Abstract
Metabolic reprogramming is the survival rule of tumor cells, and tumor cells can meet their high metabolic requirements by changing the energy metabolism mode. Metabolic reprogramming of tumor cells is an important biochemical basis of tumor malignant phenotypes. Ras‐related C3 botulinum toxin substrate 1 (Rac1) is abnormally expressed in a variety of tumors and plays an important role in the proliferation, invasion, and migration of tumor cells. However, the role of Rac1 in tumor metabolic reprogramming is still unclear. Herein, we revealed that Rac1 was highly expressed in colon cancer tissues and cell lines. Rac1 promotes the proliferation, migration, and invasion of colon cancer cells by upregulating SOX9, which as a transcription factor can directly bind to the promoters of HK2 and G6PD genes and regulate their transcriptional activity. Rac1 upregulates the expression of SOX9 through the PI3K/AKT signaling pathway. Moreover, Rac1 can promote glycolysis and the activation of the pentose phosphate pathway in colon cancer cells by mediating the axis of SOX9/HK2/G6PD. These findings reveal novel regulatory axes involving Rac1/SOX9/HK2/G6PD in the development and progression of colon cancer, providing novel promising therapeutic targets.
Keywords: glycolysis, metabolic reprogramming, pentose phosphate pathway, Rac1, SOX9
We further reveal novel regulatory axes involving Rac1/SOX9/HK2/G6PD in the development and progression of colon cancer.
Abbreviation
- 6PGD
6‐phosphogluconate dehydrogenase
- ATP
adenosine triphosphate
- DEG
differentially expressed gene
- G6P
glucose‐6‐phosphate
- G6PD
glucose‐6‐phosphate dehydrogenase
- GSEA
Gene Set Enrichment Analysis
- HK2
hexokinase
- IDH
isocitrate dehydrogenase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDHA
lactate dehydrogenase A
- PKM
pyruvate kinase
- PPP
pentose phosphate pathway
- qRT‐PCR
quantitative real‐time PCR
- Rac1
Ras‐related C3 botulinum toxin substrate 1
- SOX9
sex‐determining region Y (SRY)‐box 9
1. INTRODUCTION
Colon cancer is one of the common lower gastrointestinal tumors, which poses a great threat to people's health. Due to its high morbidity and mortality, and the characteristics of the onset age tending to be younger, it has gradually attracted attention. 1 Traditional treatments for colon cancer have limited efficacy on the vast majority of patients with advanced colorectal cancers. The occurrence of poor prognosis in colon cancer is more common in the late stage of the disease, which is associated with extensive tumor metastasis and chemoresistance. 2 , 3 Molecular targeted therapies have been shown to improve survival in patients with metastatic colon cancer, but the underlying mechanisms of these treatments are not fully understood, resulting in unsatisfactory outcomes. Therefore, it has become an urgent scientific problem to comprehensively expound the pathogenesis of colon cancer and explore the effective therapeutic targets of colon cancer.
Rac1 (Ras‐related C3 botulinum toxin substrate 1) is a key molecule of the Rho GTPase family. Rac1 protein can switch between inactive and active structures by hydrolyzing and binding GTP nucleotides. Activated Rac1 acts as an intracellular signal converter involved in control of several important cellular functions, such as cytoskeletal remodeling, cell adhesion, cell motility, and angiogenesis. 4 , 5 , 6 , 7 High expression or aberrant activation of Rac1 is mostly found in various cancer cells, such as lung, gastric, breast, and colon cancers, while mutant forms of Rac1 occur in melanoma. 7 , 8 , 9 , 10 , 11 , 12 , 13 Rac1 is involved in the development and progression of cancer by regulating multiple important signaling pathways to affect the cycle, proliferation, apoptosis, invasion, migration, and epithelial–mesenchymal transition of cancer cells. 4 , 14 , 15 , 16 Rac1 is considered to be a star target for cancer therapy such as melanoma and breast cancer, but is also found to be involved in the regulation of tumor therapy resistance. 17 Our previous study showed that Rac1 was significantly highly expressed in colon cancer and positively correlated with metastasis, clinical stage, and poor prognosis of colon cancer. 13 However, the specific molecular mechanisms of Rac1 in regulation of colon cancer invasion and metastasis remains to be further elucidated.
Metabolic reprogramming is one of the top 10 characteristics of tumor cells, including glucose metabolism, lipid metabolism, and nucleotide metabolism reprogramming. 18 As the energy source and metabolic intermediates of living cells, glucose catabolism includes glycolysis and mitochondrial oxidative phosphorylation. Abnormal glucose metabolism of tumor cells mainly showed abnormal activation of glycolysis, active glutamine metabolism, enhancement of the PPP, and changes in oxidative phosphorylation. 19 As glucose metabolism reprogramming plays an important role in tumor development, the molecular mechanism of its regulation has been a hot topic in tumor research. Abnormal function changes of metabolic key enzymes often lead to the occurrence of tumors. For example, the high expression of key enzymes, such as HK2 and PKM in tumor cells not only effectively promotes the glycolysis pathway, but has also been found to play an important role in the stemness maintenance and growth of a variety of tumor cells. 20 , 21 , 22 With the accumulation of the metabolite lactate, the acidified tumor microenvironment enhances the invasiveness of tumor cells. 23 Moreover, G6PD/G6PDH, the rate‐limiting enzyme of PPP, and NADPH, the final product, are considered to be favorable biomarkers for distant metastasis and prognosis of tumors. 24 , 25 Therefore, metabolic reprogramming provides a new strategy and direction for cancer prevention and treatment.
In the current study, we found that Rac1 could increase tumor growth and invasion by increasing the expression of SOX9, which as a transcription factor that can directly bind to the promoters of metabolic enzymes HK2 and G6PD and regulate their transcriptional activity to promote glycolysis and the activation of the PPP. In addition, we have further confirmed that Rac1 upregulates the expression of SOX9 through the PI3K/AKT signaling pathway. Our work elucidates a Rac1/SOX9/HK2/G6PD axis in the tumorigenesis and progression of colon cancer and provides prognostic indicators as well as a promising therapeutic target for colon patients.
2. MATERIALS AND METHODS
2.1. Data collection and analysis
GSE78093 were collected from the Gene Expression Omnibus datasets (https://www.ncbi.nlm.nih.gov/geo). The DEGs in GSE78093 were identified by a fold change of ≥1.5, or ≤−1.5 and p value <0.05, determined by Student's t‐test. Hierarchical clustering of DEGs was carried out using the package R software and expressed by heatmap. Volcano maps were drawn using the ggplot2 packages. The colon cancer cases in the Molecular Signature Database were stratified into high and low Rac1 groups, based on the median value in the GSE78093 database. The DEGs in gene sets named c2.kegg.v6.0.symbols.gmt between the sh‐Rac1 and control groups were analyzed by GSEA software (the Broad Institute, http://www.broadinstitute.org/gsea/index.jsp). The potential functions of DEGs in Rac1 silencing SW480 cells were analyzed by KEGG.
2.2. Patient samples
A total of 184 surgical colon cancer tissues and 40 adjacent nontumor tissues were collected from the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University from 2007 to 2011. The demographic and clinical characteristics of the patients are shown in Table S1. None of the patients received preoperative radiotherapy or chemotherapy. Written informed consent was obtained from individual patients. The experimental protocols were approved by the Joint Ethics Committee of the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University and Hunan Cancer Hospital.
2.3. Cell lines and cell culture
Human normal colonic epithelial cells NCM460 and five colon cancer cell lines (SW480, SW620, HT‐29, HCT116, and DLD1) are all stocks of our laboratory. The NCM460, SW620, HT‐29, and DLD1 cell lines were cultured in RPMI‐1640 (Gibco, Life Technologies), and SW480 and HCT116 cell lines were cultured in DMEM (Gibco, Life Technologies). All cell line culture media are supplemented with 10% FBS (Zeta Life) and 100 U/ml penicillin–streptomycin solution (Gibco, Life Technologies). The cells were cultured at 37°C under a humidified atmosphere of 5% CO2 and 95% air.
2.4. Transfection of plasmids
Three Rac1 shRNAs (shRac1‐1, shRac1‐2, and shRac1‐3) and three SOX9 shRNAs (shSOX9‐1, shSOX9‐2, and shSOX9‐3) were purchased from GeneChem. The Lipofectamine 3000 Transfection Reagent (Invitrogen) was used for plasmid transfection according to the manufacturer's protocols. The overexpression and knockdown effects were confirmed by quantitative real‐time PCR and western blot analysis.
