Key Points
Cytokine-producing, monocyte-derived macrophages are readily infected by M-CoV.
Type I IFNs curtail viral replication and sustain proinflammatory macrophages.
IFNAR deficiency leads to dysregulated immune phenotypes and immunopathology.
Abstract
Hallmarks of life-threatening, coronavirus-induced disease include dysregulated antiviral immunity and immunopathological tissue injury. Nevertheless, the sampling of symptomatic patients overlooks the initial inflammatory sequela culminating in severe coronavirus-induced disease, leaving a fundamental gap in our understanding of the early mechanisms regulating anticoronavirus immunity and preservation of tissue integrity. In this study, we delineate the innate regulators controlling pulmonary infection using a natural mouse coronavirus. Within hours of infection, the cellular landscape of the lung was transcriptionally remodeled altering host metabolism, protein synthesis, and macrophage maturation. Genetic perturbation revealed that these transcriptional programs were type I IFN dependent and critically controlled both host cell survival and viral spread. Unrestricted viral replication overshooting protective IFN responses culminated in increased IL-1β and alarmin production and triggered compensatory neutrophilia, interstitial inflammation, and vascular injury. Thus, type I IFNs critically regulate early viral burden, which serves as an innate checkpoint determining the trajectory of coronavirus dissemination and immunopathology.
Introduction
Coronaviruses have emerged as a family of ssRNA viruses causing not only mild respiratory symptoms but life-threatening infection at the root of multiple epidemics (1), including the recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced disease 2019 (COVID-19) pandemic (2, 3). Severe coronavirus disease is associated with high viral titers, exacerbated tissue injury, and dysregulated immune processes (4–6). A global effort has identified a number of innate immune phenotypes associated with disease severity, including increased monocyte proliferation and dampened Ag presentation, excessive neutrophil degranulation, impaired NK cell function, and elevated proinflammatory serum cytokines (7–12), while contradictory prognostic associations between type I IFN responses and disease outcome have been reported (13–19). Nevertheless, the relationship among these three hallmarks of disease, i.e., viral titers, immunopathology, and dysregulated immunity, remains unclear. The high, early viral burden observed in patients who go on to develop severe coronavirus-induced disease suggests either a delayed or insufficient potency of antiviral immune responses (20–22), whereas the relative absence of infectious viral particles in postmortem tissues (13, 23, 24) hints at immune responses that are sufficiently inflammatory to clear the virus and cause excessive tissue pathology. In general, for viral infections, the series of events triggered in early infection determines the outcome of late immune or pathological phenotypes and the overall course of disease (25–28). Because the sampling of symptomatic patients or dissection of postmortem tissues captures the culmination of a series of immune responses, the early regulation of inflammatory sequela balancing viral clearance and immunopathology remains largely unclear and represents a major missing link in our understanding of mechanisms driving severe coronavirus-induced disease.
The key cellular players and molecular mechanisms securing early antiviral immunity at the site of infection can be best dissected using preclinical models. The murine coronavirus (M-CoV; also known as the murine hepatitis virus) is a β-coronavirus that can induce a multiorgan disease in its natural host (29, 30) mimicking the pulmonary and extrapulmonary involvement of SARS-CoV-2 (31, 32). Genetic studies employing this mouse pathogen have identified a number of general regulators of coronavirus infection. In the context of i.p. infection, a type I IFN–driven communication axis between plasmacytoid dendritic cells (DCs) and macrophages has been shown to limit viral replication (33, 34), whereas in enteric coronavirus infection, NK and innate lymphoid cells contain M-CoV infection via the proinflammatory cytokine IL-15 (35). In contrast, functional assessment of antiviral mediators using mouse-adapted human coronaviruses or humanized mice has yielded contradicting results in terms of disease mechanisms, for example, in the case of type I IFNs (36–38). These discrepant observations may reflect the altered virulence or tropism of mouse-adapted and humanized transgenic models, which are not optimized to support the life cycle of a virus that has evolved to co-opt the replication machinery of a distinct host (29, 39). Indeed, successful viral propagation relies not only on viral entry but also uncoating, genome replication, translation of viral proteins, and new virion assembly and release from the host cell (40). Such curtailed viral propagation efficiency of mouse-adapted and humanized SARS-CoV-2 models is exemplified by the low or abrogated viral transmission in these respective models (41), in contrast with the robust transmissibility of M-CoV (42). Thus, the timely dissection of early regulators of antiviral immunity is best addressed in the context of a coronavirus model that exhibits a naturally evolved replication life cycle.
In this study, we employed the M-CoV model to dissect the early inflammatory responses mediating antiviral immunity to pulmonary coronavirus infection. To unleash viral replication, we genetically abrogated IFN responsiveness and delineated the cellular and molecular sequela preceding severe coronavirus-induced disease. Our data reveal a dynamic cellular and transcriptional remodeling of the pulmonary landscape marked by an attenuation of host cell metabolism, protein synthesis, and proinflammatory macrophage maturation, which were all dependent on type I IFN responses. Unrestricted viral replication together with impaired proinflammatory macrophage maturation triggered increased inflammation and tissue injury governed by overzealous neutrophil infiltration, as well as increased IL-1β and alarmin (S100A8/9) production. In summary, our study identifies the early threshold of viral burden as an innate checkpoint governing efficient versus pathological antiviral immunity during pulmonary coronavirus infection.
Materials and Methods
Mice
C57BL/6 mice were purchased from Charles River Laboratories, and LysM-Cre mice were purchased from Jackson Laboratory. LysM-Cre Ifnarfl/fl mice were provided by U. Kalinke (Helmholtz Center for Infection Research, Hannover, Germany) (43). β2-Microglobulin (β2M) and IFN-α receptor (IFNAR) knockout mice were maintained locally at the Institute for Laboratory Animal Sciences at the University of Zurich. All mice were on the C57BL/6 genetic background and were housed in the Institute of Immunobiology, Kantonsspital St. Gallen under specific pathogen-free conditions in individually ventilated cages. Experiments were conducted with female mice between 8 and 12 wk of age. All experiments were performed in accordance with federal and cantonal guidelines (Tierschutzgesetz) under permission numbers SG/01/19, SG/26/20, and SG/07/21 following review and approval by the Cantonal Veterinary Office (St. Gallen, Switzerland).
Viral infections and determination of viral titers
Mice were infected intranasally (i.n.) with 5 × 104 PFUs M-CoV A59 or M-CoV GFP as previously described (34, 44, 45). Mice were observed daily, and clinical disease symptoms and weights were noted. M-CoV titers were determined by standard plaque assay using L929 cells (33).
NK cell depletion
C57BL/6 mice were injected with 0.5 mg/ml anti-NK1.1 Ab i.p. 1 d before infection with 5 × 104 PFUs M-CoV. Depleting Ab was injected every second day after the first depletion. Control mice obtained 0.05 mg/ml anti-IgG Ab i.p. with the same treatment regimen.
Hematopoietic and stromal cell isolation
Cell isolation from mouse lungs was performed according to standardized protocols (46). Mice were euthanized at the indicated time points and immediately perfused with PBS. Lungs were transferred into a six-well plate containing RPMI 1640 medium (Sigma-Aldrich) supplemented with 4 U/ml elastase (Worthington), 1 U/ml Dispase II (Roche), and 10 μl/ml DNase I (AppliChem) and incubated for 45 min at 37°C. Lung tissue was then mechanically disrupted and washed in cold RPMI. Tissues were subjected to a second enzymatic digestion in RPMI 1640 medium supplemented with 25 μg/ml Liberase (Roche) and 0.25 μg/ml DNase I and incubated for 30 min at 37°C. Any larger tissue fragments were mechanically dissociated and passed through a 100-μm cell strainer. Stromal and hematopoietic cell fractions were enriched using MACS anti-CD45 microbeads (Miltenyi Biotec).
Flow cytometry
Cell characterization was performed by incubating single-cell suspensions in PBS containing 2% FCS and 2 mM EDTA (Sigma-Aldrich) for 20 min at 4 °C, using Abs against Ly6C-PerCP (HK1.4; BioLegend), CD11c-allophycocyanin-Cy7 (N418; BioLegend), I-A–I-E–AF700 (M5/114.15.2, BioLegend), CD24-Alexa (Al.) Fluor 647 (M1/69; BioLegend), CD11b-BV711 (M1/70; BioLegend), CD45-BV605 (104; BioLegend), CCR2-BV421 (SA203G11; BioLegend), CD64-PE-Cy7 (X54-5/7.1; BioLegend), Ly6G-PE (1A8; BD), CD31-PerCP (390; BioLegend), CD45-allophycocyanin-Cy7 (30-F11; BioLegend), CD24-Al. Fluor 647 (M1/69; BioLegend), I-A/I-E-BV421 (M5/114.15.2; BioLegend), CD326-PE-Cy7 (G8.8; BioLegend), podoplanin-PE (8.1.1; BioLegend), CD45-PE (104; BioLegend), and Annexin V–allophycocyanin (BioLegend). Ghost Dye Violet 510 (LubioScience) was used to identify dead cells. Ag-specific T cells were detected using PE-conjugated S598 tetramers (Sanquin Reagents) after an incubation of 20 min at 37°C. For measuring cytokine production, 106 cells were incubated with brefeldin A (5 μg/ml; Sigma) for 3.5 h at 37°C. For intracellular staining, cells were surface stained and fixed using Cytofix/Cytoperm (BD Biosciences) for 20 min. Fixed cells were incubated at 4°C for 40 min with monoclonal anti–granzyme B (eBioscience) diluted in permeabilization buffer (eBioscience). For Annexin V staining, the Annexin V–Apoptosis Staining/Detection Kit (Abcam) was used according to the manufacturer’s instructions. Samples were analyzed by flow cytometry using a FACS Fortessa flow cytometer and the FACSDiva operating program. Data were analyzed using FlowJo software (Tree Star).
