Abstract
An in vitro transformation system of carcinogen-treated Syrian hamster embryo (SHE) cell cultures represents multistep genetic and nongenetic changes that develop during the neoplastic progression of normal cells to tumor cells in vivo. During this neoplastic progression, SHE cells demonstrate an altered response to epidermal growth factor (EGF). In the present report, we examined the role of the adapter protein Gab1 (Grb2-associated binder-1) in the neoplastic progression of SHE cells. We used two asbestos-transformed SHE cell clones in different neoplastic stages: a 10W+8 clone, which is immortal and retains the ability to suppress the tumorigenicity of tumor cells in cell-cell hybrid experiments, and a 10W−1 clone, which has lost this tumor suppressor ability. 10W+8 cells expressed full-length 100-kDa Gab1 and associated 5.2-kb mRNA. Upon repeated cell passaging, 10W−1 cells showed increasing expression of a novel 87-kDa form of Gab1 as well as 4.6-kb mRNA with diminishing expression of the original 100-kDa Gab1. cDNA encoding the 87-kDa Gab1 predicts a form of Gab1 lacking the amino-terminal 103 amino acids (Gab1Δ1-103), which corresponds to loss of most of the pleckstrin homology (PH) domain. Gab1Δ1-103 retains the ability to be phosphorylated in an EGF-dependent manner and to associate with the EGF receptor and SHP-2 upon EGF stimulation. The endogenous expression of Gab1Δ1-103 in 10W−1 cells appeared closely related to EGF-dependent colony formation in soft agar. Moreover, transfection and expression of Gab1Δ1-103, but not Gab1, in 10W+8 cells enhanced their EGF-dependent colony formation in soft agar. These results demonstrate that Gab1 is a target of carcinogen-induced transformation of SHE cells and that the expression of a Gab1 variant lacking most of the PH domain plays a specific role in the neoplastic progression of SHE cells.
Many lines of evidence support the idea that neoplastic transformation is acquired by multistep genetic and nongenetic changes (1, 4, 9). Our laboratory developed a Syrian hamster embryo (SHE) cell transformation system that has been shown to be a reproducible in vitro culture system (5–8, 27, 36). In this system, normal SHE cells are isolated from 13-day-gestation fetuses and exposed to carcinogens such as asbestos, benzo[a]pyrene, or diethylstilbestrol for 24 to 48 h. During subsequent culture and passages, several defined aspects of transformation develop at different time points.
The first stage in the transformation process following carcinogen exposure is typically a morphological transformation (7). Morphologically transformed cells may still be mortal and senesce once they reach their maximum life span (16). Another important early step in the neoplastic transformation of SHE cells is the acquisition of immortality (loss of senescence) (34). Genetic instability enhances the probability of an immortal, nontumorigenic (preneoplastic) cell to acquire the additional alterations required to achieve the next step in the transformation process, tumorigenicity. These alterations include the activation of proto-oncogenes and the loss of tumor suppressor genes (9, 21). Tumorigenicity of SHE cells in nude mice is closely correlated with their growth in soft agar (6). Finally, tumorigenic SHE cells increase their ability to metastasize in vivo, to increase in the blood supply, and to avoid host tumor immunosurveillance systems (21).
Thus, in this in vitro transformation system, one can initiate intermediate or preneoplastic cells that have acquired some, but not all, of the properties necessary for tumorigenicity (5). Early-passage, chemically immortalized cells retained the ability to suppress tumorigenicity of a benzo[a]pyrene-transformed hamster fibrosarcoma cell line, BP6T, in cell-cell hybrids, thus classified as tumor suppressor gene positive phenotype (supB+). Upon subsequent passage in culture, some cell variant clones lose this tumor suppressor ability (classified as tumor suppressor gene negative phenotype [supB−]) (27). By subcloning these preneoplastic populations arising from asbestos-transformed SHE cells, two distinct clones were isolated, a supB+, 10W+8 clone and a supB−, 10W−1 clone (27). We have previously investigated signaling through the receptor for epidermal growth factor (EGF) using these two SHE cell variants. These cells were biochemically different in the modulation of ligand-induced EGF receptor (EGFR) activation by lipooxygenase products (13), dephosphorylation rate of EGFR (14), and the recruitment of signaling molecules such as SHP-2, an SH2-containing protein tyrosine phosphatase, to the activated EGFR (19).
In the present study, we investigated the expression of an adapter protein, Gab1, and its functional differences between 10W+8 and 10W−1 cells. Gab1 was first identified and cloned as a Grb2-associated protein from a human glial tumor expression library (18). Gab1 is a member of the insulin receptor substrate 1 (IRS-1) family of proteins, which includes IRS-1, IRS-2, IRS-3, Daughter of Sevenless (DOS), fibroblastic growth factor receptor substrate 2 (FRS2), Downstream of tyrosine kinase (Dok), Linker for activation of T cells (LAT), Dok-related (Dok-R), and Gab2 (10, 15, 17, 18, 23, 28, 35, 38, 44–46). The most striking homology with other IRS-1 family proteins lies within the pleckstrin homology (PH) domain in the amino terminus of Gab1 (18), which is involved in the interactions with specific phospholipids in the plasma membrane (20, 31, 33, 40). Gab1 plays a pivotal role in EGFR signaling through EGF-dependent tyrosine phosphorylation and association with SHP-2, phospholipase Cγ and phosphatidylinositol 3-kinase (PI3K) (11, 18, 29, 30, 37, 41).
Here we report that a novel form of Gab1 lacking most of the PH domain is exclusively expressed in a preneoplastic SHE cell clone of supB− phenotype. This variant form of Gab1 is associated with anchorage-independent growth of the cells in the presence of growth factors, including EGF.
MATERIALS AND METHODS
Materials.
Dulbecco's modified Institute for Biological Research medium (IBR), calcium- and magnesium-free phosphate-buffered saline (cmf-PBS), trypsin-EDTA, and gentamicin were from Life Technologies, Inc. Fetal bovine serum (FBS) was purchased from HyClone Laboratories. Nonfat dry milk was purchased from Bio-Rad. Bicinchoninic acid protein assay reagent was from Pierce. EGF and insulin were obtained from Collaborative Research Associates. The acrylamide and bisacrylamide were purchased from Amresco. The nitrocellulose membrane was obtained from Schleicher & Schuell. The immunoglobulin (Ig) conjugates, Hyperfilm, and enhanced chemiluminescence (ECL) reagents were purchased from Amersham. The anti-SHP-2 (SC-280) and antiphosphotyrosine (anti-p-Tyr) antibody (PY99) were obtained from Santa Cruz Biotechnology. Anti-Gab1 (06-579; against amino acids 664 to 694) was from Upstate Biotechnology Inc., and another anti-Gab1 (162D-124-3; against amino acids 192 to 205) was a generous gift from Richard P. DiAugustine, NIEHS. p44/42 MAP Kinase Assay kit and SAPK/JNK Assay kit were purchased from New England BioLabs, Inc. The protein A-Sepharose beads and all other reagents were obtained from Sigma.
Cell culture and transfections.
The experiments were performed with two subclones obtained from asbestos-transformed SHE fibroblast cell line clones 10W+8 and 10W−1 described above. Cells were cultured in IBR containing 10% FBS and gentamicin (10 μg/ml) at 37°C in a humidified 5% CO2, 95% air atmosphere. Trypsin-EDTA (0.05%) was used to subculture the cells. 10W+8 cells, under passages 11 to 17, were used in these experiments. 10W−1 cells under passages 11 to 17 were used as early passages and named 10W−1E, while 10W−1 cells at passages 25 to 40 were used as later passages and named 10W−1L in this study. Cell transfections were performed using the Lipofectamine Plus reagent (Gibco BRL) following the manufacturer's instruction.
Immunoprecipitation.
After reaching 50 to 60% confluence on 150-mm-diameter dishes, cells were serum deprived for 20 h to synchronize cell cycles in G0. At least 1 h after changing the medium to fresh IBR, cells were further incubated at 37°C with or without 100 ng of EGF per ml for indicated times. Then dishes were placed on ice, the cells were rinsed twice with 10 ml of ice-cold cmf-PBS, and the cells were lysed with 0.4 ml of ice-cold lysis buffer (1% Triton X-100, 50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 2 mM sodium orthovanadate, 100 μM p-nitrophenylphosphate, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 10 mM sodium phosphate, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml). After the cells were scraped, the lysate was collected, sonicated on ice for 7 s twice, and centrifuged at 2,000 × g for 10 min at 4°C. The supernatant was transferred to a new tube, and an aliquot was removed to measure the protein concentration by bicinchoninic acid protein assay reagent. Two milligrams of cellular protein from 107 cells was precleared with 5 μl of normal rabbit serum and 100 μl of protein A-Sepharose beads. The mixture was tumbled at 4°C for 1 h and centrifuged at 10,000 × g for 2 min, and the supernatant was transferred to a new tube. Specific antibodies (anti-Gab1, 2 μg; anti-SHP-2, 2 μg) were added to the supernatant, the samples were tumbled overnight at 4°C, and 100 μl of protein A-Sepharose beads was added and tumbled for an additional 2 h. After centrifugation at 10,000 × g for 2 min, the pellet was washed three times with 1 ml of 20 mM HEPES (pH 7.5)–150 mM NaCl–0.1% Triton X-100–10% glycerol. Protein sample buffer (2×) was added to the final pellet; the sample was boiled for 8 min and centrifuged at 10,000 × g for 10 min. The supernatant was immediately used or frozen at −70°C for later use with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Western blot analysis.