2.5. Quantitative real‐time PCR
Total RNA of cells was extracted with TRIzol (Invitrogen; Thermo Fisher Scientific), according to the manufacturer's protocol. The total RNA (1 μg) was reverse transcribed by Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative RT‐PCR was carried out using a Fast Start Essential DNA Green Master kit (Lifescience, Roche) in the Roche Light Cycler 96 Instrument and Instrument Software (Lifescience, Roche) to determine the relative expression levels of target genes. The sequences of the primers used for quantitative PCR are shown in Table S2. We used GAPDH as the reference and normalization control.
2.6. Western blot analysis
Cells were lysed in RIPA lysis buffer (Beyotime) supplemented with protease and phosphatase inhibitors (Life Technologies). According to the quality of the protein, the protein samples were subjected to 8%–12% SDS‐PAGE and then transferred to a PVDF membrane. The membrane was sealed with 5% skim milk for 1 h and incubated with primary Ab at 4°C overnight. The Abs used are shown in Table S3. The next day, it was washed with PBS and incubated with the secondary Abs at room temperature for 2 h. Finally, immunoblotting was undertaken according to standard procedures.
2.7. Wound healing assay
The cells were implanted in a 6‐well plate (Corning), and cultured until the fusion reached approximately 80%. The cell monolayer was scratched with a 10 μl pipette tip and washed with PBS, then 2% FBS culture medium was added. The size of the scratch gap was recorded at certain time intervals under an inverted microscope.
2.8. Cell migration and invasion assay
A Transwell chamber (8 μm, 24‐well format; Corning) was used to evaluate the migration and invasion ability of cultured cells treated with or without matrix gel. The starved cells were suspended with serum‐free medium, and 5 × 104/200 μl cells were inoculated into the chamber. After 24–48 h, the upper chamber was collected, fixed with 4% paraformaldehyde, dyed with 1% crystal violet, washed and dried, and then photographed and counted under a microscope.
2.9. Colony formation and cell proliferation assay
Two hundred cells were seeded in a 6‐well plate and cultured for 2 weeks. After clonal colony formation, they were fixed with 4% paraformaldehyde, stained with 1% crystal violet, and washed with running water. The cell suspension (2 × 103 cells/100 μl/well) was inoculated into a 96‐well plate, and after cell adhesion, 10 μl CCK‐8 solution was added to each well. After incubating in an incubator for 1–4 h, the absorbance at 450 nm was measured with a microplate reader.
2.10. Cellular metabolism assays
Processed cells and cell culture media were collected according to the protocol. Glucose levels in the culture medium were determined using the Glucose Uptake Colorimetric Assay Kit (BioVision). Lactate levels were measured using a Lactate Colorimetric Assay Kit (BioVision). Intracellular G6P level and G6PDH enzyme activity were determined using a G6P Assay Kit (Beyotime) or G6PDH Activity Assay Kit (Beyotime), respectively. In addition, the NADP+/NADPH detection kit (Beyotime) detected the NADP+/NADPH ratio.
2.11. Immunohistochemistry
The tissue slices were soaked in citrate buffer solution, and the antigen was recovered by the microwave method. The following steps follow the standard procedure of the immunohistochemistry kit. Briefly, the tissue sections were incubated with primary Abs for 1 h. The specimens were then incubated with corresponding secondary Abs conjugated with HRP. The Abs used are listed in Table S4. The samples were scored according to the staining intensity and the number of positive cells as described previously. 26
2.12. Dual luciferase assay
Cells were cotransfected with indicated vectors, reporter vector with the firefly luciferase, and Renilla luciferase expression vector pRL‐TK (Promega). After transfection for 48 h, the cells were treated with a dual‐luciferase reporter system kit. The levels of firefly and Renilla luciferase activities were measured using a Dual‐Luciferase Reporter Assay System (Promega). Relative luciferase activity = firefly luminescence / Renilla luminescence.
2.13. Animal experiment
Colon cancer cells (1 × 106 cells/mouse, five nude mice per group) were subcutaneously injected into nude mice. The weight and tumor volume of the mice were measured with Vernier calipers every other day until the end of the experiment. Tumor volumes were calculated by measuring the length (L) and width (W) of tumors using calipers. The formula tumor volume = (L × W2)/2 was used to calculate the tumor volume. This experiment was carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University.
2.14. Statistical analysis
Experiments were carried out in triplicate and repeated three times. The data obtained was graphed and analyzed with GraphPad Prism8 software. All experimental results were presented as the mean ± SD. p values less than 0.05 were considered statistically significant. *p < 0.05, **p < 0.01, ***p < 0.001; NS, not significant.
3. RESULTS
3.1. Rac1 promotes malignant biological behaviors of colon cancer
Rac1 is highly expressed in various cancers. Our previous studies have shown that Rac1 is highly expressed in colon cancer tissues and is associated with prognosis. 13 First, we analyzed the mRNA and protein levels of Rac1 in normal colon epithelial cells (NCM460) and five colon cancer cell lines (SW480, SW620, DLD1, HCT116, and HT‐29). The results showed that Rac1 expression was relatively higher in the DLD1 and SW620 cell lines than in normal colon epithelial cells, but relatively lower in the HT‐29 and HCT116 cell lines (p < 0.05, Figure 1A,B). To detect the biological function of Rac1 in colon cancer cells, we generated Rac1 stably overexpressing/silencing cell lines (p < 0.05; Figures 1C,D and S1A,B). We analyzed the growth characteristics of these colon cancer cells. Colony formation and CCK‐8 assays showed that the Rac1 overexpression promoted the colony forming and proliferation of HT‐29 cells compared to the control. In contrast, the inhibition of Rac1 reduced the colony formation and proliferation of SW620 cells (p < 0.05; Figure 1E,F). To further test the tumor‐promoting function of Rac1, we analyzed the metastasis characteristics in colon cancer cells. We found that overexpression of Rac1 promoted the migration and invasion of HT‐29 cells, while the inhibition of Rac1 significantly suppressed the migration and invasion of SW620 cells by wound healing and Transwell assays (p < 0.05; Figure 1G,H). We next investigated the role of Rac1 in colon cancer development by means of xenografts in nude mice. Colon cancer cells 1 × 106 with stably overexpressing or KO Rac1 or corresponding controls were injected into nude mice. We observed that overexpression of Rac1 significantly promoted the growth of xenograft tumors, while the silencing of Rac1 significantly inhibited the growth of xenograft tumors (p < 0.05; Figure 1I,J). These findings indicate that Rac1promotes the growth and invasion of colon cancer cells both in vitro and in vivo.
FIGURE 1.
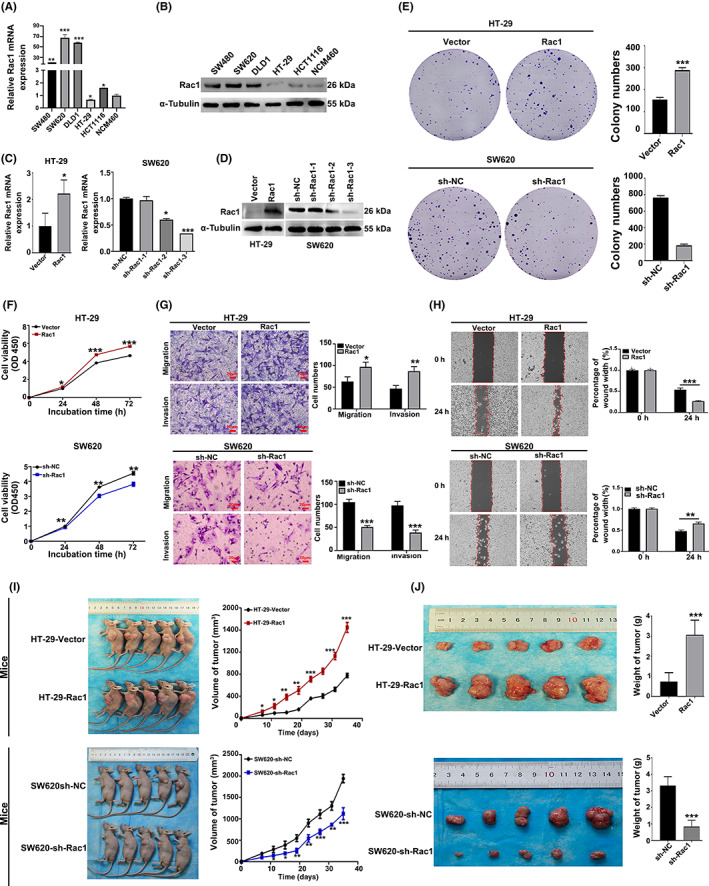
Rac1 promotes the malignant biological behaviors of colon cancer. (A) Rac1 expression in colon cancer cell lines. The mRNA and protein expression of Rac1 in normal colonic epithelial cells (NCM460) and colon cancer cells with different invasive potential (HT‐29, HCT116, SW480, SW620, and DLD1) were detected by quantitative real‐time PCR (qRT‐PCR) and western blot analysis. (B) Rac1 overexpression and knockdown validation. Rac1 protein expression levels were measured by qRT‐PCR and western blot analysis. (C–H) The effect of altered Rac1 expression on colon cancer cell migration, invasion, and proliferation. Rac1 overexpression promoted migration, invasion, and proliferation of colon cancer cells HT‐29, and Rac1 interference inhibited migration, invasion, and proliferation of colon cancer cells SW620. (I, J) Growth of subcutaneous tumors after subcutaneous injection of colon cancer cells stably overexpressing Rac1 or stably interfering with Rac1. Data are representative images or expressed as the mean ± SD of each group of samples analyzed in triplicate from three separate experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NC, negative control; NS, no statistical difference; OD, optical density.
