Abstract
Autophagy is one of the degradation pathways to remove proteins or damaged organelles in cells that plays an important role in neuroprotection. Different stages of autophagy are regulated by autophagy‐related genes, and many molecules such as transcription factor EB (TFEB) are involved. The complete autophagy process plays an important role in maintaining the dynamic balance of autophagy and is crucial to the homeostasis of intracellular substance and energy metabolism. Autophagy balance is disrupted in neurodegenerative diseases, accounting for a variety of degeneration disorders. These impairments can be alleviated or treated by the regulation of autophagy through molecules such as TFEB.
Keywords: autophagy, mitophagy, neurodegenerative, TFEB
In this manuscript, we elucidate the possible effect of autophagy and autophagy‐related molecules in neurodegenerative diseases, such as Parkinson's disease and Alzheimer's disease. Also, these molecules have huge implications for the treatment of neurodegenerative diseases.
1. INTRODUCTION
Autophagy is the process by which cells degrade proteins or organelles through lysosomes. Intracellular proteins or organelles need to be cleared or renewed by autophagy or protease systems as the cells grow or are stimulated by external stimuli. Some small molecular substances or short‐lived proteins are mainly degraded by the ubiquitin‐proteasome pathway, whereas long‐lived proteins or damaged organelles in the cytoplasm are degraded by the autophagy‐lysosomal pathway. 1 Normally, autophagy mainly eliminates senescent organelles and macromolecular proteins that are difficult to clear by the proteasome system, which plays an important role in maintaining the intracellular homeostasis of energy and material metabolism. 2 In the 1860s, Duve et al. observed autophagy in hepatocytes. Then, in the 1980s, the signal pathway of autophagy was discovered in yeast cells. 3 Since then, autophagy has gradually become the focus of research, and the liver has become the main target for studying autophagy in mammals. Owing to its special relationship with blood supply, autophagy can be observed in hepatocytes under conditions of nutrient deficiency. Thus, hepatocytes have always been an ideal material for studying autophagy under starvation conditions. Unlike the liver, the brain has priority in energy usage, which means it is difficult to observe neuronal autophagy even under starvation conditions. 4 , 5 Despite this difficulty, more and more studies have found that autophagy plays an important role in the development of nerve cells 6 and the function of synapses. 7 , 8 Abnormal levels of autophagy can cause damage to the nervous system, including autophagosome aggregation, neuronatrophy, mitochondrial depletion, and axonal and dendritic atrophy. 9 , 10 Neurodegeneration is the chronic progressive degeneration and loss of neurons in the brain and spinal cord, which can cause Parkinson's disease (PD), Alzheimer's disease (AD), Huntington's disease (HD), amyotrophic lateral sclerosis (ALS), and dementia with Lewy body (DLB). Abnormal proteins accumulate in the brain of patients with neurodegenerative disease. Studies have shown that neurodegenerative diseases are closely related to abnormal autophagy function.
2. AUTOPHAGY AND DEGRADATION
Autophagy can be divided into 3 types: microautophagy, chaperone‐mediated autophagy, and macroautophagy. Microautophagy transports small molecules into the lysosomal cavity for degradation by endocytosis or budding of the lysosome itself. 11 , 12 Chaperone‐mediated autophagy transports the target protein to lysosomes for degradation mainly by binding Hsc70 to LAMP2A on the lysosomal membrane. 13 When cells are stimulated by starvation or other stress, macroautophagy degrades most of the abnormal proteins or organelles. The autophagy described in this article refers to macroautophagy. The process of autophagy involves the formation of autophagosomes with bilayer membrane structure and the fusion and degradation with lysosomes. 6 , 14 Autophagosomes may be derived from the smooth endoplasmic reticulum and extend and mature under the action of ATG12‐ATG5 complex and ATG8/LC3, forming a bilayer membrane structure with cytoplasm inside and outside. It is degraded and acidified by the inner membrane via endocytosis, and finally fuses with the lysosome to form a monolayer membrane of autophagosomes. 15 , 16 Therefore, the observation of double‐layer or single‐layer cell membrane structure under the ultrastructure is the gold standard for the observation of autophagosomes. 17
Abnormal aggregation of proteins or mitochondrial damage 9 , 18 , 19 has been found in many neurodegenerative diseases, such as the aggregation of α‐synuclein (α‐syn) in PD, 20 and tangles of nerve fibers formed by β‐amyloid and tau proteins in AD. 21 , 22 Abnormal aggregation of proteins can be degraded by proteasome or autophagy. The ubiquitin‐proteasome system can only degrade proteins that are short‐lived, are soluble, and can expand into the proteasome, and it is difficult to eliminate protein aggregates. 6 In contrast, the autophagy‐lysosomal pathway can degrade abnormal protein aggregates, and even damaged mitochondria and other organelles. 23 , 24 , 25 When the function of proteasome or autophagy is impaired, a large number of abnormal proteins and damaged organelles accumulate in the cells, which disturbs the intracellular homeostasis of substance and energy metabolism (Figure 1). Studies have indicated that a mass of ubiquitinated protein inclusions was accumulated in the brain after the autophagy‐related gene ATG5/ATG7 was knocked out in mice. 26 , 27 More notably, the specific inhibition of autophagy in the mice brain can give rise to neurodegenerative diseases even without the accumulation of disease‐related mutant proteins.
