Abstract
The distal region (−830 to −720 bp) of the rat whey acidic protein (WAP) gene contains a composite response element (CoRE), which has been demonstrated previously to confer mammary gland-specific and hormonally regulated WAP gene expression. Point mutations in the binding sites for specific transcription factors present within this CoRE have demonstrated the importance of both nuclear factor I (NFI) and STAT5 as well as cooperative interactions with the glucocorticoid receptor (GR) in the regulation of WAP gene expression in the mammary gland of transgenic mice. This study reports the characterization of NFI gene expression during mammary gland development and the identification and cloning of specific NFI isoforms (NFI-A4, NFI-B2, and NFI-X1) from the mouse mammary gland during lactation. Some but not all of these NFI isoforms synergistically activate WAP gene transcription in cooperation with GR and STAT5, as determined using transient cotransfection assays in JEG-3 cells. On both the WAP CoRE and the mouse mammary tumor virus long terminal repeat promoter, the NFI-B isoform preferentially activated gene transcription in cooperation with STAT5A and GR. In contrast, the NFI-A isoform suppressed GR and STAT cooperativity at the WAP CoRE. Finally, unlike their interaction with the NFI consensus binding site in the adenovirus promoter, the DNA-binding specificities of the three NFI isoforms to the palindromic NFI site in the WAP CoRE were not identical, which may partially explain the failure of the NFI-A isoform to cooperate with GR and STAT5A.
Milk protein genes have been widely studied as models for understanding hormonal and developmental stage-specific gene expression in the mammary gland (46). Several transcription factors, various peptide and steroid hormones, cytokines, and cell-cell and cell-substratum interactions have been shown to influence milk protein gene expression (5, 11, 18, 24, 31, 32, 54, 56). The rat whey acidic protein (WAP) gene, the major whey protein expressed in rodents, has been employed in our laboratory as a model system to study the transcriptional regulation of milk protein genes. A mammary gland-specific and hormonally regulated DNase I-hypersensitive site at −830 to −720 bp 5′ to the site of transcription initiation of the rat WAP gene was identified previously (33). Nuclear factor I (NFI) binding at both palindromic and half-sites constituted the major DNA-protein interactions detected within this tissue-specific nuclease-hypersensitive region. Point mutations introduced into these NFI binding sites abrogated expression in transgenic mice, indicating that NFI is a key regulator of WAP gene expression.
A recognition site for STAT5, the primary mediator of prolactin-induced milk protein gene expression, was also identified immediately proximal to the NFI binding sites. Mutation of this site reduced transgene expression by approximately 90% per gene copy, but did not alter tissue specificity (34). In addition, several glucocorticoid receptor (GR) binding sites (GREs) were identified flanking the NFI sites using an in vitro DNase I footprinting assay with baculovirus-expressed GR. These sites were able to confer dexamethasone inducibility on a heterologous thymidine kinase (tk) -chloramphenicol acetyltransferase (CAT) reporter gene construct in transient cotransfection assays performed with GR in CV1 cells. These results suggest that regulation of WAP gene expression is determined by the cooperative interactions among several transcription factors that constitute a composite response element (CoRE).
The CTF/NFI family of ubiquitous transcription factors was initially discovered as part of an adenovirus DNA replication complex, but later found to be involved in the transcriptional regulation of various cellular and viral genes (38). There are four different NFI genes (NFI-A, NFI-B, NFI-C, and NFI-X). Multiple alternatively spliced isoforms of each gene have been identified from different species, and NFI isoforms exhibit a high degree of homology from chickens to humans (35, 48, 49). The 220-amino-acid N-terminal region of all the NFI proteins is highly conserved and is responsible for DNA binding, dimerization and adenovirus replication (3, 36). NFI binds to DNA as both homo- and heterodimers, and the apparent binding affinity to the consensus binding site, TTGGC(N5)GCCAA, is similar for different isoforms (21, 29).
Alternative splicing generates many variants of the proline-rich C-terminal region, which is responsible for transactivation (28). NFI proteins have been found to be important for the regulation of the genes expressed in almost every tissue and organ system, including brain (4, 16), lung (2), liver (7, 8, 13, 19, 25, 44), and chondrocytes (53). They are also important for the expression of several mammary-specific genes, such as the milk protein genes, WAP (34), and β-lactoglobulin (55) as well as other genes expressed in the mammary gland, such as mouse mammary tumor virus (MMTV) (37), testosterone-repressed prostate message (17), β1,4-galactosyltransferase (45), and carboxyl ester lipase (26).
Relatively little is known about the mechanisms by which NFI family members regulate tissue- and cell-specific gene expression, except for the observation that different NFI isoforms vary with cell type and growth conditions. In the mouse mammary gland, WAP gene expression increases at midpregnancy, is highest during lactation, and decreases rapidly at the onset of involution. Accordingly, specific NFI isoforms were cloned from mRNA isolated from mice at day 2 of lactation. These were characterized by transient transfection in JEG-3 choriocarcinoma cells, which contain undetectable levels of endogenous NFI and GR, to elucidate how these specific NFI isoforms might regulate WAP gene transcription in cooperation with GR and STAT5.
In the present study, the role of these specific NFI isoforms in regulating WAP gene expression was investigated, and transcriptional cooperativity among all three transcription factors at the WAP CoRE was demonstrated. Furthermore, the DNA-binding specificity of the three specific NFI isoforms appears in part to be correlated with their ability to activate WAP gene transcription.
MATERIALS AND METHODS
RT-PCR.
Reverse transcription (RT)-PCR was carried out with NFI-specific degenerate primers Deg1 (5′-TTCCGGATGARTTYCAYCITTYATYGARGC-3′, where R is purine [A or G], Y is pyrimidine [C or T], B is C or G, and I is inosine) and Deg2 (5′-AATCGATRTGRTGBGGCTGIAYRCAIAG-3′) as described previously by Kulkarni and Gronostajski (30). Each RT reaction was performed in a total volume of 20 μl of reaction mix containing 1 μg of total RNA isolated from different stages of mouse mammary gland development, 4 μl of 5× RT buffer, 0.2 μl of 0.1 M dithiothreitol, 1 μl of oligo(dT) (100 ng/μl), 0.5 μl of RNasin (40 U/μl), and 1 μl of Moloney murine leukemia virus RT (200 U/μl). Reactions were incubated at 37°C for 1 h followed by 97°C for 5 min. RT mix (20 μl) was added with 80 μl of PCR mix containing 8 μl of 10× PCR buffer, 8 μl of 25 mM MgCl2, 5 μl of each degenerate primer, and 2 U of Taq polymerase, and PCR was set up for 30 cycles at 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min. In the case of radiolabeled RT-PCR, 0.2 μl of [α-32P]dATP was added to the PCR mix.
Cloning of NFI isoforms.
The specific NFI isoforms were amplified by RT-PCR using NFI-specific primers from the 5′ and 3′ ends of the mRNA and 1 μg of total RNA isolated from BALB/c mice at day 2 of lactation. They were then cloned in the pGEM-T-Easy (Promega) vector and sequenced. The primers used were NFI-A3′1C (5′-GATGCTGCAACTTTTATCCCAGG-3′), NFIAE1 (5′-TGTATTCTCCGCTCTGTCTCAC-3′), NFI-BE2 (5′-GTTTTTGGCATACTACGTGCAGG-3′), NFI-B3′C (5′-TTGGGACATTTGGGACATTTGCC-3′), NFI-C5′ (5′-ATGTATTCCTCCCCGCTCTGCC-3′), NFI-C3′ (5′-TTTCCACCAAAAATGCAGGCTGG-3′), NFI-X5′ (5′-ATGTATAGCCCCGTACTGCCTC-3′), and NFI-X3′ (5′-AGGACTGAGACTCCTGTGGGATG-3′).
Plasmid constructions.
