Abstract
In addition to directing transcription initiation, core promoters integrate input from distal regulatory elements. Except for rare exceptions, it has been generally found that eukaryotic tRNA and rRNA genes do not contain TATA promoter elements and instead use protein-protein interactions to bring the TATA-binding protein (TBP), to the core promoter. Genomewide analysis revealed TATA elements in the core promoters of tRNA and 5S rRNA (Pol III), U1 to U5 snRNA (Pol II), and 37S rRNA (Pol I) genes in Schizosaccharomyces pombe. Using tRNA-dependent suppression and other in vivo assays, as well as in vitro transcription, we demonstrated an obligatory requirement for upstream TATA elements for tRNA and 5S rRNA expression in S. pombe. The Pol III initiation factor Brf is found in complexes with TFIIIC and Pol III in S. pombe, while TBP is not, consistent with independent recruitment of TBP by TATA. Template commitment assays are consistent with this and confirm that the mechanisms of transcription complex assembly and initiation by Pol III in S. pombe differ substantially from those in other model organisms. The results were extended to large-rRNA synthesis, as mutation of the TATA element in the Pol I promoter also abolishes rRNA expression in fission yeast. A survey of other organisms' genomes reveals that a substantial number of eukaryotes may use widespread TATAs for transcription. These results indicate the presence of TATA-unified transcription systems in contemporary eukaryotes and provide insight into the residual need for TBP by all three Pols in other eukaryotes despite a lack of TATA elements in their promoters.
Structural similarities shared by the RNA polymerases (Pols) of bacteria, archaea, and eukarya reflect a deep-rooted common ancestry of transcription systems in all organisms on earth. Archaea and eukaryotes exhibit greater similarity to each other in their Pol subunits, accessory transcription factors (TFs), and promoter elements than either does to bacteria (36). In eukaryotes, Pol I synthesizes large rRNA (35S to 45S, depending on the species); Pol II synthesizes mRNAs and some small nuclear (sn) RNAs, such as U1 to U5; and Pol III synthesizes mostly tRNAs and 5S rRNA, as well as U6 snRNA and a few other transcripts (53).
The core promoter orchestrates polymerase recruitment, promoter activity, and response to regulatory input (59, 66). In eukaryotes, TATA promoter elements direct transcription by Pol II of a large subset of (but not all) protein-encoding genes, but often not the far fewer snRNA genes that are transcribed by Pol II. While TATA elements are found in a minute fraction of Pol III genes, they are generally not found in the core promoter regions of Pol I genes (53). Intriguingly, despite the lack of TATA promoter elements, Pols I, II, and III all require TATA-binding protein (TBP) for initiation (17). Archaea use widespread TATA-like promoters and a TBP ortholog to direct transcription by a single Pol of all gene types, those encoding tRNA, rRNA, and mRNAs (reviewed in reference 67). Orthologs of another central initiation factor, TFIIB, cooperate with TBPs in promoter recognition in archaea and eukarya (35, 36). While TBP is shared by the three eukaryotic Pols, TFIIB and related factors exhibit polymerase and promoter specificity, such that TFIIB is used by Pol II, TFIIB-related factor (Brf) is used by Pol III for tRNA and 5S rRNA genes, and a distinct variant, BRFU/TFIIIB50, is used by Pol III for human U6 and related type 3 genes (63, 70). The TFIIB-related proteins bind adjacent to TBP on the promoter, recruit the corresponding polymerase to the transcription start site, and participate in promoter melting, an intermediate step in initiation (27, 30, 51, 58). Unlike archaeal and the eukaryal Pol II and Pol III systems, there is no apparent TFIIB homolog in the Pol I machinery (11, 84). Instead, the factor known as Rrn3p/TIF-IA bridges the core promoter-associated factors and Pol I (2, 44, 54).
Pol III promoters have historically been categorized into three major types. 5S rRNA (type 1) and tRNA (type 2) genes utilize internal TATA-less promoters, whereas U6 snRNA (type 3) promoters contain upstream TATA elements (7, 15, 28, 53, 80). For TATA-containing genes such as U6, TBP-TFIIIB can recognize the upstream DNA directly (46, 78). The tRNA promoter is composed of a proximal A box element located 10 to 20 bp downstream of the start site of transcription and a B box element at various distances farther downstream. Although the regions upstream of eukaryotic tRNA genes are generally AT rich, the sequence in this region is not conserved (33). Rather, the sequence information used to assemble a tRNA transcription complex resides in the internal promoter, which is recognized by TFIIIC. Once bound, TFIIIC recruits the initiation factor Brf and its associated TFIIIB components to the TATA-less upstream DNA (7, 28, 80). TFIIIB is an entity composed of three polypeptides, TBP, Brf, and B" (7, 28). TBP is brought to the upstream region of the tRNA gene by Brf (designated BRF/hTFIIIB90 in the human system) via stable protein-protein interactions that occur in the absence of DNA (22, 31, 75). Therefore, association with Brf provides TBP access to the TATA-less tRNA promoter (31, 40, 41, 77). In this setting, TATA is not required and the TBP in TFIIIB can bind to upstream DNA that contains stretches of only G and C residues (25).
We discovered that TATA motifs reside upstream of nearly all Schizosaccharomyces pombe tRNA and 5S rRNA genes. Here we demonstrate an obligatory role for TATA in homologous 5S rRNA and tRNA expression in S. pombe. We demonstrate differences in the mechanisms of Pol III transcription complex formation in S. pombe and Saccharomyces cerevisiae using in vitro transcription systems. Furthermore, S. pombe Brf associates with TFIIIC and Pol III in vivo, while TBP is conspicuously absent from these complexes, consistent with a TATA-dependent mechanism of TBP recruitment. The cumulative data fit a model of obligatory recruitment of TBP by the TATA element, precluding a stringent need for Brf-mediated recruitment of TBP, and lead to the proposal that this may reflect an ancient Pol III system.
Widening our search revealed TATA elements upstream of the S. pombe genes for U1 to U5 snRNAs and ≈37S rRNA genes, which do not use TATA elements in several other species. Mutation of the TATA element in the Pol I core promoter abolishes rRNA expression in vivo. The data indicate that all three Pols require TATA promoter elements for efficient expression in fission yeast. These results suggest that S. pombe may represent an ancient eukaryotic transcription system that was intermediate between that in archaea and the more diversified eukaryotic model systems that are described in textbooks.
MATERIALS AND METHODS
Construction and expression of TATA-less tRNA, 5S rRNA, and ≈37S rRNA genes and F-Ret1.
The S. cerevisiae tRNAser gene (on chromosome IX) was amplified from genomic DNA, cloned into the pJK vector, and named SctRNAserUCA. TATA was then introduced at −30; this changed the upstream sequence TATCTACAA to TATATATAA. Plasmid pFL20/18SPst5.8Si4, containing full-length S. pombe ribosomal DNA (rDNA) and 500 bp of upstream sequence producing a sequence-tagged 5.8S* rRNA (23), was left unaltered or changed at the TATAAA sequence to GGATCC by site-directed mutagenesis to create pFL20/18SPst5.8Si4-Mu2-6 (and -Mu2-7). The plasmids were used to transform S. pombe strain yAS50, and transformants were selected and grown on Edinburgh minimal medium lacking uracil. The 5S rDNA gene with flanking sequences (43) was amplified by PCR from S. pombe DNA and cloned into pGEM-T (Promega, Madison, Wis.) to create p5S-T, which was mutagenized at four nucleotide positions to create plasmid p5Smu. The 5Smu-containing fragment was subcloned into the NcoI/NdeI sites of pRep90X (21) to create pRep90X-5Smu. The TATA element of p5Smu was mutagenized and cloned into the NcoI/NdeI sites of pRep90X to create pRep90X-5Smu-Bam. These were used to transform yAS50, and transformants were selected on EMM lacking leucine. Ret1+ was amplified from genomic DNA. The product was cloned into the SalI and SmaI sites of pREP3X, resulting in pREP3X-Ret1, which was digested with PstI and BamHI and cloned into pBluescript-ura4, resulting in pBluescript-Ret1a. Ret1 5′-flanking sequence was obtained by PCR of S. pombe genomic DNA. The product was cloned into the KpnI and SalI sites of pBluescript-Ret1a, resulting in pBluescript-Ret1b. A 4.2-kb KpnI/BamHI fragment of pBluescript-Ret1b was transformed into yHL6382 (21) to create yYH3272 (h+ his3-D1 leu1-32 ura4-D18 ade6-M216 Δret1::[F-ret1, ura4+]). The genomic structure of F-ret1 in yYH3272 was confirmed by PCR (not shown). All constructs were verified by sequencing.
