Abstract
The orientation of the mitotic spindle at metaphase determines the placement of the daughter cells. Spindle orientation in animals typically relies on an evolutionarily conserved biological machine comprised of at least four proteins – called Pins, Gαi, Mud, and Dynein in flies – that exerts a pulling force on astral microtubules and reels the spindle into alignment. The canonical model for spindle orientation holds that the direction of pulling is determined by asymmetric placement of this machinery at the cell cortex. In most cell types, this placement is thought to be mediated by Pins, and a substantial body of literature is therefore devoted to identifying polarized cues that govern localized cortical enrichment of Pins. In this study we revisit the canonical model and find that it is incomplete. Spindle orientation in the Drosophila follicular epithelium and embryonic ectoderm requires not only Pins localization but also direct interaction between Pins and the multifunctional protein Discs large. This requirement can be over‐ridden by interaction with another Pins interacting protein, Inscuteable.
Keywords: discs large, Drosophila, pins, spindle orientation
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle
In the Drosophila follicular epithelium and embryonic ectoderm, spindle orientation in the apical‐basal axis relies not only on the spatial localization of Pins but also on its interaction with a partner protein.
Introduction
The orientation of cell division is implicated in cell fate, as in the asymmetric division of neural progenitor cells, and in tissue architecture, as in the expansion of the Drosophila imaginal wing disc (reviewed in Bergstralh et al, 2017). Division orientation is generally determined by the orientation of the mitotic spindle at metaphase. Spindle orientation is governed by a suite of evolutionarily conserved factors comprised of Partner of Inscuteable (Pins; vertebrate LGN; nematode GPR1/2), Gαi, Mushroom body defective (Mud; vertebrate NuMA, nematode LIN‐5), and Dynein.
A combination of genetic experiments and biochemical work has led to a generalized model in which Gαi acts as a membrane anchor for Pins, which in turn recruits Mud to the cortex. Direct binding between Pins/LGN and Mud/NuMA is mediated by tetratricopeptide repeats (TPRs) at the N‐terminus of Pins/LGN (Du et al, 2001; Siller et al, 2006). Mud and Dynein together exert a pulling force on astral microtubules that reels the spindle into alignment. This model predicts that the key determinant of division orientation should be the placement of the pulling machinery at the cortex.
In most cell types, Pins/LGN is suggested to regulate the cortical position of the pulling machinery. For that reason, multiple studies have focused on the identification and characterization of cues that regulate Pins/LGN/GPR1/2 localization (reviewed in Bergstralh et al, 2017). In Drosophila neuroblasts, which are asymmetrically dividing neural progenitor cells, this cue is thought to be a protein called Inscuteable (Kraut et al, 1996; Schaefer et al, 2000; Yu et al, 2000). In symmetrically dividing epithelial cells, proposed Pins/LGN‐positioning cues include the junctional proteins E‐Cadherin and Afadin (in mammalian culture cells) and the multifunctional scaffolding protein Discs large (in flies, cultured mammalian cells, and chick neuroepithelium; Bergstralh et al, 2013b; Saadaoui et al, 2014; Carminati et al, 2016; Chanet et al, 2017; Gloerich et al, 2017). All of these proteins are reported to interact with Pins directly. The proliferation of potential Pins‐positioning proteins, particularly in epithelial cells, presents a problem: why are multiple factors necessary, especially in the same cell type?
The function of Pins is also somewhat enigmatic. Essentially, the model holds that it is an adaptor for an adaptor, linking Mud and therefore dynein to the cortex. The inclusion of a Pins‐positioning factor like Discs large (Dlg) adds another adaptor to the mechanism. Again, there is a parsimony problem; why are so many adaptors necessary? This question is highlighted by literature showing that the number can be reduced: (i) During anaphase, NuMA anchors itself to the membrane through a cryptic motif that is uncovered by CDK1 phosphorylation (Kiyomitsu & Cheeseman, 2013; Kotak et al, 2013; Seldin et al, 2013; Zheng et al, 2014); and (ii) the planar cell polarity protein Disheveled can act as an alternative cortical anchor for Mud (Segalen et al, 2010).