NFI cDNAs were cloned in the pCH expression vector (9). To obtain the pCH-NFI-X1 construct, both pCH-NFI-X (9) and an RT-PCR-cloned NFI-X fragment (pRXh2) were digested with the Tth111I and AgeI enzymes and ligated to the 894-bp Tth111I-AgeI fragment from pRXh2 into the pCH-NFI-X vector. To generate the pWAPtk-luciferase construct, the 100-bp (−820 to −720) fragment from the WAP distal promoter region was amplified by PCR using nested primers containing KpnI and BamHI sites. The basal tk promoter (168 bp) was excised from pBLCAT2 by BamHI and XhoI digestion. The KpnI-BamHI fragment from the WAP distal promoter region was then ligated with the BamHI-XhoI fragment from the basal tk promoter. Then the KpnI-XpoI fragment of the WAP distal promoter and the basal tk promoter were ligated to the KpnI and XhoI sites of pGL2-basic vector. The orientation and correct reading frame of all plasmids were verified by sequencing. Rat STAT5A cDNA was subcloned into the pRcCMV vector (Invitrogen, Carlsbad, Calif.) described previously (58). A rat GR expression plasmid (pSTCGR3-795), described previously by Godowski et al. (20), was kindly provided by Rainer Lanz (Baylor College of Medicine) and Sandro Rusconi (University of Fribourg). The MMTV-luciferase construct (pAH Luc) was kindly provided by Sandy Grimm and Steve Nordeen. All plasmids were purified using a Qiagen DNA maxi-prep kit (Qiagen, Valencia, Calif.).
Cell culture and transfection assays.
JEG-3 choriocarcinoma cells (American Type Culture Collection) were cultured in minimal essential medium (MEM; Gibco-BRL) containing 10% fetal bovine serum. Twenty-four hours prior to transfection, cells were plated onto 60-mm dishes. Two hours before transfection, cells were cultured with fresh medium containing MEM, 10% charcoal-stripped horse serum (SHS), and insulin (5 μg/ml). Transfection was carried out using 8 to 10 μg of DNA by the calcium phosphate method. After 24 h of transfection, the cells were washed twice with phosphate-buffered saline (PBS), and then the cells were treated with fresh medium (MEM plus 10% SHS and insulin), with or without ovine prolactin (1 μg/ml) and hydrocortisone (HC) (1 μg/ml). Twenty-four hours after the treatment of hydrocortisone and prolactin, the cells were harvested, lysed with lysis buffer (Boehringer Mannheim), and assayed for luciferase and β-galactosidase activity.
RNase protection. (i) Generation of riboprobes.
Deg1 and Deg2 primers were used to amplify the 486-bp conserved DNA-binding region from each of the NFI genes. Amplified degenerate fragments were cloned in pGEM-T-Easy vectors, and the antisense strand was identified by sequencing. Antisense probes were synthesized by using the Ambion in vitro transcription kit (MAXIscript SP6/T7 kit). The pTRI-actin-mouse template was provided in the RPAIII kit (Ambion) to synthesize the mouse β-actin probe, which was used as a control, and the Century RNA marker (Ambion) was used as a molecular weight marker.
(ii) RNase protection.
Total cellular RNA was isolated by RNAzol B from different stages of mouse mammary gland development. RNase protection assays were performed using the RPAIII kit (Ambion) according to the manufacturer's protocol. Total RNA (10 μg) was used in each reaction to coprecipitate an excess amount of each NFI and β-actin probe, followed by hybridization overnight at 45°C.
EMSA.
Oligonucleotides encompassing the NFI palindromic site of the WAP CoRE (−820 to −720) and the adenovirus NFI consensus binding site were used for electrophoretic mobility shift assay (EMSA) analysis (coding strand for WAP NFI, 5′-TTGGGCACAGTGCCCAACAG-3′, and coding strand for adenovirus NFI, 5′-CTAGCTATTTTGGATTGAAGCCAATAT-3′). Equimolar concentrations of each oligonucleotide from both strands were annealed in the presence of 1× React 2 (Promega) buffer at 94°C for 10 min and then cooled to room temperature for 3 to 4 h. The double-stranded oligonucleotide was end labeled with [γ-32P]dATP using polynucleotide kinase (Gibco-BRL), and the probe was purified using p-6 Micro Bio-spin columns (Bio-Rad) followed by trichloroacetic acid precipitation to quantify the amount of labeled probe. The three NFI genes (A4, B2, and X1) were expressed in JEG-3 cells, and nuclear extracts, isolated as described previously (58), were used for the DNA-binding assays. The amount of nuclear extract required for 50% binding of the labeled probe was calculated, and then for each gene that amount was used for the competition assays. Nuclear extracts were isolated from the same passage of JEG-3 cells, and equimolar concentrations of each probe were used in all the EMSAs.
Western blotting.
Western blotting was performed using previously published protocols (58). All the NFI constructs contained a hemagglutinin (HA) epitope at their N termini. A monoclonal HA antibody (Babco, Berkeley, Calif.) was used to identify the HA-tagged NFI proteins at a dilution of 1:2,000. Biotinylated goat anti-mouse immunoglobulin G and streptavidin-horseradish peroxidase were purchased from Calbiochem (La Jolla, Calif.). A mouse monoclonal anti-GR antibody (Bu-GR2) was obtained from Affinity Bioreagents.
RESULTS
Cloning the predominant NFI isoforms expressed in the mammary gland during lactation.
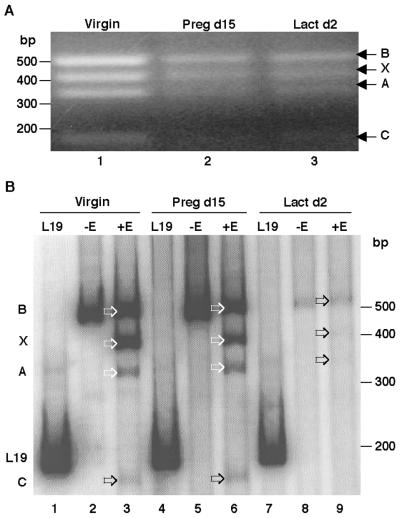
Four NFI genes (NFI-A, -B, -C, and -X) and many different alternatively spliced isoforms of these genes have been isolated from vertebrates, including mice. However, the expression patterns of these NFI isoforms at different stages of mammary gland development, especially during lactation, have not been definitively established. To initially determine the expression pattern of the four NFI genes during different stages of mammary gland development, RT-PCR was performed using total RNA isolated from virgin, pregnancy day 15, lactation day 2, and involution and degenerate primers previously shown to detect all four mouse NFI genes. As the N-terminal regions of the four NFI genes are conserved, a common band of 486 bp was generated. On the basis of their unique restriction enzyme sites, the 486-bp amplified region was digested with BamHI, BpuI1021, and NarI, and the digestion products were separated on a 2% agarose gel (Fig. 1A). The digestion products of the 486-bp RT-PCR-amplified fragment isolated from virgin, pregnancy day 15, and lactation day 2 glands are shown in Fig. 1A, lanes 1, 2, and 3, respectively. NFI-B generated a fragment of 486 bp (uncut), NFI-X one of 394 bp, NFI-A one of 327 bp, and NFI-C one of approximately 200 bp. To quantitate the levels of these individual digestion products, the same experiment was performed using radioactive PCR. After digestion the bands were separated on a 10% polyacrylamide gel, autoradiographed and quantitated using a PhosphorImager (Fig. 1B). The results from Fig. 1 suggest that all four NFI genes (A, B, C, and X) are expressed in the virgin mammary gland and during midpregnancy. However, using this approach, NFI-C was not detectable during lactation.
FIG. 1.