3.2. Rac1 promotes glycolysis and the PPP in colon cancer
To clarify the exact mechanisms through which Rac1 promoted the progression of colon cancer cells, the GSEA analysis of GSE78093 was carried out. It showed that gene sets related to glycolysis were enriched in low Rac1 expressing colon cancer cases (Figure 2A). In addition, we found 879 DEGs with a fold change of >1.5 or <−1.5 and p value of <0.01. The KEGG analysis showed that these DEGs were enriched in the PPP (Figure 2B). These data indicated that Rac1 could regulate the glycolysis and PPP in colon cancer cells.
FIGURE 2.
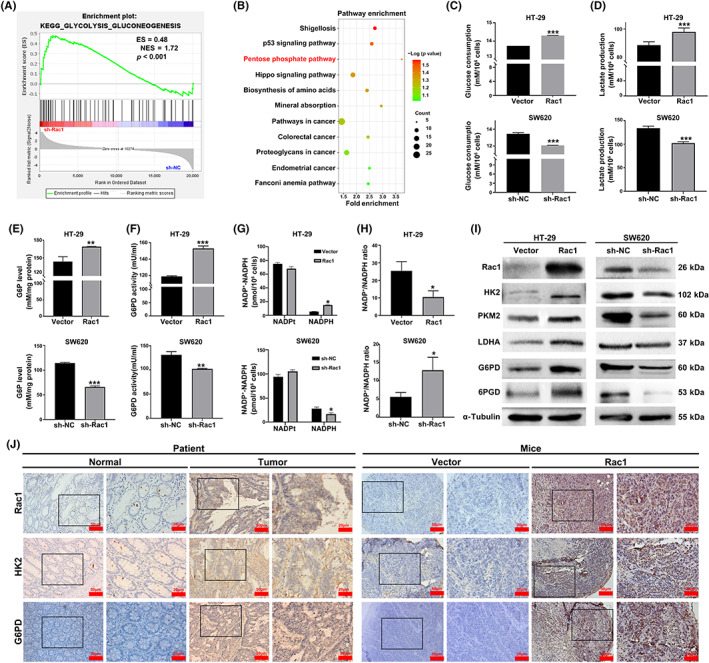
Rac1 enhances glycolysis and the pentose phosphate pathway (PPP) in colon cancer cells. (A) Gene Set Enrichment Analysis of enriched glycolysis‐related genes in high Rac1 expressing colon cancer of the GSE78093 dataset. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of 879 DEGs. (C–H) Rac1 overexpression increased glucose consumption, lactate production, intracellular glucose‐6‐phosphate (G6P) level, and glucose‐6‐phosphate dehydrogenase (G6PD) activity and decreased the ratio of NADP+/NADPH in HT‐29 cells; however, after Rac1 silencing, the results were opposite. (I) After Rac1 overexpression or silencing, Western blot was used to detect the effects of glycolysis‐related enzymes hexokinase 2 (HK2), pyruvate kinase isoenzyme 2 (PKM2), and lactate dehydrogenase A (LDHA) and PPP‐related enzymes G6PD and 6‐phosphogluconate dehydrogenase (6PGD) protein expression levels in HT‐29 and SW620 colon cancer cells. (J) Immunohistochemistry was used to detect the expression of Rac1, HK2, and G6PD in colon cancer tissues and tumor sections (magnification 200×, scale bars 50 μm; magnification 400×, scale bars 20 μm). Data are representative images or expressed as the mean ± SD of each group of samples analyzed in triplicate from three separate experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NS, no statistical difference; TK1, thymidine kinase 1.
To identify the role of Rac1 in regulation of metabolic reprogramming signaling in colon cancer, we examined the changes in glucose consumption, intracellular G6P level, and lactate production in Rac1 overexpressing/silencing colon cancer cells. Results showed that overexpression of Rac1 significantly increased glucose consumption, lactate production levels, glucose‐6‐phosphate level, and G6PD activity, whereas Rac1 silencing had the opposite effects on colon cancer cells (p < 0.05; Figure 2C–F). In addition, we detected the levels of intracellular NADPH in colon cancer cells. Compared with the controls, Rac1 overexpression decreased NADP+/NADPH ratios, while Rac1 silencing increased them in colon cancer cells (p < 0.05; Figure 2G,H). A similar pattern was observed in HCT116 and DLD1 cells (p < 0.05; Figure S1C–H). Additionally, we found that Rac1 silencing increased HK2, PKM2, LDHA, G6PD, and 6PGD levels in SW620 cells; in contrast, POU2F1 overexpression increased their expression in HT‐29 cells (Figure 2I). A similar pattern was observed in HCT116 and DLD1 cells (Figure S1I). In addition, we investigated the correlation between Rac1 and HK2/G6PD on clinical samples of colon cancer through immunohistochemical analysis. The expression of Rac1, HK2, and G6PD was significantly increased in colon cancer tissues compared with adjacent tissues (p < 0.05; Figures 2J and S2A). Furthermore, we observed that HK2 and G6PD expression positively correlated with Rac1 (HK2: r = 0.6554, p < 0.001; G6PD: r = 0.4832, p < 0.001) (Figure S2B). Similarly, HK2 and G6PD were also significantly overexpressed in the Rac1‐overexpressed group in mouse xenografts compared to the control group (Figure 2J). These results indicate that Rac1 can promote glycolysis and activate the PPP in colon cancer cells.
3.3. Rac1 upregulates expression of SOX9 in colon cancer cells
To understand how Rac1 is involved in regulating glycolysis and PPP in colon cancer, we found 879 DEGs (488 upregulated and 391 downregulated) in SW480‐sh‐Rac1 cells derived from GSE78093 (with a fold change >1.5 and p value <0.01) (Figure 3A). Among them, we found that SOX9 expression was significantly downregulated after silencing of Rac1 (Figure 3A). Previous studies have shown that SOX9, as an important transcription factor containing the HMG box sequence, is closely associated with digestive tract tumors. 27 To clarify whether Rac1 can regulate SOX9 expression, we examined SOX9 expression in Rac1 overexpressing/silencing colon cancer cells. The results showed that Rac1 upregulated the expression of SOX9, but Rac1 silencing downregulated the expression of SOX9 (p < 0.05; Figure 3B). We further examined SOX9 expression in colon cancer patient tissues and xenograft tissues (p < 0.001; Figure 3C,D), and found that SOX9 expression was positively correlated with Rac1 (r = 0.5999, p < 0.001, Figure 3C–E), suggesting that Rac1 promoted SOX9 expression in colon cancer cells.
FIGURE 3.