These questions led us to revisit the canonical model and test its predictions, starting from the most fundamental. We find that the model is incomplete. Spindle orientation relies not only on the position of Pins but also on interaction with factors that promote its activity.
Results
Cortical mud localization relies on pins
Mitotic spindles in epithelial cells tend to orient along the plane of the tissue, meaning perpendicular to the apical‐basal axis. In this way, divisions are oriented such that the two new daughter cells appear within the tissue (reviewed in Bergstralh et al, 2013a). The pulling machinery should therefore be expected to work at or adjacent to epithelial cell–cell borders (i.e. at the lateral cortex), and in agreement we have already shown that both Mud and Pins are lateral in mitotic follicle cells (Bergstralh et al, 2013b).
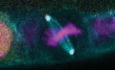
The canonical model holds that Pins acts as a cortical recruitment factor for Mud during mitosis (reviewed in Siller & Doe, 2009; Morin & Bellaïche, 2011; Werts & Goldstein, 2011; di Pietro et al, 2016; Bergstralh et al, 2017). Whether this holds true in Drosophila epithelia has not been tested outside of the pupal notum and larval wing disc, developmentally related tissues in which Mud localizes to tricellular junctions in a Pins‐independent manner (David et al, 2005; Bergstralh et al, 2016; Bosveld et al, 2016; Nakajima et al, 2019). We tested it in the Drosophila follicular epithelium (FE) and show here that cortical localization of Mud in mitotic follicle cells relies on Pins, whereas Mud localization at spindle poles is Pins‐independent (Fig 1A). This pattern was previously observed in mitotic neuroblasts (Siller et al, 2006). Unexpectedly, Mud is also cortical in interphase follicle cells, and this localization relies on Pins (Fig 1B). Pins therefore has a general, rather than mitosis‐specific, role in Mud localization in the follicular epithelium.
Figure 1. The cortical localization and spindle orienting function of Mud both rely on Pins.
-
AGFP:Mud is observed at spindle poles and at the lateral cell cortex in control mitotic follicle cells (A), but is only seen at the spindle poles in pins p62 /pins p62 cells (A'). Scale bars = 5 microns.
-
BInterphase localization of Mud at the cortex is lost in pins p62 /pins p62 follicle cells. Scale bar = 20 microns.
-
C–EFull‐length GFP:Mud shows the wild‐type pattern of localization in mud 3 /mud 4 mitotic follicle cells (C) and rescues spindle orientation (D, E). Scale bars = 5 microns. GFP:Mud lacking the Pins binding domain localizes only at spindle poles (C′) in mud 3 /mud 4 mitotic follicle cells and fails to rescue spindle orientation (D, E). Statistical significance in spindle orientation was determined using the Mann–Whitney test. ****P < 0.0001, ***P < 0.001.
The Pins‐binding domain (PBD) of Mud was previously mapped to a location between amino acids 1,928 and 1,982 (Izumi et al, 2006; Siller et al, 2006). We tested the importance of this region to spindle orientation in the FE using transgenes that encode the genomic promoter of Mud followed by either GFP‐tagged full length Mud or Mud lacking the PBD (Bosveld et al, 2016). We expressed these transgenes in mud 3/mud 4 flies, in which endogenous Mud function is strongly impaired (Yu et al, 2006). In this background, full length GFP‐Mud is observed at the cortex and at spindle poles and spindle orientation is rescued from random to the wild type distribution (Fig 1C–E). Contrastingly, GFP‐MudΔPBD is observed at spindle poles but not the cortex, and fails to rescue spindle orientation (Fig 1C–E). One caveat to interpretation is that the PBD overlaps with a region of Mud that can bind microtubules (AA1,850–2,039), meaning that cortical localization may not be the only impairment caused by the deletion (Izumi et al, 2006; Siller et al, 2006). However, taken together our results are consistent with the canonical model, in the follicle epithelium Pins is required for the cortical localization and the spindle orientation function of Mud. A corollary to this finding is that the FE is a suitable system in which to interrogate the model.