FIGURE 1.
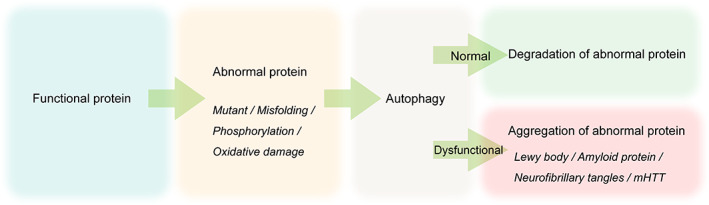
The role of autophagy in protein degradation
3. REGULATION OF AUTOPHAGY
Autophagy‐related genes (ATGs) regulate the autophagy process during different stages of autophagy. 28 Various molecules of ATG expression form different protein complexes to regulate the formation of autophagosomes, including induction of autophagy, generation of autophagosomes, expansion of autophagic vesicles, recognition and endocytosis of substrates, and clearance of autophagosomes 29 , 30 , 31 (Figure 2). In mammalian cells, mammalian rapamycin target protein complex (mTOR) can sense the level of amino acids and ATP in the cell and thus control the autophagy activity of the cell. When cells are suffering from starvation or external stimulus, mTOR is phosphorylated and autophagy is subsequently induced. 32 The formation of autophagosome is also known as the elongation stage, including the binding of ATG12 and the modification of LC3. Under the effect of E3 ligase, LC3‐I is converted into LC3‐II, binding to the forming autophagosomes. 33 Then cargos can combine with the autophagosomes through the LC3‐interacting region (LIR), such as p62. The matured autophagosomes fuse with lysosomes to form autophagolysosomes, and finally, the combined cargos are degraded. 29
FIGURE 2.
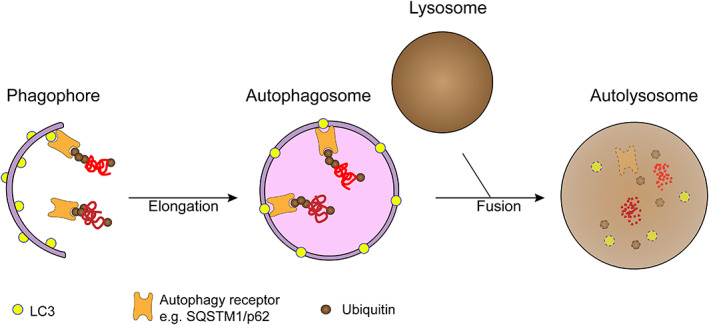
Different stages of autophagy
Some important molecules are involved in the autophagy pathway, among which the TFEB is one of the important transcription factors regulating autophagy (Figure 3). Normally, TFEB binds to 14‐3‐3 protein in the cytoplasm 34 , 35 , 36 under the action of mTORC1 or MAPK1. 34 , 35 , 36 , 37 When cells are under stress due to starvation or impaired lysosomal function, 38 TFEB is activated by dephosphorylation and enters the nucleus to regulate the expression of lysosomal and autophagy‐related molecules, thereby promoting the formation of autophagy and generation of lysosome. 36 , 39 A study found that upregulation of TFEB expression in PD can alleviate lysosomal collapse, autophagic vesicle accumulation, and the aggregation of α‐syn in dopaminergic neurons, demonstrating an obvious protective effect. 40 Another study also showed that the activation of TFEB could rescue cells from the damage of injured mitochondria and reactive oxygen species (ROS). 41
FIGURE 3.
