Abstract
Waning immunity and emerging variants necessitate continued vaccination against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Improvements in vaccine safety, tolerability, and ease of manufacturing would benefit these efforts. Here, we develop a potent and easily manufactured nanoparticle vaccine displaying the spike receptor-binding domain (RBD). Computational design to stabilize the RBD, eliminate glycosylation, and focus the immune response to neutralizing epitopes results in an RBD immunogen that resolves issues hindering the efficient nanoparticle display of the native RBD. This non-glycosylated RBD can be genetically fused to diverse single-component nanoparticle platforms, maximizing manufacturing ease and flexibility. All engineered RBD nanoparticles elicit potently neutralizing antibodies in mice that far exceed monomeric RBDs. A 60-copy particle (noNAG-RBD-E2p) also elicits potently neutralizing antibodies in non-human primates. The neutralizing antibody titers elicited by noNAG-RBD-E2p are comparable to a benchmark stabilized spike antigen and reach levels against Omicron BA.5 that suggest that it would provide protection against emerging variants.
Keywords: structure-based design, structural vaccinology, immunogen, nanoparticle, glycosylation, stabilized, SPEEDesign
Graphical abstract
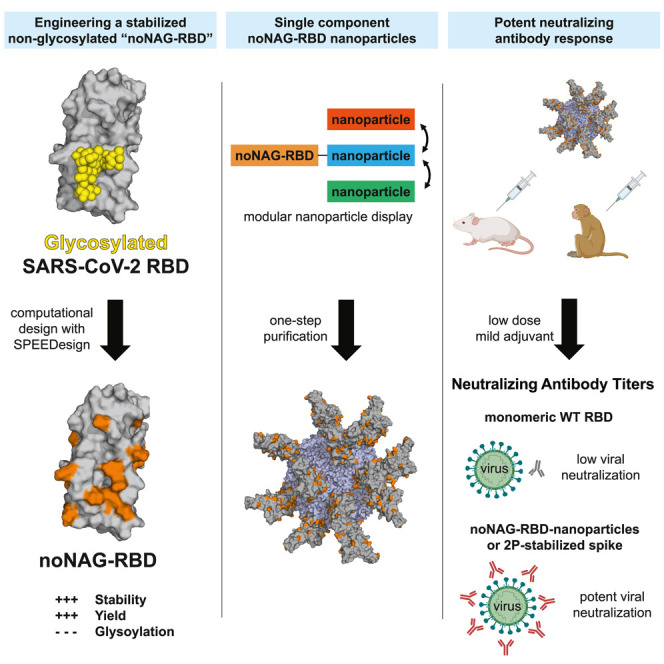
SARS-CoV-2 RBD vaccines are cost effective but require methods to increase their potency. Dickey et al. use a computational design method to create a stabilized non-glycosylated RBD that can be fused to a nanoparticle carrier, boosting its potency to levels of a gold-standard spike antigen.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines have successfully curbed disease, but, globally, thousands of people still died from COVID daily in 2022.1 These deaths largely occured in unvaccinated and at-risk populations. Increased global access to vaccines would protect those who are unvaccinated, and periodic boosters would protect at-risk populations. A vaccine that is highly effective, easily produced and distributed, low cost, and extremely safe is needed to meet this ongoing global demand for SARS-CoV-2 control. Cost and supply issues limit the distribution of the most effective vaccines, while efficacy and safety concerns limit the use of other vaccines. Furthermore, the reactogenicity of mRNA vaccines may limit their use as regular boosters.
Almost all authorized vaccines utilize a heavily glycosylated full-length spike (FL-S) protein antigen, but the receptor-binding domain (RBD) of the spike protein may be an equally efficacious and more easily produced alternative.2 , 3 The RBD binds the angiotensin-converting enzyme-2 (ACE2) receptor to mediate viral entry into the host cell. Consequently, it is the target of the most potent neutralizing antibodies against SARS-CoV-2.4 , 5 Two vaccines containing an RBD antigen produced in yeast have proven effective in clinical trials and are now authorized for use.6 , 7 Production of the recombinant RBD in yeast requires removal of the N-linked glycosylation site at Asn331.8 However, this construct retains an N-linked glycosylation site at Asn343, which could not be removed in the homologous SARS-RBD.9 The composition of the glycans at Asn343 depends on many factors, and the glycoprotein characteristics can vary between expression platforms and between batches.10 Variability in glycan composition can have a major impact on the immunogenicity, stability, solubility, safety, and efficacy of a vaccine, leading to challenges associated with the production of homogeneous reproducible glycoprotein vaccines. Removing the N-linked glycosylation site in the RBD has the potential to improve the homogeneity, reproducibility, and efficacy of a SARS-CoV-2 vaccine candidate.
The efficacy of RBD-based vaccines can also be dramatically improved through nanoparticle display.11 This concept has been demonstrated using two-component nanoparticle platforms where a purified RBD subunit is mixed with a separately purified nanoparticle subunit.12 , 13 , 14 , 15 However, two-component platforms require optimization of multiple purification steps and an assembly step, which complicate the manufacturing process and reduce the final yield of the desired product. In contrast, single-component nanoparticles do not suffer from these complications. A single-component 24-copy RBD nanoparticle can be produced, and it elicited broad and potent immunity in mice, illustrating the potential of RBD nanoparticle antigens.16 However, production of this nanoparticle required decreased production temperatures, stabilizing mutations, and stabilizing excipients, suggesting that this product may not be ideally suited for large-scale production and distribution.16
Here, we modified a ROSETTA-based computational design and screening method called SPEEDesign to produce multiple stable, single-component, non-glycosylated RBD nanoparticles.17 SPEEDesign stabilizes protein antigens and focuses the immune response to potently neutralizing epitopes.17 , 18 We identified amino acid changes that stabilize a non-glycosylated RBD, facilitating fusion to a variety of nanoparticle platforms. Immunization of mice with any of these nanoparticles elicited neutralizing antibody titers significantly higher than monomeric wild-type (WT) RBDs and at levels comparable to a benchmark spike antigen. A 60-copy nanoparticle also elicited high neutralizing antibody titers in naive non-human primates (NHPs) and effectively boosted titers in previously immunized NHPs. These non-glycosylated nanoparticles can be produced at high yields, providing a vaccine candidate capable of meeting the ongoing global demand for SARS-CoV-2 vaccines.
Results
Design of a non-glycosylated RBD immunogen using noNAG-SPEEDesign
We sought to produce a self-assembling, single-component RBD nanoparticle vaccine that would be easily produced and be highly immunogenic. Fusion of the RBD to a ferritin nanoparticle subunit, however, resulted in very low expression, consistent with previous reports (Figure S1A).16 Changing the nanoparticle carrier or the linker between the RBD and nanoparticle did not substantially increase expression (data not shown). We hypothesized that the glycosylated RBD prevents nanoparticle expression and assembly due to steric clash of bulky glycans upon high density display on a nanoparticle and that non-glycosylated RBDs would more readily assemble into nanoparticles.
Non-glycosylated RBDs cannot be produced by making single amino acid changes. The simplest manner of preventing N-linked glycosylation is to mutate the NxS/T amino acid motif recognized by the oligosaccharyltransferase complex. However, mutation of this motif at Asn343 of the RBD prevents protein expression (Figure S1B). Changing Thr345 to alanine eliminates this motif and significantly reduces expression in expi293F cells. This result is consistent with deep mutational scanning (DMS) data that show a significant decrease in yeast expression upon mutation of either residue that comprises the NxS/T motif.19 The dependence of protein expression on this motif suggests that the glycan plays a stabilizing role structurally.
We hypothesized that the decrease in protein expression upon removal of the NxS/T motif is caused by a decrease in thermodynamic stability of the RBD, and additional amino acid changes to the residues surrounding the glycan could compensate for the lost glycan to restore protein expression. We modified the previously reported SPEEDesign computational design pipeline17 by incorporating DMS data19 to create a non-glycosylated RBD immunogen (noNAG-SPEEDesign). The SPEEDesign pipeline was previously shown to focus the immune response toward neutralizing epitopes and stabilize the structure and conformation of a target antigen.17 , 18 Here, we stabilized the area around the glycan, and we focused the immune response to the ACE2-binding site because the most potent neutralizing antibodies are those that block ACE2.4 , 5
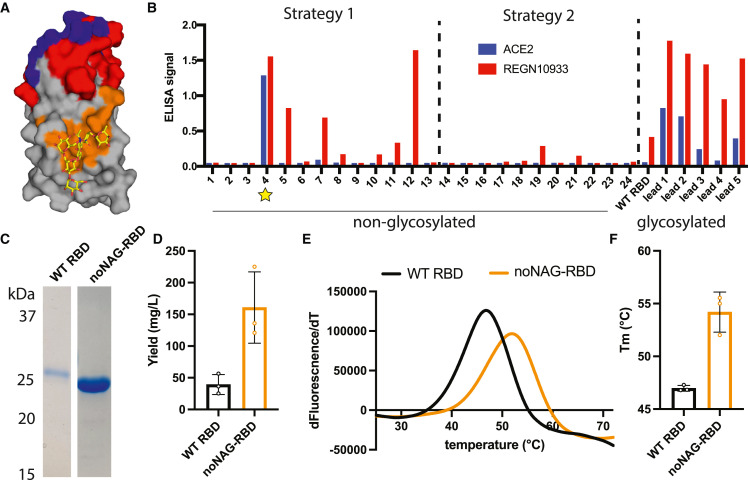
A total of 20,000 noNAG-SPEEDesign computational decoys were produced from the WA-1 RBD sequence using the two design strategies, and 24 sequences were selected to sample the top-scoring decoys in a sparse-matrix fashion (Figure 1 ). The first design strategy forced the ACE2-binding site to remain unchanged, and the second strategy expanded this definition to all known blocking epitopes (Figure 1A). Twenty-four sequences were synthesized and expressed in expi293F cell culture, and the cell-free supernatant was screened by ELISA for the presence of designed immunogens that retain structural integrity and binding to ACE2 and the neutralizing antibody REGN10933 (Figure 1B). One non-glycosylated RBD immunogen (decoy 4) bound both ACE2 and REGN10933 with reactivity far greater than WT RBDs. This construct is not the top-scoring ROSETTA decoy, which is consistent with our earlier SPEEDesign results and underscores the importance of the SPEEDesign clustering and screening process.17 , 18 The ELISA signal for decoy 4 is comparable to ELISA signals observed for optimized glycosylated immunogens previously reported (leads 1–5), suggesting that this non-glycosylated immunogen can be expressed at high levels with neutralizing epitopes intact.17
Figure 1.
