Abstract
Meiosis is a specialized cell division that halves the number of chromosomes in two consecutive rounds of chromosome segregation. In angiosperm plants is meiosis followed by mitotic divisions to form rudimentary haploid gametophytes. In Arabidopsis, termination of meiosis and transition to gametophytic development are governed by TDM1 and SMG7 that mediate inhibition of translation. Mutants deficient in this mechanism do not form tetrads but instead undergo multiple cycles of aberrant nuclear divisions that are likely caused by the failure to downregulate cyclin dependent kinases during meiotic exit. A suppressor screen to identify genes that contribute to meiotic exit uncovered a mutation in cyclin‐dependent kinase D;3 (CDKD;3) that alleviates meiotic defects in smg7 deficient plants. The CDKD;3 deficiency prevents aberrant meiotic divisions observed in smg7 mutants or delays their onset after initiation of cytokinesis, which permits formation of functional microspores. Although CDKD;3 acts as an activator of cyclin‐dependent kinase A;1 (CDKA;1), the main cyclin dependent kinase that regulates meiosis, cdkd;3 mutation appears to promote meiotic exit independently of CDKA;1. Furthermore, analysis of CDKD;3 interactome revealed enrichment for proteins implicated in cytokinesis, suggesting a more complex function of CDKD;3 in cell cycle regulation.
Keywords: Arabidopsis thaliana , cell cycle, cyclin dependent kinase, cytokinesis, meiosis
1. INTRODUCTION
The eukaryotic cell cycle consists of two major cellular events: deoxyribonucleic acid (DNA) replication in S‐phase and chromosome segregation followed by cell division in M‐phase. Alteration of these events is accomplished by executing complex molecular processes that take place in a well‐defined order to ensure faultless passage of genetic information from one cell generation to another. Cyclin‐dependent kinases (CDKs) are the master regulators of the eukaryotic cell cycle. CDKs push cells into S‐phase through activation of S‐phase specific genes and initiation of DNA replication. Further buildup of CDK activity at G2/M drives chromosome condensation, dissolution of the nuclear membrane, formation of the mitotic spindle, and other processes required for chromosome segregation and cell division. CDK activity peaks at metaphase when all chromosomes properly attach to the spindle microtubules. This triggers a rapid decline of CDK activity and the dephosphorylation of its targets, which allows for chromosome decondensation, spindle disassembly, and cytokinesis. Low CDK activity in G1 is crucial for DNA replication licensing, which assures that chromosomes are duplicated only once during the cell cycle.
The plant cell cycle is driven by two types of CDKs: The canonical cyclin‐dependent kinase A (CDKA) corresponding to the mammalian CDK1 is expressed throughout the cell cycle and the plant‐specific cyclin‐dependent kinase B (CDKB) whose expression is cell‐cycle dependent (Shimotohno et al., 2021). CDK activity and specificity are controlled by cyclins, which in Arabidopsis thaliana form a large gene family with at least 50 members (Vandepoele et al., 2002; Wang, Kong, et al., 2004). Type A, B and D cyclins are directly implicated in cell cycle, and their consecutive activities drive progression through S‐, G2‐, and M‐phases. Cyclin levels oscillate due to ubiquitin‐mediated degradation resulting in waves of CDK activities as the cell cycle progresses.
CDKA and CDKBs are further regulated by phosphorylation in their T‐loop by CDK‐activating kinases that consist of CDKDs and cyclin H (Dissmeyer et al., 2007; Harashima et al., 2007; Umeda et al., 2005; Yamaguchi et al., 1998). Plant cyclin‐dependent kinase D (CDKD) correspond to mammalian CDK7; like CDKD7, they are capable of not only activating other CDKs but also phosphorylating the C‐terminal domain (CTD) of the largest subunit of ribonucleic acid (RNA) polymerase II, thereby affecting transcription and co‐transcriptional RNA processing (Hajheidari et al., 2012; Shimotohno et al., 2003; Yamaguchi et al., 1998). Arabidopsis encodes three CDKD paralogues CDKD;1, CDKD;2, and CDKD;3 that are partially redundant, and their concomitant inactivation is lethal (Hajheidari et al., 2012; Takatsuka et al., 2015). Further functional studies showed that CDKDs regulate microtubule dynamic and meiotic cytokinesis through activation of CDKA;1 and small RNA biogenesis through phosphorylation of the CTD domain of RNA polymerase II (Hajheidari et al., 2012; Sofroni et al., 2020).
Meiosis is a specialized form of cell division that produces haploid cells from a diploid progenitor in two successive rounds of chromosome segregation without intervening S‐phase. The meiotic program substantially deviates from regular mitotic division, and its implementation requires extensive modification of chromosome segregation and cell‐cycle machinery. These include physical coupling of homologous chromosomes via recombination and synapsis in prolonged prophase I, mono‐orientation of sister kinetochores in metaphase I, protection of centromeric cohesion during anaphase I, and inhibition of DNA replication in interkinesis. These processes are brought about by specific sets of effector proteins and cell‐cycle regulators, many of which are uniquely expressed in meiosis. Despite the differences between mitosis and meiosis, the core molecular machinery driving their progression seems to be identical. For example, the key Arabidopsis mitotic kinase, CDKA;1, appears to be central also for meiosis where it orchestrates chromosome recombination and pairing, spindle organization, and cytokinesis (Sofroni et al., 2020; Wijnker et al., 2019; Yang et al., 2020). This is done in concert with several cyclins, of which some are meiosis specific, whereas others are present also in mitosis (Azumi et al., 2002; Bulankova et al., 2013; Wang, Magnard, et al., 2004).
In angiosperm plants, haploid products of meiosis undergo additional mitoses to generate rudimentary haploid gametophytes that carry germ cells. Therefore, upon completion of meiosis, the cell division machinery is reprogrammed back to mitosis. Male germ cells differentiate in pollen, a tricellular structure produced by two consecutive post‐meiotic divisions of a haploid microspore. Studies in Arabidopsis revealed a dedicated molecular mechanism that governs the termination of male meiosis and transition to post‐meiotic development. This mechanism relies on the evolutionarily conserved nonsense‐mediated RNA decay (NMD) factor SMG7 and plant specific protein TDM1 and facilitates meiotic exit through repressing translation (Cairo et al., 2022). Mutants deficient in these proteins fail to terminate meiosis and to produce pollen. In tdm1 mutants, meiosis progresses normally until telophase II, but instead of undergoing cytokinesis, the haploid nuclei attempt to further divide without replicating their DNA. Such pollen mother cells (PMCs) enter multiple cycles of chromatin condensation and spindle reassembly, which leads to unequal distribution of chromatin and formation of polyads (Bulankova et al., 2010; Cairo et al., 2022). A similar phenotype was caused by partially functional allele smg7‐6, whereas the full SMG7 inactivation halts meiotic progression in anaphase II (Capitao et al., 2021; Riehs et al., 2008). Such phenotypes are reminiscent of situations when cells fail to downregulate CDK activity at the end of M‐phase (Parry & O'Farrell, 2001; Potapova et al., 2006). Indeed, smg7 mutants arrested at anaphase II exhibit high level of T‐loop phosphorylated CDKA;1 indicating that high CDK activity persists beyond metaphase II (Bulankova et al., 2010).
To decipher molecular processes that govern meiotic exit, we performed a forward genetic screen to identify mutations that suppress phenotype of smg7‐6 plants and increase their fertility. We anticipated to recover mutations either directly affecting the TDM1/SMG7‐mediated mechanism that regulates meiotic exit or rescuing the fertility indirectly through modulation of chromosome segregation or cell cycle progression. In this study, we report on characterizing suppressor mutation in CDKD;3 and its effect on meiotic progression and exit in smg7‐6 plants.
2. RESULTS
2.1. A mutation in CDKD;3 gene restores fertility of smg7‐6 plants
Arabidopsis mutants homozygous for the hypomorphic smg7‐6 allele have reduced fertility due to 10‐fold decrease of viable pollen compared with wild type (Figure 1) (Capitao et al., 2021; Riehs‐Kearnan et al., 2012). A characteristic feature of smg7‐6 plants is that the first 15–20 flowers on the main inflorescence bolt are sterile, but later flowers regain fertility. To identify smg7‐6 suppressors, we mutagenized the seeds homozygous for smg7‐6 with ethyl‐methanesulfonate (EMS) and screened for plants with increased length of the first 20 siliques in the M2 generation, which was indicative of restored seed production (Capitao et al., 2021). One of the suppressor lines named EMS58‐1 exhibited increased fertility and about four‐fold increase in viable pollen compared with smg7‐6 parents. However, the pollen count was still lower than in wild type (Figure 1). The EMS58‐1 line did not show any obvious growth abnormalities and, with the exception of increased fertility, it was indistinguishable from smg7‐6 parents (Figure S1).
FIGURE 1.