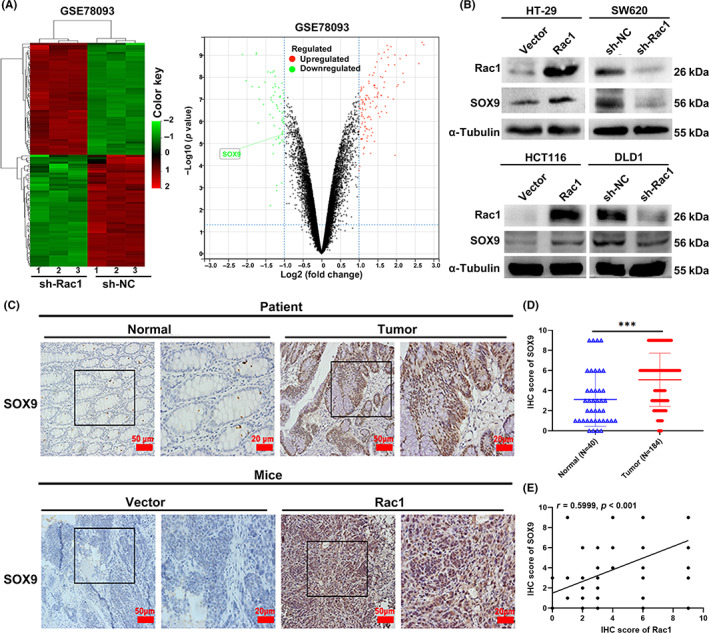
Rac1 upregulates the expression of SOX9 in colon cancer cells. (A) Volcano plot and heatmap analysis of differentially expressed genes in GSE78093. Red indicates upregulation and green indicates downregulation, and the box in the volcano figure is identified as the SOX9 gene. (B) Western blot analysis was used to detect the effect of Rac1 overexpression or silencing on SOX9 protein expression in colon cancer cell lines HT‐29, HCT116, SW620, and DLD1. (C) Immunohistochemistry (IHC) was used to detect the expression of SOX9 in colon cancer tissues and xenografts (magnification 200×, scale bars 50 μm; magnification 400×, scale bars 20 μm). (D) IHC analysis of SOX9 expression in 184 colon cancer tissues. (E) Correlation between the relative levels of Rac1 and SOX9, determined by IHC in 184 colon cancer tissues. ***p < 0.001. NC, negative control.
3.4. SOX9 promote proliferation, invasion, and migration by enhancing glycolysis and PPP in colon cancer cells
To clarify the role of SOX9 in the malignant biological phenotypes of colon cancer, we constructed HT‐29 cells with stable overexpression of SOX9 and SW620 cells with stable silencing of SOX9 (p < 0.05; Figure 4A). Using plate colony assays and CCK‐8 proliferation assays, we revealed that SOX9 overexpression significantly promoted the proliferation of HT‐29 cells, while SOX9 silencing significantly inhibited SW620 cell proliferation (p < 0.05; Figure 4B–D). We also analyzed the effects of SOX9 on the invasion and migration of colon cancer cells by Transwell and wound healing assays, and found that SOX9 overexpression significantly enhanced the invasion and migration of HT‐29 cells, whereas silencing of SOX9 expression significantly inhibited the invasion and migration of SW620 cells (p < 0.05; Figure 4 E,F). In addition, we further detected the effects of SOX9 on glucose consumption, lactate production, intracellular G6P level, G6PD activity, and NADP+/NADPH ratio in colon cancer cells. The results showed that overexpression of SOX9 increased glucose consumption, lactate production, intracellular G6P level, and G6PD activity and decreased the NADP+/NADPH ratio in HT‐29 cells, while silencing of SOX9 produced the opposite results (p < 0.05; Figure 4G–K). Western blot experiments also showed that the metabolism‐related enzymes were significantly upregulated after overexpression of SOX9, while the opposite result were observed after silencing of SOX9 (p < 0.05; Figure 4L), suggesting that SOX9 enhanced the glycolysis and PPP to promote the proliferation, invasion, and migration of colon cancer cells.
FIGURE 4.
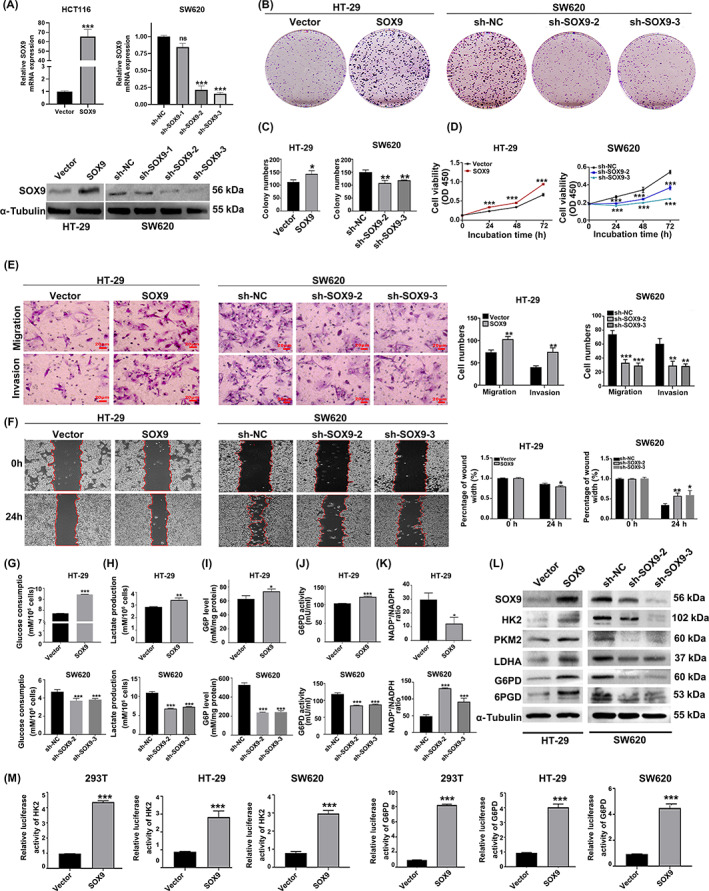
SOX9 promotes colon cancer cell proliferation, invasion, and migration, and glycolysis and the pentose phosphate pathway (PPP). (A) SOX9 overexpression and knockdown validation. SOX9 mRNA and protein expression levels were measured by quantitative real‐time PCR and western blot analysis. (B–D) SOX9 overexpression significantly enhanced the colony formation and proliferation of colon cancer cells, while SOX9 silencing significantly inhibited colon cancer cell colony formation and proliferation. (E, F) SOX9 overexpression significantly enhanced the invasion and migration of colon cancer cells, and interference with SOX9 inhibited the invasion and migration of colon cancer cells (magnification 200×, scale bars 50 μm; magnification 400×, scale bars 20 μm). (G–K) SOX9 overexpression increased glucose consumption, lactate production, intracellular glucose‐6‐phosphate (G6P) level and glucose‐6‐phosphate dehydrogenase (G6PD) activity of colon cancer cells HT‐29 and decreased the NADP+/NADPH ratio in HT‐29 cells; however, silencing SOX9 yielded the opposite results (L) Western blot analysis was used to detect the effect of SOX9 overexpression or silencing on the protein expression levels of glycolysis‐related enzymes hexokinase 2 (HK2), pyruvate kinase isoenzyme 2 (PKM2), and lactate dehydrogenase A (LDHA) and PPP‐related enzymes G6PD and 6‐phosphogluconate dehydrogenase (6PGD) in HT‐29 and SW620 colon cancer cells. (M) Regulation of SOX9 overexpression on HK2 and G6PD promoter activities was detected in 293T, HT‐29, and SW620 cells. Data are representative images or expressed as the mean ± SD of each group of samples analyzed in triplicate from three separate experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NC, negative control; NS, no statistical difference; OD, optical density.
Using Jaspar ( http://jaspar.genereg.net/), we predicted that SOX9 as a transcription factor could bind to the promoter region of HK2 and G6PD, which suggested that SOX9 could directly regulate the expression of HK2 and G6PD and thus regulate metabolism (Tables S4 and S5). We constructed luciferase reporter plasmids for HK2 and G6PD (−2000 to ~0 bp) and cotransfected them with Renilla luciferase reporter plasmid. We found that SOX9 significantly upregulated the transcriptional activities of HK2 and G6PD in 293T, HT‐29, and SW620 cells (p < 0.05; Figure 4M). Similar results were observed in HCT116 and DLD1 cells (p < 0.05; Figure S2C). Collectively, these results suggest that SOX9 promotes glycolysis and activates the PPP by enhancing the transcriptional activity of both HK2 and G6PD.