Design of a non-glycosylated RBD immunogen
(A) Design strategy for a non-glycosylated RBD immunogen in which the epitopes of ACE2-blocking antibodies (red) and/or the ACE2-binding site (blue) are fixed, while residues contacting the N-glycan (orange/yellow) are allowed to sample a diverse sequence space. Intermediate residues (gray) are constrained by DMS data.
(B) An ELISA expression screen identifies one non-glycosylated immunogen (star) that presents ACE2 and REGN10933 neutralizing epitopes and is expressed at levels comparable to previously engineered glycosylated immunogens (leads 1–5).
(C) Reducing SDS-PAGE of purified WT RBD and noNAG-RBD illustrates the increased yield and decreased size of the immunogen. Samples were run on the same gel, and intervening irrelevant lanes were removed.
(D) Purification yields for WT RBD and noNAG-RBD. Each point is a biological replicate, and bars represent mean ± standard deviation.
(E and F) Representative DSF melt curves showing the mean values of four wells for one biological replicate (E) and a summary of biological replicates (F), demonstrating an increased thermostability of noNAG-RBD relative to WT RBD. Bars represent mean ± standard deviation.
See also Figure S2.
The non-glycosylated RBD immunogen (noNAG-RBD), or decoy 4, is stabilized by changes primarily clustered around the glycan site, which presumably compensate for the absence of the structurally stabilizing glycan (Figure S2). Four of these changes (G339E, N343H, A363W, V367L) are predicted space-filling mutations in a cavity previously occupied by the N343 glycan (Figure S2C). Another six changes were made around the two helices that contact the glycan (L335P, V341I, T345E, Y365F, Y369L, A372K) (Figure S2D). Four of these changes are conservative hydrophobic amino acid changes that likely improve packing, while the other two changes introduce charged residues on the surface that may improve hydrophilicity of the molecule. There are also changes to distal surfaces that may promote stability but do not ablate neutralizing epitopes (vide infra). Only four changes in noNAG-RBD overlap with mutations in Omicron and other variants, and introduction of noNAG-RBD changes to the variant sequences should facilitate production of non-glycosylated variant RBDs as well.
Non-glycosylated RBD immunogen expression and stability exceed WT RBD
Recombinant noNAG-RBD was expressed and purified from expi293F cells to validate the biophysical characteristics of the purified protein. Purified noNAG-RBD is homogeneous by SDS-PAGE and migrates at a molecular weight consistent with a non-glycosylated RBD that is smaller than the glycosylated WT RBD (Figure 1C). Furthermore, the final purification yields for noNAG-RBD are over 150 mg/L, a greater than 4-fold increase over WT RBDs (Figure 1D). We measured the thermostability of noNAG-RBD by differential scanning fluorimetry (DSF) to determine if the stabilizing mutations fully compensated for removal of the glycan. noNAG-RBD has a melting temperature of 54°C, which is 7°C higher than WT RBD and similar to the thermostabilized glycosylated RBD immunogens previously reported (Figures 1E and 1F).17 This stabilization is likely achieved by the set of mutations described above, which include space-filling mutations that compensate for the removed glycan and mutations that improve hydrophobic packing and surface hydrophilicity. Thus, noNAG-RBD is a homogeneous immunogen with improved expression and stability.
noNAG-RBD can be assembled into single-component nanoparticles
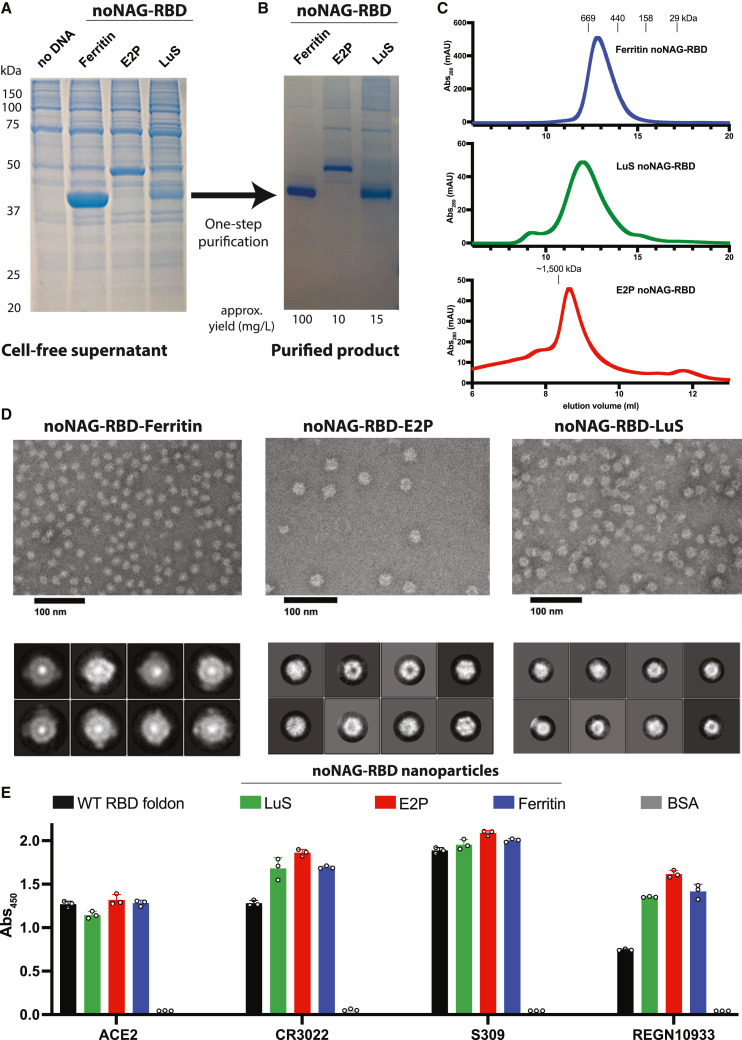
We genetically fused noNAG-RBD to three nanoparticle platforms to test our hypothesis that removal of the N-glycan promotes high-density nanoparticle display. Indeed, noNAG-RBD is highly expressed as a fusion with all nanoparticle platforms tested (Figure 2 A). These untagged fusion proteins can be purified at high yields by size-exclusion chromatography and elute at a volume consistent with the formation of intact nanoparticles (Figures 2B and 2C). Negative-stain electron microscopy established the formation of high-symmetry homogeneous nanoparticles (Figure 2D). While the symmetry cannot be directly concluded from these negative-stain images, our data are consistent with previously reported assemblies of these particles. H. pylori ferritin is a 24-copy nanoparticle with 2-, 3-, and 4-fold symmetry.20 Our construct also contains an N-terminal extension from bullfrog ferritin and mutations to eliminate N-glycosylation and preserve a salt bridge, as described previously.21 G. stearothermophilus dihydrolipoyl acetyltransferase (E2p) is a 60-copy nanoparticle with 2-, 3-, and 5-fold symmetry, and our construct is truncated to include only the oligomerization domain attached to noNAG-RBD by a 15 amino acid linker.22 , 23 , 24 A. aeolicus lumazine synthase (LuS) is also a 60-copy nanoparticles with 2-, 3-, and 5-fold symmetry, and our construct contains an engineered N-glycosylation site to improve expression, as described previously.25 , 26 The ability of noNAG-RBD to assemble on all of these platforms suggests that it is a modular antigen that can be utilized in conjunction with a variety of platforms, facilitating future optimization according to immunogenicity, production, licensing, and safety requirements.
Figure 2.
noNAG-RBD assembles into single-component nanoparticles that present neutralizing epitopes
(A) SDS-PAGE of cell-free supernatant from expi293F expression demonstrates that single-component noNAG-RBD nanoparticles have high expression levels.
(B and C) Particles can be purified at high yields be size-exclusion chromatography (B) and elute at sizes consistent with particle formation (C).
(D) Negative-stain electron microscopy (EM) images (top) and 2D class averages (bottom) of purified noNAG-RBD nanoparticles demonstrate efficient and homogeneous assembly.
(E) ELISAs on purified noNAG-RBD nanoparticles with neutralizing antibodies demonstrate the accessibility of neutralizing epitopes. Individual points are replicates, and bars represent mean ± standard deviation.
We probed several neutralizing epitopes on noNAG-RBD nanoparticles by ELISA to determine if the amino acid changes or nanoparticle assembly perturb key functional surfaces (Figure 2E). We used a sandwich ELISA with REGN10933 Fab coating the plate and REGN10933 monoclonal antibody (mAb) serving as a probe, meaning that only multivalent particles would be detected. All noNAG-RBD particles bound to REGN10933 better than a trimerized WT RBD control. ACE2 and the mAbs CR3022 and S309 were also used as probes that recognize distinct neutralizing epitopes covering the majority of the RBD surface. All these probes bind the noNAG-RBD nanoparticles, suggesting that these vaccine candidates would elicit strong neutralizing responses against SARS-CoV-2.
noNAG-RBD nanoparticles elicit high titers of neutralizing antibodies in mice
We immunized mice with noNAG-RBD nanoparticles or WT RBD monomer to determine if noNAG-RBD nanoparticles improve the antibody response against SARS-CoV-2 (Figure 3 ). We also immunized a group of mice with a trimeric FL-S antigen (FL-S-2P) similar to that used in approved vaccines to serve as a benchmark. This antigen comprises the WA-1 sequence of the complete spike ectodomain, it contains two commonly used proline mutations that promote the pre-fusion conformation, and it has a mutation that eliminates a furin protease cleavage site. We used CD-1 outbred mice to mimic the genetic diversity of humans more closely than inbred mice. Antigens were formulated in AddaVax and administered by subcutaneous injection in a prime/boost regimen with a low dose of protein (1 μg) to recapitulate real-world vaccine administration (Figure 3A).
Figure 3.
