Abstract
Cellular therapies hold enormous potential for the cure of severe hematological and oncological disorders. The forefront of innovative gene therapy approaches including therapeutic gene editing and hematopoietic stem cell transplantation needs to be processed by good manufacturing practice to ensure safe application in patients. In the present study, an effective transfection protocol for automated clinical-scale production of genetically modified hematopoietic stem and progenitor cells (HSPCs) using the CliniMACS Prodigy® system including the CliniMACS Electroporator (Miltenyi Biotec) was established. As a proof-of-concept, the enhancer of the BCL11A gene, clustered regularly interspaced short palindromic repeat (CRISPR) target in ongoing clinical trials for β-thalassemia and sickle-cell disease treatment, was disrupted by the CRISPR-Cas9 system simulating a large-scale clinical scenario, yielding 100 million HSPCs with high editing efficiency. In vitro erythroid differentiation and high-performance liquid chromatography analyses corroborated fetal hemoglobin resurgence in edited samples, supporting the feasibility of running the complete process of HSPC gene editing in an automated closed system.
Introduction
Gene editing approaches are currently used to develop therapies for the treatment of severe hematological disorders. Among other technologies, the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system has proven to be one of the most versatile and affordable technologies for gene therapy. Its applications include disruption, addition, and correction of genes of interest in a great variety of clinically relevant cells.1–6 Recently, an important improvement in the health and life quality of two patients treated by a CRISPR-based gene therapy suffering from β-thalassemia and sickle-cell disease (SCD), respectively, was reported.7 The strategy consisted in targeting BCL11A, a gene involved in the negative regulation of fetal hemoglobin (HbF), to promote the resurgence of HbF expression as previously demonstrated in preclinical studies.8–10 These results encourage the expansion of CRISPR-based protocols for the treatment of β-hemoglobinopathies and other hematological disorders.
For the implementation of these therapies, it is common to perform genetic engineering of effector cells ex vivo, before their infusion into the patient. In this context, electroporation is a valuable option for the delivery of the CRISPR-Cas9 components into the cells. The electric field exerted in the cellular membrane during the electroporation process increases the permeability and allows efficient transfer of the cargo into the cells.11,12
The CliniMACS Prodigy system (Miltenyi Biotec) offers fully automated cell culture and expansion in a closed, good manufacturing practice (GMP)-compatible system. The CliniMACS Electroporator was recently implemented into the platform design, enabling additional gene-editing protocols using a single process within one closed tubing set. In this publication, we propose an optimized transfection protocol for editing CD34+ hematopoietic stem and progenitor cells (HSPCs) at a clinical production scale that facilitates translation to patient treatment.
Materials and Methods
Ethics statement
All leukapheresis samples were obtained following standard collection procedures in accordance with the guidelines of the certified collection centers (Cytocare).
HSPC isolation and culture
CD34+ HSPCs were magnetically isolated from mobilized leukapheresis products from healthy donors using the CliniMACS Plus or CliniMACS Prodigy (Miltenyi Biotec). The isolation was performed using the CliniMACS CD34+ reagent (Miltenyi Biotec).
HSPCs were used freshly after isolation or were thawed and recovered from frozen aliquots. They were cultured at a concentration of 1 × 106 cells/mL in an HSC medium 1 day before electroporation at 37°C 5% CO2 in 24-well plates in the incubator (Heracell; Thermo Fisher) or the CliniMACS Prodigy cell culture unit (Miltenyi Biotec). The HSC medium consisted of HSC-Brew GMP Basal Medium (Miltenyi Biotec) supplemented with 1% HSC-Brew GMP Supplement (Miltenyi Biotec), 2% human serum albumin (HSA) (Octapharma), MACS® GMP Recombinant Human stem cell factor (SCF) (100 ng/mL), MACS® GMP Recombinant Human thrombopoietin (TPO) (20 ng/mL), and MACS® GMP Recombinant Human Flt-3 ligand (100 ng/mL; Miltenyi Biotec).
Small-scale HSPC electroporation
CD34+ cells were transfected in CliniMACS Electroporation Buffer (Miltenyi Biotec) at a concentration of 5 × 106 to 1.5 × 107 cells/mL using the CliniMACS Prodigy electroporator with 1 μg DsRed mRNA (in vitro transcribed as previously described).8 One hundred microliters of cell suspension was electroporated in 0.2 cm electrode distance Ingenio® electroporation cuvettes (Mirus Bio LLC) using the test cuvette adapter (TCA) and the respective software on the CliniMACS Prodigy. The used electroporation parameters are indicated in the Results section. For all experiments, two consecutive electroporation pulses with defined voltage and time were applied. In addition, interrupted (burst) as well as bipolar pulses were tested with 8 μs burst duration. After electroporation, cells were transferred to a 24-well plate for recovery in HSC medium at 37°C or 32°C, 5% CO2.
Upscale HPSC electroporation
The cuvette of the CliniMACS Prodigy EP-2 was manually filled with 600 μL of CD34+ cells in the CliniMACS Electroporation Buffer (Miltenyi Biotec) at a concentration of 5 × 106 to 1.5 × 107 cells/mL with either 30 μg/mL eGFP mRNA (Miltenyi Biotec) or 150–900 pmol ribonucleoprotein (RNP) complex per million cells. The cuvette was placed into the CliniMACS Electroporator and cells were electroporated using the TCA software on the CliniMACS Prodigy. Unless indicated otherwise, the following electroporation parameters were used:
Pulse 1 (high voltage): 600 V 104 μs burst/bipolar—8 μs burst duration.
Pulse 2 (low voltage): 200 V 5000 μs square.
After electroporation, the electroporated cell suspension was added to 6 mL of HSC medium and transferred to a 24-well plate for overnight recovery culture at 37°C or 32°C, 5% CO2.