Immunoaffinity purification of Pol III and TFIIIC complexes.
Extracts were prepared according to the method of Hamada et al. (16) with modifications. Cells were broken with a French press two times, and the lysate was prepared by adding 3.5 M (NH4)2SO4 to a final concentration of 0.4 M. The resulting precipitate was dissolved in BC100 (20 mM HEPES, pH 7.9, 20% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 100 mM KCl) and dialyzed into BC100. For immunoaffinity purification, the extract was adjusted to 0.05% NP-40. Two milligrams of extract protein was incubated with 40 μl of M2 agarose beads (Sigma) at 4°C for 4 h. The beads were washed five times with 0.5 ml of BC100) 0.05% NP-40) and eluted two times with 40 μl of BC100 containing 200 μg of FLAG peptide (Sigma)/ml.
Antisera and immunoblotting.
The following antigens were used for antiserum production: N-terminal peptides of Ret1p, spB", and Rpc39p, and a C-terminal peptide of spBrf. Anti-TBP, anti-Sfc1p, ant-Sfc3p, anti-Sfc6p, anti-Sfc4p, and anti-spLa (Sla1p) were described previously (21). Samples were separated by 4 to 20% polyacrylamide gel electrophoresis, transferred to nitrocellulose, probed with appropriate antibodies, and processed using an ECL kit (Amersham).
RESULTS
TATA elements upstream of S. pombe tRNA genes.
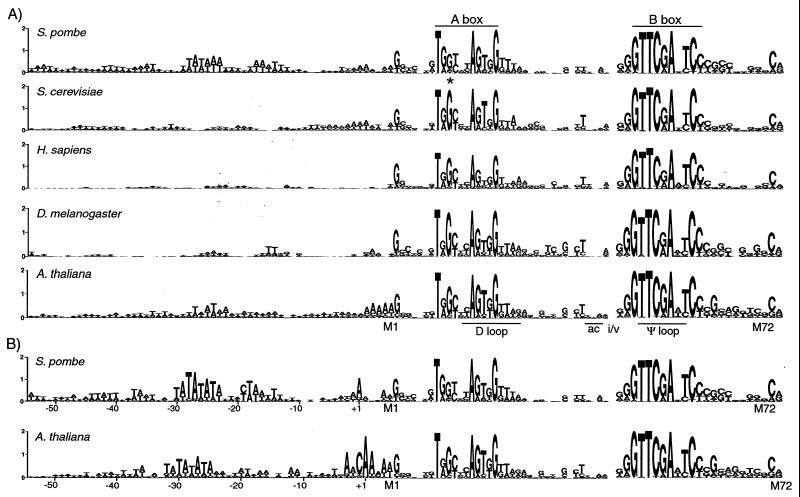
For this study, tRNAscan-SE (42) was used to identify the tRNA gene locations, and then the upstream regions were extracted from S. pombe, S. cerevisiae, Homo sapiens, Drosophila melanogaster, and Arabidopsis thaliana genomic databases. The sequence sets were then used to generate sequence Logos (Fig. 1A) (62). In the Logo display, the bases are stacked on top of each other for each position, the height of each letter being proportional to its frequency, and the letters are sorted with the most common one on top. The height of each stack is adjusted according to the information content (i.e., conservation) and plotted, in bits, on the base 2 logarithmic scale on the y axis (62).
FIG. 1.
Genomewide analysis reveals TATA motifs upstream of S. pombe tRNA genes. (A) Sequence Logos of the genomic 5′ flanking regions of tRNA sequences at 174 S. pombe, 275 S. cerevisiae, 425 D. melanogaster, 625 H. sapiens, and 600 A. thaliana loci. The positions of the A and B box promoter elements are indicated above the Logos. Note the relatively low level of sequence information content in the S. pombe A box, at the no. 10 position indicated by the asterisk, compared to that of S. cerevisiae. Although the full sets of sequences were used for the upstream regions and the first half of the tRNA for all species, for H. sapiens, A. thaliana, and D. melanogaster, the B box and downstream regions were limited to 200, 215, and 250 sequences, respectively, for practical reasons. Approximately 20% of the H. sapiens sequences were identified by tRNAscan-SE as “possible pseudogenes,” probably reflective of tRNA-like repetitive elements in the human genome (42). Approximately 1% each of the A. thaliana, S. pombe, and D. melanogaster sequences and none of the S. cerevisiae sequences were identified as possible pseudogenes. Common features of tRNA structure are indicated below the Logos. M1 indicates the first base of mature tRNA. A gap representing introns and sequences extending through the variable stem-loop (not shown) is designated i/v. ac, anticodon. (B) Sequence Logo of the S. pombe and A. thaliana sequences after prealignment of the upstream regions by the Clustal program. The numbering under the upstream region is relative to the putative consensus +1 site.
Figure 1A shows the upstream sequence (the first base of the mature tRNA is designated M1 under the A. thaliana Logo) and the first half of the tRNA sequence (ending after position 37), followed by a gap representing introns and sequences extending through the variable stem-loop (not shown), followed by the B box region and continuing to the end of the tRNA sequence (CCA is not included). The positions of the A box and B box and some features of tRNA structure are indicated. The largest amount of information content coincides with the A and B box promoter elements. In the upstream regions, the information content is higher for S. pombe than for the other species. The most concentrated upstream information appears as a TATAAA motif at a position typical of TATA promoter elements (recall that Pol III initiation occurs a variable distance [≈5 to 10 bp] upstream of the first base of the mature tRNA). This degree of information was not found in the upstream regions of the S. cerevisiae, H. sapiens, and D. melanogaster tRNA sequences (not shown, but see below). TATA-like information also appeared upstream of A. thaliana tRNA genes, although the information content was not as high nor as well defined as for S. pombe (Fig. 1A), consistent with the documented presence of the 4-nucleotide sequence TATA (13).
tRNAscan-SE used the first base of the mature tRNA sequences to isolate the upstream sequences. However, Pol III initiates at various distances (≈5 to 15 bp) upstream of the mature tRNA sequence. Prealignment of the sequences by an alignment program led to a significant increase in the information content of the TATA region in the S. pombe and, to a lesser degree, A. thaliana sequences (Fig. 1B) but not those of the other species (not shown).
To our surprise, prealignment also produced a prominent A that appeared in the A. thaliana and S. pombe sequences about six positions upstream of the mature tRNA (Fig. 1B). An increase in the information content of this position in association with the increased information content of the TATA motif that appeared in the aligned sequences (more so for S. pombe than A. thaliana), in conjunction with the 30-bp spacing of the prominent A and TATA, argued that the A represents a consensus transcription initiation site (+1). While prealignment led to a moderate increase in the TATA content of the A. thaliana sequences, a more substantial increase was observed around the predicted +1 site (underlined), which appeared in the form of ANCAA (Fig. 1B). ANCAA may be analogous to the initiator element described for certain Pol II (Inr) and Pol I (rInr) core promoters (56, 66).
Inspection of individual S. pombe sequences revealed that while 35% matched at seven or more TATATATA positions, the great majority matched at five or more positions (not shown). Thus, most, if not all, S. pombe tRNA genes appear to have at least a component of an upstream TATA element around the −30 region (not shown). Our ability to examine tRNA transcription in fission yeast led us to focus on S. pombe for the remainder of this study.
G10, which occupies the third position of the A box and is invariant in all S. cerevisiae tRNA genes and highly conserved in those of other species (14, 15), is much less conserved in S. pombe (Fig. 1A). Analysis of the subset of the S. pombe non-G10 genes revealed that they were similar to the full set of sequences in several features, including an upstream TATA, A-to-B box distance, percentage of genes with introns, +1 information, and B box consensus (not shown). We conclude that upstream TATAs are typical of the great majority of S. pombe tRNA genes. As will be described in more detail below, template commitment and other in vitro transcription assays using S. pombe extract indicate that although tRNA genes with G10 appear to compete better than non-G10 genes for a limiting TF (probably TFIIIC [see below]), these nonetheless require upstream TATA elements regardless of whether G occupies the number 10 position, i.e., even when the A and B boxes exhibit perfect matches to the S. cerevisiae consensus.
TATA promotes tRNA expression in S. pombe and in a homologous in vitro transcription system.