Identification of NFI genes expressed during different stages of mammary gland development. (A) RNA was isolated from virgin, pregnancy day 15 (Preg d15), and lactation day 2 (Lact d2) mouse mammary glands, and RT-PCR was performed with degenerate primers Deg1 and Deg2. The 486-bp amplified product was digested with BamHI, BpuI1021, and NarI and then separated on a 2% agarose gel. The bands in each lane (1, 2, and 3) represent the NFI genes in virgin, pregnancy day 15, and lactation day 2 mice, respectively. The arrows indicate the undigested 486-bp NFI-B, 394-bp NFI-X, 327-bp NFI-A, and ≈200-bp NFI-C gene fragments. The numbers on the left side represent the molecular size markers. (B) RT-PCR was performed with degenerate primers and [32P]dATP, and the BamHI-, BpuI1021-, and NarI-digested 486-bp fragments were separated on a 10% polyacrylamide gel. Lanes 1, 4, and 7 represent reactions using the L19 primers, which generated a 200-bp product, and lanes 2, 5, and 8 represent the degenerate primer-amplified 486-bp band of the conserved N-terminal region of each NFI gene (−E, not digested with enzymes). Lanes 3, 6, and 9 represent the expression of each NFI gene in virgin, pregnancy day 15, and lactation day 2 mammary glands of mice (+E, digested with enzymes after amplification). The arrows indicate the individual NFI genes. The numbers on the right side represent the molecular size markers.
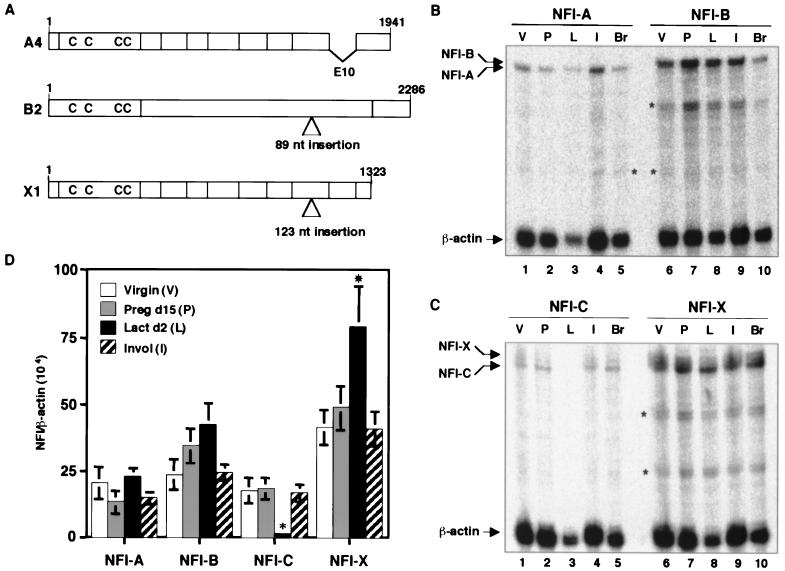
These results were confirmed using RNase protection assays (Fig. 2B, C and D). NFI-A expression was fairly constant at different stages of mammary gland development. NFI-B and -X, however, exhibited their highest levels of expression during lactation and then decreased in expression during involution. In contrast, NFI-C expression decreased significantly during lactation and then increased during involution (Fig. 2D).
FIG. 2.
RNase protection assay for identification of the expression pattern of four NFI genes at different stages of mammary gland development. RNA was isolated from mammary gland of virgin (V), pregnancy day 15 (P), lactation day 2 (L), and involution stage (I) mice and from mouse brain (Br). RNase protection assays were performed using antisense probes for each NFI gene along with a mouse β-actin control. (A) Schematic representation of the genomic structure of three NFI isoforms expressed in the mouse mammary gland during lactation. (B) Lanes 1 to 5 represent the NFI-A, and lanes 6 to 10 represent NFI-B. (C) Lanes 1 to 5 represent NFI-C, and lanes 6 to 10 represent NFI-X. Upper arrows indicate the 486-bp protected band for each NFI gene, and the asterisk indicates the nonspecific bands. The expression of each NFI gene at each developmental stage was analyzed using the PhosphorImager, and the results are graphically represented in panel D. Error bars denote the standard error of the mean (SEM) of three independent determinations. An asterisk above the error bar represents the statistical significance of the data (P < 0.05).
To clone the NFI isoforms expressed during lactation, RT-PCR was performed using primers designed on the basis of the sequence of 5′ and 3′ ends of each of the mouse NFI genes. The nucleotide sequence of several clones was determined, and the three predominant spliced NFI isoforms were identified as NFI-A4, NFI-B2, and NFI-X1 (Fig. 2A). As expected, the predicted amino acid sequences were homologous with the previously published murine NFI-A4, -B2, and -X1 proteins (28).
Cooperativity among NFI, GR, and STAT5A in the regulation of WAP gene expression.
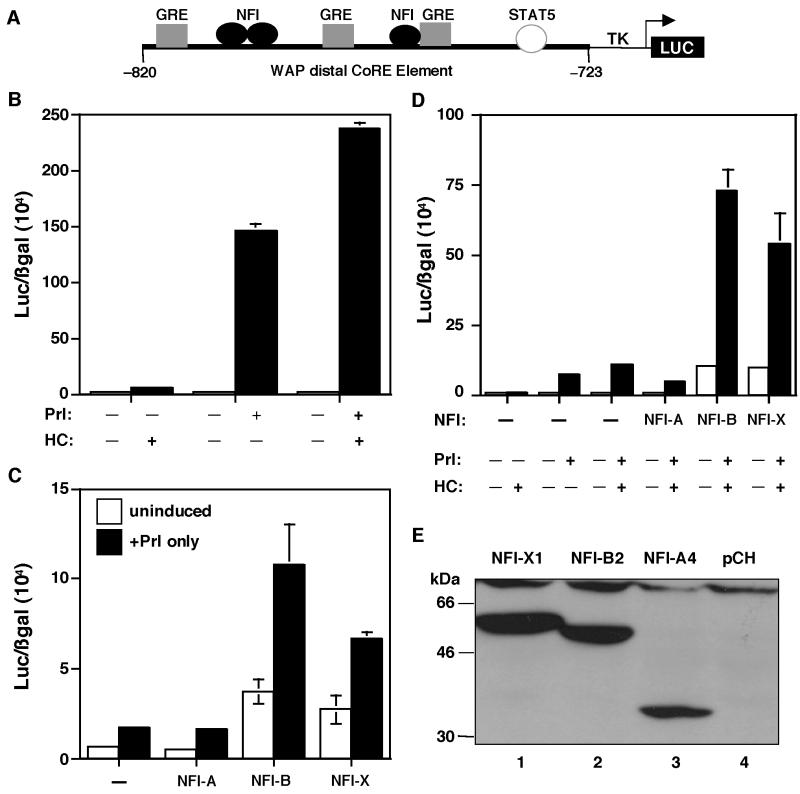
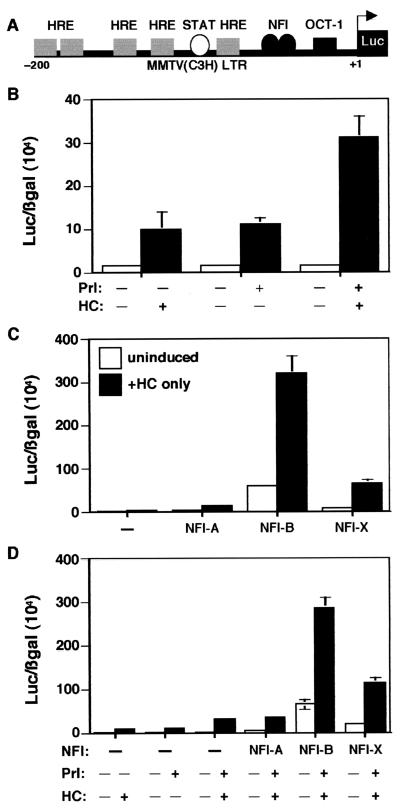
The WAP gene CoRE (−820 to −720) containing the binding sites for NFI, GR, and STAT5 is essential for the regulation of WAP gene expression (Fig. 3A). Specifically, NFI has been demonstrated to play a vital role in WAP gene regulation. To determine the role of the individual NFI isoforms in the transcriptional regulation of WAP gene expression, transient transfections were performed in JEG-3 choriocarcinoma cells, because JEG-3 cells contain low levels of endogenous NFI and GR. The three NFI isoforms were cloned in the mammalian expression vector pCH, and their activity on the pWAPtk-Luc reporter construct was determined. The prolactin receptor (PrlR), GR, and STAT5A expression constructs were cotransfected along with the different NFI isoforms as well as pRSV-β-gal to monitor transfection efficiency.