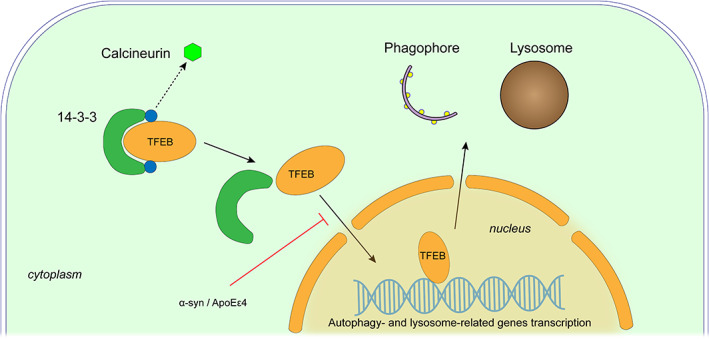
Regulation of TFEB in autophagy
4. MITOPHAGY AND NEURODEGENERATION
The damaged organelles in the cell are mainly degraded by selective autophagy, and the abnormal mitochondria in the neuron are mainly eliminated by mitophagy, to ensure the normal energy metabolism of neurons. 42 , 43 Mitophagy can be triggered by a variety of physiological or pathological factors. 6 In an animal model of PD, the mutation of PD‐related gene LRRK2 can cause mitochondrial damage and induce a mass of mitophagy in midbrain dopaminergic neurons. 44 Simultaneously, dysfunction of mitophagy is also observed, suggesting that abnormal mitophagy may play an important role in PD and other neurodegenerative diseases. 9 , 45 , 46 , 47 , 48 , 49 , 50
4.1. PINK1‐parkin pathway
The damaged mitochondria can be labeled by ubiquitin and degraded and cleared through the PINK1‐parkin pathway 51 , 52 , 53 (Figure 4). Impaired mitochondrial membrane potential decreases, leading to the accumulation of PINK1 protein in the mitochondrial outer membrane, 54 , 55 , 56 recruiting and activating the E3 ubiquitin ligase Parkin 57 by ubiquitin phosphorylation, ubiquitinating the mitochondrial outer membrane protein. It is further degraded by the ubiquitin‐proteasome and autophagy pathway. 58 , 59 The ubiquitin‐proteasome system also plays an important role in the degradation of mitochondrial outer membrane proteins, promoting the completion of PINK1‐Parkin‐mediated mitochondrial autophagy. 58 Some studies have found that TFEB is also involved in PINK1‐Parkin‐mediated mitochondrial autophagy after mitochondrial depolarization. 60 The recruitment of Parkin is regulated by TFEB, which is mTOR and ATG7 independent but requires the participation of ATG5 molecules, 60 , 61 and the specific process remains to be further studied.
FIGURE 4.