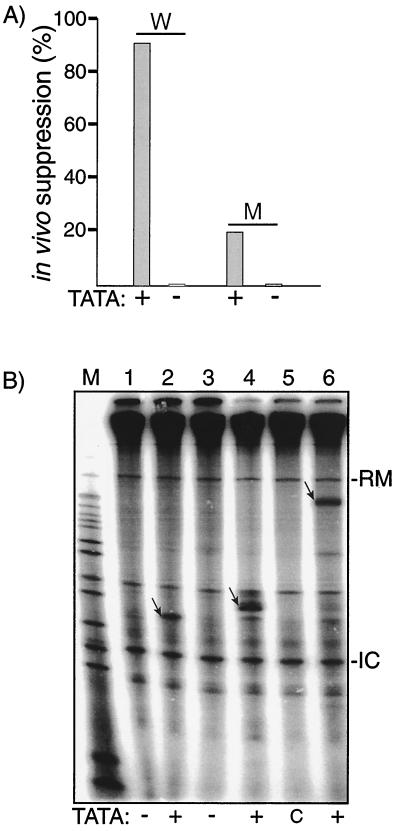
We recently characterized an in vivo suppression assay that is sensitive to the expression level of tRNAserUGA (16). Two suppressor genes were described, tRNAserUGA-W, which encodes a wild-type full-strength suppressor, andtRNAserUGA-M, which is comparably active for Pol III transcription but is less active for suppression (16). The naturally occurring TATATAAA sequence was altered in both of these genes, and their suppressor activities in S. pombe were determined (Fig. 2A). While both genes containing the wild-type TATA were active (16), the TATA-less genes were inactive.
FIG. 2.
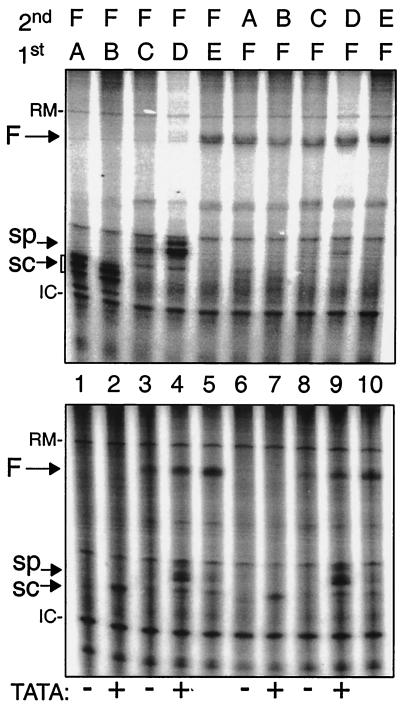
Upstream TATA is a positive determinant of tRNA expression in vivo and in a homologous in vitro S. pombe system. (A) The TATATA motifs of two opal suppressor tRNA genes, tRNAserUGA-W (W) and tRNAserUGA-M (M), were left unchanged (+) or replaced with GGATCC (−) as indicated along the horizontal axis and examined for suppressor activity in vivo as previously described (16, 24). (B) In vitro transcription in an S. pombe-derived extract was performed using three tRNA genes, containing (+) or lacking (−) upstream TATA elements, and empty plasmid control (c) as indicated below the lanes. Lanes 1 and 2, S. cerevisiae-derived tRNAserUCA gene; lanes 3 and 4, S. pombe-derived tRNAserUGA-M gene; lane 5, control plasmid containing no tRNA gene; lane 6, S. pombe-derived tRNAserUGA-M-3T gene (produces longer transcript [16]). The S. cerevisiae and S. pombe genes differ in size due to a 15-nt intron in the latter, and their A and B box elements are identical except at one position, G10 in the former and T10 in the latter. Transcript bands are indicated by arrows. -RM indicates a recovery marker added to the reactions, and -IC indicates an extract-derived internal control.
The effects of TATA on the efficiency of in vitro transcription in S. pombe-derived extract are shown in Fig. 2B. In addition to the S. pombe tRNAserUGA-M gene, we also examined an S. cerevisiae tRNAser gene that contains perfect matches to the consensus A and B box promoter elements and, as is typical of S. cerevisiae tRNA genes, contains no upstream TATA element. Specifically, the A and B box elements in the two yeasts' tRNAser genes used here are identical except for one position in the A box, T10 in the S. pombe tRNAser gene and G10 in the S. cerevisiae tRNAser gene. These genes were used for in vitro transcription (Fig. 2B). Nucleotides in the wild-type TATA-less S. cerevisiae tRNAser gene were replaced to create a TATA element. The TATA-less S. cerevisiae gene was inactive, while the substitutions that created the TATA element activated it (Fig. 2B, lanes 1 and 2). This demonstrated that a yeast tRNA gene with perfect matches to the consensus A and B box elements requires an intact upstream TATA for efficient transcription in the S. pombe system. The S. pombe tRNAserUGA-M gene whose TATA was mutated was inactive, while the gene containing wild-type TATA was active (Fig. 2B, lanes 3 and 4). A negative control, showing the products of a reaction containing a control plasmid that does not contain a tRNA gene, is shown in lane 5, and another version of the TATA-containing tRNAserUGA-M gene that differs only in the sequence at the Pol III terminator and therefore produces a distinctively longer transcript (16) is shown in lane 6.
An essential TATA element upstream of 5S rRNA genes in S. pombe.
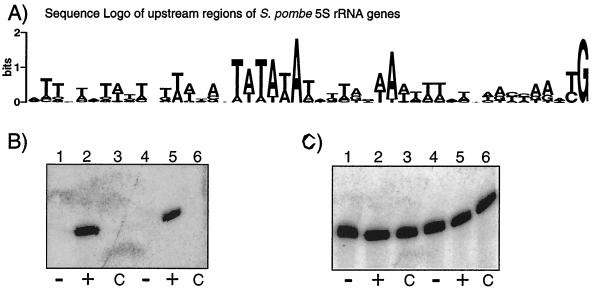
5S rRNA genes are dispersed in S. pombe, flanked by sequences that vary among copies (43). The upstream flanking regions plus the first G of the 5S rRNA sequences of multiple 5S rDNA loci are shown as a sequence Logo in Fig. 3A. Although variability is apparent at many positions, a TATA motif is prominent and typical for the majority of 5S genes.
FIG. 3.
Essential TATA motifs upstream of S. pombe 5S rRNA genes. (A) Sequence Logo of the upstream regions of multiple dispersed S. pombe 5S rDNA loci. The first base (G) of mature 5S rRNA was included as the last base on the right. (B) Northern blot analysis. A neutral sequence tag allows detection of the tagged transcript, designated 5S* rRNA, which is distinguishable from endogenous 5S rRNA. In vivo expression of plasmid-borne TATA-containing (+) and TATA-less (−) 5S* rRNA genes was monitored with a 5S*-specific probe. C, control. (C) The blot in panel B was stripped and rehybridized to detect endogenous 5S rRNA.
To investigate if the upstream TATA is important for expression, we examined 5S rRNA production from a pRep90X-derived plasmid containing an S. pombe 5S rRNA gene (43). We introduced four substitutions in the 5S sequence that correspond to nonconserved residues (derived mostly from S. cerevisiae 5S). These substitutions allow specific detection by oligonucleotide hybridization that distinguishes expression from the test gene, designated 5S*, from that of endogenous 5S rRNA and are also compatible with the predicted 5S rRNA secondary structure in this region (not shown). The wild-type upstream TATATAA was then changed to GGATCCA or left unaltered, and the resulting plasmids, which differed only at the TATA sequence, were used to transform S. pombe to a selectable leu1+ phenotype. Both plasmids yielded transformation efficiencies comparable to that of the empty control plasmid, and the transformants exhibited indistinguishable growth phenotypes (not shown). Total RNAs from duplicate sets of transformants were examined by Northern blotting (Fig. 3B). Hybridization with an antisense oligonucleotide probe revealed that 5S rRNA was expressed from the 5S* gene containing the wild-type TATA (Fig. 3B, lanes 2 and 5) but not from the TATA-less 5S* gene (lane 1 and 4) or from the control plasmid lacking an insert (lanes 3 and 6). To estimate the relative activity of the TATA-containing 5S* gene and to examine for differences in loading, the blot was stripped and rehybridized with an oligonucleotide specific for the wild-type 5S rRNA sequence under the same conditions as for the 5S* probe (Fig. 3C). Quantitation led to the estimate that 20 to 25% of the total 5S rRNA in the TATA-containing lanes was produced by the plasmid-borne 5S* rRNA gene containing the wild-type TATA element, while the TATA-less gene produced background levels of 5S* rRNA (not shown). These results indicated that the conserved TATA found upstream of S. pombe 5S rRNA genes is required for 5S rRNA expression in vivo. Sequence analysis revealed that these constructs differed only in the upstream TATA element, as expected (not shown). Therefore, the large difference in the amount of 5S* rRNA production from the TATA-containing and TATA-less 5S* rRNA genes suggests that TATA-dependent transcription represents an obligatory pathway of Pol III recruitment in S. pombe cells.