Automated generation of edited CD34+ HPSCs (Prodigy sample)
CD34+ HSPCs were manufactured using the T cell engineering (TCE) process on the CliniMACS Prodigy platform. Process parameters were defined by the operator and saved in the activity matrix of the process. The separated CD34+ HSPCs were connected to the tubing set, after evaluation of the viability and total cell number. The cells were automatically transferred to the CentriCult Unit (CCU) of the CliniMACS Prodigy TS 520 (Miltenyi Biotec) and cultivated using the same medium, temperature, and CO2 concentration used for the small-scale experiments. On day 1, CD34+ cells were electroporated using the CliniMACS Prodigy EP-2 tubing set on the CliniMACS electroporator: cells were rebuffered in the CliniMACS Electroporation Buffer (Miltenyi Biotec); the RNP was transferred to the nucleic acid bag, and electroporation was started using the optimized pulse previously described in the upscale electroporation.
After each electroporation cycle, the edited cells were automatically transferred back to the cultivation chamber and cultured in HSC medium for 24 h including shaking (shaker type 2). On day 2, the cells were formulated in NaCl supplemented with 0.5% HSA and cells were harvested in the target cell bag. The final cell product was directly used for functional analysis or frozen until further use.
CRISPR-Cas9 transfection and analysis
To prove the efficacy of the electroporation settings, a previously designed sgRNA targeting BCL11A was used (Table 1). Unless indicated otherwise, the sgRNA and Cas9 V3 ribonucleoprotein (Integrated DNA Technologies) were incubated for 20 min at room temperature at a 1:2 molar ratio, complexing 450 pmol of Cas9 and 900 pmol of sgRNA per million cells, respectively. CD34+ HSPCs were harvested 24 h postelectroporation and DNA was isolated using the NucleoSpin Tissue kit following the manufacturer's instructions (Macherey-Nagel). Polymerase chain reaction (PCR) amplification targeting the region susceptible to CRISPR-Cas9 editing was performed (primer sequences available in Table 1). After corroboration in 1% agarose gel, samples were purified using the QIAquick PCR Purification Kit (Qiagen) and Sanger sequenced (Eurofins Genomics).
Table 1.
Sequences of BCL11A gRNA and primers used in this work
| Sequence | Ref. | |
|---|---|---|
| BCL11A gRNA | CTAACAGTTGCTTTTATCAC | 8 |
| BCL11A forward primer | GTGTATGTGCTGATTGAGGGC | |
| BCL11A reverse primer | GGACAGCCCGACAGATGAAA |
The obtained results were analyzed and the InDel score was quantified by the ICE online tool (Inference of CRISPR Edits; Synthego). If applicable, Mann–Whitney nonparametric tests were performed to assess the difference in editing efficiency.
Flow cytometry
Flow cytometric measurements to monitor the cell viability, transfection efficiency, and phenotype of CD34+ HSPCs were performed on day 2 after transfection using the MACSQuant analyzer 10 (Miltenyi Biotec). To determine cell viability, 7-aminoactinomycin (Miltenyi Biotec) or propidium iodide (Miltenyi Biotec) was added. The transfection efficiency was determined by measuring the mean fluorescent intensity of the DsRed+ control cells. HSPCs were characterized by staining with two panels comprising anti-CD34-PE/Vio770 (REA1164), anti-CD133/1-APC (AC133), anti-CD45-VioBlue (5B1), or anti-CD34-PE/Vio770 (REA1164), anti-CD45RA-APC/Vio770 (T6D11), CD90-APC (REA897).
During erythroid differentiation, cells were analyzed by staining with three different panels: Panel I: CD34-VioBlue (REA1164) and CD36-APC (platelet glycoprotein 4, REA760). Panel II: CD235a-VioBlue (glycophorin A, REA175) and CD71-APC (transferrin receptor, REA902). Panel III: CD233-APC (Band3, REA368) and CD49d-FITC (α-4 integrin, MY18-24A9). All antibodies were obtained from Miltenyi Biotec. Data were analyzed using MACSQuantify software (Miltenyi Biotec).
In addition, for the automated process, cell recovery was determined for the CD34+ HPSCs by quantification of viable cells on day 2 after electroporation. Where appropriate, nonparametric Mann–Whitney tests were performed to assess the difference in cell viability.
Colony-forming unit assay
The clonal ability of the edited and nonedited CD34+ HSPCs was determined by performing a colony-forming unit (CFU) assay. Five hundred live cells resuspended in 300 μL of IMDM were added to 3 mL of StemMACS™ HSC-CFU complete with erythropoietin, human medium (Miltenyi Biotec) and equally distributed among two 35 mm wells (six-well plates). The plates were incubated for 14 days at 37°C and the resulting colonies were counted using a light microscope.
HSPC differentiation to the erythroid lineage
Erythroid in vitro differentiation of the transfected HSPCs was performed according to established protocols.8,13,14 Briefly, CD34+ HSPCs were cultured at a starting concentration of 104/mL in StemMACS HSC expansion medium supplemented with 2 mM of l-glutamine, 100 ng/mL of SCF (Miltenyi Biotec), 10 ng/mL of IL-3 (Miltenyi Biotec), 0.5 U/mL of human EPO (eBiosciences), 200 μg/mL of holo transferrin (Sigma Aldrich), and 100 U/mL of Pen/Strep for 7 days. For the second phase, the cells were seeded at a starting concentration of 105/mL in StemMACS HSC expansion medium supplemented as above, but with 3 U/mL of EPO and cultured for another 4 days. Cells were finally cultured at a starting concentration of 105/mL as in phase 1, but with 3 U/mL of EPO and 1 mg/mL of holo transferrin until day 21. Erythroid differentiation and maturation were monitored by flow cytometry on days 7, 14, and 21.
HbF quantification
For high-performance liquid chromatography (HPLC), cell pellets after 21 days of erythroid differentiation were frozen for further analysis. The frozen pellets were treated and processed as previously described.8 The analysis was performed on a LaChrom Elite HPLC-system (Merck-Hitachi) using a gradient elution mode with a bis-tris buffer system (buffer A: bis-tris 20 mM, NH4-acetate 13 mM, KCN 1 mM, and buffer B: bis-tris 20 mM, Na-acetate 38 mM, KCN 1 mM, NaCl 200 mM). Hemoglobin proteins were detected by absorbance measurements at 415 nm. Intracellular HbF was also determined on day 21 of erythroid differentiation utilizing intracellular anti-HbF-FITC staining according to the intracellular flow cytometry staining protocol (Miltenyi Biotec).