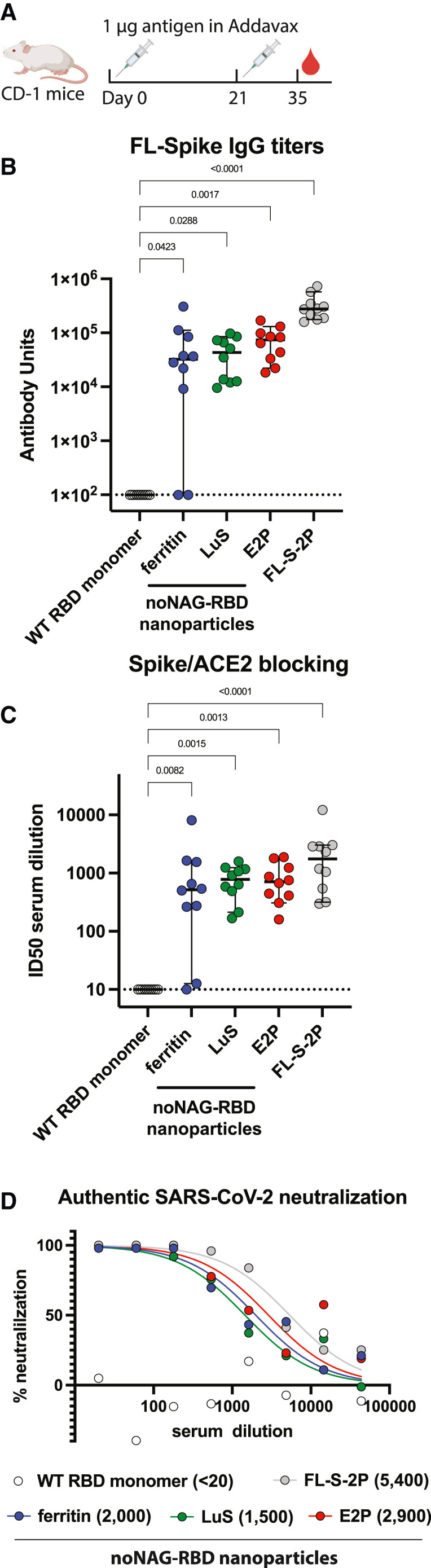
noNAG-RBD nanoparticles elicit potent neutralizing antibodies
(A) Prime/boost immunization schedule of CD-1 mice with 1-μg doses of antigen formulated in AddaVax.
(B) IgG antibody titers at day 35 against FL-spike demonstrating that noNAG-RBD nanoparticles elicit high titers while monomeric RBD does not. Dashed line indicates detection limit of assay, each point is the average of triplicate measurements of a single animal, and bars represent median titers with 95% confidence interval. Statistical comparisons were made using a Kruskal-Wallis ANOVA followed by Dunn’s comparison with WT RBD, corrected for multiple comparisons. n = 10 animals per group.
(C) Spike-/ACE2-binding inhibition assay demonstrating that noNAG-RBD nanoparticles elicit significantly higher titers of functional antibodies than monomeric WT RBD. Displayed as described for (B).
(D) Neutralization of authentic WA-1 SARS-CoV-2 by plaque assay using pooled serum. Each point is the average of duplicate wells. PRNT50 values are indicated in parentheses.
See also Figures S3 and S4.
noNAG-RBD nanoparticles elicit high titers of immunoglobulin G (IgG) antibodies that recognize FL-S-2P (Figure 3B). In contrast, the WT RBD did not elicit detectable antibody titers. The enhanced immunogenicity of the noNAG-RBD nanoparticles is even more pronounced when taking into account the RBD-specific dose administered, which is 43 pmol WT RBD compared with 23, 25, and 20 pmol noNAG-RBD on the ferritin, LuS, and E2p particles, respectively. The anti-FL-S-2P titers elicited by noNAG-RBD nanoparticles are slightly lower than those elicited by FL-S-2P, while the anti-WT-RBD titers are comparable (Figures 3B and S3A). The ratio of anti-WT RBD to anti-FL-S-2P titers elicited by the nanoparticles is significantly higher than the ratio elicited by FL-S-2P, indicating that the nanoparticles focus the immune response toward the RBD, which contains the most potently neutralizing epitopes (Figure S3B). The anti-noNAG-RBD antibody titers elicited by noNAG-RBD nanoparticles were slightly higher than those elicited by FL-S-2P, suggesting that distinct antigenic surfaces may be introduced by mutation or exposed by removing the glycan shield (Figure S4).
We measured the functional activity of these antibodies using blocking and neutralization assays. noNAG-RBD nanoparticles elicit potent spike-/ACE2-blocking antibody titers, while monomeric RBD has no detectable blocking activity (Figure 3C). Furthermore, the noNAG-RBD nanoparticle blocking antibody geometric mean titers (GMTs) are within ∼3-fold of the potent FL-S-2P antigen. All noNAG-RBD nanoparticle blocking titers are significantly higher than WT RBD, but E2p and LuS nanoparticles more consistently elicit high titers across all animals than ferritin nanoparticles. Finally, we pooled serum from each animal in a group and measured neutralization of authentic SARS-CoV-2 in a plaque reduction assay (Figure 3D). Consistent with the blocking results, the noNAG-RBD nanoparticles elicit potent neutralizing antibodies at titers comparable to FL-S-2P, while monomeric WT RBD elicits no detectable neutralizing antibodies.
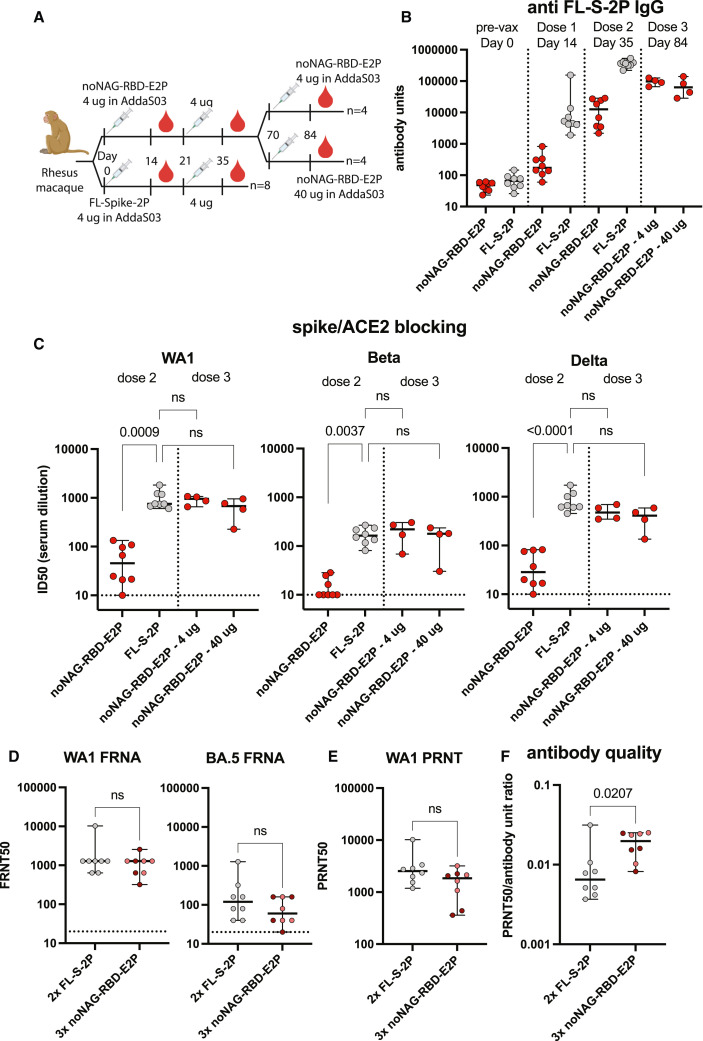
A three-dose noNAG-RBD vaccine regimen elicits potent neutralizing antibodies in NHPs
We immunized eight NHPs to further investigate the clinical viability of noNAG-RBD nanoparticle vaccines (Figure 4 A). noNAG-RBD-E2p was used as a representative particle because it elicited the highest neutralizing titers in mice and because the particle is non-glycosylated, which could confer manufacturing advantages. We used AddaS03 as an adjuvant because it is easily accessible and highly similar to the approved adjuvant AS03, which was shown to be an optimal adjuvant for other RBD nanoparticles.27 Additionally, we immunized a group of eight NHPs with FL-S-2P to serve as a benchmark for efficacy. A previous study demonstrated that two 4-μg doses of FL-S-2P adjuvanted in AS03 elicited neutralizing antibody titers sufficient for protection.28 The immunized NHPs were protected from SARS-CoV-2 challenge, and hamsters that received IgG by passive transfer from these NHPs were also protected, suggesting that the levels of neutralizing antibodies alone can serve as a benchmark for protection. This FL-S-2P vaccine has advanced to phase 3 clinical trials (ClinicalTrials.gov: NCT04904549) with positive data reported by press release, suggesting that noNAG-RBD-E2p could be clinically viable if it achieves comparable neutralizing antibody titers.
Figure 4.
Three doses of noNAG-RBD-E2p elicits a potent and focused neutralizing antibody response in NHPs
(A) Immunization schedule for rhesus macaques.
(B) Spike-binding serum antibody titers. Each point represents an individual animal measured in triplicate. Bars indicate median values and 95% confidence intervals.
(C) Spike-/ACE2-blocking antibody titers using recombinant spike protein from WA1, Beta, or Delta variants, depicted as described in (B). Horizontal dashed line indicates limit of detection. Statistical comparisons were made using a Kruskal-Wallis ANOVA followed by Dunn’s comparison with FL-S-2P, corrected for multiple comparisons. n = 8 animals for dose 2 groups, and n = 4 animals for dose 3 groups. “ns” indicates a p value > 0.05.
(D) FRNA neutralizing antibody titers against authentic WA-1 and Omicron BA.5 variants. Depicted as described for (B). Each point represents an individual animal measured in quadruplicate. The 4- and 40-μg noNAG-RBD-E2p groups are combined for analysis, with the 4-μg group depicted in light red and the 40-μg group depicted in dark red. Statistical comparisons were made using a two-tailed Mann-Whitney U test; n = 8 animals per group.
(E) PRNT neutralizing antibody titers against WA-1. Depicted as described in (D). Each point represents an individual animal measured in duplicate.
(F) Antibody quality calculated by normalizing neutralizing titers to spike-binding titers, depicted as described in (D).
See also Figures S5 and S6 and Table S1.