The samples obtained by immunoprecipitation were loaded and run on SDS-PAGE (8% acrylamide gel) and electrophoretically transferred to a nitrocellulose membrane in 25 mM Tris–192 mM glycine–20% methanol–0.1% SDS using semidry electrophoresis equipment (Hoefer). Membranes were blocked in Tris-buffered saline containing 0.1% Tween 20 (TBST) with 5% bovine serum albumin (BSA) at 4°C overnight or in 3% nonfat milk in cmf-PBS for 20 min at room temperature (for anti-Gab1). The blots were then incubated for 1 h with anti-SHP-2 antibody (1:2,000) or anti-p-Tyr antibody (1:1,000) with 1% BSA plus TBST at room temperature for 1 h or anti-Gab1 antibody (1:1,000) with 3% nonfat milk in cmf-PBS at 4°C overnight. The blots were washed five times in TBST or water (for anti-Gab1) and then incubated with horseradish peroxidase-conjugated anti-rabbit Ig (1:5,000) or anti-mouse Ig (1:5,000; for anti-p-Tyr antibody) in 1% BSA plus TBST at room temperature for 1 h or 3% nonfat milk in cmf-PBS at room temperature for 1.5 h (for anti-Gab1). The blots were again washed five times in TBST or water and once with 0.05% Tween 20 plus cmf-PBS (for anti-Gab1) and visualized using the Amersham ECL system.
Northern blot analysis.
Total RNA was isolated with Trizol Reagent (Life Technologies), and 10 μg of total RNA samples was separated by electrophoresis in 1% agarose-formaldehyde gel and transferred onto nylon membranes (Hybond-N+; Amersham) by capillary blotting. The membranes were cross-linked by UV radiation. cDNA probes of 0.4 kb corresponding to the middle portion of Gab1 cDNA sequence were amplified with 5′-CCTTTATAACCTGCCCAGGAGT-3′ and 3′-GCAG/ATCTTGAGAACTAGCATCT-5′ and labeled with [32P]dCTP with the Prime-It RT Random-Primer Labeling kit (Stratagene). Blots were prehybridized in Rapid-hyb buffer (Amersham) at 65°C for 2 h followed by hybridization at 65°C overnight. The blots were then washed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at room temperature and then twice in 0.1× SSC–0.1% SDS at 65°C. The membrane was exposed to Hyperfilm (Amersham) for 24 h.
cDNA cloning and plasmid construction.
mRNA was isolated from 10W−1L cells using mRNA Separator kit (Clontech), and a 5′ rapid amplification of cDNA ends (RACE) strategy with SMART RACE cDNA Amplification kit (Clontech) was applied for the cloning of cDNAs encoding two forms of Gab1. Two antisense primers, 3′-GTAAGAGGGAGGTAGTGTGACTGGAGAGGCTT-5′ (486R) and 3′-CAGTGGCAACGGCGTGTCTTCACTTTACGCTCTTG-5′ (2104R, which includes the translation stop codon 3′-TCA-5′ [italicized]), were used with Advantage-GC cDNA Polymerase Mix kit (Clontech). Sequence analysis of PCR products by ABI PRISM dRhodamine terminator cycle sequencing ready kit (Perkin-Elmer Applied Biosystems) was performed both directly and after a subcloning into a cytomegalovirus (CMV) promoter-driven mammalian expression vector pCR3.1 using Eukaryotic TA Cloning kit (Invitrogen). Plasmids were purified with a plasmid purification kit (Qiagen). For overexpression of hamster Gab1 of 100 kDa, a 2.1-kb cDNA product amplified by 5′-ACCATGAGCGGTGGTGAAGTGGTC-3′, which includes the translation start codon 5′-ATG-3′, and by 2104R was used. On the other hand, a cDNA product obtained by 5′-RACE with 2104R antisense primer was used for overexpression of 87-kDa Gab1/Gab1Δ1-103 (described below).
Reverse transcription (RT)-PCR.
First-strand cDNA was generated with 1 μg of total RNA and oligo(dT)18 primer using Advantage RT-for PCR kit (Clontech). RNA samples were also subjected to the reaction of cDNA synthesis without reverse transcriptase and served as negative controls. Primers used were 5′-TCGCGGCGTGCACCATGAGCGGCGGCGAAG-3′ (−14F; the translation start codon 5′-ATG-3′ is included), 5′-GAGATCTGCTGGGCTTCTCGGGGTT-3′ (LSP), and 2104R. Thermal cycling conditions were 1 cycle at 94°C for 1 min; 35 cycles at 94°C for 45 s, 68°C for 45 s, and 72°C for 2 min; and 1 cycle at 72°C for 10 min.
Cell growth in soft agar.
Soft agar assay was performed as previously described (2, 26). Briefly, 2 × 103 cells were suspended in 0.3% Bacto-agar (GIBCO) containing 0.1% Bacto Peptone and 10% FBS on a 0.6% Bacto-agar layer containing 0.1% Bacto Peptone and 10% FBS in IBR medium using six-well plates. The plates were incubated at 37°C for 14 to 21 days and scored for colonies that were greater than 60 μm in diameter (greater than 30 cells). For some assays, 50 ng of EGF per ml and/or 1 μg of insulin per ml was added to the top layer of agar.
[3H]thymidine incorporation assay.
[3H]thymidine incorporation assay was employed to measure DNA synthesis as previously described (13). Briefly, 103 cells were seeded in 96-well plates (Costar) and cultured until 60 to 70% confluence was reached. Cells were then serum deprived for 16 to 20 h and incubated in the presence of 1 μCi of [3H]thymidine (ICN Pharmaceuticals, Inc.)/well with or without 50 ng of EGF per ml at 37°C for 24 h. Cells were rinsed twice with ice-cold cmf-PBS and then treated with ice-cold 5% trichloroacetic acid for at least 30 min at 4°C. Cells were rinsed three times with water and lysed with 0.2 N NaOH–0.1% SDS at room temperature for at least 1 h. Samples were neutralized with hydrochloric acid, and radiation was quantitated in a Packard 2000CA scintillation counter. All incubations were performed in sextuplicate.
p44/42 mitogen-activated protein kinase (MAPK) activity assay.
EGF-dependent activation of p44/42 MAPK (extracellular signal-regulated kinase 1 [ERK1] and ERK2) was measured by p44/42 MAP Kinase Assay kit according to the manufacturer's instructions. Briefly, cells were stimulated with 100 ng of EGF per ml for the indicated times and lysed with cell lysis buffer. The cell lysate was subjected to immunoprecipitation of phospho-p44/42 MAPK (both Thr202 and Tyr204 phosphorylated) with an immobilized monoclonal antibody. Immunoprecipitates were washed and incubated with 200 μM ATP and 2 μg of Elk-1 fusion protein at 30°C for 30 min. Samples were analyzed by SDS-PAGE (10% acrylamide gel), followed by Western blotting with anti-phospho-Elk-1 antibody specific to Ser381-phosphorylated Elk-1.
Indirect immunofluorescence
10W+8 and 10W−1L cells (104) were plated on four-well chamber slides (Nunc, Inc.) and cultured for 3 days. After plating the slides on ice, cells were rinsed twice with ice-cold PBS and incubated with 2% paraformaldehyde in PBS on ice for 30 min. Cells were further incubated in PBS containing 50 mM ammonium chloride at room temperature, followed by washing once with PBS. After incubation with 5% BSA–0.1% Triton X-100 in PBS (BT-PBS) for 30 min, cells were treated with BT-PBS containing either 10 μg of nonimmune rabbit IgG per ml or 10 μg of anti-Gab1 per ml at 4°C overnight. Following two additional washings with PBS, cells were incubated with fluorescein isothiocyanate-conjugated anti-rabbit IgG (diluted 1:100 with BT-PBS) at room temperature for 1 h. Cells were washed five times with PBS, and fluorescence intensity was measured using a laser-emission confocal fluorescence cytometer (ACAS 570; Meridian Instruments, Inc.) as previously described (25). The threshold of fluorescence intensity representing nonspecific fluorescence was determined using the images obtained with nonimmune rabbit IgG. The images with nonimmune IgG were essentially cleared with the threshold thus obtained. Then we applied the threshold to the images with anti-Gab1 to subtract nonspecific fluorescence from the whole fluorescence.