Analysis of thawed material
Cell pellets from the resulting sample of automated large-scale gene editing and its corresponding controls were resuspended in a freezing medium composed of 90% fetal bovine serum and 10% dimethyl sulfoxide (DMSO) as a cryoprotectant. The cryovials were stored at −80°C in a liquid nitrogen tank for 6 months. Then, samples were gently thawed in a warm water bath (37°C) and were subsequently washed with StemMACS HSC expansion medium (Miltenyi Biotec) without supplements to remove DMSO. Cells were counted and seeded at 1 × 106 cells/mL of culture using a prewarmed HSC maintenance medium (StemMACS HSC expansion medium supplemented with 100 ng of SCF, 20 ng of TPO, and 100 ng of Flt-3 ligand per milliliter of medium; Miltenyi Biotec) in a 24-well plate. The plate was placed in the incubator (37°C 5% CO2) and 1 million cells' aliquots were taken at different time points for viability and genetic analyses (0, 1, 2, 4, and 24 h).
Results
Adaption of electroporation parameters for large-scale processing
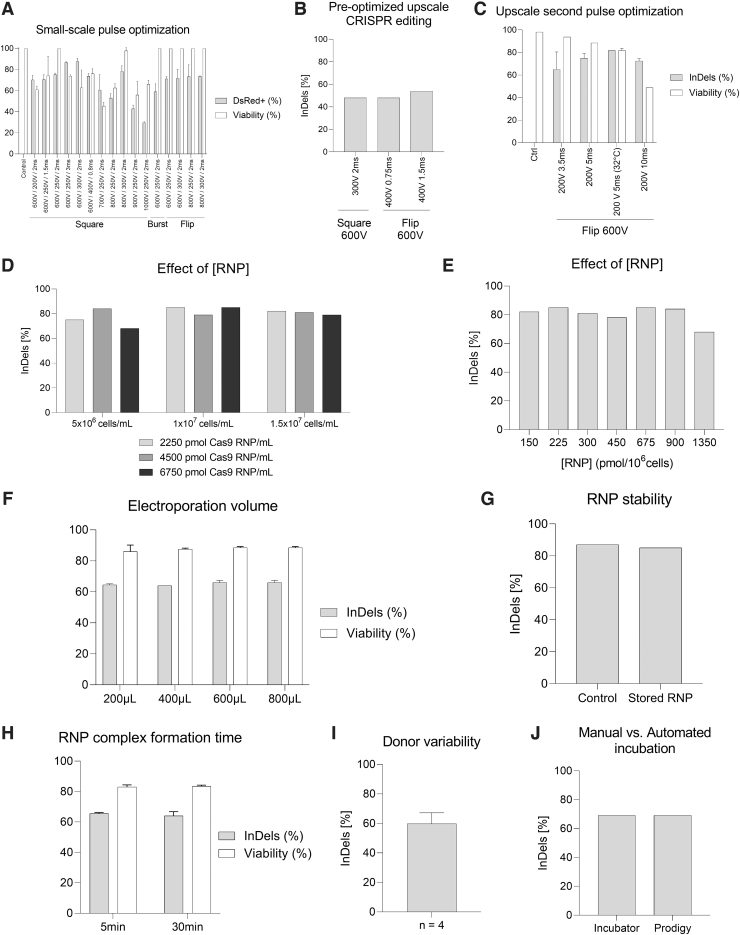
A prescreening was performed to investigate the effect of different electroporation pulses on the transfection efficiency and viability of HSPCs. Small-scale electroporations using the TCA were performed to transfect DsRed mRNA as a reporter of transfection efficiency. Three different electroporation modes were compared: square, burst, and bipolar burst (flip). The burst pulse comprises a series of short 8 μs pulses. The pulse duration is given as the sum of the individual bursts. In bipolar mode, the field directions change after each burst.
The direct comparison using 600 V and 104 μs for the first pulse and 250 V for 2 ms for the second pulse indicates the highest transfection rate of nearly 80% for the square pulse, closely followed by the flip and the burst mode. Increasing the voltage of the first pulse decreased the viability of the cells for square pulses, whereas the bipolar mode gave comparable viabilities even when applying 800 V (Fig. 1A). Increasing the time for the second pulse enhanced the transfection efficiency. Higher second pulse voltages up to 300 V also improved the efficiency, but at the cost of lower viability. However, 400 V combined with a shorter duration of 0.75 ms is also a reasonable combination (Fig. 1A).
FIG. 1.
Study and optimization of electroporation conditions using CliniMACS Prodigy electroporator. (A) Small-scale pulse optimization using DsRed mRNA to determine the transfection efficiency (gray) and viability (white), n = 2. (B) Upscale BCL11A transfection using two of the most suitable electroporation settings in small-scale optimization led to moderate InDel efficiencies, n = 1. (C) Upscale comparison of different conditions to further optimize transfection efficiency. BCL11A editing is provided in InDel rate after Sanger sequencing of the PCR product (gray). Viability was determined by flow cytometry 2 days post-transfection (white), n = 2. Nonsignificant differences were observed for editing efficiencies in Mann–Whitney tests (p > 0.05). (D) Influence of different RNP concentrations (2250 pmol/mL, light gray; 4500 pmol/mL, dark gray; 6750 pmol/mL, black) with increasing cell concentrations (5 × 106 to 1.5 × 107 cells/mL) on the BCL11A editing rate, n = 1. (E) Same data as in (D), but displayed as editing rate in relation to RNP concentration per 106 cells, n = 1. (F) Effect of the electroporation volume in CliniMACS Prodigy EP-2 cuvette on the editing performance (gray) and viability (white), n = 2. Nonsignificant differences were observed for editing efficiencies and viabilities in Mann–Whitney tests (p > 0.05). (G) RNP stability controlling freshly prepared RNP (control) versus RNP recovered after a process run and storage time of 60 min, n = 1. (H) Effect of the RNP incubation time on the editing rate (light gray) and viability (white), n = 2. Nonsignificant differences were observed for editing efficiencies and viabilities in Mann–Whitney tests (p > 0.05). (I) Average editing rate for thawed HSPCs from different donors, n = 4. (J) Comparison of BCL11A transfection efficiency after cultivation in the CliniMACS Prodigy system versus a classical cell incubator, n = 1. HSPCs, hematopoietic stem and progenitor cells; PCR, polymerase chain reaction; RNP, ribonucleoprotein.