FIG. 3.
Cooperativity of GR and STAT5A on the WAP CoRE. (A) Schematic diagram of the pWAPtk-luciferase construct. (B) GR and STAT5A cooperativity. The pWAPtk-luciferase construct (1 μg) was transiently transfected into JEG-3 cells along with GR (0.2 μg), STAT5A (1 μg), PrlR (0.3 μg), and RSV-β-gal (0.3 μg). Twenty-four hours after induction with hormones (1 μg/ml of both HC and Prl), luciferase expression was measured and transfection efficiencies were normalized to β-galactosidase activity. Cells were induced with HC, Prl, and both HC and Prl, as indicated. (C) Cooperativity of STAT5A and NFI isoforms (0.2 μg, 0.5 μg, and 1 μg of A, B, and X, respectively) for the activation of the WAP CoRE. Three NFI isoforms were transfected into JEG-3 cells along with STAT5A, PrlR, pWAPtk-luciferase, and RSV-β-gal, and the cells were induced with Prl. Cells were transfected with STAT5A and PrlR and induced with Prl and with NFI-A, -B, and -X, respectively, as shown. (D) Cooperativity of GR, STAT5A, and NFI isoforms on the WAP CoRE. Cells were transiently transfected with GR, STAT5A, the PrlR, and pWAPtk-luciferase along with RSV-β-gal. In addition to these constructs, cells were transfected with either NFI-A, -B, or -X. Cells were induced with HC, Prl, or both HC and Prl as indicated. Error bars represent the standard error of three independent determinations. (E) Expression of the three NFI isoforms in JEG-3 cells. Total proteins were isolated from the cells transfected with NFI-X1 (lane 1), NFI-B2 (lane 2), and NFI-A4 (lane 3) and from cells transfected with only the pCH vector (lane 4). These extracts were analyzed on sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (SDS–8% PAGE), and Western blots were performed using an anti-HA antibody. The molecular sizes of the protein markers are shown.
HC-activated GR induced WAP promoter-driven luciferase expression approximately 5-fold (Fig. 3B), while STAT5 with Prl induction resulted in a further 30-fold increase in activity (Fig. 3B). When STAT5 and GR were cotransfected and induced by both Prl and HC, WAP-driven luciferase expression increased almost 47-fold compared to HC alone (Fig. 3B). These results indicate that there is synergism between GR and STAT5 in the regulation of WAP gene expression, similar to that observed previously on the β-casein promoter (15).
To determine the interaction of STAT5A with the three different specific NFI isoforms in the regulation of WAP CoRE activity, STAT5A was cotransfected with the PrlR and each of the three different NFI isoforms, and the cells were then treated with Prl. As illustrated in Fig. 3C, WAP-driven luciferase activity was induced by STAT5 and Prl, but no further induction was observed when STAT5A was cotransfected with the NFI-A4 isoform. However, when STAT5A was cotransfected with NFI-B2, reporter expression increased fivefold. With NFI-X1, the increase observed was only 2.5-fold compared to STAT5A alone (Fig. 3C). Interestingly, NFI-B2 and -X1 resulted in an increase in both basal and Prl-induced expression. Thus, NFI-B2 appeared to exhibit the greatest cooperativity with STAT5A, while NFI-A4 was ineffective in the transcriptional activation of the WAP distal CoRE.
To determine how GR might influence this cooperativity between STAT5 and the three different NFI isoforms, GR was included in the transfections, and the cells were induced with both HC and Prl. As expected, the synergistic interaction of GR and STAT5 was once again observed (Fig. 3D), but surprisingly, when GR and STAT were cotransfected with NFI-A4, expression was significantly repressed (Fig. 3D). In contrast, when GR and STAT5 were cotransfected with NFI-B2, expression was increased 7.3-fold, while with NFI-X1, it increased 5.3-fold (Fig. 3D). These experiments suggest that cooperative activity among the transcription factors GR, STAT5A, and NFI-B2 results in optimal transcriptional activation of WAP gene expression. The expression levels of the three NFI isoforms in these transient transfection experiments were confirmed by Western blotting with an anti-HA antibody (Fig. 3E).
Interaction between GR and NFI in transcriptional activation of WAP CoRE.
GR plays a critical role in the transcription of several mammary gland-specific genes. The WAP distal CoRE contains three GREs, one of which is a half-GRE that overlaps the NFI binding site (Fig. 3A). An interaction between GR and different NFI isoforms has already been demonstrated on the MMTV promoter (10). When GR was cotransfected with the NFI-A4 isoform, no induction of the WAP CoRE-driven luciferase activity was observed following HC treatment (Fig. 4C). However, with NFI-B2, WAP promoter activity was induced 22-fold compared to GR alone (Fig. 4C). With NFI-X1, this induction was 16-fold (Fig. 4C). Thus, once again the NFI-B isoform exhibited a greater cooperativity with GR than the NFI-X isoform, and the NFI-A isoform was inactive.
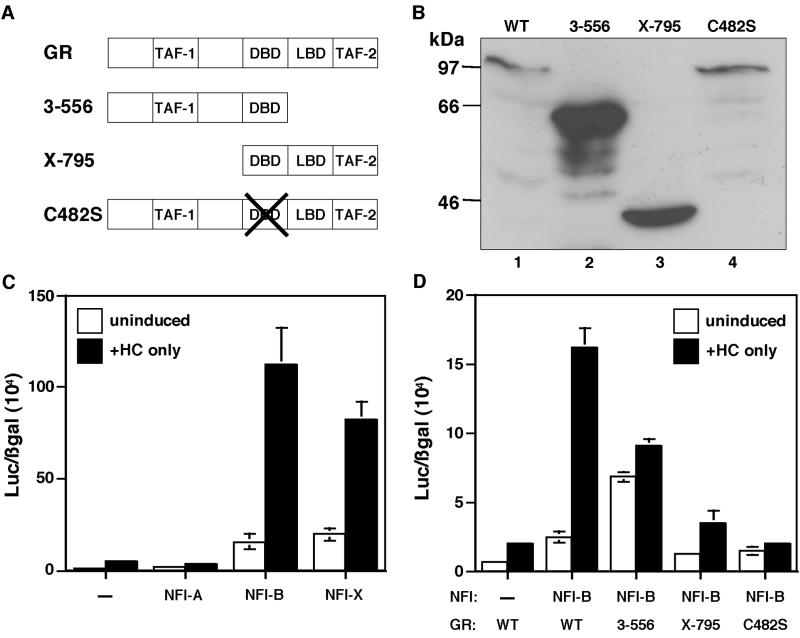
FIG. 4.
GR domains required for cooperative interactions. (A) Schematic diagram of wild-type (WT) GR and mutant GR proteins. The C-terminal mutant is designated GR 3-556, the N-terminal mutant is GR X-795, and the DNA-binding domain (DBD) mutant is GR C482S. (B) The levels of expression of GR and the different GR mutants shown in panel D determined by Western blotting. Lane 1, wild-type GR; lane 2, GR 3-556; lane 3, GR X-795; lane 4, GR C482S. Total protein isolated from transfected cells depicted in panel D was separated by SDS–8% PAGE and blotted with mouse monoclonal anti-GR antibody. The numbers on the left indicate the molecular sizes of the protein markers. (C) Cooperativity of GR with NFI isoforms for the activation of the WAP CoRE. GR was cotransfected with three NFI isoforms into JEG-3 cells along with pWAPtk-luciferase and RSV-β-gal, and the cells were induced with HC. Cells were transfected with only GR and with NFI-A, NFI-B, or NFI-X as illustrated. (D) Analysis of GR and GR mutant cooperativity with NFI-B on the WAP CoRE: cells were transfected with wild-type GR or one of the three GR mutants and NFI-B along with pWAPtk-luciferase and RSV-β-gal and induced with HC. Cells were transfected with wild-type GR or with both NFI-B and wild-type GR or GR 3-556 C-terminal, GR X-795 N-terminal, and GR C482S DBD mutants, respectively, as shown. Error bars represent the standard error of three independent determinations.