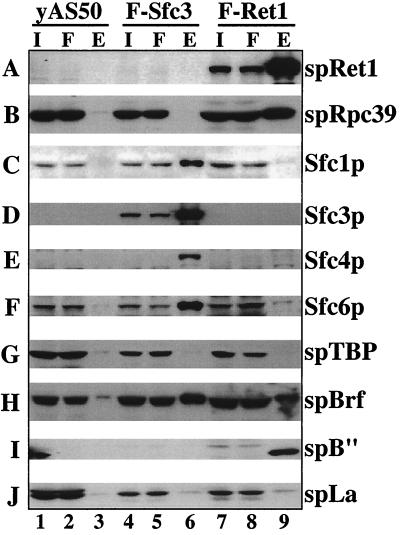
The Brf subunit of TFIIIB copurifies with fission yeast TFIIIC and Pol III with no associated TBP.
Extracts of a previously described S. pombe strain carrying FLAG-tagged Sfc3p, the S. pombe homolog of the B box-binding subunit of TFIIIC (21); a new strain carrying FLAG-tagged spRet1p, the S. pombe homolog of the second-largest subunit of Pol III; and a control strain, yAS50, were subjected to immunoaffinity purification using monoclonal anti-FLAG antibody (M2) cross-linked to agarose (Fig. 4). The input, flowthrough, and affinity-eluted materials were subjected to immunoblotting to detect the proteins indicated to the right of each panel. The spBrf homolog was found associated with Sfc1p, Sfc4p, Sfc6p, and the FLAG-tagged Sfc3p (Fig. 4C to F and H, lanes 6), while two other TFIIIB homologs, spTBP and spB", were not associated (Fig. 4G and I, lanes 6). The relative amounts of Brf in the negative control (lanes 3) and the Sfc3p-TFIIIC complex (lanes 6) are comparable to the ratio seen for Sfc6p, a genuine TFIIIC component (lanes 3 and 6). Quantitative analysis using known amounts of recombinant proteins indicated that a substantial fraction of spTFIIIC is specifically associated with Brf (not shown). The Pol III subunit homologs, spRet1p and spRpc39p were not associated with the S. pombe TFIIIC complex (Fig. 4A and B, lanes 6), nor was the Pol III nascent transcript-binding protein, spLa (Fig. 4J, lane 6), further indicating specificity of the Brf association. The same profile was also reproducible when a strain carrying epitope-tagged Sfc6p was used for purification (not shown). These results indicate that in S. pombe, Brf is specifically associated with a substantial fraction of TFIIIC, and most significantly, this occurs in the absence of TBP.
FIG. 4.
Fission yeast Brf and B" associate with Pol III TF complexes in the absence of stably associated TBP. Extracts prepared from a wild-type strain, the FLAG-Sfc3 strain, and a FLAG-spRet1p strain were incubated with anti-FLAG immunoglobulin G (M2)-agarose. After incubation, the supernatants were collected as the flowthrough, the agarose was washed five times with buffer containing 250 mM NaCl, and the bound material was eluted. The input (I), flowthrough (F), and eluate (E) of the M2-agarose from the three extracts were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting using antisera to the proteins as indicated on the right.
Analysis of the spRet1p complex revealed the presence of the associated Pol III subunit, spRpc39p, as expected (Fig. 4A and B, lanes 9). In this case, spBrf and spB" homologs were reproducibly found associated with the complex, while spTBP, spLa, and four spTFIIIC subunits were not associated (Fig. 4C to J, lanes 9). The specificity of the Brf association with the TFIIIC and Pol III complexes was reproducible in multiple experiments and was observed with affinity-purified antibody raised against a terminal peptide of spBrf (Fig. 4), as well as with two antisera raised against recombinant spBrf (not shown). The same profile of associated and nonassociated proteins was observed when a strain carrying the spRPC53p subunit of Pol III was epitope tagged and used for purification (not shown; as will be described elsewhere, FLAG-tagged Pol III is active for promoter-dependent transcription, as well as efficient termination and recycling on preassembled transcription complexes isolated on immobilized template DNA).
It was noted that TFIIIB components could not be detected when the epitope-tagged TFIIIC220 human counterpart of Sfc3p was used to isolate TFIIIC complexes from HeLa cells (74), suggesting that the Brf-TFIIIC association may be a unique characteristic of S. pombe. TBP together with Brf was readily found associated with epitope-tagged Pol III isolated from S. cerevisiae (7). spTBP could not be detected in our Brf-containing spTFIIIC complex or the Brf-containing Pol III complex, even though TBP is found tightly associated with Brf (in the absence of DNA) in other systems (20, 22, 31, 45, 55, 75). Thus, compared to the TFIIIB subunit associations in other systems, the S. pombe TFIIIB subunits appear to be arranged differently, although in a manner that is appropriate for the unique promoter architecture of S. pombe tRNA genes (see below). Furthermore, we have been unable to demonstrate association between TBP and Brf in S. pombe extracts by using direct immunoprecipitation or epitope-tagging approaches (Y. Huang, M. Weindel, and R. Maraia, unpublished data). The apparent weak association of TBP and Brf would appear to reflect a functional feature of the S. pombe Pol III system: principally TATA-mediated, rather than principally Brf-mediated, recruitment of TBP to the core promoter regions of tRNA genes.
Requirement for an upstream TATA element to program a functional Pol III preinitiation complex.
Since tRNA expression in S. pombe involves the widespread use of upstream TATA elements, we wanted to examine another hallmark feature of class III genes, their ability to program stable transcription complexes (15). While the interaction of TFIIIC with certain tRNA (and VA1) genes generates a complex that is stable on subsequent challenge with orthologous promoter DNA, this is not true for other tRNA genes, in large part due to variances in the A and/or B box elements (1, 12, 29, 37). However, inclusion of TFIIIB in the TFIIIC-DNA complex does program stable complex formation (33, 37). According to the assembly pathway elucidated using S. cerevisiae components, TFIIIC binds to the internal promoter and then recruits TFIIIB to the upstream DNA through contacts made principally to Brf (6, 57, 60). Thus, on TATA-less promoters, TFIIIC recruits and induces Brf and associated TBP to bind upstream DNA (6, 29, 47, 57, 60; reviewed in reference 7). This assembly pathway leads to a preinitiation complex that is stable upon challenge by homologous promoters and is consistent with the assemblages elucidated for human, Drosophila, and Xenopus (3, 30, 37, 59, 76). Using currently available activities, we can examine complex assembly and stability with a template commitment assay (3, 37, 61). In this assay, the first template is allowed to assemble with TFs and a second template is then added to challenge the stability of the first transcription complex. Because several templates were compared in S. cerevisiae and S. pombe extracts (Fig. 5), multiple observations regarding cis-acting elements and trans-acting factors are noteworthy.
FIG. 5.
An upstream TATA is required to program a functional Pol III preinitiation complex on an A box- and B box-containing tRNA gene in S. pombe. Transcription complexes were monitored using template exclusion assays performed in parallel in S. cerevisiae (top) and S. pombe (bottom) in vitro transcription systems. The tRNA genes used for Fig. 2B were tested for the ability to form stable transcription complexes. After incubation of the first template (the order of addition is indicated above the lanes) with transcription buffer (previously determined to be optimal at 20 min), nucleoside triphosphates and [α-32P]GTP were added along with the second template (indicated above the lanes), and transcription was allowed to proceed. The reaction was stopped, and RNA was extracted and separated on 6% polyacrylamide–8 M urea. The templates were as follows: A, sc-tRNASer −TATA; B, sc-tRNASer +TATA; C, sp-tRNASer −TATA; D, sp-tRNASer +TATA; E, pJK148 (empty vector); F, sp-tRNASer-3T +TATA. The arrows point to the different transcripts synthesized.