Both antigens elicit antibodies that recognize FL-S-2P after one immunization in NHPs, and titers are further increased after a second immunization 3 weeks later (Figure 4B). The anti-FL-S-2P IgG titers elicited by noNAG-RBD-E2p are lower than those elicited by FL-S-2P, consistent with the removal of domains and epitopes outside the RBD. The anti-WT-RBD titers elicited by noNAG-RBD-E2p are also lower than those elicited by FL-S-2P, which could be attributed to lower immunogenicity of noNAG-RBD-E2p or the mutation of RBD epitopes outside of the retained ACE2-binding site (Figure S5A). The ratio of WT-RBD/FL-S-2P titers elicited by noNGA-RBD-E2p is significantly higher than that elicited by FL-S-2P, demonstrating that noNAG-RBD-E2p focuses the immune response to the RBD (Figure S5B). Interestingly, the homologous anti-noNAG-RBD titers elicited by noNAG-RBD-E2p are slightly lower than the titers elicited by FL-S-2P (Figure S6A). This confirms that noNAG-RBD-E2p is less immunogenic than FL-S-2P and suggests that the response could be improved by optimizing the vaccination regimen. Consistent with the IgG titers, the blocking antibody GMTs elicited by two doses of noNAG-RBD-E2p in NHPs are more than 10-fold lower than FL-S-2P (Figure 4C). This is much greater than the 2- to 3-fold difference seen in mice, which could be attributed to differences in species, dosing, and/or adjuvant.
A third dose of noNAG-RBD-E2p administered 7 weeks after the second dose significantly increased functional antibody titers to levels comparable to FL-S-2P (Figures 4B–4E). We tested both 4- and 40-μg doses and saw that both groups elicited blocking titers that were not significantly different from FL-S-2P (Figure 4C). To improve the statistical power of our comparisons with FL-S-2P, we combined the 4- and 40-μg groups for further analysis. We measured neutralizing antibody titers against the WA-1 variant of SARS-CoV-2 using both the fluorescent reduction neutralization assay (FRNA) and the plaque reduction neutralization test (PRNT) (Figures 4D and 4E). Three doses of noNAG-RBD-E2p elicited FRNT50s and PRNT50s that are not significantly different from FL-S-2P. Neutralizing titers against the Omicron variant BA.5 were also not significantly different between the two groups (Figure 4D). Calculated GMTs are within 2-fold, and with the limited number of animals, we have 95% confidence that the noNAG-RBD-E2p PRNT50 GMT is within 5.3-fold of FL-S-2P. Together, these data suggest that three doses of noNAG-RBD-E2p would be expected to achieve a similar efficacy to two doses of FL-S-2P (Figure 4D). Interestingly, the total spike-binding antibody titers elicited by three doses of noNAG-RBD-E2p remain lower than FL-S-2P despite having comparable neutralizing titers, suggesting that noNAG-RBD-E2p focuses the antibody response to neutralizing epitopes (Figure 4B). These results are consistent with the ELISA data that show that noNAG-RBD-E2p focuses the antibody response to the RBD (Figures S5A and S5B). The neutralizing antibody quality can be quantified by calculating the ratio of neutralizing antibodies to binding antibodies, demonstrating a significantly higher-quality antibody response elicited by noNAG-RBD-E2p (Figure 4F).
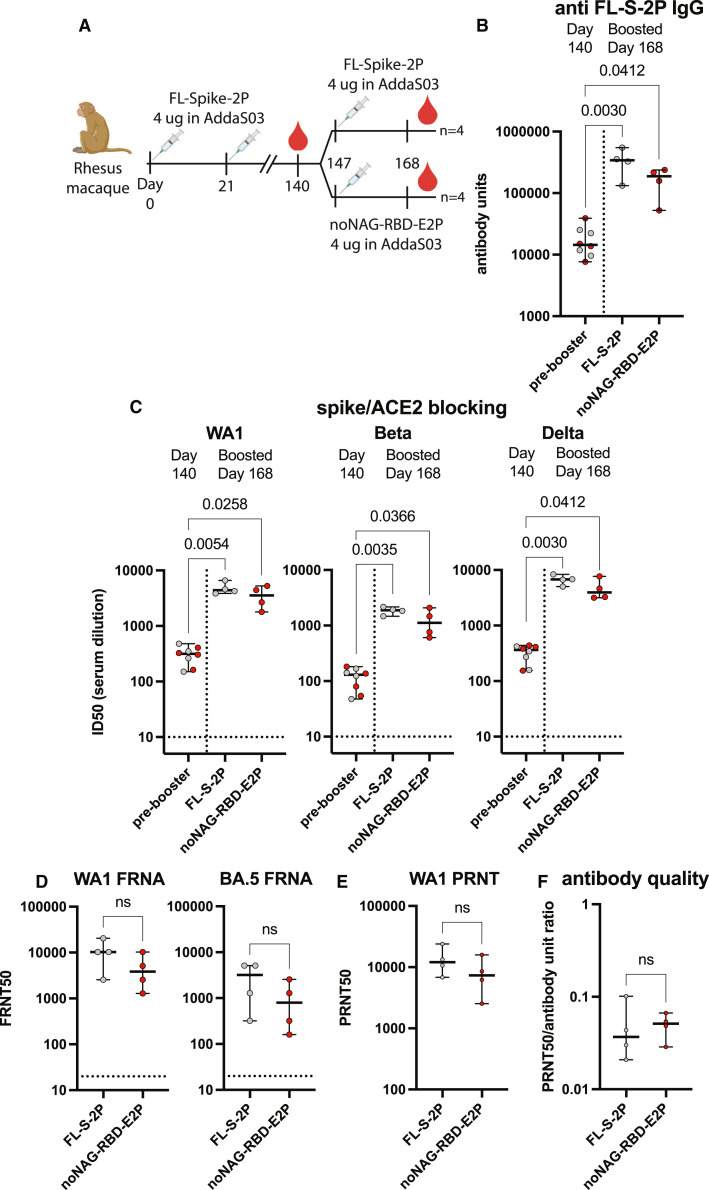
noNAG-RBD effectively boosts titers of animals previously immunized with FL-S-2P
Future SARS-CoV-2 vaccination strategies are still being debated, but it is likely that high-risk individuals will continue to receive periodic boosters to increase protective antibody titers. To determine if noNAG-RBD-E2p could serve as a booster vaccine candidate, we immunized four NHPs that had previously received two doses of FL-S-2P (Figure 5 A). We immunized the other four NHPs with a third dose of FL-S-2P, as a comparator. Boosters were administered 4 months (day 147) after the second FL-S-2P immunization when binding antibody GMTs had decayed 22-fold from peak values on day 35 (Figures 4B and 5B). Interestingly, blocking antibody GMTs were only 3-fold lower than peak values, suggesting that binding and blocking antibodies decay at different rates (Figures 4C and 5C).
Figure 5.
noNAG-RBD-E2p is an effective booster following FL-S-2P immunization
(A) Immunization schedule for primary immunization series and booster.
(B) Spike-binding serum antibody titers. Each point represents an individual animal measured in triplicate. Bars indicate median values and 95% confidence intervals. Gray circles in the pre-booster sample are animals subsequently boosted with FL-S-2P and red circles were subsequently boosted with FL-S-2P. Statistical comparisons were made using a Kruskal-Wallis ANOVA followed by Dunn’s comparison with the pre-booster group, corrected for multiple comparisons. n = 8 animals for pre-booster group, and n = 4 animals for boosted groups.
(C) Spike-/ACE2-blocking antibody titers depicted as described in (B).
(D) FRNA neutralizing antibody titers against authentic WA-1 and Omicron BA.5 variants, depicted as described in (B). Each point represents an individual animal measured in quadruplicate. Statistical comparisons were made using a two-tailed Mann-Whitney U test; n = 4 animals per group. “ns” indicates a p value > 0.05.
(E) PRNT neutralizing antibody titers against WA-1. Depicted as described in (D). Each point represents an individual animal measured in duplicate.
(F) Antibody quality calculated by normalizing neutralizing titers to spike-binding titers, depicted as described in (D).
See also Figures S5 and S6 and Table S1.
After boosting with noNAG-RBD-E2p or FL-S-2P, binding and blocking antibody titers increased significantly against all variants tested (Figures 5B and 5C). The FL-S-2P booster increased WA-1 FL-S-2P binding antibody GMT 19-fold and blocking antibody GMT 11-fold, suggesting a similar effect on both neutralizing and non-neutralizing antibodies. Conversely, the noNAG-RBD-E2p booster increased WA-1 FL-S-2P binding antibody GMT 9-fold and blocking antibody GMT 17-fold, suggesting a more targeted increase in functional antibodies. The targeted nature of the noNAG-RBD-E2p booster is also apparent in the ratio of RBD/FL-S-2P antibodies, which is significantly increased by the noNAG-RBD-E2p booster but not the FL-S-2P booster (Figures S5C and S5D). There was no significant difference in binding or blocking titers between the FL-S-2P- and noNAG-RBD-E2p-boosted groups.
The neutralizing antibody titers elicited by a noNAG-RBD-E2p booster are comparable to an FL-S-2P booster. Neutralizing antibody GMTs were not significantly different between the two antigens against both WA-1 and Omicron BA.5 (Figures 5D and 5E). Calculated GMTs were within 3-fold, and with the limited number of animals, there is 95% confidence that the noNAG-RBD-E2p PRNT50 GMT is within 8.2-fold of FL-S-2P. All animals had higher neutralizing titers after the booster (day 168) than after the second immunization (day 35), except for one animal, which had the highest titers on day 35 (Table S1). Together, these results demonstrate that a noNAG-RBD-E2p booster increases functional antibody titers to a similar extent as FL-S-2P. Thus, noNAG-RBD-E2p can serve as a booster vaccine following a primary immunization series with a conventional FL-S-2P antigen.
