Abstract
RNA editing in kinetoplastid mitochondria occurs by a series of enzymatic steps that is catalyzed by a macromolecular complex. Four novel proteins and their corresponding genes were identified by mass spectrometric analysis of purified editing complexes from Trypanosoma brucei. These four proteins, TbMP81, TbMP63, TbMP42, and TbMP18, contain conserved sequences to various degrees. All four proteins have sequence similarity in the C terminus; TbMP18 has considerable sequence similarity to the C-terminal region of TbMP42, and TbMP81, TbMP63, and TbMP42 contain zinc finger motif(s). Monoclonal antibodies that are specific for TbMP63 and TbMP42 immunoprecipitate in vitro RNA editing activities. The proteins are present in the immunoprecipitates and sediment at 20S along with the in vitro editing, and RNA editing ligases TbMP52 and TbMP48. Recombinant TbMP63 and TbMP52 coimmunoprecipitate. These results indicate that these four proteins are components of the RNA editing complex and that TbMP63 and TbMP52 can interact.
RNA editing in trypanosomes posttranscriptionally inserts and deletes uridylates (U's) at multiple sites in most mitochondrial pre-mRNAs to produce mature mRNAs. U insertion and deletion are directed by guide RNAs (gRNAs) and are catalyzed by a macromolecular complex. Editing occurs by a series of enzymatic steps that include endoribonuclease, 3′ terminal uridylyl transferase (TUTase), 3′ exouridylylase, and RNA ligase activities (reviewed in references 3, 8, 21, and 23). Although editing can be extensive, with the insertion and deletion of numerous U's, it is also very specific. The characteristics of the enzymatic activities contribute to this specificity (7), but noncatalytic proteins may be required for editing and may contribute to the specificity.
RNA editing is catalyzed by a 20S ribonucleoprotein complex (2, 15), and identification of its components and the composition of the fully functional complex is at an early stage. Initial studies estimated that a complex that can catalyze at least some of the steps of editing in vitro contains 7 to 20 polypeptides (11, 14, 16). Two related proteins, TbMP52 and TbMP48, were identified by mass spectrometric analysis of purified editing complexes (14), and TbMP52 was shown to be essential for RNA editing and for survival of bloodstream forms in vivo and in vitro (19). In addition, TbMP52 and TbMP48 correspond to the larger and smaller adenylatable proteins in the RNA editing complex, respectively, and were found to be RNA ligases (12, 17, 19). TbMP52 corresponds to T. brucei V and T. brucei p52 and TbMP48 corresponds to T. brucei IV and T. brucei p48 (12, 17). The fully functional editing complex, which may consist of a catalytic core complex and accessory and regulatory factors may contain numerous proteins. Several other candidate proteins, some of which have RNA binding activities, have been described and may play a role in RNA editing (6, 9, 11, 13, 25). However, except for mHel61p (13), none of these proteins have been shown to play a direct role in editing.
In this study, we describe the identification of four additional proteins that are present in the RNA editing complex by immunoprecipitation, mass spectrometric, and/or gradient sedimentation analyses. These four proteins have sequence similarities to each other and the three largest contain one or two C2H2 zinc finger motif(s). One protein was also shown to interact in vitro with the TbMP52 editing RNA ligase.
MATERIALS AND METHODS
In vitro editing and adenylation assays.
Deletion and insertion editing were assayed in vitro using 3′-labeled A6-U5 pre-mRNA substrate with gA6[14]Δ16G gRNA and precleaved 5′CL18 and 3′CL13pp substrates with gPCA6-2A RNAs, respectively, as previously described (7, 20). The reaction products were resolved on polyacrylamide-urea gels and visualized on Storm PhosphorImager screens (Molecular Dynamics). Adenylation of the editing RNA ligases was determined as previously described (18) using 15-min incubations at 28°C with 2.5 μCi of [α-32P]ATP in 25 mM HEPES (pH 7.9)–10 mM Mg(OAc)2–50 mM KCl–0.5 mM dithiothreitol–10% dimethyl sulfoxide. The proteins were resolved on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) gels, and the radiolabeled proteins were detected by phosphorimaging.
Protein and gene identification.
Trypanosoma brucei procyclic cells (strain IsTaR 1.7a) were grown to log phase in vitro (22), and the mitochondrial vesicles were isolated (5). The editing complex was isolated by sequential ion-exchange (SP- and Q-Sepharose) and gel filtration (Superose 6) column chromatography (14). The proteins in the peak deletion editing fractions from Superose 6 column were separated on an SDS–10% PAGE gel and stained with Coomassie brilliant blue. The protein bands were excised, digested in-gel with trypsin, and analyzed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) and T. brucei nucleotide database searches (reference 14 and references cited therein). Complete open reading frames (ORFs) were determined when significant peptide matches were found to genomic sequences for which the complete ORF was not available. The DNA fragment was amplified by PCR using primers designed based on database analysis, was cloned, and was sequenced, and the ORF was identified by multiple-sequence alignment using the Seqman program (DNASTAR, Inc.). Details of how proteins TbMP81, TbMP63, TbMP42, and TbMP18 were identified are described in the Results. The National Center for Biotechnology Information nonredundant protein database was searched for homologs to the predicted protein sequences by using the BLAST algorithm, and the PROSITE, BLOCKS, and CDD databases were searched for the presence of known motifs and domains in the predicted proteins. Mitochondrial targeting signals were predicted using the PSORT II algorithm (http://psort.ims.u-tokyo.ac.jp/form2.html), and an amphiphilic helix was predicted at the N terminus of the proteins by using Gene Runner (Hastings Software, Inc.). Amino acid repeats in the predicted protein sequence were identified by Dotplot analysis using MegAlign software, and a hydrophobic region was predicted by Protean analysis (DNASTAR, Inc). Multiple sequences were aligned using the ClustalW algorithm (http://www.ebi.ac.uk/clustalw/).
Immunoprecipitation of editing complex.
