Abstract
Prostate cancer (PCa) continues to threaten men’s health, and treatment targeting the androgen receptor (AR) pathway is the major therapy for PCa patients. Several second-generation androgen receptor inhibitors (SG-ARIs), including enzalutamide (ENZ), apalutamide (APA) and darolutamide (DARO), have been developed to better block the activity of AR. Unavoidably, emergence of resistance to these novel drugs still persists. Herein, we identified glutathione S-transferase Mu 2 (GSTM2) as an important determinant in the acquisition of resistance to SG-ARIs. Elevated GSTM2 was detected in enzalutamide-resistant (ENZ-R) PCa, and overexpression of GSTM2 in naïve enzalutamide-sensitive (ENZ-S) cells effectively transformed them to ENZ-R PCa. Aryl hydrocarbon receptor (AhR), the upstream transcription factor, was implicated in the overexpression of GSTM2 in ENZ-R cells. Mechanistically, GSTM2 antagonized the effect of ENZ by rescuing cells from oxidative stress-associated damage and activation of p38 MAPK pathway. Surprisingly, high GSTM2 levels also associated with cross-resistance to APA and DARO. Taking together, these results provide new insight to ameliorate resistance to SG-ARIs and improve treatment outcome.
INTRODUCTION
According to the latest statistics, prostate cancer (PCa) is the third leading diagnosed type of cancer and the sixth leading cause of death [1], thus remaining to be a major obstacle to human well-being. Androgen receptor (AR) plays a pivotal role in PCa and serves as the basis of disease progression. As a result, current therapy for PCa focuses mainly on pharmaceutical inhibition of AR activity. Despite the initial effectiveness, most patients gradually develop resistance to the first-line treatment and progress to castration-resistant prostate cancer (CRPC) [2]. Established evidence shows that CRPC is characterized with androgen insensitivity accompanied with overexpression of AR. As such, even at the CRPC stage, AR remains a valid target for treatment [3]. To better target the AR pathway, several second-generation AR inhibitors (SG-ARIs) have been developed. Enzalutamide (ENZ) was the early approved SG-ARI to treat CRPC. Following that, apalutamide (APA) and darolutamide (DARO) were greenlighted as well [4]. Based on data obtained from large-scale clinical trials, these novel AR inhibitors possessed much higher potency to block AR signaling and consistently delivered better therapeutic effect [5–7]. Although patients’ welfare has been significantly improved, subsequent acquisition of resistance to SG-ARIs greatly impairs the treatment outcome. Since ENZ was early approved to treat CRPC, resistance to it has been extensively studied, whereas research regarding resistance to APA and DARO is still lacking. Therefore, more experiments are needed to explore the mechanism of resistance to SG-ARIs.
Sufficient evidence from both experimental and clinical studies indicates that oxidative stress (OS) is inherent and might play a critical role in prostate carcinogenesis [8, 9]. Marked by the level of reactive oxygen species (ROS), OS can be either beneficial or harmful to the cells. Under the condition of low ROS, several growth factor pathways are activated to enhance cell proliferation and promote the progression of cancer. However, if the level of ROS reaches a threshold, unrestricted OS leads to damage and triggers cell death [10]. Thus, it is critical for cells to develop preemptive mechanism to protect them from OS-associate damage while still enjoying the benefit of ROS. This tug-of-war is kept in balance by the cellular antioxidants and salvage system, which include ROS scavengers that directly eliminate excessive ROS and regulatory components that antagonize the activation of death pathways [11, 12]. Considering the double-edged effect of OS, tilting the redox balance toward the harmful edge can be a novel treatment modality for PCa.
Glutathione S-Transferase Mu 2 (GSTM2) is a phase II metabolizing enzyme that conjugates glutathione to electrophilic compounds. This process is very important to detoxify harmful chemicals [13]. In addition, GSTM2 has been implicated in cellular anti-OS defense to protect cells from OS-associated damage and cell death [14]. Considering the pivotal role of OS in tumorigenesis, it is not surprising that GSTM2 is closely related to cancer progression, and there have been several publications regarding the function of GSTM2 in drug resistance [15–19]. Nonetheless, these data do not always convey consistent results and are sometimes interpreted diametrically. In addition, little is known about the role of GSTM2 in PCa progression and drug resistance, which needs to be further uncovered. Here we identified that GSTM2 was overexpressed in enzalutamide-resistant (ENZ-R) PCa and sustained the nature of drug resistance. Inhibition or depletion of GSTM2 effectively overwhelmed resistance to ENZ by counteracting the treatment-induced OS and activation of cell death. Surprisingly, this relation was not restricted to a single drug, as GSTM2 also contributed to general resistance to SG-ARIs through similar mechanism. In sum, our findings likely benefit PCa patients by enhancing the potency of current treatment regimen.
RESULTS
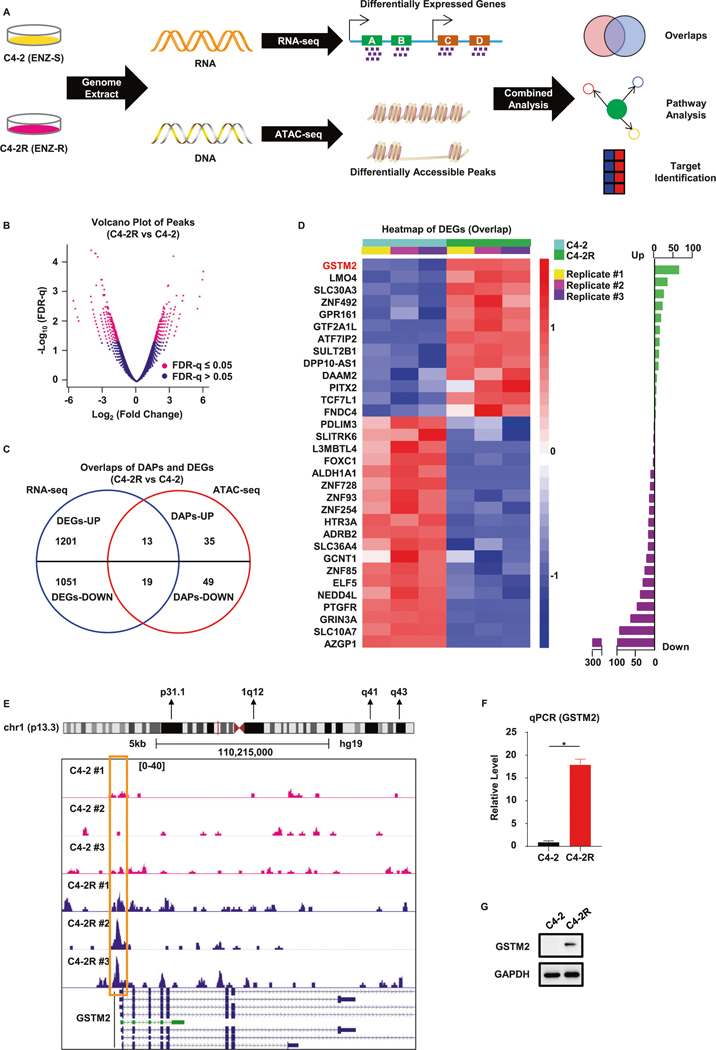
Integrative analysis identifies overexpression of GSTM2 in ENZ-R cells
To investigate a potential causal link between the genetic profile and resistance to SG-ARIs, we used C4–2 and C4–2R cell lines as our starting point. Whereas C4–2 is an ENZ-sensitive (ENZ-S) cell line, C4–2R is the ENZ-R derivative from C4–2 through long-term drug selection. We extracted the genomic contents and performed Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), which detects the open chromatin regions of genome [20]. Together with the RNA-seq we reported previously [21], we sought to integrate two datasets to see whether there were any overlapping genes and pathways that could serve as a promising target (Fig. 1A). To access the reliability of the ATAC-seq results, we first performed hierarchical clustering analysis and principal component analysis. The results indicated that each replicate was closely correlated with others with minimal variation, validating the quality of ATAC-seq (Fig. S1A, B). A total of 37,328 peaks were identified in ATAC-seq, of which 11,400 were shared between C4–2 and C4–2R cells (Fig. S1C). A total of 963 differentially accessible peaks (DAPs) were identified, as shown in the volcano plot (Fig. 1B and Supplementary Table 1). We then drew the average density plot and corresponding heatmap of each DAP. Compared with C4–2, 403 and 560 DAPs were gained or lost in C4–2R, respectively (Fig. S1D, E). After mapping them to the whole genome, we noticed that the DAPs were distributed among various functional gene elements located at different position (Fig. S1F). To investigate whether there were any consensus gene signals between the two sequencings, we made an overlap of differentially expressed genes (DEGs) in RNA-seq and DAPs in ATAC-seq. The result indicated that 114 genes were consistently recognized in both sequencings (Fig. S1G). Of note, although ATAC-seq does not tell us the information of gene expression, DAPs of promoter regions often imply the transcription capability and gene expression indirectly, as an open status of promoter regions facilitates the access of transcription machinery. In light of this notion, we further overlapped DEGs with DAPs of promoter regions and identified 32 consensus genes (Fig. 1C). We generated the heatmap and showed that a total of 13 and 19 genes were up-regulated or down-regulated in C4–2R, respectively (Fig. 1D and Supplementary Table 2). Specifically, we were interested in GSTM2, as it was the most remarkably up-regulated gene in ENZ-R C4–2R, highlighting it as a potential therapeutic target to overcome drug resistance. Indeed, a closer look at GSTM2 loci confirmed strong peak signals around the promoter region and transcription start site in C4–2R, suggesting active transcription of this gene in ENZ-R cells (Fig. 1E). Detection of the mRNA and protein expression in C4–2 and C4–2R confirmed the elevated level of GSTM2 in C4–2R (Fig. 1F, G).