RESULTS
Expression of a novel 87-kDa form of Gab1 in 10W−1 cells.
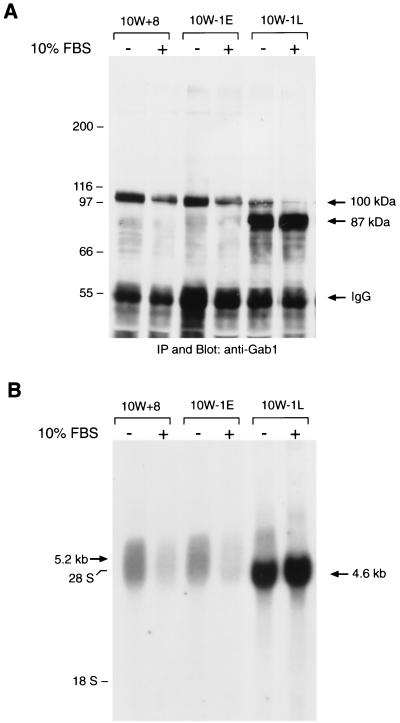
First, we examined the expression of Gab1 in preneoplastic SHE cell variants 10W+8 (passages 11 to 17), 10W−1E (passages 11 to 17), and 10W−1L (passages 25 to 40). Cell lysates were subjected to immunoprecipitation with specific antibodies against the carboxy terminus of human Gab1, followed by SDS-PAGE (8% acrylamide) and anti-Gab1 blotting (Fig. 1A). All of these variants showed the expression of Gab1 with an apparent molecular mass of 100 kDa, although its expression was minimal in 10W−1L cells. Instead, an intense band at 87 kDa was observed in 10W−1L cells, which was not detected in 10W+8 and slightly detectable in 10W−1E. Both 100- and 87-kDa proteins were also detected by another anti-Gab1 antibody raised against a different epitope (amino acids 192 to 205, data not shown), which strongly suggests that the 87-kDa protein is a variant form of Gab1. We analyzed the expression of these two forms of Gab1 following serum starvation and stimulation and found that these two forms were oppositely regulated. When stimulated with 10% FBS, the expression of the 100-kDa Gab1 expression decreased while the expression of the 87-kDa Gab1 increased.
FIG. 1.
Increasing expression of a novel 87-kDa form of Gab1 protein and 4.6-kb mRNA in 10W−1 cells during passages. (A) 10W+8, 10W−1E, and 10W−1L cells were cultured with 10% FBS for 48 to 72 h, and some of the cells were serum starved for 20 h before harvest (10% FBS − lanes). Gab1 was immunoprecipitated (IP) from the lysates of 10W+8, 10W−1E, and 10W−1L cells, analyzed by SDS–8% PAGE followed by anti-Gab1 blotting, and visualized by ECL. Signals at 100 kDa commonly observed among those cells and signals at 87 kDa clearly detected in 10W−1L cells are indicated by arrows as well as signals representing IgG. (B) 10W+8, 10W−1E, and 10W−1L cells were cultured with 10% FBS, and some of the cells were serum starved for 20 h before harvest (10% FBS − lanes). Ten micrograms of total RNA was loaded onto each lane and analyzed for Gab1 mRNA expression. A broad signal around 5.2 kb observed in all cells and an intense signal at 4.6 kb clearly detected in 10W−1L cells are indicated by arrows.
To determine whether the difference between the 100- and 87-kDa products occurs before or after translation, we performed a Northern blot analysis using a 32P-labeled specific probe for Gab1 (Fig. 1B). 10W+8 cells showed a broad signal of mRNA around 5.2 kb. The expression of mRNA around 5.2 kb decreased with 10% FBS. On the other hand, 10W−1L cells intensely expressed a 4.6-kb mRNA as well as a limited signal around 5.2 kb. 10W−1E cells typically showed similar results with 10W+8, although a modest expression of the 4.6-kb mRNA band was often detected. Moreover, 10% FBS stimulation significantly increased the expression of the 4.6-kb mRNA. Based on these results, it is very likely that 100- and 87-kDa proteins are translated from the 5.2- and 4.6-kb mRNAs, respectively. Interestingly, the expression of 100-kDa Gab1 as well as 5.2-kb mRNA in 10W−1L cells continuously decreased by further passaging and became hardly detectable typically at passages 25 to 30 (data not shown).
Time courses of Gab1 expression in 10W+8 and 10W−1L cells during cell growth.
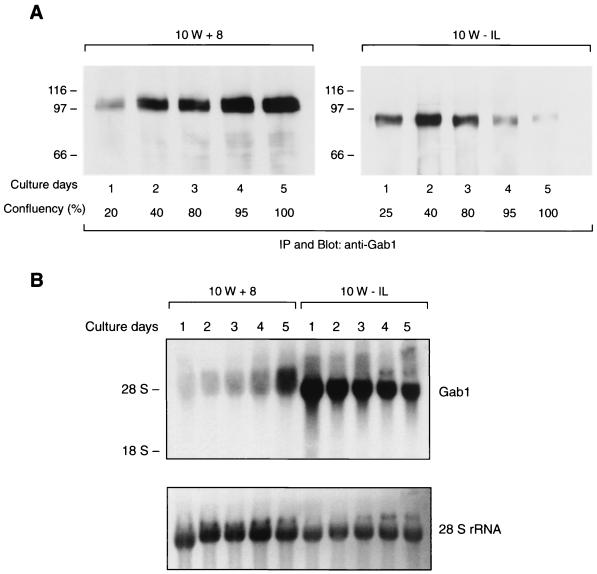
We also examined the time course of expression of 100-kDa Gab1 in 10W+8 and that of 87-kDa Gab1 in 10W−1L cells (Fig. 2A). The expression of 100-kDa Gab1 in 10W+8 was low on day 1 of culture (the next day after cell plating) and gradually increased with cell growth. The highest expression of 100-kDa Gab1 was observed when the cells reached full confluence on day 5. In contrast, the expression of 87-kDa protein in 10W−1L was intense on days 1 to 3 and dramatically decreased after the cells became confluent. Concordant with the results of protein expression, Northern analysis showed increasing expression of 5.2-kb mRNA in 10W+8 and the decreasing expression of 4.6-kb mRNA in 10W−1L during the growth of the cells (Fig. 2B). These results further support the hypothesis that 100- and 87-kDa forms of Gab1 are translated from 5.2- and 4.6-kb mRNA, respectively.
FIG. 2.
Time course of protein and mRNA expression of two forms of Gab1 during cell growth. 10W+8 or 10W−1L cells (105) were plated on 150-mm-diameter dishes and cultured with 10% FBS for 5 days. The degrees of cell confluence, 20 to 25% on day 1 to 100% on day 5, are indicated (A). On each day, cells were simultaneously harvested both for protein analysis (A) and RNA analysis (B). To examine the Gab1 protein expression, cell lysates containing 1 mg of protein from 10W+8 and 10W−1L cells were subjected to immunoprecipitation (IP) of Gab1 and analyzed by anti-Gab1 blotting (A). The 100-kDa form of Gab1 and the 87-kDa form of Gab1 were detected in 10W+8 and 10W−1L, respectively, with different time courses during cell growth. For Northern analyses, 20 μg of total RNA from 10W+8 cells and 10 μg from 10W−1L were examined for Gab1 mRNA expression and visualized with ethidium bromide staining (B, lower panel).
The 87-kDa form of Gab1 shares pivotal functions in EGFR signaling with 100-kDa Gab1.