To mimic a clinical treatment scenario, the electroporation cuvette included in the CliniMACS Prodigy® EP-2 tubing set was manually filled with magnetically isolated CD34+ HSPCs (94.2% ± 2% viable CD34+ HSPCs among all viable white blood cells [WBCs]) to test the upscale of the electroporation conditions. In this context, square and flip settings with 600 V as the first pulse were selected to transfect HSPCs with BCL11A RNP as they proved to be highly efficient in the small-scale optimization (Fig. 1A). However, the observed editing performance was moderate (48–54%, Fig. 1B), indicating that further optimization was needed to achieve higher InDel rates. To proceed with the upscale screening of the most efficient electroporation settings, the flip protocol was set as it previously led to the highest BCL11A editing (54% InDel rate, Fig. 1B). For the first pulse, 600 V 104 μs burst/bipolar—8 μs burst, was applied.
To promote suitable viability, it was decided to decrease the voltage of the second pulse to 200 V since higher voltages, such as 300 V, have been shown to reduce viability (Fig. 1A). In addition, it was considered to increase the duration of the second pulse to promote efficient gene editing. By increasing the length of the second 200 V pulse from 3.5 to 5 ms, the efficiency of BCL11A editing was boosted (InDel rate from 65% ± 15.6% to 75% ± 4.3%, Fig. 1C). Further improvement was achieved by postincubation of the electroporated cells at 32°C (InDel score 82%, Fig. 1C). This “cold shock” has proven to increase InDels derived from nonhomologous end joining repair.15 Further increase of the second pulse to 10 ms drastically harmed the viability of the samples (from ≈80% to 50%, Fig. 1C).
BCL11A editing is effective in a wide range of RNP and cell concentrations
Different RNP and cellular concentrations were tested to identify the most suitable composition during electroporation (Fig. 1D, E). Cells in the electroporation buffer were mixed with RNP and manually filled into the cuvette of the CliniMACS Prodigy EP-2 tubing set to simulate upscale conditions. The editing rate appeared robust within the range of tested RNP and cell concentrations with a slight tendency to lower editing rates for the low cell concentration of 5 × 106 cells/mL (Fig. 1D). Considering the RNP-to-cell ratio, as depicted in Figure 1E, comparable InDel rates of 78–85% were observed for 150–900 pmol of RNP per million cells. For the high RNP-to-cell ratio (1350 pmol per million cells), respectively, a cell concentration of 5 × 106 cells/mL combined with a high RNP concentration of 6750 pmol/mL, the editing rate dropped (InDel rate 68%, Fig. 1E).
A cell concentration of 1 × 107 cells/mL and an RNP concentration of 2250–6750 pmol/mL (225–675 pmol per million cells) during electroporation were depicted as the most effective condition for further experiments (Fig. 1D, E).
The selected transfection conditions work for different electroporation volumes
The typical filling volume during automated large-scale electroporation in the CliniMACS Prodigy electroporator is 600–650 μL. To investigate the potential impact of different electroporation volumes, a range of 200–800 μL was tested. Independent of the volume, the editing efficiency was consistent between all tested samples (InDel rate 64–66%, Fig. 1F).
CRISPR RNP complex stability is not a limiting factor in the editing process
The CliniMACS Prodigy electroporator sequentially electroporates 600–650 μL of cells per cycle with a cycle time of ≈30 s. The duration of the electroporation process will depend on the total volume to be processed. More specifically, we used 25 mL of cell suspension for the large-scale electroporation with an approximate duration of 40 min. We considered that the RNP might be degraded over time. To investigate the effect of storage at a defined room temperature (22°C), the remaining RNP complex of a large-scale electroporation was used in a small-scale electroporation using the TCA after a storage time of 1 h. The InDel rate was comparable with freshly prepared RNP (85% InDel rate, Fig. 1G).
In a different experiment, the RNP complex was additionally incubated at room temperature (22°C) for 5 and 30 min, showing similar results in the editing performance (InDel rate 65.5% ± 0.5% and 64% ± 2%, respectively, Fig. 1H). Within the tested time scale of up to 1 h, storage of the RNP complex has no impact on the editing efficiency.
Comparable editing rates can be reached for different donors
Cells from different donors can affect the experimental outcome. To assess the effect of individual variability of cellular fitness along the process, CD34+ HSPCs from four different donors were thawed and processed. Although slightly different efficiencies can be observed, the editing was consistent in all samples (BCL11A InDel rate: 60 ± 7.3, Fig. 1I).
Culture conditions can be scaled up without impacting the cell product
The post-transfection recovery is crucial for gene editing and cell survival. The manual cultivation in the incubator was compared with the cultivation in the CCU of the CliniMACS Prodigy system. The editing performance was equivalent, indicating optimal conditions of the automated culture for transfection efficiency (69% InDel rate, Fig. 1J).
