Regulation of cell morphology and cytochrome P450 expression in human hepatocytes by extracellular matrix and cell-cell interactions. Cell Tissue Res 306, 85–99. 10.1007/s004410100429 [DOI] [PubMed] [Google Scholar]
- 64.Hoffmaster KA et al. (2004) P-glycoprotein expression, localization, and function in sandwich-cultured primary rat and human hepatocytes: relevance to the hepatobiliary disposition of a model opioid peptide. Pharm Res 21, 1294–1302. 10.1023/b:pham.0000033018.97745.0d [DOI] [PubMed] [Google Scholar]
- 65.Khetani SR and Bhatia SN (2008) Microscale culture of human liver cells for drug development. Nat Biotechnol 26, 120–126. 10.1038/nbt1361 [DOI] [PubMed] [Google Scholar]
- 66.Cho CH et al. (2010) Layered patterning of hepatocytes in co-culture systems using microfabricated stencils. Biotechniques 48, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee SA et al. (2013) Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte-hepatic stellate cell interactions and flow effects. Lab Chip 13, 3529–3537. 10.1039/c3lc50197c [DOI] [PubMed] [Google Scholar]
- 68.Toh YC et al. (2009) A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip 9, 2026–2035. 10.1039/b900912d [DOI] [PubMed] [Google Scholar]
- 69.Kang YB et al. (2015) Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol Bioeng 112, 2571–2582. 10.1002/bit.25659 [DOI] [PubMed] [Google Scholar]
- 70.Tan K et al. (2019) A high-throughput microfluidic microphysiological system (PREDICT-96) to recapitulate hepatocyte function in dynamic, re-circulating flow conditions. Lab Chip 19, 1556–1566 [DOI] [PubMed] [Google Scholar]
- 71.Sung JH and Shuler ML (2009) A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip 9, 1385–1394. 10.1039/b901377f [DOI] [PubMed] [Google Scholar]
- 72.Chen P-Y et al. (2021) Liver-on-a-chip platform to study anticancer effect of statin and its metabolites. Biochem Engineer J 165, 107831 [Google Scholar]
- 73.Skardal A et al. (2017) Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep 7, 8837. 10.1038/s41598-017-08879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin F et al. (2021) HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs. Lab Chip 21, 571–581 [DOI] [PubMed] [Google Scholar]
- 75.Kostrzewski T et al. (2017) Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J Gastroenterol 23, 204–215. 10.3748/wjg.v23.i2.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skardal A et al. (2016) A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng 113, 2020–2032. 10.1002/bit.25950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J et al. (2020) Three-Dimensional Human Liver-Chip Emulating Premetastatic Niche Formation by Breast Cancer-Derived Extracellular Vesicles. ACS Nano 14, 14971–14988. 10.1021/acsnano.0c04778 [DOI] [PubMed] [Google Scholar]
- 78.Dvir T et al. (2009) Prevascularization of cardiac patch on the omentum improves its therapeutic outcome. Proceedings of the National Academy of Sciences of the United States of America 106, 14990–14995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liau B et al. (2011) Pluripotent stem cell-derived cardiac tissue patch with advanced structure and function. Biomaterials 32, 9180–9187. 10.1016/j.biomaterials.2011.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stevens KR et al. (2009) Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue Eng Part A 15, 1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Radisic M et al. (2003) High-density seeding of myocyte cells for cardiac tissue engineering. Biotechnol Bioeng 82, 403–414 [DOI] [PubMed] [Google Scholar]
- 82.Radisic M et al. (2004) Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proceedings of the National Academy of Sciences of the United States of America 101, 18129–18134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimmermann WH et al. (2000) Three-dimensional engineered heart tissue from neonatal rat cardiac myocytes. Biotechnology and Bioengineering 68, 106–114 [PubMed] [Google Scholar]
- 84.Boudou T et al. (2012) A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A 18, 910–919. 10.1089/ten.TEA.2011.0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Naito H et al. (2006) Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation 114, I72–78 [DOI] [PubMed] [Google Scholar]