Pins may act as a cortical anchor for mud, but this function is not obligate
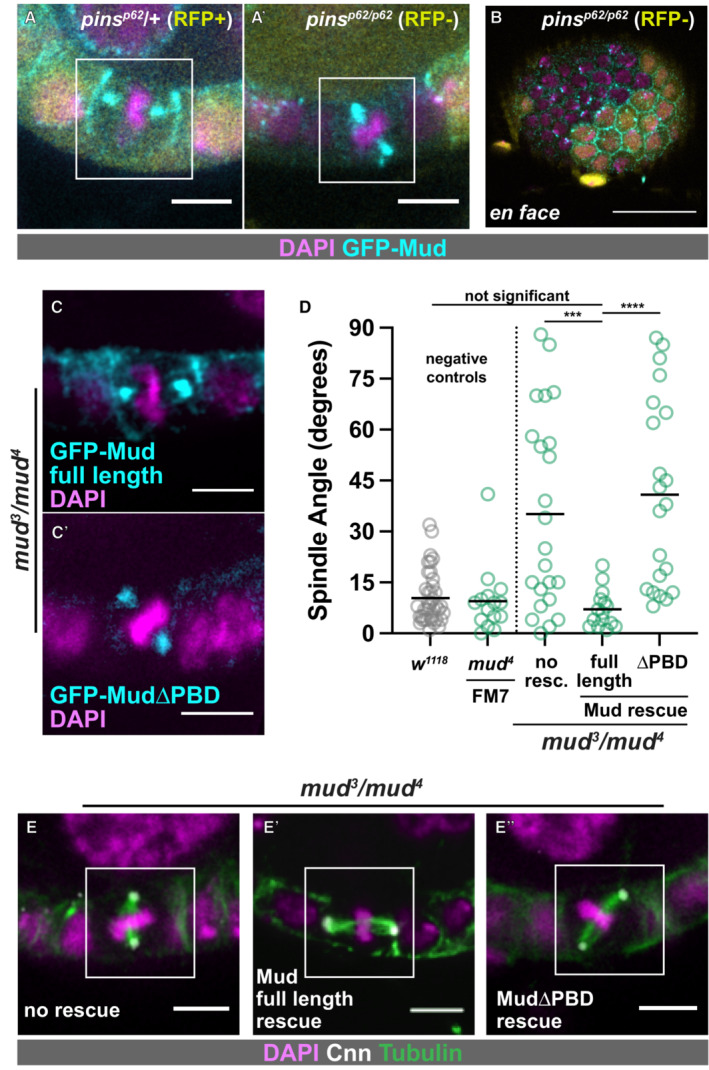
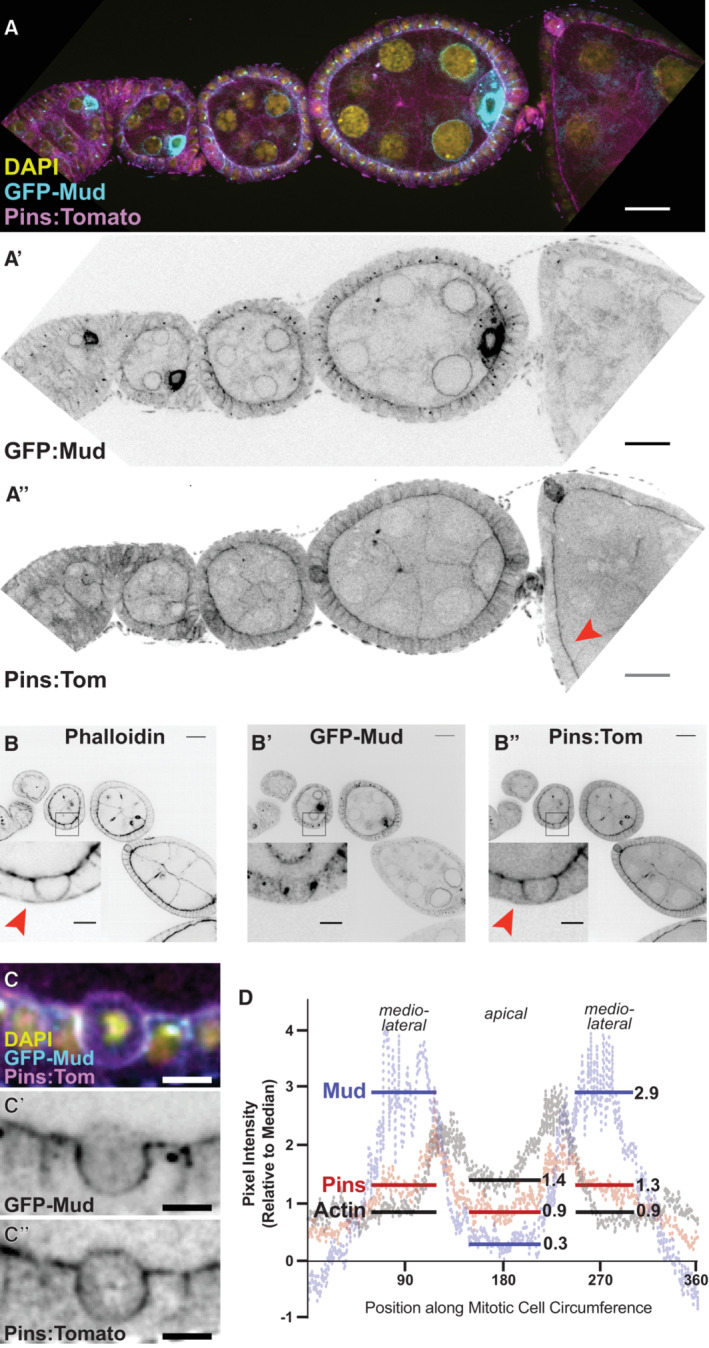
We next characterized the subcellular localization of Pins in the FE. We generated transgenic flies expressing Pins:Tomato (Pins:Tom) under the control of the Pins genomic promoter. Pins:Tom is observed in all cell types in the egg chamber during the developmental stages at which follicle cells divide (prior to developmental Stage 7), and unlike Mud it can be detected past that point (Fig 2A). Pins:Tom overlaps with cortical actin; both are observed at the border between the FE and the interior germline cells and are more weakly detected at follicle cell–cell borders (Fig 2A). Unlike actin, however, Pins:Tom is not obvious at the basal cell surface (Fig 2B). We considered the possibility that Pins signal at the follicle cell‐germline border is contributed by the germline cells, but clonal expression of Ubi‐Pins‐YFP shows that Pins localizes to the apical surface of follicle cells and accounts for the great majority of the signal at the border (Fig EV1A). In addition, although UAS‐Pins‐GFP is difficult to detect (a problem encountered by previous researchers) it is apically enriched when expressed in the FE but not the germline (Fig EV1B) (Chanet et al, 2017). Together these results agree with previous work showing that the vertebrate ortholog of Pins (LGN) is apical in interphase epithelial cells, though we can also find Pins along lateral surfaces (Hao et al, 2010).
Figure 2. Expression and localization of Pins:Tom and GFP:Mud in interphase and mitotic follicle cells.
-
APins:Tom and GFP:Mud both appear enriched at the apical surface of follicle cells, and can be weakly detected at lateral surfaces. Enrichment of Mud at the oocyte nucleus has been reported previously (Zhao et al, 2012). Unlike GFP:Mud, Pins:Tom can be seen past Stage 6, the developmental point at which follicle cell division ceases. The red arrow points to a later (~ Stage 8) egg chamber. Scale bars = 20 microns.