Monoclonal antibodies (MAbs) generated against the purified native editing complex were conjugated to anti-mouse immunoglobulin G (IgG)-coated Immunomagnetic beads (Dynabeads M-450; DYNAL), and immunoprecipitation from the mitochondrial 20S fraction was performed as previously described (14). The samples bound to the beads were directly assayed for deletion editing and precleaved insertion editing in vitro, adenylation activity, and editing-associated enzymatic activities (14). The primary and secondary antibodies were cross-linked as described by the manufacturer (DYNAL) for MS analysis of the immunoprecipitated proteins. The editing complex was then immunoprecipitated from mitochondrial lysate (14), and the proteins were eluted from the antibody complex with 0.5 M acetic acid, pH 2.6, and neutralized with NaOH. The eluted proteins were concentrated using Centricon-YM10 membrane (Amicon) and desalted with 10 mM Tris (pH 7.2), and approximately 2 μg of the proteins was digested with 40 ng of trypsin and analyzed by LC-MS/MS.
Cloning and expression of TbMP63 and TbMP42.
DNA corresponding to the TbMP63 ORF was amplified from T. brucei genomic DNA with primers 3316 (TGG ATC CTC AAG ACG ATG TAC C) and 3317 (AAG GGA ATT CGG TAT TCT CC) (restriction sites are italicized). The amplified DNA was cloned into the pGEM-T Easy vector (Promega), and its sequence was confirmed. The insert was released by digestion with BamHI and EcoRI and cloned into a pRSET C vector (Invitrogen) cut with the same enzymes. Similarly, the DNA corresponding to the TbMP42 ORF was PCR amplified with primers 3528 (AGG ATC CAC TTG TGT TGT CGC) and 3529 (TGA ATT CTC ACA CCT TCA ACA C) and cloned into the pRSET B vector at the BamHI-EcoRI site. Amino acids 1 to 12 of the predicted TbMP42 protein sequence were omitted in the resultant Tb08-pRSET construct. The plasmids were transformed into BL21 DE3-LysS cells, and recombinant proteins were expressed at 30°C with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induction. Proteins from recombinant Escherichia coli cells were separated by SDS–10% PAGE, transferred to nitrocellulose filters, and reacted with MAbs. E. coli cells transformed with vector only were used as a negative control.
In vitro expression of proteins.
DNA corresponding to the TbMP63 ORF was PCR amplified from T. brucei genomic DNA using primers 3602 (CTG AAT TCG ATG TAC CGC A) and 3603 (CTG GAT CCG GTA TTC TCC CCT). The amplified DNA was digested with EcoRI and BamHI and cloned into the EcoRI-BamHI site of the pSG1 vector. DNA corresponding to the TbMP42 ORF was amplified with primers 3604 (GGA ATT CAT GAA GCG TGT TAC) and 3605 (CGG GAT CCT CAC ACC TTC AAC A) and cloned into the pSG1 vector at the EcoRI-BamHI site. Recombinant TbMP63 (rTbMP63) and TbMP42 (rTbMP42) proteins were produced in vitro using a cell-free, coupled transcription-translation system (TNT; Promega) according to the manufacturer's instructions. The proteins were labeled by incorporation of [35S]methionine during translation. Recombinant TbMP52 (rTbMP52) was expressed under similar conditions by using the plasmid pSG-TbMP52 (19).
Protein-protein interaction assay.
Protein-protein interactions were investigated by coimmunoprecipitation of in vitro-translated recombinant proteins using MAbs that were coupled to immunomagnetic beads as described above. 35S-labeled and unlabeled recombinant TbMP63 (rTbMP63), rTbMP52, and rTbMP42 were produced in vitro. The labeled and unlabeled recombinant proteins were mixed together in various combinations and immunoprecipitated with MAbs P1H3-D7 (anti-TbMP63) or P3C1-G2 (anti-TbMP52) by using in vitro-expressed 35S-labeled luciferase protein as negative control. The incubations and washes were carried out at high stringency using IP500 buffer (10 mM Tris [pH 7.5], 5 mM EDTA, 500 mM KCl, 0.5% Triton X-100, 1% bovine serum albumin). The immunoprecipitated samples were digested and resolved on SDS–10% PAGE gels, and the labeled proteins were detected by phosphorimaging.
Nuclease treatment and Western analysis.
Fifty-microliter samples of pooled glycerol gradient fractions from the peak of in vitro editing activity were treated for 30 min at 25°C with either 20 μg of RNase A or a mixture of RNases (DNase-free RNase; Boehringer Mannheim). Mock treatment was carried out with buffer alone (10 mM Tris [pH 7.2], 10 mM MgCl2, 5 mM CaCl2, 50 mM KCl). Following the treatment, the samples were immunoprecipitated with anti-TbMP63 MAb in immunoprecipitation (IP) buffer (10 mM Tris [pH 7.2], 10 mM MgCl2, 200 mM KCl, 0.1% Triton X-100). These immunoprecipitates were resolved on SDS–10% PAGE gels, transferred to polyvinylidene difluoride membranes, and examined by Western analyses using MAbs specific for TbMP81, TbMP63, TbMP52, and TbMP42 that were generated against the native editing complex as previously described (14). Total mitochondrial lysates, glycerol gradient (10 to 30%) fractions of these lysates, and recombinant proteins were subjected to similar Western analyses.
Nucleotide sequence accession numbers.
The nucleotide sequences have been submitted to GenBank with accession numbers AF382333, AF382334, AF382335, and AF382336.
RESULTS
Identification of TbMP81, TbMP63, TbMP42, and TbMP18.
Four proteins and their corresponding genes were identified by high-performance LC-MS/MS analysis of protein bands from purified editing complexes (14). Fractions containing the peak editing activities from sequential SP-Sepharose, Q-Sepharose, and Superose 6 column chromatography were concentrated and separated on an SDS–10% PAGE gel. Coomassie blue-stained bands that correspond to those in most purified fraction (14) (see Fig. 2) were excised and analyzed by LC-MS/MS. Comparison of the MS/MS data from the ∼99-kDa protein band to data in T. brucei DNA sequence databases identified 15 tryptic peptides that match those in a protein encoded by a 2,289-nucleotide ORF in The Institute of Genomic Research (TIGR) T. brucei database [positions 12616 to 14904, GenBank accession no. (gb) AC008031.3, chromosome II, clone RPCI93-25N14]. The predicted preprocessed protein has 762 amino acids and a mass of 81 kDa; hence, the protein was designated TbMP81.