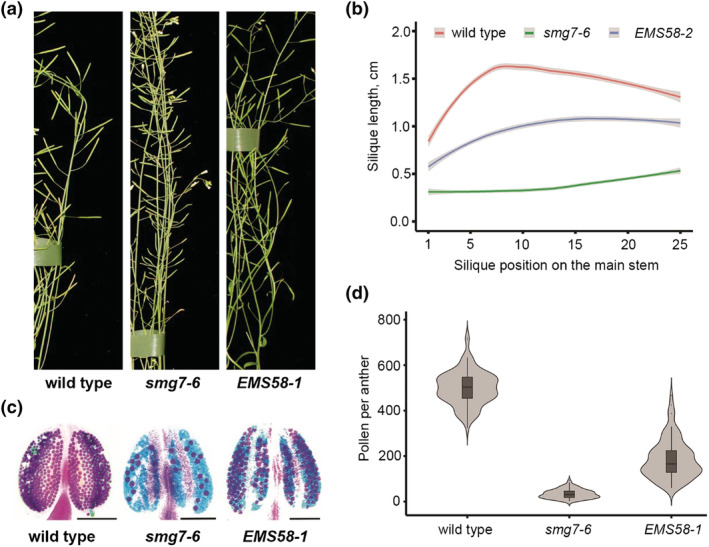
Fertility of the EMS58‐1 suppressor line. (a) Details of inflorescence bolt with siliques from 6 week old plants depicted in Figure S1. (b) Quantification of silique length along the main inflorescence bolt in wild type (n = 25), smg7‐6 (n = 37), and EMS58‐1 (n = 48). The trend lines of the data were plotted by locally weighted scatterplot smoothing (LOESS) function (colored lines; shaded area represents 95% confidence intervals). (c) Anthers of indicated lines after Alexander staining. Scale bar = 10 μm. (d) Violin plots showing viable pollen per anther (wild type n = 96, smg7‐6 n = 97, and EMS58‐1 n = 97)
We used association mapping to identify the mutation restoring fertility in the EMS58‐1 line. The increased fertility phenotype segregated as a recessive trait. The associated de novo mutations were identified by comparative whole genome sequencing of fertile B2 plants with smg7‐6 parents. This analysis revealed de novo mutations that associated with fertility on chromosome 1 between 5.5 and 7 Mb (Figure S2, Table S1). Polymerase chain reaction (PCR)‐based genotyping of several selected mutations in this region in 106 B2 plants showed the highest association between phenotype and mutation in the At1g18040 locus coding for CDKD;3. The mutation represents a C to T transition in the third exon of the gene and causes a proline to leucine substitution (P137L) at amino acid position 137 (Figure 2a). The P137L mutation is located in the evolutionary highly conserved kinase active site and likely impairs CDKD;3 activity (Figure 2b). We refer to this new allele as cdkd;3‐3. Transformation of EMS58‐1 plants with a wild copy of the CDKD;3 gene reduced fertility in the majority of T1 transformants back to the level observed in smg7‐6, confirming the causality of the mutation (Figure S3). When introgressed to wild type Col‐0 background, cdkd;3‐3 mutants did not exhibit any obvious phenotype and were fully fertile (Figure S4).
FIGURE 2.
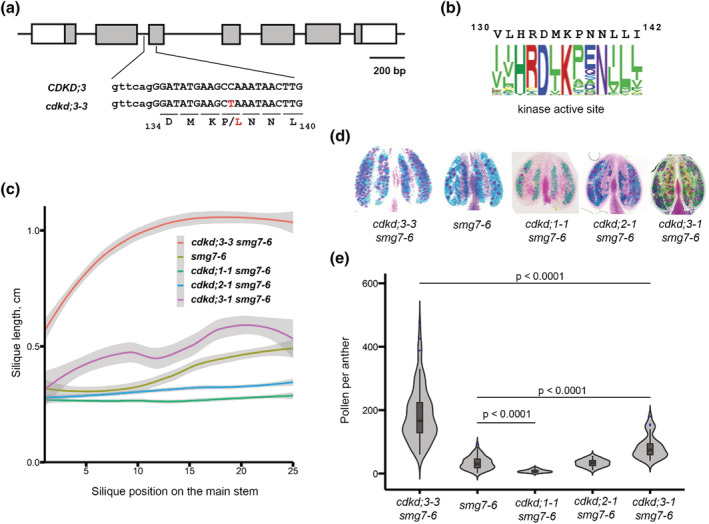
Effect of CDKDs on fertility of smg7‐6 plants. (a) Diagram of CDKD;3 gene with exons marked as boxes. DNA sequence surrounding the de novo mutation (depicted in red) in the cdkd;3‐3 allele is shown; capital letters denote exon sequence. (b) The conserved amino acid sequence present in the kinase active site of CDKD;3 is represented as web logo. A total of 131,130 kinase sequences were aligned with CDKD;3 active kinase site (amino acid residues 130 to 142). (c) Quantification of silique length along the main inflorescence bolt in wild type and indicated mutants (smg7‐6 n = 25, cdkd;1‐1 smg7‐6 n = 20, cdkd;2‐1 smg7‐6 n = 20, and cdkd;3‐1 smg7‐6 n = 11). The trend lines of the data were plotted by LOESS function (colored lines; shaded area represents 95% confidence intervals). (d) Anthers of indicated mutants after Alexander staining. (e) Violin plots showing viable pollen per anther. Significance of the difference is indicated (two tailed t‐test; cdkd;3‐3 smg7‐6 ‐ 97 anthers/10 plants, smg7‐6 ‐ 94 anthers/10 plants, cdkd;1‐1 smg7‐6 ‐ 70 anthers/6 plants, cdkd;2‐1 smg7‐6 ‐ 35 anthers/3 plants, and cdkd;3‐1 smg7‐6‐36 anthers in 4 plants).
Because Arabidopsis CDKD;3 paralogues exhibit functional redundancy (Hajheidari et al., 2012; Takatsuka et al., 2015), we next examined the impact of each paralogue on the fertility of smg7‐6 plants. We crossed T‐DNA insertion alleles cdkd;1‐1, cdkd;2‐1, and cdkd;3‐1 individually with smg7‐6 and examined fertility in the respective double mutants (Figure S5). Only cdkd;3‐1 smg7‐6 plants showed partially improved fertility and pollen count (Figure 2c–e). Inactivation of the other two CDKDs further decreased fertility of smg7‐6 with cdkd;1‐1 smg7‐6 plants producing no pollen. These data suggest partial functional divergence of the Arabidopsis CDKD paralogues. Interestingly, the cdkd;3‐1 allele improved pollen yield to a much lesser extent than cdkd;3‐3 allele (Figure 2f). We suspect that the CDKD;3P137L variant produced from cdkd;3‐3 sequesters cyclin H and other subunits in a nonproductive complex, therefore exerting a bigger effect on CDKD activity than full disruption of the gene in cdkd;3‐1 plants.
2.2. Meiotic progression in cdkd;3‐3 smg7‐6 plants
PMCs in smg7‐6 mutants do not conclude meiosis by cytokinesis. Instead, haploid nuclei undergo multiple rounds of chromatin condensation/decondensation and spindle assembly/disassembly (referred here to as meiosis III, meiosis IV, etc.; Capitao et al., 2021). This results in random distribution of chromatids and formation of polyads in more than 95% of PMCs (Figure 3a,b). In contrast, we observed that 20%–80% of PMCs in individual anther lobes in cdkd;3‐3 smg7‐6 plants formed tetrads. This result was further reaffirmed by crossing cdkd;3‐3 smg7‐6 with qrt1 plants that are deficient in pectin methylesterase and do not separate pollen derived from the same PMC (Francis et al., 2006). The fraction of tetrads containing four viable pollen rose from 21% in smg7‐6 qrt1 to 52% in cdkd;3‐3 smg7‐6 qrt1 pants (Figure 3c). These data suggest that a substantial portion of PMCs in cdkd;3‐3 smg7‐6 properly terminate meiosis and do not enter aberrant postmeiotic chromosome segregation.
FIGURE 3.
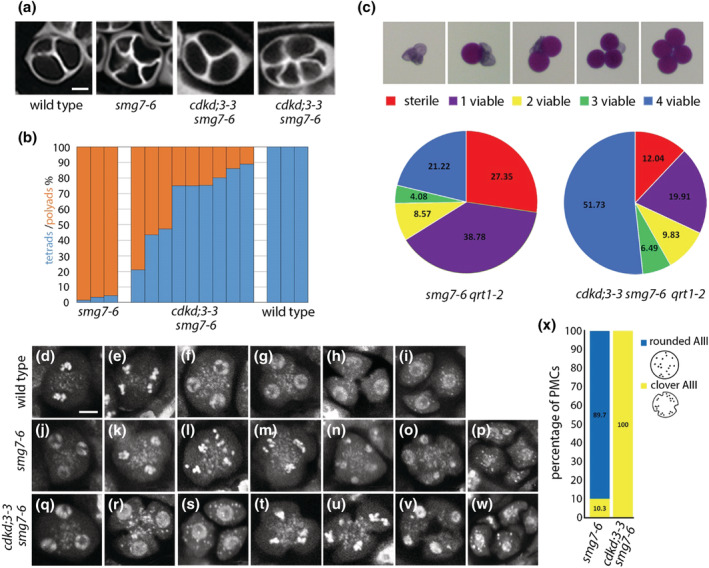
Male meiosis in cdkd;3‐3 smg7‐6. (a) Tetrads and polyads in indicated mutants visualized by callose staining with SCRI Renaissance 2200 (SR2200). Scale bar = 5 μm. (b) Frequency of tetrads and polyads in individual anthers. (c) Viability of pollen derived from the same tetrad assessed by Alexander staining. Pie charts show proportion of tetrads with indicated number of viable pollens in smg7‐6 qrt1‐2 (n = 245 tetrads) and cdkd;3‐3 smg7‐6 qrt1‐2 (n = 1219). (d–w) Pollen mother cells (PMCs) and tetrads stained by DAPI. Labels (k–p) and (t–w) show aberrant post‐meiotic divisions resulting in polyads, whereas labels (g–h) and (q–s) represent regular cytokinesis and tetrad formation. Scale bar = 5 μm; (d) metaphase II, (e) anaphase II, (m, t, u) clover anaphase III, and (l) rounded anaphase III. (x) Frequency of rounded and clover anaphases III in smg7‐6 (n = 183) and cdkd;3‐3 smg7‐6 mutants (n = 74)
Interestingly, we also noticed that a portion of tetrads harboring two or three viable pollen is higher relative to tetrads with no or a single pollen in cdkd;3‐3 smg7‐6 qrt1 compared with smg7‐6 qrt1 (Figure 3c). We assume that PMCs producing 1 to 3 viable pollen represent situations where cells entered aberrant postmeiotic segregation, but the original set(s) of chromosomes did not fully separate and remained clustered forming functional haploid nuclei (Capitao et al., 2021). A higher frequency of tetrads with 2 to 3 viable pollen in cdkd;3‐3 smg7‐6 plants indicated increase propensity of chromosomes to stay together during aberrant divisions.
Cytogenetic analysis showed two populations of PMCs in cdkd;3‐3 smg7‐6 plants: ones that terminated meiosis normally and formed tetrads (Figure 3d–i and q–s) and the other ones that entered aberrant chromosome segregation (Figure 3t–w) typical for smg7‐6 mutants (Figure 3j–p). However, there was a difference between smg7‐6 and cdkd;3‐3 smg7‐6 PMCs undergoing aberrant segregations. Whereas smg7‐6 PMCs progressing through meiosis III were mostly rounded (Figure 3l,x), the majority of cdkd;3‐3 smg7‐6 meiocytes in anaphase III were pinched inwards as if undergoing cytokinesis (referred to as “clover” PMCs; Figure 3t,u,x). This observation indicated that the onset of cytokinesis precedes the completion of aberrant chromosome segregation in cdkd;3‐3 smg7‐6 plants.