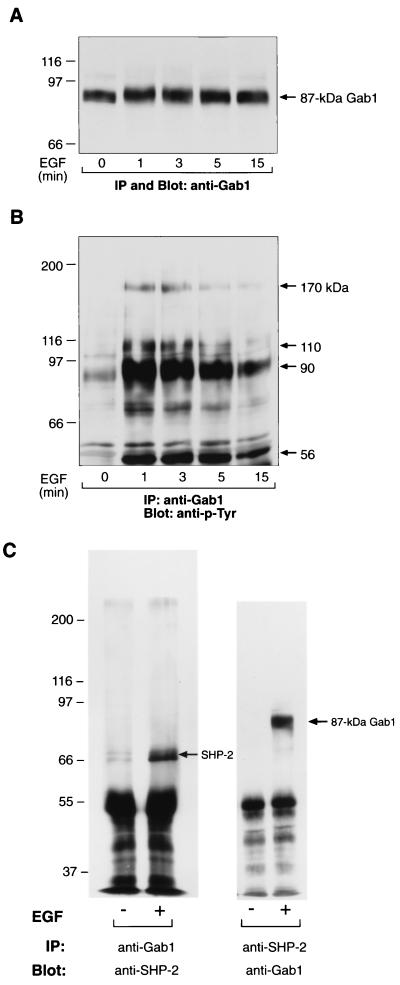
Next, we examined whether the 87-kDa Gab1 is a functional protein involved in EGFR signaling as is the 100-kDa Gab1. We investigated the phosphorylation of 87-kDa Gab1 and its association with other proteins upon EGF stimulation. The incubation of 10W−1L cells with 100 ng of EGF per ml for 1 min resulted in the mobility shift of 87-kDa Gab1 to 90 kDa, which was sustained over 15 min (Fig. 3A). Anti-p-Tyr blot analysis of Gab1 immunoprecipitants revealed that 87-kDa Gab1 was tyrosine phosphorylated upon EGF stimulation and became associated with EGFR (170 kDa) and other tyrosine-phosphorylated proteins (Fig. 3B). In addition, the 87-kDa Gab1 showed an EGF-dependent association with SHP-2 (Fig. 3C). These results demonstrated that 87-kDa Gab1 retains some shared functions with 100-kDa Gab1 in EGFR signaling.
FIG. 3.
The 87-kDa form of Gab1 is a functional protein involved in EGFR signaling in 10W−1 cells. (A and B) 10W−1L cells were serum starved for 20 h and left unstimulated or stimulated with 100 ng of EGF/ml for 1, 3, 5, and 15 min. Cells were lysed and subjected to immunoprecipitation (IP) of Gab1, followed by SDS–8% PAGE and anti-Gab1 (A) or anti-p-Tyr (B) blotting. (C) Gab1 (left) or SHP-2 (right) was immunoprecipitated (IP) from the lysates of 10W−1L cells left unstimulated or stimulated by 100 ng of EGF/ml for 1 min and analyzed by anti-SHP-2 (left) and anti-Gab1 (right) blottings, respectively.
Growth factor-dependent growth of 10W−1L in soft agar.
Previous works in our laboratory showed that 10W−1 clones exhibited significant growth in soft agar only when stimulated with growth factors like EGF, insulin, and platelet-derived growth factor (2, 26). On the other hand, 10W+8 clones did not grow in soft agar even with the combination treatment of EGF, insulin, and platelet-derived growth factor. Therefore, we examined the possible relationship between the expression of 87-kDa Gab1 variant and the growth factor-dependent colony formation in soft agar. 10W+8 cells did not show any colony formation with 10% FBS alone and exhibited a limited growth in soft agar (3.9% colony-forming efficiency [CFE] in the presence of both EGF [50 ng/ml] and insulin [1 μg/ml] (Table 1). 10W−1E showed a slightly increased CFE of 7.4% in the presence of both EGF and insulin. Interestingly, 10W−1L cells were found to grow in the presence of either EGF or insulin, with EGF (CFE, 13.6%) proving more potent than insulin (CFE, 7.7%). A combined stimulation of 10W−1L with EGF and insulin showed a synergistic increase of CFE up to 26.4%. Moreover, 10W−1L, but not 10W+8 or 10W−1E, formed colonies large enough to be observed without microscopic magnification after staining with 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyltetrazolium chloride (data not shown). A benzo[a]pyrene-transformed hamster fibrosarcoma cell line, BP6T, was cultured as a positive control and grew well in soft agar in the absence of additional growth factors. Thus, 10W−1 cells showed a remarkable increase in growth factor-dependent colony formation in soft agar with the development of 87-kDa Gab1 expression.
TABLE 1.
Colony-forming efficiency of carcinogen-transformed SHE cells in soft agar and effects of additional growth factor stimulation on it
| Cell | Growth factor(s) added to 10% serum | Colony-forming efficiency (%) |
|---|---|---|
| 10W+8 | None | 0.0 |
| EGFa + Insulinb | 3.9 | |
| 10W−1E | None | 0.0 |
| EGF | 1.3 | |
| Insulin | 0.0 | |
| EGF + Insulin | 7.4 | |
| 10W−1L | None | 0.2 |
| EGF | 13.6 | |
| Insulin | 7.7 | |
| EGF + Insulin | 26.4 | |
| BP6T | None | 63.4 |
EGF concentration, 50 ng/ml.
Insulin concentration, 1 mg/ml.
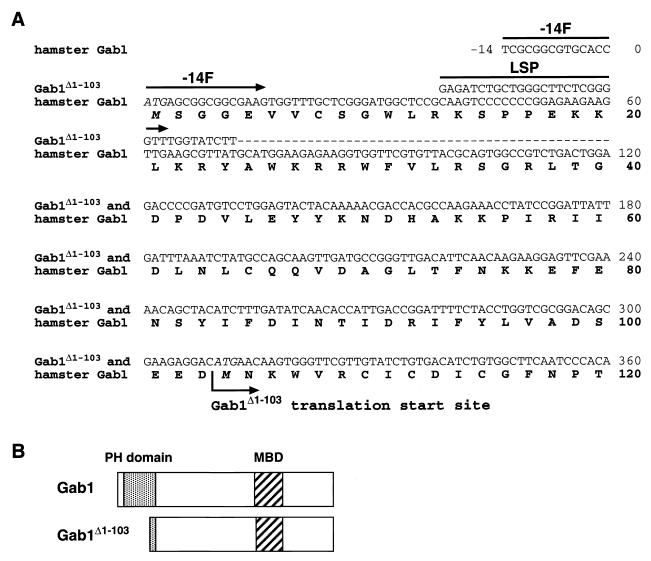
Cloning of cDNA encoding the 87-kDa Gab1 revealed that it lacks the amino-terminal 103-amino-acid sequence corresponding to most of the PH domain.
In order to further characterize the 87-kDa Gab1, we attempted to determine its cDNA. We used two different antisense primers (486R and 2104R) in 5′-RACE of cDNA from 10W−1L, and both PCRs gave us two products with an ≈0.4-kb difference in their sizes, the shorter products being much more predominant (data not shown). Sequence analysis showed that the longer product is the same as the cDNA obtained from nontransformed SHE 83-9 cells. This cDNA encodes hamster Gab1 composed of 694 amino acids (GenBank accession number AF307847) with 92% homology to human (GenBank accession number U43885) and mouse (GenBank accession number AJ250669) Gab1 (18, 24, 43). On the other hand, the shorter product lacked the 5′-untranslated region of ≈0.4 kb found in the longer product. When we compared the nucleotide sequence of the shorter product with the hamster Gab1 sequence, the first 72-nucleotide sequence of coding region was missing in the shorter form and it was replaced by an uncharacterized nucleotide sequence, 5′-GAGATCTGCTGGGCTTCTCGGGGTTTGGTATCTT-3′ (sense), followed by a sequence identical to the original Gab1 sequence to the 3′ end (Fig. 4A). Protein translation analysis revealed that the shorter form should be translated from 310ATG, which encodes the second methionine of the original 100-kDa hamster Gab1 (104M), thus Gab1Δ1-103. Gab1Δ1-103 lacks most of the PH domain (amino acids 14 to 116) and retains the Met-binding domain (MBD) and binding sites for PI3K and SHP-2, all of which are clustered in the carboxy-terminal portion of Gab1 (Fig. 4B). These results are consistent with the EGF-dependent association of 87-kDa Gab1 with EGFR (through MBD) and SHP-2 (40).
FIG. 4.
Comparison of two cDNA sequences and deduced protein sequences cloned from 10W−1L cells. (A) The nucleotide sequence of cDNA encoding Gab1 was cloned by 5′- and 3′-RACE strategy. Although hamster Gab1 is composed of 694 amino acids (2,085 nucleotide bases for the coding region), only the amino-terminal sequence of 120 amino acids (up to 360A in the nucleotide sequence) is shown here. Two cDNAs were cloned from 10W−1L, one of which (the minor product) was identical with cDNA from nontransformed SHE 83-9. A cDNA product specific to 10W−1L lacks the upstream sequence to 73G of the coding region of the original hamster Gab1 cDNA, and that sequence has been replaced by an uncharacterized sequence of at least 34 nucleotide bases, 5′-GAGATCTGCTGGGCTTCTCGGGGTTTGGTATCTT-3′ (sense), followed by the common sequence to the 3′ end. Therefore, translation of the product specific to 10W−1 starts from 310ATG (M104), instead of 1ATG (M1). A 5′ primer named −14F (5′-TCGCGGCGTGCACCATGAGCGGCGGCGAAG-3′; sense) is specific to cDNA encoding Gab1, while another 5′ primer named LSP (5′-GAGATCTGCTGGGCTTCTCGGGGTT-3′; sense) is specific to cDNA encoding Gab1Δ1-103. Dashes, nucleotide bases of Gab1Δ1-103 identical with those of hamster Gab1. (B) Comparison of the protein structures of Gab1 and Gab1Δ1-103. Gab1Δ1-103 lacks most of the amino-terminal PH domain (amino acids 14 to 116) observed in Gab1 and shares downstream structure including the MBD with Gab1.