Single-cell RNA sequencing analysis
Cell sorting was performed on a BD FACS Melody cell sorter using BD Chorus software. For discrimination between live and dead cells, 7-aminoactinomycin D (Sigma-Aldrich) was added before sorting. Sorted cells were passed through the 10× Chromium (10× Genomics) system, and cDNA libraries were generated according to the manufacturer’s recommendations (Chromium Single Cell 3′ Reagent Kit [version 3 chemistry]). Sequencing of libraries was done using the NovaSeq 6000 Illumina sequencing system at the Functional Genomic Center Zürich. Initial processing and gene expression estimation were performed using Cell Ranger (version 5.0.1) with the Ensembl GRCm38.94 release as a reference, as well as the M-CoV-A59 nucleocapsid gene sequence (NC_001846.1) to build index files for alignments. Subsequent quality control was performed with the Scater (version 1.20.1) (47) and SingleCellExperiment (version 1.10.1) (48) R/Bioconductor packages in R version 4.0.1. Cells with particularly high or low numbers of detected genes or unique molecular identified counts (more than two median absolute deviations from the median across all cells), as well as cells with a large fraction of mitochondrial genes (more than two median absolute deviations above the median across all cells), were removed to avoid contamination with damaged cells. After quality control and removal of contaminants, in total 35,616 cells were retained for further processing using the Seurat package (version 4.0.1) (49), which corresponded to 8196 cells from naive controls, 14,646 cells from infected control mice, and 12,774 cells from infected LysM-Cre Ifnarfl/fl mice. For transcriptional analysis of macrophages, a total of 10,678 cells were re-embedded, corresponding to 686 cells from naive mice, 7558 cells from infected control mice, and 2434 cells from infected LysM-Cre Ifnarfl/fl mice. Data were analyzed using functions from the Seurat package for normalization, scaling, dimensional reduction with principal-component analysis and t-distributed stochastic neighbor embedding (t-SNE), and graph-based clustering. Resulting clusters were characterized based on expression pattern of canonical marker genes and cluster markers calculated with the FindMarkers function of the Seurat package. Differentially expressed genes were conceived from Wilcoxon test as implemented in the FindMarkers function to compare expression profiles across conditions. To investigate functional differences between inflammatory macrophages in LysM-Cre Ifnarfl/fl and control mice, we tested differentially expressed genes in macrophage clusters for enrichment in gene ontologies using the ‘enrichGO’ function from the clusterProfiler R/Bioconductor package (version 3.18.1) (50). Top significant ontologies (q < 0.05) were visualized. Functional gene signatures were summarized from top differentially expressed genes based on their reported functions and projected on a diffusion map (51) by running the ‘DiffusionMap’ function from the destiny R package (3.4.0) (52).
Immunofluorescence
Murine lungs were perfused and fixed overnight at 4°C in 4% paraformaldehyde (Merck Millipore) under agitation. Tissues were embedded and oriented in 4% low-melting agarose (Invitrogen) in PBS and cut into 60- to 80-μm sections using a vibratome (VT-1200; Leica). Sections were blocked in PBS containing 10% FCS, 1 mg/ml anti-Fcγ receptor (BD Biosciences), and 0.1% Triton X-100 (Sigma). Sections were stained overnight with the following conjugated Abs against M-CoV–nucleocapsid–Al. Fluor 546 (J3.3, kindly provided by S. Siddell, University of Würzburg, Germany), CD45-Biotin (104; BioLegend), CD45-FITC (104; BioLegend), activated caspase 3–Al. Fluor 647 (92-605; BD), and smooth muscle actin–eFluor660 (1A4; Thermo Fischer Scientific) and unconjugated Abs against surfactant protein C (SFTPC)-rabbit (Invitrogen), podoplanin-hamster (8.1.1; BioLegend), and GFP-chicken (BD). Unconjugated Abs were detected with the following secondary Abs by incubation for 1 h at room temperature: anti-chicken Al. Fluor 488 (Thermo Fischer Scientific), goat anti–syrian hamster-D649 (Jackson Immunoresearch), donkey anti-rabbit-Cy3 (Jackson Immunoresearch), and streptavidin-Cy3 (BioLegend). Nuclei were stained with DAPI dihydrochloride (Life Technologies), and lung sections were mounted on glass microscopy slides using Fluorescence Mounting Medium (Dako). Imaging was performed with an LSM 980 confocal microscope (Carl Zeiss), and microscopy data were processed with ZEN software (Zeiss) and Imaris version 9 software (Bitplane). Overview images of lungs were acquired using tile scans at 200× magnification and overlapping tiles were fused together using ZEN software (Zeiss). For histological quantification of cells coexpressing caspase 3 and M-CoV nucleocapsid protein, cells were counted individually and expressed as a ratio over total M-CoV nucleocapsid–stained cells per field.
M-CoV infection of bone marrow–derived macrophages
Bone marrow was isolated from bones from LysM-Cre− littermate control (LMC), LysM-Cre Ifnarfl/fl, and Ifnar-deficient mice. Murine bone marrow–derived macrophages (BMDMs) were generated after culture with GM-CSF–containing supernatant. After 7 d, 105 BMDMs were plated into 48-well plates and on the following day incubated with 0.01 multiplicity of infection of M-CoV at 37°C. After 1 h, the virus-containing supernatant was replaced with medium, and supernatants were collected after 6 or 12 h. M-CoV titers in the supernatant were determined by standard plaque assay.
Cytokine quantification
For cytokine measurements, lung homogenate supernatants from day 2–infected mice and uninfected mice were used. IL-1β and S100A8/9 levels were measured using the IL-1β ELISA kit (Invitrogen) and Mouse S100A8/S100A9 ELISA kit (Invitrogen) according to the manufacturer’s instructions. Absorbances were measured at 450 nm on a microplate reader (Tecan). TNF-α and MCP-1 cytokine levels were quantified using the Cytometric bead assay Mouse inflammation kit (BD Biosciences) according to the manufacturer’s instructions and measured by flow cytometry using a FACS Fortessa flow cytometer.
Vascular permeability
Vascular permeability assays were performed as per standardized protocols (53). A total of 40 kDa of FITC-Dextran (Invitrogen) was injected i.v. into mice. One hour later, mice were sacrificed, and blood was collected by cardiac puncture. Serum was diluted 1:10 in PBS, and the fluorescence intensity was measured at 528 nm on a microplate reader (Tecan). Alternatively, 200 µl of 0.5% Evans blue (Sigma) was injected i.v. into mice. One hour later, mice were sacrificed and perfused, and lungs were stored in 4% paraformaldehyde.
Statistical analyses
Statistical analyses were performed with GraphPad Prism 8.0 with Mann–Whitney U test, and longitudinal comparison between different groups was performed with one- or two-way ANOVA with Tukey’s posttest or Kruskal–Wallis test with Dunn’s posttest. Statistical analyses are indicated in the figure legends. Statistical significance was defined as p < 0.05.
Data and code availability
The single cell RNA-sequencing (scRNA-seq) data generated in this study have been deposited in the BioStudies database (https://www.ebi.ac.uk/biostudies/) under accession code E-MTAB-11781. The R code will be made available from the authors on request.
Results
Type I IFN–responsive macrophages control acute pulmonary M-CoV infection
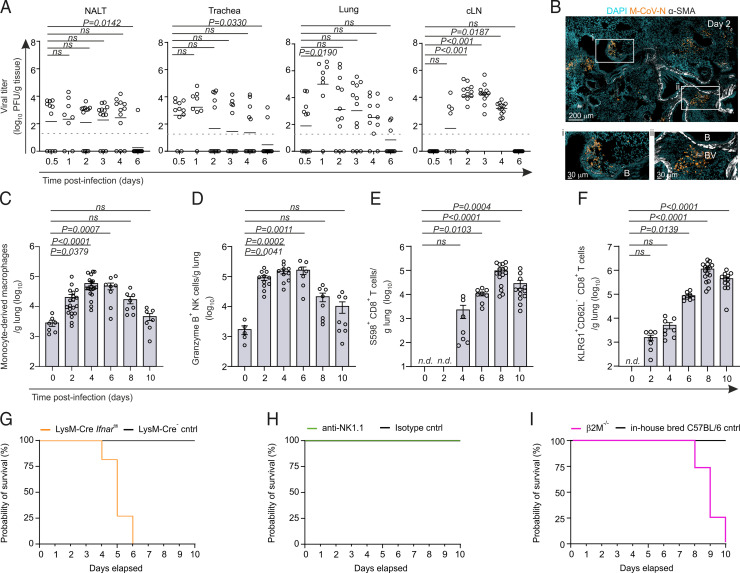
To elucidate the trajectory of viral replication of the natural mouse coronavirus, we infected C57BL/6 mice with 5 × 104 PFUs of M-CoV via the i.n. route and measured viral titers along the upper and lower airways and draining lymphoid tissues within the first 6 d postinfection (p.i.). All mice used for viral titer analysis showed the presence of virus in at least one organ (Supplemental Fig. 1A). i.n. infection of mice led to detectable viral titers in the nasopharyngeal-associated lymphoid tissue, the trachea, and lung already 12 h p.i. (Fig. 1A). Viral titers in the lungs peaked between days 1 and 2 p.i. Histological analysis of day 2–infected mouse lungs revealed that clusters of virus-infected cells expressing the M-CoV nucleocapsid protein consistently appeared in proximity to bronchioles, in line with an infection trajectory along the airways (Fig. 1B). By day 6 p.i., most mice had cleared the virus in the upper and lower airways. In cervical lymph nodes, replicating virus was first detectable 1 d p.i., with complete clearance by day 6. Pulmonary M-CoV infection triggered a rapid influx of myeloid cells, especially CCR2+Ly6C+CD64+ monocyte-derived macrophages and granzyme B–expressing NK cells as early as day 2 p.i. (Fig. 1C, 1D; Supplemental Fig. 1B–E). The numbers of M-CoV spike-specific, S598 tetramer-binding CD8+ T cells and activated KLRG1+CD62L−CD8+ T cells peaked in the lungs on day 8 p.i. (Fig. 1E, 1F, Supplementary Fig. 1F–H), highlighting the canonical immune processes leading to elimination of virus-infected cells (44).