3.5. Rac1 promotes malignant behaviors of colon cancer through SOX9
To determine whether Rac1 promotes colon malignant biological behaviors through SOX9, we induced SOX9 overexpression in Rac1 silencing cells or SOX9 silencing in Rac1overexpressing cells. The results of Figure 5A–G indicate that SOX9 silencing also significantly decreased their proliferation, colony formation, migration, and invasion in Rac1 overexpressing HT‐29 cells, whereas SOX9 overexpression had dramatically opposite effects on Rac1 silencing SW620 cells (p < 0.05). A similar pattern was observed in measurement of glucose consumption, lactate production, G6P level, and G6PD activity (p < 0.05; Figures 5H–K and S2D–G). Furthermore, SOX9 silencing also rescued the Rac1 overexpression‐decreased NADP+/NADPH ratios (p < 0.05; Figures 5L and S2H). Western blot assays revealed that Rac1 overexpression enhanced HK2, PKM2, LDHA, G6PD, and 6PGD expression, which were mitigated or abrogated by SOX9 silencing (Figures 5M and S2I). Together, these data indicated that Rac1 enhances the glycolysis and PPP activity to promote colon cancer cell proliferation and metastasis through upregulating SOX9 expression.
FIGURE 5.
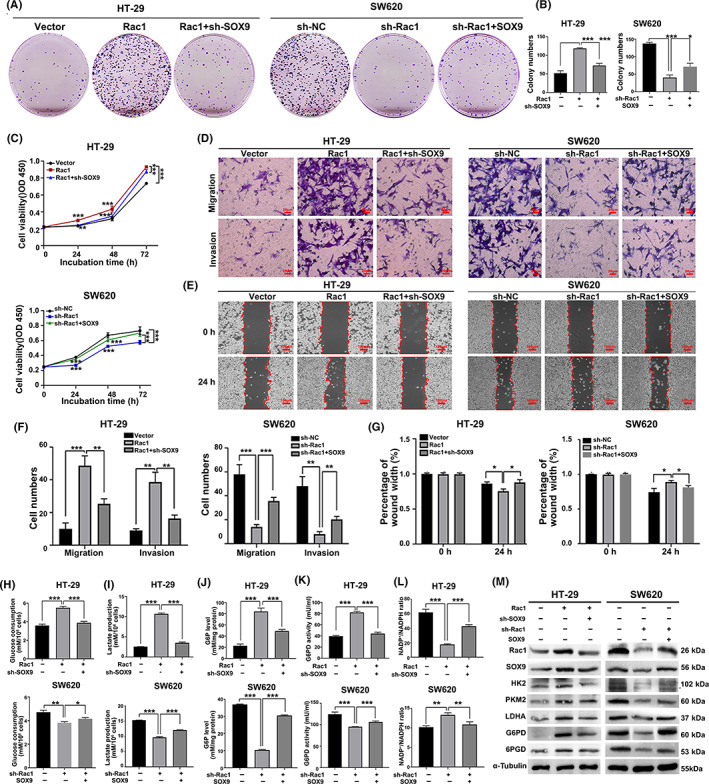
Rac1 promotes colon cancer malignant biological behaviors through SOX9. (A–C) SOX9 overexpression partially reversed the inhibitory effect of Rac1 silencing on colon cancer cell colony formation and proliferation. (D–G) SOX9 overexpression partially reversed the inhibitory effect of Rac1 silencing on invasion and migration of colon cancer cells (magnification 200×, scale bars 50 μm; magnification 400×, scale bars 20 μm). (H–L) Overexpression of SOX9 significantly restored glucose consumption, lactate production, increased intracellular glucose‐6‐phosphate (G6P) level and glucose‐6‐phosphate dehydrogenase (G6PD) activity, and decreased NADP+/NADPH ratio in Rac1‐silenced colon cancer cells. (M) Western blot analysis showed that Rac1 overexpression enhanced the expression of glycolysis‐related enzymes hexokinase 2 (HK2), pyruvate kinase isoenzyme 2 (PKM2), and lactate dehydrogenase A (LDHA), and pentose phosphate pathway‐related enzymes G6PD and 6‐phosphogluconate dehydrogenase (6PGD) in HT‐29 cells, which were attenuated or abolished by SOX9 silencing. Data are representative images or expressed as the mean ± SD of each group of samples analyzed in triplicate from three separate experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NC, negative control; NS, no statistical difference; OD, optical density.
3.6. Rac1 upregulates expression of SOX9 through the PI3K/AKT signaling pathway
We further investigated the mechanism through which Rac1 regulates SOX9 expression. Previous studies have reported that SOX9 is a downstream target gene of the PI3K/AKT signaling pathway. 28 , 29 Interestingly, Rac1 can activate the PI3K/AKT signaling pathway. 30 , 31 , 32 We thus speculated that Rac1 upregulates SOX9 expression by activating the PI3K/AKT signaling pathway. Indeed, we found that Rac1 overexpression significantly upregulated the expression of PI3K, p‐AKT (Ser473), and SOX9, whereas Rac1 interference significantly downregulated the protein expression of PI3K, p‐AKT, and SOX9 (Figure 6A). We cotreated Rac1‐overexpression/silencing colon cancer cells with10 μg/ml LY294002 (a PI3K/AKT pathway inhibitor) or 20 μg/ml 740Y‐P (an agonist of the PI3K/AKT signaling pathway). We found that LY294002 significantly inhibited the increased protein levels of SOX9 caused by Rac1 overexpression, while 740Y‐P significantly increased the decreased protein levels of SOX9 caused by Rac1 silencing (Figure 6B), suggesting that Rac1 promotes the expression of SOX9 protein depending on the activation of the PI3K/AKT signaling pathway.
FIGURE 6.
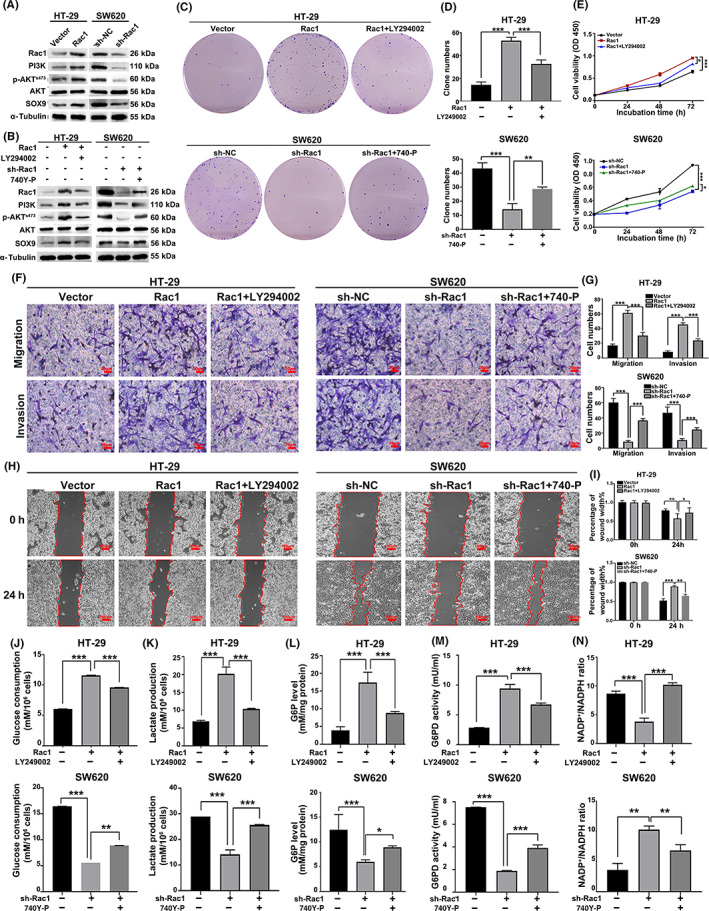
Rac1 upregulated the expression of SOX9 through the PI3K/AKT signaling pathway. (A) Western blot analysis was used to detect the effect of Rac1 overexpression or silencing on PI3K, p‐AKT (Ser473), and SOX9 protein expression. (B) Rac1 overexpressing/silencing colon cancer cells were treated with LY294002 (10 μg/ml), an inhibitor of the PI3K/AKT signaling pathway, or 740Y‐P (20 μg/ml), an agonist, to further detect PI3K, AKT, p‐AKT, and SOX9 protein expression. (C–I) Overexpression of Rac1 reversed the inhibitory effects of PI3K inhibitor LY294002 on colon cancer cell proliferation, invasion, and migration, whereas silencing Rac1 weakened the promoting effects of PI3K agonist 740Y‐P on colon cancer cell proliferation, invasion, and migration (magnification 200×, scale bars 50 μm; magnification 400×, scale bars 20 μm). (J–N) Overexpression of Rac1 partially reversed the inhibition of glucose consumption, lactate production, intracellular glucose‐6‐phosphate (G6P) level, glucose‐6‐phosphate dehydrogenase (G6PD) activity and the promoting effect on the intracellular NADP+/NADPH ratio by the PI3K inhibitor LY294002. Data are representative images or expressed as the mean ± SD of each group of samples analyzed in triplicate from three separate experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NC, negative control; NS, no statistical difference.