We next performed live cell imaging of PMCs to assess meiotic progression in cdkd;3‐3 smg7‐6. We used two experimental setups: In the first one, we measured the duration of stages when chromatin is condensed (meiosis I, meiosis II, and meiosis III) and decondensed (interkinesis I and interkinesis II) using HTA10:TagRFP chromatin marker (Figure 4a, Movies [Link], [Link], [Link]). In the second setup, we measured the time interval between spindle disassembly either in meiosis II or in meiosis III and cytokinesis using the TagRFP:TUB4 reporter for microtubules (Figure 4b, Movies [Link], [Link], [Link]). Together, these markers allowed us to assess the duration of meiosis from diakinesis/metaphase I to cytokinesis. cdkd;3‐3 smg7‐6 meiocytes that formed regular tetrads were compared with wild type, whereas cdkd;3‐3 smg7‐6 meiocytes undergoing meiosis III were compared with smg7‐6. This analysis revealed that all stages of meiosis were delayed in cdkd;3‐3 smg7‐6 plants compared with the respective controls (Figure 4c–e). Nevertheless, the most pronounced difference was observed in interkinesis II and cytokinesis in PMCs undergoing meiosis III, both of which lasted ~1 h longer in cdkd;3‐3 smg7‐6 than in smg7‐6 plants (Figure 4e). We further noticed that chromatids in cdkd;3‐3 smg7‐6 PMCs undergoing anaphase III tend to spread less than chromatids in smg7‐6 (Figure S6).
FIGURE 4.
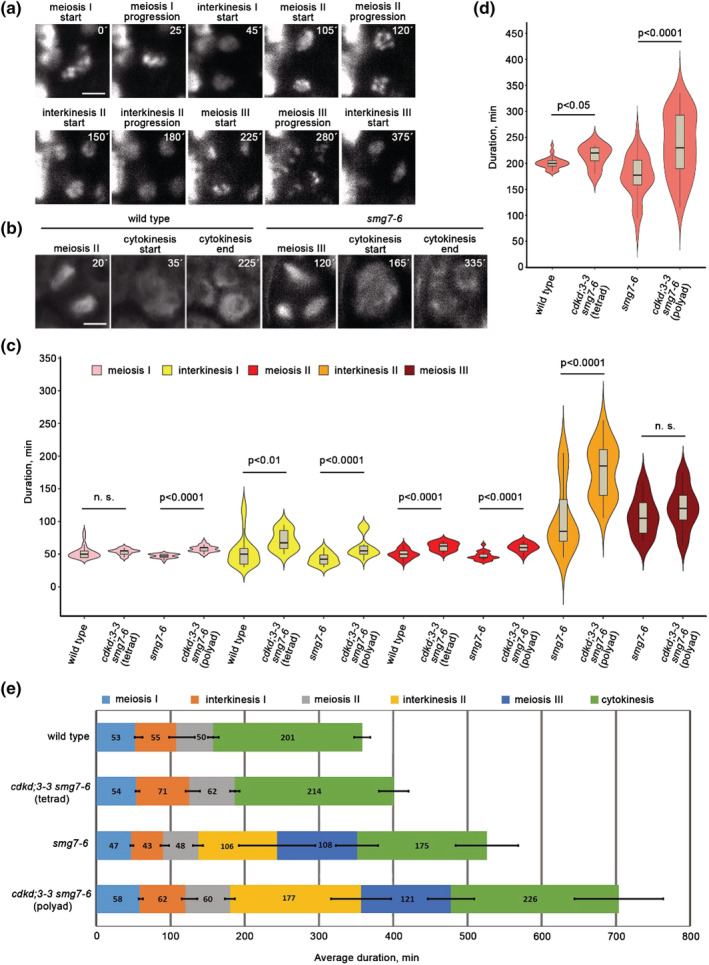
Live imaging of meiotic progression in cdkd;3‐3 smg7‐6. (a) Time‐lapse series of smg7‐6 meiocyte with HTA10:TagRFP labeled chromatin. Time points relate to metaphase I. Scale bar = 5 μm. (b) Time‐lapse series of wild type and smg7‐6 meiocytes containing microtubules labeled with TagRFP:TUB4 reporter. Time points relate to the formation of meiosis I spindle. Scale bar = 5 μm. (c) Violin plots showing duration of individual meiotic stages assessed from live cell imaging of pollen mother cells (PMCs) with labeled chromatin. Significance of the difference is indicated (two tailed t‐test; wild type n = 25, cdkd;3‐3 smg7‐6 (tetrad) n = 16, smg7‐6 n = 18, and cdkd;3‐3 smg7‐6 (polyad) n = 23). (d) Violin plots showing duration of cytokinesis assessed from live cell imaging of PMCs with labeled microtubules. Significance of the difference is indicated (two tailed t‐test; wild type n = 35, cdkd;3‐3 smg7‐6 (tetrad) n = 5, smg7‐6 n = 40, and cdkd;3‐3 smg7‐6 (polyad) n = 35). (e) Graphical representation of meiotic duration based on data from (c) and (d). Error bars represent standard deviations (SDs) for the duration of each meiotic stage.
Together, these data indicate two mechanisms that contribute to the increased fertility and pollen production in cdkd;3‐3 smg7‐6 plants. First, a portion of meiocytes does not enter aberrant postmeiotic divisions and forms normal tetrads after meiosis II. In the remaining PMCs is meiosis III substantially delayed and coincides with the onset of cytokinesis. Cytokinesis restricts the movement of chromatids, which increases the chance for assembly of haploid nuclei with the full set of chromosomes.
2.3. Downregulation of CDKA;1 does not restore meiotic exit in smg7‐6 mutants
Arabidopsis CDKDs were described as the CDKA;1 activating kinases, and CDKA;1 appears to be a key driver of meiotic progression. In Arabidopsis, cytokinesis occurs simultaneously after meiosis is completed (De Storme & Geelen, 2013). Inhibition of cytokinesis after meiosis I is mediated by CDKA;1, and combined reduction of CDKD;3 and CDKA;1 activities led to sequential cytokinesis or premature meiotic exit after meiosis I, depending on the extent of the reduction (Sofroni et al., 2020). Our previous work hinted that aberrant meiotic exit in SMG7 deficient plants may be caused by the failure to downregulate CDKA;1 (Bulankova et al., 2010). Therefore, identification of CDKD;3 in the suppressor screen is consistent with the scenario where CDKD insufficiency leads to a lower activity of CDKA;1, which in turn promotes cytokinesis and meiotic exit in smg7‐6 plants.
To directly test this scenario, we combined smg7‐6 mutation with a line where endogenous CDKA;1 was substituted with its hypomorphic CDKA;1T14V;Y15F variant (CDKA;1VF) that exhibits lower activity (Dissmeyer et al., 2009). The CDKA;1 VF plants have partially reduced fertility and pollen count (Figure 5a–c), which is consistent with the key role of CDKA;1 in meiotic progression. Cytogenetic analysis showed that many CDKA;1VF PMCs initiate cell wall formation prior to the tetrad stage (Figure 5d–f). This leads to partial separation of interkinesis nuclei, which permits more flexibility in spindle orientation during meiosis II, resulting in tetrad configurations reflecting tetrahedral (regular), parallel, or perpendicular divisions (Figure 5d–f). Surprisingly, CDKA;1VF did not suppress the smg7‐6 phenotype, and meiocytes in CDKA;1 VF smg7‐6 plants progressed into aberrant post‐meiotic divisions and formed polyads (Figure 5d–f). Furthermore, smg7‐6 prevented the partial cytokinesis observed in CDKA;1 VF plants (Figure 5e). These data indicate that suppression of aberrant meiosis and promotion of cytokinesis in EMS58‐1 plants are mediated through a CDKA;1 independent mechanism.
FIGURE 5.