FIGURE 1.
Type I IFN–responsive macrophages control pulmonary M-CoV infection. (A) M-CoV viral titer in the nasal-associated lymphoid tissue (NALT), trachea, lungs, and cervical lymph node at the indicated time points p.i. (B) Representative immunofluorescence image of the localization of virus-infected cells as demarcated by M-CoV nucleocapsid (M-CoV N) staining in the lungs of infected mice 48 h p.i. Smooth muscle actin (SMA) demarcates the bronchioles (B) and blood vessels (BV). (C–F) Flow cytometric cell enumeration of CCR2+ Ly6C+ CD64+ monocyte-derived macrophages (C), Granzyme B+ NK cells (D), S598+CD8+ T cells (E), and KLRG1+CD62L−CD8+ T cells (F) in the lungs of M-CoV–infected mice at the indicated times. (G–I) Survival curves after M-CoV infection of LysM-Cre Ifnarfl/fl mice compared with LysM-Cre–negative controls (G), anti-NK1.1–depleted mice compared with IgG isotype-treated controls (H), and β2M−/− compared with in-house bred C57BL/6 mice (I). Data in (A) are representative of three independent experiments with n = 11 (day 0.5 p.i.), n = 8 (day 1 p.i.), and n = 12 (days 2–6 p.i.); means are indicated. Dotted lines indicate the detection limit of the assay. Data in (B) are representative of at least six mice. Data in (C)–(F) are representative of three independent experiments with n = 5–10 naive mice, n = 7–20 for day 2 p.i., n = 7–20 for day 4 p.i., n = 7–8 for day 6 p.i., n = 8–16 for day 8 p.i., and n = 7–12 for day 10 p.i. Means ± SEM are indicated. In (E) and (F), T cell counts are normalized to day 0. Data in (G)–(I) are representative of two independent experiments each, with n = 7–9 mice per group. Statistical analysis in (A) and (C)–(F) was performed using Kruskal–Wallis test with Dunn’s multiple comparisons test. n.d., not detected.
Previous work has demonstrated the requirement for type I IFN responsiveness in myeloid cells, as well as a role for NK cells in controlling early M-CoV infection in the peritoneum and gastrointestinal tract, respectively (34, 35). Given the rapid increase in both of these cell populations in the lungs of M-CoV–infected mice, we sought to delineate the relevant cell types mediating viral containment during pulmonary M-CoV infection. To this end, we assessed the survival of mice lacking the gene encoding the IFNAR in LysM-Cre–targeted myeloid cells (54), as well as in mice treated with the NK cell–depleting anti-NK1.1 Ab. Consistent with i.p. M-CoV infection (34), mice lacking IFNAR in myeloid-lineage cells exhibited critical infection compared with LysM-Cre− LMCs (Fig. 1G), whereas NK cell depletion had no effect on survival (Fig. 1H, Supplemental Fig. 1I, 1J). As anticipated, CD8+ T cell–deficient, β2M-knockout mice exhibited a high probability of succumbing to infection between days 8 and 10 p.i. (Fig. 1I). Thus, pulmonary M-CoV infection was accompanied by increased inflammatory macrophage and NK cell infiltration, although type I IFN responsiveness in myeloid cells crucially controlled early M-CoV infection.
Transcriptional remodeling of the pulmonary landscape by M-CoV
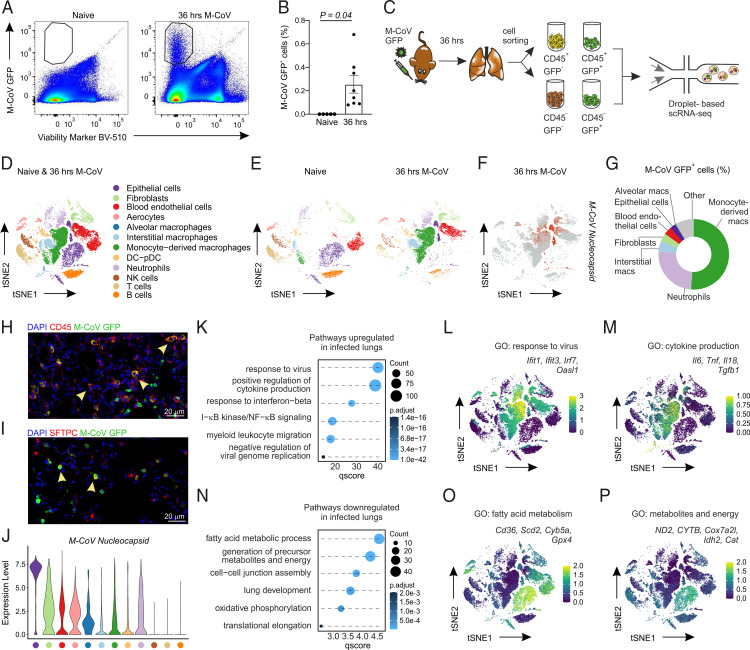
Discrepant observations regarding the protective or pathogenic role for type I IFNs are reported within the COVID-19 patient population (13–19, 55), in part because of the differences in sampling bronchoalveolar lavage (BAL) or peripheral blood at various time points over the course of symptomatic disease. To define the early molecular changes that accompany the rapid pulmonary influx of innate immune cells into the lungs during natural M-CoV infection, we sought to elucidate the transcriptional reprogramming of the pulmonary landscape at the peak of viral infection. Given the cytopathic nature of the coronavirus life cycle, we anticipated that virus-infected cells would account for a small percentage of overall cells in the lung. To detect and enrich for a sufficient number of live, virus-infected cells, we infected mice with the fluorescent M-CoV GFP virus. Consistent with the dynamics of viral replication in the lungs (Fig. 1A), we observed the highest proportion of M-CoV GFP+–infected cells at 48 h p.i. (Supplemental Fig. 2A, 2B). We reasoned that at the peak of the infection, cells would be more susceptible to cell death and sensitive to sorting for downstream scRNA-seq analysis. To achieve an optimal balance between infected cell yield and viability, we selected the 36 h time point p.i. for cell sorting experiments (Fig. 2A, 2B). To characterize both infected and noninfected cells, we sorted for GFP+CD45+/CD45− and GFP−CD45+/CD45– cells from M-CoV GFP–infected lungs at 36 h p.i. and for CD45+/CD45− cells from the lungs of naive mice (Fig. 2C). After quality control and processing, we obtained 22,842 cells that captured the desired spectrum of hematopoietic and nonhematopoietic cells in the lungs (Fig. 2D). Cell types were assigned based on known marker genes and comprised epithelial cells (Sftpc, Krt8, Epcam); fibroblastic cells (Col3a1, Pdgfra, Pdgfrb); blood endothelial cells (Cdh5, Pecam1, Cldn5), including a specialized population of aerocytes (Car4, Apln); alveolar macrophages (Fabp1, Mrc1, Marco, Siglecf); interstitial macrophages (Fcgr4, Treml4, Fabp4, Cx3cr1); monocyte-derived macrophages (Ccr2, Ly6c); a joint cluster of DCs and plasmacytoid DCs (Batf3, H2-Ab1, Siglech); neutrophils (S100a9, Ly6g); NK cells (Ncr1, Nkg7); T cells (Cd3e, Trbc2); and B cells (Cd79a, Cd19) (Supplemental Fig. 2C) (56–59). All cell types were present in lungs of both naive and M-CoV–infected mice, although the altered two-dimensional distribution of cells in t-SNE plots suggested significant transcriptional changes p.i. (Fig. 2E). Many of these transcriptional shifts were observed in infected cells, as identified by the expression of M-CoV nucleocapsid transcripts, including macrophages, neutrophils, epithelial cells, fibroblasts, and blood endothelial cells (Fig. 2F). To determine the relative abundance of infected cells, we quantified the composition of M-CoV GFP+ cells in the lungs of infected mice by flow cytometry (Fig. 2G, Supplemental Fig. 2D, 2E). Monocyte-derived macrophages, which matched the phenotype of Ly6C+ inflammatory macrophages, were the most readily infected cell type, comprising 51.2% ± 3.3% of GFP+ cells. Neutrophils were the second most infected cell type (24.9% ± 1.1%), followed by interstitial macrophages (4.9% ± 0.5%). Cells of the nonhematopoietic compartment, including epithelial cells, comprised between 2.7% and 3.8% of M-CoV GFP+ cells. The infection of CD45+ hematopoietic and SFTPC-expressing alveolar type II epithelial cells was confirmed by confocal microscopy (Fig. 2H, 2I). Intriguingly, despite the relatively low abundance of recovered infected epithelial cells, this cell population expressed the highest M-CoV nucleocapsid transcript levels, suggestive of host cell type–specific differences in supporting viral replication (Fig. 2J).
FIGURE 2.