To further determine whether RAC1 can regulate the malignant biological behaviors of colon cancer through the PI3K/AKT signaling pathway, we treated colon cells with agonists or inhibitors of the PI3K/AKT signaling pathway. The results showed that LY294002 attenuated the promoting effect of Rac1 overexpression on proliferation, invasion, and migration of colon cancer cells, whereas 740Y‐P significantly reversed the inhibition of Rac1 silencing on colon cancer cell proliferation, invasion, and migration (p < 0.05; Figure 6C–I). In addition, LY294002 partially reversed the promotion of Rac1overexpression on glucose consumption, lactate production, intracellular G6P level, and G6PD activity and the promoting effect on intracellular NADP+/NADPH ratio, but 740Y‐P partially enhanced glucose consumption, lactate production, intracellular G6P level, and G6PD activity and decreased the intracellular NADP+/NADPH ratio in Rac1‐silencing colon cancer cells (p < 0.05; Figure 6J–N). Collectively, these results suggest that Rac1 increased SOX9 expression by activating the PI3K/AKT signaling pathway, and then promoted glycolysis and the PPP in colon cancer cells.
4. DISCUSSION
Colon cancer is one of the most common gastrointestinal malignancies with a high mortality rate. 33 Studies have shown that the main reason affecting the poor clinical prognosis of patients with colon cancer is metastasis, and the existing treatment can only achieve clinical cure in 10%–25% of patients with colon cancer metastasis. 34 Therefore, it is of great practical significance to deeply reveal the molecular mechanisms related to colon cancer metastasis and explore molecular biomarkers that can predict colon cancer metastasis for the prevention and treatment of colon cancer.
Rac1 acts as a “molecular switch” and is essential for cell behavior and regulation of multiple signaling pathways. Rac1 is abnormally activated in tumors, such as breast cancer, non‐small‐cell lung cancer, colon cancer, and liver cancer, and is involved in the regulation of the malignant biological behaviors of tumor cells. Metabolic reprogramming has been found to play an important role in the mechanisms through which Rac1 regulates tumor malignant proliferation, invasion, and migration and drug resistance. For example, Rac1 induces cisplatin resistance in esophageal squamous carcinoma cells by enhancing AKT/FOXO3a signaling and glycolytic processes. 35 In addition, it has been found that silencing of Rac1 can inhibit the nonoxidative pentose phosphate pathway, thereby overcoming chemoresistance in breast cancer. 36 , 37 Targeting the rapamycin (Rictor)/Rac1 pathway can inhibit IDH mutations, resulting in malignant biological behaviors due to metabolic reprogramming in gliomas. 38 Recent reports also indicated that Rac1 is involved in the regulation of glucose uptake in multiple ways. 39 , 40 Although many studies have reported that Rac1 is involved in the regulation of metabolic reprogramming in tumor cells, the specific mechanism remains unclear. Therefore, it is of great theoretical significance and clinical value to elaborate the regulation of Rac1 on metabolic reprogramming in tumor cells.
We have previously confirmed that Rac1 as an oncogene can promote epithelial–mesenchymal transition, invasion, and migration of colon cancer, but the specific mechanism needs further elucidation. 13 In this study, we investigated the relationship between Rac1 and the malignant biological phenotypes of colon cancer cells from the perspective of metabolic reprogramming. SOX9 is an important transcription factor closely related to digestive tract tumors, and encodes a protein with highly conserved HMG box sequence characteristics. 41 , 42 SOX9 is abnormally highly expressed in various malignancies, such as osteosarcoma, lung cancer, liver cancer, and breast cancer, and has a significant correlation with disease grade, stage, and prognosis. 27 , 43 SOX9 plays an oncogenic role in tumors and is involved in the regulation of important events, such as tumorigenesis, proliferation, migration, and chemoresistance by regulating signaling pathways, such as Wnt/β‐catenin, Hippo/YAP, and transforming growth factor‐β. 44 , 45 , 46 , 47 Hexokinase and G6PD, as the rate‐limiting enzymes of glycolysis and the PPP, respectively, can affect the occurrence and development of tumors by regulating the reprogramming of glucose metabolism in tumor cells. Herein, we confirmed that SOX9 upregulates the expression of HK2 and G6PD by binding to HK2 and G6PD promoters to enhance their transcriptional activity, but its specific binding site and regulatory mechanisms need to be further explored.
However, this article still has many deficiencies and shortcomings, which need further study. First, in the immunohistochemical analysis of HK2 in patient specimens, we observed that stromal cells are positively stained rather than cancer cells. Recent data indicated that cancer‐associated fibroblasts with upregulated HK2 expression could promote tumor progression and increase the maximum standard uptake value (SUVmax) of 18F‐FDG. 48 Various types of stromal cells (fibroblasts, lymphocytes, macrophages, and endothelial cells) with high expression of HK2 can establish a tumor microenvironment that promotes tumor angiogenesis and immune escape, 49 , 50 , 51 suggesting that the increased expression of HK2 in stromal cells might play an important role in tumor progression. Furthermore, we observed that SOX9 overexpression could increase the Rac1 expression in Rac1‐silencing colon cancer cells, suggesting that there is a mutual regulation relationship between Rac1 and SOX9. In this article, we initially showed that silencing Rac1 downregulated SOX9 expression by inactivating the PI3K/AKT pathway. However, as a transcription factor, SOX9 overexpression could translocate into the nucleus through the role of transporters and regulate the transcriptional expression of the Rac1 gene, ultimately increasing Rac1 protein expression. Whether SOX9 transcriptionally regulates Rac1 expression gives us a promising research direction, and exploring the positive feedback regulation loop of the Rac1‐SOX9 axis is necessary in further study.
In summary, this study found that Rac1 upregulated the expression of SOX9 through the PI3K/AKT signaling pathway, which in turn promoted glycolysis and the PPP by upregulating the expression of HK2 and G6PD, thereby promoting the development and progression of colon cancer (Figure 7). In recent years, targeting the relevant molecules in tumor metabolic reprogramming has become a new direction for the research and development of new antitumor drugs, that is, inhibiting the malignant proliferation, invasion, and metastasis of tumor cells by interfering with abnormal metabolic processes. 52 For example, inhibitors targeting IDH mutants are approved for the treatment of acute myelogenous leukemia with IDH mutations, 53 and various specific small molecule inhibitors such as HK2, PKM2, and LDHA are in clinical trials. 54 Therefore, given the relationship between Rac1 and SOX9 in this study, it is expected to provide an experimental basis for the clinical development of metabolic drugs against Rac1 and SOX9.
FIGURE 7.