Exclusive expression of mRNA encoding Gab1Δ1-103 in 10W−1 cells.
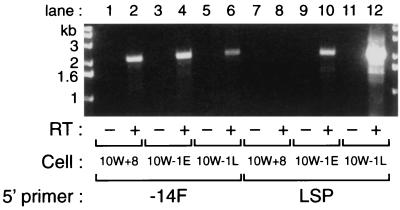
To confirm that the mRNA encoding Gab1Δ1-103 is exclusively expressed in 10W−1 cells, we performed RT-PCR with two sense primers: 5′-TCGCGGCGTGCACCATG AGCGGCGGCGAAG-3′ (−14F), which is specific to the original Gab1 cDNA, and 5′-GAGATCTGCTGGGCTTCTCGGGGTT-3′ (LSP), which is specific to the novel sequence observed in Gab1Δ1-103 cDNA (Fig. 4A). As an antisense primer, 2104R, corresponding to the terminal portion of the coding region, was used. A single product of 2.1 kb arose from PCR amplifications using −14F sense primer with all cells examined (Fig. 5, lanes 2, 4, and 6). However, it is noteworthy that the intensity of the PCR product with 10W−1L (lane 6) was significantly decreased compared to those with 10W+8 (lane 2) or 10W−1E (lane 4) cells. In contrast, when we performed PCR with LSP sense primer, 10W+8 cells gave us no products (lane 8) and 10W−1E showed a small amount of 2.1-kb product (lane 10), which dramatically increased with 10W−1L cells (lane 12). These results demonstrate that cDNA encoding Gab1Δ1-103 corresponds to the 4.6-kb mRNA observed in Fig. 1B and 2B while cDNA encoding full-length Gab1 containing the −14F primer sequence corresponds to the 5.2-kb mRNA. In addition, the expression of the 4.6-kb mRNA seems to develop at the stage of the loss of tumor suppressor gene function (supB−) in the neoplastic progression of SHE cells.
FIG. 5.
Exclusive expression of Gab1 mRNA led by LSP sequence in 10W−1, especially in 10W−1L cells. One microgram of RNA from 10W+8, 10W−1E, and 10W−1L cells was subjected to reverse transcription reaction using oligo(dT)18 primer. For some samples, reverse transcriptase (RT) was not included in the reaction (lanes 1, 3, 5, 7, 9, and 11). cDNA, in turn, was provided for PCR with either −14F or LSP as 5′ primer and 2104R as 3′ primer. PCR products were analyzed by 1.2% agarose gel and visualized with ethidium bromide staining and UV exposure.
Different subcellular localization between Gab1 and Gab1Δ1-103.
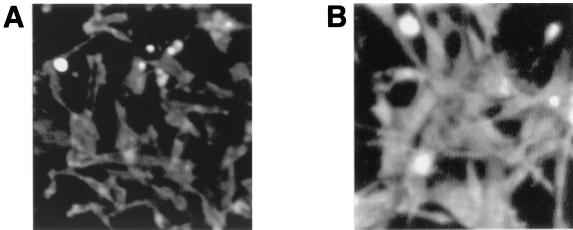
The results reported by Maroun et al. (32, 33) prompted us to examine the subcellular localization of Gab1 in 10W+8 cells and that of Gab1·Δ1-103 in 10W−1L cells, respectively. As shown in Fig. 6, Gab1 is preferably localized at cell membranes, while Gab1Δ1-103 is diffusely expressed in the cytoplasm. These results were similar to those with human wild-type Gab1 and Gab1 lacking the PH domain (32, 33).
FIG. 6.
Subcellular localization of Gab1 in 10W+8 cells (A) and Gab1Δ1-103 in 10W−1L cells (B). Cells were cultured in 10% FBS-containing medium for 3 days and subjected to the analysis of the cellular expression of Gab1/Gab1Δ1-103 using indirect immunofluorescence.
Overexpression of Gab1 and Gab1Δ1-103 in 10W+8 resulted in the enhanced mitogenic response to EGF stimulation.
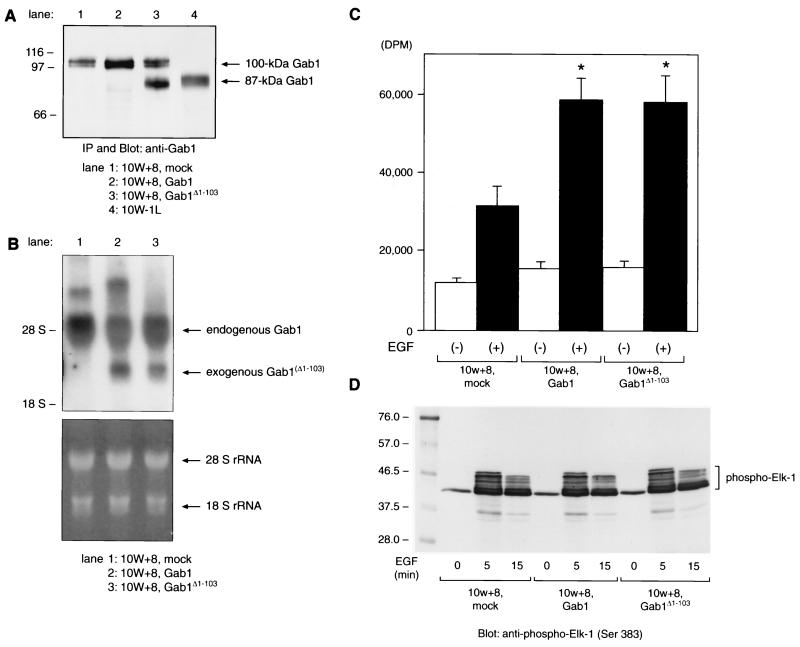
We cloned cDNAs encoding hamster Gab1 and Gab1Δ1-103 into a CMV promoter-containing mammalian expression vector pCR 3.1 and constructed 10W+8 cells overexpressing either Gab1 or Gab1Δ1-103 as well as mock-transfected cells. 10W+8 cell pools stably expressing exogenous Gab1 or Gab1Δ1-103 were obtained (Fig. 7A and B). 10W+8 cells with exogenous expression of 100-kDa Gab1 (lane 2) as well as its endogenous expression showed approximately twofold overall expression of 100-kDa Gab1 compared to mock-transfected (lane 1) 10W+8 (Fig. 7A). Similarly, the amount of exogenous expression of Gab1Δ1-103 was comparable to that of endogenous 100-kDa Gab1 expression (lane 3). The exogenously expressed Gab1Δ1-103 showed a migration on SDS-PAGE identical to that of the 87-kDa Gab1 from 10W−1L, which appears to confirm that the 87-kDa Gab1 is Gab1Δ1-103. Northern analysis (Fig. 7B) enabled us to distinguish the exogenous expression of Gab1 or Gab1Δ1-103 mRNA (2.7 kb) from endogenous Gab1 mRNA (5.2 kb).
FIG. 7.