Cell type–dependent infection and transcriptional remodeling of the pulmonary landscape. (A) Representative flow cytometry plots showing the presence of M-CoV GFP+ cells in the lungs of infected, but not naive, mice at 36 h p.i. (B) Quantification of the relative abundance of GFP+ cells in naive and 36 h p.i. mouse lungs. (C) Schematic representation of the experimental approach from cell isolation to droplet-based scRNA-seq. (D and E) Dimensional reduction t-SNE plots demonstrating the cell types in the naive and M-CoV–infected lungs merged (D) or separated per naive and infected condition (E). (F) Feature plot representing the expression pattern of the M-CoV nucleocapsid transcript. (G) Circle plot depicting the relative proportion of M-CoV GFP+ cells after 36 h p.i. as quantified by flow cytometry. (H and I) Representative confocal microscopy images of M-CoV GFP–infected CD45+ hematopoietic cells (H) and SFTPC+ alveolar epithelial type II cells (I). (J) Violin plot of M-CoV nucleocapsid expression across cell types in M-CoV–infected lungs. (K) Significantly enriched gene sets in the lungs of mice 36 h p.i. compared with naive mice. (L and M) Expression profiles of select genes belonging to the indicated gene sets projected onto t-SNE plots of merged M-CoV–infected and naive lungs. (N) Significantly downregulated gene sets in the lungs of mice 36 h p.i. compared with naive mice. (O and P) Average expression of select genes belonging to the indicated gene sets projected onto t-SNE plots of merged M-CoV–infected and naive lungs. Data in (A) and (B) are representative of three independent experiments, with n = 5 naive mice and n = 8 M-CoV–infected mice. In (B), statistical analysis was performed using unpaired two-tailed Student t test; means ± SEM are indicated. Data in (D)–(F) and (J)–(P) are representative of 22,842 cells. Data in (G) are representative of three independent experiments with n = 8 mice. Data in (H) and (I) are representative of at least four mice.
Given the breadth of infected cell types and evident transcriptional shifts associated with viral infection, we sought to define the transcriptional reprogramming of the cellular landscape in the lungs at this early time point. Analysis of the gene sets enriched in infected compared with naive mouse lungs revealed an upregulation of pathways associated with antiviral defense mechanisms, such as response to virus or cytokine signaling (Fig. 2K). To examine whether transcriptional changes were induced to a similar extent across different cell types, we projected the genes found in enriched pathways on t-SNE plots of merged naive and infected lungs. Although the upregulation of IFN-stimulated genes (ISGs) was evident across cell types in M-CoV–infected lungs, macrophages and neutrophils showed the strongest level of ISG expression (Fig. 2L), with proinflammatory cytokine transcripts most abundant in monocyte-derived macrophages from infected mice (Fig. 2M). Gene sets downregulated in infected lungs were associated with energy production, oxidative phosphorylation, and cellular translation (Fig. 2N), consistent with known and emerging type I IFN–mediated effector mechanisms limiting pathogen replication (60–64). Projection of genes involved in these pathways demonstrated the infection-associated downregulation of metabolic programs within nonhematopoietic cell types, including epithelial cells, blood endothelial cells, and fibroblasts (Fig. 2O, 2P). In turn, myeloid cells expressed low levels of transcripts involved in fatty acid metabolism and energy production already in the naive condition. In summary, although M-CoV was observed to infect multiple cell types in murine lungs, cell type–specific differences were observed pertaining to the relative abundance of infected cells, level of viral transcript expression, and infection-induced transcriptional reprogramming. In particular, the induction of ISGs and proinflammatory cytokine transcripts was most pronounced in myeloid cells, which appeared to employ these pathways to critically control early pulmonary M-CoV infection.
Type I IFN signaling curtails viral replication and host cell death
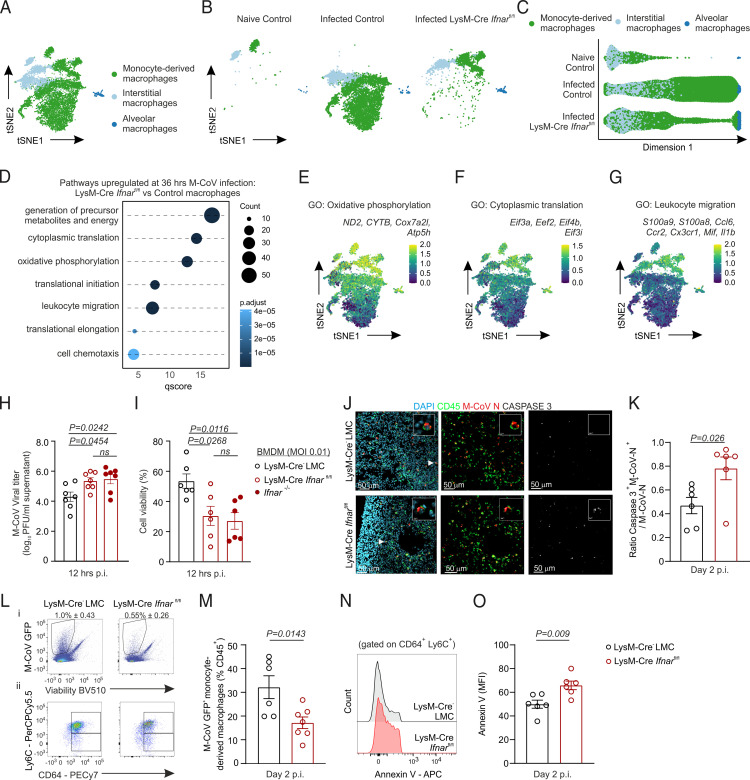
Because the sensing of type I IFNs by myeloid cells was observed to critically protect from pulmonary coronavirus infection (Fig. 1G), we next sought to address the spectrum of type I IFN–mediated transcriptional reprogramming of macrophages. To this end, we performed scRNA-seq of hematopoietic and nonhematopoietic cells from the lungs of LysM-Cre Ifnarfl/fl mice at 36 h p.i. All cell populations previously identified in Ifnar-competent mice were recapitulated in the lungs of conditionally Ifnar-deficient mice, although the distribution of cells in t-SNE plots already suggested an altered transcriptional identity of macrophage populations (Supplementary Fig. 3A). To first delineate type I IFN–induced transcriptional reprogramming of LysM-Cre–targeted cells, we re-embedded monocyte-derived, interstitial, and alveolar macrophages from Ifnar-sufficient, uninfected, and M-CoV–infected mice and from Ifnar-deficient, M-CoV–infected mice (Fig. 3A). Separation of macrophages according to infected condition and genotype, as well as visualization using diffusion maps as an alternative nonlineal dimensionality reduction technique (51), suggested that macrophages from LysM-Cre Ifnarfl/fl mice adopted an intermediate molecular state compared with those from Ifnar-sufficient uninfected and M-CoV–infected counterparts (Fig. 3B, 3C). In contrast with the infection-induced downregulation of gene sets involved in metabolism and energy production (Fig. 2N–P), macrophages from LysM-Cre Ifnarfl/fl mice retained an elevated expression of genes regulating metabolite and energy generation, oxidative phosphorylation, cytoplasmic translation, and cell chemotaxis (Fig. 3D–G).
FIGURE 3.
IFNAR signaling restricts metabolic resources and cytoplasmic translation machinery to curtail viral replication and host cell death. (A and B) Dimensional reduction t-SNE plot of pulmonary macrophages from Ifnar-sufficient naive and M-CoV–infected control mice and infected LysM-Cre Ifnarfl/fl mice either merged (A) or shown per individual condition and genotype (B). (C) Diffusion map of macrophage subsets from naive and M-CoV–infected control mice and infected LysM-Cre Ifnarfl/fl mice. (D) Significantly enriched gene sets in lung macrophages of infected LysM-Cre Ifnarfl/fl compared with infected control mice. (E–G) Average expression of selected genes belonging to the indicated gene sets projected onto t-SNE plots of naive and M-CoV–infected control macrophages and LysM-Cre Ifnarfl/fl macrophages. (H and I) Quantification of M-CoV titers (H) and cell viability (I) in BMDM cultures from LysM-Cre− LMC and LysM-Cre Ifnarfl/fl and Ifnar−/− mice at 12 h after infection (multiplicity of infection 0.1). (J) Representative immunofluorescence images of apoptotic M-CoV nucleocapsid–stained, virus-infected cells as demarcated by caspase 3 staining in the lungs of LysM-Cre Ifnarfl/fl and LysM-Cre− LMC mice 48 h p.i. Arrowheads demarcate the magnified cell in the inset image. (K) Histological quantification of the ratio of caspase 3 and M-CoV nucleocapsid costained cells per total M-CoV nucleocapsid–demarcated cells in lungs of LysM-Cre Ifnarfl/fl and LysM-Cre− LMC mice 48 h p.i. (L) Representative flow cytometric plots of M-CoV GFP+ live cells (Li) and M-CoV GFP+ monocyte-derived macrophages (Lii) from lungs of LysM-Cre− LMC and LysM-Cre Ifnarfl/fl M-CoV–infected mice. (M) Quantification of the percent abundance of LysM-Cre− GFP+ monocyte-derived macrophages from CD45+ cells in the lungs of LysM-Cre Ifnarfl/fl and LysM-Cre− LMC mice. (N and O) Representative histograms (N) and quantification (O) of the mean fluorescent intensity (MFI) of Annexin V among monocyte-derived macrophages in the lungs of LysM-Cre Ifnarfl/fl and LysM-Cre− LMC mice. scRNA-seq data in (A)–(G) are representative of 10,678 macrophages. Data in (H) and (I) are representative of four independent experiments with n = 6–7 repeats for each genotype. p values as per one-way ANOVA with Tukey’s multiple comparisons test; means ± SEM are depicted. Data in (L) and (M) are representative of two independent experiments with n = 6 LysM-Cre− LMC and n = 7 LysM-Cre Ifnarfl/fl mice. Data in (J), (K), (N), and (O) are representative of two independent experiments with n = 6 LysM-Cre− LMC and n = 6 LysM-Cre Ifnarfl/fl mice. p values as per unpaired two-tailed Student t test; means ± SEM are indicated.