- 86.Radisic M et al. (2007) Pre-treatment of synthetic elastomeric scaffolds by cardiac fibroblasts improves engineered heart tissue. J Biomed Mater Res A, [DOI] [PMC free article] [PubMed]
- 87.Mastikhina O et al. (2020) Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials 233, 119741. 10.1016/j.biomaterials.2019.119741 [DOI] [PubMed] [Google Scholar]
- 88.Sadeghi AH et al. (2017) Engineered 3D Cardiac Fibrotic Tissue to Study Fibrotic Remodeling. Advanced healthcare materials 6, 10.1002/adhm.201601434. 10.1002/adhm.201601434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang EY et al. (2022) Design and Fabrication of Biological WiresBiological wires for Cardiac FibrosisFibrosis Disease Modeling. In Cardiac Tissue Engineering: Methods and Protocols (Coulombe, K.L.K. and Black Iii L.D, eds), pp. 175–190, Springer; US [Google Scholar]
- 90.Zimmermann WH et al. (2002) Tissue engineering of a differentiated cardiac muscle construct. Circulation Research 90, 223–230 [DOI] [PubMed] [Google Scholar]
- 91.Zimmermann WH et al. (2006) Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med 12, 452–458 [DOI] [PubMed] [Google Scholar]
- 92.Shen S et al. (2022) Physiological calcium combined with electrical pacing accelerates maturation of human engineered heart tissue. Stem Cell Reports 17, 2037–2049. 10.1016/j.stemcr.2022.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee EK et al. (2017) Machine Learning of Human Pluripotent Stem Cell-Derived Engineered Cardiac Tissue Contractility for Automated Drug Classification. Stem Cell Reports 9, 1560–1572. 10.1016/j.stemcr.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shadrin IY et al. (2017) Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat Commun 8, 1825. 10.1038/s41467-017-01946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sidorov VY et al. (2017) I-Wire Heart-on-a-Chip I: Three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta biomaterialia 48, 68–78. 10.1016/j.actbio.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huebsch N et al. (2016) Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Sci Rep 6, 24726. 10.1038/srep24726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schaaf S et al. (2011) Human Engineered Heart Tissue as a Versatile Tool in Basic Research and Preclinical Toxicology. PloS one 6, e26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mathur A et al. (2015) Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep 5, 8883. 10.1038/srep08883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.King O et al. (2022) Functional microvascularization of human myocardium in vitro. Cell Rep Methods 2, 100280. 10.1016/j.crmeth.2022.100280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ng R et al. (2021) Shortening Velocity Causes Myosin Isoform Shift in Human Engineered Heart Tissues. Circulation Research 128, 281–283. doi: 10.1161/CIRCRESAHA.120.316950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nunes SS et al. (2013) Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nature methods 10, 781–787. 10.1038/nmeth.2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grosberg A et al. (2011) Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab on a chip 11, 4165–4173. 10.1039/c1lc20557a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Agarwal A et al. (2013) Microfluidic heart on a chip for higher throughput pharmacological studies. Lab on a chip 13, 3599–3608. 10.1039/c3lc50350j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ahn S et al. (2018) Mussel-inspired 3D fiber scaffolds for heart-on-a-chip toxicity studies of engineered nanomaterials. Anal Bioanal Chem 410, 6141–6154. 10.1007/s00216-018-1106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Williams RR et al. (2000) NHLBI family blood pressure program: methodology and recruitment in the HyperGEN network. Hypertension genetic epidemiology network. Annals of epidemiology 10, 389–400 [DOI] [PubMed] [Google Scholar]
- 106.Mastikhina O et al. (2020) Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials 233, 119741. 10.1016/j.biomaterials.2019.119741 [DOI] [PubMed] [Google Scholar]