FIG. 2.
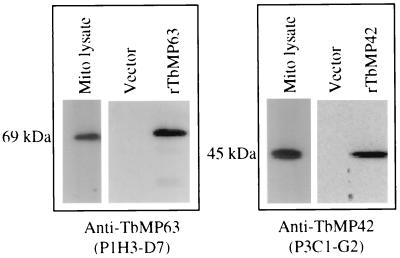
Western analysis showing the reactions of MAb P1H3-D7 with native and recombinant TbMP63 and of MAb P3C12-D8 with native and recombinant TbMP42. E. coli cells transformed with vector alone were used as negative control. Recombinant TbMP63 and TbMP42 containing an N-terminal His6 tag were expressed in E. coli cells.
Similar analysis of the ∼69-kDa protein band did not identify a complete ORF but revealed MS/MS matches to several DNA sequences, allowing us to identify and sequence the complete gene as described below. The analysis identified three tryptic peptides, (K)PLHQPDR, (R)TFVDEEER, and (K)LHLQQAHGGDGSVEAGPGPGVEASSVNVVNTPVPS PISPVER, that match peptides predicted from T. brucei genomic DNA clone G654 (gb B07464). It also identified tryptic peptides (R)LHPVYEASNNK in genomic DNA clone G432 (gb B07352), (K)PLQPPQPK in genomic DNA clone 37L1.TR (TIGR database, gb AQ950249), and (R)LEGNFVHSTMVMK and (K)PLQPPQPK in genomic DNA clone 16E22.TF (TIGR database, gb AQ650728). Furthermore, the nucleotide sequence of clone G654 matched that of EST clone T651 (EMBL accession no. T26253). cDNA clone T651 was completely sequenced and found to overlap with the sequences of the above genomic DNA clones, suggesting that the identified peptides correspond to a single protein. The 5′ region of the ORF, which was not in the DNA sequence databases, was amplified by PCR, cloned, and sequenced. It was amplified using two primers (GCT CGC TTG TGC ATG TTG and CGC AGC AAA CAC CTT CAT CAC) that were based on 5′- and 3′-end sequences from DNA clone 16E22. The complete ORF sequence was assembled using these sequences and additional matching genomic DNA sequences (110F8.TR [gb AZ215909], 115A2.TF [gb AZ214294], 115A2.TR [gb AZ214297]) that were identified in the databases. The complete ORF was then PCR amplified from T. brucei genomic DNA, cloned, and sequenced. The 1,767-nucleotide ORF predicts a 588-amino-acid protein with a mass of 63 kDa, and the protein was designated TbMP63.
LC-MS/MS analysis of the ∼45-kDa protein identified two tryptic peptides, (R)LINALNHHIMTK and (K)LEEVNPEEIK, corresponding to genomic DNA clone 34C8.TF (TIGR database, gb AQ943880) and two other tryptic peptides, (R)VFDSDYSSR and (R)DGEWFLVTGR, corresponding to genomic DNA clone 35I8.TF (TIGR database, gb AQ941226). Further database analysis showed that the sequences of these two clones overlap and that they represent a single ORF. The sequences overlapped with four other genomic DNA clones from the TIGR database [105A1.TR, 70B2.TR, 25O15.TR (gb AQ655403), 11 M2.TR (gb AQ661213)]. The complete ORF sequence (1,182 nucleotides long) was assembled from these sequences. The ORF was predicted to encode a 393-amino-acid protein with a mass of 42 kDa, which was designated TbMP42.
Analysis of the ∼22-kDa protein band identified four tryptic peptides, (R)CFGELFSAEVK, (K)EGNVVCVNGR, (R)LSPQLEPSCNK, and (R)TVPAAVNPAVEDIK, corresponding to genomic DNA clone 15K6.TR (TIGR database, gb AQ660117). This clone overlaps with another genomic DNA clone, 15 M18.TR (TIGR database, gb AQ649021). This identified a 495-nucleotide ORF that encodes a 164-amino-acid, 18-kDa protein, which was designated TbMP18. The identity of TbMP18 was further confirmed by N-terminal amino acid sequencing. Microsequencing of the first 23 amino acids of native TbMP18 identified peptide sequence XKSVNSVTLVGVVHDIQSGFVYE, which matches amino acids 19 to 41 of the predicted TbMP18 sequence. This suggests that the first 18 amino acids form the mitochondrial import signal.
Sequence comparisons.