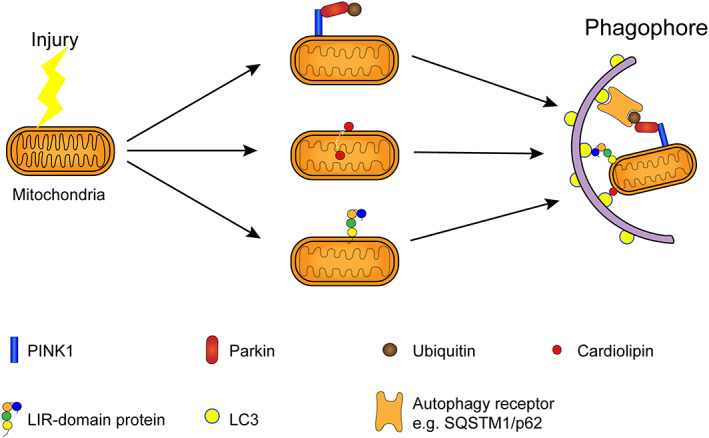
Mitochondrial autophagy mediated by different pathways
4.2. Other mitochondrial autophagy pathways
A variety of transmembrane receptors, including BNIP3L (Nix), 62 , 63 FUNDC1, 64 , 65 and FKBP8, 66 can mediate mitophagy through the non‐PINK1‐Parkin pathway (Figure 2). These transmembrane receptor proteins contain LIR domains that can bind to LC3 and induce mitochondrial autophagy. 6 In cortical neurons and neuroblastomas, the researchers observed mitophagy in the non‐PINK1‐Parkin pathway mediated by cardiolipin. 46 When the mitochondria are damaged, cardiolipin is exposed to the mitochondrial outer membrane from the inner membrane and directly interacts with LC3 to degrade the abnormal mitochondria through the autophagy pathway 46 , 67 (Figure 2). In addition to cardiolipin, ceramides and steroids also participate in the regulation of mitophagy. 68 , 69
Thus, mitophagy‐related molecules may play an important role in neurodegeneration. The dysfunction of mitophagy can induce neuronal damage. On the other hand, the regulation of these mitophagy‐related molecules may attenuate the impact of mitochondrial dysfunction. Moreover, mitophagy‐related molecules can be used to construct animal models of neurodegeneration disease.
5. AUTOPHAGY HOMEOSTASIS AND NEURODEGENERATION
5.1. The transport of autophagosomes in neurons
The entire autophagy process includes the formation, transport, and degradation of autophagosomes, and any abnormality in any of these steps will lead to autophagy dysfunction. Autophagosomes in neurons are mainly formed at the axon ends that are growing or have synaptic connections, 70 , 71 which is closely related to the synthesis and metabolism of synaptic vesicles. 5 The newly generated autophagic vesicles rapidly fuse with syntaxin‐17 to obtain endolysosome markers LAMP1 and Rab17. 72 , 73 The vesicles are dependent on dynein to transport substances from the axon terminal to the nucleus via microtubules 74 and are degraded by fusion with lysosomes. 75 , 76 Studies have shown many neurodegenerative diseases to be associated with lysosomal dysfunction. 77 , 78 , 79 Researchers found a mass of lysosome accumulation in AD models, 73 , 80 suggesting that abnormal transport or degradation of autophagosomes and lysosomes may play an important role in degenerative diseases. Besides, TFEB has been shown to play an important role in the axonal transport of autophagosomes and lysosomes. Moreover, activated TFEB can transcribe lysosomal transmembrane proteins, which bind to dynein and exert a lasting effect on lysosomal transport. 76
5.2. Homeostasis in the process of autophagy
In normal autophagy process, the synthesis and degradation of autophagosomes are in a dynamic balance, 38 whereas in neurodegenerative diseases, multiple intermediate processes of autophagy process may be abnormal. 3 , 38 , 68 , 81 , 82 The decrease in transport, fusion, and degradation efficiency of autophagosomes and the increase in autophagy demand caused by continuous external stimulation all lead to an increase of intracellular autophagy pressure. Once the dynamic balance between autophagy synthesis and degradation is broken, autophagy stress will occur, 38 and induce abnormal autophagy. Thus, a large number of substances need to be transported between the neuron body and axon through microtubules. However, neurons are sensitive to energy changes. Even at a normal level of autophagy, energy depletion or Becin1/Bcl‐2 imbalance may cause cell death when autophagy pressure increases. 83 The degeneration of dopaminergic neurons in the nigrostriatum of the midbrain in PD may be due to the autophagy stress resulting from damage to the neuron body or synapses. 38 , 44
5.3. Interference of abnormal proteins with autophagy homeostasis
On the other hand, abnormal proteins may interfere with autophagy homeostasis in some neurodegenerative diseases. In normal circumstances, the autophagy process can eliminate abnormally aggregated proteins or damaged organelles in cells, and play a protective role in nerve cells. However, cells are damaged or even killed by autophagy stress when the normal autophagy process is disturbed or inhibited. 38 At the same time, mutated LRRK2 or α‐syn was shown to interfere with chaperone‐mediated autophagy in some PD models. 84 , 85 The Jnk‐Bcl‐2 pathway was inhibited by E46K mutation, 86 which impaired the clearance of abnormal α‐syn and damaged nerve cells. Besides, α‐syn is structurally similar to the 14–3‐3 protein, the chaperone of TFEB, 87 which binds to TFEB competitively and blocks TFEB entry into the nucleus, interfering with the entire autophagy process. 88 , 89 In AD, ApoEε4 competes with TFEB for the lysosomal protein promoter SQSTM, MAP1LC3B, LAMP2, leading to abnormal autophagy. 90 , 91 In neurodegenerative diseases such as PD and AD, the mutated molecules induce autophagy imbalance while causing structure damage, resulting in a vicious cycle and eventually leading to cell death. Therefore, the complete autophagy process plays an important role in maintaining the homeostasis of autophagy and is essential for the homeostasis of intracellular substances and energy metabolism.