BCL11A knockout in CD34+ HSPCs is efficient in a large-scale scenario with clinically relevant cell numbers
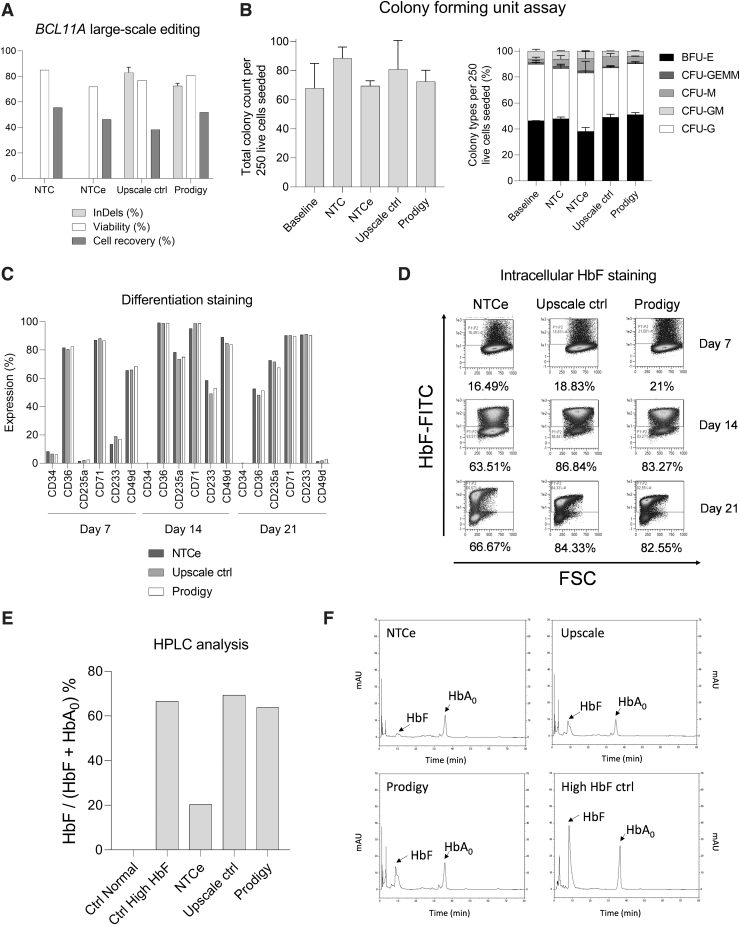
After the identification of the most suitable electroporation parameters and conditions for efficient BCL11A editing, a large-scale run was performed. CD34+ HSPCs were isolated from a mobilized leukapheresis using the CliniMACS CD34 enrichment. 2.1 × 108 CD34+ HSPCs (94.7% viable CD34+ HSPCs among all viable WBCs) were further processed using the TCE process on the CliniMACS Prodigy. On day 2, 1.1 × 108 cells were harvested (52% recovery, Fig. 2A) and further analyzed. The editing efficiency of the upscale control and large-scale samples was 80–86% and 71–74%, respectively, with a viability of 76.9% and 80.8%, which was similar to 85% viable cells observed for nontransfected control (NTC) (Fig. 2A).
FIG. 2.
Large-scale BCL11A editing of HSPCs using CliniMACS Prodigy system with electroporator compared with NTC, NTCe, and upscale controls using the CliniMACS Prodigy EP-2 cuvette. (A) BCL11A editing at the genomic level (n = 1 with technical replicates), cellular viability, and recovery at day 2 after electroporation (n = 1). (B) CFU assay of large-scale samples compared with upscale samples. Total colonies counted for 250 seeded HSPCs (left) and proportion of different colonies, n = 1 with technical replicates. (C) Erythroid differentiation staining on days 7, 14, and 21. Positive rate by flow cytometry for CD34, CD36, CD235a, CD71, CD233, and CD49d, mock electroporated cells (NTCe) (dark gray), upscale control electroporation (light gray), and cells electroporated by the CliniMACS Prodigy process (white), n = 1. (D) HbF levels of electroporated samples measured by flow cytometry on days 7, 14, and 21. (E) HbF/(HbF+HbA0) ratio as determined by HPLC analysis of normal control cells, HbF expressing control cells, and processed cells after additional erythroid differentiation: mock electroporated cells (NTCe), upscale control electroporated cells, and cells electroporated by the CliniMACS Prodigy process, n = 1. (F) HPLC chromatograms of HbF expressing control cells and electroporated samples after in vitro erythroid differentiation. BFU-E, burst-forming unit-erythroid; CFU, colony-forming unit; CFU-G, CFU-granulocyte; CFU-GEMM, CFU-granulocyte erythrocyte macrophage megakaryocyte; CFU-GM, CFU-granulocyte macrophage; CFU-M, CFU-macrophage; HbF, fetal hemoglobin; HPLC, high-performance liquid chromatography; NTC, nontransfected controls; NTCe, electroporated nontransfected controls.
The differentiation potential of CD34+ HSPCs is not compromised after BCL11A knockout and HbF resurgence is independent of the production scale
CFU assays were conducted to determine the proliferation and differentiation potential of the electroporated cells and therefore assess whether the cells remained functional. Cells were seeded immediately after separation (baseline, day 0) and compared with the samples with cells after electroporation and cultivation in the CliniMACS Prodigy on day 2 (Fig. 2B). The cells developed into the typical myeloid colonies exhibiting very similar total colony counts, as well as distributions of colony types among the samples.
To corroborate that the differentiation potential of the edited HSPCs is not compromised and to investigate the induced expression of HbF in the edited cells, an in vitro erythroid differentiation protocol was performed. The expression of relevant differentiation markers was assessed during the differentiation on days 7, 14, and 21 (Fig. 2C). Whereas the purity of CD34+ cells was typically more than 95% after separation and 2 days of culture, the expression decreased during the differentiation below the detection limit at day 14 (Fig. 2C and Supplementary Fig. S1A). Along the differentiation, CD36 (an early marker for erythroid differentiation)16 increased from day 7 to 14 and decreased until day 21. CD233 and CD235a erythrocyte markers17 increased their positive population from below 20% and 5% to roughly 90% and 75% on day 21 as described during erythropoiesis (Fig. 2C and Supplementary Fig. S1B, C).