Exogenous expression of Gab1 or Gab1Δ1-103 in 10W+8 cells enhances the mitogenic response to EGF stimulation without an upregulated p44/42 MAPK activation. (A) Immunoprecipitation (IP) and Western analysis of Gab1- or Gab1Δ1-103-overexpressing 10W+8. Gab1Δ1-103 expressed in 10W+8 (lane 3) comigrated with 87-kDa Gab1 from 10W−1L (lane 4). (B) Northern analysis of 10W+8 cells exogenously expressing Gab1 or Gab1Δ1-103. Exogenously expressed mRNA of Gab1 (lane 2, 2.7 kb) or Gab1Δ1-103 (lane 3, 2.7 kb) lacking most of the 5′- and 3′-untranslated region was distinct from endogenous Gab1 mRNA (5.2 kb). The lower panel shows equal RNA loading onto each lane and the integrity of RNA samples by visualization of 28S/18S rRNA with ethidium bromide staining and UV exposure. (C) [3H]thymidine incorporation assay was performed using mock-transfected 10W+8 and Gab1/Gab1Δ1-103-overexpressing 10W+8 cells. Cells were serum starved for 20 h and incubated with or without 50 ng of EGF/ml for a further 24 h in the presence of 1 μCi of [3H]thymidine/well. Both Gab1 and Gab1Δ1-103 overexpression significantly potentiated the mitogenic response to EGF stimulation. ∗, P < 0.05 versus mock-transfected 10W+8. (D) EGF-dependent activation of p44/42 (ERK1/2) MAPK in 10W+8 cells was not significantly modulated by Gab1 or Gab1Δ1-103 overexpression. Cells were serum starved for 20 h and left unstimulated or stimulated with 100 ng of EGF/ml for 5 and 15 min. Cell lysates containing 200 μg of protein were incubated with anti-phospho-MAPK antibody. Immunoprecipitated phospho-MAPK was then reacted in vitro with Elk-1 as a substrate. MAPK-phosphorylated Elk-1 was analyzed by SDS–10% PAGE and anti-phospho-Elk-1 blotting. Some of the Ser381-phosphorylated Elk-1 showed various degrees of delayed migration on SDS-PAGE.
Next we examined the effect of exogenous expression of Gab1 or Gab1Δ1-103 on EGF-dependent mitogenic activity of 10W+8 cells assessed by the [3H]thymidine incorporation assay. As shown in Fig. 7C, exogenous expression of Gab1Δ1-103, as well as Gab1, significantly enhanced the mitogenic response to EGF stimulation, up to twofold compared to that in mock-transfected 10W+8. To elucidate the molecular mechanism of the increased mitogenic response to EGF in Gab1- or Gab1Δ1-103-overexpressing cells, we compared the activation levels of MAPK, p44/ERK1, and p42/ERK2 among these cells by a MAPK activity assay using Elk-1 as a substrate (Fig. 7D). Mock-transfected 10W+8 cells showed EGF-dependent MAPK activation assessed by Elk-1 phosphorylation, which peaked at 5 min after EGF stimulation and gradually decayed. Unexpectedly, overexpression of either Gab1 or Gab1Δ1-103 did not enhance the EGF-dependent MAPK activation measured by Elk-1 phosphorylation at any time point examined including 60 min after EGF stimulation (data not shown). These results indicate the involvement of other signaling pathways in the enhanced mitogenic response to EGF by exogenous expression of Gab1 or Gab1Δ1-103 in 10W+8 cells.
Exogenous Gab1Δ1-103, but not Gab1, expression potentiated growth of 10W+8 cells in soft agar in response to EGF stimulation.
Finally we performed a soft agar assay using 10W+8 cells exogenously expressing Gab1 or Gab1Δ1-103 to determine whether the enhanced CFE of growth factor-stimulated 10W−1L cells (Table 1) can be attributed to the development of 87-kDa Gab1/Gab1Δ1-103 expression. Mock-transfected 10W+8 showed 7.8% of CFE when stimulated with 50 ng of EGF per ml, which was higher than that of parental 10W+8 (Table 2). This increase might be related to the transfection procedure including Lipofectamine treatment and many passages during antibiotic selection of stably expressing cells. Therefore, cells with identical passage numbers were used in this comparative study. Additional expression of Gab1 did not enhance the CFE of 10W+8 but rather slightly decreased it. In contrast, exogenous expression of Gab1Δ1-103 increased the CFE up to threefold (22.2%) the value observed with mock-transfected cells when stimulated with EGF. None of these cells showed significant growth in soft agar when cultured with 10% FBS alone. All of these results taken together indicate that 10W−1 cells have acquired the phenotype of growth factor-dependent, anchorage-independent growth, at least in part, by developing Gab1Δ1-103 expression.
TABLE 2.
Effects of exogenous expression of Gab1 and Gab1Δ1-103 on colony-forming efficiency of 10W+8 cells in soft agar
| Cell | EGF concn (ng/ml) added to 10% serum | Colony-forming efficiency (%) |
|---|---|---|
| 10W+8, mock | 0 | 0.0 |
| 50 | 7.8 | |
| 10W+8, Gab1 | 0 | 0.0 |
| 50 | 3.1 | |
| 10W+8, Gab1Δ1-103 | 0 | 0.0 |
| 50 | 22.2 |
DISCUSSION
In the present report, we demonstrated the expression of a novel 87-kDa Gab1 during the neoplastic progression of carcinogen-transformed SHE cells. The cloning and expression of the cDNA encoding 87-kDa Gab1 revealed that the protein lacks the amino-terminal 103 amino acids of Gab1, hence the designation Gab1Δ1-103. Gab1Δ1-103 lacks most of the amino-terminal PH domain of Gab1 (amino acids 14 to 116). Therefore, the differences between Gab1 and Gab1Δ1-103 are most likely to be attributed to the presence or absence of the functional PH domain.
The PH domains were first identified as regions that share homology with an internal repeat of pleckstrin, a major substrate of protein kinase C in platelets (31, 39). These domains are composed of ≈100 amino acids with seven β strands, which form two antiparallel β sheets, with a carboxy-terminal α helix. PH domains are believed to mediate intermolecular interactions, primarily protein-phospholipid interactions. The Gab1 PH domain preferentially binds to phosphatidylinositol 3,4,5-triphosphate, which is a product of phosphatidylinositol 3-kinase (20, 33, 40). While the present work was in progress, several reports that examined the roles of the Gab1 PH domain using artificial deletion mutants of the whole PH domain or site-directed mutants of Trp26 and Arg29 residues were published (32, 33, 40). These reports showed several observations similar to ours, as follows: (i) Gab1 lacking the functional PH domain was phosphorylated upon hepatocyte growth factor or EGF stimulation, although its phosphorylation seemed to be decreased and/or altered compared to that of Gab1 (32, 33, 40); and (ii) the Gab1 PH domain is not required for its ability to associate with the Met receptor and SHP-2 following activation of the Met receptor (32). Likewise, 87-kDa Gab1/Gab1Δ1-103 showed an EGF-dependent phosphorylation determined by a modest mobility shift on SDS-PAGE (Fig. 3A) and anti-p-Tyr blotting (Fig. 3B), as well as the EGF-dependent association with EGFR (Fig. 3B) and SHP-2 (Fig. 3C). Other characteristic features of Gab1 lacking a functional PH domain reported so far include the following: (i) it failed to play a role in Met-dependent branching tubulogenesis (32, 33); (ii) the functional Gab1 PH domain is required for the localization of Gab1 to sites of cell-cell contact (32, 33); and (iii) PH domain-deleted Gab1 did not activate Jun kinase (JNK) by EGF stimulation (40). Furthermore, the Gab1 PH domain was able to act as a dominant-negative mutant of Gab1 and inhibited the EGF-stimulated increase in JNK activity (40).
The analysis of genomic DNA sequence encoding hamster Gab1 identified an exon-intron junction between 72T and 73G (H. Kameda and T. E. Eling, unpublished data). Therefore, mRNA encoding Gab1Δ1-103 is likely to be an alternatively spliced form. Moreover, the opposite, complementary regulations of Gab1 and Gab1Δ1-103 expression (for example, serum starvation and cell confluence increase the expression of 5.2-kb mRNA and decrease the 4.6-kb mRNA expression) suggest that the transcription of these two mRNAs may be driven by distinct promoter regions. In addition, such a complementary expression pattern between two forms of Gab1 indicates their distinct roles in cell proliferation and cell-cell interactions. In this context, we have analyzed the subcellular localization of wild-type Gab1 in 10W+8 cells and Gab1Δ1-103 in 10W−1L cells. We found that wild-type Gab1 was preferably localized at cell membranes, especially at the site of cell-cell contact, while Gab1Δ1-103 was predominantly expressed in the cytoplasm (Fig. 6). These observations were consistent with recent reports (32, 33) and may explain why the expression of 100-kDa Gab1 increases after confluence, while Gab1Δ1-103 expression decreases.
There may exist a mutational hot spot in genomic DNA encoding Gab1 because Gab1 expression has been lost in an asbestos-transformed tumorigenic SHE cell clone 10W2T (24). Therefore, at least two distinct alterations in Gab1 expression have been demonstrated during the neoplastic progression of SHE cells. And when we consider that most proto-oncogenes and tumor suppressor genes are involved in the regulation of cellular growth (1, 9), the roles of the altered expression of Gab1 in the signaling pathways required for the neoplastic progression of SHE cells seem to be much more complicated.
The expression of mRNA encoding Gab1Δ1-103 was always clearly observed in 10W−1E cells by RT-PCR (Fig. 5), although Northern analysis (Fig. 1B) and immunoprecipitation and Western analysis (Fig. 1A) seem to be less sensitive than RT-PCR. In addition, the expression amount of Gab1Δ1-103 dramatically increased during passages. Therefore, genetic changes leading to the development of Gab1Δ1-103 are likely to follow, or at least occur along with, the loss of tumor suppressor gene(s).