To better define the interaction between GR and the NFI-B isoform, three GR mutants (3-556, a C-terminal deletion mutant; X-795, an N-terminal deletion mutant; and C482S, a DNA-binding mutant) (depicted schematically in Fig. 4A) were employed in transient transfection assays together with the NFI-B isoform. The C-terminal mutant of GR can activate transcription in a ligand-independent manner, and accordingly, both the HC-induced activation and uninduced activation of WAP CoRE-driven luciferase expression were significantly less than that observed with wild-type GR (Fig. 4D). The N-terminal and DNA-binding mutants also failed to increase the WAP promoter expression compared to wild-type GR (Fig. 4D). These results indicate that the C-terminal, N-terminal, and DNA-binding regions of GR are all required for optimal regulation of WAP promoter activity. Western blot analysis indicated that the differences observed were not due to decreased levels of expression of the different GR mutants compared to wild-type GR (Fig. 4B)
Cooperative activation of GR, STAT, and NFI on the MMTV LTR promoter.
The MMTV long terminal repeat (LTR) is perhaps the most widely used promoter for studying effects of GR on gene regulation. The composition of the transcription factor binding sites of the MMTV LTR promoter is related to that of the WAP promoter, but the former contains several consensus GREs (Fig. 5A). MMTV expression is also hormonally regulated during mammary gland development (1). Functional interactions between STAT5 and GR have already been shown on the MMTV LTR by transient transfection experiments performed in COS cells (50). However, unlike the cooperativity observed between these transcription factors on WAP CoRE activity in JEG-3 cells, STAT5 has been reported to antagonize GR activity on the MMTV LTR. In order to compare directly the regulation of the MMTV LTR to that of the WAP CoRE, GR and STAT5A were cotransfected into JEG-3 cells and then induced by lactogenic hormones. Unlike the WAP CoRE, the induction of MMTV-luciferase by only HC was similar to that observed for Prl alone (Fig. 5B). However, with both HC and Prl, expression was increased approximately another threefold (Fig. 5B). In contrast to the results reported by Stocklin et al. (50) in COS cells, STAT5 and GR appeared to act cooperatively on the MMTV LTR in a manner similar to that observed on the WAP distal promoter.
FIG. 5.
Cooperative interactions on the MMTV promoter. (A) Schematic diagram of MMTV-luciferase construct. (B) Cooperativity of STAT and GR on the MMTV promoter. MMTV-Luc was transfected with only GR, with only STAT5A, and with both GR and STAT into JEG-3 cells, and the cells were induced with HC, with Prl, and with both hormones as illustrated. (C) Cooperative activation of GR with NFI isoforms on the MMTV promoter. MMTV-Luc and RSV-β-gal were cotransfected into JEG-3 cells along with GR alone, with both GR and NFI-A, with both GR and NFI-B, and with both GR and NFI-X, as shown. The cells were induced with HC. (D) Cooperativity among GR, STAT5A, and NFI isoforms required for the activation of MMTV. MMTV-Luc was transiently transfected into JEG-3 cells with GR, with STAT5A, with both GR and STAT5A, with GR, STAT5A, and NFI-A, with GR, STAT5A, and NFI-B, and with GR, STAT5A, and NFI-X, as illustrated. The cells were induced with HC, with Prl, and with both hormones, as depicted.
To determine the interactions between GR and specific NFI isoforms on the MMTV LTR, GR was cotransfected with the individual NFI isoforms followed by treatment with HC. Similar to WAP CoRE-driven constructs, when GR was cotransfected with the NFI-B isoform, expression was increased to a much greater extent than with the NFI-A and NFI-X isoforms (Fig. 5C). In fact, this effect of the NFI-B isoform appeared even more selective than that observed on the WAP CoRE-driven reporter construct.
Finally, synergism was also observed between the NFI-B and -X isoforms, GR, and STAT5 on the MMTV LTR. The NFI-B isoform, when cotransfected with GR and STAT5, resulted in almost a ninefold induction compared to GR and STAT activation on the MMTV LTR (Fig. 5D). Once again, the NFI-X isoform resulted in less activation (only fourfold) compared to GR and STAT, and no induction was observed with the NFI-A isoform (Fig. 5D).
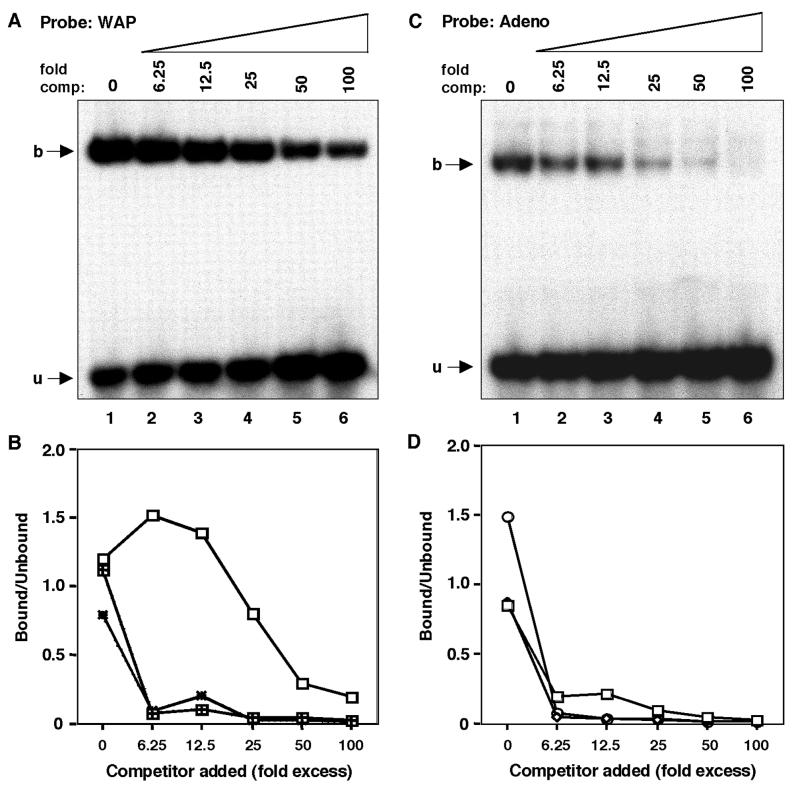
DNA-binding affinity of the NFI isoforms to the palindromic site in the WAP CoRE.
The CoRE of the rat WAP gene (Fig. 3A) contains one palindromic and one NFI half-site, and these binding sites are highly conserved in both the mouse and rabbit WAP genes. In previous studies, the palindromic site, designated FP2, was shown to bind NFI with a higher affinity in vitro and demonstrated a greater mammary gland-specific occupancy in vivo than the half-site, designated FP1. However, the FP2 interaction was considerably weaker than that observed with the consensus adenovirus NFI binding site (34). The transient transfection results reported herein indicate that the NFI-B isoform exhibited greater cooperativity with GR and STAT5A than NFI-X and, surprisingly, that NFI-A displayed no cooperativity. EMSAs were therefore performed to determine if these three lactation-specific NFI isoforms might exhibit different binding affinities to the FP2 binding site.