Three tRNA gene templates, in TATA-containing and TATA-less versions, were examined simultaneously in S. cerevisiae and S. pombe extracts (Fig. 5, top and bottom, respectively). The S. cerevisiae tRNAser gene contains A and B elements that match the consensus, while the A and B box elements of the S. pombe tRNAser genes differ from those of the S. cerevisiae gene at one position only, T at position 10. Both the TATA-containing and TATA-less versions of the S. cerevisiae tRNAser gene (Fig. 5, templates A and B) were efficiently transcribed and excluded transcription of the challenging tRNA gene (template F) in S. cerevisiae extract (top, lanes 1 and 2). In contrast to this, while either of the S. cerevisiae tRNA genes could exclude the second template in S. pombe extract, only the TATA-containing gene (template B) was active for transcription while the TATA-less gene (template A) was inactive (bottom, lane 1 and 2). Most significantly, this indicates that although the tRNA gene is able to interact stably with limiting S. pombe TFs, this alone is not sufficient to program a functional transcription complex in the absence of a TATA element.
Both versions of the S. pombe tRNAserUGA gene excluded transcription from the second template in S. cerevisiae extract (Fig. 5, top, lanes 3 and 4), although the TATA-containing gene (template D) was transcribed more efficiently than the TATA-less gene (template C). Prior data suggest that the relatively low-level transcription of the S. pombe gene in S. cerevisiae extract is almost certainly due to the single nonconsensus nucleotide, T10 (48) (Fig. 5, top, compare lanes 1 and 3).
In contrast to the stability observed in the S. cerevisiae system with the S. pombe tRNAserUGA gene, this gene did not exclude transcription from the second gene (template F) in the S. pombe system (Fig. 5, lanes 3 and 4). Because the promoters of the S. cerevisiae and S. pombe genes differ at only one position in the internal promoter (G10 versus T10, respectively), which is a significant determinant of transcription (48), and because prior studies indicate that interaction of TFIIIC with the A box can be a determinant of template exclusion (1, 37, 61, 65), it is reasonable to deduce from the data in Fig. 5 that S. pombe TFIIIC stably interacts with the consensus promoter (templates A and B, lanes 1 and 2) but not with the nonconsensus promoter (templates C and D, lanes 3 and 4) and that this is the basis of template exclusion in the former but not the latter. Moreover, Fig. 5 provides additional information that is relevant to a major conclusion of this study: even in the case where a stable complex is formed in the S. pombe system (Fig. 5, bottom, lanes 1 and 2), presumably reflecting a stable interaction of TFIIIC with the consensus promoter (37), this is not sufficient to program a functional preinitiation complex in the absence of a TATA element. By contrast, stable TFIIIC binding is sufficient to recruit functional TFIIIB in the S. cerevisiae system (29, 30, 33, 61, 65) (Fig. 5, top). The cumulative data in Fig. 5 argue strongly that a TATA element is required for the functional programming of an S. pombe transcription complex even when all of the necessary activities are present.
The results support the model in which the TFIIIC-mediated placement of TFIIIB, which leads to a functional transcription complex in S. cerevisiae, does not appear to occur efficiently, if at all, in the S. pombe system (Fig. 5, bottom, lanes 1 and 2). A second point that is suggested by the data is that the trans-acting factors that recognize TATA to promote transcription need not be recruited into a highly stable complex in the S. pombe system, since even when a functional complex is assembled, it is not stable on challenge by the orthologous promoter (Fig. 5, bottom, lane 4). This unexpected observation suggests that S. pombe TFIIIB interacts less stably with tRNA transcription complexes than does S. cerevisiae TFIIIB (see Discussion). The demonstrable differences in the two transcription systems were confirmed and strengthened by results obtained with control templates and were also observed when the order of addition of the genes was switched (lanes 5 to 10). The results provide convincing biochemical evidence that the S. pombe and S. cerevisiae transcription systems interact differently with tRNA genes but in a manner that is consistent with the associations of TFIIIB subunits shown in Fig. 4.
A TATA element in the S. pombe Pol I core promoter.
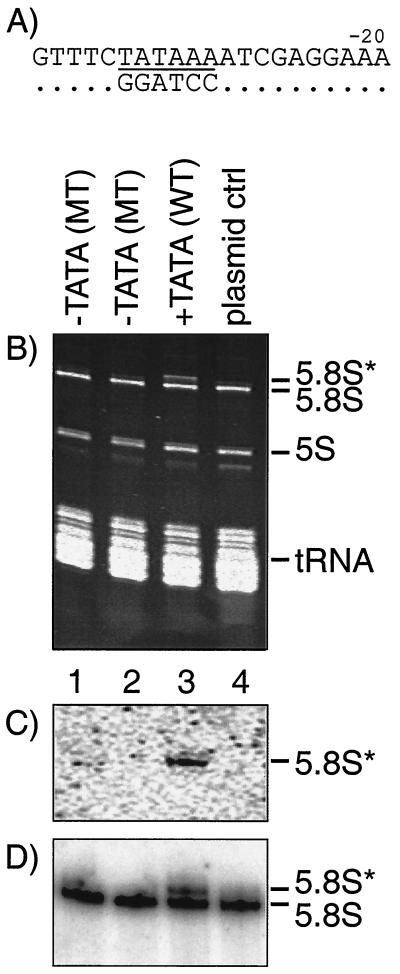
TATAAAA was also found in the region upstream of the transcription initiation sites of the genes that encode large rRNAs. In this promoter, TATAAAA overlaps the −30 region of the previously determined Pol I start site (Fig. 6A) (8). We examined 5.8S rRNA expression from a plasmid that harbors the TATA-containing wild-type 37S rRNA gene of S. pombe. The 5.8S rRNA produced from this plasmid is marked with a 4-nucleotide sequence tag that allows its transcript (designated 5.8S*) to be distinguished from endogenous 5.8S rRNA by slower mobility on polyacrylamide gels (23). The upstream TATAAA was changed to GGATCC (Fig. 6A) or left unaltered, and the resulting plasmids were used to transform ura4-D18 S. pombe cells to the ura4+ phenotype. The two plasmids yielded comparable transformation efficiencies, and the transformants exhibited indistinguishable growth phenotypes (not shown). Total RNA was prepared from the transformants and examined by polyacrylamide gel electrophoresis and ethidium bromide staining (Fig. 6B). As described, the tagged 5.8S* rRNA was visible as a clear band with slower mobility than 5.8S rRNA (23), produced from the wild-type TATA-containing plasmid (lane 3). Cells containing plasmids bearing the TATA-less promoter (lanes 1 and 2) and the control plasmid lacking the rRNA gene (lane 4) did not express the 5.8S* rRNA. As a control, the TATA-less promoter was converted back to a TATA-containing promoter by site-directed mutagenesis, and this rescued 5.8S* rRNA (not shown). Hybridization with an antisense probe confirmed that the tagged 5.8S* rRNA was expressed from the gene containing the wild-type TATA (Fig. 6C, lane 3) but not from the TATA-less genes or the control plasmid (lanes 1, 2, and 4). An oligonucleotide complementary to wild-type 5.8S and 5.8S* rRNAs revealed comparable loading in all lanes (Fig. 6D); again as expected, 5.8S* was expressed only from the TATA-containing plasmid (Fig. 6D, lane 3). These results indicated that the TATAAA sequence that is naturally found upstream of large rRNA genes in S. pombe is important for rRNA expression in vivo.
FIG. 6.
Pol I-transcribed large-rRNA genes use an upstream TATA motif in the core promoter region. (A) Sequence of promoter region (−40 through −20 relative to +1), with TATAAA motif underlined, of S. pombe rDNA (top line) (8) and TATA mutant (bottom line); dots indicate identical residues. (B) Ethidium bromide-stained polyacrylamide gel. Total RNA was isolated from S. pombe strains that had been transformed with plasmids containing 37S rRNA genes in which the 5.8S rRNA sequence contained a unique sequence tag (5.8S*; see the text). Lanes 1 and 2, two independent isolates in which the TATA element was mutated by site-directed substitution; lane 3, wild-type TATA; lane 4, control plasmid containing no rRNA gene. (C) Northern blot analysis using oligonucleotide probe conditions specific for the tagged 5.8* rRNA; the lanes are the same as in panel B. (D) The Northern blot in panel B was stripped and hybridized using an oligonucleotide probe that recognizes both wild-type (endogenous) 5.8S rRNA and 5.8* rRNA; the lanes are the same as in panel B.