Fig. 1. Integrative analysis of RNA-seq and ATAC-seq indicates critical role of GSTM2 in ENZ-R PCa.
A Schematic illustration of the methodology used to explore enzalutamide-resistant (ENZ-R) and enzalutamide-sensitive (ENZ-S) prostate cancer (PCa). B Volcano plot of all peaks in the assay for transposase-accessible chromatin using sequencing (ATAC-seq). A total of 963 differentially accessible peaks (DAPs) are identified, marked with log2(fold change) ≥2 and FDR-q ≤ 0.05. The X-axis indicates the ATAC-seq signal differences between C4–2R and C4–2. C Venn diagram of differentially expressed genes (DEGs) in RNA-seq and DAPs of promoter regions in ATAC-seq. A total of 32 genes have the same trend in both cases. D Differential expression analysis of the overlap genes identified in C. Left panel: heatmap of 32 DEGs with normalized gene expression (TPM) from RNA-seq. Right panel: barplot of 32 DEGs preranked with −log10(FDR-q). Up-regulated genes in C4–2R are marked with green and down-regulated genes are marked with purple. E Genome browser tracks of ATAC-seq signal density at the GSTM2 loci in C4–2R and C4–2 cell lines. The rectangle indicates the promoter region of GSTM2. F, G qPCR and immunoblot (IB) to verify the overexpression of GSTM2 in ENZ-R C4–2R. qPCR data are normalized to GAPDH, then normalized to C4–2 and shown as mean ± SD (n = 6). *p < 0.05.
Inhibition of GSTM2 overcomes resistance to ENZ
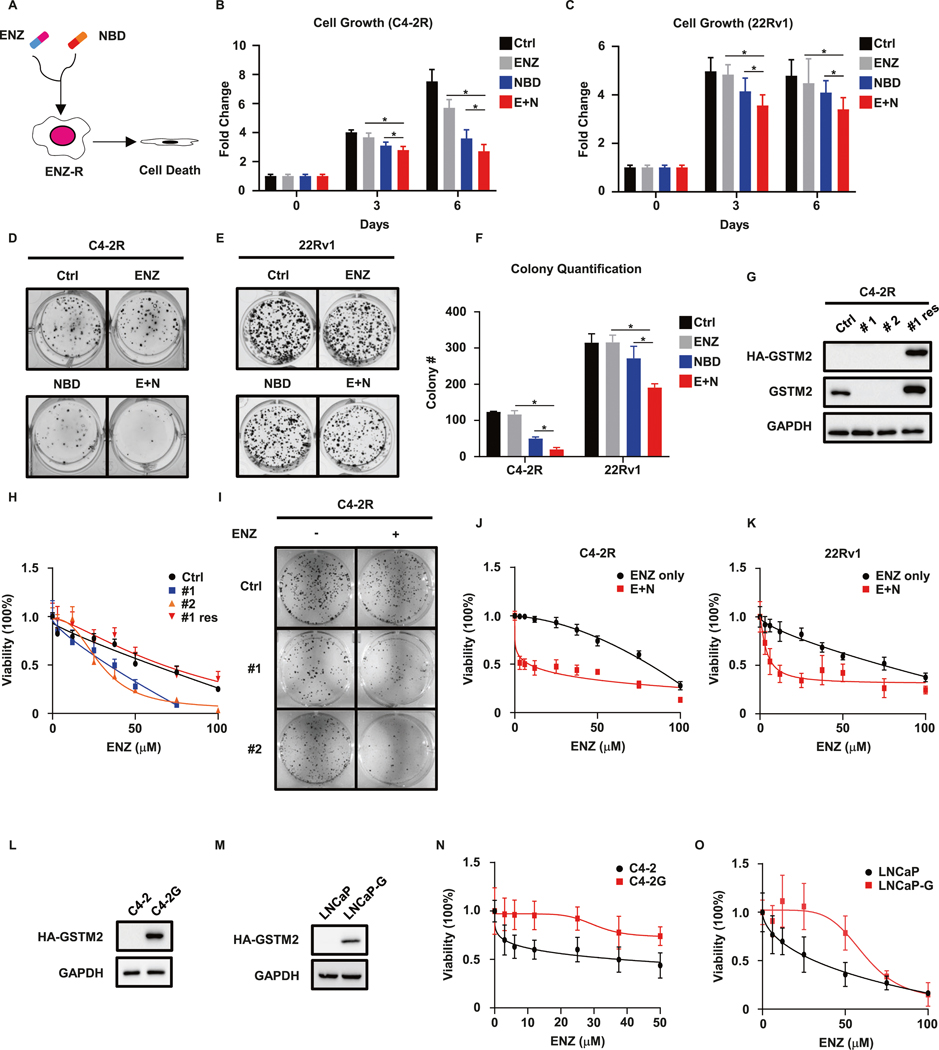
The elevated GSTM2 in ENZ-R C4–2R was predicted to be critical for the acquisition of resistance to ENZ, so we hypothesized that GSTM2 inhibitor would be a potential candidate to overcome resistance to ENZ. Although there are no commercially available inhibitors of GSTM2, the discovery of substitute was kind of serendipity when we searched literatures of other isoforms of glutathione S-transferases (GST), of which we were especially interested in GST Pi 1 (GSTP1) and its specific inhibitor NBDHEX (NBD) [22]. Although NBD was initially designed for GSTP1, it displays much higher potency toward GSTM2 (Fig. S2A, B). Since NBD also targeted GSTP1, and it has been reported that GST Mu 1 (GSTM1) shares high similarity with GSTM2 [23], we examined the expression levels of these GST isoforms in our cell lines to rule out the off-target effect of NBD and possible functional compensation. We also included 22Rv1 cell line as it is the well-known ENZ-R cells that was previously demonstrated to have negligible expression of GSTM1 and GSTP1 [24, 25]. Luckily, none of them expressed GSTM1 and GSTP1, and only GSTM2 could be detected in C4–2R and 22Rv1 (Fig. S2C). These results made us confident that we could test NBD in ENZ-R cells to see whether it could disrupt drug resistance (Fig. 2A). While ENZ per se had minimal effect on ENZ-R cells, pharmacological inhibition of GSTM2 by NBD sensitized them to ENZ treatment as indicated by proliferation assays (Fig. 2B, C). This result was further supported by the decreased clonogenic abilities when combining ENZ and NBD (Fig. 2D, F). To further validate this hypothesis, we depleted GSTM2 in C4–2R, then tested cells’ sensitivity to ENZ (Fig. 2G). As expected, knockdown of GSTM2 made cells vulnerable to ENZ treatment as indicated by decreased viability and clonogenic ability, which was reversed by add-back of GSTM2 (Fig. 2H, I). The results mentioned above indicated that combination of ENZ and NBD displayed synergy to inhibit ENZ-R PCa. To numerically measure this synergistic effect, we calculated the 50% combination index (CI50%) of ENZ and NBD using their IC50 values of NBD (Fig. S2D, E), as described elsewhere [26]. As predicted, the CI50% for ENZ in C4–2R and 22Rv1 were 0.575 and 0.607 (Fig. 2J, K and Tables 1 and 2), respectively, confirming synergism between NBD and ENZ. Next, we asked whether overexpression of GSTM2 would be sufficient to confer resistance to ENZ upon ENZ-S C4–2 and LNCaP cells. When GSTM2 was introduced into these cells (Fig. 2L, M), they exhibited resistant features as shown by viability assays (Fig. 2N, O). Taken all together, these results reiterated the indispensable role of GSTM2 in the acquisition of resistance to ENZ and validated its potential as a target to overcome this resistance.
Fig. 2. Targeting GSTM2 overcomes resistance to ENZ.
A Treatment strategy in ENZ-R cells that combines enzalutamide (ENZ) and the GSTM2 inhibitor NBDHEX (NBD). B, C Growth assays in ENZ-R C4–2R and 22Rv1 treated with DMSO (Ctrl), ENZ, NBD or both (E + N). The concentrations of the two drugs are 10 μM for ENZ and 200 nM for NBD. Data are normalized to day 0 and denoted as mean ± SD (n = 8). D, E Clonogenic assays of C4–2R and 22Rv1 cells treated with the indicated drugs for up to 14 days. The concentrations of the drugs are 5 μM for ENZ and 100 nM for NBD. F Quantification of D and E. Data are mean ± SD of three independent replicates. G IB to show the shRNA depletion of endogenous GSTM2 (#1 and #2) and restoration of exogenous HA-tagged GSTM2 in C4–2R (#1 res). H, I Viability assay and clonogenic assay of cells in G. Data of viability are scaled into percentage and normalized to the untreated group (ENZ = 0), then shown as mean ± SD (n = 8). J, K Viability assays in C4–2R and 22Rv1 treated with ENZ only or ENZ plus NBD (400 nM). Data are scaled into percentage and normalized to the untreated group, then shown as mean ± SD (n = 8). L, M Overexpression of GSTM2 in ENZ-S C4–2 (C4–2G) and LNCaP (LNCaP-G) cell lines. Viability assays of cells in N and O. Data are scaled into percentage and normalized to the untreated group shown as mean ± SD (n = 8). *p < 0.05.
Table 1.