Competition EMSAs were performed using oligonucleotides containing the WAP FP2 site and the consensus NFI site of adenovirus replication origin region. Nuclear extracts were prepared from JEG-3 cells following transfection of NFI-A, -B, and -X, as described in Materials and Methods. Relative binding specificities were then estimated by measuring the amount of complex formation between the three NFI isoforms and DNA as a function of probe DNA concentration using increasing amounts of the unlabeled probe as a competitor (Fig. 6A and C). The results of these studies are summarized in Fig. 6B and D. These experiments indicate that the NFI-A isoform exhibited less specific binding to the palindromic site in the WAP promoter, in agreement with its lack of demonstrated cooperativity. However, the binding specificities of the NFI-B and -X isoforms were quite similar, perhaps accounting in part for their ability to cooperatively transactivate the WAP CoRE. In contrast, the binding specificities of all three NFI isoforms to the adenovirus NFI site were quite similar. Therefore, the DNA-binding specificities of these two isoforms were not sufficiently different to account for decreased activity of NFI-X versus NFI-B on the WAP CoRE. This most likely reflects differences in protein-protein interactions between these two NFI isoforms and possibly STAT5 and GR.
FIG. 6.
DNA-binding specificities of different NFI isoforms. (A) DNA-binding specificity of NFI-A to the palindromic motif present on the WAP distal promoter estimated by competition EMSA. The specific DNA-protein complexes (described in Materials and Methods) were made to compete with the same palindromic NFI oligonucleotide (homologous competitor) using increasing concentrations as shown (lanes 2 to 8; no competitor in lane 1). (B) Comparison of DNA-binding specificity of the three NFI isoforms to the WAP palindromic motif. The DNA-binding specificities of NFI-B and -X were also determined along with NFI-A (A) by competition EMSA, and the DNA-protein complex and free probe were quantitated by PhosphorImager. Binding specificities were determined by plotting the percentage of bound/unbound versus competitor added. Open squares represent NFI-A, squares filled with the + sign represent NFI-B, and dotted rectangles represent NFI-X. (C) DNA-binding specificity of NFI-A isoform to the palindromic motif on the adenovirus origin of replication region estimated by competition EMSA. The specific DNA-protein complexes (described in Materials and Methods) were made to compete with the same palindromic NFI oligonucleotide (homologous competitor) using increasing concentrations as shown (lanes 2 to 8; no competitor in lane 1). (D) Comparison of DNA-binding specificity of the three NFI isoforms to the adenovirus palindromic motif. The DNA-binding specificities of NFI-B and -X were also determined along with NFI-A (C) by competition EMSA, and the DNA-protein complex and free probe were quantitated by PhosphorImager. Binding specificities were determined by plotting the percentage of bound/unbound versus competitor added. Open squares represent NFI-A, open circles represent NFI-B, and the open rectangles represent NFI-X.
DISCUSSION
To elucidate the mechanisms by which NFI, STAT5, and GR might act in a cooperative or synergistic fashion to regulate WAP gene expression, specific NFI isoforms were identified and then studied using transient transfection assays in JEG-3 cells in combination with STAT5 and GR. Both STAT5 and GR are critical regulators of WAP gene expression, and cooperative interactions between these factors now have been demonstrated for the first time using transient transfection assays in JEG-3 cells. STAT5 and GR cooperativity has been demonstrated previously for the activation of β-casein gene expression in cotransfection experiments in several different cell types (14, 50, 58). This appears to be due to several mechanisms, including prolonged activation and DNA binding of STAT5, contribution of the GR transactivation domain, and coactivator recruitment (22, 58). Transcriptional cooperativity between GR and STAT along with C/EBPβ has recently also been shown to be important for the regulation of β-casein gene expression (57). In contrast, the interaction of STAT5 and GR has been reported to suppress the GR induction of the MMTV LTR promoter in COS 7 cells (50). This was clearly not the case for GR and STAT5 in JEG-3 cells, which appear to better reflect the known cooperativity between Prl and glucocorticoids in the regulation of MMTV gene expression during mammary gland development (43). These cell type differences most likely reflect the levels of expression of these different transcription factors and coactivators. Differences in STAT5 and GR cooperativity observed between the β-casein and MMTV LTR may also result from the presence of consensus palindromic GREs, compared to half-sites and incomplete palindromic sites, in the MMTV LTR and the β-casein promoters, respectively. The context of these sites may also play an important role in the degree of cooperativity observed.
The functional interaction between STAT5 and NFI in transfection experiments has not been described previously. Both STAT5 and NFI have been suggested to be involved in the developmental stage-specific expression of mammary gland gene expression (27, 34), but this has been difficult to demonstrate in vivo using mouse knockouts due to the presence of closely related family members.
Among the specific NFI isoforms identified in this study, NFI-B2 exhibited the best cooperativity with STAT5 at the WAP distal promoter in transient transfection assays (Fig. 3C). Interestingly, the same NFI-B2 isoform also displayed the best cooperativity with GR in regulating both WAP distal CoRE-driven (Fig. 4C) and MMTV LTR-driven reporter activity (Fig. 5C). Studies of NFI-B2 with three different GR mutants indicated that the N-terminal, C-terminal, and DNA-binding regions of GR were all necessary for transcriptional cooperativity with NFI-B2 at the WAP CoRE (Fig. 4D).
These studies suggest that the NFI-B2 isoform might interact more efficiently with GR and STAT than other lactation-specific isoforms. Preliminary studies using chimeras between NFI-A and -B have suggested that the C-terminal domain of the NFI-B isoform may help facilitate these protein-protein interactions, either by physical interaction or via coactivator complexes, but this remains to be established (data not shown). Cooperativity between GR and NFI has also been demonstrated previously for the transcriptional activation of other promoters, e.g., at a simple promoter containing a single NFI binding site upstream of the adenovirus major late promoter or the MMTV promoter, suggesting the differential activation of simple versus complex NFI-responsive promoters by NFI isoforms (10).
The relative expression levels of the four different NFI genes at different stages of mammary gland development were determined using both RNase protection and RT-PCR assays (Fig 2D). The overall expression of NFI-X appeared to be slightly greater than that of NFI-B throughout development, while the expression of NFI-A appeared approximately equivalent to that of NFI-B. There does appear to be a difference in the specificity of DNA binding of the different NFI isoforms for the nonconsensus palindromic FP2 binding site present in the WAP distal CoRE. Specifically, the NFI-A isoform displayed a much reduced binding specificity to this site compared to the B2 and X1 isoforms (Fig. 6A and B). Thus, the decreased binding efficiency of the NFI-A4 isoform appears to correlate with its reduced transcriptional activity at the WAP CoRE.
Using just the NFI DNA-binding domains from the four different NFI genes expressed as glutathione S-transferase fusions in Escherichia coli, Osada et al. have reported that the DNA-binding affinities of NFI-A and NFI-X are slightly greater (two- to threefold) than those of NFI-B and NFI-C using the NFI binding site from the adenovirus replication origin (42). Binding affinities also varied from 1.3- to 5-fold among the NFI binding sites of four different promoters. In the present studies, the dissociation constants of the individual NFI isoforms could not be determined directly because nuclear extracts made from JEG-3 cells transiently transfected with different NFI genes were compared. Presumably, posttranslational modifications of the NFI proteins also might influence their ability to bind DNA, and these modifications are not present in the NFI DNA glutathione S-transferase fusions expressed in E. coli. These posttranslational modifications may play a significant role in NFI function (12), and it will be important to determine how they might influence WAP gene expression and interactions with GR, STAT5, and coactivators.