Discussion
We have developed a generalizable design procedure to eliminate unwanted glycans in antigens while increasing their production yield and stability. Successful application of this approach to the RBD of SARS-CoV-2 enabled the development of multiple versatile noNAG-RBD nanoparticles. Of these nanoparticles, noNAG-RBD-E2p elicited potent neutralizing antibodies in NHPs as a primary vaccine or as a booster vaccine. Three doses of noNAG-RBD-E2p elicited WA-1 neutralizing GMTs of 987 and 1,311, as measured by FRNA and PRNT, respectively. As a booster following an FL-S-2P primary series, noNAG-RBD-E2p elicited even higher neutralizing GMTs of 3,620 and 6,795 for FRNA and PRNT, respectively. These titers are comparable to previously reported neutralizing titers in NHPs against authentic WA-1 SARS-CoV-2, which range from 800 to 4,000 for approved mRNA vaccines, RBD nanoparticle vaccines, and recombinant RBD and FL-S-2P vaccines.27 , 28 , 29 , 30 , 31 , 32 Each of these studies demonstrated protection of NHPs upon challenge with SARS-CoV-2, meaning that the titers elicited by noNAG-RBD-E2p are also likely to be protective. While this comparison of neutralizing titers between publications is informative, there are potential artifacts due to differences in assays. To control for these artifacts, we included an internal control group immunized with FL-S-2P. Previously, this vaccine was shown to confer protection with a neutralizing GMT of 801.28 We measured similar neutralizing GMTs for FL-S-2P of 1,396 and 2,624 by FRNA and PRNT, respectively, suggesting a successful reproduction of the previous results. noNAG-RBD-E2p neutralizing GMTs were 987/1,311 after three immunizations and 3,620/6,795 after boosting. These results indicate that RBD-noNAG-E2p will likely provide potent protection as a vaccine or booster.
noNAG-RBD-E2p also elicits neutralizing antibodies against BA.5, suggesting that a booster would confer protection against future immune escape variants. Humans with three doses of mRNA vaccine have approximately 10-to 20-fold lower neutralizing titers against BA.5 than WA-1.33 , 34 , 35 , 36 Similarly, noNAG-RBD-E2p and FL-S-2P have approximately 10-fold lower neutralizing antibody titers against BA.5 than WA-1 (Figures 4D and 5D). The titers elicited by two doses of FL-S-2P against WA-1 can again be used as a benchmark for protection (FRNT50 GMT of 1,396). Three doses of noNAG-RBD-E2p elicit an FRNT50 GMT of 67.3, suggesting that a primary series of noNAG-RBD-E2p may confer limited protection against BA.5. A noNAG-RBD-E2p booster, however, elicits an FRNT50 GMT of 640 against BA.5. This value is comparable to the benchmark, suggesting that a noNAG-RBD-E2p booster would confer protection against BA.5.
It is notable that these high neutralizing GMTs were achieved with a low 4-μg dose of noNAG-RBD-E2p nanoparticle and a mild adjuvant. This dosing regimen was optimal for an FL-S-2P immunogen, but we found that noNAG-RBD-E2p was slightly less immunogenic under this regimen. Increasing the noNAG-RBD-nanoparticle dose from 4 μg or optimizing the adjuvant could further improve outcomes based on observations from other nanoparticle studies that used 50- to 100-μg doses.27 , 31 , 37
noNAG-RBD was created using a modified version of the SPEEDesign method and appears to have achieved the goals of stabilizing the protein antigen and focusing the immune response to the potently neutralizing ACE2-binding site.17 noNAG-RBD has purification yields approximately 4-fold higher than WT RBDs and thermostability 5°C higher (Figures 1D–1F). This enhanced stability is even more significant compared with the non-glycosylated WT RBD, which cannot be expressed. noNAG-RBD-E2p also elicits a higher proportion of neutralizing antibodies than FL-S-2P, consistent with a focused response toward neutralizing epitopes (Figure 4F). Together, these results support the ability of SPEEDesign to stabilize antigens, remove unwanted glycans, and focus the immune response toward neutralizing epitopes.
The removal of the N-glycan on noNAG-RBD has a relatively minor effect on the quality of the immune response, which is offset by the benefits of nanoparticle display. Some neutralizing antibodies, like S309, contact the glycan at Asn343. Therefore, removal of the N-glycan in noNAG-RBD could eliminate some neutralizing epitopes and reduce the quality of the immune response. However, these antibodies do not block ACE2 binding and are less potently neutralizing than those that do.4 , 5 , 38 noNAG-RBD retains the ACE2-binding site and, consequently, the most potently neutralizing epitopes. Indeed, the anti-WT-RBD IgG titers elicited by noNAG-RBD nanoparticles are lower than the titers elicited by FL-S-2P, potentially due to removal of glycan-containing epitopes (Figure S5A). However, noNAG-RBD retains binding to S309 (Figure 2E) and elicits a potent neutralizing antibody response that likely includes both potently neutralizing mAbs that block ACE2 binding and less potent mAbs targeting other epitopes (Figures 3, 4, and 5). One limitation of our study is that we cannot compare noNAG-RBD-E2p with a glycosylated RBD nanoparticle to carefully deconvolute the impact of the glycan. However, comparison with a monomeric glycosylated WT RBD clearly demonstrates that nanoparticle display of noNAG-RBD offsets any potential decrease in antibody quality caused by removing the glycan (Figure 3D).
noNAG-RBD-E2p is a non-glycosylated, self-assembling, single-component nanoparticle with manufacturing advantages over antigens that elicit similar neutralizing antibody titers. The removal of N-linked glycosylation sites improves consistency across manufacturing platforms. In comparison, FL-S-2P is a heavily glycosylated protein, requiring production in insect or mammalian cell culture. The unmodified RBD can be produced in yeast, but it also retains at least one N-glycan.8 The composition of these glycans changes depending on expression platform and many factors of the expression process. The composition of the glycans can impact vaccine homogeneity, stability, safety, and efficacy, meaning that glycoprotein production must be carefully controlled and monitored. noNAG-RBD-E2p does not require these measures, simplifying the manufacturing process. The single-component nature of noNAG-RBD particles also simplifies the purification process compared with multi-component RBD nanoparticles.12 , 13 , 14 , 15 , 37 , 39 , 40 Multi-component particles require independent cGMP production and purification processes for each individual component, in addition to the final assembled product, dramatically increasing the complexity and cost of production. In contrast, noNAG-RBD particles can be expressed and purified in a single process. Perhaps the most comparable antigen to noNAG-RBD particles is a single-component RBD-ferritin particle.16 This particle also required stabilizing mutations to the RBD to enable production, and these mutations are distinct from those in noNAG-RBD. However, the RBD-ferritin particle remained relatively unstable, requiring a decreased expression temperature and stabilizing excipients. Some multi-component nanoparticles also require stabilizing excipients.15 , 27 noNAG-RBD-ferritin can be expressed at 37°C and achieves purification yields approximately 4-fold higher than RBD-ferritin without excipients or purification tags (Figures 2A and 2B). The modular nature of noNAG-RBD particles also allowed us to create single-component 60-copy RBD particles (E2p and LuS). We expect that noNAG-RBD would be compatible with all other nanoparticle platforms as well. The stabilizing amino acid changes in noNAG-RBD are also likely to be compatible with emerging variants because the vast majority do not overlap with naturally occurring mutations. Thus, noNAG-RBD particles are versatile, stable, and effective antigens that can be easily manufactured.
Limitations of the study
This study is limited to an analysis of the systemic antibody response elicited by these vaccine candidates. Other immune responses, such as T cell responses, may play a role in vaccine efficacy. However, the neutralizing antibody response we have characterized is necessary and sufficient for protection in animal models and correlates very well with protection in humans.28 , 41 , 42 Another limitation of this study is the analysis of only one adjuvant and one booster dose due to the number of animals available. These animals had complex immunization histories with antigens from Leishmania major, malaria, Ebola, Epstein-Barr virus, and/or cytomegalovirus (Table S2). None of these pathogens are closely related to SARS-CoV-2, suggesting they would have little impact on our study, and the anti-SARS-CoV-2 IgG titers at day 0 of our study reveal no pre-existing immunity (Figure 4B). Furthermore, animals with various histories are distributed between groups of this study and are therefore unlikely to contribute to the conclusions we report. These complex histories may in fact represent a diverse human population more closely than naive animals, increasing the robustness of our findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| REGN10933 | Hansen et al.43 | N/A |
| CR3022 | ter Meulen et al.44 | RRID:AB_2848080 |
| S309 | Pinto et al.45 | N/A |
| Peroxidase-AffiniPure Goat Anti-Human IgG | Jackson ImmunoResearch Laboratories, Inc. | Cat.# 109-035-098; RRID:AB_2337586 |
| Peroxidase-AffiniPure Goat Anti-Mouse IgG | Jackson ImmunoResearch Laboratories, Inc. | Cat.# 115-035-164; RRID:AB_2338510 |
| SARS-CoV Nucleoprotein Antibody | Sino Biologicals | Cat# 40143-R001; RRID:AB_2827974 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor™ 647 | Thermo Fisher Scientific | Cat# A-21245; RRID:AB_2535813 |
| Bacterial and virus strains | ||
| SARS-CoV-2 | Centers for Disease Control and Prevention [CDC], Atlanta, GA | GenBank: MN985325.1 |
| Chemicals, peptides, and recombinant proteins | ||
| noNAG-RBD | This paper | N/A |
| no-NAG-RBD-E2p | This paper | N/A |
| no-NAG-RBD-LuS | This paper | N/A |
| no-NAG-RBD-ferritin | This paper | N/A |
| WT RBD | Dickey et al.17 | N/A |
| full-length spike (FL-S-2P) | Wrapp et al.46 | N/A |
| ACE2-Fc(IgG1) fusion | This paper | N/A |
| biotinylated ACE2-Fc fusion | This paper | N/A |
| Beta variant FL-S-2P | This paper | N/A |
| Delta variant FL-S-2P | This paper | N/A |
| REGN10933 Fab | This paper | N/A |
| AddaVax | Invivogen | Cat#vac-adx-10 |
| AddaS03 | Invivogen | Cat#vac-as03-10 |
| Streptavidin Protein, HRP | Thermo Fisher Scientific | Cat#21127 |
| Critical commercial assays | ||
| Ni-NTA HisSorb Plates | Qiagen | Cat#35061 |
| Protein Thermal Shift Dye Kit | Thermo Fisher Scientific | Cat#4461146 |
| Experimental models: Cell lines | ||
| expi293F cells | Thermo Fisher Scientific | Cat#A14527; RRID:CVCL_D615 |
| Vero E6 cells stably expressing TMPRSS2 | Liu et al.47 | N/A |
| VeroE6/TMPRSS2cells | Nao et al.48 | RRID:CVCL_YQ49 |
| Experimental models: Organisms/strains | ||
| CD-1 mice | Charles River Laboratories | RRID:IMSR_CRL:022 |
| Recombinant DNA | ||
| pHL-sec plasmid | Addgene plasmid # 9984549 | RRID:Addgene_99845 |
| Software and algorithms | ||
| ROSETTA 3.10 | https://www.rosettacommons.org/home | RRID:SCR_015701 |
| CD-HIT 2007 | http://weizhong-lab.ucsd.edu/cd-hit/ | RRID:SCR_007105 |
| Unicorn 7 | Cytiva | N/A |
| Protein Thermal Shift Software v1.4 | Thermo Fisher Scientific | N/A |
| Relion 3.0 | Zivanov et al.50 | RRID:SCR_016274 |
| Prism 9 | GraphPad | N/A |
| PyMOL v2.3 | Schrodinger LLC | N/A |
| Other | ||
| Superose 6 Increase 10/300 GL column | Cytiva | Cat#29091596 |
| SRT SEC-1000 column | Sepax | Cat#215100-7830 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Niraj Tolia (niraj.tolia@nih.gov).