Calculation of 50% combination index (CI50%) in C4–2R (ENZ + NBD).
| ENZ + NBD (C4–2R) | |||
|---|---|---|---|
| IC50 | *IC50 | *IC50/IC50 | |
| ENZ | 80 μM | 6 μM | 0.075 |
| NBD | 0.8 μM | / | / |
| CI50%(ENZ) = 0.075 + 0.500 = 0.575 < 1 | |||
IC50 original IC50 of ENZ and NBD,
IC50 conditioned IC50 of ENZ when half IC50 of NBD was set as the background.
Table 2.
Calculation of 50% combination index (CI50%) in 22Rv1 (ENZ + NBD).
| ENZ + NBD (22Rv1) | |||
|---|---|---|---|
| IC50 | *IC50 | *IC50/IC50 | |
| ENZ | 75 μM | 8 μM | 0.107 |
| NBD | 0.8 μM | / | / |
| CI50%(ENZ) = 0.107 + 0.500 = 0.607 < 1 | |||
IC50 original IC50 of ENZ and NBD,
IC50 conditioned IC50 of ENZ when half IC50 of NBD was set as the background.
AhR-GSTM2 axis is involved in resistance to ENZ
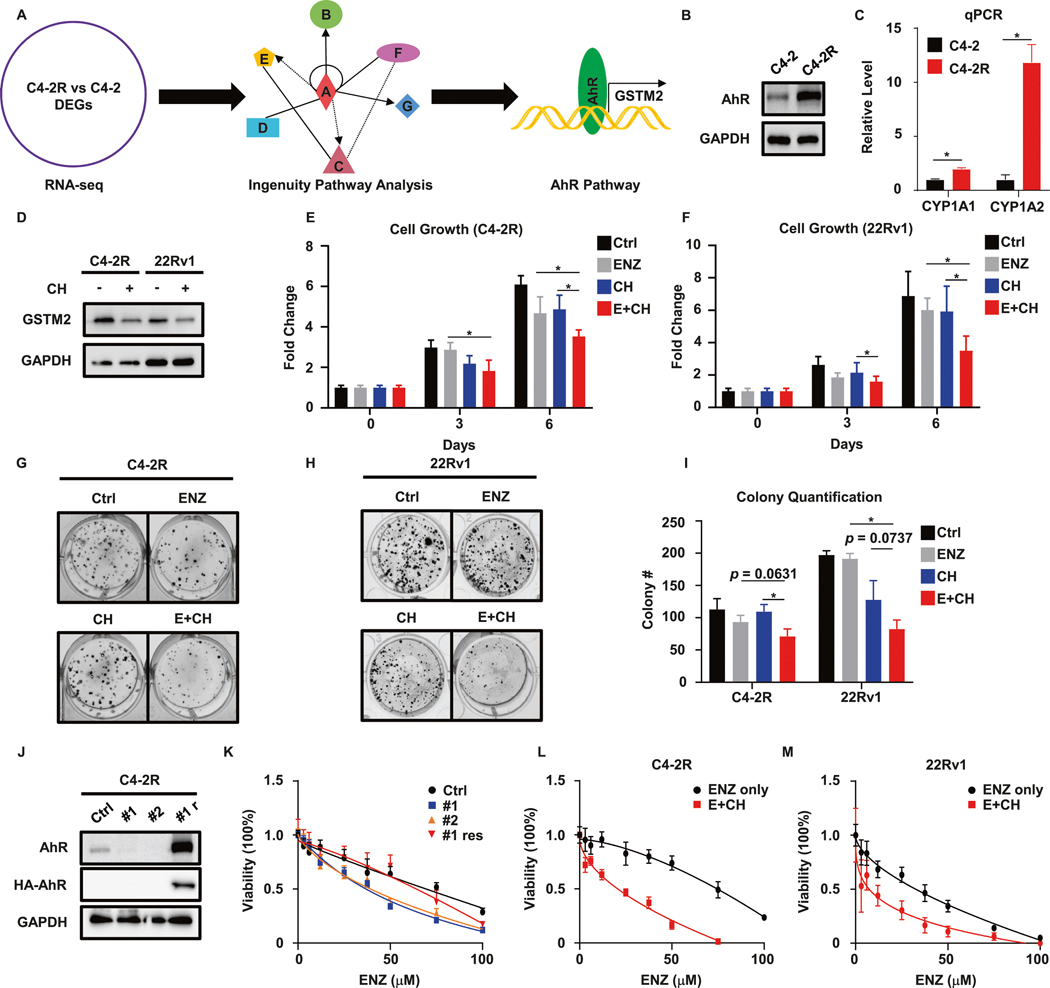
To dissect how GSTM2 was regulated, we ran the web-based Ingenuity Pathway Analysis (IPA) based on the DEGs of RNA-seq. The IPA unraveled that the aryl hydrocarbon receptor (AhR) pathway was activated in C4–2R (Fig. 3A). Of note, AhR is a well-known master transcription factor for GSTM2 [27]. Based on this preliminary analysis, we suspected that the AhR pathway would be involved in GSTM2-incurred resistance to ENZ. To test this conjecture, we first compared the expression of AhR between ENZ-R and ENZ-S cells and found that the protein level of AhR was indeed elevated in C4–2R (Fig. 3B). Further detection of CYP1A1 and CYP1A2, which are two canonical downstream targets of AhR [28], and luciferase reporter assay assured the active status of this pathway in C4–2R and 22Rv1 (Figs. 3C and S2F, 22Rv1 data not shown). To authenticate this upstream regulation, we treated C4–2R and 22Rv1 with the AhR-specific inhibitor CH-223191 (CH) and observed a decreased level of GSTM2 (Fig. 3D), providing direct evidence of this regulatory axis. Having established the involvement of AhR, we designed similar experiments to ask whether targeting AhR could recapitulate the results acquired by GSTM2 inhibitor. In agreement with NBD treatment, addition of the AhR inhibitor remarkably augmented the effect of ENZ on resistant cells (Fig. 3E–I). In addition, we depleted endogenous AhR in C4–2R, followed by restoration of its expression to see whether modulation of the AhR level affected the cells’ sensitivity to ENZ. Consistently, abolishment of AhR sensitized cells to ENZ, and this was reverted by re-expression of AhR (Fig. 3J, K). Again, we calculated CI50% after we acquired the IC50 of CH (Fig. S2G–I). Since the 3-day IC50 of CH in 22Rv1 was too high and hard to utilize (Fig. S2H), we calculated 3-day CI50% and 6-day CI50% in C4–2R and 22Rv1, respectively. The results showed that combination of ENZ and CH synergistically inhibited cell growth (Fig. 3L, M and Tables 3 and 4). In short, these results established the regulation of GSTM2 by AhR and their contributions to resistance to ENZ.
Fig. 3. AhR is associated with ENZ-R phenotype caused by GSTM2.
A Illustration of Ingenuity Pathway Analysis (IPA), which identifies the AhR pathway that regulates GSTM2. B, C IB and qPCR to confirm that AhR signaling is active in ENZ-R cells. qPCR results are normalized to GAPDH, then normalized to C4–2 and shown as mean ± SD (n = 6). D IB to detect GSTM2 after treated with 20 μM AhR inhibitor CH-223191 (CH) for 48 h. E, F Growth assays in C4–2R and 22Rv1 treated with DMSO (Ctrl), ENZ, CH or both (E + CH). The concentrations of the two drugs are both 10 μM. Data are normalized to day 0 and shown as mean ± SD (n = 8). G, H Clonogenic assay of C4–2R and 22Rv1 with the indicated treatments for up to 14 days. The concentrations of the two drugs are both 5 μM. I Quantification of G and H. Data are mean ± SD of three independent replicates. J IB to detect the efficacy of AhR knockdown by shRNA (#1 and #2) and restoration of exogenous HA-tagged AhR in C4–2R (#1 res). K Viability assay of cells in J. Results are scaled into percentage and shown as mean ± SD (n = 8). L, M Viability assays in C4–2R and 22Rv1 treated with ENZ only or ENZ plus CH. Due to the high 3-day IC50 of CH in 22Rv1, the assay for 22Rv1 is cultured up to 6 days. The concentrations of CH are 50 μM for C4–2R and 5 μM for 22 Rv1. Data are scaled into percentage and shown as mean ± SD (n = 8). *p < 0.05.
Table 3.
Calculation of 50% combination index (CI50%) in C4–2R (ENZ + CH).
| ENZ + CH (C4–2R) | |||
|---|---|---|---|
| IC50 | *IC50 | *IC50/IC50 | |
| ENZ | 80 μM | 20 μM | 0.250 |
| CH | 100 μM | / | / |
| CI50%(ENZ) = 0.250 + 0.500 = 0.750 < 1 | |||
IC50 original IC50 of ENZ and CH,
IC50 conditioned IC50 of ENZ when half IC50 of CH was set as the background.
Table 4.
Calculation of 50% combination index (CI50%) in 22Rv1 (ENZ + CH).
| ENZ + CH (22Rv1) | |||
|---|---|---|---|
| IC50 | *IC50 | *IC50/IC50 | |
| ENZ | 33 μM | 10 μM | 0.303 |
| CH | 10 μM | / | / |
| CI50%(ENZ) = 0.303 + 0.500 = 0.803 < 1 | |||
IC50 original IC50 of ENZ and CH,
IC50 conditioned IC50 of ENZ when half IC50 of CH was set as the background.