-
BPins and actin overlap extensively, though actin is observed at the basal surface (red arrow in B) whereas Pins is not (red arrow in B″). Scale bars = 5 microns.
-
C, DPins:Tom and GFP:Mud have overlapping but not identical localization patterns in mitotic follicle cells. Representative pictures (C) and quantification from 9 cells (D) are shown. Scale bars = 5 microns.
Figure EV1. Pins signal at the border between interior germline and exterior follicle cells reflects apical localization in follicle cells—Related to Fig 2 .
- Clonal expression of Ubi‐Pins‐YFP shows that most signal at the border between germline and follicle cells originates in the follicle epithelium. The arrow marks the clone border.
- Pins‐GFP appears at the apical surface when expressed only in the follicle epithelium.
- Clonal expression of Ubi‐Pins‐YFP shows that signal at the border between the germline and a mitotic follicle cell originates primarily in the mitotic cell. In this field of view, only the mitotic cell lacks expression of Ubi‐Pins‐YFP.
Data information: scale bars = 5 microns.
The canonical model describes Pins as a recruitment factor for Mud, suggesting that the two proteins should localize together at the cortex. During interphase, GFP:Mud shows a similar pattern to Pins:Tom, though the signal is not strong enough to confirm colocalization (Fig 2A). Consistent with our prior work, we find that both proteins appear along the basolateral cortex in mitotic cells, with an enrichment just below the apical surface (we term this position “mediolateral”) and a decrease toward the basal surface (Fig 2B and C; Bergstralh et al, 2013b). However, unlike Mud, Pins:Tom can also be distinguished at the apical cell surface (Fig 2C and D). Clonal expression of Ubi‐Pins‐YFP confirms that this signal originates primarily in the mitotic cell rather than the adjacent germline cell (Fig EV1C). Actin shows a similar pattern to Pins:Tom, but stronger enrichment at the apical surface (Fig 2C and D). A likely explanation is that this is at least in part because of cortical actin in the adjacent germline cell.
Together, these results show that in the follicular epithelium, the locations of Pins and Mud do not always correspond. This has also been shown in the wing disc and pupal notum, in which Pins is cortically localized but does not control the location of Mud (David et al, 2005; Bergstralh et al, 2016; Bosveld et al, 2016). Our findings do not contradict a role for Pins in cortical anchoring of Mud. However, they suggest that this function is not obligate even in cells in which Mud relies on Pins for its cortical location.
Relocalization of pins at mitosis does not require a pins‐specific cue
We next addressed the question of how Pins relocalizes from the apical to the lateral cortex at mitosis. Previous work, including our own, has suggested that this an active change mediated by a cortical polarity cue (Hao et al, 2010; Bergstralh et al, 2013b). Another model to consider is that Pins relocalization occurs passively. During mitosis, the composition and material properties of the actin cortex undergo changes associated with cell rounding (Chugh & Paluch, 2018). The possibility that the change in Pins localization simply reflects these changes is suggested by the following observations: (i) Pins:Tom overlaps closely with actin throughout the cell cycle (Figs 2B and EV2A); (ii) both proteins are enriched at the mitotic cortex (Fig 2B); and (iii) the unrelated transmembrane protein Basigin, a marker for follicle cell boundaries, is also cortically enriched at mitosis (Figs 2B and EV2B) (Bergstralh et al, 2015).
Figure EV2. Localization of cortical proteins in the follicular epithelium—Related to Fig 3 .