High expression of CD49d was reported on day 14, but it dropped below 5% on day 21 as expected in the late stage of erythroid differentiation (Fig. 2C and Supplementary Fig. S1C).14 More than 80% of the cells expressed CD71 on day 7, maintaining high expression levels until day 21. This observation correlates with the development of proerythroblasts to basophilic and polychromatophilic erythroblasts and finally late-stage orthochromatic erythroblasts17 (Fig. 2C and Supplementary Fig. S1). Both upscale and large-scale samples reported similar receptor expression levels to those observed in the NTC. CD34−, CD235a+, CD71+ cells indicate hemoglobin-expressing, mature differentiated erythrocytes as required for the analysis of HbF expression.18
Concerning HbF resurgence after BCL11A disruption, flow cytometry and HPLC analyses revealed an upregulation of HbF in upscale and large-scale samples (Fig. 2D–F). The HbF staining and flow cytometry analyses indicate HbF induction in the differentiation control sample, increasing from 16% on day 7 to 64% and 67% on days 14 and 21, respectively (Fig. 2D). However, the rate of HbF expressing cells appeared higher for the edited cells with 19%, 97%, and 84% for the upscale control sample and 21%, 83%, and 83% for the large-scale sample processed on the CliniMACS Prodigy (Fig. 2D).
The HPLC analysis corroborates the cytometry data with an HbF to HbF plus HbA ratio of 20.4% for the electroporated but nontransduced control, 69.3% for the upscale, and 63.8% for the large-scale electroporation sample (Fig. 2E, F). The HbF expression, besides the spontaneous or differentiation-induced HbF expression in the control sample, resembles the editing rate on the genomic level.
Freeze and thawing of edited HSPCs decreased the BCL11A knockout population
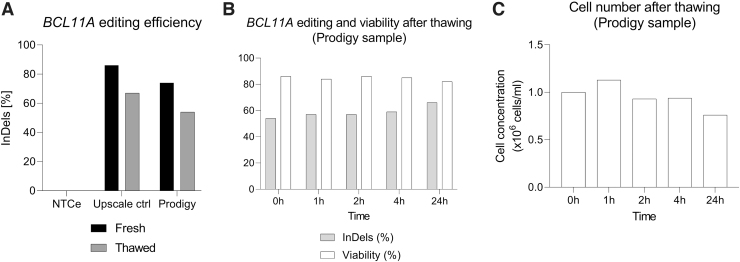
After the large-scale run, samples were frozen and stored at −80°C for 6 months to analyze the InDel score and evaluate the recovery of the cells after thawing. Unexpectedly, there was a reduction in BCL11A editing for both upscale control and large-scale electroporation samples (from 86% to 67% and 74% to 54%, respectively, Fig. 3A). Genetic analyses were performed with the large-scale sample at different time points after thawing to assess this observation, revealing a similar InDel score (54–59% at 0–4 h, Fig. 3B) that was moderately increased after 24 h (66% InDels, Fig. 3B). The thawed cells showed high viability (82–86%, Fig. 3B) and consistent cellular numbers during the first few hours after thawing (9.4 × 105–1.12 × 106 cells/mL at 0–4 h, Fig. 3C), whereas a decline was observed after 24 h (7.6 × 105 cells/mL, Fig. 3C).
FIG. 3.
Effect of freezing/thawing cycle in gene editing and viability. (A) BCL11A editing efficiency comparison of freshly edited HSPCs versus thawed edited HSPCs after freezing storage at −80°C for 6 months, n = 1. (B) Time course analysis of BCL11A editing efficiency and cellular viability after thawing for large-scale Prodigy sample. (C) Time course analysis of cell number after thawing for large-scale Prodigy sample.
Discussion
Hemoglobinopathies are among the most common human genetic disorders worldwide.19 Besides hematopoietic stem cell transplantation (HSCT), there has been no curative transfusion-dependent β-thalassemia (TDT) and SCD. However, gene therapies either using viral vectors or genome editing strategies could overcome this shortcoming. In this context, the first gene therapy based on ex vivo transduced HPSCs to integrate a modified form of the β-globin gene has just recently been approved by the FDA.20
We investigated different gene editing approaches and considered BCL11A a promising target.8 In a proof-of-principle investigation, editing of the BCL11A locus has shown clinical benefit for β-thalassemia (TDT) and SCD patients.7 As for other individualized cell therapies, safe and cost-efficient production is crucial for the future availability of these treatments. Therefore, in parallel to clinical developments, we were aiming to improve traditional GMP gene editing approaches and implement the whole cell engineering protocol in an automated closed system. Predefined electroporation pulses are often used and the respective parameters such as voltage or pulse duration cannot be adapted. As shown in this work, optimizing the used conditions enables to balance transfection efficiency and cell survival.
Traditionally, different devices are used, for example, for cell concentration, culture, and electroporation. Accordingly, the respective instruments need to be operated individually and cells have to be transferred in between. Reducing manual and open steps is important to minimize risks during production. In this way, the impact of different operators is also diminished and reproducibility can be improved.21
Apart from that, scalability will be a prerequisite to serving patients, especially for prevalent diseases such as hemoglobinopathies. As discussed during the annual ISCT meeting, the availability of trained staff can be a challenge for sophisticated cell products.22 Simplified procedures and automation will relieve some of the tasks, allowing to focus on monitoring the production processes. As personnel costs have been addressed for up to 47% of the total costs,23 lowering the workload is expected to also substantially contribute to cost-effectiveness.
Another considerable cost factor is the necessary clean room facilities. In this study, using a closed system for the production of cellular therapies could overcome the limitation of a single-cell product manufactured per clean room.23 Thereby, the fixed costs for the facility could be reduced by parallel production. Considering reliable production procedures and cost-effectiveness is not only relevant for late clinical trials or commercial manufacturing, but can be critical already during early developments.
With our work, we tackled some challenges related to different scales and automations. To achieve reasonable throughput, the electroporation volume is increased compared with the manual electroporation systems. The raised conductivity bears the risks of impaired transfection efficiency. To address this issue, we adapted the electroporation pulse (Fig. 1C) and confirmed the reproducible performance for increasing electroporation volumes (Fig. 1F).
The electroporation process on the CliniMACS Prodigy system performs cyclic electroporations until all cells are processed. The volume of cells that can be processed during electroporation (25–200 mL) with a recommended concentration of up to 5 × 107 cells/mL represents a feasible scale for clinical applications. The system has been designed to store the material to be electroporated, that is, the RNP, in a separate bag. Thereby, the risk of RNA degradation is minimized as the two fractions are mixed for each cycle just before the electroporation. However, deviations in the mixing ratio could impair the editing efficiency. We have shown that the CRISPR editing efficiency is sustained in a wide range of RNP and cellular concentrations (Fig. 1D, E).