ENZ-R cells are immune to treatment-induced OS-associate damage and cell death
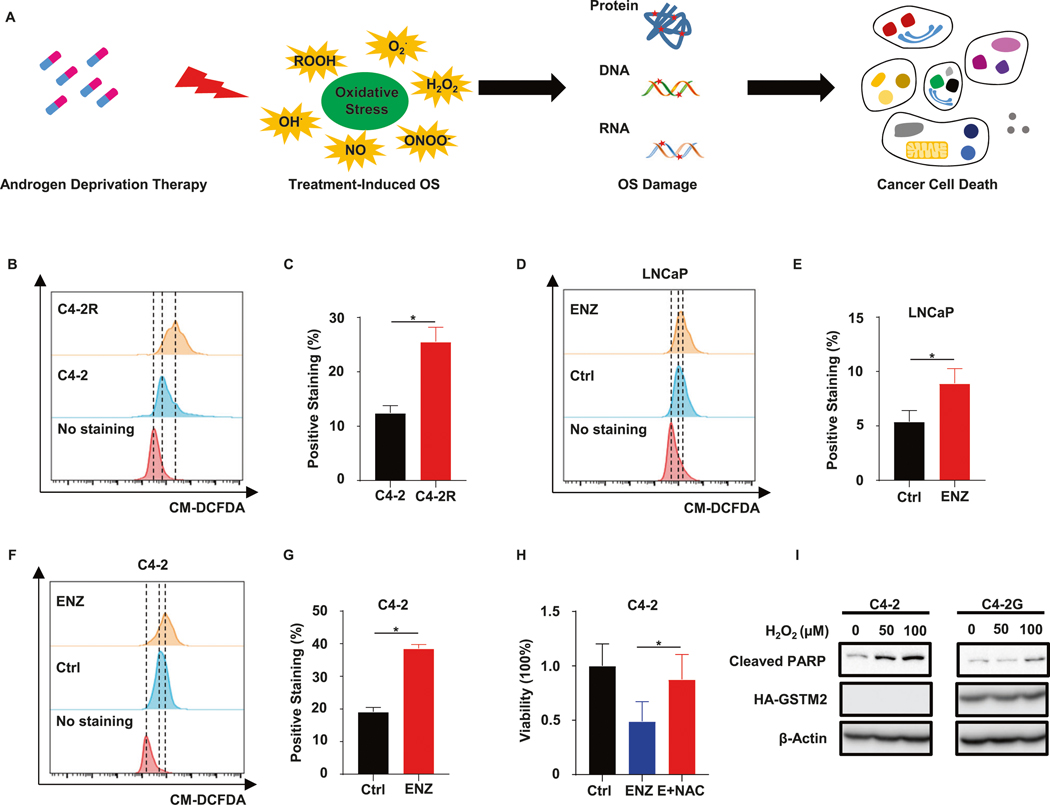
Having confirmed the liaison between GSTM2 and resistance to ENZ, we investigated the mechanism underlying this finding. Of note, androgen deprivation therapy (ADT) has been known to induce OS [29]. As described before, the dichotomous consequence of OS could be skewed to the dark side to suppress cancer progression (Fig. 4A). Because one of the major functions of GSTM2 is to antagonize OS, we performed a series of experiments to inspect whether this function was essential for resistance to ENZ. First, we performed flow cytometry analyses to measure intracellular ROS level in C4–2 and C4–2R. Intriguingly, ENZ-R C4–2R exhibited a much higher level of intracellular ROS in comparison to C4–2 cells (Fig. 4B, C), which was accompanied by the down-regulation of general antioxidant genes as shown by the plot of Gene Set Enrichment Analysis (GSEA) of RNA-seq (Fig. S3A). Second, we treated LNCaP and C4–2 with ENZ, and corroborated that ENZ did indeed induce intracellular ROS and worsened the condition of OS (Fig. 4D–G), which might be partially due to the decrease of general ROS scavengers such as SOD1 (Fig. S3B). Third, we treated C4–2 with ENZ or ENZ plus the general antioxidant n-acetylcysteine (NAC). Although NAC did not have an observable effect on cell viability, addition of it to ENZ treatment dramatically rescued cells from the cytotoxic effect of ENZ (Figs. S3C and 4H). Since ENZ enhanced the intracellular ROS level and rendered cells susceptible to OS-associate damage, it was astonishing that C4–2R, which already tolerated severe OS, still survived well upon ENZ treatment. This unconventional event implicated that ENZ-R cells had developed a remedy to crush this harmful condition. Taking account of the phenomena described above, we assumed that ENZ-R cells protected themselves from OS-associated damage through up-regulation of GSTM2, which reversed the cell fate that initially doomed to death. To justify this assumption, we overexpressed GSTM2 in C4–2, then challenged cells with hydrogen peroxide (H2O2). Upon overexpression of GSTM2, cells became recalcitrant to H2O2-induced apoptosis, as indicated by decrease of apoptotic marker cleaved PARP (Fig. 4I). In sum, these data convinced us that ENZ-R cells escaped treatment-induced OS-associated damage and cell death by elevation of GSTM2.
Fig. 4. ENZ-R cells rely on endogenous remedy to survive OS-associated damage incurred by treatment.
A Iconograph of oxidative stress (OS), which is induced by androgen deprivation therapy, and its impact on cell fate. B Flow cytometry analyses of intracellular reactive oxygen species (ROS) in C4–2 and C4–2R. C Quantification of intracellular ROS signal in B. Data are mean ± SD (n = 3). D, F Flow cytometry analyses of intracellular ROS in LNCaP and C4–2 upon treated with DMSO or 10 μM ENZ for 24 h. E, G Quantification of intracellular ROS levels in D and F. Data are mean ± SD (n = 3). H Viability assay in C4–2 treated with DMSO, ENZ or ENZ plus antioxidant N-acetylcysteine (E + NAC) for 6 days. The concentrations of chemicals are: 10 μM for ENZ and 5 mM for NAC. Data are normalized to Ctrl and scaled into percentage, then shown as mean ± SD (n = 6). I IB to show apoptosis in C4–2 and C4–2G treated with the indicated concentrations of hydrogen peroxide (H2O2) for 24 h. *p < 0.05.
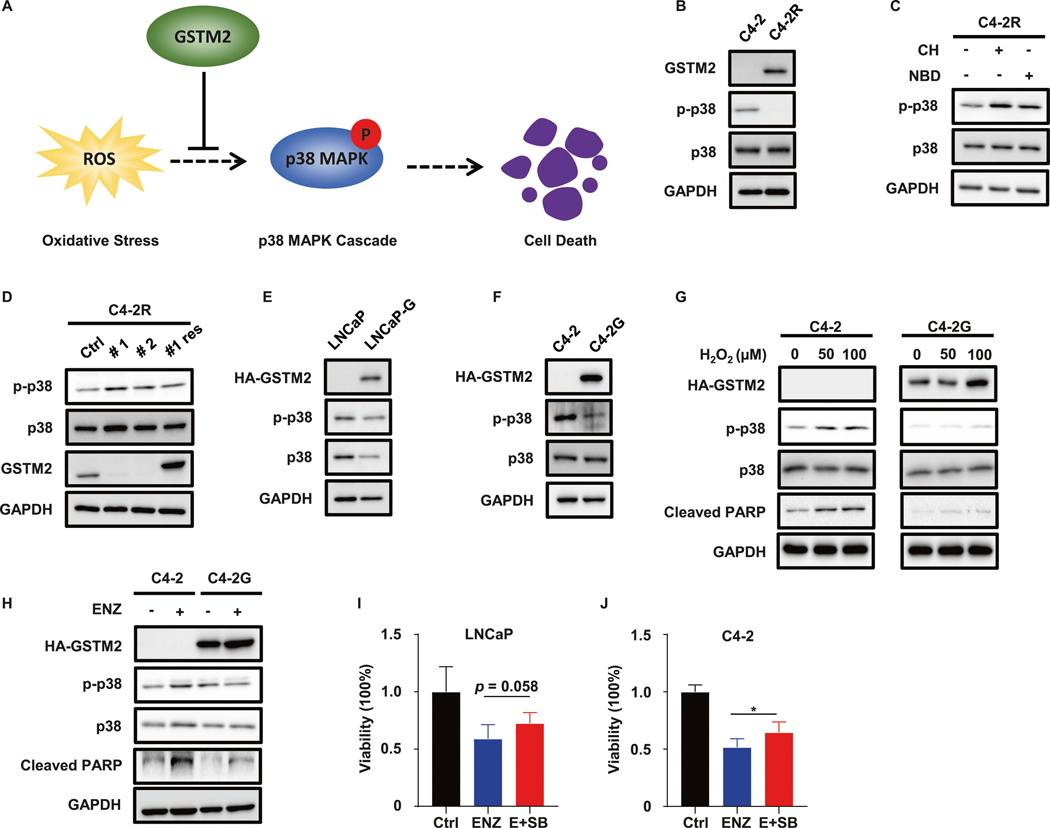
GSTM2 safeguards cells by counteracting the p38 MAPK cascade
Next, we aimed to further shed light on the precise mechanism of how GSTM2 exerted its anti-OS function during acquisition of resistance to ENZ. Although GSTM2 is not a direct ROS scavenger, it has been proven that GSTM2 negates activation of p38 MAPK pathway [30]. Without GSTM2, p38 MAPK initiates cell death pathways whenever cells cannot tolerate outrageous ROS (Fig. 5A) [31]. We speculated that this salvage system would be responsible for the anti-OS function of GSTM2. Preliminary examination of genes in the p38 MAPK cascade suggested the disparate status of this pathway between C4–2R and C4–2, albeit it did not recognize which cell line had robust activation (Fig. S3D). Considering the contrasting level of GSTM2, it was very likely that the salvage system functioned in ENZ-R C4–2R to silence activation of p38 MAPK. To test our speculation, we compared phospho-p38 MAPK (p-p38) in C4–2 and C4–2R. Not surprised, the level of p-p38 was lower in C4–2R which also contained a higher level of GSTM2 (Fig. 5B). Treatment with either CH or NBD raised p-p38 epitope, indicating the functional regulatory axis in C4–2R (Fig. 5C). In addition, depletion of GSTM2 in C4–2R provoked an increase of the p-p38 epitope, which was abrogated by replenishment of GSTM2 (Fig. 5D). Moreover, once GSTM2 was introduced into LNCaP and C4–2, the p-p38 signals decreased in response to the expression of GSTM2 (Fig. 5E, F). These results were congruent with the established model, so we were confident that this salvage system operated in ENZ-R cells. To further substantiate this model, we overexpressed GSTM2 in C4–2 and exposed cells to H2O2. Although H2O2 activated p38 MAPK pathway and triggered cell death as marked by the uptrend of p-p38 and Cleaved PARP, overexpression of GSTM2 rendered the cells insensitive to this effect (Fig. 5G). Likewise, ENZ treatment also induced the p-p38 epitope and cell death, which were saved by GSTM2 overexpression (Fig. 5H). Indeed, despite the fact that the p38 MAPK inhibitor SB203580 (SB) did not affect cell viability by itself (Fig. S3E), the effect of ENZ was partially offset by the addition of SB as seen in LNCaP and C4–2 (Fig. 5I, J). These results strongly argued for the concept that ENZ-induced OS and activation of p38 MAPK pathway could be alleviated by GSTM2, which served as a salvage system to safeguard ENZ-R cells.