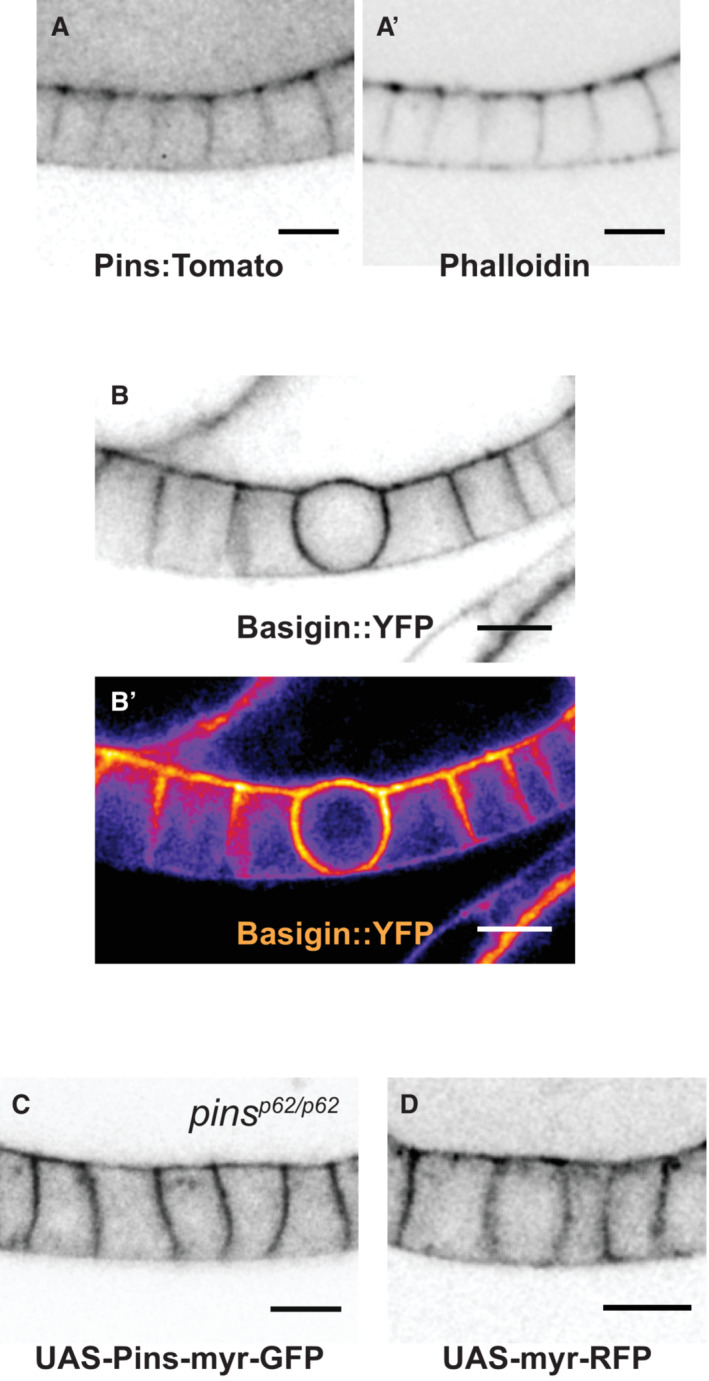
-
APins:Tom and actin overlap at the apical and lateral cortex during interphase.
-
BBasigin::YFP is enriched in a mitotic cell. Both a b/w and heat map image are shown to highlight enrichment. The protein can be found at the borders of germline and follicle cells.
-
C, DUAS‐myr‐Pins‐GFP (C) and UAS‐myr‐RFP localization (D) show similar localization in interphase follicle cells. Scale bars = 5 microns.
We used UAS‐Pins‐myr‐GFP, a myristoylated Pins variant developed by the Martin lab, to test between the active and passive models for Pins relocalization at mitosis (Chanet et al, 2017). We expressed Pins.myr‐GFP in the FE and examined its localization and impact on spindle orientation. The active model predicts that the myristoyl modification might circumvent mechanisms that control asymmetric localization of Pins at the cortex, and that Pins‐myr should therefore localize broadly around the plasma membrane. It does not. During interphase, Pins‐myr is observed at the apical surface and lateral borders, though unlike Pins:Tom, Pins‐myr signal extends evenly along the length of the epithelial cell–cell border rather than concentrating towards the apical side (Fig EV2A and C). During mitosis Pins‐myr is enriched at the lateral cell–cell border and more weakly observed at the apical surface, a pattern very similar if not identical to Pins:Tom (Fig 3A and C). Myristoylated‐RFP, a negative control, demonstrates similar localization to Pins:Tom and Pins‐myr (Figs 3B and C, and EV2D). These results are consistent with the passive model. We also find that Pins‐myr expression rescues spindle orientation in pins null tissue, indicating that it is able to substitute for wild‐type Pins (Fig 3D–F). Together, our results indicate that the apparent relocalization of Pins from the apical to the lateral surface at mitosis—from which position it drives spindle orientation—can largely be explained as the placement of a membrane‐anchored protein.