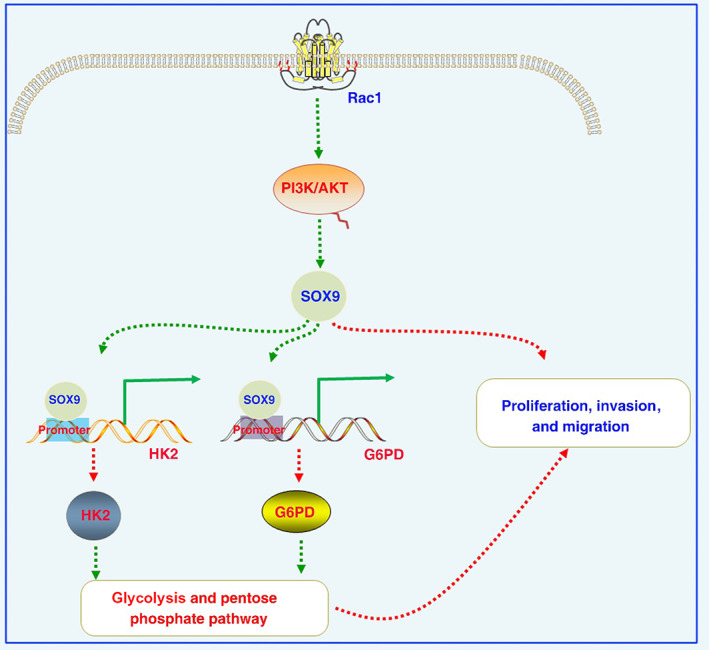
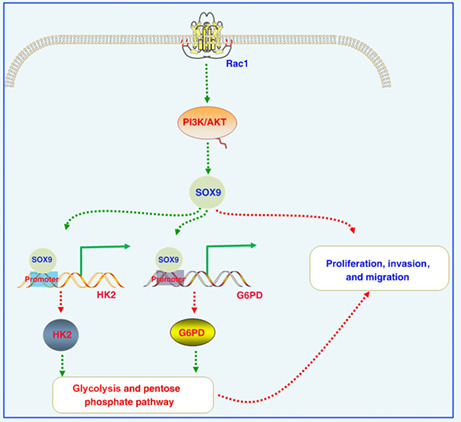
Increased levels of Rac1 activate the PI3K/AKT signaling pathway, thereby increasing SOX9 expression in colon cancer cells. Increased SOX9 expression activates the hexokinase (HK2) and glucose‐6‐phosphate dehydrogenase (G6PD) promoter and promotes the expression of HK2 and G6PD at the transcriptional level, then enhances glycolysis and the pentose phosphate pathway to enhance proliferation, invasion, and migration. We further revealed novel regulatory axes involving Rac1/SOX9/HK2/G6PD in the development and progression of colon cancer, providing novel promising therapeutic targets.
AUTHOR CONTRIBUTIONS
Q. Liao and Y. Zhou conceived and designed all experiments. J. Liang and L. Xia performed experiments, collected and analyzed the data, and wrote the manuscript. Q. Liu, L. Oyang, Y. Tang, M. Su, N. Wu, and X. Jiang collected and analyzed the data. Q. Liu, J. Lin, S. Tan, Q. Peng, X. Xu, X. Luo, and Y. Yang performed experiments. Q. Liu provided the clinical samples and reagents. All authors critically reviewed and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an institutional review board: The use of human tissue and experimental animals in this study was approved by the Joint Ethics Committee of the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University and Hunan Cancer Hospital.
Informed consent: Informed consent was obtained from all participants.
Registry and registration no. of the study/trial: N/A.
Animal studies: The use of experimental animals in this study was approved by the Joint Ethics Committee of the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University and Hunan Cancer Hospital. The ethical approval number is KYJJ‐2018‐019.
CONSENT FOR PUBLICATION
Not applicable.
Supporting information
Figure S1.
Figure S2.
Appendix S1.
ACKNOWLEDGMENTS
This work was supported in part by grants from the following sources: the National Natural Science Foundation of China (82173142, 81972636, 81872281), the Natural Science Foundation of Hunan Province (2020JJ5336, 2019JJ40175, 2019JJ40183), the Research Project of Health Commission of Hunan Province (202204085344, 202204085195, 202109031837, 202109032010, 20201020), China Hunan Provincial Science and Technology Department (2020TP1018), Ascend Foundation of National cancer center (NCC201909B06, NCC2018b68), Hunan Cancer Hospital Climb Plan (ZX2020001‐3, YF2020002), and the Fundamental Research Funds for the Central Universities of Central South University (2019zzts832, 2019zzts833).
Liang J, Liu Q, Xia L, et al. Rac1 promotes the reprogramming of glucose metabolism and the growth of colon cancer cells through upregulating SOX9 . Cancer Sci. 2023;114:822‐836. doi: 10.1111/cas.15652
Jiaxin Liang, Qiang Liu, and Longzheng Xia contributed equally to this work.
Contributor Information
Qianjin Liao, Email: march-on@126.com.
Yujuan Zhou, Email: zhouyujuan@hnca.org.cn.
DATA AVAILABILITY STATEMENT
The public datasets analyzed during the current study are available in the repositories listed below: Gene Expression Omnibus GSE78093: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78093
REFERENCES
- 1. Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713‐732. doi: 10.1038/s41575-019-0189-8 [DOI] [PubMed] [Google Scholar]
- 2. Goss PE, Strasser‐Weippl K, Lee‐Bychkovsky BL, et al. Challenges to effective cancer control in China, India, and Russia. Lancet Oncol. 2014;15(5):489‐538. doi: 10.1016/S1470-2045(14)70029-4 [DOI] [PubMed] [Google Scholar]
- 3. Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33(16):1787‐1796. doi: 10.1200/JCO.2014.60.0213 [DOI] [PubMed] [Google Scholar]
- 4. Benitah SA, Valeron PF, van Aelst L, Marshall CJ, Lacal JC. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim Biophys Acta. 2004;1705(2):121‐132. doi: 10.1016/j.bbcan.2004.10.002 [DOI] [PubMed] [Google Scholar]
- 5. Bosco EE, Mulloy JC, Zheng Y. Rac1 GTPase: a "Rac" of all trades. Cell Mol Life Sci. 2009;66(3):370‐374. doi: 10.1007/s00018-008-8552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509‐514. doi: 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- 7. Recouvreux MV, Commisso C. Macropinocytosis: a metabolic adaptation to nutrient stress in cancer. Front Endocrinol (Lausanne). 2017;8:261. doi: 10.3389/fendo.2017.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leng R, Liao G, Wang H, Kuang J, Tang L. Rac1 expression in epithelial ovarian cancer: effect on cell EMT and clinical outcome. Med Oncol. 2015;32(2):329. doi: 10.1007/s12032-014-0329-5 [DOI] [PubMed] [Google Scholar]
- 9. Lin Y, Fu F, Lv J, et al. Identification of potential key genes for HER‐2 positive breast cancer based on bioinformatics analysis. Medicine (Baltimore). 2020;99(1):e18445. doi: 10.1097/MD.0000000000018445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopez‐Cortes A, Paz YMC, Guerrero S, et al. OncoOmics approaches to reveal essential genes in breast cancer: a panoramic view from pathogenesis to precision medicine. Sci Rep. 2020;10(1):5285. doi: 10.1038/s41598-020-62279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Venugopal SV, Caggia S, Gambrell‐Sanders D, Khan SA. Differential roles and activation of mammalian target of rapamycin complexes 1 and 2 during cell migration in prostate cancer cells. Prostate. 2020;80(5):412‐423. doi: 10.1002/pros.23956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Y, Zhao Y, Huan L, et al. An LTR retrotransposon‐derived long noncoding RNA lncMER52A promotes hepatocellular carcinoma progression by binding p120‐catenin. Cancer Res. 2020;80(5):976‐987. doi: 10.1158/0008-5472.CAN-19-2115 [DOI] [PubMed] [Google Scholar]
- 13. Xia L, Lin J, Su J, et al. Diallyl disulfide inhibits colon cancer metastasis by suppressing Rac1‐mediated epithelial‐mesenchymal transition. Onco Targets Ther. 2019;12:5713‐5728. doi: 10.2147/OTT.S208738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merajver SD, Usmani SZ. Multifaceted role of rho proteins in angiogenesis. J Mammary Gland Biol Neoplasia. 2005;10(4):291‐298. doi: 10.1007/s10911-006-9002-8 [DOI] [PubMed] [Google Scholar]
- 15. Toyama Y, Kontani K, Katada T, Shimada I. Conformational landscape alternations promote oncogenic activities of Ras‐related C3 botulinum toxin substrate 1 as revealed by NMR. Sci Adv. 2019;5(3):eaav8945. doi: 10.1126/sciadv.aav8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang FS, Wang CJ, Chen YJ, et al. Ras induction of superoxide activates ERK‐dependent angiogenic transcription factor HIF‐1alpha and VEGF‐A expression in shock wave‐stimulated osteoblasts. J Biol Chem. 2004;279(11):10331‐10337. doi: 10.1074/jbc.M308013200 [DOI] [PubMed] [Google Scholar]
- 17. Arnold CR, Abdelmoez A, Thurner G, et al. Rac1 as a multifunctional therapeutic target to prevent and combat cancer metastasis. Onco Targets Ther. 2014;1(8):513‐521. doi: 10.18632/oncoscience.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:6487. doi: 10.1126/science.aaw5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. 2019;95(7):912‐919. doi: 10.1080/09553002.2019.1589653 [DOI] [PubMed] [Google Scholar]