6. TREATMENT OF NEURODEGENERATIVE DISEASES BY REGULATING AUTOPHAGY
Abnormal autophagy is closely related to neurodegenerative diseases, and many studies have attempted to alleviate or treat neurodegenerative diseases by regulating the level of autophagy. Studies found that regulating the level of autophagy can alleviate or treat neurodegenerative diseases such as PD, AD, and HD. 6 In cell or animal models of HD, drug‐induced autophagy reduces huntingtin accumulation and alleviates HD symptoms in mice and Drosophila. 92 In the APP transgenic mouse model of AD, it was found that injection of the lentivirus expressing Beclin1 into the brain significantly reduced the accumulation of amyloid in the brain of mouse models of early AD. 93 At the same time, Becn1 with F121A mutation attenuated the inhibition of beclin1 by BCL‐2, which reduced amyloid deposition and improved survival rate and cognitive function in APP/PS1 transgenic mice. 94 Moreover, upregulation of ATG7 expression in α‐syn‐overexpressing transgenic mice can decrease α‐syn levels. 93
Neuroprotection can be achieved by increasing the level of autophagy without causing autophagy stress. TFEB is involved in the regulation of autophagosome synthesis, transport, fusion, and lysosomal functions within the autophagy process, and may play an important role in the neuroprotective effects of autophagy. Therefore, TFEB is an ideal target for the treatment of neurodegenerative diseases. 39 Studies have found that upregulation of TFEB expression in tau‐overexpressing rTg4510 model mice can reduce nerve fiber tangling and synaptic injury, and ameliorate neural behavior abnormalities. 94 In the human neuroblastoma cell BE‐M17, upregulation of TFEB attenuates lysosomal collapse in the cytoplasm and reduces autophagic vesicles and α‐syn accumulation simultaneously. 40 In vitro and in some rodent models, the therapeutic effect of TFEB on various neurodegenerative diseases such as PD and AD has attracted increasing attention. However, the protective effect of TFEB has not been confirmed in non‐human primates. Thus, further research on TFEB is needed on the therapeutic effects of neurodegenerative diseases.
7. CONCLUSION
Autophagy homeostasis is essential for the maintenance of normal cell function and is closely associated with the development of neurodegenerative diseases. As an important autophagy‐regulatory protein, TFEB is of great interest. More research on TFEB‐related pathways and drug targets is needed.
9. AUTHOR CONTRIBUTIONS
Changsong Dou wrote the paper. Professor Chuan Qin, Yu Zhang and Ling Zhang reviewed and edited the manuscript.
8. ACKNOWLEDGMENTS
The work was supported by National Natural Science Foundation of China Grant (31970510, 81941012), CAMS Innovation Fund for Medical Sciences (CIFMS) grant (2016‐I2M‐2‐006, 2016‐I2M‐1‐010). Grateful for the suggestions and correction of Professor Chuan Qin, Yu Zhang and Ling Zhang.
Dou C, Zhang Y, Zhang L, Qin C. Autophagy and autophagy‐related molecules in neurodegenerative diseases. Anim Models Exp Med. 2023;6:10‐17. doi: 10.1002/ame2.12229
Funding information
National Natural Science Foundation of China grant (31970510, 81941012), CAMS Innovation Fund for Medical Sciences (CIFMS) grant (2016‐I2M‐2‐006, 2016‐I2M‐1‐010).