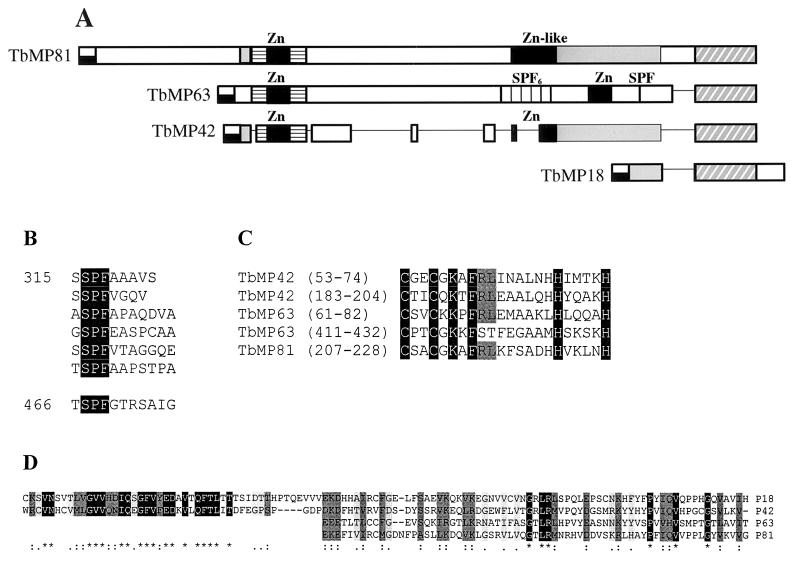
The proteins TbMP81, TbMP63, TbMP42, and TbMP18 are predicted to localize in mitochondria by PSORT II analysis and since the amino acids at the N terminus in all these four proteins can fold into an amphiphilic helix (results not shown). BLAST searches to National Center for Biotechnology Information nonredundant protein database revealed no proteins related to TbMP81, TbMP63, TbMP42, and TbMP18, indicating that they are novel. Motif and domain analyses identified one zinc finger motif in TbMP81 (amino acids 207 to 228), two in TbMP63 (amino acids 61 to 82 and 411 to 432) and two in TbMP42 (amino acids 53 to 74 and 183 to 204) (Fig. 1A). No known motifs or domains were identified in TbMP18. Dotplot analysis of TbMP63 identified six serine-proline-phenylalanine (SPF) tripeptide repeats, each separated by five to eight amino acids (amino acids 316 to 368), and another SPF sequence (amino acids 467 to 469) separated from the first cluster by 90 amino acids which contain a zinc finger motif (Fig. 1A and B). The repeat region predicts a hydrophobic domain in the protein. All of the zinc finger motifs have the signature C2H2 zinc ligands (Fig. 1C). As illustrated in Fig. 1A, all four proteins have sequence similarity between them in various regions. TbMP18 has considerable similarity to the C-terminal region of TbMP42 with 41% identity and 60% similarity over 108 amino acids (Fig. 1D), but TbMP18 contains an additional 34-amino-acid C-terminal sequence. All four proteins, TbMP81, TbMP63, TbMP42, and TbMP18, have sequence similarity in the C-terminal portion (65- to 67-amino-acid region), with six amino acid positions being identical. However, 15 amino acids are identical in TbMP81, TbMP42, and TbMP18, which are more closely related to each other than to TbMP63. Pairwise sequence comparisons showed additional sequence relationships between the three largest proteins. All three proteins have a domain near the N terminus which contains a zinc finger motif that is related, although the ones between TbMP81 and TbMP42 are more closely related than those between TbMP63 and these other two proteins. TbMP81, however, has an additional N-terminal sequence. The C-terminal region of TbMP42 that is similar to TbMP81 contains a zinc finger motif, while TbMP81 has a zinc finger-like motif with an insertion of 22 amino acids in this region. TbMP63 contains a zinc finger motif in its C-terminal region. The central regions of TbMP81 and TbMP63 have a low level of sequence similarity.
FIG. 1.
Sequence relationships among TbMP81, TbMP63, TbMP42, and TbMP18 proteins. (A) Diagram showing N-terminal amphipathic helices that are likely mitochondrial targeting signals (black and white) and conserved C-terminal domains (cross-hatched). C2H2 Zn finger and Zn finger like motifs (black boxes) and SPF repeats (vertical hatched) are labeled accordingly. The thin lines indicate gaps in the sequence. Shaded regions in TbMP18, TbMP42, and TbMP81 show sequence similarity, and horizontal hatched boxes represent similar regions between TbMP81, TbMP63, and TbMP42 sequences. The central regions of TbMP81 and TbMP63 have a low level of sequence similarity. (B) SPF tripeptide repeats in TbMP63. The six-unit repeat begins with amino acid 315, and the single repeat begins with amino acid 466. (C) Shown are sequences of the C2H2 Zn finger motifs, and the locations of the protein sequence are in parentheses. (D) Amino acid sequence alignment of TbMP18 (amino acids 19 to 131), TbMP42 (amino acids 286 to 393), TbMP63 (amino acids 523 to 588), and TbMP81 (amino acids 695 to 762). Indicated are amino acid identity (∗), conservation (:), and similarity(.).
Association of TbMP81, TbMP63, TbMP42, and TbMP18 with RNA editing complex.
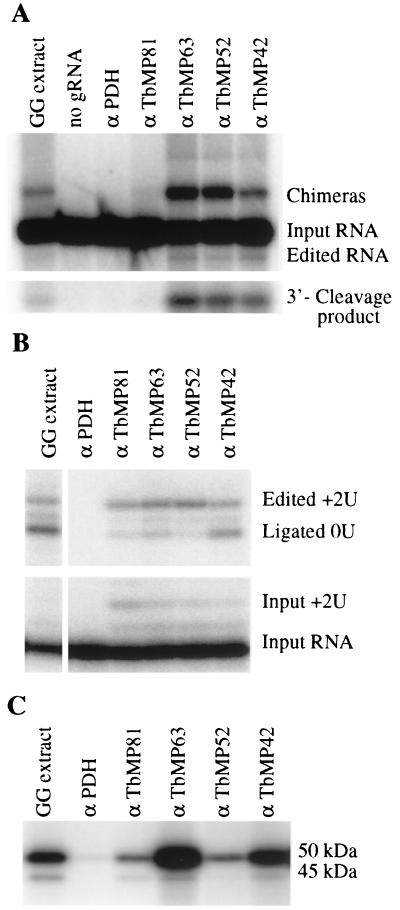
The association of the proteins with the editing complex was assessed by the ability of MAbs against specific proteins to precipitate editing activities from mitochondrial lysate and detection of these proteins in the affinity-purified editing complex. MAbs P1H3-D7 and P3C12-D8, respectively, which were generated against the isolated editing complex (14), specifically reacted with both native and recombinant TbMP63 and TbMP42 proteins (Fig. 2). The MAbs did not cross-react with any other proteins in the mitochondrial lysate. Both antibodies immunoprecipitated in vitro deletion editing, precleaved insertion editing, and adenylation activities from the mitochondrial 20S fraction (Fig. 3). A MAb that is specific for TbMP52, an RNA ligase that has been shown to be essential for editing (19), also immunoprecipitated these activities. The lower adenylation activity with the anti-TbMP52 MAb relative to that of the anti-TbMP63 and anti-TbMP42 MAbs reflected inhibition of the adenylation by the anti-TbMP52 MAb. In contrast, MAbs that are specific for the E2 subunit of pyruvate dehydrogenase (MAb 58) and ATP synthase α (MAb P7D9-A12) did not precipitate editing activities (14). Thus, TbMP63 and TbMP42 are tightly associated with and are likely components of the editing complex.