Fig. 5. GSTM2 rescues cell fate by inhibiting the p38 MAPK pathway.
A Simplified model of how GSTM2 rescues cell death by negatively regulating the p38 MAPK pathway. B IB to compare the GSTM2 and phospho-p38 MAPK (p-p38) level between C4–2 and C4–2R. C IB to detect p-p38 level in C4–2R treated with either GSTM2 inhibitor or AhR inhibitor. The concentrations of chemicals are 20 μM for CH and 400 nM for NBD. D IB to detect p-p38 level after shRNA depletion of endogenous GSTM2 (#1 and #2) and restoration of exogenous HA-tagged GSTM2 in C4–2R (#1 res). E, F IB to detect the p-p38 level in LNCaP, C4–2 and their derivative cells with overexpression of GSTM2. G IB to detect p-p38 level in C4–2 and C4–2G challenged with the indicated concentrations of H2O2 for 12 h. H IB to detect p-p38 level in C4–2 and C4–2G treated with 20 μM ENZ for 48 h. I, J Viability assays in LNCaP and C4–2 treated with DMSO, ENZ or ENZ plus p38 inhibitor SB203580 (E + SB) for 3 days. The concentrations of chemicals are: 10 μM for ENZ and 2 μM for SB. Data are normalized to Ctrl and scaled into percentage, then shown as mean ± SD (n = 6 for I and n = 8 for J). *p < 0.05.
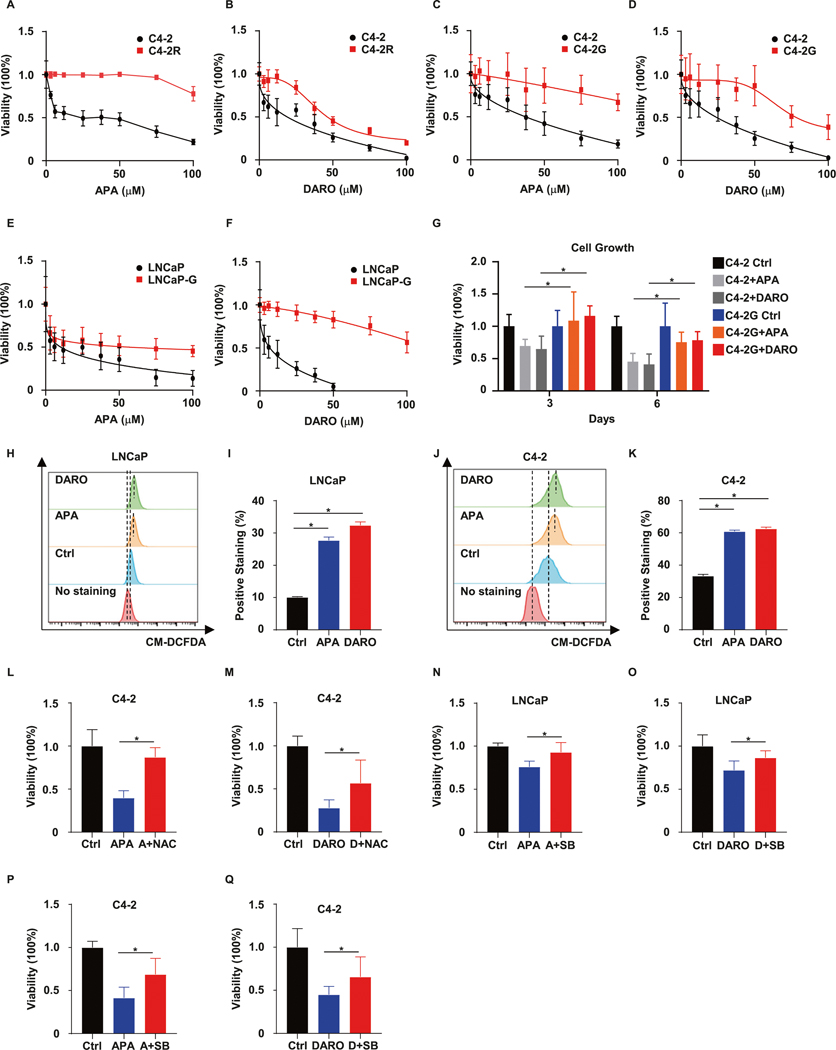
GSTM2 determines cross-resistance to SG-ARIs
Since ADT-induced OS is a universal consequence of this treatment regime, we extrapolated our explanation of resistance to ENZ to APA and DARO, which belong to the same class of SG-ARIs. We proposed that elevated GSTM2 would also herald cross-resistance to APA and DARO. To evaluate this corollary, we first compared the sensitivity of C4–2 and C4–2R to APA and DARO. The results specified that ENZ-R C4–2R was cross-resistant to APA and DARO (Fig. 6A, B). To verify that this was attributable to GSTM2, we overexpressed GSTM2 in LNCaP and C4–2, followed by examining their responsiveness to APA and DARO treatment. As anticipated, overexpression of GSTM2 rendered these cells less responsive to APA and DARO (Fig. 6C–F). In addition, C4–2 cells with an elevated level of GSTM2 persisted even after long-term exposure to treatment (Fig. 6G). These data revealed that GSTM2 prompted cross-resistance to APA and DARO. To inquire whether GSTM2 led to cross-resistance in a similar way as for ENZ, we exposed LNCaP and C4–2 to APA and DARO and then examined the ROS levels. In accordance with the results acquired from ENZ, application of APA and DARO slightly abolished general ROS scavengers and boosted ROS generation (Figs. 6H–K and S3F), suggesting that cross-resistance to SG-ARIs entailed a conserved tactic. Furthermore, the repressive effect of APA and DARO could be substantially eliminated by NAC treatment in C4–2 (Fig. 6L, M). Finally, the p38 MAPK inhibitor SB discontinued the inhibition effect of APA and DARO in these two cell lines as it did for ENZ treatment (Fig. 6N–Q). In brief, these results provided strong evidence for our extrapolation that GSTM2 and a related anti-OS salvage system were prerequisites for cross-resistance to SG-ARIs.
Fig. 6. GSTM2 and cross-resistance to SG-ARIs.
A, B Viability assays in C4–2 and C4–2R treated with the indicated concentrations of the two second-generation androgen receptor inhibitors (SG-ARIs), apalutamide (APA) and darolutamide (DARO). Viability are normalized to the untreated group (APA/DARO = 0) and scaled into percentage, then shown as mean ± SD (n = 8). Viability assays in C4–2 (C, D) and LNCaP (E, F) treated with the indicated concentrations of APA and DARO, in the context of GSTM2 overexpression or not. Data are scaled into percentage and normalized to the untreated group, then expressed as mean ± SD (n = 8). G Growth assays in C4–2 and C4–2G treated with DMSO, 10 μM APA or DARO for 6 days. Data are scaled into percentage and normalized to Ctrl of day 3 and day 6, then shown as mean ± SD (n = 8). H, J Flow cytometry analyses of intracellular ROS in LNCaP and C4–2 upon treated with DMSO, 10 μM APA or DARO for 24 h. I, K Quantification of intracellular ROS levels in H and J. Data are mean ± SD (n = 3) of one representative. L, M Viability assays in C4–2 treated with DMSO, APA, APA plus NAC (A + NAC), DARO or DARO plus NAC (D + NAC) for 3 days. The concentrations of the chemicals are: 10 μM for APA and DARO; 5 mM for NAC. Results are normalized to Ctrl and scaled into percentage, then shown as mean ± SD (n = 8). Viability assays in LNCaP (N, O) and C4–2 (P, Q) treated with DMSO, APA, APA plus SB (A + SB), DARO or DARO plus SB (D + SB) for 3 days. The concentrations of chemicals are: 10 μM for APA and DARO; 2 μM for SB. Results are normalized to Ctrl and scaled into percentage, then shown as mean ± SD (n = 8). *p < 0.05.