The reason why the overexpression of Gab1Δ1-103, but not of the full-length Gab1, resulted in the upregulated soft agar colony formation of 10W+8 cells in response to EGF stimulation remains to be clarified. Gab1 is implicated in the transforming signaling through Tpr-Met, the oncogenic version of the Met receptor in which the 5′ sequence of Tpr derived from chromosome 1 forms a hybrid with the 3′ sequence of Met located on chromosome 7 (3, 12). Moreover, two Gab1-overexpressing (8- and 13-fold over control) NIH 3T3 cell clones were shown to exhibit significant colony formation in soft agar in the presence of additional growth factors, with EGF being more potent than insulin (18). To avoid possible clonal deviations (cellular factors other than Gab1 overexpression) contributing to the cell growth in soft agar, we used transfected-cell pools instead of single-cell clones. In these experiments, we did not observe any increase in the anchorage-independent cell growth of 10W+8 cells by Gab1 overexpression. In contrast, we did observe an enhanced colony formation in soft agar by overexpression of Gab1Δ1-103. The expression amounts of the exogenous Gab1 and Gab1Δ1-103 were comparable and did not explain the exclusive effect of Gab1Δ1-103 (Fig. 7A and B). And we have confirmed the expression of Gab1 in 10W+8 cells and Gab1Δ1-103 in 10W−1L cells cultured under anchorage-independent (suspension) conditions (data not shown). The exogenous expression of Gab1 and Gab1Δ1-103 is likely to be regulated similarly by the CMV promoter-driven expression system. Therefore, we could not explain the specific effect of Gab1Δ1-103 expression on EGF-dependent, anchorage-independent cell growth by the overall amount of Gab1/Gab1Δ1-103 expression in SHE cells. Moreover, the transforming ability of Gab1Δ1-103 was found to be closely related to Gab1Δ1-103 expression when we examined several single-cell clones (data not shown). To address a dominant effect of the Gab1Δ1-103 expression on EGF-dependent colony formation in soft agar over the wild-type Gab1 expression, we constructed 10W−1L cells exogenously expressing wild-type Gab1. The expression of wild-type Gab1 did not significantly inhibit the EGF-dependent colony formation of 10W−1L cells (10W−1L-mock showed 13.9% of CFE with EGF stimulation; 10W−1L-Gab1 showed 11.1% of CFE with EGF stimulation). These results were consistent with the fact that 10W−1 cells at around passage 23, which expressed both Gab1 and Gab1Δ1-103, showed a colony formation comparable to that of 10W−1L cells but not of 10W−1E cells (data not shown). It is noteworthy that both 10W−1L cells and exogenously Gab1Δ1-103-expressing 10W+8 cells require growth factor stimulation in addition to 10% serum for their colony formation in soft agar. Therefore, another step, such as the overexpression of growth factor receptor or autocrine production of growth factors, may be needed for 10W−1 cells before the acquisition of tumorigenicity. Indeed, 10W−1L cells did not form tumors when subcutaneously injected into nude mice (Kameda and Eling, unpublished).
The two Gab1-overexpressing NIH 3T3 clones investigated by Holgado-Madruga et al. showed an attenuated EGF-dependent MAPK (p42/ERK2) activation compared to that in control cells (18). However, several subsequent studies demonstrated the upregulated MAPK (p44/ERK1 and p42/ERK2) activation upon growth factor and cytokine stimulation, including EGF, by Gab1 overexpression (40, 42, 43). Moreover, Gab1-deficient mouse embryonic fibroblasts showed a reduced MAPK activation (22). In our study, although exogenous expression of either Gab1 or Gab1Δ1-103 resulted in an enhanced mitogenic activity upon EGF stimulation as assessed by [3H]thymidine incorporation, EGF-dependent MAPK activation was not modulated by the exogenous expression of Gab1/Gab1Δ1-103. Thus, a multiple signaling pathway from EGFR through Gab1 to nuclear events may exist, and factors other than p44/42 MAPKs may be involved in these pathways. We examined the involvement of JNK in our cells, but we could not detect any JNK activity.
In conclusion, we demonstrated the expression of a novel 87-kDa Gab1/Gab1Δ1-103 in carcinogen-transformed SHE cells at a specific stage of neoplastic progression. The expression of Gab1Δ1-103 showed a transforming ability measured by cell growth in soft agar upon stimulation by growth factors, including EGF. These observations suggest that Gab1 is a key element in EGF signal transduction and that its transforming potential may be activated by the loss of its amino-terminal PH domain.
ACKNOWLEDGMENTS
We thank John P. O'Bryan and Cynthia A. Afshari for reading of the manuscript and helpful comments and Julie Angerman-Stewart, Mark Geller, and Leigh Wilson for laboratory assistance.
REFERENCES
- 1.Aaronson S A. Growth factors and cancer. Science. 1991;254:1146–1153. doi: 10.1126/science.1659742. [DOI] [PubMed] [Google Scholar]
- 2.Afshari C A, Barrett J C. Negative regulation of mitogen-stimulated, anchorage-independent cell growth by a tumor-suppressor gene function. Mol Carcinog. 1993;7:249–256. doi: 10.1002/mc.2940070407. [DOI] [PubMed] [Google Scholar]
- 3.Bardelli A, Longati P, Gramaglia D, Stella M C, Comoglio P M. Gab1 coupling to the HGF/Met receptor multifunctional docking site requires binding of Grb2 and correlates with the transforming potential. Oncogene. 1997;15:3103–3111. doi: 10.1038/sj.onc.1201561. [DOI] [PubMed] [Google Scholar]
- 4.Barrett J C. Mechanisms of multistep carcinogenesis and carcinogen risk assessment. Environ Health Perspect. 1993;100:9–20. doi: 10.1289/ehp.931009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett J C. A preneoplastic stage in the spontaneous neoplastic transformation of Syrian hamster embryo cells in culture. Cancer Res. 1980;40:91–94. [PubMed] [Google Scholar]
- 6.Barrett J C, Crawford B D, Mixter L O, Schechtman L M, Ts'o P O P, Pollack R. Correlation of in vitro growth properties and tumorigenicity of Syrian hamster cell lines. Cancer Res. 1979;39:1504–1510. [PubMed] [Google Scholar]
- 7.Barrett J C, Ts'o P O P. Evidence for the progressive nature of neoplastic transformation in vitro. Proc Natl Acad Sci USA. 1978;75:3761–3765. doi: 10.1073/pnas.75.8.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett J C, Wong A, McLachlan J A. Diethylstilbestrol induces neoplastic transformation without measurable gene mutation at two loci. Science. 1981;212:1402–1404. doi: 10.1126/science.6262919. [DOI] [PubMed] [Google Scholar]
- 9.Boyd J A, Barrett J C. Genetic and cellular basis of multistep carcinogenesis. Pharmacol Ther. 1990;46:469–486. doi: 10.1016/0163-7258(90)90028-z. [DOI] [PubMed] [Google Scholar]
- 10.Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B. p62dok: a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 11.Deb T B, Wong L, Salomon D S, Zhou G, Dixon J E, Gutkind J S, Thompson S A, Johnson G R. A common requirement for the catalytic activity and both SH2 domains of SHP-2 in mitogen-activated protein (MAP) kinase activation by the ErbB family of receptors. A specific role for SHP-2 in MAP, but not c-Jun amino-terminal kinase activation. J Biol Chem. 1998;273:16643–16646. doi: 10.1074/jbc.273.27.16643. [DOI] [PubMed] [Google Scholar]
- 12.Fixman E D, Holgado-Madruga M, Nguyen L, Kamikura D M, Fournier T M, Wong A J, Park M. Efficient cellular transformation by the Met oncoprotein requires a functional Grb2 binding site and correlates with phosphorylation of the Grb2-associated proteins, Cbl and Gab1. J Biol Chem. 1997;272:20167–20172. doi: 10.1074/jbc.272.32.20167. [DOI] [PubMed] [Google Scholar]
- 13.Glasgow W C, Afshari C A, Barrett J C, Eling T E. Modulation of the epidermal growth factor mitogenic response by metabolites of linoleic and arachidonic acid in Syrian hamster embryo fibroblasts. Differential effects in tumor suppressor gene (+) and (−) phenotypes. J Biol Chem. 1992;267:10771–10779. [PubMed] [Google Scholar]