Another concern had been the storage of the RNP during the processing time. Material loss, for example, by nonspecific binding to the storage bag or dissociation of the RNP complex could lead to a decreased efficiency over time. The editing efficiency was shown to be robust after the common duration of clinical-scale electroporation (storage time of 1 h, Fig. 1G).
Besides the electroporation itself, also the culture conditions might influence the results. In a direct comparison, we observed no differences between the automated process in the CentriCult Unit (CCU) of the CliniMACS Prodigy and cells cultured under standard incubation conditions in an incubator (Fig. 1J).
To assess the effect of individual variability of cellular fitness along the process, frozen HSPCs from four different healthy donors were tested. The editing efficiencies were very similar (Fig. 1I). Moderate editing could be a result of the use of thawed cellular material, which is also observable for the thawed cell samples used in other experiments reported in this publication (Fig. 1F, H, J) and is consistent with previous research conducted in the immunotherapy field.8
After the optimization screenings, we performed a clinical-scale production run. For this purpose, purified HSPCs (94.7% viable CD34+ HSPCs among all viable WBCs) were processed in the CliniMACS Prodigy with the electroporator system using the CliniMACS Prodigy TS 520 and CliniMACS Prodigy EP-2 set. Posterior genomic studies revealed high disruption in BCL11A enhancer (Fig. 2A). Normal HSPC functionality and increased HbF levels were observed after CRISPR treatment, thereby proving the success of the strategy and feasibility of the overall process (Fig. 2B–F). With a yield of 52% (Fig. 2A), a clinically relevant cell number of about 1 × 108 cells were harvested from the isolated CD34+ cells.
According to the American Society for Blood and Marrow Transplant (ASBMT), the minimum recommended stem cell dose for autologous HSCT is 2 × 106 CD34+ cells/kg.24 Accordingly, the cell product of 1 × 108 CD34+ cells would be sufficient for the treatment of a 50 kg patient. Taking the recommended stem cell collection target of 3–5 × 106 CD34+ cells per kilogram into account, we demonstrated the feasibility of the process for future clinical use.24 As in our experiments, the cell number is typically limited by mobilization of HSPCs and the apheresis. Due to the additional manipulation steps for cell engineering and the related cell losses, mobilization, apheresis, and the production process need to be well balanced with the potential cell doses. The clinical-scale run had been intended as proof-of-principle experiment and further technical runs will be needed to confirm the reproducibility of the performance and suitability for routine use.
Finally, one of the most critical processes in HSPC gene therapy lies in the logistics from the manufacturing to the clinical site and the maintenance of the so-called cryochain.25 As common clinical procedures involve gene editing, expansion, and freezing of the cellular material in specialized centers for subsequent transportation, thawing, and administration to the patient in the clinic,7 the large-scale processed sample was frozen for 6 months and thawed for genetic and viability analyses up to 24 h. While cell numbers were properly maintained with high viability during the first hours, it was observed a reduction in the BCL11A knockout population right after thawing in comparison with freshly edited cells (Fig. 3).
We hypothesize that this result could be explained by the superior cellular resilience of unedited cells compared with the CRISPR-edited ones against freezing stress. More runs would be needed to assess this potential negative effect derived from freezing and thawing cycles in clinical-scale gene editing efficiency. In addition, the freezing and thawing process was performed with conventional laboratory protocols, and thereby a different outcome can be expected following clinical standard procedures. Nonetheless, the use of thawed material has been associated with higher risks of cell loss during therapy development and graft failure.25–28 On-site automated production by CliniMACS Prodigy can facilitate the usage of fresh material, suitably bypassing the challenging cryochain and favoring timely treatment availability. Still, it is important to remark that the use of fresh material can interfere with necessary quality controls and gene editing assessments as it would require a much faster application to the patient than cryopreserved cells.
As a consequence, the use of fresh cells for human therapy is mainly suspended in Europe since 2007 due to the adoption of the European Union Tissues and Cells Directive (EUTCD).28 The routine use of fresh material, despite being highly desirable, still needs the optimization of quality assessments and GMP-compliant protocols to make it feasible in the clinic.
Conclusions
The large-scale run results indicate that efficient editing could be obtained at the clinical scale using CRISPR-Cas9 transfection in the CliniMACS Prodigy system with electroporator. Gene-edited HSPC generation and the subsequent cultivation for the treatment of β-hemoglobinopathies can be performed in a closed and automated system, enabling feasible on-site production that could potentially translate into clinical trials similar to NCT03655678 and NCT03745287. In addition, the generated protocol can be transferred to other diseases whose treatment is based on HSCT. In this way, we hope to contribute to accelerating cellular gene therapy accessibility in the near future and support the development of novel treatments for patient care.
Supplementary Material
Authors' Contributions
G.U.-B.: Conceptualization, investigation, visualization, data curation, writing—original draft, and writing—review and editing (equal). M.B.: Conceptualization, investigation, visualization, data curation, writing—original draft, and writing—review and editing (equal). T.G.: Investigation. F.A. Investigation. J.Q.: Investigation. D.K.: Investigation. A.D.-M.: Investigation. A.L.-C.: Investigation. T.E.: Investigation, resources, and supervision. R.H.: Conceptualization, resources, and supervision. S.W.: Conceptualization, resources, supervision, and writing—review and editing (equal). M.M.: Conceptualization, resources, supervision, and writing—review and editing (equal).
Author Disclosure Statement
M.B., T.G., F.A., J.Q., D.K., and S.W. are employed at Miltenyi Biotec. The rest of the authors declare no conflict of interest.
Funding Information
This work was supported by Miltenyi Biotec, Clinician Scientist Program (No. 440-0-0), Stefan Morsch Stiftung, and the University Children's Hospital of Tübingen.