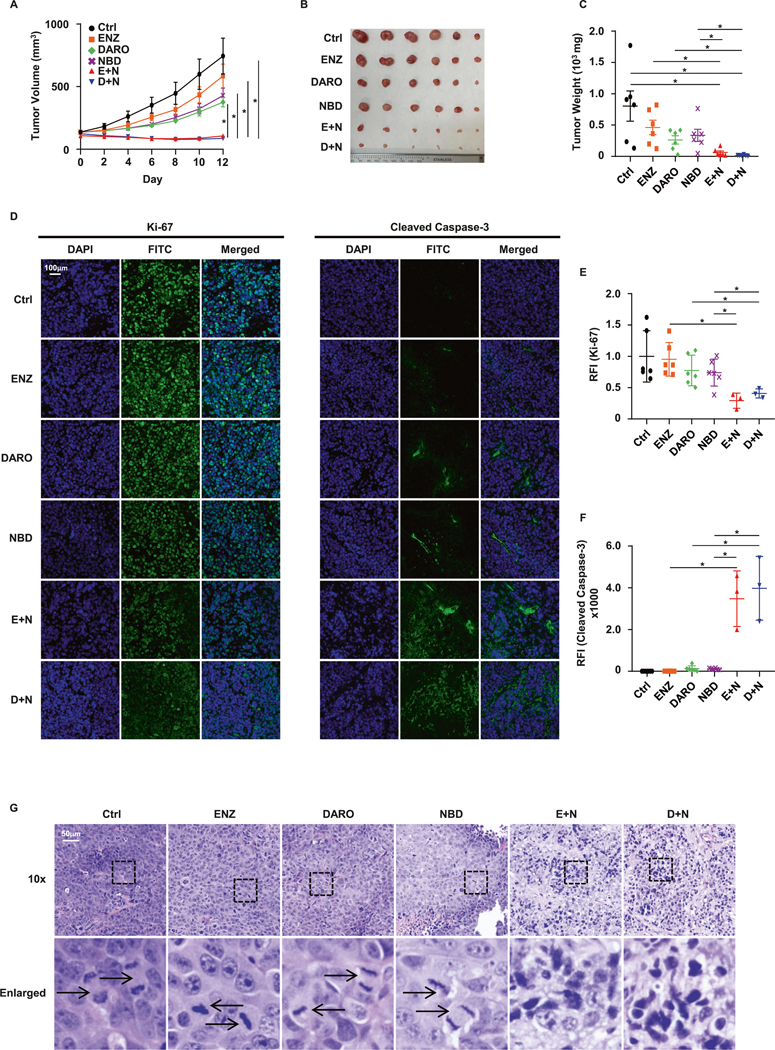
Targeting GSTM2 enhances the efficacy of SG-ARIs in 22Rv1 xenograft
To validate the crucial function of GSTM2 in resistance to SG-ARIs, we performed an in vivo xenograft experiment utilizing the 22Rv1 cell line because of its strong ability to form tumors and verified nature of resistance to all three SG-ARIs [32]. Whereas APA is an analog of ENZ and shares some similarities [33], DARO was developed independently with distinct features from ENZ and APA [34]. Considering these facts, we designed two separate drug combinations by administering ENZ plus NBD, or DARO plus NBD in immunodeficient mice harboring 22Rv1 tumors. In keeping with the in vitro results, even though monotherapy showed little ability to suppress tumor growth, combination therapies effectively decelerated this process and resulted in lower tumor weights (Fig. 7A–C). In addition, the body weight of the mice over treatment period, as well as the final body weight, showed no difference among all treatment groups, suggesting that the treatment regimen had minor toxicity and was well tolerated in vivo (Fig. S4A, B). Detection of GSTM2 in protein extracts of tumors verified the expression of GSTM2 in all treatment groups, confirming the specificity of NBD in vivo to target GSTM2 (Fig. S4C). To better visualize the effect of combination therapies, we stained the tumor slides with Cleaved Caspase-3 and Ki-67 antibodies, which are apoptotic and proliferative indicators, respectively. In line with our expectations, the combination therapies obviously inhibited cell growth and stimulated apoptosis (Fig. 7D–F). We also stained tumor slides with hematoxylin and eosin (Fig. 7G). The images showed that tumors from the monotherapy mice displayed stereotypical architecture and cell shapes, highlighted by a considerable number of mitotic figures. In contrast, this hallmark was not observed in tumors from the combination treatment mice, which instead contained large areas of apoptotic cells marked by condensed nuclei. Therefore, these results illustrated that targeting GSTM2 was an effective strategy to overcome resistance to SG-ARIs in vivo.
Fig. 7. In vivo 22Rv1 xenograft experiment.
A Tumor growth of 22Rv1 xenograft in nude mice over 2 weeks. Results are shown as mean ± SEM (n = 6). B Picture of tumors upon harvest. C Final weight of tumors. Results are shown as mean ± SD (n = 6). D Immunofluorescent staining of tumor nuclei (DAPI) and Ki-67 & Cleaved Caspase-3 (FITC). Images are exemplary results of each group. E, F Quantification of Ki-67 and Cleaved Caspase-3 signals in D. Results are normalized to Ctrl and shown as mean ± SD of 6 mice per group, except for E + N and D + N groups, which only have three mice undergoing paraffin sectioning. RFI relative fluorescent intensity. G H&E staining of tumor slides. Representative images of each group are shown. Arrows indicate mitotic cells. *p < 0.05.
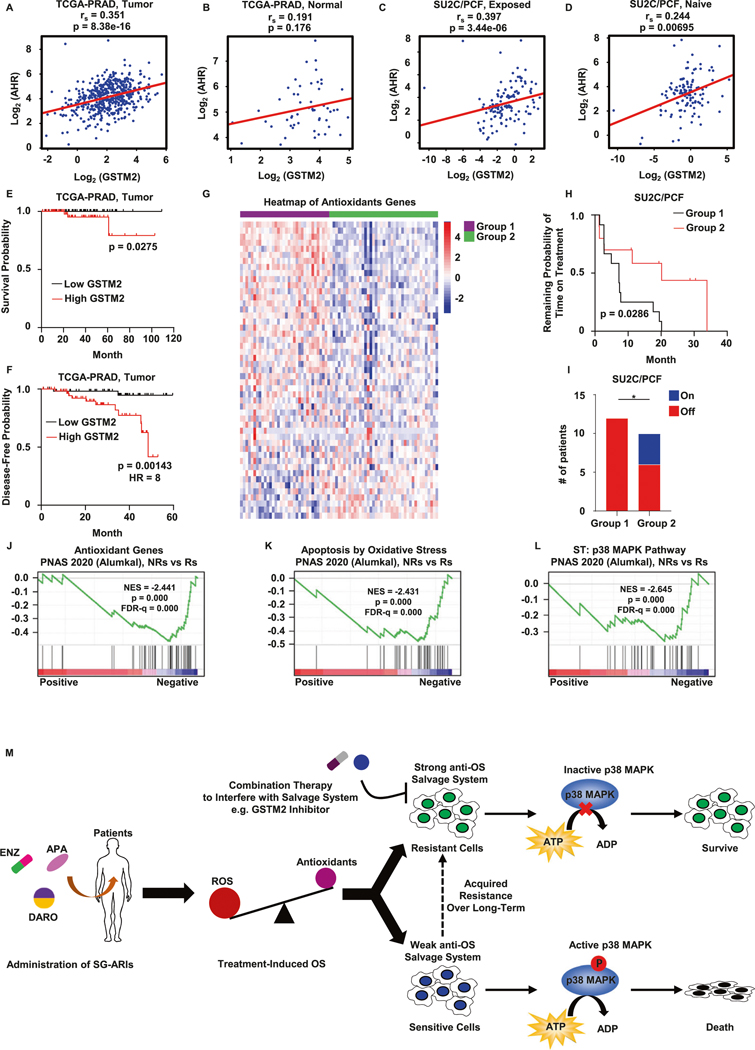
Clinical evidence of GSTM2 and anti-OS salvage system in resistance to SG-ARIs
To uncover potential translational value of our findings, we performed several analyses based on patients’ data. First, we checked the public TCGA database of primary PCa [35], and found that the level of GSTM2 was positively correlated with AhR and its downstream target CYP1B1 in PCa samples (Figs. 8A and S4D). In contrast, paired normal tissues did not have such a relationship (Figs. 8B and S4E). This phenomenon was double confirmed by another available PCa dataset (Fig. S4F–I) [36]. Furthermore, this correlation persisted in late stage PCa patients medicated with SG-ARIs (Figs. 8C and S4J), but was less prominent in patients who were naïve to SG-ARIs (Figs. 8D and S4K), as denoted by data of metastatic CRPC samples collected from recent genomic landscape research [37]. In brief, these results supported the positive regulation of GSTM2 by AhR in clinical scenarios of resistance to SG-ARIs, as indicated in our cell studies. Second, we separated patients from TCGA into two groups based on median level of GSTM2. Surprisingly, although the 10-year survival rate is close to 99% [38], a high level of GSTM2 still was associated with early death of PCa (Fig. 8E). Also, elevated GSTM2 forecasted early recurrence of PCa (Fig. 8F). In addition, we analyzed 75 patients with longitudinal data of SG-ARIs treatment from the aforementioned CRPC collection [37]. Of note, samples of these patients were sequenced before SG-ARIs treatment and thus could be used to predict the efficacy of treatment. To better visualize individual differences, we sorted these patients with a predefined antioxidant gene signature [39]. Clearly, these samples could be categorized into two groups. Patients in Group 1 displayed abundant expression of anti-OS components and a potentially strong salvage system, whereas those in Group 2 retained only scarce anti-OS modules and a potentially weak salvage system (Fig. 8G). Although there was no obvious difference among all patients (Fig. S4L), the subcohort of patients treated with ENZ in Group 2 exhibited a longer duration of treatment (Fig. 8H). Furthermore, all patients in Group 1 eventually quit by the end of the effective monitoring period, whereas a notable number of patients in Group 2 continued the current treatment (Fig. 8I). We did not observe any distinction in the overall survival of these patients, suggesting that other factors might interfere with the survival of patients after they shifted to other treatment plans (Fig. S4M). Finally, we collected patients’ data from another shared dataset and performed GSEA [40]. Among those patients, 7 were confirmed non-responders (NRs) to ENZ and 18 were responders (Rs). It was noteworthy that samples of these patients were sequenced only after exposure to ENZ, thus were useful to evaluate the consequence of treatment. We noticed that the levels of antioxidant genes generally diminished in the NRs group, indicating that these patients bore higher OS and were technically predisposed to OS-induced p38 activation and cell death (Fig. 8J). Unfortunately, this was not true in the NRs group, as the analyses proved that OS-induced cell death and the p38 MAPK pathway were repressed in this group (Figs. 8K, L and S4N, O). Of note, we observed a trend of AhR activation in the NRs group, although it was not statistically significant (Fig. S4P). Taken together, these results served as important complements and provided some translational value to our study.