REFERENCES
- 1. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xie W, Zhou J. Aberrant regulation of autophagy in mammalian diseases. Biol Lett. 2018;14(1):20170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27(5–6):503‐519. [DOI] [PubMed] [Google Scholar]
- 4. Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36(12):2491‐2502. [DOI] [PubMed] [Google Scholar]
- 5. Pan PY, Zhu Y, Shen Y, et al. Crosstalk between presynaptic trafficking and autophagy in Parkinson's disease. Neurobiol Dis. 2019;122:64‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu CT. Mechanisms of selective autophagy and mitophagy: implications for neurodegenerative diseases. Neurobiol Dis. 2019;122:23‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Plowey ED, Chu CT. Synaptic dysfunction in genetic models of Parkinson's disease: a role for autophagy? Neurobiol Dis. 2011;43(1):60‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lieberman OJ, McGuirt AF, Tang G, et al. Roles for neuronal and glial autophagy in synaptic pruning during development. Neurobiol Dis. 2019;122:49‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cherra SR, Steer E, Gusdon AM, et al. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol. 2013;182(2):474‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernandez D, Torres C A, Setlik W, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron, 2012, 74(2): 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li WW, Li J, Bao JK. Microautophagy: lesser‐known self‐eating. Cell Mol Life Sci. 2012;69(7):1125‐1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oku M, Sakai Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays. 2018;40(6):e1800008. [DOI] [PubMed] [Google Scholar]
- 13. Kaushik S, Cuervo AM. Chaperone‐mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ejlerskov P, Ashkenazi A, Rubinsztein DC. Genetic enhancement of macroautophagy in vertebrate models of neurodegenerative diseases. Neurobiol Dis. 2019;122:3‐8. [DOI] [PubMed] [Google Scholar]
- 15. Shelburne JD, Arstila AU, Trump BF. Studies on cellular autophagocytosis. The relationship of autophagocytosis to protein synthesis and to energy metabolism in rat liver and flounder kidney tubules in vitro. Am J Pathol. 1973;73(3):641‐670. [PMC free article] [PubMed] [Google Scholar]
- 16. Eskelinen EL. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1(1):1‐10. [DOI] [PubMed] [Google Scholar]
- 17. Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17(3):897‐908. [DOI] [PubMed] [Google Scholar]
- 18. Plowey ED, Johnson JW, Steer E, et al. Mutant LRRK2 enhances glutamatergic synapse activity and evokes excitotoxic dendrite degeneration. Biochim Biophys Acta. 2014;1842(9):1596‐1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verma M, Callio J, Otero PA, et al. Mitochondrial calcium dysregulation contributes to dendrite degeneration mediated by PD/LBD‐associated LRRK2 mutants. J Neurosci. 2017;37(46):11151‐11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalia LV, Lang AE. Parkinson's disease. Lancet. 2015;386(9996):896‐912. [DOI] [PubMed] [Google Scholar]
- 21. Pistollato F, Sumalla CS, Elio I, et al. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr Rev. 2016;74(10):624‐634. [DOI] [PubMed] [Google Scholar]
- 22. Tomita T. Aberrant proteolytic processing and therapeutic strategies in Alzheimer disease. Adv Biol Regul. 2017;64:33‐38. [DOI] [PubMed] [Google Scholar]
- 23. Rideout HJ, Lang‐Rollin I, Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol. 2004;36(12):2551‐2562. [DOI] [PubMed] [Google Scholar]
- 24. Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med. 2015;47:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rubinsztein DC. The roles of intracellular protein‐degradation pathways in neurodegeneration. Nature. 2006;443(7113):780‐786. [DOI] [PubMed] [Google Scholar]
- 26. Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885‐889. [DOI] [PubMed] [Google Scholar]
- 27. Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880‐884. [DOI] [PubMed] [Google Scholar]
- 28. Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self‐digestion. Nature. 2008;451(7182):1069‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maiuri MC, Zalckvar E, Kimchi A, et al. Self‐eating and self‐killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8(9):741‐752. [DOI] [PubMed] [Google Scholar]
- 30. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22(2):140‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125‐156. [DOI] [PubMed] [Google Scholar]
- 33. Roczniak‐Ferguson A, Petit CS, Froehlich F, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):a42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429‐1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation‐induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu CT. Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol. 2006;65(5):423‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473‐477. [DOI] [PubMed] [Google Scholar]