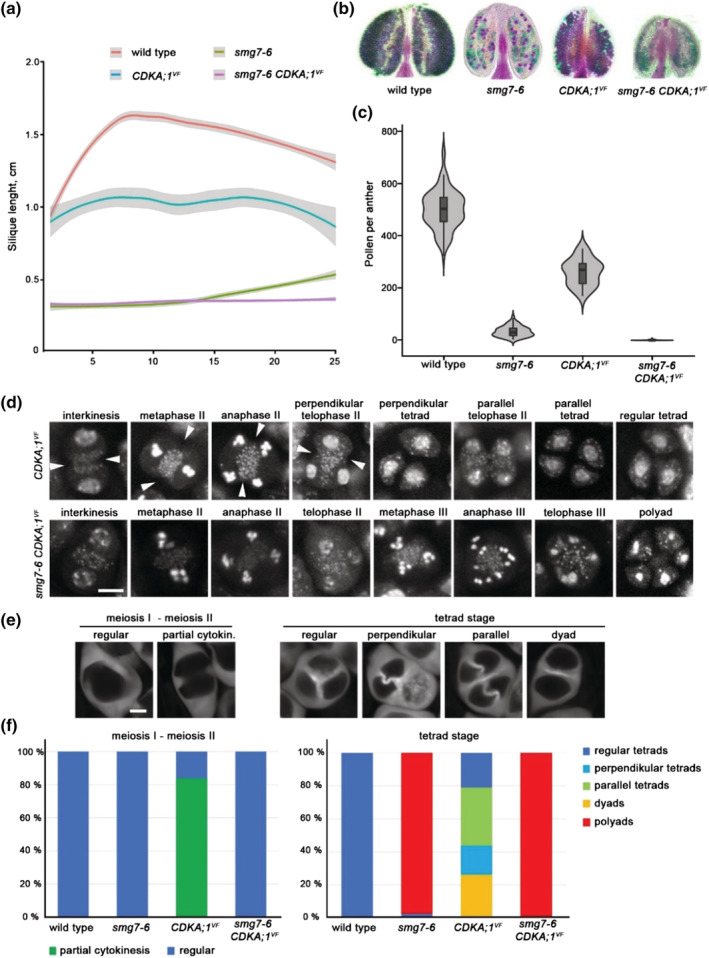
Genetic interaction between CDKA;1 VF and smg7‐6. (a) Quantification of silique length along the main inflorescence bolt in wild type and indicated mutants (wild type n = 25, smg7‐6 n = 25, CDKA;1 VF n = 9, and CDKA;1 VF smg7‐6 n = 23 plants). The trend lines of the data were plotted by LOESS smooth function (colored lines; shaded area represents 95% confidence intervals). (b) Anthers of indicated mutants after Alexander staining. (c) Violin plots showing viable pollen per anther (wild type n = 96, smg7‐6 n = 94, CDKA;1 VF n = 30, and CDKA;1 VF smg7‐6 n = 116). (d) Cytogenetic analysis of meiosis II and cytokinesis in DAPI‐stained pollen mother cells (PMCs). Arrowheads point to cell wall invaginations prior to completion of meiosis II. Scale bar = 5 μm. (e) Callose staining of CDKA;1 VF PMCs with SR2200 dye to visualize cell wall. Scale bar = 5 μm. (f) Quantification of PMCs undergoing partial cytokinesis prior to completion of chromosome segregation (left panel; wild type n = 86, smg7‐6 n = 166, CDKD;1 VF n = 155, and smg7‐6 CDKD;1 VF n = 95) and tetrad configurations after cytokinesis (right panel; wild type n = 200, smg7‐6 n = 524, CDKD;1 VF n = 256, and smg7‐6 CDKD;1 VF n = 246)
To further explore the role of CDKD;3 in meiotic cytokinesis, we analyzed its genetic interaction with tardy asynchronous meiosis (TAM) (Wang, Magnard, et al., 2004). TAM is an A‐type cyclin expressed during meiosis I whose inactivation results in meiotic exit after meiosis I and formation of diploid microspores. Due to the absence of meiosis II, the pollen count in tam mutants is reduced to half compared with wild type (Figure 6a,b). Live cell imaging revealed that tam mutants also form ectopic spindle‐like and phragmoplast‐like structures in the cytoplasm of prophase I PMCs (Prusicki et al., 2019). Consistent with these data, we observed that PMCs in TAM deficient plants are often pinched and form bud‐like structures that indicate attempts of ectopic and premature cytokinesis (Figure 6c,d). The ectopic cytokinesis was substantially reduced in cdkd;3‐3 tam double mutants (Figure 6e). Thus, the effect of cdkd;3‐3 mutation on cytokinesis is context‐dependent: While it reduces the premature cytokinesis during prophase I in TAM‐null plants, it promotes cytokinesis after meiosis II in smg7‐6 mutants.
FIGURE 6.
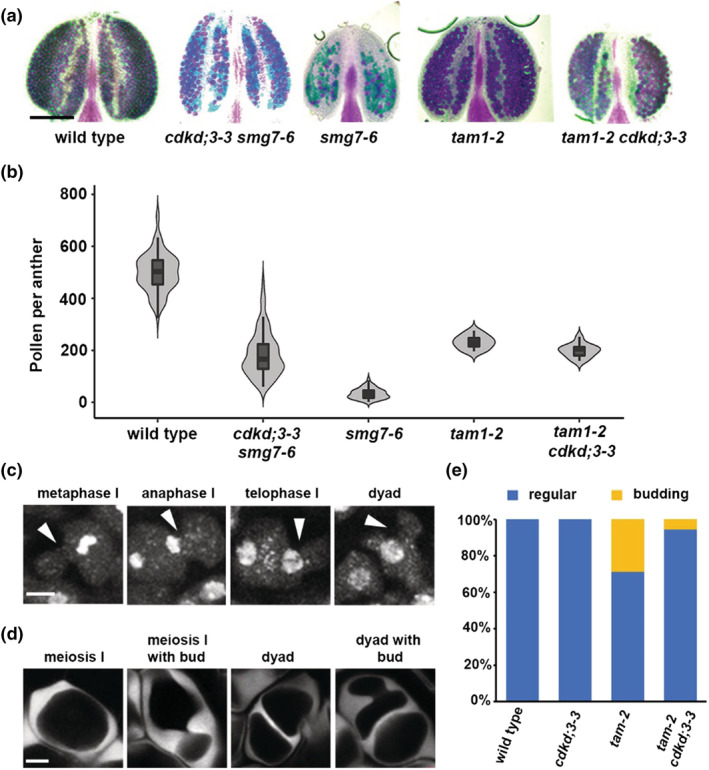
Genetic interaction between tam and cdkd;3‐3. (a) Anthers of indicated mutants after Alexander staining. (b) Violin plots showing viable pollen per anther (wild type n = 96, cdkd;3‐3 smg7‐6 n = 97, smg7‐6 n = 94, tam‐2 n = 10, and tam‐2 cdkd;3‐3 n = 13). (c) Cytogenetic analysis of meiosis I in DAPI‐stained pollen mother cells (PMCs) of tam. Arrowheads point to ectopic cell wall invaginations that form buds during metaphase I. Scale bar = 5 μm. (d) Callose staining of tam PMCs with SR2200 dye to visualize cell wall. Scale bar = 5 μm. (e) Quantification of PMCs exhibiting ectopic cell wall formation and budding (wild type n = 386, cdkd;3‐3 n = 455, tam‐2 n = 187, and tam‐2 cdkd;3‐3 n = 101)
2.4. Exploration of alternative mechanisms of CDKD;3 action by transcriptomics and proteomics
Our data indicate that cdkd;3‐3 may restore the fertility of smg7‐6 plants independently of the CDKD function in activating CDKA;1. The CTD domain of RNA PolI is so far the only other known substrate of CDKDs besides CDKA;1. Therefore, to assess the impact of CDKD;3 on transcription, we compare RNA‐seq data obtained from rosette leaves of wild type, cdkd;3‐1, and smg7‐6 mutants as well as of cdkd;3‐1 smg7‐6 double mutants. Interestingly, cdkd;3‐1 caused deregulation of only 114 genes compared with wild type with 105 genes upregulated and nine genes downregulated (Figure 7a; Table S2). The vast majority of these genes (91/114) were also altered in smg7‐6 mutants, where in total 656 genes were deregulated. This is a striking result, as it indicates that CDKD3 targets almost exclusively genes also regulated by SMG7.
FIGURE 7.
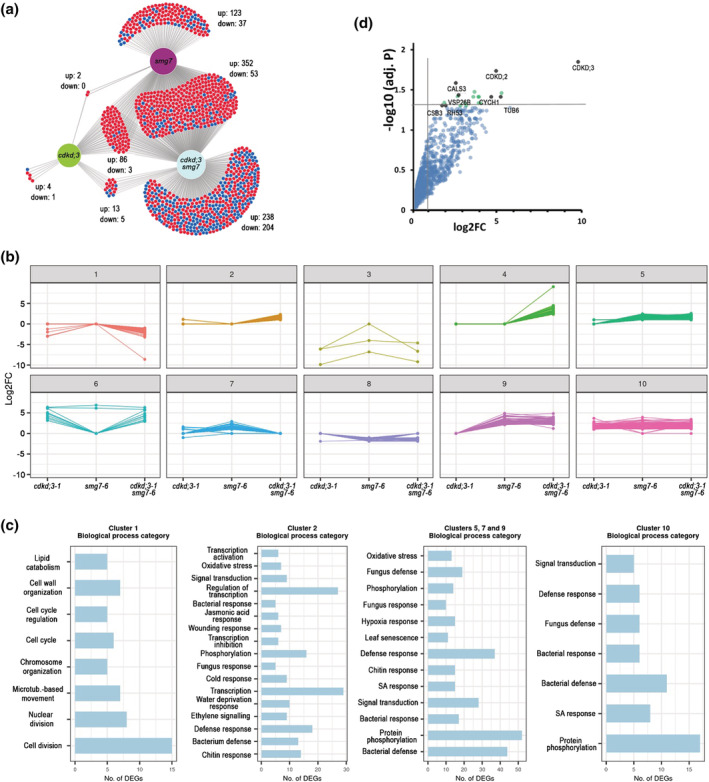
Transcriptomics and proteomics analysis of CDKD;3. (a) Venn diagram showing overlaps between differentially expressed genes relative to wild type in cdkd;3‐1, smg7‐6, and cdkd;3‐1 smg7‐6 mutants. Numbers of up‐ and down‐regulated genes are indicated. (b) K‐means clustering of differentially expressed genes. Clustering was performed on log2FC to build expression patterns in 10 clusters for 1125 differentially expressed protein‐coding genes. (c) Subsequent gene ontology term (GO‐term) enrichment analysis of selected clusters. Genes in the clusters were analyzed with regard to their biological function by GO enrichment analysis (p‐value <.05; Benjamini–Hochberg test). Most enriched GO categories are shown for representative categories of clusters. (d) Dot plot of proteins identified by LC–MS in pull‐downs from wild type and CDKD;3:YFP expressing plants using Green fluorescent protein (GFP) antibody. Proteins with the highest confidence of enrichment are indicated as black dots. Fold change (log2FC) and significance of the difference between wild type and CDKD;3:YFP samples are plotted.
To further analyze the expression datasets, we clustered genes based on their differential expression in different mutant combinations in 10 clusters and performed gene ontology term (GO‐term) enrichment analysis (Figure 7b,c). Genes upregulated in all three mutant combinations including cdkd;3‐1 (cluster 10) were almost exclusively enriched for GO‐terms associated with pathogen defense response. Pathogen defense and oxidative stress were also enriched in clusters 5, 7, and 9, which include genes upregulated in smg7‐6 and cdkd;3‐1 smg7‐6, as well as in cluster 2 encompassing genes upregulated only in double mutants (Figure 7b,c). SMG7 controls pathogen response via suppressing plant intracellular immune receptors, and its inactivation leads to a strong autoimmune response (Gloggnitzer et al., 2014; Riehs‐Kearnan et al., 2012). This analysis indicates that even the relatively mild smg7‐6 allele with respect to NMD causes substantial upregulation of pathogen defense genes. CDKD;3 inactivation also leads to pathogen response, though much milder than in smg7‐6. Moreover, cdkd;3‐1 further enhances pathogen and other stress responses when combined with smg7‐6 (cluster 2). The cdkd;3‐1 smg7‐6 mutants also show a large portion of downregulated genes (Figure 7a). Their GO‐term analysis revealed strong enrichment for genes involved in cell cycle and division (cluster 1, Figure 7b,c).