DISCUSSION
A major conclusion of this study is that transcription in S. pombe involves widespread use of TATA promoter elements. This is significant not so much because of the number of promoters involved but more so because TATA use is widespread in the Pol I and III systems. It has generally been found that, except for rare exceptions (see below), eukaryotic tRNA gene promoters are TATA-less. The results presented here should extend the known diversity of gene structure and promoter function in eukaryotes (59, 66). Although, as detailed below, scattered reports indicate TATA elements upstream of some tRNA and 5S rRNA genes, this is the first report that indicates TATA function for the great majority of tRNA, 5S rRNA, and large-rRNA genes. Moreover, for these genes, TATA-mediated transcription is an obligatory pathway of expression, indicating that functional use of TATA elements is the rule for tRNA transcription in S. pombe rather than the exception (see below). The inability to detect expression from TATA-less tRNA and 5S rRNA genes in S. pombe provides strong evidence to suggest that the mechanism to bring TFIIIB activity to the core promoter in the absence of a TATA element that has been elucidated for other model organisms is not available to and/or not used in S. pombe. This further suggests a fundamental difference between fission yeast and other model systems in the mechanisms that bring the central TF, TBP, to the core promoter. The presence of TATA elements upstream of a substantial fraction of A. thaliana tRNA genes suggests that plants and perhaps other branches of the evolutionary tree share this feature with fission yeast. Thus, this study provides a basis to suggest that, contrary to what has been generally accepted, a significant portion of eukaryotes may use TATA-dependent mechanisms for the great majority of their Pol III transcription.
S. pombe Pol III promoters: TATA elements are the rule.
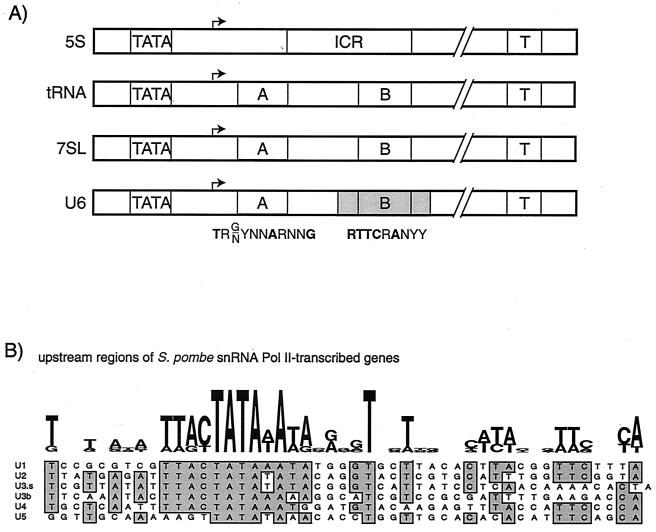
As derived from Fig. 1, the consensus sequences 8TRG/NYNNARNNG18 and 53RTTCRANYY62 represent the A and B boxes of S. pombe tRNA genes. For the S. pombe 7SL and U6 RNA genes, sequence matches to the A box begin 13 and 17 bp, respectively, downstream of their first nucleotide. These genes also match the B box consensus sequence beginning at positions 60 and 69 of 7SL and U6, respectively. Substitution of conserved B box residues in the S. pombe U6 gene rendered this template inactive in the S. pombe in vitro transcription system (M. Hamada and R. Maraia, unpublished data). Thus, in S. pombe, tRNA, U6, and 7SL genes exhibit similar promoter architectures, in which TATA appears as one of multiple promoter elements (Fig. 7A).
FIG. 7.
Conserved TATA motifs in the promoters of small RNA genes in S. pombe. (A) Pol III-transcribed genes. The upstream TATA, internal control region (ICR) (for 5S only), A box and B box (for tRNAs and U6 and 7SL RNAs), and terminators (T) are shown. The bent arrows indicate the +1 positions (as previously determined). Consensus sequences for the A and B box elements that fit all tRNA genes, as well as U6 and 7SL genes, are shown below the schematics; boldface letters are invariant or very highly conserved. The U6 B box resides in an intron (shaded; see the text) (B) Upstream regions of Pol II-transcribed snRNA genes are presented both in Logo (above) and as individual aligned sequences, ending at +1.
The data in Fig. 5 suggest that although tRNA genes with A box residues other than G at the number 10 position represent a significant fraction of genes, these may interact with TFIIIC in a less stable manner than those with G at this position. Attempts to identify other features of these genes that distinguished them from the main set of tRNA genes was not successful. It is also interesting in this regard that the A box sequences of the S. pombe U6 and 7SL genes contain a G at the corresponding position of their A box sequences. In considering the potential implications of these findings, it may be important to recall that both the G10 and the non-G10 tRNA genes required an upstream TATA for expression in our assays. While the results suggest a potential for effects of the A box residue corresponding to G10 related to transcription complex formation and/or stability, the physiological significance of this, if any, remains an open question.
Species specificity of Pol III-dependent tRNA transcription.
It is important to note that the data indicate that the role of TATA is not to provide simple additive transcriptional activity to the internal promoters of tRNA and 5S rRNA genes in S. pombe; rather, the homologous transcription machinery is highly dependent on and functionally adapted to the TATA element, even when the internal promoter is strong (Fig. 2, bottom, lanes 1 and 2).
The requirement for an upstream TATA may explain, at least in part, observations that indicate differential expression of tRNA genes in the two yeasts such that while S. pombe tRNA genes are efficiently expressed in S. cerevisiae, the TATA-less tRNA genes from S. cerevisiae were not expressed in S. pombe (34, 79).
Intriguingly, the data in Fig. 1 reveal an inverse correlation between the prevalence of guanosine at position 10 and the presence of an upstream TATA motif, most readily observable for S. cerevisiae, S. pombe, and A. thaliana. Since G10 is invariant or highly conserved in species that rely on TATA-less transcription, the data suggest that G10 may be a significant determinant of TATA-less transcription. This is consistent with the data in Fig. 5 (top). For templates bearing T10 but no TATA, transcription activity is low relative to the same template with TATA (compare lanes 3 and 4), while for templates containing G10, the presence or absence of TATA makes little if any difference (compare lanes 1 and 2).
TATA elements have been noted upstream of tRNA genes in A. thaliana, and a TATA element has been reported to effect reinitiation of tRNA transcription in tobacco (13, 81). A CAA motif was noted to be a transcription initiation site for plant tRNA (81). The analysis in Fig. 1B suggests that this motif may be extended to ANCAANA/T (the underlining reflects a +1 site), reminiscent of initiator elements (Inr) found in some Pol I and Pol II genes (56, 66). However, several differences distinguish the patterns of sequence information content in S. pombe and A. thaliana tRNA genes (Fig. 1B): (i) the information content is higher at TATA for S. pombe, (ii) the information content is higher at position 10 (greater propensity for G10) in A. thaliana, and (iii) the information content is higher around +1 for A. thaliana. The A. thaliana pattern is consistent with data that show partial or little effect of TATA mutations but larger effects of CAA mutations on plant tRNA expression (9, 81). Thus, the cumulative evidence indicates that although fission yeasts and plants exhibit TATA elements upstream of their tRNA genes, S. pombe is more obligatorily dependent on the TATA promoter element for transcription.
TATA and tRNA transcription: exceptional cases in conventional model organisms.
Although in certain cases TATA elements direct Pol III transcription of tRNAs, these appear to be unusual instances. The transcription start site for the gene encoding Xenopus laevis selenocysteine tRNA[ser]sec is dictated by an upstream promoter that includes a TATA element (5, 39). However, this gene utilizes a unique pathway of expression, as evidenced by the Pol III initiation site, which occurs at a highly unusual position for a tRNA, coinciding with the 5′ end of the mature tRNA (38). The sequence of tRNA[ser]sec does not contain an A box promoter element, and start site selection is instead mediated by the TATA element (5, 52). An upstream TATA has also been shown to control expression of a tRNA gene in the insect Bombix mori, in which two genes contribute to silk gland-specific tRNAAla synthesis in a tissue-regulated manner. In this case, an upstream TATA directs expression of the more active of the two tRNAAla genes in a TBP-dependent manner (50). This tRNAAla gene was also analyzed in D. melanogaster cells, where its transcription was stimulated by increased TBP levels (71). Figure 1A demonstrates that TATA elements are not found upstream of most tRNA genes in the insect D. melanogaster, for which it was recently reported that TBP-related factor, TRF, rather than TBP itself, directs tRNA transcription (68). The cumulative data are consistent with a model in which TATA and TBP may be used for special cases of tRNA expression in insects and other organisms. In summary, prior to the present report, TATA elements had been characterized as promoters of tRNA transcription in exceptional cases.