The sustained expression of genes encoding for host cytoplasmic translational machinery and the failure to upregulate ISGs that directly inhibit viral replication suggested that cells lacking IFNAR may be better able to support new virion assembly. To assess the cell-intrinsic requirement for IFN-responsiveness on viral replication in an isolated system, we infected BMDMs from LysM-Cre Ifnarfl/fl mice, LMCs, and Ifnar-knockout mice. Compared with Ifnar-sufficient macrophages, Ifnar-deficient macrophages supported increased viral replication (Fig. 3H) and exhibited reduced cell viability at 12 h after M-CoV infection (Fig. 3I). Consistent with the observed increase in cell death in in vitro assays, histological assessment of apoptotic cells in situ revealed that lungs of LysM-Cre Ifnarfl/fl mice exhibited a significantly increased proportion of M-CoV nucleocapsid–demarcated cells expressing caspase 3 compared with LMCs (Fig. 3J, 3K). Given that monocyte-derived macrophages were observed to be the most readily infected cell type in murine lungs (Fig. 2G), and that the absence of Ifnar culminated in increased susceptibility of infected cells in vitro, we next examined whether infected, monocyte-derived macrophages were underrepresented in the lungs of LysM-Cre Ifnarfl/fl mice. Consistent with the observed reduction of macrophages bearing M-CoV nucleocapsid transcripts (Supplemental Fig. 3B), the proportion of M-CoV GFP+ monocyte-derived macrophages was reduced in the lungs of LysM-Cre Ifnarfl/fl mice compared with Ifnar-sufficient mice at day 2 p.i. (Fig. 3L, 3M). To determine to what extent the reduced abundance of this macrophage population was due to cell death, we examined the relative expression of Annexin V, an early apoptotic marker. Indeed, monocyte-derived macrophages from LysM-Cre Ifnarfl/fl mice expressed higher levels of Annexin V compared with Ifnar-sufficient controls (Fig. 3N, 3O), suggesting that type I IFNs protect and sustain monocyte-derived macrophages. In summary, the impaired sensing of type I IFNs unleashed viral replication while reducing host cell survival, biological effects that were concomitant with a failure to dampen transcriptional programs involved in metabolism, oxidative phosphorylation, and protein translation.
Type I IFNs sustain proinflammatory macrophage maturation
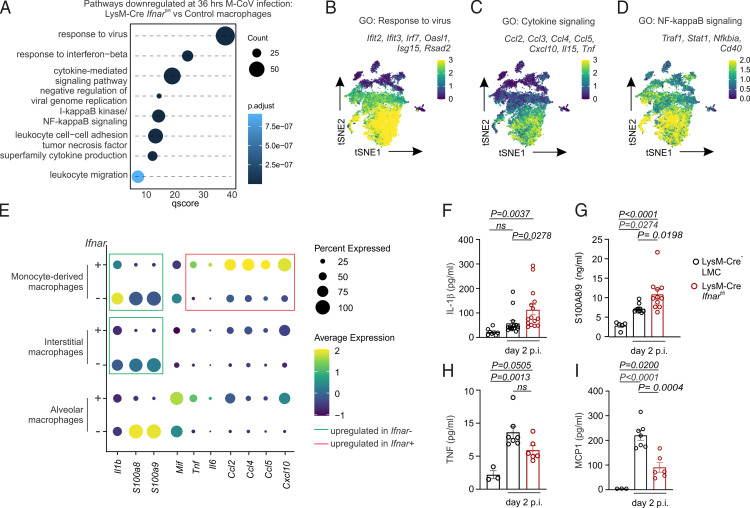
In the COVID-19 patient population, both dampened IFN responses and dysfunctional innate immunity correlate with worse disease outcome (8–12, 15–19). Nevertheless, the underlying mechanisms driving these phenotypes and whether they are linked remains unclear and difficult to address in humans given the rapid induction and resolution of type I IFNs. Enrichment analysis of gene sets preferentially expressed at a high level in Ifnar-deficient compared with Ifnar-sufficient macrophages highlighted an IFN-induced antiviral state associated with translational and metabolic reprogramming (Fig. 3D). To assess whether the production of proinflammatory cytokines by monocyte-derived macrophages (Fig. 2M) was also dependent on type I IFNs, we examined the pathways that failed to be upregulated in Ifnar-deficient macrophages on M-CoV infection. Gene set charting suggested that the inflammation-induced maturation of macrophages expressing inflammatory mediators such as Tnf, Ccl2, and Cxcl10 was dependent on IFNAR signaling (Fig. 4A–D) and restricted to LysM-Cre–targeted myeloid cells (Supplemental Fig. 3C–E). This appeared in contrast with the IFN-independent expression of leukocyte migration genes (Fig. 3G). Comparison of these two cytokine and chemokine expression profiles revealed a dichotomy between Tnf, Il6, Ccl2, and Cxcl10 expression in Ifnar-sufficient macrophages versus Il1b, S100a8, and S100a9 expression in Ifnar-deficient macrophages (Fig. 4E). We next assessed the expression profile of these cytokines and chemokines in macrophages relative to the overall cellular landscape of infected lungs in control and LysM-Cre Ifnarfl/fl mice. Neutrophils expressed the highest transcript levels of Il1b, S100a8, and S100a9, and these gene expression profiles were elevated in the lungs of conditionally Ifnar-deficient mice compared with Ifnar-sufficient mice (Supplemental Fig. 3F). Protein-level validation of these IFN-dependent and -independent gene expression changes was confirmed in lung homogenate supernatants from M-CoV–infected mice, with lungs from Ifnar-deficient mice exhibiting elevated IL-1β and S100A8/9 production and dampened amounts of MCP-1 (encoded by Ccl2) and TNF on day 2 p.i. (Fig. 4F–I). Collectively, these data revealed an IFN-dependent maturation of proinflammatory macrophages producing cytokines and chemokines such as TNF and CCL2, respectively. Strikingly, an increase in IL-1β and alarmins appeared as a compensatory process in the context of Ifnar deficiency.
FIGURE 4.
IFN-responsiveness dictates early proinflammatory macrophage maturation in the lungs of M-CoV–infected mice. (A) Significantly downregulated gene sets in lung macrophages of M-CoV–infected LysM-Cre Ifnarfl/fl compared with Ifnar-sufficient control mice. (B–D) Average expression of selected genes belonging to the indicated gene sets is projected onto t-SNE plots of macrophages from naive and M-CoV–infected control mice and infected LysM-Cre Ifnarfl/fl mice. (E) Average expression of inflammatory cytokines and chemokines in control and LysM-Cre–targeted, Ifnar-deficient macrophage clusters. (F–I) Cytokine levels of IL-1β (F), S100A8/9 (G), TNF (H), and MCP-1 (I) in the lung homogenate supernatant of LysM-Cre− LMC and LysM-Cre Ifnarfl/fl mice 2 days p.i. and naive LysM-Cre− LMC mice; means ± SEM are indicated. scRNA-seq data in (A)–(E) are representative of 10,678 macrophages. Data in (F) are representative of four independent experiments with n = 8 naive LysM-Cre− LMC mice, n = 15 M-CoV–infected LysM-Cre− LMC mice, and n = 15 M-CoV–infected LysM-Cre Ifnarfl/fl mice. Data in (G) are representative of two independent experiments with n = 5 naive LysM-Cre− LMC mice, n = 10 M-CoV–infected LysM-Cre− LMC mice, and n = 11 M-CoV–infected LysM-Cre Ifnarfl/fl mice. Data in (H) and (I) are representative of two independent experiments with n = 3 naive LysM-Cre− LMC mice, n = 7 M-CoV–infected LysM-Cre− LMC mice, and n = 6 M-CoV–infected LysM-Cre Ifnarfl/fl mice. p values in (F)–(I) as per one-way ANOVA with Tukey’s multiple comparisons test.