In addition to lactogenic hormones, extracellular matrix (ECM)-epithelial cell interactions play a key role in determining the fully differentiated mammary epithelial cell phenotype (34, 47, 52) and ensuring epithelial cell survival (51, 52). Furlong et al. (17) have nicely demonstrated that the expression of a 74-kDa NFI isoform is associated with mammary epithelial cell apoptosis and that its expression is suppressed when the cells are overlaid with ECM and cell death is inhibited. Similarly, Streuli et al. (52) have shown matrix-dependent regulation of transcription factors NFI and Sp1 in the mammary cells. Nebl et al. (39) have shown that a Ha-v-ras-mediated downregulation of both CTF/NFI and NFI-X mRNA levels in mouse mammary cell lines occurs through a decrease in the stability of the NFI transcripts. Destabilization of mRNA may also be due to ECM-mediated regulation of NFI at the transcript level (21). Similarly, in this study the expression of NFI-C was below the level of detection during lactation by both RNase protection assay and RT-PCR. However, this does not preclude differences in the relative levels of the different NFI isoforms that may exist at the protein level. Decreased NFI-C expression during lactation may facilitate milk protein gene expression by the NFI-B isoform, since the C-terminal domain of NFI-C has been implicated in the repression of the MMTV promoter (23). The NFI-A4 isoform also repressed GR- and STAT-mediated transcriptional activity at both the WAP CoRE and MMTV LTR. A similar repressor-like activity of NFI-A has been shown on the rat glutathione transferase P, as well as in the human metallothionein IIA promoters (40, 41).
Despite the demonstration that GR, STAT5, and specific NFI isoforms cooperatively activate WAP distal promoter transcription, it is still not clear precisely how or whether these multiple transcriptional complexes regulate tissue- and developmental stage-specific WAP gene expression. A working hypothesis is that the accumulation of particular NFI isoforms coupled with the Prl-mediated activation of STAT5 at specific times during mammary gland development is critical for the appropriate temporal and spatial regulation of WAP gene expression. In addition, GR may play a critical role in nucleosome reorganization to facilitate the binding of these transcription factors.
Burdon et al. (6), using organ explant cultures from virgin and pregnant mice, have shown that Prl and glucocorticoids are both required for the synergistic activation of WAP gene expression, and they have also suggested that neither signaling pathway by itself is sufficient to induce WAP transcription. The results of the transient transfection assays also suggest that each of these factors is important in the regulation of WAP CoRE activity, but none of these factors alone is sufficient to induce maximal expression. The precise mechanisms by which GR, NFI, and STAT5 enhance transcription are also not known, but presumably these entail either a direct interaction with the basal transcription complex or the maintenance of an open chromatin structure through interaction with coactivators. Although the distal CoRE region is critical for enhanced WAP gene expression, the involvement of other complexes or transcription factors in regulating tissue- and stage-specific WAP gene expression should not be excluded.
ACKNOWLEDGMENTS
These studies were supported by National Institutes of Health grant CA 16303 (to J.M.R.) and DK 58401 (to R.M.G.).
We thank Sandy Grimm and Michele Kallesen for critiquing the manuscript and for help in preparing the figures.
REFERENCES
- 1.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. . (Erratum, 256:161.) [DOI] [PubMed] [Google Scholar]
- 2.Bachurski C J, Kelly S E, Glasser S W, Currier T A. Nuclear factor I family members regulate the transcription of surfactant protein-C. J Biol Chem. 1997;272:32759–32766. doi: 10.1074/jbc.272.52.32759. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay S, Starke D W, Mieyal J J, Gronostajski R M. Thioltransferase (glutaredoxin) reactivates the DNA-binding activity of oxidation-inactivated nuclear factor I. J Biol Chem. 1998;273:392–397. doi: 10.1074/jbc.273.1.392. [DOI] [PubMed] [Google Scholar]
- 4.Bedford F K, Julius D, Ingraham H A. Neuronal expression of the 5HT3 serotonin receptor gene requires nuclear factor 1 complexes. J Neurosci. 1998;18:6186–6194. doi: 10.1523/JNEUROSCI.18-16-06186.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum J L, Zeigler M E, Wicha M S. Regulation of rat mammary gene expression by extracellular matrix components. Exp Cell Res. 1987;173:322–340. doi: 10.1016/0014-4827(87)90274-6. [DOI] [PubMed] [Google Scholar]
- 6.Burdon T, Wall R J, Shamay A, Smith G H, Hennighausen L. Overexpression of an endogenous milk protein gene in transgenic mice is associated with impaired mammary alveolar development and a milchlos phenotype. Mech Dev. 1991;36:67–74. doi: 10.1016/0925-4773(91)90073-f. [DOI] [PubMed] [Google Scholar]
- 7.Cardinaux J R, Chapel S, Wahli W. Complex organization of CTF/NF-I, C/EBP, and HNF3 binding sites within the promoter of the liver-specific vitellogenin gene. J Biol Chem. 1994;269:32947–32956. [PubMed] [Google Scholar]
- 8.Cereghini S, Raymondjean M, Carranca A G, Herbomel P, Yaniv M. Factors involved in control of tissue-specific expression of albumin gene. Cell. 1987;50:627–638. doi: 10.1016/0092-8674(87)90036-5. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhry A Z, Lyons G E, Gronostajski R M. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhry A Z, Vitullo A D, Gronostajski R M. Nuclear factor I (NFI) isoforms differentially activate simple versus complex NFI-responsive promoters. J Biol Chem. 1998;273:18538–18546. doi: 10.1074/jbc.273.29.18538. [DOI] [PubMed] [Google Scholar]
- 11.Chen L H, Bissell M J. A novel regulatory mechanism for whey acidic protein gene expression. Cell Regul. 1989;1:45–54. doi: 10.1091/mbc.1.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke D W, Lane M D. The transcription factor nuclear factor I mediates repression of the GLUT4 promoter by insulin. J Biol Chem. 1999;274:12917–12924. doi: 10.1074/jbc.274.18.12917. [DOI] [PubMed] [Google Scholar]
- 13.Corthesy B, Claret F X, Wahli W. Estrogen receptor level determines sex-specific in vitro transcription from the Xenopus vitellogenin promoter. Proc Natl Acad Sci USA. 1990;87:7878–7882. doi: 10.1073/pnas.87.20.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doppler W, Groner B, Ball R K. Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc Natl Acad Sci USA. 1989;86:104–108. doi: 10.1073/pnas.86.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doppler W, Hock W, Hofer P, Groner B, Ball R K. Prolactin and glucocorticoid hormones control transcription of the beta-casein gene by kinetically distinct mechanisms. Mol Endocrinol. 1990;4:912–919. doi: 10.1210/mend-4-6-912. [DOI] [PubMed] [Google Scholar]
- 16.Elder G A, Liang Z, Snyder S E, Lazzarini R A. Multiple nuclear factors interact with the promoter of the human neurofilament M gene. Brain Res Mol Brain Res. 1992;15:99–107. doi: 10.1016/0169-328x(92)90156-6. [DOI] [PubMed] [Google Scholar]
- 17.Furlong E E, Keon N K, Thornton F D, Rein T, Martin F. Expression of a 74-kDa nuclear factor 1 (NF1) protein is induced in mouse mammary gland involution. Involution-enhanced occupation of a twin NF1 binding element in the testosterone-repressed prostate message-2/clusterin promoter. J Biol Chem. 1996;271:29688–29697. doi: 10.1074/jbc.271.47.29688. [DOI] [PubMed] [Google Scholar]