- 107.Wang EY et al. (2021) An organ-on-a-chip model for pre-clinical drug evaluation in progressive non-genetic cardiomyopathy. J Mol Cell Cardiol 160, 97–110. 10.1016/j.yjmcc.2021.06.012 [DOI] [PubMed] [Google Scholar]
- 108.Kuzmanov U et al. (2020) Mapping signalling perturbations in myocardial fibrosis via the integrative phosphoproteomic profiling of tissue from diverse sources. Nature Biomedical Engineering 4, 889–900. 10.1038/s41551-020-0585-y [DOI] [PubMed] [Google Scholar]
- 109.Ma Z et al. (2018) Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat Biomed Eng 2, 955–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takebe T et al. (2017) Synergistic Engineering: Organoids Meet Organs-on-a-Chip. Cell Stem Cell 21, 297–300. 10.1016/j.stem.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 111.Tonon F et al. (2019) In vitro metabolic zonation through oxygen gradient on a chip. Sci Rep 9, 13557. 10.1038/s41598-019-49412-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.kang YB et al. (2018) Metabolic patterning on a chip – towards in vitro liver zonation of primary rat and human hepatocytes. Sci Rep 8. 10.1038/s41598-018-27179-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tonon F et al. (2019) In vitro metabolic zonation through oxygen gradient on a chip. Sci Rep 9. 10.1038/s41598-019-49412-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Michaels YS et al. (2022) DLL4 and VCAM1 enhance the emergence of T cell–competent hematopoietic progenitors from human pluripotent stem cells. Sci Adv 8, eabn5522. doi: 10.1126/sciadv.abn5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ronaldson-Bouchard K et al. (2022) A multi-organ chip with matured tissue niches linked by vascular flow. Nat Biomed Eng 6, 351–371. 10.1038/s41551-022-00882-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahrens JH et al. (2022) Programming Cellular Alignment in Engineered Cardiac Tissue via Bioprinting Anisotropic Organ Building Blocks. Advanced Materials 34, 2200217. 10.1002/adma.202200217 [DOI] [PubMed] [Google Scholar]
- 117.Ma X et al. (2018) Rapid 3D bioprinting of decellularized extracellular matrix with regionally varied mechanical properties and biomimetic microarchitecture. Biomaterials 185, 310–321. 10.1016/j.biomaterials.2018.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.(2021–2022). FDA Modernization Act of 2021. 117th Congress
- 119.Nerem RM (2010) Regenerative medicine: the emergence of an industry. J R Soc Interface 7 Suppl 6, S771–775. 10.1098/rsif.2010.0348.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vacanti JP and Langer R (1999) Tissue engineering: the design and fabrication of living replacement devices for surgical reconstruction and transplantation. The Lancet 354, st32–st34 [DOI] [PubMed] [Google Scholar]
- 121.Xia Y and Whitesides GM (1998) Soft Lithography. Angewandte Chemie International Edition 37, 550–575. [DOI] [PubMed] [Google Scholar]
- 122.Radisic M et al. (2006) Biomimetic approach to cardiac tissue engineering: oxygen carriers and channeled scaffolds. Tissue Eng 12, 2077–2091. 10.1089/ten.2006.12.2077 [DOI] [PubMed] [Google Scholar]
- 123.Radisic M et al. (2005) Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers. American journal of physiology. Heart and circulatory physiology 288, H1278–1289. 10.1152/ajpheart.00787.2004 [DOI] [PubMed] [Google Scholar]
- 124.Lee J et al. (2021) Organoid Model in Idiopathic Pulmonary Fibrosis. Int J Stem Cells 14, 1–8. 10.15283/ijsc20093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shin YJ et al. (2021) 3D bioprinting of mechanically tuned bioinks derived from cardiac decellularized extracellular matrix. Acta biomaterialia 119, 75–88. 10.1016/j.actbio.2020.11.006 [DOI] [PubMed] [Google Scholar]
- 126.Lin Y et al. (2018) Soft lithography based on photolithography and two-photon polymerization. Microfluidics and Nanofluidics 22, 97. 10.1007/s10404-018-2118-5 [DOI] [Google Scholar]
- 127.Lind JU et al. (2017) Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nature materials 16, 303–308. 10.1038/nmat4782 [DOI] [PMC free article] [PubMed] [Google Scholar]