Fig. 8. Clinical evidence of GSTM2 and the salvage system of OS damage in the acquisition of resistance to SG-ARIs.
Spearman correlations of AhR and GSTM2 in prostate adenocarcinoma samples (A, n = 495) and paired normal prostate tissues (B, n = 52) from TCGA-PRAD database (https://www.cancer.gov/tcga). rs, Spearman’s rank correlation coefficient. Spearman correlations of AhR and GSTM2 in castration-resistant prostate cancer (CRPC) samples that are exposed (C, n = 128) or naïve (D, n = 121) to second-generation ARIs from SU2C/PCF Dream Team database (Abida et al. [37], PMID: 31061129). E, F Kaplan–Meier curves of survival and 5-year disease-free probability of PCa patients from TCGA-PRAD subcohort iCluster 3. Patients are separated into low GSTM2 (n = 69) and high GSTM2 (n = 69) based on median. HR hazard radio. G Heatmap to show the expression of antioxidant genes of all patients from SU2C/PCF before treated with SG-ARIs (n = 75, 50 abiraterone, 22 ENZ and 3 others). Patients are separated into two groups by K-Means algorithm. Gene set is from MSigDB-GSEA (http://www.gsea-msigdb.org/gsea/index.jsp, M5938). H Kaplan–Meier curve of remaining probability of time on treatment of patients that are medicated with ENZ (n = 22) in G. I Histogram of patients’ outcome in H. Patients that continue ENZ treatment at the observation point are counted as On, whereas those who stop treatment are counted as Off. J–L Pathways by Gene Set Enrichment Analysis (GSEA) of 7 non-responders (NRs) and 18 responders (Rs) treated with ENZ from PNAS 2020 (Alumkal et al., PMID: 32424106). NES normalized enrichment score. All gene sets are from MSigDB-GSEA, and individual systematic name is: M5938 (J), M22556 (K) and M12012 (L). M Putative working model of resistance to SG-ARIs as interpreted by this study. *p < 0.05.
DISCUSSION
The data conveyed above support a divergent anti-OS route in response to SG-ARIs treatment (Fig. 8M). Theoretically, treatment-induced OS jeopardizes cells by activation of p38 MAPK and subsequent cell death as observed in sensitive cells. However, resistant cells have adopted a salvage system to brake the p38 MAPK cascade and evade catastrophe. Counteracting this system in resistant cells through a combination of drugs is a promising strategy to overcome resistance to SG-ARIs, and chemicals such as GSTM2 inhibitors are good candidates to be leveraged. Gradual transition of sensitive cells to their resistant counterparts is the established fact, however the precise mechanism of this process is still elusive, more likely involving extensive epigenetic reprogramming of the whole genomic landscape as reported previously [41–44]. In that case, combining RNA-seq with other sequencing approaches that detect epigenetic changes will be the standard way to conduct similar researches in the field. In fact, despite the fact that GSTM2 is regulated by AhR in resistant cells, overexpression of AhR in sensitive cells does not elevate GSTM2 as expected (data not shown). The failure of GSTM2 induction implies the distinct chromatin accessibility at GSTM2 loci between resistant and sensitive cells, which cannot be achieved by simple introduction of its regulator. Further studies will need to address this discrepancy and identify important epigenetic modulators underlying the elevation of GSTM2 in resistant cells.
As a potential hallmark of cancer, modulation of OS and ROS has long been discussed as a therapeutic strategy [10]. Since the ROS is the major driver of OS, understanding the source and generation of ROS is critical for managing OS-related events. So far, the major source of ROS is considered to be mitochondria, where ROS is the by-product of oxidative phosphorylation. Thus, enhanced mitochondrial respiration activity accompanies the increase of ROS production. Interestingly, we do observe enhanced oxidative phosphorylation capacity in ENZ-R C4–2R (data not shown), which may account for the higher level of ROS in C4–2R. Moreover, extramitochondrial sources of ROS cannot be omitted, and whether they contribute to ROS production needs to be further explored. We also show that SG-ARIs treatment decreases expression of several ROS scavengers, which is partially responsible for the treatment-induced OS. However, other concurrent events may happen to exacerbate the process. The major metabolites of APA and ENZ are their demethylated forms [45, 46], but it has been reported that they can also react with glutathione in mouse and human hepatocytes [47]. Although it has not yet been determined whether this reaction happens in our PCa cells, it is worthwhile to investigate this possibility, as one of the major reasons for OS is glutathione depletion. The consequence of these events is to cause imbalance of ROS production and elimination, which can be taken advantage of for cancer treatment.
Since the discovery of SG-ARIs, the drug resistance issue has been investigated for a long time. The recent genomic study of drug-exposed patients illuminates a novel direction and renders important information about resistance to SG-ARIs. Regardless of significant progress, we are still short of clinical samples from patients treated with SG-ARIs, as the majority of patients are not treated with any of the SG-ARIs mentioned in our study. For example, most patients are medicated with abiraterone, which belongs to another class of SG-ARIs to inhibit de novo synthesis of androgen. However, our results indicate that response to abiraterone is unique and does not share the same mechanism, therefore the anti-OS system has very limited value to predict its efficacy (Fig. S4K). This observation is consistent with a previous report about resistance to SG-ARIs [44]. Also, another question that needs to be addressed is why a high level of GSTM2 coincides with early recurrence and death only in patients categorized into iCluster 3. Patients in this category may have distinct genetic changes that facilitate disease progression together with GSTM2. Hence, future studies need to address these issues and provide more detailed genomic blueprints, especially in patients treated with the three SG-ARIs tested in our study. Nonetheless, the conclusion is that targeting inherent anti-OS defense is a feasible approach to overcome resistance to SG-ARIs.
EXPERIMENTAL PROCEDURES
Cell culture
All cells were cultured at 37 °C in RPMI 1640 medium with 10% FBS and 5% penicillin-streptomycin in 5% CO2. LNCaP and 22Rv1 cell lines were purchased from ATCC. ENZ-R C4–2R and paired C4–2 were kindly provided by Dr. Allen Gao [48]. LNCaP and 22Rv1 were authenticated before experiments. All cell lines were tested negative for mycoplasma contamination. The concentration of ENZ to maintain C4–2R was 20 μM. Before any experiment, C4–2R cells were cultured in medium without ENZ for 48 h to remove any residual effect of ENZ.
Cell proliferation and viability assay
In total, 3 × 103 cells were seeded per well onto 96-well plates in medium without phenol red and incubated for overnight. On the next day (day 0) and day 3, the medium was refreshed (no phenol red) with the indicated concentration of chemicals. AquaBluer (MultiTarget Pharmaceuticals LLC, 6015) reagent was added to the medium, then incubated for 4 h. The fluorescent intensity at 540ex/590em was read by GloMax Discover plate reader (Promega). All experiments were repeated three times and one was shown.
Clonogenic assay
In total, 1 × 103 cells were seeded onto plates and cultured overnight. On the next day, cells were treated with the indicated concentration of chemicals for up to 14 days. Cells were fixed with 10% formalin for 10 min and washed once with clean PBS, then 0.5% crystal violet was applied to stain colonies for 30 min, followed by washing with tap water until the wells were clear. After air drying, images were taken with ChemiDoc Imaging System (Bio-Rad). All experiments were repeated three times and one was shown.
RNAi and transfection
For RNAi-based gene depletion, cells were infected with the pLKO.1-puro empty vector or vectors containing shRNAs. In total, 2 ug/ml puromycin was used to select infected cells.
HA-GSTM2 and HA-AhR were constructed using their coding sequencing onto pLV-EF1a-IRES-Hygro (Addgene, 85134), which was a gift from Tobias Meyer [49]. Transfection was performed using jetPRIME® Versatile DNA/siRNA transfection reagent (Poly-plus-transfection, 114–15) according to the manufacturer’s instructions.