Materials availability
Plasmids and other unique and stable reagents generated in this study can be provided upon completion of a material transfer agreement.
Experimental model and subject details
Mammalian cell lines
Expi293F cells were used for protein expression (Thermo Fisher Scientific RRID: CVCL_D615). This is a transformed cell line derived from human female kidney cells. Authentication was not performed after purchase. Cells were cultured at 37°C, >80% humidity, 8% CO2, suspended in Expi293 Expression Medium by shaking at 125 rpm (Thermo Fisher Scientific cat#A1435103).
Mouse studies
Mouse studies were performed in an AAALAC-accredited facility under the guidelines and approval of the Institutional Animal Care and Use Committee (IACUC) at the National Institutes of Health. Ten naive 5-6 week old female CD-1 mice (Charles River Laboratories) per group (50 mice total) were immunized with 1 μg antigen each. Mice were randomly assigned to each group. Experimental units comprised single animals and groups of 5 mice receiving the same immunization were co-housed. Mice were maintained in a specific pathogen-free vivarium, housed in a climate-controlled room on a 12-hour day/night cycle, and had access to food and water ad libitum.
Non-human primate studies
NHP studies were performed in an AAALAC-accredited facility under the guidelines and approval of the Institutional Animal Care and Use Committee (IACUC) at the National Institutes of Health. For naïve immunization studies, 16 healthy male rhesus macaques (Macaca mulatta), were randomized into two groups of 8 based on prior immunization history (Table S2). Animals ranged from 5-9 years of age and 7.49-13.55 kg at the beginning of the study. The eight animals that received two doses of noNAG-RBD-E2p were randomized into two groups of four by immunization history and antibody titers at day 35 to receive a third dose of either 4 ug or 40 ug noNAG-RBD-E2p. The eight animals that received two doses of FL-S-2P were later immunized in a booster study. These animals were randomized into two groups of 4 based on previous immunization history and antibody titers at day 35. Experimental units comprised single animals and groups of 2-4 NHPs were co-housed. A high protein monkey diet was provided, supplemented by fresh fruits, vegetables and nuts, with water ad libitum. Animals were provided with hanging and floor toys, with perches in the cages allowing for exercise. The facility was maintained at a temperature between 73°F and 79°F, a humidity of less than 70%, and a 12-hour day/night cycle.
Method details
noNAG-SPEEDesign
The SPEEDesign pipeline categorizes each residue as unchanged, intermediate, or deep search to define the amino acids identities sampled at each position during the computational design process. The original SPEEDesign pipeline limits the amino acids sampled at intermediate residues to those found in homologous protein sequences, thereby identifying changes compatible with the RBD and related structures.17 In noNAG-SPEEDesign, we modified this approach to leverage DMS data.19 This modified approach limits the computational search at intermediate residues to only those amino acid changes that have positive effects on expression in yeast.19 However, this deep mutational scanning data was collected on glycosylated proteins and does not identify changes that will compensate for the removal of the glycan. Therefore, we defined the NxS/T motif and all residues that contact the glycan as deep search, allowing them to sample all amino acids, except cysteine (Figure 1A). Finally, we defined the ACE2-binding site as unchanged to maintain the most potent neutralizing epitopes while allowing the computational design process to find an optimally stabilized solution (Figure 1A). We also used a second design strategy in which the epitopes of all known antibodies that block ACE2 binding were unchanged. These amino acid constraints were used to create a ResFile for each strategy that was used by the FastDesign module in ROSETTA.
Decoy clustering was performed as described previously.17 For each computational strategy, decoys with scores in the 95th percentile were clustered by sequence similarity using CD-HIT and the top scoring decoy form each cluster was selected as a representative sequence.51 The number of clusters was selected based on the sequence diversity produced in each computational strategy.
noNAG-SPEEDesign in vitro screening
Immunogen screening was performed as described previously.17 Synthetic DNA coding for secreted RBD immunogens was cloned (GenScript) into a customized pHL-sec expression plasmid. pHL-sec was a gift from Edith Yvonne Jones (Addgene plasmid # 99845 ; http://n2t.net/addgene:99845 ; RRID:Addgene_99845).49 Plasmid was transfected into human expi293F cells and grown in a 96-well plate according to manufacturer instructions (ThermoFisher Scientific, RRID:CVCL_D615). Cell-free supernatant was harvested after 4 days of expression.
Cell-free supernatant was diluted in PBST + 2% BSA and added to Ni-NTA HisSorb Plates (Qiagen) to capture His-tagged immunogens. After incubation for 1 hour at room temperature, plates were washed three times with PBST. Neutralizing epitopes were probed using an ACE2-Fc(IgG1) fusion (0.2 ug/well) or a human IgG1 antibody containing the REGN10933 variable domain (0.05 ug/well). After incubation for 1 hour at room temperature, plates were washed three times with PBST and 200 μl 1:5000 peroxidase-conjugated anti-human IgG was added (Jackson ImmunoResearch Laboratories, Inc. Cat.# 109-035-098, RRID:AB_2337586). Plates were incubated 30 minutes at room temperature and washed three times with PBST. Finally, 70 μl Tetramethylbenzidine (TMB) (MilliporeSigma) was added and incubated 5 minutes at room temperature before quenching with 70 μl 0.16 M H2SO4. Absorbance at 450 nm was measured using a Biotek Synergy H1 plate reader.
RBD and FL-S-2P expression and purification
Recombinant RBD antigens were expressed in expi293F cells, as described above. FL-S-2P was also expressed in expi293F cells using a modified pHL-sec vector. The construct contains amino acids 1-1208 of the spike protein followed by a foldon trimerization domain and a 6-His tag. This construct also contains the “2P” stabilizing mutations at K986P and V987P and mutation of the furin cleavage site (682-685 GSAS to RRAR).46
Cell-free supernatant was harvested after 4 days and His-tagged antigens were purified by gravity chromatography using Ni Sepharose excel resin according to manufacturer instructions (Cytiva). Antigens were further purified by size-exclusion chromatography using a Superdex 75 Increase 10/300 GL column (RBD) or Superose 6 Increase 10/300 GL column (FL-S-2P) equilibrated in 1x PBS. Fractions corresponding to trimeric FL-spike or monomeric RBD were pooled and the concentration for immunization was determined by measuring Abs280 and using extinction coefficients calculated by the ExPASy ProtParam tool.52 Aliquots of protein were snap frozen in liquid nitrogen and stored at -80°C.
Transfection, expression, and purification was performed in triplicate on three separate days to calculate RBD immunogen purification yields. Each replicate consisted of a 30 mL culture, and yields were calculated by integrating the area under the monomeric peak on the Abs280 chromatogram during size-exclusion chromatography. These yields closely matched yields calculated from pooled fractions. Extinction coefficients were calculated using the ExPASy ProtParam tool.52
Nanoparticle expression and purification
noNAG-RBD nanoparticles were created by genetically fusing nanoparticle carriers to the C-terminus of noNAG-RBD. There is a 15-amino acid linker (GGGGS)3 between noNAG-RBD and E2p/LuS and a GSGGGG linker between noNAG-RBD and ferritin. The LuS construct contains an engineered N-glycan at position N71.25 Additionally, a potential N-glycosylation site was removed by mutation of S104A. The ferritin construct is a previously described engineered construct from bullfrog (Rana catesbeiana) and H. pylori ferritin.21 The E2p construct contains amino acids 185-426 from G. stearothermophilus 23 with a S276A mutation to remove a potential N-glycosylation site. All constructs were created without additional purification or solubility tags.
Proteins were expressed in expi293F cells, as described above. Cell-free supernatant was harvested after 4 days of expression at 37°C and concentrated using 100,000 kDa molecular weight cutoff Amicon centrifugal filter units. Concentrated supernatant was purified by size-exclusion chromatography using a Superose 6 Increase 10/300 GL column (ferritin and LuS) or a Sepax SRT SEC-1,000 column (E2p) equilibrated in 1x PBS. Fractions corresponding to nanoparticles were pooled, snap frozen in liquid nitrogen, and stored at -80°C. Reported yields were calculated from final products using extinction coefficients calculated by the ExPASy ProtParam tool.52
Antibody/ACE2 expression and purification
Antibodies for ELISAs were created by fusing the variable regions for the indicated antibody to the human IGHG∗01, IGKC∗01, or IGLC2∗02 constant regions and cloning into the pHL-sec plasmid (GenScript). Heavy and light chain plasmids were mixed in equal amounts and transfected into expi293F cells according to manufacturer instructions and cell-free supernatant was harvested after 4 days of expression.
Cell-free supernatant was batch incubated with protein A agarose resin (GoldBio) for 1 hour at room temperature. Resin was collected and washed with 10 column volumes (CV) protein A IgG binding buffer (ThermoFisher Scientific). Protein was eluted with 10 CV IgG elution buffer (ThermoFisher Scientific) and neutralized with 1 CV 1 M Tris pH 9.0. Antibodies were concentrated and buffer exchanged into PBS using an Amicon centrifugal filter (MilliporeSigma).
Fab for sandwich ELISAs was produced by fusing the REGN10933 variable region to a His-tagged human IGHG∗01 CH1 domain and cloning into pHL-sec (GenScript). Heavy chain Fab plasmid was co-transfected 1:1 with light chain in expi293F cells according to manufacturer instructions and cell-free supernatant was harvested after 4 days of expression (ThermoFisher Scientific). His-tagged Fab was purified from cell-free supernatant by gravity chromatography using Ni Sepharose excel resin according to manufacturer instructions (Cytiva). Fab was further purified by size-exclusion chromatography using a Superdex 75 increase 10/300 GL column (Cytiva) equilibrated in 1x PBS.