CDKD;3 might also phosphorylate additional substrates besides CDKA;1 and RNA PolII. To identify putative CDKD3;1 targets, we generated an Arabidopsis line expressing the CDKD;3::CDKD;3:YFP construct and performed immunoprecipitation coupled with mass spectrometry from floral bud protein extracts. The most significantly enriched proteins were CDKD;3, CDKD;2, and cyclin H, which reaffirmed the specificity of the immunoprecipitation protocol (Figure 7d). The other proteins with the highest confidence enrichment in the immunoprecipitated fraction were callose synthase 3 (CALS3; At5g13000), tubulin beta‐6 (TUBB6, At5g12250), and components of vesical trafficking systems coatomer subunit beta‐3 (CSB3, At3g15980) and vacuolar protein sorting‐associated protein 26B (VPS26B, At4g27690). Notably, the molecular functions of these proteins can be related to cytokinesis. While CDKD;3 resides most of the cell cycle in nuclei (Sofroni et al., 2020), it is released into the cytoplasm during the M‐phase (Movie S7), and therefore, it has an opportunity to interact with these non‐nuclear proteins.
3. DISCUSSION
Arabidopsis deficient in SMG7 exhibits an unusual meiotic phenotype that is characterized by a failure to complete meiosis and proceed to gametophytic development. Plants lacking SMG7 function arrest in meiotic anaphase II, whereas hypomorphic smg7‐6 mutants expressing SMG7 N‐terminus form haploid nuclei that undergo multiple cycles of aberrant chromosome segregation without DNA replication (Capitao et al., 2021; Riehs et al., 2008). These additional meiotic cycles resemble meiosis II, where chromatid segregation also proceeds without DNA duplication. The key mechanism driving exit from the M‐phase is CDK downregulation via anaphase promoting complex (APC/C) mediated degradation of M‐type cyclins. This results in dephosphorylation of CDK substrates reverting molecular processes that led to the M‐phase and allowing for chromosome decondensation, spindle disassembly, formation of nuclei, and eventually cytokinesis (Lopez‐Aviles et al., 2009; Potapova et al., 2006). Further decrease of CDK activity in G1 creates permissive conditions for licensing of origins of replication, which ensures the alternation of DNA replication and chromosome segregation. Therefore, the gradual downregulation of CDK activity provides order and directionality to molecular processes that occur during transition from M to G1. However, M‐phase exit is not irreversible. Pharmacological manipulation of CDK activity during mitotic exit in human cells expressing nondegradable cyclin B could revert cells back to M‐phase (Potapova et al., 2006). This principle is utilized in interkinesis between meiosis I and II when a partial inhibition of APC/C prevents full CDK inactivation and leads to reversion to M‐phase prior to licensing of replication origins (Cromer et al., 2012; Izawa et al., 2005).
The Arabidopsis smg7 phenotypes indicate insufficient downregulation of CDK activity at the end of meiosis. In support of this hypothesis, failure to fully activate APC/C and to downregulate CDK activity during meiotic exit in Drosophila and budding yeast has been reported to result in phenotypes similar to the ones described in smg7 Arabidopsis (Chu et al., 2001; Page & Orr‐Weaver, 1996; Wang et al., 2020). Thus, it is expected that mutations in genes that contribute to CDK activity would alleviate meiotic defects caused by SMG7 dysfunction. Indeed, we identified cdkd;3‐3 in the suppressor screen designed to uncover such mutations. The cdkd;3‐3 allele restores fertility of smg7‐6 plants by promoting orderly cytokinesis and tetrad formation, or, in PMCs that enter meiosis III by delaying the onset of aberrant chromosome segregation after initiation of cytokinesis (Figure 2t,u). Because high CDK activity prevents transition to cytokinesis in plants (Sasabe et al., 2011), these results are consistent with the idea that CDKD;3 is a CDK activator. CDKD;3 was reported to phosphorylate CDKA;1 and act as CDKA;1 activating kinase (Sofroni et al., 2020; Yamaguchi et al., 1998). However, we failed to mimic the cdkd;3‐3 effect on meiotic exit by the hypomorphic CDKA;1VF, which exhibits low CDK activity (Dissmeyer et al., 2009). This indicates that meiosis II is governed by another CDK, perhaps one of the CDKBs that were implicated to act together with B1 cyclins in microtubule organization during mitosis (Romeiro Motta et al., 2022).
The observation that CDKA;1VF does not promote meiotic exit in smg7‐6 mutants opens up alternative scenarios, in which CDKD;3 affects meiotic exit through regulating other substrates than CDKs. Plant CDKDs are orthologous to human CDK7 kinases that are implicated in regulating transcription and RNA processing through phosphorylation of RNA Pol II (Fisher, 2019; Joubes et al., 2000). Whereas CDKD;3 has been reported to phosphorylate RNA Pol II in vitro (Hajheidari et al., 2012; Shimotohno et al., 2003; Yamaguchi et al., 1998), our transcriptomics analysis in cdkd;3‐1 mutant revealed only negligible impact on transcriptome likely due to functional redundancy between Arabidopsis CDKDs. Interestingly, the majority of upregulated transcripts were associated with pathogen response and almost perfectly overlapped with transcripts deregulated in smg7‐6 mutants. We observed that the combination of cdkd;3‐1 and smg7‐6 further enhanced upregulation of pathogen response and led to the downregulation of genes involved in cell cycle and division.
CDKD;3 may also phosphorylate additional proteins and affect the meiotic exit through direct regulation of proteins executing cell division. Remarkably, the top candidates we co‐immunoprecipitated with CDKD;3 can be linked to cytokinesis. One such protein, TUB6, is part of the β‐tubulin family that forms microtubules. Microtubules play a central role in mitosis and cytokinesis, and phosphorylation of β‐tubulin was proposed to regulate microtubule dynamics in human (Fourest‐Lieuvin et al., 2006). Another candidate, CALS3, belongs to a family of callose synthases that mediate callose deposition in a wide range of cell types including meiocytes. Some members of the family have also been implicated in cytokines and microspore formation (Chen et al., 2009; De Storme & Geelen, 2013). We have also identified two proteins involved in endosomal trafficking important for the transport and turnover of plasma membrane proteins and cell wall biosynthetic enzymes. VPS26B is a subunit of the retromer complex that mediates protein recycling between Golgi apparatus, plasma membrane, and vacuoles (Paez Valencia et al., 2016). CSB3 is a subunit of COPI coatomer complex responsible for Golgi vesicle trafficking, and its suppression results in aberrant cell plate formation during cytokinesis in tobacco cells (Ahn et al., 2015). To conclude, besides CDKA;1 and RNA Pol II, CDKD;3 may interact with and phosphorylate proteins involved in cytokinesis.
Our current work presents a more nuanced picture of the CDKD;3's role in meiosis. In addition to dictating the simultaneous rather than sequential meiotic cytokinesis typical for dicots by inhibiting cell wall formation during interkinesis (Sofroni et al., 2020), CDKD;3 can also affect meiotic exit by influencing the rate of meiotic progression and onset of cytokinesis. These abilities are likely facilitated through regulation of not only CDKA;1 but also additional protein substrates involved in cell division.
4. EXPERIMENTAL PROCEDURES
4.1. Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col‐0) and mutant seeds were grown on soil in growth chambers at 21°C at 50%–60% humidity with 16/8 h light/dark illumination. EMS58‐1 plants characterized in Figure 1 are derived from B3 seeds generated from the backcross between a M2 plant derived from the mutagenized smg7‐6 plants and non‐mutagenized parent. To generate reporter lines for live cell imaging or double mutants, the B3 plants were crossed with mutant/reporter lines, and F3 or F4 plants from these crosses were analyzed in Figures 3, 4, and 6. The following mutant lines were used in this study: smg7‐6 (Riehs‐Kearnan et al., 2012), tam‐2 (Bulankova et al., 2010), cdka;1‐1 CDKA;1VF (Dissmeyer et al., 2009), cdkd;3‐1, cdkd;2‐1, and cdkd;1‐1 (Hajheidari et al., 2012; Shimotohno et al., 2006). Mutations were assessed by PCR genotyping using primers described in Table S3. Plants used for live‐cell imaging were generated by crossing plants containing reporter constructs HTA10:RFP (Valuchova et al., 2020) and pRPS5A::TagRFP:TUB4 (Prusicki et al., 2019) with the parental smg7‐6 and the EMS58‐1 line.
4.2. Generation of transgenic lines
For the complementation study, a PCR amplified CDKD;3 gene including 471 bp region upstream of the start codon and the 450 bp downstream of stop codon was generated using the primers provided in Table S3. The amplicon was cloned into the vector pENTR™/D‐TOPO®Vector (Invitrogen) and transferred by Gateway LR reaction into the binary pGWB601 vector (Nakagawa et al., 2007). The construct was transformed by the floral dipping method into the EMS58‐1 line, and T1 transformants were selected with Basta. The CDKD;3:YFP (yellow fluorescent protein) reporter line was generated the same Gateway cloning strategy by introducing the CDKD;3 gene fragment devoid of stop codon PCR amplified with primers indicated in Table S3 into the destination vector pGWB640 containing C terminus YFP tag. The resulting construct was transformed into wild‐type Col‐0 plants. Transformants were Basta selected and propagated up to the T3 generation.
4.3. Assessment of plant fertility
Pollen viability was determined by Alexander staining (Alexander, 1969). The cell count plugin on FIJI (Schindelin et al., 2012) was used to count viable pollen. Silique length was assessed from images of main stems scanned by an Epson scanner and the siliques were measured using the FIJI Analyze/Measure function.
4.4. Genetic mapping and complementation
EMS58‐1 seeds were obtained by the suppressor screening in smg7‐6 background described in Capitao et al. (2021). Mutations associated with improved fertility were identified by whole genome sequencing of pooled fertile plants using ArtMAP (Javorka et al., 2019). The identified cdkd;3‐3 mutation was genotyped by the high‐resolution melting (HRM) qPCR‐based method using primers described in Table S3 using Lightcycler96 thermocycler (Roche) using the inbuilt HRM profile.
4.5. Cytology
Staining of PMCs in whole anthers was performed as previously described (Capitao et al., 2021) using 2 μg/ml 4′,6‐diamidino‐2‐phenylindole (DAPI) stock solution for DNA staining or .1% (v/v) solution of SR2200 (Renchem) for callose staining. Images were acquired by LSM700 confocal microscope (1024p resolution, emission/absorption wavelengths of 405/435, 1 AU, 35% laser power).
4.6. Live cell imaging
Live imaging was performed as previously described (Valuchova et al., 2020). Floral buds .3 to .7 mm long were selected from the main inflorescence. The reproductive organs were exposed by removing the sepals and placed into glass capillaries (size 4, Zeiss) containing 1/2 Murashige and Skoog (MS) medium (5% sucrose, pH 5.8) with 1% low melting point agarose (Sigma Aldrich). The inflorescence embedded in solidified MS was pushed out of the capillary and then placed into the capillary holder for the Z.1 ZEISS light‐sheet microscope. After the holder is inserted in the microscope chamber, the remaining space is filled with liquid 1/2 MS medium (5% sucrose, pH 5.8). Images were taken every 5 min with a 10× objective (detection optics 10×/.5), single illumination (illumination optics 10×/.2), 561‐nm laser (15% intensity). The large raw data files were processed by ZEN Blue software (Zeiss).
4.7. Transcriptome analysis
Leaf tissue was collected from three week‐old plants, and total RNA was extracted using RNA Blue (Top‐Bio), following the manufacturer's protocol. For each sample, 10 μg of total RNA was treated with DNase I (Roche). Strand‐specific libraries were prepared from ribosomal RNA‐depleted total RNA using ScriptSeq complete kit for plant leaf (Epicentre; BPL1224) following manufacturer's protocol. After assessing the quality and quantity of RNASeq libraries by fragment analyzer, libraries were 125/150 bp paired‐end sequenced on Illumina HiSeqv4/NextSeq. Low‐quality reads and adapters were trimmed from the total read pairs using cutadapt v2.5. Then, the RNA‐Seq data were aligned on reference AtRTD2 transcript dataset (Zhang et al., 2017) and simultaneously quantified with quasi‐mapping‐based algorithm using Salmon v1.5 (Patro et al., 2017). The downstream data normalization and differential expression analysis was done using 3D RNA‐Seq (Calixto et al., 2018). A gene was significantly differentially expressed (DE) in a contrast group if it had adjusted p‐value <.05 (Benjamini–Hochberg [BH]‐corrected) (Benjamini & Yekutieli, 2001) and absolute log2FC ≥ 1. The transcript overlaps among datasets were visualized by a custom Venn diagram tool available at https://divenn.tch.harvard.edu/. Further, functional annotation and GO enrichment were done online via DAVID bioinformatic database (https://david.ncifcrf.gov/summary.jsp; Sherman et al., 2022). Next, to categorize the DE genes with similar expression patterns, k‐means clustering was performed, and plots were generated using R packages.
4.8. Immunoprecipitation and mass spectrometry
Meiotic buds from ~150 inflorescences from CDKD;3:YFP and wild type control were grinded in liquid nitrogen and processed using the ChromoTek GFP‐Trap® Magnetic Agarose kit (Chromotek). Briefly, the sample was mixed with 200 μl of RIPA buffer (NEB) supplemented with 2.5 μl of DNAse I (NEB), 10 μl of 50 mM MgCl2, and 3.5 μl of protease and phosphatase inhibitor cocktail (Halt) and incubated on ice for 45 min. Lysates were sedimented by centrifugation, and the supernatant was transferred to a new 1.5 ml tube containing 450 μl ice‐cold dilution buffer included in the kit and 6.5 μl of protease and phosphatase inhibitor cocktail. Two hundred micrograms of the extracted proteins was added to 25 μl of equilibrated magnetic agarose beads (Chromotek) and mixed in a rotating shaker for 45 min at 4°. The supernatant was mixed with equilibrated ChromoTek GFP‐Trap® Magnetic Agarose beads GFP‐Trap MA beads and incubated for 90 min at 4 °C. After washing with the dilution buffer, the bead bound protein complexes were digested directly on beads by addition of .5 μg (1 μg/μl) of trypsin (sequencing grade, Promega) in 50 mM NaHCO3 buffer and incubated at 37°C for 18 h. Resulting peptides were extracted into liquid chromatography mass spectrometry (LC–MS) vials by 2.5% formic acid and acetonitrile with addition of polyethylene glycol (20,000; final concentration .001%) and concentrated in a SpeedVac concentrator (Thermo Fisher Scientific).
LC–MS/MS analyses of all peptide mixtures were done using Ultimate 3000 RSLCnano system connected to Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). Prior to LC separation, tryptic digests were online concentrated and desalted using trapping column (100 μm × 30 mm, 3.5 μm particles, X‐Bridge BEH 130 C18 sorbent; Waters, Milford, MA, USA; temperature of 40 °C). After washing of trapping column with .1% formic acid (FA), the peptides were eluted (flow rate—500 nl/min) from the trapping column onto an analytical column (Acclaim Pepmap100 C18, 3 μm particles, 75 μm × 500 mm; column compartment temperature of 40 °C, Thermo Fisher Scientific) by 135 min nonlinear gradient program (1%–56% of mobile phase B; mobile phase A: .1% FA in water; mobile phase B: .1% FA in 80% acetonitrile). The analytical column outlet was directly connected to the Digital PicoView 550 (New Objective) ion source with sheath gas option and SilicaTip emitter (New Objective; FS360‐20‐15‐N‐20‐C12) utilization. ABIRD (Active Background Ion Reduction Device, ESI Source Solutions) was installed.
MS data were acquired in a data‐dependent strategy with defined number of scans based on precursor abundance with survey scan (m/z 350–2000). The resolution of the survey scan was 120,000 (at m/z 200) with a target value of 4 × 105 ions and maximum injection time of 100 ms. Higher energy collisional dissociation (HCD) MS/MS (30% relative fragmentation energy, normal mass range) spectra were acquired with a target value of 5.0 × 104 and resolution of 15,000 (at m/z 200). The maximum injection time for MS/MS was 22 ms. Dynamic exclusion was enabled for 30 s after one MS/MS spectra acquisition. The isolation window for MS/MS fragmentation was set to 1.2 m/z.
The analysis of the mass spectrometric RAW data files was carried out using the MaxQuant software (version 1.6.2.0) using default settings unless otherwise noted. MS/MS ion searches were done against modified cRAP database (based on http://www.thegpm.org/crap) containing protein contaminants like keratin and trypsin and UniProtKB protein database for Arabidopsis thaliana (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/reference_proteomes/Eukaryota/UP000006548_3702.fasta.gz; downloaded May 2018, version 2018/05, number of protein sequences: 27,582). Oxidation of methionine and proline, deamidation (N and Q) and acetylation (protein N‐terminus) as optional modification, and trypsin/P enzyme with 2 allowed missed cleavages were set. Peptides and proteins with false discovery rate threshold <.01 and proteins having at least one unique or razor peptide were considered only. Match between runs was set among all analyzed samples. Protein abundance was assessed using protein intensities calculated by MaxQuant. Protein intensities reported in proteinGroups.txt file (output of MaxQuant) were further processed using the software container environment (https://github.com/OmicsWorkflows), version 3.7.2a.
CONFLICT OF INTEREST
The Authors did not report any conflict of interest.
AUTHOR CONTRIBUTIONS
S.T., A.S. and K.R. designed the work. S.T., N.S., A.C. and C.C. performed the experiments. N.S. analyzed transcriptomics data. P.M. and S.V. contributed to bioimaging. R.S.G. and Z.Z. performed the mass spectrometry. V.K.R. contributed to data analysis. K.R. and S.T. wrote the manuscript. All authors revised the manuscript.
Supporting information
Movie S1. Time‐lapse microscopy of meiotic divisions in wild type PMCs from diakinesis till telophase II in 5 min increments. Chromatin was labeled with HTA10:TagRFP reporter. Scale bar = 20 μm.
Movie S2. Time‐lapse microscopy of meiotic divisions in smg7‐6 PMCs from diakinesis till telophase III in 5 min increments. Chromatin was labeled with HTA10:TagRFP reporter. Scale bar = 20 μm.
Movie S3. Time‐lapse microscopy of meiotic divisions in cdkd;3‐3 smg7‐6 PMCs from diakinesis till telophase III in 5 min increments. Chromatin was labeled with HTA10:TagRFP reporter. Scale bar = 20 μm.
Movie S4. Time‐lapse microscopy of meiotic divisions in wild type PMCs from interkinesis till the tetrad stage in 5 min increments. Microtubules were labeled with GFP:TUA5 reporter. Scale bar = 20 μm.
Movie S5. Time‐lapse microscopy of meiotic divisions in smg7‐6 PMCs from interkinesis, through meiosis II and meiosis III till the polyad stage in 5 min increments. Microtubules were labeled with GFP:TUA5 reporter. Scale bar = 20 μm.
Movie S6. Time‐lapse microscopy of meiotic divisions in cdkd;3‐3 smg7‐6 PMCs from interkinesis, through meiosis II and meiosis III till the polyad stage in 5 min increments. Note that some PMCs do not proceed in meiosis III and form regular tetrads. Microtubules were labeled with GFP:TUA5 reporter. Scale bar = 20 μm.
Movie S7. Time‐lapse microscopy of CDKD;3 localization in PMCs from late prophase I till telophase II s in 5 min increments. CDKD;3 and chromatin were visualized using CDKD;3:YFP (green) and HTA10:TagRFP (magenta) reporters, respectively.
Figure S1. Arabidopsis cdkd and smg7‐6 mutants. Approximately 6‐weks old plants are shown. Longer inflorescence bolt in the smg7‐6 mutant is caused by reduced fertility.
Figure S2. Mapping mutations associated with the rescued fertility in EMS58‐1 by whole genome sequencing. The causative mutation is recessive and is, therefore, expected to fully associate with the rescued phenotype. The charts indicate positions of de novo mutations on individual Arabidopsis chromosomes (x axis) and their frequency in population of B2 plants exhibiting increased fertility. Colored diamonds on chromosome 1 mark de novo mutations with the highest association. This region is zoomed in the lower left graph with indicated AGI codes for the mutated genes.
Figure S3. Genetic complementation of EMS58‐1 plants with the CDKD;3 genic construct causes infertility in transformed T1 plants. The chart shows average silique length in individual T1 transformants (error bars indicate SD; n = 25). Data for smg7‐6 are shown in red, for EMS58‐1 in gray, for complemented T1 transformants in blue and for non‐complemented transformants in green.
Figure S4. Fertility of cdkd;3‐3 mutants. (a) Quantification of silique length along the main inflorescence bolt in wild type (n = 10) and cdkd;3‐3 (n = 5). The trend lines of the data were plotted by LOESS smooth function (colored line; shaded area represents 95% confidence intervals). (b) Anthers from wild type and cdkd;3‐3 plants after Alexander staining. (c) Violin plots showing viable pollen per anther. Significance of the difference is indicated (two tailed t‐test; wild type n = 20; cdkd;3‐3 = 5). (d) Tetrads in anther lobes stained with DAPI.
Figure S5. Approximately six weeks old mutant plants.
Figure S6. Time‐lapse series of smg7‐6 and EMS58‐1 meiotic nuclei undergoing meiosis II and meiosis III. Nuclei derived from the same PMC are encircled with identical color. Note that chromatid tends to spread over larger area during anaphase III in smg7‐6 mutants. Chromatin is visualized with HTA10:TagRFP marker. Scale bar = 5 μm.
Table S1: De novo mutations detected in EMS58‐1.
Table S2: List of differentially expresssed genes relative to wild type.
Table S3. List of primers.
ACKNOWLEDGMENTS
We acknowledge the support from CEITEC MU Core facilities Plant Sciences, Proteomics, and CELLIM, supported by the Czech‐Bioimaging (No. LM2018129) and CIISB (No. LM2018127) infrastructure projects funded by MEYS CZ. The genome sequencing was performed by the Next Generation Sequencing Facility at Vienna BioCenter Core Facilities (VBCF), member of the Vienna BioCenter (VBC), Austria. This work was supported by the Ministry of Education, Youth, and Sports of the Czech Republic, the European Regional Development Fund‐Project ‘REMAP’ (No. CZ.02.1.01/.0/.0/15_003/0000479 to K.R.), the Czech Science Foundation (grant 21‐25163J to K.R.) and German Research Foundation (grant 452003411 to A.S.).
Tanasa, S. , Shukla, N. , Cairo, A. , Ganji, R. S. , Mikulková, P. , Valuchova, S. , Raxwal, V. K. , Capitao, C. , Schnittger, A. , Zdráhal, Z. , & Riha, K. (2023). A complex role of Arabidopsis CDKD;3 in meiotic progression and cytokinesis. Plant Direct, 7(3), e477. 10.1002/pld3.477
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ahn, H. K. , Kang, Y. W. , Lim, H. M. , Hwang, I. , & Pai, H. S. (2015). Physiological functions of the COPI complex in higher plants. Molecules and Cells, 38, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technology, 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Azumi, Y. , Liu, D. , Zhao, D. , Li, W. , Wang, G. , Hu, Y. , & Ma, H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin‐like protein. The EMBO Journal, 21, 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics, 29, 1165–1188. 10.1214/aos/1013699998 [DOI] [Google Scholar]
- Bulankova, P. , Akimcheva, S. , Fellner, N. , & Riha, K. (2013). Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genetics, 9, e1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankova, P. , Riehs‐Kearnan, N. , Nowack, M. K. , Schnittger, A. , & Riha, K. (2010). Meiotic progression in Arabidopsis is governed by complex regulatory interactions between SMG7, TDM1, and the meiosis I‐specific cyclin TAM. Plant Cell, 22, 3791–3803. 10.1105/tpc.110.078378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo, A. , Vargova, A. , Shukla, N. , Capitao, C. , Mikulkova, P. , Valuchova, S. , Pecinkova, J. , Bulankova, P. , & Riha, K. (2022). Meiotic exit in Arabidopsis is driven by P‐body‐mediated inhibition of translation. Science, 377, 629–634. [DOI] [PubMed] [Google Scholar]
- Calixto, C. P. G. , Guo, W. , James, A. B. , Tzioutziou, N. A. , Entizne, J. C. , Panter, P. E. , Knight, H. , Nimmo, H. G. , Zhang, R. , & Brown, J. W. S. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell, 30, 1424–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitao, C. , Tanasa, S. , Fulnecek, J. , Raxwal, V. K. , Akimcheva, S. , Bulankova, P. , Mikulkova, P. , Crhak Khaitova, L. , Kalidass, M. , Lermontova, I. , Mittelsten Scheid, O. , & Riha, K. (2021). A CENH3 mutation promotes meiotic exit and restores fertility in SMG7‐deficient Arabidopsis. PLoS Genetics, 17, e1009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. Y. , Liu, L. , Lee, E. , Han, X. , Rim, Y. , Chu, H. , Kim, S. W. , Sack, F. , & Kim, J. Y. (2009). The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiology, 150, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, T. , Henrion, G. , Haegeli, V. , & Strickland, S. (2001). Cortex, a Drosophila gene required to complete oocyte meiosis, is a member of the Cdc20/fizzy protein family. Genesis, 29, 141–152. [DOI] [PubMed] [Google Scholar]
- Cromer, L. , Heyman, J. , Touati, S. , Harashima, H. , Araou, E. , Girard, C. , Horlow, C. , Wassmann, K. , Schnittger, A. , De Veylder, L. , & Mercier, R. (2012). OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genetics, 8, e1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme, N. , & Geelen, D. (2013). Cytokinesis in plant male meiosis. Plant Signaling & Behavior, 8, e23394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer, N. , Nowack, M. K. , Pusch, S. , Stals, H. , Inze, D. , Grini, P. E. , & Schnittger, A. (2007). T‐loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell, 19, 972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer, N. , Weimer, A. K. , Pusch, S. , De Schutter, K. , Kamei, C. L. , Nowack, M. K. , Novak, B. , Duan, G. L. , Zhu, Y. G. , De Veylder, L. , & Schnittger, A. (2009). Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell, 21, 3641–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. P. (2019). Cdk7: A kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription, 10, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourest‐Lieuvin, A. , Peris, L. , Gache, V. , Garcia‐Saez, I. , Juillan‐Binard, C. , Lantez, V. , & Job, D. (2006). Microtubule regulation in mitosis: Tubulin phosphorylation by the cyclin‐dependent kinase Cdk1. Molecular Biology of the Cell, 17, 1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, K. E. , Lam, S. Y. , & Copenhaver, G. P. (2006). Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology, 142, 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer, J. , Akimcheva, S. , Srinivasan, A. , Kusenda, B. , Riehs, N. , Stampfl, H. , Bautor, J. , Dekrout, B. , Jonak, C. , Jimenez‐Gomez, J. M. , Parker, J. E. , & Riha, K. (2014). Nonsense‐mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host & Microbe, 16, 376–390. [DOI] [PubMed] [Google Scholar]
- Hajheidari, M. , Farrona, S. , Huettel, B. , Koncz, Z. , & Koncz, C. (2012). CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C‐terminal domain of Arabidopsis RNA polymerase II. Plant Cell, 24, 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima, H. , Shinmyo, A. , & Sekine, M. (2007). Phosphorylation of threonine 161 in plant cyclin‐dependent kinase A is required for cell division by activation of its associated kinase. The Plant Journal, 52, 435–448. [DOI] [PubMed] [Google Scholar]
- Izawa, D. , Goto, M. , Yamashita, A. , Yamano, H. , & Yamamoto, M. (2005). Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature, 434, 529–533. [DOI] [PubMed] [Google Scholar]
- Javorka, P. , Raxwal, V. K. , Najvarek, J. , & Riha, K. (2019). artMAP: A user‐friendly tool for mapping ethyl methanesulfonate‐induced mutations in Arabidopsis. Plant Direct, 3, e00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes, J. , Chevalier, C. , Dudits, D. , Heberle‐Bors, E. , Inze, D. , Umeda, M. , & Renaudin, J. P. (2000). CDK‐related protein kinases in plants. Plant Molecular Biology, 43, 607–620. [DOI] [PubMed] [Google Scholar]
- Lopez‐Aviles, S. , Kapuy, O. , Novak, B. , & Uhlmann, F. (2009). Irreversibility of mitotic exit is the consequence of systems‐level feedback. Nature, 459, 592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. , Kawai, T. , Tanaka, K. , Niwa, Y. , Watanabe, Y. , Nakamura, K. , Kimura, T. , & Ishiguro, S. (2007). Improved gateway binary vectors: High‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology, and Biochemistry, 71(2095–2), 100. [DOI] [PubMed] [Google Scholar]
- Paez Valencia, J. , Goodman, K. , & Otegui, M. S. (2016). Endocytosis and endosomal trafficking in plants. Annual Review of Plant Biology, 67, 309–335. [DOI] [PubMed] [Google Scholar]
- Page, A. W. , & Orr‐Weaver, T. L. (1996). The Drosophila genes grauzone and cortex are necessary for proper female meiosis. Journal of Cell Science, 109(Pt 7), 1707–1715. [DOI] [PubMed] [Google Scholar]
- Parry, D. H. , & O'Farrell, P. H. (2001). The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Current Biology, 11, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R. , Duggal, G. , Love, M. I. , Irizarry, R. A. , & Kingsford, C. (2017). Salmon provides fast and bias‐aware quantification of transcript expression. Nature Methods, 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova, T. A. , Daum, J. R. , Pittman, B. D. , Hudson, J. R. , Jones, T. N. , Satinover, D. L. , Stukenberg, P. T. , & Gorbsky, G. J. (2006). The reversibility of mitotic exit in vertebrate cells. Nature, 440, 954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusicki, M. A. , Keizer, E. M. , van Rosmalen, R. P. , Komaki, S. , Seifert, F. , Muller, K. , Wijnker, E. , Fleck, C. , & Schnittger, A. (2019). Live cell imaging of meiosis in Arabidopsis thaliana . eLife, 8, e42834. 10.7554/eLife.42834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehs, N. , Akimcheva, S. , Puizina, J. , Bulankova, P. , Idol, R. A. , Siroky, J. , Schleiffer, A. , Schweizer, D. , Shippen, D. E. , & Riha, K. (2008). Arabidopsis SMG7 protein is required for exit from meiosis. Journal of Cell Science, 121, 2208–2216. 10.1242/jcs.027862 [DOI] [PubMed] [Google Scholar]
- Riehs‐Kearnan, N. , Gloggnitzer, J. , Dekrout, B. , Jonak, C. , & Riha, K. (2012). Aberrant growth and lethality of Arabidopsis deficient in nonsense‐mediated RNA decay factors is caused by autoimmune‐like response. Nucleic Acids Research, 40, 5615–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeiro Motta, M. , Zhao, X. , Pastuglia, M. , Belcram, K. , Roodbarkelari, F. , Komaki, M. , Harashima, H. , Komaki, S. , Kumar, M. , Bulankova, P. , Heese, M. , Riha, K. , Bouchez, D. , & Schnittger, A. (2022). B1‐type cyclins control microtubule organization during cell division in Arabidopsis . EMBO Reports, 23, e53995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe, M. , Boudolf, V. , De Veylder, L. , Inze, D. , Genschik, P. , & Machida, Y. (2011). Phosphorylation of a mitotic kinesin‐like protein and a MAPKKK by cyclin‐dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proceedings of the National Academy of Sciences of the United States of America, 108, 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J. Y. , White, D. J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. , & Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, B. T. , Hao, M. , Qiu, J. , Jiao, X. , Baseler, M. W. , Lane, H. C. , Imamichi, T. , & Chang, W. (2022). DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Research, 50, W216–W221. 10.1093/nar/gkac194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno, A. , Aki, S. S. , Takahashi, N. , & Umeda, M. (2021). Regulation of the plant cell cycle in response to hormones and the environment. Annual Review of Plant Biology, 72, 273–296. [DOI] [PubMed] [Google Scholar]
- Shimotohno, A. , Matsubayashi, S. , Yamaguchi, M. , Uchimiya, H. , & Umeda, M. (2003). Differential phosphorylation activities of CDK‐activating kinases in Arabidopsis thaliana . FEBS Letters, 534, 69–74. [DOI] [PubMed] [Google Scholar]
- Shimotohno, A. , Ohno, R. , Bisova, K. , Sakaguchi, N. , Huang, J. , Koncz, C. , Uchimiya, H. , & Umeda, M. (2006). Diverse phosphoregulatory mechanisms controlling cyclin‐dependent kinase‐activating kinases in Arabidopsis. The Plant Journal, 47, 701–710. [DOI] [PubMed] [Google Scholar]
- Sofroni, K. , Takatsuka, H. , Yang, C. , Dissmeyer, N. , Komaki, S. , Hamamura, Y. , Bottger, L. , Umeda, M. , & Schnittger, A. (2020). CDKD‐dependent activation of CDKA;1 controls microtubule dynamics and cytokinesis during meiosis. The Journal of Cell Biology, 219, e201907016. 10.1083/jcb.201907016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka, H. , Umeda‐Hara, C. , & Umeda, M. (2015). Cyclin‐dependent kinase‐activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis thaliana . The Plant Journal, 82, 1004–1017. [DOI] [PubMed] [Google Scholar]
- Umeda, M. , Shimotohno, A. , & Yamaguchi, M. (2005). Control of cell division and transcription by cyclin‐dependent kinase‐activating kinases in plants. Plant & Cell Physiology, 46, 1437–1442. 10.1093/pcp/pci170 [DOI] [PubMed] [Google Scholar]
- Valuchova, S. , Mikulkova, P. , Pecinkova, J. , Klimova, J. , Krumnikl, M. , Bainar, P. , Heckmann, S. , Tomancak, P. , & Riha, K. (2020). Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy. eLife, 9, e52546. 10.7554/eLife.52546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K. , Raes, J. , De Veylder, L. , Rouze, P. , Rombauts, S. , & Inze, D. (2002). Genome‐wide analysis of core cell cycle genes in Arabidopsis. Plant Cell, 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Zhang, R. , Feng, W. , Tsuchiya, D. , Ballew, O. , Li, J. , Denic, V. , & Lacefield, S. (2020). Autophagy of an amyloid‐like translational repressor regulates meiotic exit. Developmental Cell, 52(141–151), e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Kong, H. , Sun, Y. , Zhang, X. , Zhang, W. , Altman, N. , DePamphilis, C. W. , & Ma, H. (2004). Genome‐wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin‐like proteins. Plant Physiology, 135, 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Magnard, J. L. , McCormick, S. , & Yang, M. (2004). Progression through meiosis I and meiosis II in Arabidopsis anthers is regulated by an A‐type cyclin predominately expressed in prophase I. Plant Physiology, 136, 4127–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker, E. , Harashima, H. , Muller, K. , Parra‐Nunez, P. , de Snoo, C. B. , van de Belt, J. , Dissmeyer, N. , Bayer, M. , Pradillo, M. , & Schnittger, A. (2019). The Cdk1/Cdk2 homolog CDKA;1 controls the recombination landscape in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 116, 12534–12539. 10.1073/pnas.1820753116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M. , Umeda, M. , & Uchimiya, H. (1998). A rice homolog of Cdk7/MO15 phosphorylates both cyclin‐dependent protein kinases and the carboxy‐terminal domain of RNA polymerase II. The Plant Journal, 16, 613–619. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Sofroni, K. , Wijnker, E. , Hamamura, Y. , Carstens, L. , Harashima, H. , Stolze, S. C. , Vezon, D. , Chelysheva, L. , Orban‐Nemeth, Z. , Pochon, G. , Nakagami, H. , Schlogelhofer, P. , Grelon, M. , & Schnittger, A. (2020). The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. The EMBO Journal, 39, e101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Calixto, C. P. G. , Marquez, Y. , Venhuizen, P. , Tzioutziou, N. A. , Guo, W. , Spensley, M. , Entizne, J. C. , Lewandowska, D. , Have, S. T. , Frey, N. F. , Hirt, H. , James, A. B. , Nimmo, H. G. , Barta, A. , Kalyna, M. , & Brown, J. W. S. (2017). A high quality Arabidopsis transcriptome for accurate transcript‐level analysis of alternative splicing. Nucleic Acids Research, 45, 5061–5073. 10.1093/nar/gkx267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Time‐lapse microscopy of meiotic divisions in wild type PMCs from diakinesis till telophase II in 5 min increments. Chromatin was labeled with HTA10:TagRFP reporter. Scale bar = 20 μm.
Movie S2. Time‐lapse microscopy of meiotic divisions in smg7‐6 PMCs from diakinesis till telophase III in 5 min increments. Chromatin was labeled with HTA10:TagRFP reporter. Scale bar = 20 μm.
Movie S3. Time‐lapse microscopy of meiotic divisions in cdkd;3‐3 smg7‐6 PMCs from diakinesis till telophase III in 5 min increments. Chromatin was labeled with HTA10:TagRFP reporter. Scale bar = 20 μm.
Movie S4. Time‐lapse microscopy of meiotic divisions in wild type PMCs from interkinesis till the tetrad stage in 5 min increments. Microtubules were labeled with GFP:TUA5 reporter. Scale bar = 20 μm.
Movie S5. Time‐lapse microscopy of meiotic divisions in smg7‐6 PMCs from interkinesis, through meiosis II and meiosis III till the polyad stage in 5 min increments. Microtubules were labeled with GFP:TUA5 reporter. Scale bar = 20 μm.
Movie S6. Time‐lapse microscopy of meiotic divisions in cdkd;3‐3 smg7‐6 PMCs from interkinesis, through meiosis II and meiosis III till the polyad stage in 5 min increments. Note that some PMCs do not proceed in meiosis III and form regular tetrads. Microtubules were labeled with GFP:TUA5 reporter. Scale bar = 20 μm.
Movie S7. Time‐lapse microscopy of CDKD;3 localization in PMCs from late prophase I till telophase II s in 5 min increments. CDKD;3 and chromatin were visualized using CDKD;3:YFP (green) and HTA10:TagRFP (magenta) reporters, respectively.
Figure S1. Arabidopsis cdkd and smg7‐6 mutants. Approximately 6‐weks old plants are shown. Longer inflorescence bolt in the smg7‐6 mutant is caused by reduced fertility.
Figure S2. Mapping mutations associated with the rescued fertility in EMS58‐1 by whole genome sequencing. The causative mutation is recessive and is, therefore, expected to fully associate with the rescued phenotype. The charts indicate positions of de novo mutations on individual Arabidopsis chromosomes (x axis) and their frequency in population of B2 plants exhibiting increased fertility. Colored diamonds on chromosome 1 mark de novo mutations with the highest association. This region is zoomed in the lower left graph with indicated AGI codes for the mutated genes.
Figure S3. Genetic complementation of EMS58‐1 plants with the CDKD;3 genic construct causes infertility in transformed T1 plants. The chart shows average silique length in individual T1 transformants (error bars indicate SD; n = 25). Data for smg7‐6 are shown in red, for EMS58‐1 in gray, for complemented T1 transformants in blue and for non‐complemented transformants in green.
Figure S4. Fertility of cdkd;3‐3 mutants. (a) Quantification of silique length along the main inflorescence bolt in wild type (n = 10) and cdkd;3‐3 (n = 5). The trend lines of the data were plotted by LOESS smooth function (colored line; shaded area represents 95% confidence intervals). (b) Anthers from wild type and cdkd;3‐3 plants after Alexander staining. (c) Violin plots showing viable pollen per anther. Significance of the difference is indicated (two tailed t‐test; wild type n = 20; cdkd;3‐3 = 5). (d) Tetrads in anther lobes stained with DAPI.
Figure S5. Approximately six weeks old mutant plants.
Figure S6. Time‐lapse series of smg7‐6 and EMS58‐1 meiotic nuclei undergoing meiosis II and meiosis III. Nuclei derived from the same PMC are encircled with identical color. Note that chromatid tends to spread over larger area during anaphase III in smg7‐6 mutants. Chromatin is visualized with HTA10:TagRFP marker. Scale bar = 5 μm.
Table S1: De novo mutations detected in EMS58‐1.
Table S2: List of differentially expresssed genes relative to wild type.
Table S3. List of primers.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