Figure 3. Pins does not require a Pins‐specific cue to relocalize at mitosis.
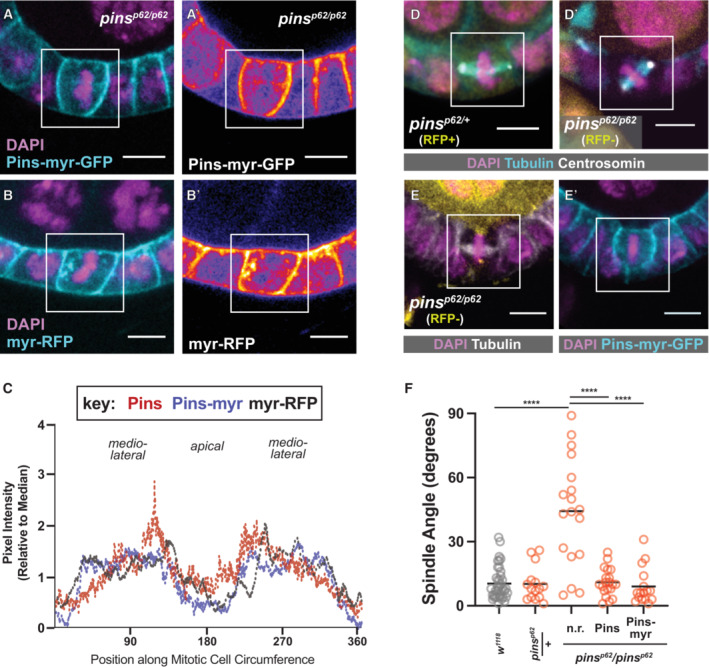
-
A–CPins‐myr‐GFP and myr‐RFP show a similar localization pattern to Pins:Tom during mitosis. Representative pictures are shown (A, B); a heatmap lookup table (A', B′) emphasizes the relative enrichments of Pins‐myr‐GFP and myr‐RFP along the cortex. Quantification (Pins‐myr: 4 cells, myr‐RFP: 2 cells) (C).
-
D–FRandom spindle orientation in pins p62 /pins p62 null mutant follicle cells is rescued by the expression of full‐length Pins or Pins‐myr‐GFP. Representative pictures (D, E) and quantification (F) are shown. Scale bars = 5 microns. Statistical significance in spindle orientation was determined using the Mann–Whitney test. ****P < 0.0001.
Pins is cortical in the absence of Dlg
The finding that membrane anchoring is sufficient to explain Pins localization at mitosis presents a challenge to our previous work, in which we suggested that relocalization relies on the epithelial polarity factor Discs large (Bergstralh et al, 2013b). Subsequent studies in the early Drosophila embryo, chick neuroepithelium, and unpolarized cultured cells indicated that Dlg is not just a cue but an anchor for Pins at the cell boundary (Saadaoui et al, 2014; Chanet et al, 2017). However, Pins/LGN is thought to be linked to the membrane by Gαi‐GDP, raising the question of why a second anchor would be required (reviewed in Siller & Doe, 2009; Morin & Bellaïche, 2011; Werts & Goldstein, 2011; Bergstralh et al, 2017). We therefore revisited the question of how Dlg participates in spindle orientation.