FIG. 3.
Immunoprecipitation analyses of editing complex. (A) In vitro deletion editing assay of immunoprecipitates from the mitochondrial 20S fraction using MAbs against (α) TbMP81, TbMP63, TbMP52, and TbMP42. Input pre-mRNA and the edited RNA, chimeras, and 3′-cleavage product that result from editing activities are indicated. Anti-pyruvate dehydrogenase subunit E2 MAb (α PDH) was used as negative control antibody, and the 20S fraction from glycerol gradient-separated mitochondrial extract with (GG extract) and without (no gRNA) gRNA served as positive and negative controls, respectively, for the editing assay. (B) In vitro-precleaved insertion editing assays of samples and controls (except for no gRNA) as in panel A. The input 5′-labeled 5′ substrate RNA and the product with two added Us are ligation products of the input substrate RNA and unlabeled 3′ substrate RNA and the accurately edited RNA product. Products with one added U and the product of its ligation with the 3′ substrate RNA are not labeled. (C) Adenylation assay of samples and controls as in panel B. The adenylated native 50-kDa TbMP52 and 45-kDa TbMP48 RNA ligases are labeled. The 50-kDa adenylated protein band in the α PDH negative control lane was also detected with other nonspecific antibodies, and this was due to nonspecific binding.
MAb P4D8-F6, which was also generated using native editing complexes, is specific for TbMP81. It specifically reacts with the native protein (Fig. 4A and B, top band) and recombinant protein which was expressed in vitro after cloning into the eukaryotic expression vector pSG1 (data not shown), since we were unable to express the protein in E. coli cells. This MAb immunoprecipitated precleaved insertion editing and adenylation activities from the 20S fraction of the mitochondrial lysate (Fig. 3B and C). The lower level of adenylation activity relative to the anti-TbMP63 and anti-TbMP42 MAb precipitates is due to relatively less protein in the precipitate as determined by Western analysis (results not shown). Neither in vitro deletion RNA editing nor endonucleolytic cleavage of the input pre-mRNA substrate was detected in the immunoprecipitate (Fig. 3A). The presence of both the 50- and 45-kDa adenylatable proteins in the immunoprecipitate (Fig. 3C), which correspond to TbMP52 and TbMP48, respectively (12, 19), provides evidence that TbMP81 is present in the editing complex along with TbMP63 and TbMP42. Comparable studies were not possible for TbMP18 since no suitable MAb is available.
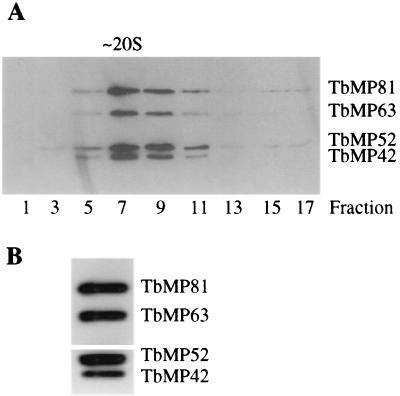
FIG. 4.
Western analyses of sedimented and immunoprecipitated editing complexes. (A) Analysis of glycerol gradient fractions from cleared mitochondrial lysate. The top of the gradient is fraction 1. (B) Analysis of editing complexes that were immunoaffinity purified from the 20S fraction by using a MAb specific for TbMP63.
Additional studies showed that TbMP81, TbMP63, TbMP42, and TbMP18 cosediment and/or coimmunoprecipitate with each other, with editing complex ligases, and with in vitro editing activity. Western analysis of glycerol gradient-fractionated mitochondrial lysate showed that the bulk of the native TbMP81, TbMP63, and TbMP42 proteins cosediment together at ∼20S along with TbMP52 (Fig. 4A), which has been shown to be an essential part of the editing complex (14, 19). Anti-TbMP63 MAb (P1H3-D7) was used to precipitate the editing complex from either crude or glycerol gradient-fractionated mitochondrial lysate. Western analysis identified TbMP81, TbMP63, TbMP52, and TbMP42 proteins in the immunoprecipitated sample (Fig. 4B). In addition to these proteins, MS analysis of the immunoaffinity-purified complex identified TbMP48 (RNA ligase) and TbMP18 in the complex.
Nuclease treatment of complex.
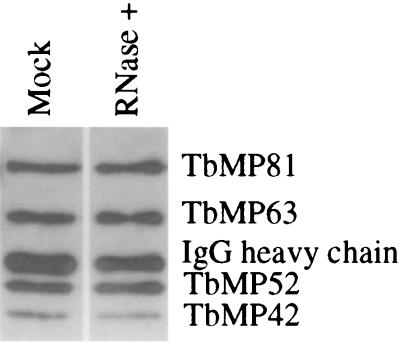
The mitochondrial 20S fraction from glycerol gradients was treated with RNases and the complex was immunoprecipitated with anti-TbMP63 MAb as described in Materials and Methods. Western analysis of the precipitated sample showed that the three zinc finger proteins were still associated with a complex following RNase treatment, as was RNA editing ligase TbMP52. No significant quantitative difference was observed between RNase-treated and mock-treated sample (Fig. 5). Despite extensive washing traces of RNase remained in the precipitated sample, which upon incubation with editing substrates degraded most of the RNA. Hence, we were unable to assess whether the RNase treatment affected editing associated enzymatic activities.
FIG. 5.
Western analysis of RNase treated editing complexes, using the panel of MAbs that are specific for the indicated proteins. The complexes were immunoprecipitated with a MAb specific for TbMP63. Pooled 20S fractions of cleared mitochondrial lysate were treated with RNase or mock treated as described in Materials and Methods. The anti-TbMP63 MAb that was used for immunoprecipitation and which reacts with the second antibody in the Western analysis is indicated.