The biotinylated ACE2-Fc fusion was created by fusing ACE2 (amino acids 19-615) to the human IGHG∗01, followed by an HRV 3C protease site, a biotin ligase (BirA) recognition site (AviTag), and a 6x-His tag. Plasmid encoding BirA was mixed at a 1:9 ratio with the ACE2 expression plasmid and transfected into expi293F cells. 100 μM biotin was immediately added to the media and cell-free supernatant was harvested after 4 days of expression. Protein was purified from cell-free supernatant by gravity chromatography using Ni Sepharose excel resin according to manufacturer instructions (Cytiva) and further purified by size-exclusion chromatography using a Superdex 200 increase 10/300 GL column (Cytiva) equilibrated in 1x PBS.
Differential scanning fluorimetry (DSF)
DSF was performed using the Protein Thermal Shift Dye Kit according to manufacturer instructions (ThermoFisher Scientific). Final reactions contained 0.4 mg/mL purified immunogen, 1x Protein Thermal Shift buffer, 1x Thermal Shift Dye, and 0.63x PBS. Fluorescence was monitored using a 7500 Fast Real-Time PCR system (ThermoFisher Scientific) as the temperature was increased from 25°C to 95°C at a ramp rate of 1%. Melting temperature (Tm) was calculated as the peak of the derivative of the melt curve. DSF reactions were performed in technical quadruplicate on each plate and in biological triplicate using three different protein preps on 3 separate days. Technical replicates were averaged to calculate the Tm for a biological replicate, and the three biological replicates were averaged to calculate the reported Tm.
Negative-stain electron microscopy
Purified nanoparticles (0.01mg/ml) were adsorbed on a glow discharged 300 mesh carbon-coated copper grids (EMD Science) for 30s followed by NanoW (Nanoprobes) staining. Raw Micrographs were recorded in Thermo Scientific Tecnai T20 microscope equipped with a charge-coupled device (CCD) camera. Particles were auto-picked using Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) and 2D classes were generated using RELION 3.0.50
Sandwich ELISA of nanoparticle epitopes
Nunc MaxiSorp plates (ThermoFisher Scientific) were coated with 100 μl 0.01 mg/mL purified REGN10933 Fab diluted in 50 mM Na-carbonate pH 9.5. Plates were coated overnight at 4°C then washed three times with PBST. Plates were blocked 1 hour at room temperature with 2% BSA in PBST then washed three times with PBST. Purified nanoparticles were diluted to 0.01 mg/mL in blocking buffer and 100 μl was added to the plate. Plates were incubated at room temperature for one hour then washed three times with PBST. 100 μl primary antibody was added to each well at the indicated concentration: ACE2 – 15.6 ng/mL, REGN10933, CR3022, and S309 – 5 ng/mL. Primary antibody was incubated 1 hour at room temperature then plates were washed three times with PBST and 200 μl 1:5000 peroxidase-conjugated anti-human IgG was added (Jackson ImmunoResearch Laboratories, Inc. Cat.# 109-035-098, RRID:AB_2337586). Plates were incubated 30 minutes at room temperature and washed three times with PBST. Finally, 70 μl Tetramethylbenzidine (TMB) (MilliporeSigma) was added and incubated 5 minutes at room temperature before quenching with 70 μl 2 M H2SO4. Absorbance at 450 nm was measured using a Biotek Synergy H1 plate reader.
Mouse immunizations
Antigen was purified, as described above, snap frozen in liquid nitrogen, and stored at -80. On the day of immunization, antigen was thawed and formulated as a 1:1 ratio in AddaVax, according to manufacturer recommendations (Invivogen). 100 μl formulated antigen was delivered by subcutaneous injection. Blood was collected on day 35 and serum was separated and stored at -80°C.
NHP immunizations
Vaccines were administered intramuscularly with a 500 μl injection into alternating biceps. Antigen was purified, as described above, snap frozen in liquid nitrogen, and stored at -80. On the day of immunization, antigen was thawed and formulated as a 1:1 ratio in AddaS03, according to manufacturer recommendations (Invivogen). Blood was collected at indicated time points and serum was separated and stored at -80°C. All serum samples were blinded and randomized for analysis.
Serum antibody titer ELISAs
Nunc MaxiSorp plates (ThermoFisher Scientific) were coated with 100 μl 0.01 mg/mL purified FL-S-2P, WT RBD, or noNAG-RBD diluted in 50 mM Na-carbonate pH 9.5. Plates were incubated overnight at 4°C then washed three times with PBST. Plates were blocked 1 hour at room temperature with 2% BSA in PBST then washed three times with PBST. Serum was diluted in 2% BSA in PBST and 100 μl was added to each well. After 1 hour incubation at room temperature, plates were washed three times with PBST and 200 μl 1:5000 peroxidase-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc. Cat.# 115-035-164, RRID:AB_2338510) or 1:8,000 peroxidase-conjugated anti-monkey IgG (Southern Biotech anti-monkey IgG-HRP clone SB108a, RRID:AB_2796069) was added. Plates were incubated 30 minutes at room temperature and washed three times with PBST. Finally, 70 μl Tetramethylbenzidine (TMB) (MilliporeSigma) was added and incubated 20 minutes at room temperature before quenching with 70 μl 0.16 M H2SO4. Absorbance at 450 nm was measured using a Biotek Synergy H1 plate reader.
Pooled serum from mice or NHPs immunized with FL-S-2P was used as a standard curve on each plate to calculate the antibody titers of individual animals in all groups. One antibody unit (AU) was defined as the dilution of the standard serum required to achieve an Abs450 value of 1. Each plate included triplicate 2-fold serial dilutions of the standard serum from 20 to 0.01 AU. Serum from each animal was diluted such that the Abs450 fell in the informative portion of the standard curve between 0.1 and 2.0. The Abs450 values for the standard curve were fit to a 4-parameter logistic curve, which was used to convert Abs450 values to AU for each individual animal. AU values for each individual animal were measured in triplicate on separate plates and the average was reported.
Spike/ACE2 blocking assay
Nunc MaxiSorp plates (ThermoFisher Scientific) were coated with 100 μl 0.1 μg/mL purified FL-S-2P diluted in 50 mM Na-carbonate pH 9.5. Plates were incubated overnight at 4°C then washed three times with PBST. Plates were blocked 1 hour at room temperature with 2% BSA in PBST then washed three times with PBST. Serum was diluted in 2% BSA in PBST in a 3-fold dilution series starting at 1:10 or 1:30. 50 μl serum was mixed with 10 μl 60 nM biotinylated ACE2-Fc or 10 μl buffer as a background control. 50 μl serum mixture was added to FL-S-2P-coated plates and incubated 1 hour at room temperature. Plates were washed three times with PBST and 200 μl 1:10,000 streptavidin-HRP was added (Thermo Fisher). Plates were incubated 30 minutes at room temperature and washed three times with PBST. Finally, 70 μl Tetramethylbenzidine (TMB) (MilliporeSigma) was added and incubated 20 minutes at room temperature before quenching with 70 μl 0.16 M H2SO4. Absorbance at 450 nm was measured using a Biotek Synergy H1 plate reader.
Spike/ACE2 binding inhibition was calculated by first subtracting the Abs450 values of the background controls lacking biotinylated ACE2-Fc. Four wells without serum were used to calculate the maximum signal. Inhibition was calculated using the following formula:
where X is the Abs450 of a well after background-subtraction and max is the average of the 4 samples without serum after background-subtraction.
% inhibition values were measured for each serum dilution in triplicate and average values were plotted in GraphPad Prism 9. Data were fit using a normalized dose response curve with a variable slope:
where X is the serum dilution, Y is the % inhibition, and HillSlope and ID50 are calculated parameters corresponding to the slope of the curve and the dilution at which 50% inhibition occurs, respectively.
SARS-CoV-2 plaque assay (PRNT)
Vero E6 cells stably expressing TMPRSS2 were plated in a 24-well plate one day prior to infection.47 Serum was heat inactivated at 56°C for 30 minutes. Serum was diluted 1:200 in infection media (DMEM, 2% heat-inactivated FBS, 1x Glutamax (ThermoFisher), 0.25 mg/mL hygromycin), followed by 3-fold serial dilutions. Diluted serum was mixed 1:1 with WA-1 variant SARS-CoV-2 (GenBank: MN985325.1) and incubated 30 minutes at 37°C. Approximately 30 PFU virus was added to each well of Vero-TMPRSS2 cells and incubated 30 minutes at 37°C before overlay with 0.65% methyl cellulose 400 cP. Cells were incubated 72 hr at 37°C before staining with 0.2% crystal violet in 10% neutral buffered formalin.
% neutralization was calculated relative the average of 16 wells without serum. % neutralization values were measured for each serum dilution in duplicate and average values were plotted in GraphPad Prism 8. Data were fit using a normalized dose response curve:
where X is the serum dilution, Y is the % neutralization, and ID50 is calculated corresponding to the dilution at which 50% inhibition occurs.
Fluorescent reduction neut. assay (FRNA)
The assay was performed similarly to previously described.53 Vero E6 TMPRSS2 cells (JCRB1819, National Institutes of Biomedical Innovation, Health, and Nutrition, RRID:CVCL_YQ49) were seeded into 384-well tissue culture treated assay plates at 6,000 per well in 30 μL DMEM (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA) the day prior to infection. Heat-inactivated serum was serially diluted in 384-well dilution blocks to generate a twelve-point dose curve for each sample, replicated in quadruplicate (n = 4) in 30 μL per well. Under biocontainment conditions, virus was diluted in DMEM with 10% FBS to a concentration of 6,000 plaque forming units (PFU) per 30 μL (2e5 PFU/mL) and 30 μL per well was stamped into the dilution blocks using an Integra Viaflo384 semi-automated liquid handling robot. Following incubation at 37C for one hour, 30 μL was transferred from the dilution block to a corresponding assay plate. Both WA-1 and BA.5 plates were fixed in 10% neutral buffered formalin 12 hours post-infection. Upon removal from biocontainment, cells were stained with a SARS-CoV/SARS-CoV-2 nucleoprotein-specific primary antibody (Sino Biologicals, RRID:AB_2827974) followed by secondary antibody conjugated to Alexa647 fluorophore (Invitrogen, RRID:AB_2535813) and Hoechst dye (Invitrogen) was added for detection of cell nuclei. Fluorescence readout was quantitated using a PerkinElmer Operetta high-content imaging system. Half-maximal inhibitory neutralization concentration (FRNT50) was calculated as previously described54 using GraphPad Prism Software (La Jolla, CA). Z' factor scores were assessed as quality-control parameters for each plate.