The spindle‐orienting function of Dlg is thought to be mediated by direct interaction between the inactive C‐terminal Guanylate Kinase (GUK) domain of Dlg and an evolutionarily conserved Pins phosphoserine (Drosophila S436, human S408; Johnston et al, 2009; Hao et al, 2010; Zhu et al, 2011; Saadaoui et al, 2014). We confirmed this in the follicle epithelium in two ways: Firstly, we tested the role of Dlg. The Dlg 1P20 allele encodes a truncated protein that is missing approximately one‐third of its GUK domain and cannot bind to Pins in vitro (Bergstralh et al, 2016). We showed previously that spindles are misoriented in Dlg 1P20 FE tissue, though apico‐basal polarity is maintained (Bergstralh et al, 2013b). That analysis was performed using mitotic clones, raising the possibility that the spindle orientation defect could be caused by a second mutation on the Dlg 1P20 chromosome. We tested this possibility using flies heterozygous for Dlg 1P20 and Dlg 2 , a strong temperature‐sensitive hypomorphic allele. We find that follicle cell spindles in Dlg 1P20/2 egg chambers are misoriented at restrictive temperature (29°C; Fig 4A). These results confirm that disruption of the Dlg C‐terminus causes spindle misorientation. Secondly, we examined the consequence of Pins phosphorylation. We generated transgenic flies encoding Pins, Pins‐S436D (phosphomimetic), or Pins‐S436A (unphosphorylatable) under UAS control and expressed these variants in pins null clones. We find that Pins and Pins‐S436D both rescue spindle orientation, while Pins‐S436A does not, in agreement with previous work (Fig 4B and C) (Johnston et al, 2009; Hao et al, 2010). These results are consistent with a model in which Dlg participates in spindle orientation through interaction with phosphorylated Pins.
Figure 4. Dlg and Pins interact to promote spindle orientation in the follicle epithelium, but Dlg is not required for cortical localization of Pins.
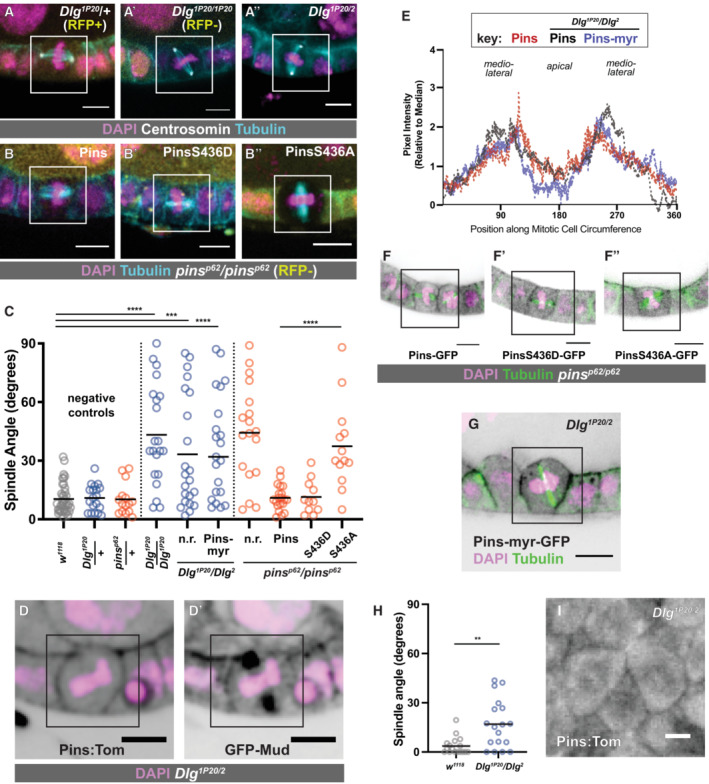
- Spindle orientation is randomized in Dlg 1P20 /Dlg 1P20 and Dlg 1P20 /Dlg 2 mutant follicle cells.
- Random spindle orientation in pins p62 /pins p62 null mutant follicle cells is rescued by the expression of full‐length Pins or phosphomimetic (S436D) Pins. