Abstract
This study investigates the fibril nanostructure of fresh celery samples by modeling the anisotropic behavior of the transverse relaxation time (T2) in nuclear magnetic resonance (NMR). Experimental results are interpreted within the framework of a previously developed theory, which was successfully used to model the nanostructures of several biological tissues as a set of water filled nanocavities, hence explaining the anisotropy the T2 relaxation time in vivo. An important feature of this theory is to determine the degree of orientational ordering of the nanocavities, their characteristic volume, and their average direction with respect to the macroscopic sample. Results exhibit good agreement between theory and experimental data, which are, moreover, supported by optical microscopic resolution. The quantitative NMR approach presented herein can be potentially used to determine the internal ordering of biological tissues noninvasively.
Keywords: Relaxation anisotropy, Dipole–dipole interactions, Transverse relaxation time, T2, Nanocavity, Ordering degree, Nanostructure, NMR, Quantitative MRI
1. Introduction
Deep understanding of the function of biological tissues requires mapping their internal structure. Nuclear magnetic resonance (NMR) spectroscopy is one of the efficient tools for mapping the structure in biological samples at the morphological, molecular, and atomic levels [1]. NMR has two main applications in biology and medicine: NMR spectroscopy for the determination of the chemical composition of macromolecules, and magnetic resonance imaging (MRI) for mapping anatomy, physiology, and metabolism functions [1-3]. These applications require development of specialized magnets, radio frequency (RF) pulses, and signal processing techniques.
It has long been known that the second moment of the NMR signal [4] and the transverse relaxation time (T2) [5] of liquids exhibit anisotropic properties when contained in fibril tissues. Namely, the values of these properties depend on the orientation of the tissue relative to the external magnetic field. The anisotropy of the T2 relaxation time was previously explored, via its dependency on the orientation of bound water molecules along hydrophilic tissue fibrils [4-9].
The phenomenon of relaxation anisotropy in collagenous tissues is usually attributed to non-averaged dipolar couplings of water bound to the collagen [4-13]. Alternative models for this phenomenon include the formation of strong water bridges [11] and bound water molecules along hydrophilic tissue fibrils [4-9,13]. These theories, however, did not consider the residual inter-molecular dipole–dipole interactions in water entrapped in the cavity itself.
Recently, a method based on modeling fibril’s internal structure as composed of water filled nanocavities has been successfully used to explain the anisotropy of the transverse and longitudinal relaxation times observed in biological tissues [14-18]. The developed model [14-18] is based on consideration of the averaged Hamiltonian for water entrapped in nanocavities and the bivariate normal distribution of the polar and azimuthal angles, which determine the fibril direction. As such, this model links the water relaxation time’ anisotropy, and the degree of orientational ordering of fibrils.
For an adequate description of a spin system bounded by a cavity, the Hamiltonian of the dipole–dipole interaction must be averaged over the coordinate space. Let us estimate the characteristic size l of a nanocavity that contain water. The Hamiltonian of spins in this water pool can be represented by its non-zero average shape due to the limited molecular motion in the nanocavity. The characteristic size lch of a cavity in which the dipolar Hamiltonian (3) does not average to zero is estimated from the condition: , where D ≈ 3 · 106 nm2/ms is the diffusion coefficient of water [19] and tmag ≈ 2.74 · 10−2 ms is the typical NMR time scale for protons which characterizes the flip-flop transition due to the dipole–dipole interaction [20]. Estimation gives that the characteristic size should be less than 810 nm.
The relaxation time anisotropy can thus be used as an indirect and sensitive probe of the three-dimensional (3D) tissue morphology at the microscopic and nanoscopic length-scales. Herein we further establish the correlation between the anisotropy of the transverse relaxation time (T2) and the orientation and degree of ordering of fibrils in plant samples. The manuscript is organized as follows: in Section 2, for convenience of the readers, we briefly show that the angular dependency of the transverse relaxation time results from residual dipole–dipole interaction in liquid entrapped in nanocavities. We then consider the transverse relaxation time for various sets of nanocavities with different average directions relative to the external magnetic field, and different angle distributions. In Section 3, we describe the tested samples and experimental procedures. In Section 4, the theoretical predictions and the experimental data are compared, and in the last section, we conclude that the transverse relaxation time is strongly correlated with the internal degree of nanofibril ordering in biological tissues.
2. Theory
In this section we recapitulate, for the convenience of the readers, the mechanisms of the transverse relaxation in a single nanocavity containing liquid (e.g., water), and then consider the influence of internal orientational ordering of the nanocavities on the transverse relaxation times.
2.1. Anisotropy of transverse relaxation time
In liquids entrapped in nanocavities transverse relaxation may be the result of several types of interactions with different relative weights. The main contribution, however, originates from dipole-dipole (DD) interaction of the nuclear spins [3-5,10,21-27]. Due to restricted motion of water molecules in nanocavities the inter-molecular DD interactions are not averaged completely to zero and the averaged Hamiltonian of the DD interaction takes the following form [28]:
| (1) |
Here, is the angular momentum operators of the j-th nuclear spin, Izj is the projection of the spin operator onto the z-axis where j = 1,2,…,N. Gd in Eq. (1) is the space-averaged coupling constant for a pair of the nuclear spins, given by.
| (2) |
where γ is the gyromagnetic ratio of nucleus, V is the volume of nanocavity, P2 is the second Legendre polynomial, i.e., the angle θn between the main axis Zn of the n-th nanocavity and the external magnetic field (see Fig. 1), and F is the form-factor [14-18,28].
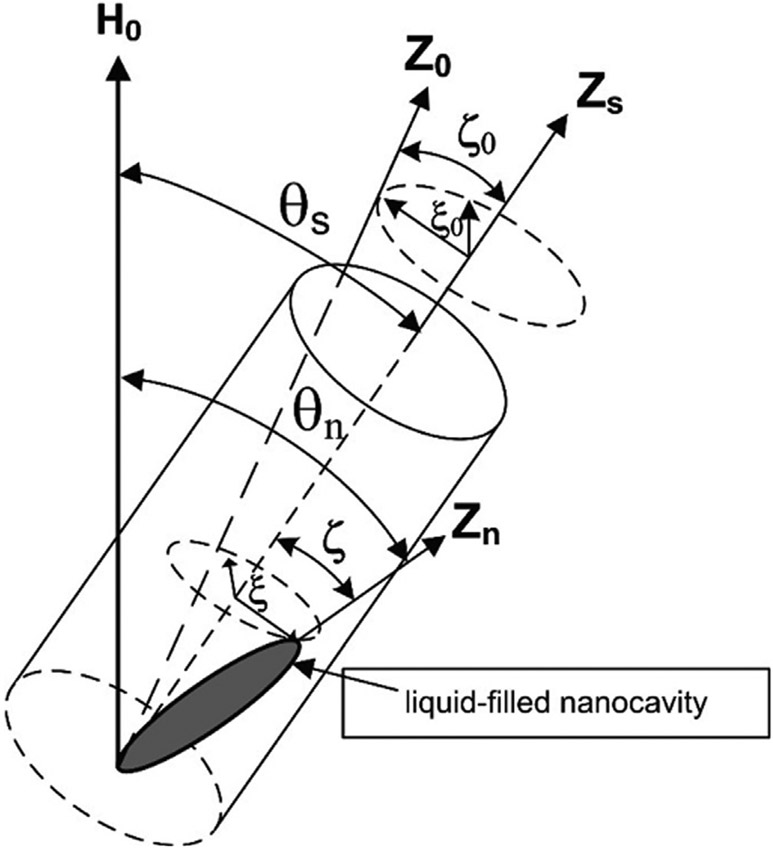
Fig. 1.
Model of a sample containing liquid entrapped in nanocavities. θs is the angle between the external magnetic field and the sample main axis (Zs). θn denotes the angle between the external magnetic field H0 and the axis of a n-th nanocavity (Zn). Z0 axis represents the averaged orientation of the ensemble of all nanocavities. The polar ζ and azimuthal ξ angles reflect the deviation of the main axis of a specific nanocavity (Zn) from the main Zs axis. Lastly, ζ0 and ξ0 are the polar and azimuthal angles which determine the deviation of the Z0-axis from the Zs-axis.
Using Eqs. (1) and (2) the transverse relaxation rate R2 = 1/T2 can be expressed by [18].
| (3) |
where ρ is the proton density in the tissue. According to Eq. (3), R2 is proportional to the square root of the concentration of the liquid contained in the nanocavity and is inversely proportional to the square root of the volume of the nanocavity. Importantly, it also depends on the orientation of the main axis of the nanocavity (Zn) relative to the external magnetic field, causing R2 to vanish at an angle equal to the magic angle where no relaxation occurs and T2 → ∞.
2.2. Orientational distribution of nanocavities
Eq. (3) describes the orientation dependency of R2 for a single nanocavity. If all nanocavities are oriented along the sample Zs axis, then Eq. (3) describes the angular anisotropy of R2 for the entire sample. In actual biological samples, however, a deviation from this ideal ordering is usually observed. Moreover, the volume and form factors may also vary across different nanocavities, and the main axis of each nanocavity can then be thought of as distributed around a single averaged orientation.
To expand Eq. (3) to account for a range of orientations (rather than a single one) it is necessary to average R2 over an ensemble of nanocavities, each having a different orientation (ζ, ξ) relative to the main sample axis Zs (see Fig. 1). The measured signal is usually modeled by a decaying exponential function, which, for an ensemble of nanocavities can be expressed by:
| (5) |
where n is an index running over all nanocavities. Assuming t ⪡ T2, the averaged relaxation rate can be expressed as.
| (6) |
where An is the relative fraction of each nanocavity.
The form factor of elongated axis-symmetrical nanocavities, a >> b = c is practically independent of the cavity size [12]. In biological and plant fibril tissues this condition is met, allowing to replace Vn and Fn with average volume V and form factor F, which characterize the tissue’s nanostructure,
| (7) |
where
| (8) |
To average all orientations, we define the deviation of each nanocavity from the main axis of the sample (Zs) by the polar ζ and azimuthal ξ angles (Fig. 1). Several previous studies have shown that a Gaussian distribution is good approximation for the angular distribution of nanocavities [14-18], showing good agreement between theory and experimentally measured spin–lattice and transverse relaxation rates R1 and R2. Similarly, we assume here that the distribution of nanocavities over both polar ζ and azimuthal ξ angles is given by a bivariate normal distribution function, with mean orientation denoted by Z0. The sum in Eq. (7) can be now expressed as integration over all possible values of ζ = 0, …, π and ξ = 0, …, 2π, yielding,
| (9) |
Φ(ζ, ξ) is the probability density function given by.
| (10) |
with σζ and σξ denoting the standard deviations (SD) around the average orientation Z0, defined using ζ0 and ξ0 relative to the Zs axis (Fig. 1). Since we do not know a priori the values of Vand F, it is useful to define a normalized relaxation rate which does not include these parameters according to:
| (11) |
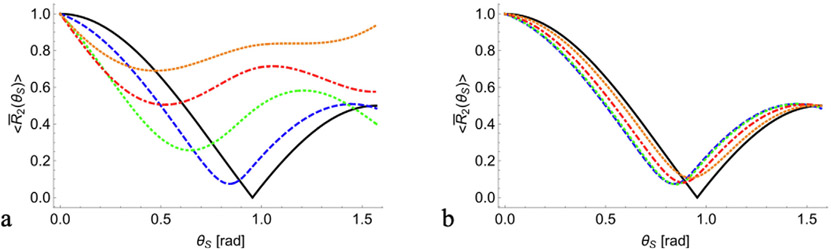
Examining Eq. (11) we note that this operation eliminates the dependency of the normalized transverse relaxation rate on the volume and form factor, thereby isolating the dependency on the orientation of sample’s main axis θs. Fig. 2 presents computer simulations of the angular dependency of for several choices for the SD of ζ and ξ around Z0. As can be seen R2 vanishes at the magic angle only when all nanocavities are perfectly aligned oriented along the sample Zs axis (solid black curves), while for non-zero SD R2 minimum moves away from zero. The difference between the maximum and minimum of the function decreases as SD values increase, making the effect of anisotropy less noticeable. Accordingly, an increase of σζ causes the minimum of the function to monotonically shifts towards θs = 0 (Fig. 2a). An increase of σζ, on the other hand, does not exhibit monotonic trend monotonic and the function’s minimum moves towards θs = 0 and then away from zero angle. Comparison of Fig. 2a and Fig. 2b shows that variations in the SD of the polar angle σζ causes significantly stronger changes in the angular dependence of R2 than variations in the SD of the azimuthal angle σξ.
Fig. 2.
Angular dependency of the normalized transverse relaxation rate on the SD of the polar and azimuthal angles ζ and ξ around the averaged orientation ζ0 = 0 and ξ0 = 0. The solid black curves are calculated according to Eq. (3) for an ensemble of nanocavities, which are all perfectly oriented along the sample axis Zs (i.e., zero SD). (a) for fixed SD σξ = 0.1 of the azimuthal angle, and varying SD of the polar angle σζ = 0.1, 0.3, 0.5, 0.7 (blue dashed, green dotted, red dash-dotted, and light brown dotted lines respectively). (b) for fixed SD σζ = 0.1 of the polar angle, and varying SD of the azimuthal angle σξ = 0.1, 0.4, 1.0, 1.7 (blue dashed, green dotted, red dash-dotted, and light brown dotted lines respectively). Note that the dashed blue line in (a) and in (b) was calculated using the same parameter values. All curves were calculated using Eq. (11).
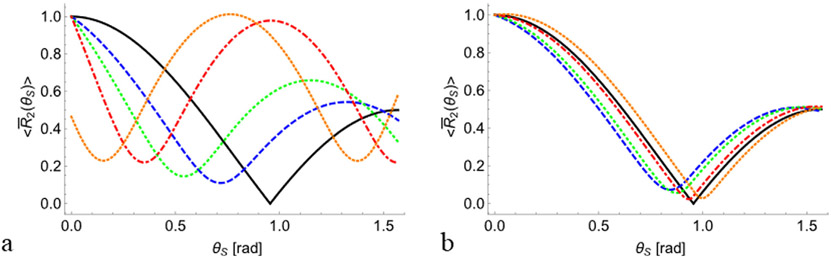
The angular dependency of R2 is determined not only by the degree of internal ordering of nanocavities (i.e., σζ and σξ), but also by the averaged direction of all nanoparticles Z0, determined by the polar ζ0 and azimuthal ξ0 angles (Fig. 1). Fig. 3 shows this dependency of R2 on the deviation of Z0 from the sample main axis Zs. As can be seen, even small changes in the polar angle ζ0 lead to significant changes in R2 anisotropy, while changes in the azimuthal angle ξ0 have a much smaller effect on the relaxation rate. The analysis above shows how the transverse relaxation rate depends on the nanoscale ordering of a set of nanocavities, and specifically the average orientation and the standard deviation around this orientation (Figs. 2 and 3). To consider the effect of the orientational distributions on the relaxation time T2, we use Eq. (9) to calculate the dimensionless value ηT2 according to,
| (12) |
Fig. 3.
Angular dependency of the normalized transverse relaxation rate on the deviation of the averaged orientation Z0 from the main sample axis Zs, for fixed distribution pattern given by σξ = 0.1 and σζ = 0.1. The solid black lines are calculated according to Eq. (3) for an ensemble of nanocavities, which are perfectly oriented along the sample axis Zs (i.e., zero SD, ξ0 = 0, and ζ0 = 0). (a) for fixed ξ0 = 0, and varying ζ0 equal to 0.2, 0.4, 0.6, and 0.8 (blue dashed, green dotted, red dash-dotted, and light brown dotted lines respectively). (b) for fixed ζ0 = 0, and varying ξ0 set to 0.2, 0.8, 1.4, 2.0, and 0.8 (blue dashed, green dotted, red dash-dotted, and light brown dotted lines respectively). All curves were calculated using Eq. (11).
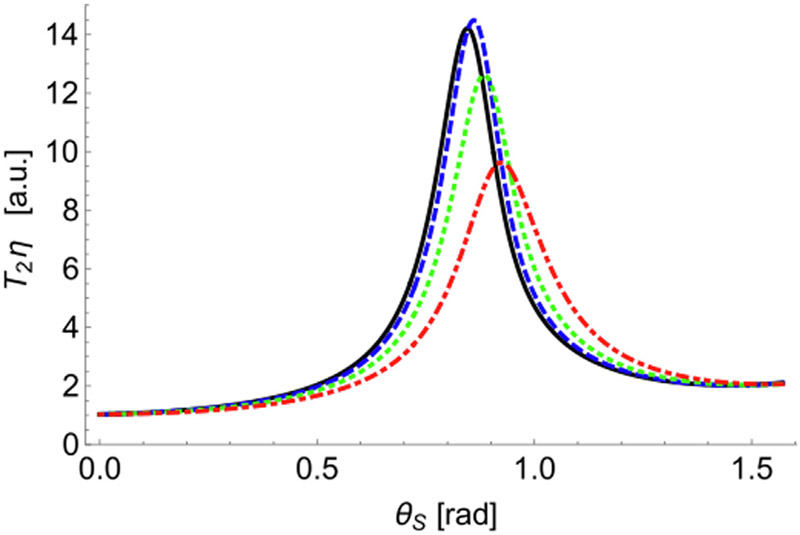
Fig. 4 shows the dependency of ηT2 on the standard deviations σζ and σξ. The maximum of the relaxation time shifts to higher values of θs, and decreases by a factor of ~1.5 as the standard deviation of the azimuthal angle σξ increases.
Fig. 4.
Relaxation time T2 multiplied by the factor η as a function of the SD of the azimuthal angle and fixed SD of the polar angle σζ = 0.1. Curves correspond to σξ = 0.3, 0.6, 1.0, and 1.7 (black solid, blue dashed, green dotted, and red dot-dashed curves). All curves were calculated using Eq. (12).
3. Methods
3.1. Preparation of celery samples
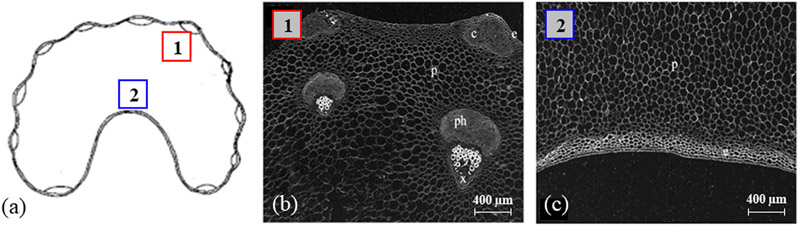
Fig. 5a presents a schematic view of an axial slice of a fresh celery stem, marked with two regions of interest (ROIs). Fig. 5b and 5c contain light microscopy images of a transverse section of a fresh celery stem, showing the structural variations of celery at 2 μm2 pixel resolution. Each image in Fig. 5b and 5c covers a tissue region of 2.8 × 2.0 mm2.
Fig. 5.
Transverse vibratome-cut section of a stem celery (a). The areas from which samples were cut out. Sample #1, outer (lateral) part of the stem celery (red inset); sample #2, core part of the stem celery (blue inset). (b-c) Microscopic images of sample ROIs #1 & #2 respectively [x, xylems; ph, phloems; c, collenchyma; c, parenchyma; e, epidermis].
Four types of tissue structure can be identified inside of a celery stem: the vascular bundles that include water-transporting xylems (marked by ‘x’ in Fig. 5b); nutrient-transporting phloems (marked by ‘ph’ in Fig. 5b); collenchyma that provides support for rigidity of the plant (marked by ‘c’ in Fig. 5b); and parenchyma that is isodiametric ground tissue (marked by ‘p’ in Fig. 5b and 5c). The outmost layer of a celery stem is the epidermis (skin, marked by ‘e’ in Fig. 5b and 5c). The vessels of the xylem and phloem are formed by overlapping cells that form hollow cavities connected into one long micrometer-diameter tube. Collenchyma consists of elongated living cells filled with water and is located along the outer edges of the stem and tissue of the vascular bundle, being in a longitudinal orientation. Collenchyma cells do not divide as much as the surrounding parenchyma cells. Importantly, the cell walls have a fibril structure and consist of nanofibers with a diameter in the range of 6–25 nm [28-31]. The parenchyma consists of non-oriented living cells filled with water and cell fluid, which are larger in diameter and often shorter than that of collenchyma [29]. Water is also part of the fibril structure and is located between microfibrils, presumably serving to stabilize them due to hydrogen bonds. Since a fresh celery is very hydrated and contains up to 95% water, the NMR signal comes mainly from water molecules.
Two test samples were used in this study, both cut from fresh celery purchased from local supermarkets.
3.1.1. Celery sample #1
Sample #1 was cut out from the outer (lateral) part of the celery stem in the form of a small piece of about 1 cm long and 2 × 2 mm2 cross-section [33]. The sample consisted mainly of collenchyma, xylem, and phloem (Fig. 5b), with minimal parenchyma. Thus, Sample #1 contains mainly the oriented fibril tissues without the epidermis (i.e., without the skin, marked by ‘e’ in Fig. 5b). The sample was placed in an empty glass tube before scanning.
3.1.2. Celery sample #2
Sample #2 was cut out from the inner (core) part of the celery stem. Its dimensions were about 2.5 cm long and 0.9 × 0.9 mm2 cross-section and consisted of mainly parenchyma without the epidermis (i.e., without the skin, marked by e in Fig. 5c). These parenchyma cells consist of cellulose microfibrils, which are characterized by uniform orientation, and has the potential to show a magic angle effect [29-31]. The celery sample was placed in an Eppendorf tube filled with Fluorinert, which does not produce any MR signal. This helped in keeping the external magnetic field as homogeneous as possible, while ensuring that the only signal arrives from water molecules inside the celery.
3.2. NMR scans
3.2.1. Sample #1 scans
1H NMR scans were carried out at ambient temperature (293 ± 0.5 K) using a Tecmag pulse solid state NMR spectrometer, a home-built NMR probe and an Oxford superconducting magnet of 8.0 T . The spin–spin, transverse, relaxation times T2 were measured using the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence [32]. Remaining experimental details are presented in [33].
3.2.2. Sample #2 scans
MRI experiments on Sample #2 were performed on the Bruker Biospec 7 T MRI magnet, running a Paravision 6.0 software. Scans consisted of a high-resolution morphologic scan (RARE), eleven multi echo spin echo (MESE) scans to measure T2 at different angles, and a diffusion tensor imaging (DTI) scan used to correct diffusion related bias of T2 measurements [34,35].
A 360° rotating bed was used to rotate the sample while inside the MRI scanner. The bed is shown in Fig. 6 and consists of a circular steering wheel located outside the scanner, which is connected through a cogwheel rack to a rotating test tube base located inside the scanner. This allowed to scan the sample at different angles, without the need to extract and re-insert the sample between scans, thereby largely reducing inter-scan variability which might result from repeating the matching, tuning, and shimming calibration steps. The sample was centered within a test Eppendorf tube in order to maintain the field of view (FOV) as small as possible.
Fig. 6.
The 3D printed rotating bed device used in this study, allowing rotation of the sample while inside the MRI scanner. The ability to image at different angles without the need to extract and reinsert the sample reduces spurious variability due to the need to repeat the processes of matching, tuning, and shimming between scans.
MESE scans parameters were: NEchoes = 32, TE = 18 ms, Nslices = 9, slice-thickness = 0.8 mm, in-plane resolution = 0.375 × 0.375 mm2, FOV = 36 × 36 mm2, and a total scan time of 4:48 min. Sample main axis orientation with respect to B0 was set to 0°, 9°, 18°, 27°, 36°, 45°, 54°, 63°, 72°, 81°, 90° degrees. DTI scan parameters included TE = 18 ms, TR = 3000 ms, Naverages = 3, Nslices = 5, slice-thickness = 0.5 mm, in-plane resolution = 0.5 × 0.5 mm2, and FOV = 32 × 32 mm2. Diffusion encoding used four b values = 0, 200, 500, 1000 sec/mm2 along 3 orthogonal directions and a total scan time was 11:42 min.
4. Results and discussion
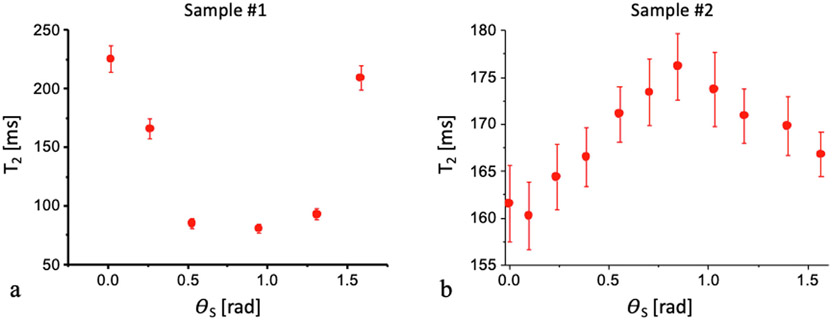
Fig. 7 shows the dependence of the relaxation time T2 on the angle between the main sample axis and H0 for the two assayed samples. A clear dependency was observed with extremum values at different values of the angle θs.
Fig. 7.
Transverse relaxation time T2 (ms) as a function of the rotation angle (rad) for sample #1 (a) and sample #2 (b) (panel (a) is reproduced with permission from [33]
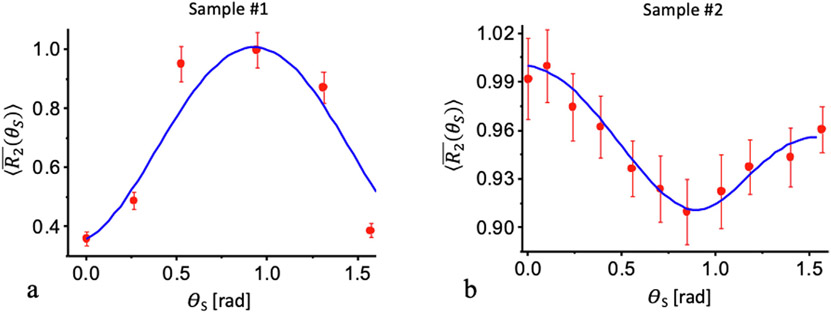
The model resented herein allows to search for the angular distribution and average direction of nanocavities which best fits the experimental measurements of the relaxation rates. Fitting to the model yielded SD and average directions of σζ = 0.33; σξ = 0.1; ζ0 = 2.3; ξ0 = 0.1 for sample #1 and σζ = 1.18; σξ = 0.8; ζ0 = 0.36; ξ0 = 1.5 for sample #2 (both ζ0 and ξ0 are in unit of radians). Fig. 8 presents the averaged transverse relaxation rates calculated according to Eq. (11) overlaid on the experimentally measured data.
Fig. 8.
Angular dependence of the transverse relaxation rate in the celery. Red circles mark the experimentally measured relaxation rate value, while the blue solid line represent the theoretically fitted model. (a) Normalized relaxation rate, measured using a CPMG pulse sequence for sample #1 (data reproduced with permission from [33]). (b) values for sample #2, measured using an imaging MESE protocol and fitted using the echo modulation curve (EMC) algorithm. The solid blue curves are calculated according to Eq. (8) with the parameters of σζ = 0.33; σξ = 0.1; ζ0 = 2.3; ξ0 = 0.1 for sample #1 and σζ = 1.18; σξ = 0.8; ζ0 = 0.36; ξ0 = 1.5 for sample #2.
Good agreement is achieved for both assayed samples, which exhibit different orientational dependency. To interpret these results, we consider the microscopic and morphological structures of the celery. According to Fig. 1, the polar and azimuthal angles (ζ0 and ξ0) determine the deviation of the average orientation of all nanocavities Z0 from the main sample axis ZS. For the polar angles, the model fits the experimental data best when ζ0 = 2.3 and σζ = 0.33 for sample #1, and ζ0 = 0.36 and σζ = 1.18 for sample #2. The value of ζ0 for Sample #1 (2.3 rad, 131.8°) means that the average orientation of the nanocavities is almost perpendicular to ZS. Sample #2 on the other hand produced ζ0 = 0.36 rad, or 20.6° meaning that the average axis of nanocavities is almost in parallel to ZS. This illustrates the different in the nanoscopic morphology of the two specimens, which is congruent with the microstructural differences between the two samples as shown in Fig. 5b-c. In addition, a much smaller variability was observed in the polar angle of sample #1 (σζ = 0.33) compared to sample #2 (σζ = 1.18), meaning that the first sample exhibited higher degree of internal ordering.
With regard to the azimuthal angle ξ0, the model fits best when ξ0 = 0.1 and σ ξ = 0.1 for sample #1, and ξ0 = 1.5 and σξ = 0.8 for sample #2. These values also reflect the morphological differences between the two samples: sample #1 contains mostly the organized nanocavities with small baseline and variability of the azimuthal angle, while sample #2 contains mostly isodiametric ground tissue (parenchyma), characterized by large azimuthal angle and large standard deviations around the mean.
To summarize, good agreement is achieved between the theoretical and experimental values, indicating that the nanoscale ordering in sample #1 is higher than that in sample #2. The best agreement of calculations is achieved with the following values: in the sample #1 the average polar and azimuthal angles equal to ζ0 = 2.3 and ξ0 = 0.1, while for the sample #2 these angles came out to be ζ0 = 0.3 and ξ0 = 1.5, respectively. These angles determine the average directions of the nanocavities relative to the probe axes. On one hand various orientations of the samples inside the probes can explain different angular positions of the maxima and minima of the relaxation rates (Fig. 8). On the other hand, the average direction Z0 relative to the sample axis ZS can be considered as one of the structural ordering parameters of the fibril tissue.
As can be seen in Fig. 5, the physical dimensions of the anatomic structures in celery are in tens of microns (from 60 to 80 μm in parenchyma, to 8–10 μm in collenchyma). Hence the water molecules inside each biological structure (e.g., parenchyma) should have an isotropic motion. However, the walls of parenchyma, which consist of layers of cellulose microfibrils with uniform orientation, are much smaller in dimension and highly anisotropic, which can be used to explain the marked differences in the polar and azimuthal angles in samples #1 and #2.
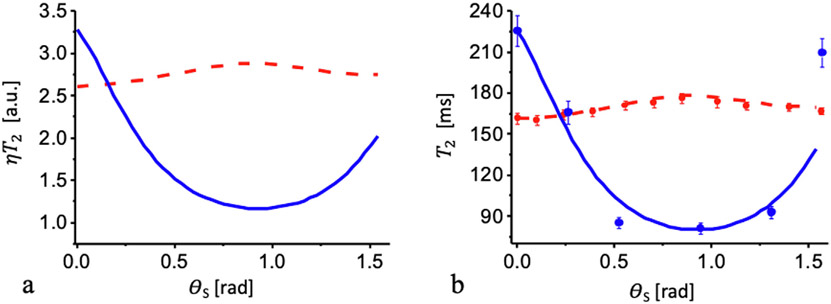
Significant differences in the experimentally obtained angular dependence of relaxation times from different parts of the sample (Fig. 8) can be explained based on the examples shown previously in Figs. 2 and 3. Fig. 9 presents the results of calculation multiplied by factor η according to Eq. (12) with the parameters obtained for fitting the experimental results for the relaxation rates above: σζ = 0.33; σξ = 0.1; ζ0 = 2.3; ξ0 = 0.1 for sample #1 and σζ = 1.18; σξ = 0.8; ζ0 = 0.3; ξ0 = 1.5 for sample #2. The factor can be found comparing the theoretical (Fig. 9a) and experimental (Fig. 7) relaxation times T2: η = (1/67.8) = 1.46 × 10−2 for sample #1 (Fig. 9b), and η = (1/61.8) = 1.62 × 10−2 for sample #2 (Fig. 9b). These values can now be used to estimate the characteristic volume for a nanocavity according to Eq. (8). Assuming nanocavities having elongated ellipsoidal shape where a⪢b the form factor is equal to F ≈ 2π/3 [36], the spin density for water is ρ = 66 spin/nm3, and the term γ2h is equal to 2π × 0.120 nm3/ms. The resulting volumes are 7.7 × 105 nm3 for sample #1 and 6.0 × 105 nm3 for sample #2. For b ≈ 10 nm [29], the length of the nanocavity for the first and second samples will be about 1430 nm and 1840 nm, respectively. A precise verification of these modeling results awaits further experiments.
Fig. 9.
Angular dependency of the transverse relaxation time T2. (a) T2 multiplied by the factor η. (b) The transverse relaxation time T2: blue dot – experimental data for sample #1; red dot – for sample #2. Solid blue and dashed red curves are calculated according to Eq. (12) for sample #1 and sample #2, respectively. Values of the factors are given in the text.
5. Conclusions
In this study we performed experimental and theoretical investigation of the angular dependence of water transverse relaxation time in a celery plant stem. Experimental results were interpreted within the framework of the previously developed theory [14-18], in which the fibril structure of biological tissues is represented as a set of aqueous nanocavities containing water, while the transverse relaxation time is determined by the dipole–dipole spin interaction which are affected by the restricted tumbling and rotational motion of water molecule inside the nanocavities.
This model is successfully applied to explain the anisotropy of R2 relaxation rate in fibril tissues. Good agreement was achieved between the calculations and experimental data with the use of few fitting parameters: the standard deviations of the bivariate Gaussian distribution of the nanocavities, and the polar and azimuth angles of their averaged direction. The approach allows the use of NMR data for quantitative determination of the nanostructural ordering of biological tissues non-invasively and non-destructively. We have shown that dependence of the relaxation time on the angle between the magnetic field and the main axis of the sample makes it possible to identify the nanoscale properties of fibrillar tissues [37].
An important feature of this study is that we were able to determine the degree of orientational ordering of nanocavities in the celery and the average direction of nanocavities with respect to the orientation of the sample. The obtained values of standard deviations (σζ = 0.33 and σξ = 0.1 for sample #1 and σζ = 1.18 and σξ = 0.8 for sample #2) indicate a noticeable ordering of the cavities in the first sample and less ordering in the second sample, which agree with the microscopic images of celery stem.
Our approach showed that NMR can be used to study the microscopic vascular structure of plants. Variations in these structures can be associated with the economic characteristics of plants, for example, high / low yields of food crops or long / short growth times of vegetables [38,39]. Since NMR and MRI methods are completely non-invasive and non-destructive, they can be used to study the fine vascular structures of living organisms both in situ and in vivo, as well as to diagnose human pathology that involves changes to the tissues’ nanostructure, e.g., around collagen rich tissues like tendons and cartilage [40].
Acknowledgment
This research was supported by the Israel Binational Science Foundation (BSF No. 2019033), and by a grant from the National Institutes of Health (NIH, AR069047).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
No experiments or investigations were performed involving human participants or living animals.
References
- [1].Hausser KH, Kalbitzer HR, NMR in Medicine and Biology, Springer-Verlag, Berlin Heidelberg, 1989. [Google Scholar]
- [2].Callaghan PT, Principles of Nuclear Magnetic Resonance Microscopy, Oxford Clarendon Press, 1991. [Google Scholar]
- [3].Xia Y, Momot K, Biophysics and Biochemistry of Cartilage by NMR and MRI, The Royal Society of Chemistry, Cambridge UK, 2016. [Google Scholar]
- [4].Berendsen HJ, Nuclear Magnetic Resonance Study of Collagen Hydration, J. Chem. Phys 16 (1962) 3297–3305. [Google Scholar]
- [5].Fullerton GD, Cameron IL, Ord VA, Orientation of tendons in the magnetic field and its effect on T2 relaxation times, Radiology 155 (1985) 433–435. [DOI] [PubMed] [Google Scholar]
- [6].Wang N, Xia Y, Anisotropic analysis of multi-component T2 and T1r relaxations in Achilles tendon by NMR spectroscopy and microscopic MRI, JMRI 38 (2013) 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shao H, Pauli C, Li S, Ma Y, Tadros AS, Kavanough A, Chang EY, Tang G, Du J, Magic angle effect plays a major role in both T1rho and T2 relaxation in articular cartilage, Osteoarthritis Cartilage 25 (2017) 2022–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xia Y, Moody JB, Alhadlaq H, Orientational dependence of T2 relaxation in articular cartilage: a microscopic MRI (mMRI) study, Magn. Reson. Med 48 (2002) 460–469. [DOI] [PubMed] [Google Scholar]
- [9].Navon G, Eliav U, Demco DE, Blümich B, Study of order and dynamic processes in tendon by NMR and MRI, J. Magn. Reson. Imaging 25 (2007) 362–380. [DOI] [PubMed] [Google Scholar]
- [10].Peto S, Gillis P, Henri VP, Structure and Dynamics of Water in Tendon from NMR Relaxation Measurements, Biophys. J 57 (1990) 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fullerton GD, Rahal A, Collagen Structure: The Molecular Source of the Tendon Magic Angle Effect, J. Magn. Reson. Imaging 25 (2007) 345–361. [DOI] [PubMed] [Google Scholar]
- [12].Momot KI, Pope JM, Wellard RM, Anisotropy of Spin Relaxation of Water Protons in Cartilage and Tendon, NMR Biomed. 23 (2010) 313–324. [DOI] [PubMed] [Google Scholar]
- [13].Karjalainen J, Henschel H, Nissi MJ, Nieminen MT, Hanni M, Dipolar Relaxation of Water Protons in the Vicinity of a Collagen-like Peptide, J. Phys. Chem. B 126 (2022) 2538–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Furman GB, Goren SD, Meerovich VM, Sokolovsky VL, Anisotropy of spin–spin and spin–lattice relaxation times in liquids entrapped in nanocavities: Application to MRI study of biological systems, J. Magn. Reson 263 (2016) 71–78. [DOI] [PubMed] [Google Scholar]
- [15].Furman GB, Goren SD, Meerovich VM, Sokolovsky VL, Correlation of transverse relaxation time with structure of biological tissue, J. Magn. Reson 270 (2016) 7–11. [DOI] [PubMed] [Google Scholar]
- [16].Furman G, Meerovich V, Sokolovsky V, Xia Y, Spin locking in liquid entrapped in nanocavities: Application to study connective tissues, J. Magn. Reson 299 (2019) 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Furman G, Meerovich V, Sokolovsky V, Xia Y, Spin-lattice relaxation in liquid entrapped in a nanocavity, J. Magn. Reson 311 (2020) 106669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Furman G, Kozyrev A, Meerovich V, Sokolovsky V, Xia Y, Dynamics of Zeeman and dipolar states in the spin locking in a liquid entrapped in nanocavities: application to study of biological systems, J. Magn. Reson 325 (2021) 106933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Holz M, Heil SR, Sacco A, Temperature-dependent self-diffusion coefficients of water and six selected molecular liquids for calibration in accurate 1H NMRPFG measurements, PCCP 2 (2000) 4740. [Google Scholar]
- [20].Furman GB, Goren SD, Meerovich VM, Sokolovsky VL, Multiple-pulse spin locking in nanofluids, RSC Adv. 5 (2015) 44247–44257. [Google Scholar]
- [21].Freeman DM, Bergman G, Glover G, Short TE MR microscopy: accurate measurement and zonal differentiation of normal hyaline cartilage, Magn. Reson. Med 37 (1997) 72. [DOI] [PubMed] [Google Scholar]
- [22].Xia Y, Farquhar T, Burton-Wurster N, Lurst G, Origin of cartilage laminae in MRI, J. Magn. Reson. Imaging 7 (1997) 887. [DOI] [PubMed] [Google Scholar]
- [23].Migchelson C, Berendsen HJC, Proton exchange and molecular orientation of water in hydrated collagen fibers. An NMR study of H2O and D2O, J. Chem. Phys 59 (1973) 296. [Google Scholar]
- [24].Henkelman RM, Stainz GJ, Kim JK, Bronskill MJ, Anisotropy of NMR properties of tissues, Magn. Reson. Med 32 (1994) 592. [DOI] [PubMed] [Google Scholar]
- [25].Tadimalla S, Momot KI, Effect of partial H2O–D2O replacement on the anisotropy of transverse proton spin relaxation in bovine articular cartilage, PLoS ONE 9 (2014) e115288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Eliav U, Navon G, A study of dipolar interactions and dynamic processes of water molecules in tendon by 1H and 2H homonuclear and heteronuclear multiple-quantum-filtered nMr spectroscopy, J. Magn. Reson 137 (1999) 295–310. [DOI] [PubMed] [Google Scholar]
- [27].Xia Y, Moody JB, Burton-Wurster N, Lust G, Quantitative In Situ Correlation Between Microscopic MRI and Polarized Light Microscopy Studies of Articular Cartilage, Osteoarthritis Cartilage 9 (2001) 393–406. [DOI] [PubMed] [Google Scholar]
- [28].Fel’dman EB, Rudavets MG, Nonergodic nuclear depolarization in nanocavities, J. Exp. Theor. Phys 98 (2004) 207–219. [Google Scholar]
- [29].Thimm JC, Burritt DJ, Ducker WA, Melton LD, Celery (Apium graveolens L.) parenchyma cell walls examined by atomic force microscopy, Planta 212 (2000) 25–32. [DOI] [PubMed] [Google Scholar]
- [30].Leroux O, Collenchyma: a versatile mechanical tissue with dynamic cell walls, Ann. Bot 110 (2012) 1083–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC, Structure of cellulose microfibrils in primary cell walls from collenchyma, Plant Physiol. 161 (1) (2013. Jan) 465–476, 10.1104/pp.112.206359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Meiboom S, Gill D, Modified Spin-Echo Method for Measuring Nuclear Relaxation Times, Rev. Sci. Instrum 29 (1958) 688–691. [Google Scholar]
- [33].Furman G, Goren S, Meerovich V, Panich A, Sokolovsky V, Xia Y, Anisotropy of transverse and longitudinal relaxations in liquids entrapped in nano- and micro-cavities of a plant stem, J. Magn. Reson 331 (2021) 107051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bnaiahu N, Wilczynski E, Levy S, Omer N, Blumenfeld-Katzir T, Ben-Eliezer N. Compensating diffusion bias of quantitative T2 mapping on high-field MRI scanners. In: Proceedings of the 28th Annual meeting of ISMRM, Montreal, Canada, vol. 5437, 2019. https://archive.ismrm.org/2020/3415.html. [Google Scholar]
- [35].Radunsky D, Stern N, Nassar J, Tsarfaty G, Blumenfeld-Katzir T, Ben-Eliezer N, Quantitative platform for accurate and reproducible assessment of transverse (T2) relaxation time, NMR Biomed. 34 (2021) e4537. [DOI] [PubMed] [Google Scholar]
- [36].Furman GB, Goren SD, Meerovich VM, Sokolovsky VL, Dipole-dipole interactions in liquids entrapped in confined space, J. Mol. Liq 272 (2018) 468–473. [Google Scholar]
- [37].Furman G, Sokolovsky V, Ben-Eliezer N, Invention “Determining characteristics of nanoporous materials by NMR measurements: internal orientation, degree of ordering, and volume of nanocavity”, Provisional Patent Application 63/329,431 was filed on 10 April 2022. [Google Scholar]
- [38].Jenner CF, Xia Y, Eccles CD, Callaghan PT, Circulation of water within wheat grain revealed by nuclear magnetic resonance micro-imaging, Nature 336 (1988) 399. [Google Scholar]
- [39].Xia Y, Sarafis V, Campbell EO, Callaghan PT, Non-invasive imaging of water flow in plants by NMR microscopy, Protoplasma 173 (1993) 170–176. [Google Scholar]
- [40].Ben-Eliezer N, Raya JG, Babb JS, Youm T, Sodickson DK, Lattanzi R, A new method for cartilage evaluation of femoroacetabular impingement using quantitative T2 Magnetic Resonance Imaging: preliminary validation against arthroscopic findings, Cartilage 13 (2021) 1315S–1323S. [DOI] [PMC free article] [PubMed] [Google Scholar]