- 14.Glasgow W C, Hui R, Everhart A L, Jayawickreme S P, Angerman-Stewart J, Han B-B, Eling T E. The linoleic acid metabolite, (13S)-hydroperoxyoctadecadienooic acid, augments the epidermal growth factor receptor signaling pathway by attenuation of receptor dephosphorylation. Differential response in Syrian hamster embryo tumor suppressor phenotypes. J Biol Chem. 1997;272:19269–19276. doi: 10.1074/jbc.272.31.19269. [DOI] [PubMed] [Google Scholar]
- 15.Gu H, Pratt J C, Burakoff S J, Neel B G. Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol Cell. 1998;2:729–740. doi: 10.1016/s1097-2765(00)80288-9. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick L. The cell biology of human aging. N Engl J Med. 1976;295:1302–1308. doi: 10.1056/NEJM197612022952308. [DOI] [PubMed] [Google Scholar]
- 17.Herbst R, Carroll P M, Allard J D, Schilling J, Raabe T, Simon M A. Daughter of sevenless is a substrate of the phosphotyrosine phosphatase Corkscrew and functions during sevenless signaling. Cell. 1996;85:899–909. doi: 10.1016/s0092-8674(00)81273-8. [DOI] [PubMed] [Google Scholar]
- 18.Holgado-Madruga M, Emlet D R, Moscatello D K, Godwin A K, Wong A J. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature (London) 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 19.Hui R, Kameda H, Risinger J I, Angerman-Stewart J, Han B, Barrett J C, Eling T E, Glasgow W C. The linoleic acid metabolite, 13-HpODE augments the phosphorylation of EGF receptor and SHP-2 leading to their increased association. Prostaglandins Leukot Essent Fatty Acids. 1999;61:137–143. doi: 10.1054/plef.1999.0083. [DOI] [PubMed] [Google Scholar]
- 20.Isakoff S J, Cardozo T, Andreev J, Li Z, Ferguson K M, Abagyan R, Lemmon M A, Aronheim A, Skolnik E Y. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isfort R J, LeBoeuf R A. The Syrian hamster embryo (SHE) cell transformation system: a biologically relevant in vitro model—with carcinogen predicting capabilities—of in vivo multistage neoplastic transformation. Crit Rev Oncogene. 1995;6:251–260. doi: 10.1615/critrevoncog.v6.i3-6.30. [DOI] [PubMed] [Google Scholar]
- 22.Itoh M, Yoshida Y, Nishida K, Narimatsu M, Hibi M, Hirano T. Role of Gab1 in heart, placenta, and skin development and growth factor- and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol Cell Biol. 2000;20:3695–3704. doi: 10.1128/mcb.20.10.3695-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones N, Dumont D J. The Tek/Tie2 receptor signals through a novel Dok-related docking protein, Dok-R. Oncogene. 1998;17:1097–1108. doi: 10.1038/sj.onc.1202115. [DOI] [PubMed] [Google Scholar]
- 24.Kameda H, Risinger J I, Han B-B, Baek S J, Barrett J C, Glasgow W C, Eling T E. Identification of epidermal growth factor receptor-Grb2-associated binder-1-SHP-2 complex formation and its functional loss during neoplastic cell progression. Cell Growth Diff. 2001;12:307–318. [PubMed] [Google Scholar]
- 25.Kameda H, Morita I, Handa M, Kaburaki J, Yoshida T, Mimori T, Murota S-I, Ikeda Y. Reexpression of functional P-selectin molecules on the endothelial cell surface by repeated stimulation with thrombin. Br J Haematol. 1997;97:348–355. doi: 10.1046/j.1365-2141.1997.522700.x. [DOI] [PubMed] [Google Scholar]
- 26.Koi M, Afshari C A, Annab L A, Barrett J C. Role of a tumor-suppressor gene in the negative control of anchorage-independent growth of Syrian hamster cells. Proc Natl Acad Sci USA. 1989;86:8773–8777. doi: 10.1073/pnas.86.22.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koi M, Barrett J C. Loss of tumor-suppressive function during chemically induced neoplastic progression of Syrian hamster embryo cells. Proc Natl Acad Sci USA. 1986;83:5992–5996. doi: 10.1073/pnas.83.16.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 29.Laffargue M, Raynal P, Yart A, Peres C, Wetzker R, Roche S, Payrastre B, Chap H. An epidermal growth factor receptor/Gab1 signaling pathway is required for activation of phosphoinositide 3-kinase by lysophosphatidic acid. J Biol Chem. 1999;274:32835–32841. doi: 10.1074/jbc.274.46.32835. [DOI] [PubMed] [Google Scholar]
- 30.Lehr S, Kotzka J, Herkner A, Klein E, Siethoff C, Knebel B, Noelle V, Brüning J C, Klein H W, Meyer H E, Krone W, Müller-Wieland D. Identification of tyrosine phosphorylation sites in human Gab-1 protein by EGF receptor kinase in vitro. Biochemistry. 1999;38:151–159. doi: 10.1021/bi9818265. [DOI] [PubMed] [Google Scholar]
- 31.Lemmon M A, Ferguson K M, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 32.Maroun C R, Holgado-Madruga M, Royal I, Naujokas M A, Fournier T M, Wong A J, Park M. The Gab1 PH domain is required for localization of Gab1 at sites of cell-cell contact and epithelial morphogenesis downstream from the Met receptor tyrosine kinase. Mol Cell Biol. 1999;19:1784–1799. doi: 10.1128/mcb.19.3.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maroun C R, Moscatello D K, Naujokas M A, Holgado-Madruga M, Wong A J, Park M. A conserved inositol phospholipid binding site within the pleckstrin homology domain of the Gab1 docking protein is required for epithelial morphogenesis. J Biol Chem. 1999;274:31719–31726. doi: 10.1074/jbc.274.44.31719. [DOI] [PubMed] [Google Scholar]
- 34.Newbold R F, Overell R W, Connell J R. Induction of immortality is an early event in malignant transformation of mammalian cells by carcinogens. Nature (London) 1982;299:633–635. doi: 10.1038/299633a0. [DOI] [PubMed] [Google Scholar]
- 35.Nishida K, Yoshida Y, Itoh M, Fukada T, Ohtani T, Shirogane T, Atsumi T, Takahashi-Tezuka M, Ishihara K, Hibi M, Hirano T. Gab-family adapter proteins act downstream of cytokine and growth factor receptors and T- and B-cell antigen receptors. Blood. 1999;93:1809–1816. [PubMed] [Google Scholar]
- 36.Oshimura M, Hesterberg T W, Tsutsui T, Barrett J C. Correlation of asbestos-induced cytogenetic effects with cell transformation of Syrian hamster embryo cells in culture. Cancer Res. 1984;44:5017–5022. [PubMed] [Google Scholar]
- 37.Qu C-K, Yu W-M, Azzarelli B, Feng G-S. Genetic evidence that Shp-2 tyrosine phosphatase is a signal enhancer of the epidermal growth factor receptor in mammals. Proc Natl Acad Sci USA. 1999;96:8528–8533. doi: 10.1073/pnas.96.15.8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raabe T, Riesgo-Escovar J, Liu X, Bausenwein B S, Deak P, Maröy P, Hafen E. DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between sevenless and Ras1 in Drosophila. Cell. 1996;85:911–920. doi: 10.1016/s0092-8674(00)81274-x. [DOI] [PubMed] [Google Scholar]
- 39.Rebecchi M J, Scarlata S. Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues G A, Falasca M, Zhang Z, Ong S H, Schlessinger J. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol Cell Biol. 2000;20:1448–1459. doi: 10.1128/mcb.20.4.1448-1459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Z-Q, Yu D-H, Park M, Marshall M, Feng G-S. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol Cell Biol. 2000;20:1526–1536. doi: 10.1128/mcb.20.5.1526-1536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi-Tezuka M, Yoshida Y, Fukada T, Ohtani T, Yamanaka Y, Nishida K, Nakajima K, Hibi M, Hirano T. Gab1 acts as an adapter molecule linking the cytokine receptor gp130 to ERK mitogen-activated protein kinase. Mol Cell Biol. 1998;18:4109–4117. doi: 10.1128/mcb.18.7.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weidner K M, Di Cesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature (London) 1996;384:173–176. doi: 10.1038/384173a0. [DOI] [PubMed] [Google Scholar]
- 44.Yamanashi Y, Baltimore D. Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 45.Yenush L, White M F. The IRS-signalling system during insulin and cytokine action. Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- 46.Zhao C, Yu D-H, Shen R, Feng G-S. Gab2, a new pleckstrin homology domain-containing adapter protein, acts to uncouple signaling from ERK kinase to Elk-1. J Biol Chem. 1999;274:19649–19654. doi: 10.1074/jbc.274.28.19649. [DOI] [PubMed] [Google Scholar]