- 38. Dehay B, Bove J, Rodriguez‐Muela N, et al. Pathogenic lysosomal depletion in Parkinson's disease. J Neurosci. 2010;30(37):12535‐12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhuang XX, Wang SF, Tan Y, et al. Pharmacological enhancement of TFEB‐mediated autophagy alleviated neuronal death in oxidative stress‐induced Parkinson's disease models. Cell Death Dis. 2020;11(2):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum Mol Genet. 2010;19(R1):R28‐R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soubannier V, McLelland GL, Zunino R, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22(2):135‐141. [DOI] [PubMed] [Google Scholar]
- 42. Guzman JN, Ilijic E, Yang B, et al. Systemic isradipine treatment diminishes calcium‐dependent mitochondrial oxidant stress. J Clin Invest. 2018;128(6):2266‐2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chinta SJ, Mallajosyula JK, Rane A, et al. Mitochondrial alpha‐synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486(3):235‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197‐1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dagda RK, Cherra SR, Kulich SM, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284(20):13843‐13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dagda RK, Zhu J, Kulich SM, et al. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy. 2008;4(6):770‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Osellame LD, Duchen MR. Defective quality control mechanisms and accumulation of damaged mitochondria link Gaucher and Parkinson diseases. Autophagy. 2013;9(10):1633‐1635. [DOI] [PubMed] [Google Scholar]
- 48. Zhu JH, Horbinski C, Guo F, et al. Regulation of autophagy by extracellular signal‐regulated protein kinases during 1‐methyl‐4‐phenylpyridinium‐induced cell death. Am J Pathol. 2007;170(1):75‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hamacher‐Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73(4):775‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McWilliams TG, Muqit MM. PINK1 and parkin: emerging themes in mitochondrial homeostasis. Curr Opin Cell Biol. 2017;45:83‐91. [DOI] [PubMed] [Google Scholar]
- 51. Yin XM, Ding WX. The reciprocal roles of PARK2 and mitofusins in mitophagy and mitochondrial spheroid formation. Autophagy. 2013;9(11):1687‐1692. [DOI] [PubMed] [Google Scholar]
- 52. Kawajiri S, Saiki S, Sato S, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584(6):1073‐1079. [DOI] [PubMed] [Google Scholar]
- 53. Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits parkin to damaged mitochondria and activates latent parkin for mitophagy. J Cell Biol. 2010, 2;189:211‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol. 2010;8(1):e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shiba‐Fukushima K, Imai Y, Yoshida S, et al. PINK1‐mediated phosphorylation of the parkin ubiquitin‐like domain primes mitochondrial translocation of parkin and regulates mitophagy. Sci Rep. 2012;2:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chan NC, Salazar AM, Pham AH, et al. Broad activation of the ubiquitin‐proteasome system by parkin is critical for mitophagy. Hum Mol Genet. 2011;20(9):1726‐1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoshii SR, Kishi C, Ishihara N, et al. Parkin mediates proteasome‐dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286(22):19630‐19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nezich CL, Wang C, Fogel AI, et al. MiT/TFE transcription factors are activated during mitophagy downstream of parkin and Atg5. J Cell Biol. 2015;210(3):435‐450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vincow ES, Merrihew G, Thomas RE, et al. The PINK1‐parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110(16):6400‐6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11(1):45‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu L, Sakakibara K, Chen Q, et al. Receptor‐mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24(7):787‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu L, Feng D, Chen G, et al. Mitochondrial outer‐membrane protein FUNDC1 mediates hypoxia‐induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14(2):177‐185. [DOI] [PubMed] [Google Scholar]