- 20. Zhou Y, Zhou Y, Shingu T, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286(37):32843‐32853. doi: 10.1074/jbc.M111.260935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang W, Zheng Y, Xia Y, et al. ERK1/2‐dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295‐1304. doi: 10.1038/ncb2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang W, Xia Y, Cao Y, et al. EGFR‐induced and PKCepsilon monoubiquitylation‐dependent NF‐kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48(5):771‐784. doi: 10.1016/j.molcel.2012.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Estrella V, Chen T, Lloyd M, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73(5):1524‐1535. doi: 10.1158/0008-5472.CAN-12-2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen J, Ye C, Dong J, et al. Metabolic classification of circulating tumor cells as a biomarker for metastasis and prognosis in breast cancer. J Transl Med. 2020;18(1):59. doi: 10.1186/s12967-020-02237-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo Q, Ren XH, Zhao CP, Zhang BH. MicroRNA‐206 inhibits tumor metastasis of nasopharyngeal carcinoma through targeting G6PD. J Biol Regul Homeost Agents. 2020;34(2):479. doi: 10.23812/20-36A [DOI] [PubMed] [Google Scholar]
- 26. Zhou Y, Liao Q, Han Y, et al. Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non‐small cell lung cancer. J Cancer. 2016;7(14):2100‐2109. doi: 10.7150/jca.16198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sashikawa Kimura M, Mutoh H, Sugano K. SOX9 is expressed in normal stomach, intestinal metaplasia, and gastric carcinoma in humans. J Gastroenterol. 2011;46(11):1292‐1299. doi: 10.1007/s00535-011-0443-5 [DOI] [PubMed] [Google Scholar]
- 28. Cheng CC, Uchiyama Y, Hiyama A, Gajghate S, Shapiro IM, Risbud MV. PI3K/AKT regulates aggrecan gene expression by modulating Sox9 expression and activity in nucleus pulposus cells of the intervertebral disc. J Cell Physiol. 2009;221(3):668‐676. doi: 10.1002/jcp.21904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suryo Rahmanto A, Savov V, Brunner A, et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J. 2016;35(20):2192‐2212. doi: 10.15252/embj.201693889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qu H, Sun H, Wang X. Neogenin‐1 promotes cell proliferation, motility, and adhesion by up‐regulation of zinc finger E‐box binding Homeobox 1 via activating the Rac1/PI3K/AKT pathway in gastric cancer cells. Cell Physiol Biochem. 2018;48(4):1457‐1467. doi: 10.1159/000492255 [DOI] [PubMed] [Google Scholar]
- 31. Yang Y, Du J, Hu Z, et al. Activation of Rac1‐PI3K/Akt is required for epidermal growth factor‐induced PAK1 activation and cell migration in MDA‐MB‐231 breast cancer cells. J Biomed Res. 2011;25(4):237‐245. doi: 10.1016/S1674-8301(11)60032-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin CH, Cheng HW, Ma HP, Wu CH, Hong CY, Chen BC. Thrombin induces NF‐kappaB activation and IL‐8/CXCL8 expression in lung epithelial cells by a Rac1‐dependent PI3K/Akt pathway. J Biol Chem. 2011;286(12):10483‐10494. doi: 10.1074/jbc.M110.112433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benson AB, Venook AP, Al‐Hawary MM, et al. NCCN guidelines insights: colon cancer, version 2.2018. J Natl Compr Canc Netw. 2018;16(4):359‐369. doi: 10.6004/jnccn.2018.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andre T, Boni C, Mounedji‐Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350(23):2343‐2351. doi: 10.1056/NEJMoa032709 [DOI] [PubMed] [Google Scholar]
- 35. Zeng RJ, Zheng CW, Gu JE, et al. RAC1 inhibition reverses cisplatin resistance in esophageal squamous cell carcinoma and induces downregulation of glycolytic enzymes. Mol Oncol. 2019;13(9):2010‐2030. doi: 10.1002/1878-0261.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ganapathy‐Kanniappan S. Rac1 repression reverses chemoresistance by targeting tumor metabolism. Cancer Biol Ther. 2020;21(10):888‐890. doi: 10.1080/15384047.2020.1809923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Q, Qin T, Bi Z, et al. Rac1 activates non‐oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun. 2020;11(1):1456. doi: 10.1038/s41467-020-15308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y, Lu Y, Li A, et al. mTORC2/Rac1 pathway predisposes cancer aggressiveness in IDH1‐mutated glioma. Cancers (Basel). 2020;12(4):787. doi: 10.3390/cancers12040787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Audzeyenka I, Rogacka D, Rachubik P, et al. The PKGIalpha‐Rac1 pathway is a novel regulator of insulin‐dependent glucose uptake in cultured rat podocytes. J Cell Physiol. 2021;236(6):4655‐4668. doi: 10.1002/jcp.30188 [DOI] [PubMed] [Google Scholar]
- 40. Yue Y, Zhang C, Zhang X, et al. An AMPK/Axin1‐Rac1 signaling pathway mediates contraction‐regulated glucose uptake in skeletal muscle cells. Am J Physiol Endocrinol Metab. 2020;318(3):E330‐E342. doi: 10.1152/ajpendo.00272.2019 [DOI] [PubMed] [Google Scholar]
- 41. Grimm D, Bauer J, Wise P, et al. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2020;67(Pt 1):122‐153. doi: 10.1016/j.semcancer.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 42. Zhou T, Wu L, Ma N, et al. SOX9‐activated FARSA‐AS1 predetermines cell growth, stemness, and metastasis in colorectal cancer through upregulating FARSA and SOX9. Cell Death Dis. 2020;11(12):1071. doi: 10.1038/s41419-020-03273-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nouri M, Massah S, Caradec J, et al. Transient Sox9 expression facilitates resistance to androgen‐targeted therapy in prostate cancer. Clin Cancer Res. 2020;26(7):1678‐1689. doi: 10.1158/1078-0432.CCR-19-0098 [DOI] [PubMed] [Google Scholar]
- 44. Jana S, Madhu Krishna B, Singhal J, et al. SOX9: the master regulator of cell fate in breast cancer. Biochem Pharmacol. 2020;174:113789. doi: 10.1016/j.bcp.2019.113789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qi G, Li L. Long non‐coding RNA PVT1 contributes to cell growth and metastasis in non‐small‐cell lung cancer by regulating miR‐361‐3p/SOX9 axis and activating Wnt/beta‐catenin signaling pathway. Biomed Pharmacother. 2020;126:110100. doi: 10.1016/j.biopha.2020.110100 [DOI] [PubMed] [Google Scholar]
- 46. Zhou H, Li G, Huang S, Feng Y, Zhou A. SOX9 promotes epithelial‐mesenchymal transition via the hippo‐YAP signaling pathway in gastric carcinoma cells. Oncol Lett. 2019;18(1):599‐608. doi: 10.3892/ol.2019.10387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chao M, Liu N, Sun Z, et al. TGF‐beta signaling promotes glioma progression through stabilizing Sox9. Front Immunol. 2020;11:592080. doi: 10.3389/fimmu.2020.592080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim BG, Sung JS, Jang Y, et al. Compression‐induced expression of glycolysis genes in CAFs correlates with EMT and angiogenesis gene expression in breast cancer. Commun Biol. 2019;2:313. doi: 10.1038/s42003-019-0553-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo D, Tong Y, Jiang X, et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2‐mediated phosphorylation of IkappaBalpha. Cell Metab. 2022;34(9):1312‐24 e6. doi: 10.1016/j.cmet.2022.08.002 [DOI] [PubMed] [Google Scholar]
- 50. Meng YM, Jiang X, Zhao X, et al. Hexokinase 2‐driven glycolysis in pericytes activates their contractility leading to tumor blood vessel abnormalities. Nat Commun. 2021;12(1):6011. doi: 10.1038/s41467-021-26259-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dunn J, Ferluga S, Sharma V, et al. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine. 2019;40:77‐91. doi: 10.1016/j.ebiom.2018.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Munger J, Bennett BD, Parikh A, et al. Systems‐level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26(10):1179‐1186. doi: 10.1038/nbt.1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waitkus MS, Diplas BH, Yan H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell. 2018;34(2):186‐195. doi: 10.1016/j.ccell.2018.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30(7):671‐678. doi: 10.1038/nbt.2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Appendix S1.
Data Availability Statement
The public datasets analyzed during the current study are available in the repositories listed below: Gene Expression Omnibus GSE78093: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78093