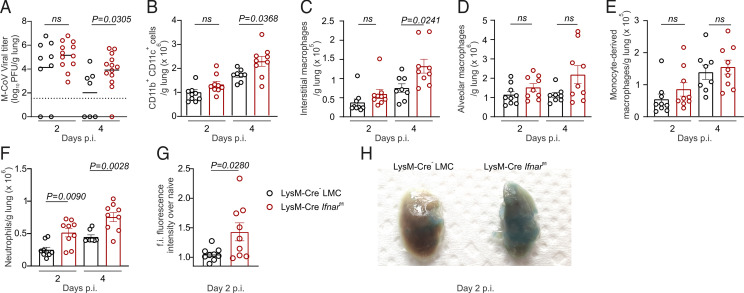
Compensatory neutrophilia and vascular permeability exacerbate lung pathology
Given the observed impact of type I IFNs in driving the transcriptional remodeling and inflammatory maturation of cells in M-CoV–infected lungs, we next investigated the physiological relevance of these programs on viral clearance and pulmonary inflammation. Consistent with the observed elevation of viral titers after in vitro infection, mice with a conditional Ifnar deficiency showed significantly impaired viral clearance in the lungs at day 4 p.i. (Fig. 5A). In addition to bearing higher viral concentration in the lungs, Ifnar-deficient mice exhibited uncontrolled viral spread to the trachea, spleen, and liver by 4 d p.i. (Supplemental Fig. 4A–C). Impaired viral containment in LysM-Cre Ifnarfl/fl mice was accompanied by a concomitant increase in the number of CD11b+CD11c+ myeloid cells in the lungs (Fig. 5B). Flow cytometry analysis revealed that the numbers of interstitial, but not alveolar or monocyte-derived, macrophages were significantly increased in LysM-Cre Ifnarfl/fl mice on day 4 p.i. (Fig. 5C, 5D, Supplemental Fig. 4D, 4E). The observed preservation of overall numbers of monocyte-derived macrophages in the lungs of Ifnar-deficient, M-CoV–infected mice, together with the heightened susceptibility of this macrophage subset to cytopathic cell death (Fig. 3K), suggested a higher turnover of these cells compared with other tissue-resident macrophage populations. In contrast with interstitial macrophages that were present at higher numbers only on day 4 p.i., uniquely the absolute number of neutrophils was observed to be significantly increased in the lungs of LysM-Cre Ifnarfl/fl mice already at day 2 p.i., preceding systemic viral spread (Fig. 5F, Supplemental Fig. 4F). Consistent with the known role for neutrophils in promoting edema and leukocyte infiltration (65), the lungs of LysM-Cre Ifnarfl/fl mice showed significantly higher vascular permeability as measured by serum FITC-Dextran fluorescence intensity on day 2 p.i. compared with Ifnar-sufficient controls (Fig. 5G) and confirmed by the accumulation of Evans blue in the lung tissue (Fig. 5H). Because increased vascular permeability could serve as a portal for leukocyte infiltration into the lungs, we performed H&E staining to determine whether vascular injury was associated with increased lung pathology. Indeed, lungs from day 2–infected LysM-Cre Ifnarfl/fl mice revealed an increased thickening of alveolar walls compared with Ifnar-sufficient controls (Supplemental Fig. 4G), indicative of exacerbated interstitial inflammation (66). Taken together, heightened viral burden and impaired IFN-dependent, proinflammatory macrophage maturation were counteracted by higher monocyte turnover, early neutrophilia, and increased pulmonary injury, which preceded systemic coronavirus dissemination.
FIGURE 5.
Neutrophilia-associated lung pathology and vascular permeability in Ifnar-deficient mice. (A) Viral titers in the lungs of LysM-Cre− LMC and LysM-Cre Ifnarfl/fl mice at days 2 and 4 after M-CoV infection. (B–F) Flow cytometric cell enumeration of CD11b+CD11c+ myeloid cells (B), interstitial macrophages (C), alveolar macrophages (D), monocyte-derived macrophages (E), and neutrophils (F) in the lungs of M-CoV–infected LysM-Cre Ifnarfl/fl mice compared with LysM-Cre− LMC mice at the indicated time points after infection. (G) Fold increase (f.i.) of the fluorescence intensity of serum FITC-Dextran in LysM-Cre− LMC and LysM-Cre Ifnarfl/fl mice infected 2 days earlier with M-CoV. (H) Representative images of lungs from LysM-Cre– LMC and LysM-Cre Ifnarfl/fl mice injected with Evans blue i.v. 2 days after M-CoV infection. Data in (A) are representative of three independent experiments with n = 9 LysM-Cre− LMC mice on day 2 p.i., n = 7 LysM-Cre− LMC mice on day 4 p.i., n = 12 LysM-Cre Ifnarfl/fl mice on day 2 p.i., and n = 13 LysM-Cre Ifnarfl/fl mice on day 4 p.i. Statistical analysis was performed using unpaired, two-tailed Student t test; mean is indicated. Data in (B)–(F) are representative of three independent experiments with n = 9 LysM-Cre− LMC mice and LysM-Cre Ifnarfl/fl mice on day 2 p.i., n = 8 LysM-Cre− LMC mice on day 4 p.i., and n = 9 LysM-Cre Ifnarfl/fl on day 4 p.i. p values as per one-way ANOVA with Tukey’s multiple comparisons test; means ± SEM are indicated. Data in (G) are representative of two independent experiments with n = 9 LysM-Cre− LMC and LysM-Cre Ifnarfl/fl mice each. Data in (H) are representative of two independent experiments with three mice per genotype. p values as per unpaired, two-tailed Student t test; means ± SEM are indicated.
Discussion
Dysregulated immunity is currently seen as a key determinant of immunopathological tissue damage associated with severe coronavirus-induced disease (15, 67). However, the relationships between the dynamics of viral replication, (dys)functional antiviral immunity, and resultant tissue damage remain unclear. In this article, we demonstrate that high, early viral replication overshooting protective type I IFN responses is the primary trigger of a compensatory innate immune cascade that culminates in excessive immunopathology. By genetically perturbing a front-line, innate defense mechanism, we delineate the impact of viral burden and IFN responses as regulators of excessive inflammation during pulmonary coronavirus infection. Several hallmarks of immune dysregulation observed in individuals with severe COVID-19 were recapitulated in exacerbated, natural M-CoV infection, including increased monocyte turnover, elevated IL-1β and S100A8/9 expression, and neutrophilia (8–10, 12, 68). In contrast with the debated pathological impact of type I IFNs in clinical and preclinical studies (10, 13–19, 34, 36–38), our data demonstrate a critical role for IFNs in controlling early viral dissemination, both by directly restricting viral replication and by securing next-in-line, proinflammatory innate immune responses. Thus, the threshold of early viral burden was found to be a key innate checkpoint that, if insufficiently controlled by IFN-mediated defenses, triggered dysregulated immune responses, increased tissue pathology, and severe coronavirus-induced disease.
Genetic mutations in the IFN signaling pathway have been documented in the context of severe COVID-19 but are extremely rare in the population (69–71). Instead, our data propose that a high viral infection dose drives excessive disease severity, a finding corroborated by anecdotal evidence in the patient population (72) and high viral titers at hospital admission (20–22). Indeed, early M-CoV burden has previously been shown to correspond to the magnitude of tissue damage, with higher peak viral loads exacerbating tissue pathology compared with lower peak viral titers (73). Our observed requirement of type I IFN–dependent control of viral replication is at odds with studies using adeno-associated virus–mediated overexpression of human entry receptors (38), likely because of the indeterminate viral tropism of such models. Indeed, we demonstrate that macrophages readily propagate M-CoV replication, a feature reported of SARS-CoV-2 (12, 23, 74). Our data now provide mechanistic insight into the mediators of coronavirus-induced immunopathology, demonstrating that impaired IFN responsiveness culminates in a preferential ablation of proinflammatory monocyte-derived macrophages and a compensatory, pathological IL-1β–alarmin–neutrophil axis concomitant with unrestricted viral replication. Although early neutrophil-mediated vascular remodeling crucially promotes leukocyte infiltration into infected tissues (75), excessive neutrophilia can cause life-threatening edema and loss of organ function (76). The concomitant increase in S100A8/9 expression may further promote neutrophil degranulation (77–79), exacerbating local tissue injury. Although the temporary dampening of proinflammatory mediators such as CCL2 and TNF observed in early M-CoV infection may appear as a disconnect from the serological hallmarks associated with severe clinical symptoms, the increased systemic viral dissemination and elevated IL-1β levels are expected to rapidly reverse this pattern. In individuals infected with a high viral dose, it is likely that exponential viral replication outpaces IFN responses, triggering such a compensatory, immunopathological inflammatory cascade. This rationale is supported by modeling studies that predict a proportionally increased requirement for IFN responses to meet increasing viral replication rates (26). Thus, high viral loads induce exacerbated and less efficient innate immune responses, culminating in immunopathology and severe coronavirus-induced disease.
Type I IFNs are the first line of innate immune defense induced within hours of infection to attenuate viral replication and prevent new virion assembly, budding, and release (80). Our data demonstrate that in addition to interfering with such canonical type I IFN–induced antiviral mechanisms, the IFN-driven attenuation of fatty acid metabolism, mitochondrial electron transport, and oxidative phosphorylation was associated with reduced viral replication, consistent with systemic metabolic changes observed after SARS-CoV-2 infection (81–83). However, we also demonstrated that such IFN-dependent metabolic changes were associated with the proinflammatory maturation of monocyte-derived macrophages. A switch from oxidative phosphorylation to glycolysis is involved in the activation of proinflammatory macrophages (84, 85), suggesting that in addition to restricting energy stores for viral replication, the observed type I IFN–dependent dampening of oxidative phosphorylation may enable the maturation of key innate immune mediators. Together, such early, IFN-dependent proinflammatory immune responses efficiently constrain coronavirus replication, preventing viral titers from exceeding a threshold that triggers dysregulated, immunopathological inflammatory sequela.
It is generally considered that cells in a local tissue microenvironment respond ubiquitously to type I IFNs. However, IFN-induced gene signatures depend, among other factors, on cell type– and activation state–associated transcriptional regulation (86) that can act as an additional determinant of viral replication and tropism (87, 88). Our comprehensive analysis of the molecular landscape of M-CoV–infected lungs revealed cell type–specific differences in the transcriptional reprogramming of antiviral mechanisms. This is evidenced by the almost complete dampening of fatty acid metabolism and extensive upregulation of ISGs in infected macrophages compared with the relatively modest fine-tuning of these responses in epithelial cells. Epithelial cells are widely considered the primary target of human coronaviruses (31, 89, 90), despite an abundance of virus-infected macrophages detected in COVID-19 patient BAL (12, 74) or postmortem tissue (23). Our data demonstrate that, although epithelial cells constitute a relatively small proportion of M-CoV–infected cells in comparison with monocyte-derived macrophages, viral transcripts were higher expressed in epithelial cells than in macrophages. A higher proportion of infected macrophages bearing lower SARS-CoV-2 genome transcript levels relative to epithelial cells has also been observed in COVID-19 patient BAL (12). Thus, it is conceivable that such cell-intrinsic differences in IFN-regulated energy supply and ISG signatures could explain either a higher replication capacity of coronaviruses in epithelial cells or an increased susceptibility of macrophages to M-CoV infection. Additional studies examining the dependence of coronavirus tropism on cell type–dependent metabolic regulation and ISG induction are warranted.