A functional TATA element in a Pol I core promoter.
Our data demonstrate that substitution of the TATA element that overlaps the −30 region of the S. pombe Pol I promoter abolishes rRNA expression in vivo. Although our results do not address the mechanism by which TATA promotes expression by Pol I in S. pombe, they nonetheless indicate an important role of TATA in rRNA expression in fission yeasts. The location of the TATAAA sequence, positioned 35 bp upstream of the previously determined start site of Pol I transcription, suggests that TATA-TBP functions as part of the core promoter in S. pombe.
TATA elements are also more widespread in the Pol II-transcribed genes in S. pombe than in other species.
Although TATA promoter elements are a recognized hallmark of protein-encoding genes, not all Pol II promoters contain TATA elements, as some mRNA-encoding genes use other core promoter elements for transcription (4, 66). Another class of genes, those that produce snRNAs (e.g., U1 to U5), are transcribed by Pol II from TATA-less promoters in yeast, human, and Drosophila (18, 73, 82). In contrast to this, TATA motifs have been noted in the promoter regions of some S. pombe snRNA genes that are transcribed by Pol II (reference 83 and references therein). Alignment of the upstream regions of the U1 to U5 snRNA genes of S. pombe revealed a prominent TATA motif in addition to an upstream TTAC sequence (Fig. 7B).
Evolutionary implications.
It seems reasonable to consider S. pombe a tentative link to an ancient ancestor that used ubiquitous TATA elements to promote transcription of a wide range of genes, if not of all genes. The ubiquitous TATA-like elements of an archaeonlike organism would have remained after divergence of the three nuclear Pols. Thus, while S. pombe may have retained TATA elements as essential components of the Pol I, II, and III systems, many genes of other organisms would have lost their TATAs. This scenario is consistent with a residual requirement for TBP and/or TBP-related factors by all three Pols, even though most of their target genes do not use TATA elements in many species. It is also consistent with the use of TBP-related factors in metazoans (19, 68, 69, 72). Although plant tRNA genes are also flanked by upstream TATAs (13) (Fig. 1), we are unaware of a simple line of inheritance between plants and S. pombe that could account for this common feature. 5S rRNA genes are also dispersed in Neurospora crassa, and transcription was also reported to require a TATA at −29 (64). TATA elements upstream of a substantial fraction of plant and other species' 5S and tRNA genes suggests that fission yeasts are not unique in the structure of their Pol III promoters.
The transcription system characterized here suggests that Pol III initiation in S. pombe may be comparable to type 3 gene transcription and other systems that use TATA elements to recruit TBP. Although for tRNA transcription, the yeast and mammalian TFIIIB subunits appear to be completely orthologous, the situation is more complex for U6 snRNA expression in metazoa (e.g., type 3 genes), as a distinct variant of BRF known as BRFU/TFIIIB50 characterizes the TFIIIB activity that mediates type 3 gene transcription (63, 70). The BRFU/TFIIIB50 variant that acts at the U6 TATA-containing promoter differs from Brf in its lack of a C-terminal domain that in Brf has been shown to interact with TBP (10, 26, 32), and consistent with this, it may not stably bind TBP, since association of TFIIIB with TBP was notably weak (70). This suggests principal recruitment of TBP by the TATA element, which may obfuscate a requirement for stable interaction between TBP and BRFU in the absence of DNA (70). In this regard, the S. pombe Pol III initiation system may be comparable to Pol II, in which TBP and TFIIB do not form a stable association in the absence of DNA. Instead, association of TBP with the TATA element appears to create a binding site for TFIIB, in part due to the specific bending effect of TBP on TATA DNA (49). While our results indicate a significant role for TATA in tRNA transcription in S. pombe, they also leave open the possibility that Brf-mediated recruitment of TBP occurs at some level in vivo and that some tRNA genes are less dependent on the TATA element for TBP recruitment than others. However, the cumulative data suggest that TATA-mediated recruitment of TBP plays a major role in Pol III transcription in S. pombe, while Brf may be brought to the upstream region in part by TFIIIC and in part by recognition of the TATA-TBP complex. In any case, S. pombe has revealed a novel Pol III system that should be useful for studying eukaryotic transcription initiation and the function of the core promoter.
ACKNOWLEDGMENTS
We are grateful to W. Makalowski for alignments and for suggesting Logo. We thank R. Intine for pFL20/18SPst5.8Si4 and technical advice, E. P. Geiduschek, N. Hernandez, R. Roeder, D. Setzer, and D. Reinberg for discussions, and I. Willis for encouragement and expert advice.
Mitsuhiro Hamada and Ying Huang contributed equally to this work.
REFERENCES
- 1.Baker R E, Hall B D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984;3:2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodem J, Dobreva G, Hoffmann-Rohrer U, Iben S, Zentgraf H, Delius H, Vingron M, Grummt I. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 2000;1:171–175. doi: 10.1093/embo-reports/kvd032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogenhagen D F, Wormington W M, Brown D D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982;28:413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 4.Burke T W, Willy P J, Kutach A K, Butler J E, Kadonaga J T. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harbor Symp Quant Biol. 1998;63:75–82. doi: 10.1101/sqb.1998.63.75. [DOI] [PubMed] [Google Scholar]
- 5.Carbon P, Krol A. Transcription of the Xenopus laevis selenocysteine tRNA(Ser)Sec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. EMBO J. 1991;10:599–606. doi: 10.1002/j.1460-2075.1991.tb07987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 7.Chedin S, Ferri M L, Peyroche G, Andrau J C, Jourdain S, Lefebvre O, Werner M, Carles C, Sentenac A. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp Quant Biol. 1998;63:381–389. doi: 10.1101/sqb.1998.63.381. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Guo A, Pape L. An immunoaffinity purified Schizosaccharomyces pombe TBP-containing complex directs correct initiation of the S. pombe rRNA gene promoter. Nucleic Acids Res. 1997;25:1633–1640. doi: 10.1093/nar/25.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choisne N, Carneiro V T, Pelletier G, Small I. Implication of 5′-flanking sequence elements in expression of a plant tRNA(Leu) gene. Plant Mol Biol. 1998;36:113–123. doi: 10.1023/a:1005988004924. [DOI] [PubMed] [Google Scholar]
- 10.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 11.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 12.Dean N, Berk A J. Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol Cell Biol. 1988;8:3017–3025. doi: 10.1128/mcb.8.8.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieci G, Percudani R, Giuliodori S, Bottarelli L, Ottonello S. TFIIIC-independent in vitro transcription of yeast tRNA genes. J Mol Biol. 2000;299:601–613. doi: 10.1006/jmbi.2000.3783. [DOI] [PubMed] [Google Scholar]
- 14.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 15.Geiduschek E P, Tocchini-Valentini G P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 16.Hamada M, Sakulich A L, Koduru S B, Maraia R. Transcription termination by RNA polymerase III in fission yeast: a genetic and biochemically-tractable model system. J Biol Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 19.Holmes M C, Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh Y J, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Hamada M, Maraia R J. Isolation and cloning of four subunits of a fission yeast TFIIIC complex that includes an ortholog of the human regulatory protein TFIIIC-beta. J Biol Chem. 2000;275:31480–31487. doi: 10.1074/jbc.M004635200. [DOI] [PubMed] [Google Scholar]
- 22.Huet J, Conesa C, Manaud N, Chaussivert N, Sentenac A. Interactions between yeast TFIIIB components. Nucleic Acids Res. 1994;22:2282–2288. doi: 10.1093/nar/22.12.2282. . (Corrected and republished, 22:3433–3439.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Intine R V, Good L, Nazar R N. Essential structural features in the Schizosaccharomyces pombe pre-rRNA 5′ external transcribed spacer. J Mol Biol. 1999;286:695–708. doi: 10.1006/jmbi.1998.2502. [DOI] [PubMed] [Google Scholar]
- 24.Intine R V A, Sakulich A L, Koduru S B, Huang Y, Pierstorrf E, Goodier J L, Phan L, Maraia R J. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol Cell. 2000;6:339–348. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 25.Jaozeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 26.Kassavetis G, Bardeleben C, Kumar A, Ramirez E, Geduschek E P. Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol Cell Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kassavetis G, Letts G, Geiduschek E P. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J. 2001;20:2823–2834. doi: 10.1093/emboj/20.11.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 107–126. [Google Scholar]