Quantification and statistical analysis
All statistical tests are indicated in figure legends. These include Kruskal-Wallis ANOVA followed by Donn’s comparison, corrected for multiple comparisons or a two-tailed Mann-Whitney U test when comparing two groups. P-values <0.05 were considered significant. All statistical tests were performed in Prism 9 (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
We thank all the members of LMIV COVID group for discussions of results. Reed Johnson and Nicole Lackemeyer at the NIAID SARS-CoV-2 virology core (SVC) provided training and resources for PRNT experiments. This study used the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID; Bethesda, MD, USA). This work was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health.
Author contributions
Conceptualization, N.H.T. and T.H.D.; data analysis, N.H.T. and T.H.D.; software, N.H.T. and T.H.D.; validation, P.P.; methodology, T.H.D., N.H.T., and E.E.; formal analysis, T.H.D., N.H.T., J.L.K., and M.M.; investigation, T.H.D., R.M., B.B., H.M., T.O., S.O.-G., and B.E.; resources, P.P. and N.D.S.; visualization, T.H.D. and N.H.T.; funding acquisition, N.H.T. and T.H.D.; project administration, N.H.T., N.D.S., L.E.L., and T.H.D.; supervision, N.H.T., L.E.L., and M.R.H.; writing – original draft, T.H.D., and N.H.T.; writing – review & editing, T.H.D., and N.H.T.
Declaration of interests
N.H.T. and T.H.D. are inventors on a patent application related to this work.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: March 6, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112266.
Supplemental information
Data and code availability
-
•
All NHP data are included in Table S1. All other source data will be provided upon request.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 6.Toledo-Romani M.E., Garcia-Carmenate M., Silva C.V., Baldoquin-Rodriguez W., Pérez M.M., Gonzalez M.C.R., Moreno B.P., Hernández I.C.M., Romero R.G.-M., Tabio O.S., et al. Efficacy and safety of SOBERANA 02, a COVID-19 conjugate vaccine in heterologous three-dose. combination. 2021 doi: 10.1016/j.lana.2022.100423. [DOI] [Google Scholar]
- 7.Thuluva S., Paradkar V., Turaga K., Gunneri S., Yerroju V., Mogulla R., Suneetha P.V., Kyasani M., Manoharan S.K., Adabala S., et al. Immunogenic superiority and safety of Biological E’s CORBEVAX™ vaccine compared to COVISHIELD™ (ChAdOx1 nCoV-19) vaccine studied in a phase III, single blind, multicenter, randomized clinical trial. medRxiv. 2022 doi: 10.1101/2022.03.20.22271891. Preprint at. [DOI] [Google Scholar]
- 8.Pollet J., Chen W.H., Versteeg L., Keegan B., Zhan B., Wei J., Liu Z., Lee J., Kundu R., Adhikari R., et al. SARSCoV-2 RBD219-N1C1: a yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. Hum. Vaccines Immunother. 2021;17:2356–2366. doi: 10.1080/21645515.2021.1901545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W.H., Du L., Chag S.M., Ma C., Tricoche N., Tao X., Seid C.A., Hudspeth E.M., Lustigman S., Tseng C.T.K., et al. Yeast-expressed recombinant protein of the receptor-binding domain in SARS-CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum. Vaccines Immunother. 2014;10:648–658. doi: 10.4161/hv.27464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Zhao W., Mao Y., Chen Y., Wang S., Zhong Y., Su T., Gong M., Du D., Lu X., et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol. Cell. Proteomics. 2021;20:100058. doi: 10.1074/mcp.RA120.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen B., Tolia N.H. Protein-based antigen presentation platforms for nanoparticle vaccines. npj Vaccines. 2021;6:70. doi: 10.1038/s41541-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang Y.F., Sun C., Zhuang Z., Yuan R.Y., Zheng Q., Li J.P., Zhou P.P., Chen X.C., Liu Z., Zhang X., et al. Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano. 2021;15:2738–2752. doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- 13.Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.e9. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan T.K., Rijal P., Rahikainen R., Keeble A.H., Schimanski L., Hussain S., Harvey R., Hayes J.W.P., Edwards J.C., McLean R.K., et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021;12:542. doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce M.G., Chen W.H., Sankhala R.S., Hajduczki A., Thomas P.V., Choe M., Martinez E.J., Chang W.C., Peterson C.E., Morrison E.B., et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021;37:110143. doi: 10.1016/j.celrep.2021.110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickey T.H., Tang W.K., Butler B., Ouahes T., Orr-Gonzalez S., Salinas N.D., Lambert L.E., Tolia N.H. Design of the SARS-CoV-2 RBD vaccine antigen improves neutralizing antibody response. Sci. Adv. 2022;8:eabq8276. doi: 10.1126/sciadv.abq8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickey T.H., Gupta R., McAleese H., Ouahes T., Orr-Gonzalez S., Ma R., Muratova O., Salinas N.D., Hume J.C.C., Lambert L.E., et al. Design of a stabilized non-glycosylated Pfs48/45 antigen enables a potent malaria transmission-blocking nanoparticle vaccine. npj Vaccines. 2023;8:20. doi: 10.1038/s41541-023-00619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hempstead P.D., Yewdall S.J., Fernie A.R., Lawson D.M., Artymiuk P.J., Rice D.W., Ford G.C., Harrison P.M. Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol. 1997;268:424–448. doi: 10.1006/jmbi.1997.0970. [DOI] [PubMed] [Google Scholar]
- 21.Kanekiyo M., Bu W., Joyce M.G., Meng G., Whittle J.R.R., Baxa U., Yamamoto T., Narpala S., Todd J.P., Rao S.S., et al. Rational design of an Epstein-Barr virus vaccine targeting the receptor-binding site. Cell. 2015;162:1090–1100. doi: 10.1016/j.cell.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen M.D., Perham R.N. The catalytic domain of dihydrolipoyl acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Expression, purification and reversible denaturation. FEBS Lett. 1997;413:339–343. doi: 10.1016/s0014-5793(97)00932-0. [DOI] [PubMed] [Google Scholar]
- 23.Domingo G.J., Orru S., Perham R.N. Multiple display of peptides and proteins on a macromolecular scaffold derived from a multienzyme complex. J. Mol. Biol. 2001;305:259–267. doi: 10.1006/jmbi.2000.4311. [DOI] [PubMed] [Google Scholar]
- 24.Izard T., Aevarsson A., Allen M.D., Westphal A.H., Perham R.N., de Kok A., Hol W.G. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA. 1999;96:1240–1245. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B., Chao C.W., Tsybovsky Y., Abiona O.M., Hutchinson G.B., Moliva J.I., Olia A.S., Pegu A., Phung E., Stewart-Jones G.B.E., et al. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 2020;10:18149. doi: 10.1038/s41598-020-74949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Meining W., Fischer M., Bacher A., Ladenstein R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 2001;306:1099–1114. doi: 10.1006/jmbi.2000.4435. [DOI] [PubMed] [Google Scholar]
- 27.Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- 28.Francica J.R., Flynn B.J., Foulds K.E., Noe A.T., Werner A.P., Moore I.N., Gagne M., Johnston T.S., Tucker C., Davis R.L., et al. Protective antibodies elicited by SARS-CoV-2 spike protein vaccination are boosted in the lung after challenge in nonhuman primates. Sci. Transl. Med. 2021;13:eabi4547. doi: 10.1126/scitranslmed.abi4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 31.Cohen A.A., van Doremalen N., Greaney A.J., Andersen H., Sharma A., Starr T.N., Keeffe J.R., Fan C., Schulz J.E., Gnanapragasam P.N.P., et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science. 2022;377:eabq0839. doi: 10.1126/science.abq0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pino M., Abid T., Pereira Ribeiro S., Edara V.V., Floyd K., Smith J.C., Latif M.B., Pacheco-Sanchez G., Dutta D., Wang S., et al. A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Sci. Immunol. 2021;6:eabh3634. doi: 10.1126/sciimmunol.abh3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hachmann N.P., Miller J., Collier A.R.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowen J.E., Addetia A., Dang H.V., Stewart C., Brown J.T., Sharkey W.K., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377:890–894. doi: 10.1126/science.abq0203. eabq0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He L., Lin X., Wang Y., Abraham C., Sou C., Ngo T., Zhang Y., Wilson I.A., Zhu J. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 2021;7:eabf1591. doi: 10.1126/sciadv.abf1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 46.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X., Luongo C., Matsuoka Y., Park H.S., Santos C., Yang L., Moore I.N., Afroz S., Johnson R.F., Lafont B.A.P., et al. A single intranasal dose of a live-attenuated parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in hamsters. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2109744118. e2109744118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nao N., Sato K., Yamagishi J., Tahara M., Nakatsu Y., Seki F., Katoh H., Ohnuma A., Shirogane Y., Hayashi M., et al. Consensus and variations in cell line specificity among human metapneumovirus strains. PLoS One. 2019;14:e0215822. doi: 10.1371/journal.pone.0215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 50.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 52.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett R.S., Postnikova E.N., Liang J., Gross R., Mazur S., Dixit S., Kocher G., Yu S., Georgia-Clark S., Gerhardt D., et al. Scalable, micro-neutralization assay for assessment of SARS-CoV-2 (COVID-19) virus-neutralizing antibodies in human clinical samples. Viruses. 2021;13:893. doi: 10.3390/v13050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Covés-Datson E.M., Dyall J., DeWald L.E., King S.R., Dube D., Legendre M., Nelson E., Drews K.C., Gross R., Gerhardt D.M., et al. Inhibition of Ebola virus by a molecularly engineered banana lectin. PLoS Negl. Trop. Dis. 2019;13:e0007595. doi: 10.1371/journal.pntd.0007595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All NHP data are included in Table S1. All other source data will be provided upon request.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.