In summary, our study delineates the inflammatory sequela underlying dysregulated innate immune phenotypes associated with severe pulmonary coronavirus infection. Using a natural mouse coronavirus, we unveil an early immune checkpoint that regulates the balance between efficient, proinflammatory immune responses and immunopathology according to viral burden. The latter is triggered when viral replication outpaces critical type I IFN–mediated reprogramming of antiviral and proinflammatory defense mechanisms. Thus, type I IFN and proinflammatory responses proportional to the early viral burden secure resolution of pulmonary coronavirus infection by limiting innate immunopathology and viral dissemination.
Supplementary Material
This article is featured in Top Reads, p. 697
Footnotes
This work was supported by the Swiss National Science Foundation (Grant 180011), the Novartis Foundation for Medical-Biological Research, and Stiftung Experimentelle Biomedizin Peter Hans Hofschneider Professorship grants awarded to N.B.P. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The online version of this article contains supplemental material.
The single-cell RNA sequencing data presented in this article have been submitted to the BioStudies database (https://www.ebi.ac.uk/biostudies/) under accession number E-MTAB-11781.
- Al.
- Alexa
- BAL
- bronchoalveolar lavage
- BMDM
- bone marrow–derived macrophage
- COVID-19
- severe acute respiratory syndrome coronavirus 2–induced disease 2019
- DC
- dendritic cell
- IFNAR
- IFN-α receptor
- i.n.
- intranasal
- ISG
- IFN-stimulated gene
- LMC
- littermate control
- β2M
- β2-microglobulin
- M-CoV
- murine coronavirus
- p.i.
- postinfection
- SARS-CoV-2
- severe acute respiratory syndrome coronavirus 2
- scRNA-seq
- single-cell RNA sequencing
- SFTPC
- surfactant protein C
- t-SNE
- t-distributed stochastic neighbor embedding
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Adachi, S., Koma T., Doi N., Nomaguchi M., Adachi A.. 2020. Commentary: origin and evolution of pathogenic coronaviruses. Front. Immunol. 11: 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu, N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang, X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., et al. 2020. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 8: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nicholls, J. M., Poon L. L., Lee K. C., Ng W. F., Lai S. T., Leung C. Y., Chu C. M., Hui P. K., Mak K. L., Lim W., et al. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361: 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ng, D. L., Al Hosani F., Keating M. K., Gerber S. I., Jones T. L., Metcalfe M. G., Tong S., Tao Y., Alami N. N., Haynes L. M., et al. 2016. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am. J. Pathol. 186: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Witkowski, M., Tizian C., Ferreira-Gomes M., Niemeyer D., Jones T. C., Heinrich F., Frischbutter S., Angermair S., Hohnstein T., Mattiola I., et al. 2021. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature 600: 295–301. [DOI] [PubMed] [Google Scholar]
- 8. cOvid-19 Multi-omics Blood aTlas (COMBAT) Consortium . 2022. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell 185: 916–938.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silvin, A., Chapuis N., Dunsmore G., Goubet A. G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., et al. 2020. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 182: 1401–1418.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schulte-Schrepping, J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Deutsche COVID-19 OMICS Initiative (DeCOI) . 2020. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182: 1419–1440.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang, J., Jiang M., Chen X., Montaner L. J.. 2020. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 108: 17–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bost, P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., et al. 2020. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 181: 1475–1488.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nienhold, R., Ciani Y., Koelzer V. H., Tzankov A., Haslbauer J. D., Menter T., Schwab N., Henkel M., Frank A., Zsikla V., et al. 2020. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 11: 5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lucas, C., Wong P., Klein J., Castro T. B. R., Silva J., Sundaram M., Ellingson M. K., Mao T., Oh J. E., Israelow B., et al. Yale IMPACT Team . 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hadjadj, J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang, J. Y., Wang X. M., Xing X., Xu Z., Zhang C., Song J. W., Fan X., Xia P., Fu J. L., Wang S. Y., et al. 2020. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 21: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 17. Lee, J. S., Park S., Jeong H. W., Ahn J. Y., Choi S. J., Lee H., Choi B., Nam S. K., Sa M., Kwon J. S., et al. 2020. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 5: eabd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blanco-Melo, D., Nilsson-Payant B. E., Liu W. C., Uhl S., Hoagland D., Møller R., Jordan T. X., Oishi K., Panis M., Sachs D., et al. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181: 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wauters, E., Van Mol P., Garg A. D., Jansen S., Van Herck Y., Vanderbeke L., Bassez A., Boeckx B., Malengier-Devlies B., Timmerman A., et al. CONTAGIOUS collaborators . 2021. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 31: 272–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magleby, R., Westblade L. F., Trzebucki A., Simon M. S., Rajan M., Park J., Goyal P., Safford M. M., Satlin M. J.. 2021. Impact of severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin. Infect. Dis. 73: e4197–e4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. El Zein, S., Chehab O., Kanj A., Akrawe S., Alkassis S., Mishra T., Shatta M., El-Hor N., Salimnia H., Chandrasekar P.. 2021. SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic. pLoS One 16: e0255981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller, E. H., Zucker J., Castor D., Annavajhala M. K., Sepulveda J. L., Green D. A., Whittier S., Scherer M., Medrano N., Sobieszczyk M. E., et al. 2021. Pretest symptom duration and cycle threshold values for severe acute respiratory syndrome coronavirus 2 reverse-transcription polymerase chain reaction predict coronavirus disease 2019 mortality. Open Forum Infect. Dis. 8: ofab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delorey, T. M., Ziegler C. G. K., Heimberg G., Normand R., Yang Y., Segerstolpe Å., Abbondanza D., Fleming S. J., Subramanian A., Montoro D. T., et al. 2021. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schurink, B., Roos E., Radonic T., Barbe E., Bouman C. S. C., de Boer H. H., de Bree G. J., Bulle E. B., Aronica E. M., Florquin S., et al. 2020. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe 1: e290–e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bocharov, G., Ludewig B., Bertoletti A., Klenerman P., Junt T., Krebs P., Luzyanina T., Fraser C., Anderson R. M.. 2004. Underwhelming the immune response: effect of slow virus growth on CD8+-T-lymphocyte responses. [Published erratum appears in 2014 J. Virol. 78: 6079.] J. Virol. 78: 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bocharov, G., Züst R., Cervantes-Barragan L., Luzyanina T., Chiglintsev E., Chereshnev V. A., Thiel V., Ludewig B.. 2010. A systems immunology approach to plasmacytoid dendritic cell function in cytopathic virus infections. pLoS Pathog. 6: e1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ou, R., Zhou S., Huang L., Moskophidis D.. 2001. Critical role for alpha/beta and gamma interferons in persistence of lymphocytic choriomeningitis virus by clonal exhaustion of cytotoxic T cells. J. Virol. 75: 8407–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crouse, J., Kalinke U., Oxenius A.. 2015. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 15: 231–242. [DOI] [PubMed] [Google Scholar]
- 29. Grabherr, S., Ludewig B., Pikor N. B.. 2021. Insights into coronavirus immunity taught by the murine coronavirus. Eur. J. Immunol. 51: 1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sariol, A., Perlman S.. 2020. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity 53: 248–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu, J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., et al. 2021. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta, A., Madhavan M. V., Sehgal K., Nair N., Mahajan S., Sehrawat T. S., Bikdeli B., Ahluwalia N., Ausiello J. C., Wan E. Y., et al. 2020. Extrapulmonary manifestations of COVID-19. Nat. Med. 26: 1017–1032. [DOI] [PubMed] [Google Scholar]
- 33. Cervantes-Barragan, L., Züst R., Weber F., Spiegel M., Lang K. S., Akira S., Thiel V., Ludewig B.. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109: 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cervantes-Barragán, L., Kalinke U., Züst R., König M., Reizis B., López-Macías C., Thiel V., Ludewig B.. 2009. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 182: 1099–1106. [DOI] [PubMed] [Google Scholar]
- 35. Gil-Cruz, C., Perez-Shibayama C., Onder L., Chai Q., Cupovic J., Cheng H. W., Novkovic M., Lang P. A., Geuking M. B., McCoy K. D., et al. 2016. Fibroblastic reticular cells regulate intestinal inflammation via IL-15-mediated control of group 1 ILCs. Nat. Immunol. 17: 1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Channappanavar, R., Perlman S.. 2017. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 39: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mahlakõiv, T., Ritz D., Mordstein M., DeDiego M. L., Enjuanes L., Müller M. A., Drosten C., Staeheli P.. 2012. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 93: 2601–2605. [DOI] [PubMed] [Google Scholar]
- 38. Israelow, B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M. M., Wei J., Dong H., Homer R. J., et al. 2020. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 217: e20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gordon, D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O’Meara M. J., Rezelj V. V., Guo J. Z., Swaney D. L., et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. V’kovski, P., Kratzel A., Steiner S., Stalder H., Thiel V.. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19: 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chu, H., Chan J. F., Yuen K. Y.. 2022. Animal models in SARS-CoV-2 research. Nat. Methods 19: 392–394. [DOI] [PubMed] [Google Scholar]
- 42. Rehg, J. E., Blackman M. A., Toth L. A.. 2001. Persistent transmission of mouse hepatitis virus by transgenic mice. Comp. Med. 51: 369–374. [PubMed] [Google Scholar]
- 43. Barchet, W., Cella M., Odermatt B., Asselin-Paturel C., Colonna M., Kalinke U.. 2002. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 195: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cupovic, J., Onder L., Gil-Cruz C., Weiler E., Caviezel-Firner S., Perez-Shibayama C., Rülicke T., Bechmann I., Ludewig B.. 2016. Central nervous system stromal cells control local CD8(+) T cell responses during virus-induced neuroinflammation. Immunity 44: 622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Züst, R., Cervantes-Barragán L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V.. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. pLoS Pathog. 3: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakano, H., Nakano K., Cook D. N.. 2018. Isolation and purification of epithelial and endothelial cells from mouse lung. Methods Mol. Biol. 1799: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCarthy, D. J., Campbell K. R., Lun A. T., Wills Q. F.. 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Amezquita, R. A., Lun A. T. L., Becht E., Carey V. J., Carpp L. N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C., et al. 2020. Orchestrating single-cell analysis with Bioconductor. [Published erratum appears in 2020 Nat. Methods 17: 242.] Nat. Methods 17: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. 3rd, Hao Y., Stoeckius M., Smibert P., Satija R.. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu, G., Wang L. G., Han Y., He Q. Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haghverdi, L., Buettner F., Theis F. J.. 2015. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31: 2989–2998. [DOI] [PubMed] [Google Scholar]
- 52. Angerer, P., Haghverdi L., Büttner M., Theis F. J., Marr C., Buettner F.. 2016. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32: 1241–1243. [DOI] [PubMed] [Google Scholar]
- 53. Adil, M. S., Somanath P. R.. 2021. Vascular permeability assays in vivo. Methods Mol. Biol. 2367: 165–175. [DOI] [PubMed] [Google Scholar]
- 54. Clausen, B. E., Burkhardt C., Reith W., Renkawitz R., Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8: 265–277. [DOI] [PubMed] [Google Scholar]
- 55. Domizio, J. D., Gulen M. F., Saidoune F., Thacker V. V., Yatim A., Sharma K., Nass T., Guenova E., Schaller M., Conrad C., et al. 2022. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature 603: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Joshi, N., Watanabe S., Verma R., Jablonski R. P., Chen C. I., Cheresh P., Markov N. S., Reyfman P. A., McQuattie-Pimentel A. C., Sichizya L., et al. 2020. A spatially restricted fibrotic niche in pulmonary fibrosis is sustained by M-CSF/M-CSFR signalling in monocyte-derived alveolar macrophages. Eur. Respir. J. 55: 1900646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schyns, J., Bai Q., Ruscitti C., Radermecker C., De Schepper S., Chakarov S., Farnir F., Pirottin D., Ginhoux F., Boeckxstaens G., et al. 2019. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat. Commun. 10: 3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gibbings, S. L., Thomas S. M., Atif S. M., McCubbrey A. L., Desch A. N., Danhorn T., Leach S. M., Bratton D. L., Henson P. M., Janssen W. J., Jakubzick C. V.. 2017. Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 57: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gillich, A., Zhang F., Farmer C. G., Travaglini K. J., Tan S. Y., Gu M., Zhou B., Feinstein J. A., Krasnow M. A., Metzger R. J.. 2020. Capillary cell-type specialization in the alveolus. Nature 586: 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou, Q. D., Chi X., Lee M. S., Hsieh W. Y., Mkrtchyan J. J., Feng A. C., He C., York A. G., Bui V. L., Kronenberger E. B., et al. 2020. Interferon-mediated reprogramming of membrane cholesterol to evade bacterial toxins. Nat. Immunol. 21: 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. York, A. G., Williams K. J., Argus J. P., Zhou Q. D., Brar G., Vergnes L., Gray E. E., Zhen A., Wu N. C., Yamada D. H., et al. 2015. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 163: 1716–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Raniga, K., Liang C.. 2018. Interferons: reprogramming the metabolic network against viral infection. Viruses 10: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Olson, G. S., Murray T. A., Jahn A. N., Mai D., Diercks A. H., Gold E. S., Aderem A.. 2021. Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 35: 109195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McNab, F., Mayer-Barber K., Sher A., Wack A., O’Garra A.. 2015. Type I interferons in infectious disease. Nat. Rev. Immunol. 15: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wedmore, C. V., Williams T. J.. 1981. Control of vascular permeability by polymorphonuclear leukocytes in inflammation. Nature 289: 646–650. [DOI] [PubMed] [Google Scholar]
- 66. Recalde-Zamacona, B., García-Tobar L., Argueta A., Álvarez L., De Andrea C. E., Fernández Alonso M., Ezponda A., Carmona Torre F., Jordán Iborra C., Quiroga J. A., et al. 2020. Histopathological findings in fatal COVID-19 severe acute respiratory syndrome: preliminary experience from a series of 10 Spanish patients. Thorax 75: 1116–1118. [DOI] [PubMed] [Google Scholar]
- 67. Merad, M., Blish C. A., Sallusto F., Iwasaki A.. 2022. The immunology and immunopathology of COVID-19. Science 375: 1122–1127. [DOI] [PubMed] [Google Scholar]
- 68. Mann, E. R., Menon M., Knight S. B., Konkel J. E., Jagger C., Shaw T. N., Krishnan S., Rattray M., Ustianowski A., Bakerly N. D., et al. CIRCO . 2020. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci. Immunol. 5: eabd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang, Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I. K. D., Hodeib S., Korol C., et al. COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group . 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370: eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Khanmohammadi, S., Rezaei N., Khazaei M., Shirkani A.. 2022. A case of autosomal recessive interferon alpha/beta receptor alpha chain (IFNAR1) deficiency with severe COVID-19. J. Clin. Immunol. 42: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Abolhassani, H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., Aranda-Guillén M., Sardh F., Zuo F., Zhang P., et al. 2022. Inherited IFNAR1 deficiency in a child with both critical COVID-19 pneumonia and multisystem inflammatory syndrome. J. Clin. Immunol. 42: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schrock, J. M., Ryan D. T., Saber R., Benbow N., Vaught L. A., Reiser N., Velez M. P., Hsieh R., Newcomb M., Demonbreun A. R., Mustanski B., McNally E. M., ’’Aquila R., McDade T. W.. 2021. Cohabitation with a known coronavirus disease 2019 case is associated with greater antibody concentration and symptom severity in a community-based sample of seropositive adults. Open Forum Infect. Dis. 8: ofab244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eriksson, K. K., Cervantes-Barragán L., Ludewig B., Thiel V.. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1’’-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 82: 12325–12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grant, R. A., Morales-Nebreda L., Markov N. S., Swaminathan S., Querrey M., Guzman E. R., Abbott D. A., Donnelly H. K., Donayre A., Goldberg I. A., et al. NU SCRIPT Study Investigators . 2021. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 590: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kolaczkowska, E., Kubes P.. 2013. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13: 159–175. [DOI] [PubMed] [Google Scholar]
- 76. Cui, L. L., Zhang Y., Chen Z. Y., Su Y. Y., Liu Y., Boltze J.. 2020. Early neutrophil count relates to infarct size and fatal outcome after large hemispheric infarction. CNS Neurosci. Ther. 26: 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Simard, J. C., Girard D., Tessier P. A.. 2010. Induction of neutrophil degranulation by S100A9 via a MAPK-dependent mechanism. J. Leukoc. Biol. 87: 905–914. [DOI] [PubMed] [Google Scholar]
- 78. Simard, J. C., Cesaro A., Chapeton-Montes J., Tardif M., Antoine F., Girard D., Tessier P. A.. 2013. S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-κB(1.). pLoS One 8: e72138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang, S., Song R., Wang Z., Jing Z., Wang S., Ma J.. 2018. S100A8/A9 in Inflammation. Front. Immunol. 9: 1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. MacMicking, J. D. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12: 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Thomas, T., Stefanoni D., Reisz J. A., Nemkov T., Bertolone L., Francis R. O., Hudson K. E., Zimring J. C., Hansen K. C., Hod E. A., et al. 2020. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 5: e140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Valdés, A., Moreno L. O., Rello S. R., Orduña A., Bernardo D., Cifuentes A.. 2022. Metabolomics study of COVID-19 patients in four different clinical stages. Sci. Rep. 12: 1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pang, Z., Zhou G., Chong J., Xia J.. 2021. Comprehensive meta-analysis of COVID-19 global metabolomics datasets. Metabolites 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kelly, B., O’Neill L. A.. 2015. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Viola, A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A.. 2019. The metabolic signature of macrophage responses. Front. Immunol. 10: 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ivashkiv, L. B., Donlin L. T.. 2014. Regulation of type I interferon responses. Nat. Rev. Immunol. 14: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fay, E. J., Aron S. L., Macchietto M. G., Markman M. W., Esser-Nobis K., Gale M. Jr., Shen S., Langlois R. A.. 2020. Cell type- and replication stage-specific influenza virus responses in vivo. pLoS Pathog. 16: e1008760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ida-Hosonuma, M., Iwasaki T., Yoshikawa T., Nagata N., Sato Y., Sata T., Yoneyama M., Fujita T., Taya C., Yonekawa H., Koike S.. 2005. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J. Virol. 79: 4460–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hou, Y. J., Okuda K., Edwards C. E., Martinez D. R., Asakura T., Dinnon K. H. III, Kato T., Lee R. E., Yount B. L., Mascenik T. M., et al. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182: 429–446.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chua, R. L., Lukassen S., Trump S., Hennig B. P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M. T., et al. 2020. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38: 970–979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell RNA-sequencing (scRNA-seq) data generated in this study have been deposited in the BioStudies database (https://www.ebi.ac.uk/biostudies/) under accession code E-MTAB-11781. The R code will be made available from the authors on request.