Immunoblotting
Cells were harvested and washed with ice-cold PBS. 1x RIPA buffers containing 50x protease inhibitor cocktail and 100x phosphatase inhibitor were used for lysis. Protein concentration was quantified with Pierce BCA Protein Assay Kit (Thermo, 23225). In total, 15 μg proteins were loaded onto SDS-PAGE gels for electrophoresis, followed by transfer to PVDF membranes and blocking with skim milk. Primary antibodies in 1x TBST buffer were incubated overnight. On the next day, membranes were washed with 1x TBST buffer three times and HRP-linked secondary antibodies were incubated in TBST buffer for another hour. After repeating the washing step, the protein bands were probed with SuperSignal West Dura Extended Duration Substrate (Thermo, 34076) and visualized with ChemiDoc Imaging System. Immunoblots were repeated three times and representative images were shown.
Statistical analysis
Unless denoted elsewhere, results were analyzed by two-tailed unpaired Student’s t test (equal SD) or Welch’s t-test (unequal SD) assuming normal distribution, except for the one-way ANOVA analysis of the mice body weight. The analysis software was GraphPad Prism 8. A p value < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
The research is generously supported by NIH R01 CA157429 (XL), R01 CA196634 (XL), R01 CA264652 (XL), R01 CA256893 (XL). This research is also supported by the Biospecimen Procurement & Translational Pathology, Biostatistics and Bioinformatics, Redox Metabolism, and Flow Cytometry and Immune Monitoring Shared Resources of the University of Kentucky Markey Cancer Center (P30CA177558). We thanks Dr. Alumkal Joshi for the generous sharing of the RNA-seq raw data, and Eleanor Erikson for the critical reading and editing of the manuscript.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41388-022-02444-1.
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. [DOI] [PubMed] [Google Scholar]
- 2.Teo MY, Rathkopf DE, Kantoff P. Treatment of advanced prostate cancer. Annu Rev Med. 2019;70:479–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coutinho I, Day TK, Tilley WD, Selth LA. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr Relat Cancer. 2016;23:T179–t197. [DOI] [PubMed] [Google Scholar]
- 4.Enzalutamide Higano C., apalutamide, or darolutamide: are apples or bananas best for patients?. Nat Rev Urol. 2019;16:335–6. [DOI] [PubMed] [Google Scholar]
- 5.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. [DOI] [PubMed] [Google Scholar]
- 6.Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN, et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med. 2018;378:1408–18. [DOI] [PubMed] [Google Scholar]
- 7.Fizazi K, Shore N, Tammela TL, Ulys A, Vjaters E, Polyakov S, et al. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N Engl J Med. 2019;380:1235–46. [DOI] [PubMed] [Google Scholar]
- 8.Shiota M, Yokomizo A, Naito S. Oxidative stress and androgen receptor signaling in the development and progression of castration-resistant prostate cancer. Free Radic Biol Med. 2011;51:1320–8. [DOI] [PubMed] [Google Scholar]
- 9.Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–85. [DOI] [PubMed] [Google Scholar]
- 10.Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem. 2017;86:715–48. [DOI] [PubMed] [Google Scholar]
- 12.Neha K, Haider MR, Pathak A, Yar MS. Medicinal prospects of antioxidants: a review. Eur J Med Chem. 2019;178:687–704. [DOI] [PubMed] [Google Scholar]
- 13.Allocati N, Masulli M, Di C. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou SG, Wang P, Pi RB, Gao J, Fu JJ, Fang J, et al. Reduced expression of GSTM2 and increased oxidative stress in spontaneously hypertensive rat. Mol Cell Biochem. 2008;309:99–107. [DOI] [PubMed] [Google Scholar]
- 15.Han I, Jeong SJ, Lee HJ, Koh W, Lee HJ, Lee EO, et al. Proteomic analysis of mesenchymal stem-like cells derived from ovarian teratoma: potential role of glutathione S-transferase M2 in ovarian teratoma. Proteomics. 2011;11:352–60. [DOI] [PubMed] [Google Scholar]
- 16.Guo E, Wei H, Liao X, Wu L, Zeng X. Clinical significance and biological mechanisms of glutathione S-transferase mu gene family in colon adenocarcinoma. BMC Med Genet. 2020;21:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andonova IE, Justenhoven C, Winter S, Hamann U, Baisch C, Rabstein S, et al. No evidence for glutathione S-transferases GSTA2, GSTM2, GSTO1, GSTO2, and GSTZ1 in breast cancer risk. Breast Cancer Res Treat. 2010;121:497–502. [DOI] [PubMed] [Google Scholar]
- 18.Tang SC, Wu CH, Lai CH, Sung WW, Yang WJ, Tang LC, et al. Glutathione S-transferase mu2 suppresses cancer cell metastasis in non-small cell lung cancer. Mol Cancer Res. 2013;11:518–29. [DOI] [PubMed] [Google Scholar]
- 19.Peng L, Zhuang L, Lin K, Yao Y, Zhang Y, Arumugam T, et al. Downregulation of GSTM2 enhances gemcitabine chemosensitivity of pancreatic cancer in vitro and in vivo. Pancreatology. 2021;21:115–23. [DOI] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Lanman NA, Kong Y, He D, Mao F, Farah E, et al. Inhibition of the erythropoietin-producing receptor EPHB4 antagonizes androgen receptor overexpression and reduces enzalutamide resistance. J Biol Chem. 2020;295:5470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricci G, De Maria F, Antonini G, Turella P, Bullo A, Stella L, et al. 7-Nitro-2,1,3-benzoxadiazole derivatives, a new class of suicide inhibitors for glutathione S-transferases. Mechanism of action of potential anticancer drugs. J Biol Chem. 2005;280:26397–405. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharjee P, Paul S, Banerjee M, Patra D, Banerjee P, Ghoshal N, et al. Functional compensation of glutathione S-transferase M1 (GSTM1) null by another GST superfamily member, GSTM2. Sci Rep. 2013;3:2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagar YK. Does glutathione transferase loading into exosomes from prostate cancer cells influence progression? 2016. https://eprints.qut.edu.au/98015/. [Google Scholar]
- 25.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L, Au JL, Wientjes MG. Comparison of methods for evaluating drug-drug interaction. Front Biosci. 2010;2:241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes JD, Dinkova-Kostova AT, McMahon M. Cross-talk between transcription factors AhR and Nrf2: lessons for cancer chemoprevention from dioxin. Toxicol Sci. 2009;111:199–201. [DOI] [PubMed] [Google Scholar]
- 28.Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 2019;19:184–97. [DOI] [PubMed] [Google Scholar]
- 29.Shiota M, Yokomizo A, Tada Y, Inokuchi J, Kashiwagi E, Masubuchi D, et al. Castration resistance of prostate cancer cells caused by castration-induced oxidative stress through Twist1 and androgen receptor overexpression. Oncogene. 2010;29:237–50. [DOI] [PubMed] [Google Scholar]
- 30.Dolado I, Swat A, Ajenjo N, De Vita G, Cuadrado A, Nebreda AR. p38alpha MAP kinase as a sensor of reactive oxygen species in tumorigenesis. Cancer Cell. 2007;11:191–205. [DOI] [PubMed] [Google Scholar]
- 31.Canovas B, Nebreda AR. Diversity and versatility of p38 kinase signalling in health and disease. Nat Rev Mol Cell Biol. 2021;22:346–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, Ning S, Lou W, Yang JC, Armstrong CM, Lombard AP, et al. Cross-resistance among next-generation antiandrogen drugs through the AKR1C3/AR-V7 axis in advanced prostate cancer. Mol Cancer Ther. 2020;19:1708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res. 2012;72:1494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moilanen AM, Riikonen R, Oksala R, Ravanti L, Aho E, Wohlfahrt G, et al. Discovery of ODM-201, a new-generation androgen receptor inhibitor targeting resistance mechanisms to androgen signaling-directed prostate cancer therapies. Sci Rep. 2015;5:12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25. https://pubmed.ncbi.nlm.nih.gov/26544944/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med. 2016;375:1415–24. [DOI] [PubMed] [Google Scholar]
- 39.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci USA. 2020;117:12315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, Wei T, Ye Z, Orme JJ, Lin D, Sheng H, et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun. 2021;12:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjöström M, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet. 2020;52:778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies A, Nouruzi S, Ganguli D, Namekawa T, Thaper D, Linder S, et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat Cell Biol. 2021;23:1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z, Zhou C, Li X, Barnes SD, Deng S, Hoover E, et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell. 2020;37:584–.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbons JA, Ouatas T, Krauwinkel W, Ohtsu Y, van der Walt JS, Beddo V, et al. Clinical pharmacokinetic studies of enzalutamide. Clin Pharmacokinet. 2015;54:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Vries R, Jacobs F, Mannens G, Snoeys J, Cuyckens F, Chien C, et al. Apalutamide absorption, metabolism, and excretion in healthy men, and enzyme reaction in human hepatocytes. Drug Metab Dispos. 2019;47:453–64. [DOI] [PubMed] [Google Scholar]
- 47.Ji C, Guha M, Zhu X, Whritenour J, Hemkens M, Tse S, et al. Enzalutamide and apalutamide: in vitro chemical reactivity studies and activity in a mouse drug allergy model. Chem Res Toxicol. 2020;33:211–22. [DOI] [PubMed] [Google Scholar]
- 48.Liu C, Lou W, Zhu Y, Yang JC, Nadiminty N, Gaikwad NW, et al. Intracrine androgens and AKR1C3 activation confer resistance to enzalutamide in prostate cancer. Cancer Res. 2015;75:1413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, et al. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol. 2016;18:1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.