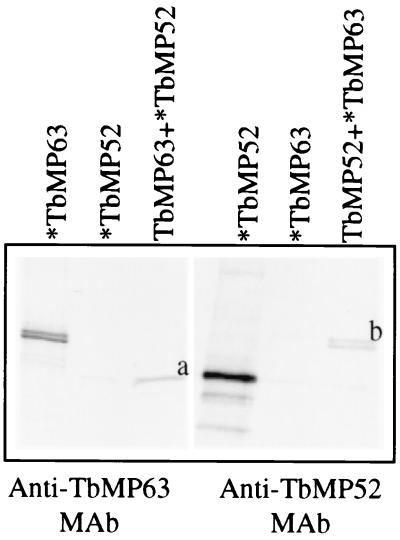
TbMP63 binds to TbMP52 in vitro.
Coimmunoprecipitation analyses showed that TbMP63 and TbMP52 can interact. Anti-TbMP63 MAb precipitated rTbMP52 only in the presence of rTbMP63, and similarly, anti-TbMP52 MAb specifically precipitated rTbMP63 upon coincubation with rTbMP52 (Fig. 6), indicating these two proteins bind to each other. Anti-TbMP63 MAb did not precipitate TbMP52 when present alone nor did anti-TbMP52 MAb precipitate TbMP63 alone. The binding reactions and washes were carried out under highly stringent conditions, and the luciferase control did not coprecipitate with these proteins nor did anti-His6 MAb pull down the proteins (results not shown), indicating that the interaction between TbMP52 and TbMP63 was specific. Under similar conditions TbMP81 did not coprecipitate with these proteins and no other specific interactions were observed among TbMP42, TbMP48, TbMP52, and TbMP63 (results not shown).
FIG. 6.
Autoradiogram of SDS-PAGE analysis of coimmunoprecipitation analyses of TbMP63 and TbMP52 recombinant proteins. The protein that was radiolabeled in each reaction is indicated (∗), and the 52-kDa (a) and 63-kDa (b) proteins that were coimmunoprecipitated are shown.
Editing complex subunit or fragment.
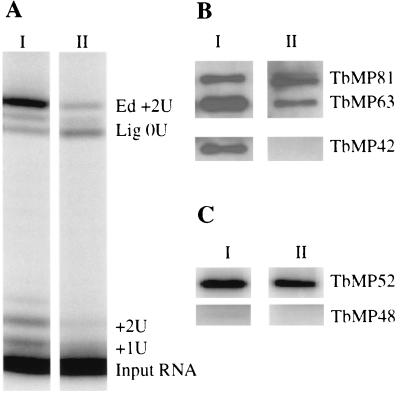
In vitro editing and most of the associated catalytic activities elute at ∼1,600 kDa (peak I) from Superose 6 columns during purification of the editing complex, but some of the activities elute at 450 to 500 kDa (peak II) (11, 14). The peak I sediments at 20S (14) and peak II sediments have not been tested due to low abundance but may sediment at less than 20S. In vitro deletion editing was not detected in material that was immunoprecipitated with anti-TbMP63 MAb from peak II although it was detected in peak I immunoprecipitates (data not shown). In addition, there was less precleaved insertion editing activity and U addition activity but a greater ratio of ligated-to-edited RNA in peak II than in peak I immunoprecipitates (Fig. 7A). Thus, the peak II immunoprecipitate contains TUTase and ligase activities although apparently less than in immunoprecipitates from peak I. Western analysis of these samples showed that while the TbMP81, TbMP63, and TbMP42 zinc finger proteins are present in the immunoprecipitate from peak I, the former two but not TbMP42 was detected in immunoprecipitates from peak II (Fig. 7B). In addition, while adenylatable TbMP52 and TbMP48 RNA ligases were present in immunoprecipitates from peak I, adenylatable TbMP48 was not detected in immunoprecipitates from peak II (Fig. 7C). Thus, peak II appears to contain some but not all of the components of the editing complexes that are present in peak I.
FIG. 7.
Analyses of complexes with different molecular masses. Complexes were immunoprecipitated with anti-TbMP63 MAb from Superose 6 column fractions that correspond to 1,600 kDa (I) and 500 kDa (II) after sequential chromatography with SP Sepharose and Q Sepharose columns. (A) In vitro precleaved insertion editing assay of immunoprecipitates labeled as in Fig. 2B. (B) Western analysis of the immunoprecipitates using MAbs specific for the proteins indicated. Note the absence of TbMP42 in II. (C) Adenylation analysis of the immunoprecipitates revealing the lack of adenylated TbMP48 in II.
DISCUSSION
This study describes the identification of four proteins from purified RNA editing complexes of T. brucei and the corresponding genes which encode preprocessed proteins with masses of 81, 63, 42, and 18 kDa. All four proteins contain a similar C-terminal domain, and the 42- and 18-kDa proteins have considerable sequence similarity. In addition, the 81-kDa protein contains a single zinc finger motif while the 63- and 42-kDa proteins each contain two zinc finger motifs. All four proteins were shown, by a combination of Western and MS analyses, to be present along with two editing RNA ligases in functional editing complexes. These complexes were specifically immunoprecipitated from total mitochondrial lysate, the 20S fraction of glycerol gradients, and the 1,600-kDa fraction from Superose 6 columns. These immunoprecipitation studies used MAbs that were prepared with native complex proteins and are specific for the 81-, 63-, and 42-kDa proteins, as well as the 52-kDa editing RNA ligase. Coimmunoprecipitation studies using these MAbs showed that the 63- and 52-kDa editing RNA ligase recombinant proteins interact. We conclude that these four proteins along with the 48- and 52-kDa RNA ligases are components of the RNA editing complex of T. brucei and that the 52- and 63-kDa proteins may be in contact in the complex.
The proteins were identified by MS, which has become an important proteomic tool (reviewed in reference 4). Identification of the proteins and their corresponding genes in the public databases by computational means was possible despite the fact that the genome sequencing of T. brucei is not yet complete but is predominantly composed of end sequences of clones from genomic DNA libraries. This was accomplished by reiterative comparisons among mass spectrometric and DNA sequence data and, finally, PCR amplification, cloning, and sequencing of the complete gene. The confirmation of gene identity in all cases illustrates the reliability of this approach. Gene identity was confirmed by specific reaction of native and recombinant 81-, 63-, and 42-kDa proteins with MAbs that were generated against the isolated complex and by the match between the N-terminal sequences of the 18-kDa protein sequence and the gene identified by LC-MS/MS.
Several lines of evidence lead to the conclusion that the four proteins identified in this study are components of the RNA editing complex along with the TbMP52 and TbMP48 RNA ligases. These proteins cofractionate with in vitro deletion and insertion editing, their component catalytic activities, and the 52- and 48-kDa editing RNA ligases, all of which ultimately elute from Superose 6 columns at 1,600 kDa and sediment at 20S in glycerol gradients (14) (Fig. 4A). In addition, all six proteins were detected by Western or MS/MS analysis in complexes that were immunoprecipitated with MAbs specific for these proteins and for the 52-kDa editing RNA ligase (Fig. 3 and 4). They were not immunoprecipitated by MAbs specific for mitochondrial proteins from the ATP synthase and pyruvate dehydrogenase complexes that did not cosediment with the RNA editing complex. The immunoprecipitations were performed and washed under stringent conditions (i.e., in buffer containing up to 400 mM KCl), which should dissociate adsorbed proteins from the immunoprecipitated complexes (14). RNase treatment, which would have eliminated proteins present as a result of RNA binding proteins, did not eliminate immunoprecipitations of proteins that could have been assayed with the available MAbs.
The potential functions of the four proteins identified in this study are not evident from homology searches. The C2H2 zinc finger motifs found in the 81-, 63-, and 42-kDa proteins imply that these proteins may have roles in nucleic acid or protein binding. Such motifs are common in eukaryotes and can function in DNA binding for gene regulation (1, 26) or protein-RNA and protein-protein interactions (10). These motifs may bind the pre-mRNAs and gRNAs that are present in purified editing complexes (11, 14) and that associate with the complex during editing and/or bind proteins to the editing complex. The demonstration in this study that TbMP63 can bind to TbMP52 (Fig. 6) is suggestive of a protein binding role. The finding that nuclease treatment did not dissociate the zinc finger or 52-kDa editing RNA ligase from an immunoprecipitable complex (Fig. 5A) suggests that RNA that is accessible to the nuclease degradation (some RNA resists degradation [24]) is not essential for overall complex integrity. Thus, the zinc finger proteins may play a role in the structural organization, integration of catalytic functions, and/or regulatory functions of the editing complex. The SPF tripeptide repeats in the 63-kDa protein are the only other evident motif which may be important for the structure and thereby the function of this protein.
The four editing complex proteins identified here may have a variety of functions in addition to molecular binding, including catalytic activity. The MAb specific for the 81-kDa protein immunoprecipitated precleaved insertion editing, revealing TUTase and RNA ligase activities, but the RNA substrate was not cleaved, suggesting the absence of endoribonuclease activity in the immunoprecipitate (Fig. 3). Preliminary results by Salavati and Drozdz et al. (unpublished results) suggest that the TbMP81 protein is associated with endonuclease activity. The relationship between the 81-kDa protein and the other three proteins implies that they may have comparable roles, and the substantial similarity between the C-terminal portion of the 42-kDa protein and the 18-kDa proteins implies that they may have substantially similar functions. It is uncertain from our results if all four proteins are derived from an ancestral protein by sequence divergence following gene duplication or if some are derived by modification via domain switching. Genetic studies are in progress to assess which of the proteins are essential to editing and to identify their functions.
The total composition of the fully functional editing complex is uncertain. Functional editing complexes have been biochemically prepared from T. brucei by different methods (11, 14, 16). Purification was monitored by tracking RNA ligase adenylation (11, 16) or in vitro editing plus associated editing activities, including RNA ligase adenylation (14). The complexes prepared in the Sollner-Webb lab (16) appear to lack RNA and contain seven major proteins while those prepared for this study (14) and in the Hajduk lab (11) contain mRNA and gRNA and 20 or more major proteins. Proteins designated IV and V by the Sollner-Webb lab correspond, respectively, to the TbMP52 (T. brucei p52) and TbMP48 (T. brucei p48) editing RNA ligases (12, 17, 19). Based on electrophoretic mobility, proteins II, III, and VII may correspond to the 81-, 63-, and 18-kDa proteins identified here, respectively, and proteins I and VI may correspond to previously observed 99- and 44-kDa proteins (14) that we also detected in MAb affinity-purified complexes (24). The proteins in complexes purified in the Hajduk lab (11) have similar electrophoretic mobilities but different relative silver staining intensities compared to those used here. This suggests differences in relative abundance and/or presence of various proteins, including contaminants in the preparations. Consequently, except for editing ligases, we could not identify likely corresponding proteins.
The purified complex preparations described above are able to edit a single site as specified by a gRNA. However, fully functional in vivo complexes edit numerous sites and use multiple gRNAs, indicating the need for additional functions, such as pre-mRNA translocation and gRNA replacement. It seems likely that there is a core catalytic complex with the endonuclease, TUTase, exo-Uase, and RNA ligase catalysts. As shown here, the 81-, 63-, and 52-kDa (RNA ligase), 48-kDa (RNA ligase), and 42- and 18-kDa proteins are stable components of a complex that catalyzes gRNA-directed editing at a single site. These proteins, along with the 99-kDa protein (24), may correspond to the complexes purified in the Sollner-Webb lab (16) which catalyze deletion and insertion editing. While this suggests that the five non-RNA ligase proteins have the endonuclease, exoUase, and TUTase activities, the presence of other proteins with these activities in low abundance cannot yet be excluded. In addition, 61-kDa (RNA helicase) and 55-, 53-, 47-, 44-, and 24-kDa proteins are also associated with the complex (references 14 and 24 and unpublished data). These latter six proteins may vary in the stability of their association with the complex, and this may be affected by the presence of RNA. The Simpson lab has identified a gene for a 1,120-amino-acid protein (∼130 kDa) with TUTase activity from L. tarentolae and its homologous gene from T. brucei (L. Simpson, personal communication). Since a protein of this size was not detected in the complexes described here, this may indicate that there may have been multiple editing TUTases, that TUTase was present in low abundance in our preparations, or that it was not the editing TUTase but perhaps an enzyme that added U tails to gRNA.
Functional analyses of the components of the editing complex are at an early stage. An RNA helicase, mHel61p, appears to have a nonessential role in editing since null mutants edit, but with reduced activity (13). Perhaps there is another editing RNA helicase that compensates for mHel61p in the null mutants. The TbMP52 RNA ligase is essential to editing (19) while the related TbMP48 is not essential to editing (24). Thus, TbMP52 may compensate for the loss of TbMP48 but TbMP48 may not compensate for the loss of TbMP52. The significance of this functional redundancy is unclear. The presence of four related proteins, two of which have a region of substantial similarity, is a further indication of such potential redundancy. Furthermore, the tetrameric organization of 20S complexes from the 1,600-kDa Superose 6 peak of an editing complex preparation (24) implies a higher order of functional redundancy in which the monomers may contain proteins with similar functions, although not necessarily the same proteins. The 500-kDa complexes from Superose 6 (14) contain TUTase and ligase activities and the 81- and 63-kDa zinc finger proteins, but not 42-kDa zinc finger protein and perhaps not adenylatable TbMP48 RNA ligase (Fig. 7). These complexes may represent subunits, assembly intermediates, or dissociated fragments of the editing complex. The definitive identification of components of the RNA editing complex with clear roles in editing promises to accelerate progress in determining the composition of the fully functional complex and the functions of its components. This information along with an understanding of the structural organization of the complex will be essential to elucidate how pre-mRNAs undergo extensive but accurate sequence remodeling by RNA editing.
ACKNOWLEDGMENTS
We thank Barbara Morach, Brian Panicucci, Jeff O'Rear, Micaiah Evans, and RoseMary Reed for technical help, Maciek Drozdz for communicating unpublished results, Peter Myler for helpful suggestions, Elizabeth Wayner for assistance in MAb production, Sandy Kielland for N-terminal microsequencing, John Donelson for clone T651, and Najib El Sayed for providing several genomic clones. Sequence data were obtained from The Institute for Genomic Research (http://www.tigr.org) and The Sanger Centre (http://www.sanger.ac.uk) websites that were supported by NIAID and The Wellcome Trust as part of the Trypanosoma Genome Network that was coordinated with support from the TDR section of the World Health Organization.
This work was supported by NIH Postdoctoral Fellowship AI10312 to R.I., NIH grant HG00041 to S.G., grants IR33 CA84698 and RO1 A141109-01 to R.A., NIH grants AI14102 and GM4218 to K.S., and HFSPO grant RG0316/1997 M.
REFERENCES
- 1.Berg J M, Godwin H A. Lessons from zinc-binding peptides. Annu Rev Biophys Biomol Struct. 1997;26:357–371. doi: 10.1146/annurev.biophys.26.1.357. [DOI] [PubMed] [Google Scholar]
- 2.Corell R A, Read L K, Riley G R, Nellissery J K, Allen T E, Kable M L, Wachal M D, Seiwert S D, Myler P J, Stuart K D. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol Cell Biol. 1996;16:1410–1418. doi: 10.1128/mcb.16.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estévez A M, Simpson L. Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene. 1999;240:247–260. doi: 10.1016/s0378-1119(99)00437-0. [DOI] [PubMed] [Google Scholar]
- 4.Gygi S P, Aebersold R. Mass spectrometry and proteomics. Curr Opin Chem Biol. 2000;4:489–494. doi: 10.1016/s1367-5931(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 5.Harris M E, Moore D R, Hajduk S L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- 6.Hayman M L, Read L K. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J Biol Chem. 1999;274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- 7.Igo R P, Jr, Palazzo S S, Burgess M L K, Panigrahi A K, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kable M L, Heidmann S, Stuart K D. RNA editing: getting U into RNA. Trends Biochem Sci. 1997;22:162–166. doi: 10.1016/s0968-0004(97)01041-4. [DOI] [PubMed] [Google Scholar]
- 9.Köller J, Müller U, Schmid B, Missel A, Kruft V, Stuart K, Göringer H U. Trypanosoma brucei gBP21: an arginine rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- 10.Mackay J P, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 11.Madison-Antenucci S, Sabatini R S, Pollard V W, Hajduk S L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 1998;17:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McManus M T, Shimamura M, Grams J, Hajduk S L. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Missel A, Souza A E, Norskau G, Göringer H U. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panigrahi A K, Gygi S, Ernst N, Igo R P, Jr, Palazzo S S, Schnaufer A, Weston D, Carmean N, Salavati R, Aebersold R, Stuart K D. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard V W, Harris M E, Hajduk S L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusché L N, Cruz-Reyes J, Piller K J, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusché L N, Huang C E, Piller K J, Hemann M, Wirtz E, Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabatini R, Hajduk S L. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J Biol Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 19.Schnaufer A, Panigrahi A K, Panicucci B, Igo R P, Jr, Salavati R, Stuart K D. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 20.Seiwert S D, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 21.Stuart K, Allen T E, Heidmann S, Seiwert S D. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart K, Gobright E, Jenni L, Milhausen M, Thomashow L, Agabian N. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J Parasitol. 1984;70:747–754. [PubMed] [Google Scholar]
- 23.Stuart K, Panigrahi A K, Salavati R. RNA editing in kinetoplastid mitochondria. In: Bass B L, editor. RNA editing: frontiers in molecular biology. Oxford, United Kingdom: Oxford University Press; 2000. pp. 1–19. [Google Scholar]
- 24.Stuart K D, Panigrahi A K, Schnaufer A, Drozdz M, Clayton C, Salavati R. Composition of the editing complex of Trypanosoma brucei. 2001. Philos. Trans. R. Soc. Lond., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaïli N, Göringer U, Benne R, Pays E. Trypanosoma brucei TBRGG1: a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J Biol Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe S A, Nekludova L, Pabo C O. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]