Supplementary Material
References
- 1. González-Romero E, Martínez-Valiente C, García-Ruiz C, et al. CRISPR to fix bad blood: A new tool in basic and clinical hematology. Haematologica 2019;104:881–893; doi: 10.3324/haematol.2018.211359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liang X, Potter J, Kumar S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 2015;208:44–53; doi: 10.1016/j.jbiotec.2015.04.024 [DOI] [PubMed] [Google Scholar]
- 3. Grote S, Ureña-Bailén G, Chan KC-H, et al. In vitro evaluation of CD276-CAR NK-92 functionality, migration and invasion potential in the presence of immune inhibitory factors of the tumor microenvironment. Cells 2021;10:1020; doi: 10.3390/cells10051020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mandal PK, Ferreira LMR, Collins R, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 2014;15:643–652; doi: 10.1016/j.stem.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brunetti L, Gundry MC, Kitano A, et al. Highly efficient gene disruption of murine and human hematopoietic progenitor cells by CRISPR/Cas9. J Vis Exp 2018134:57278; doi: 10.3791/57278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu X, Zhang Y, Cheng C, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res 2017;27:154–157; doi: 10.1038/cr.2016.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frangoul H, Altshuler D, Cappellini MD, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med 2020;384:252–260; doi: 10.1056/NEJMoa2031054 [DOI] [PubMed] [Google Scholar]
- 8. Lamsfus-Calle A, Daniel-Moreno A, Antony JS, et al. Comparative targeting analysis of KLF1, BCL11A, and HBG1/2 in CD34+ HSPCs by CRISPR/Cas9 for the induction of fetal hemoglobin. Sci Rep 2020;10:10133; doi: 10.1038/s41598-020-66309-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daniel-Moreno A, Lamsfus-Calle A, Raju J, et al. CRISPR/Cas9-modified hematopoietic stem cells—Present and future perspectives for stem cell transplantation. Bone Marrow Transplant 2019;54:1940–1950; doi: 10.1038/s41409-019-0510-8 [DOI] [PubMed] [Google Scholar]
- 10. Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: Prospects for new therapies for the β-globin disorders. Blood 2012;120:2945–2953; doi: 10.1182/blood-2012-06-292078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lino CA, Harper JC, Carney JP, et al. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv 2018;25:1234–1257; doi: 10.1080/10717544.2018.1474964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glass Z, Lee M, Li Y, et al. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol 2018;36:173–185; doi: 10.1016/j.tibtech.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dever DP, Bak RO, Reinisch A, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 2016;539:384–389; doi: 10.1038/nature20134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dulmovits BM, Appiah-Kubi AO, Papoin J, et al. Pomalidomide reverses γ-globin silencing through the transcriptional reprogramming of adult hematopoietic progenitors. Blood 2016;127:1481–1492; doi: 10.1182/blood-2015-09-667923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo Q, Mintier G, Ma-Edmonds M, et al. ‘Cold shock’ increases the frequency of homology directed repair gene editing in induced pluripotent stem cells. Sci Rep 2018;8:2080; doi: 10.1038/s41598-018-20358-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu J, Liu J, Xue F, et al. Isolation and functional characterization of human erythroblasts at distinct stages: Implications for understanding of normal and disordered erythropoiesis in vivo. Blood 2013;121:3246–3253; doi: 10.1182/blood-2013-01-476390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen K, Liu J, Heck S, et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A 2009;106:17413–17418; doi: 10.1073/pnas.0909296106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Romero Z, Urbinati F, Geiger S, et al. β-globin gene transfer to human bone marrow for sickle cell disease. J Clin Invest 2013;123:3317–3330; doi: 10.1172/JCI67930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mussolino C, Strouboulis J. Recent approaches for manipulating globin gene expression in treating hemoglobinopathies. Front Genome Ed 2021;3:618111; doi: 10.3389/fgeed.2021.618111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. FDA Approves First Cell-Based Gene Therapy to Treat Adult and Pediatric Patients with Beta-thalassemia Who Require Regular Blood Transfusions. Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-first-cell-based-gene-therapy-treat-adult-and-pediatric-patients-beta-thalassemia-who (Last accessed: August 25, 2022).
- 21. Sarah Callens JH, McCoy R, Ward S. Cell and gene therapy manufacturing: The necessity for a cost-based development approach. Cell Gene Ther Insights 2016;2:115–120; doi: 10.18609.cgti.2016.014 [Google Scholar]
- 22. ISCT provided post-pandemic venue for the entire cell and gene therapy sector to generate solutions to the major barriers to providing therapies to patients. Available from: https://www.isctglobal.org/telegrafthub/blogs/audrey-le/2022/07/24/isct-publishes-conclusions-and-consensus-from-its [Last accessed: November 1, 2022].
- 23. ten Ham RMT, Hövels AM, Hoekman J, et al. What does cell therapy manufacturing cost? A framework and methodology to facilitate academic and other small-scale cell therapy manufacturing costings. Cytotherapy 2020;22:388–397; doi: 10.1016/j.jcyt.2020.03.432 [DOI] [PubMed] [Google Scholar]
- 24. Giralt S, Costa L, Schriber J, et al. Optimizing autologous stem cell mobilization strategies to improve patient outcomes: Consensus guidelines and recommendations. Biol Blood Marrow Transplant 2014;20:295–308; doi: 10.1016/j.bbmt.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 25. Meneghel J, Kilbride P, Morris GJ. Cryopreservation as a key element in the successful delivery of cell-based therapies—A review. Front Med (Lausanne) 2020;7:59222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dholaria B, Malki MMA, Artz A, et al. Securing the graft during pandemic: Are we ready for cryopreservation for all? Biol Blood Marrow Transplant 2020;26:e145–e146; doi: 10.1016/j.bbmt.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hornberger K, Yu G, McKenna D, et al. Cryopreservation of hematopoietic stem cells: Emerging assays, cryoprotectant agents, and technology to improve outcomes. Transfus Med Hemother 2019;46:188–196; doi: 10.1159/000496068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cottle C, Porter AP, Lipat A, et al. Impact of cryopreservation and freeze-thawing on therapeutic properties of mesenchymal stromal/stem cells and other common cellular therapeutics. Curr Stem Cell Rep 2022;8:72–92; doi: 10.1007/s40778-022-00212-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.