- 18.Ganguly R, Ganguly N, Mehta N M, Banerjee M R. Absolute requirement of glucocorticoid for expression of the casein gene in the presence of prolactin. Proc Natl Acad Sci USA. 1980;77:6003–6006. doi: 10.1073/pnas.77.10.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil G, Smith J R, Goldstein J L, Slaughter C A, Orth K, Brown M S, Osborne T F. Multiple genes encode nuclear factor 1-like proteins that bind to the promoter for 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci USA. 1988;85:8963–8967. doi: 10.1073/pnas.85.23.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godowski P J, Rusconi S, Miesfeld R, Yamamoto K R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987;325:365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- 21.Goyal N, Knox J, Gronostajski R M. Analysis of multiple forms of nuclear factor I in human and murine cell lines. Mol Cell Biol. 1990;10:1041–1048. doi: 10.1128/mcb.10.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groner B, Gouilleux F. Prolactin-mediated gene activation in mammary epithelial cells. Curr Opin Genet Dev. 1995;5:587–594. doi: 10.1016/0959-437x(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 23.Gronostajski R M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 24.Hobbs A A, Richards D A, Kessler D J, Rosen J M. Complex hormonal regulation of rat casein gene expression. J Biol Chem. 1982;257:3598–3605. [PubMed] [Google Scholar]
- 25.Jackson D A, Rowader K E, Stevens K, Jiang C, Milos P, Zaret K S. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol Cell Biol. 1993;13:2401–2410. doi: 10.1128/mcb.13.4.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannius-Janson M, Lidberg U, Hulten K, Gritli-Linde A, Bjursell G, Nilsson J. Studies of the regulation of the mouse carboxyl ester lipase gene in mammary gland. Biochem J. 1998;336:577–585. doi: 10.1042/bj3360577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazansky A V, Raught B, Lindsey S M, Wang Y F, Rosen J M. Regulation of mammary gland factor/Stat5a during mammary gland development. Mol Endocrinol. 1995;9:1598–1609. doi: 10.1210/mend.9.11.8584036. [DOI] [PubMed] [Google Scholar]
- 28.Kruse U, Sippel A E. The genes for transcription factor nuclear factor I give rise to corresponding splice variants between vertebrate species. J Mol Biol. 1994;238:860–865. doi: 10.1006/jmbi.1994.1343. [DOI] [PubMed] [Google Scholar]
- 29.Kruse U, Sippel A E. Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Lett. 1994;348:46–50. doi: 10.1016/0014-5793(94)00585-0. [DOI] [PubMed] [Google Scholar]
- 30.Kulkarni S, Gronostajski R M. Altered expression of the developmentally regulated NFI gene family during phorbol ester-induced differentiation of human leukemic cells. Cell Growth Differ. 1996;7:501–510. [PubMed] [Google Scholar]
- 31.Lee E Y, Parry G, Bissell M J. Modulation of secreted proteins of mouse mammary epithelial cells by the collagenous substrata. J Cell Biol. 1984;98:146–155. doi: 10.1083/jcb.98.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine J F, Stockdale F E. Cell-cell interactions promote mammary epithelial cell differentiation. J Cell Biol. 1985;100:1415–1422. doi: 10.1083/jcb.100.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Rosen J M. Distal regulatory elements required for rat whey acidic protein gene expression in transgenic mice. J Biol Chem. 1994;269:14235–14243. [PubMed] [Google Scholar]
- 34.Li S, Rosen J M. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol Cell Biol. 1995;15:2063–2070. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisterernst M, Rogge L, Foeckler R, Karaghiosoff M, Winnacker E L. Structural and functional organization of a porcine gene coding for nuclear factor I. Biochemistry. 1989;28:8191–8200. doi: 10.1021/bi00446a034. [DOI] [PubMed] [Google Scholar]
- 36.Mermod N, O'Neill E A, Kelly T J, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 37.Mink S, Hartig E, Jennewein P, Doppler W, Cato A C. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992;12:4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagata K, Guggenheimer R A, Enomoto T, Lichy J H, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci USA. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nebl G, Mermod N, Cato A C. Posttranscriptional down-regulation of expression of transcription factor NF1 by Ha-ras oncogene. J Biol Chem. 1994;269:7371–7378. [PubMed] [Google Scholar]
- 40.Osada S, Daimon S, Ikeda T, Nishihara T, Yano K, Yamasaki M, Imagawa M. Nuclear factor 1 family proteins bind to the silencer element in the rat glutathione transferase P gene. J Biochem (Tokyo) 1997;121:355–363. doi: 10.1093/oxfordjournals.jbchem.a021595. [DOI] [PubMed] [Google Scholar]
- 41.Osada S, Ikeda T, Xu M, Nishihara T, Imagawa M. Identification of the transcriptional repression domain of nuclear factor 1-A. Biochem Biophys Res Commun. 1997;238:744–747. doi: 10.1006/bbrc.1997.7382. [DOI] [PubMed] [Google Scholar]
- 42.Osada S, Matsubara T, Daimon S, Terazu Y, Xu M, Nishihara T, Imagawa M. Expression, DNA-binding specificity and transcriptional regulation of nuclear factor 1 family proteins from rat. Biochem J. 1999;342:189–198. [PMC free article] [PubMed] [Google Scholar]
- 43.Pauley R J, Rosen J M, Socher S H. Mammary tumour virus and casein gene transcription during mouse mammary development. Nature. 1978;275:455–457. doi: 10.1038/275455a0. [DOI] [PubMed] [Google Scholar]
- 44.Quinn P G, Wong T W, Magnuson M A, Shabb J B, Granner D K. Identification of basal and cyclic AMP regulatory elements in the promoter of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1988;8:3467–3475. doi: 10.1128/mcb.8.8.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajput B, Shaper N L, Shaper J H. Transcriptional regulation of murine beta1,4-galactosyltransferase in somatic cells. Analysis of a gene that serves both a housekeeping and a mammary gland-specific function. J Biol Chem. 1996;271:5131–5142. doi: 10.1074/jbc.271.9.5131. [DOI] [PubMed] [Google Scholar]
- 46.Rosen J M, Wyszomierski S L, Hadsell D. Regulation of milk protein gene expression. Annu Rev Nutr. 1999;19:407–436. doi: 10.1146/annurev.nutr.19.1.407. [DOI] [PubMed] [Google Scholar]
- 47.Roskelley C D, Srebrow A, Bissell M J. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rupp R A, Kruse U, Multhaup G, Gobel U, Beyreuther K, Sippel A E. Chicken NFI/TGGCA proteins are encoded by at least three independent genes: NFI-A, NFI-B and NFI-C with homologues in mammalian genomes. Nucleic Acids Res. 1990;18:2607–2616. doi: 10.1093/nar/18.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santoro C, Mermod N, Andrews P C, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 50.Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 51.Streuli C H, Bailey N, Bissell M J. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Streuli C H, Schmidhauser C, Bailey N, Yurchenco P, Skubitz A P, Roskelley C, Bissell M J. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabo P, Moitra J, Rencendorj A, Rakhely G, Rauch T, Kiss I. Identification of a nuclear factor-I family protein-binding site in the silencer region of the cartilage matrix protein gene. J Biol Chem. 1995;270:10212–10221. doi: 10.1074/jbc.270.17.10212. [DOI] [PubMed] [Google Scholar]
- 54.Topper Y J, Freeman C S. Multiple hormone interactions in the developmental biology of the mammary gland. Physiol Rev. 1980;60:1049–1106. doi: 10.1152/physrev.1980.60.4.1049. [DOI] [PubMed] [Google Scholar]
- 55.Watson C J, Gordon K E, Robertson M, Clark A J. Interaction of DNA-binding proteins with a milk protein gene promoter in vitro: identification of a mammary gland-specific factor. Nucleic Acids Res. 1991;19:6603–6610. doi: 10.1093/nar/19.23.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiens D, Park C S, Stockdale F E. Milk protein expression and ductal morphogenesis in the mammary gland in vitro: hormone-dependent and -independent phases of adipocyte-mammary epithelial cell interaction. Dev Biol. 1987;120:245–258. doi: 10.1016/0012-1606(87)90122-9. [DOI] [PubMed] [Google Scholar]
- 57.Wyszomierski S L, Rosen J M. Cooperative effects of STAT5 (signal transducer and activator of transcription 5) and C/EBPbeta (CCAAT/enhancer-binding protein-beta) on beta-casein gene transcription are mediated by the glucocorticoid receptor. Mol Endocrinol. 2001;15:228–240. doi: 10.1210/mend.15.2.0597. [DOI] [PubMed] [Google Scholar]
- 58.Wyszomierski S L, Yeh J, Rosen J M. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol Endocrinol. 1999;13:330–343. doi: 10.1210/mend.13.2.0232. [DOI] [PubMed] [Google Scholar]