- 29.Kassavetis G A, Bartholomew B, Blanco J A, Johnson T E, Geiduschek E P. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc Natl Acad Sci USA. 1991;88:7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 31.Kassavetis G A, Joazeiro C A P, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 32.Kassavetis G A, Kumar A, Ramirez E, Geiduschek E P. Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol Cell Biol. 1998;18:5587–5599. doi: 10.1128/mcb.18.9.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krupp G, Thurianx P, Willis I, Gamulin V, Soll D. First identification of an amber nonsense mutation in Schizosaccharomyces pombe: major differences in the efficiency of homologous versus heterologous yeast suppressor tRNA genes. Mol Gen Genet. 1985;201:82–87. doi: 10.1007/BF00397990. [DOI] [PubMed] [Google Scholar]
- 35.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassar A B, Martin P L, Roeder R G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 38.Lee B J, de la Pena P, Tobian J A, Zasloff M, Hatfield D. Unique pathway of expression of an opal suppressor phosphoserine tRNA. Proc Natl Acad Sci USA. 1987;84:6384–6388. doi: 10.1073/pnas.84.18.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B J, Kang S G, Hatfield D. Transcription of Xenopus selenocysteine tRNA Ser (formerly designated opal suppressor phosphoserine tRNA) gene is directed by multiple 5′ extragenic regulatory elements. J Biol Chem. 1989;264:9696–9702. [PubMed] [Google Scholar]
- 40.Lobo S M, Tanaka M, Sullivan M L, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-De-Leon A, Librizzi M, Puglia K, Willis I M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 42.Lowe T M, Eddy S R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao J, Appel B, Schaack J, Sharp S, Yanada H, Soll D. The 5S RNA genes of Schizosaccharomyces pombe. Nucleic Acids Res. 1982;10:487–500. doi: 10.1093/nar/10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller G, Panov K I, Friedrich J K, Trinkle-Mulcahy L, Lamond A I, Zomerdijk J C. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittal V, Hernandez N. Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science. 1997;275:1136–1140. doi: 10.1126/science.275.5303.1136. [DOI] [PubMed] [Google Scholar]
- 47.Moir R D, Sethy-Coraci I, Puglia K, Librizzi M D, Willis I M. A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol Cell Biol. 1997;17:7119–7125. doi: 10.1128/mcb.17.12.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nichols M, Bell J, Klekamp M S, Weil P A, Soll D. Multiple mutations of the first gene of a dimeric tRNA gene abolish in vitro tRNA-gene transcription. J Biol Chem. 1989;264:17084–17090. [PubMed] [Google Scholar]
- 49.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 50.Ouyang C, Martinez M J, Young L S, Sprague K U. TATA-binding protein-TATA interaction is a key determinant of differential transcription of silkworm constitutive and silk gland-specific tRNA (Ala) genes. Mol Cell Biol. 2000;20:1329–1343. doi: 10.1128/mcb.20.4.1329-1343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 52.Park J M, Choi I S, Kang S G, Lee J Y, Hatfield D L, Lee B J. Upstream promoter elements are sufficient for selenocysteine tRNA[Ser]Sec gene transcription and to determine the transcription start point. Gene. 1995;162:13–19. doi: 10.1016/0378-1119(95)00340-c. [DOI] [PubMed] [Google Scholar]
- 53.Paule M R, White R J. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poon D, Weil P A. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J Biol Chem. 1993;268:15325–15338. [PubMed] [Google Scholar]
- 56.Radebaugh C A, Gong X, Bartholomew B, Paule M R. Identification of previously unrecognized common elements in eukaryotic promoters. A ribosomal RNA gene initiator element for RNA polymerase I. J Biol Chem. 1997;272:3141–3144. doi: 10.1074/jbc.272.6.3141. [DOI] [PubMed] [Google Scholar]
- 57.Rameau G, Puglia K, Crowe A, Sethy I, Willis I. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol Cell Biol. 1994;14:822–830. doi: 10.1128/mcb.14.1.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinberg D, Orphanides G, Ebright R, Akoulitchev S, Carcamo J, Cho H, Cortes P, Drapkin R, Flores O, Ha I, Inostroza J A, Kim S, et al. The RNA polymerase II general transcription factors: past, present, and future. Cold Spring Harbor Symp Quant Biol. 1998;63:83–103. doi: 10.1101/sqb.1998.63.83. [DOI] [PubMed] [Google Scholar]
- 59.Roeder R G. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- 60.Ruth J, Conesa C, Dieci G, Lefebvre O, Dusterhoft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 61.Schaack J, Sharp S, Dingermann T, Soll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983;258:2447–2453. [PubMed] [Google Scholar]
- 62.Schneider T D, Stephens R M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schramm L, Pendergrast P S, Sun Y, Hernandez N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000;14:2650–2663. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selker E U, Morzycka-Wroblewska E, Stevens J N, Metzenberg R L. An upstream signal is required for in vitro transcription of Neurospora 5S RNA genes. Mol Gen Genet. 1986;205:189–192. doi: 10.1007/BF02428052. [DOI] [PubMed] [Google Scholar]
- 65.Sharp S, Dingermann T, Schaack J, DeFranco D, Soll D. Transcription of eukaryotic tRNA genes in vitro. I. Analysis of control regions using a competition assay. J Biol Chem. 1983;258:2440–2446. [PubMed] [Google Scholar]
- 66.Smale S T, Jain A, Kaufmann J, Emami K H, Lo K, Garraway I P. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harbor Symp, Quant Biol. 1998;63:21–31. doi: 10.1101/sqb.1998.63.21. [DOI] [PubMed] [Google Scholar]
- 67.Soppa J. Transcription initiation in Archaea: facts, factors and future aspects. Mol Microbiol. 1999;31:1295–1305. doi: 10.1046/j.1365-2958.1999.01273.x. [DOI] [PubMed] [Google Scholar]
- 68.Takada S, Lis J T, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 69.Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait B T, Roeder R G. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci USA. 1999;96:13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teichmann M, Wang Z, Roeder R G. A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci USA. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trivedi A, Young L S, Ouyang C, Johnson D L, Sprague K U. A TATA element is required for tRNA promoter activity and confers TATA-binding protein responsiveness in Drosophila Schneider-2 cells. J Biol Chem. 1999;274:11369–11375. doi: 10.1074/jbc.274.16.11369. [DOI] [PubMed] [Google Scholar]
- 72.Veenstra G J, Weeks D L, Wolffe A P. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Stumph W E. Identification and topological arrangement of Drosophila proximal sequence element (PSE)-binding protein subunits that contact the PSEs of U1 and U6 small nuclear RNA genes. Mol Cell Biol. 1998;18:1570–1579. doi: 10.1128/mcb.18.3.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z, Roeder R G. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;89:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Z, Roeder R G. TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White R J, Jackson S P. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell. 1992;71:1041–1053. doi: 10.1016/0092-8674(92)90398-v. [DOI] [PubMed] [Google Scholar]
- 78.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 79.Willis I, Nichols M, Chisholm V, Soll D, Heyer W D, Szankasi P, Amstutz H, Munz P, Kohli J. Functional complementation between mutations in a yeast suppressor tRNA gene reveals potential for evolution of tRNA sequences. Proc Natl Acad Sci USA. 1986;83:7860–7864. doi: 10.1073/pnas.83.20.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Willis I M. RNA polymerase III genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 81.Yukawa Y, Sugita M, Choisne N, Small I, Sugiura M. The TATA motif, the CAA motif and the poly(T) transcription termination motif are all important for transcription re-initiation on plant tRNA genes. Plant J. 2000;22:439–447. doi: 10.1046/j.1365-313x.2000.00752.x. [DOI] [PubMed] [Google Scholar]
- 82.Zamrod Z, Tyree C M, Song Y, Stumph W E. In vitro transcription of a Drosophila U1 small nuclear RNA gene requires TATA box-binding protein and two proximal cis-acting elements with stringent spacing requirements. Mol Cell Biol. 1993;13:5918–5927. doi: 10.1128/mcb.13.9.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou D, Lobo-Ruppert S M. Transcription of the Schizosaccharomyces pombe U2 gene in vivo and in vitro is directed by two essential promoter elements. Nucleic Acids Res. 2001;29:2003–2011. doi: 10.1093/nar/29.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zomerdijk J C, Beckmann H, Comai L, Tjian R. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science. 1994;266:2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]