- 63. Lv M, Wang C, Li F, et al. Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy. Protein Cell. 2017;8(1):25‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bhujabal Z, Birgisdottir AB, Sjottem E, et al. FKBP8 recruits LC3A to mediate parkin‐independent mitophagy. EMBO Rep. 2017;18(6):947‐961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kagan VE, Jiang J, Huang Z, et al. NDPK‐D (NM23‐H4)‐mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23(7):1140‐1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22(3):367‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ivatt RM, Sanchez‐Martinez A, Godena VK, et al. Genome‐wide RNAi screen identifies the Parkinson disease GWAS risk locus SREBF1 as a regulator of mitophagy. Proc Natl Acad Sci U S A. 2014;111(23):8494‐8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30(1):71‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vanhauwaert R, Kuenen S, Masius R, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017;36(10):1392‐1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng XT, Zhou B, Lin MY, et al. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol. 2015;209(3):377‐386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's‐like axonal dystrophy. J Neurosci. 2011;31(21):7817‐7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McKenney RJ, Huynh W, Tanenbaum ME, et al. Activation of cytoplasmic dynein motility by dynactin‐cargo adapter complexes. Science. 2014;345(6194):337‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jongsma ML, Berlin I, Wijdeven RH, et al. An ER‐associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166(1):152‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Willett R, Martina JA, Zewe JP, Wills R, Hammond GRV, Puertollano R. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun. 2017;8(1):1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lie P, Nixon RA. Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol Dis. 2019;122:94‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fraldi A, Klein AD, Medina DL, et al. Brain disorders due to lysosomal dysfunction. Annu Rev Neurosci. 2016;39:277‐295. [DOI] [PubMed] [Google Scholar]
- 77. Lloyd‐Evans E, Haslett LJ. The lysosomal storage disease continuum with ageing‐related neurodegenerative disease. Ageing Res Rev. 2016;32:104‐121. [DOI] [PubMed] [Google Scholar]
- 78. Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno‐electron microscopy study. J Neuropathol Exp Neurol. 2005;64(2):113‐122. [DOI] [PubMed] [Google Scholar]
- 79. Cherra SR, Chu CT. Autophagy in neuroprotection and neurodegeneration: a question of balance. Future Neurol. 2008;3(3):309‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2011;8(2):108‐117. [DOI] [PubMed] [Google Scholar]
- 81. Pattingre S, Tassa A, Qu X, et al. Bcl‐2 antiapoptotic proteins inhibit Beclin 1‐dependent autophagy. Cell. 2005;122(6):927‐939. [DOI] [PubMed] [Google Scholar]
- 82. Martinez‐Vicente M, Talloczy Z, Kaushik S, et al. Dopamine‐modified alpha‐synuclein blocks chaperone‐mediated autophagy. J Clin Invest. 2008;118(2):777‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tanik SA, Schultheiss CE, Volpicelli‐Daley LA, et al. Lewy body‐like alpha‐synuclein aggregates resist degradation and impair macroautophagy. J Biol Chem. 2013;288(21):15194‐15210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yan JQ, Yuan YH, Gao YN, et al. Overexpression of human E46K mutant alpha‐synuclein impairs macroautophagy via inactivation of JNK1‐Bcl‐2 pathway. Mol Neurobiol. 2014;50(2):685‐701. [DOI] [PubMed] [Google Scholar]
- 85. Ostrerova N, Petrucelli L, Farrer M, et al. Alpha‐synuclein shares physical and functional homology with 14‐3‐3 proteins. J Neurosci. 1999;19(14):5782‐5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Decressac M, Mattsson B, Weikop P, et al. TFEB‐mediated autophagy rescues midbrain dopamine neurons from alpha‐synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110(19):E1817‐E1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Siddiqui A, Bhaumik D, Chinta SJ, et al. Mitochondrial quality control via the PGC1alpha‐TFEB signaling pathway is compromised by parkin Q311X mutation but independently restored by rapamycin. J Neurosci. 2015;35(37):12833‐12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parcon PA, Balasubramaniam M, Ayyadevara S, et al. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement. 2018;14(2):230‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Reddy K, Cusack CL, Nnah IC, et al. Dysregulation of nutrient sensing and CLEARance in presenilin deficiency. Cell Rep. 2016;14(9):2166‐2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585‐595. [DOI] [PubMed] [Google Scholar]
- 91. Pickford F, Masliah E, Britschgi M, et al. The autophagy‐related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118(6):2190‐2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rocchi A, Yamamoto S, Ting T, et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer's disease. PLoS Genet. 2017;13(8):e1006962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Crews L, Spencer B, Desplats P, et al. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha‐synucleinopathy. PLoS One. 2010;5(2):e9313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94. Polito VA, Li H, Martini‐Stoica H, et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med. 2014;6(9):1142‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]


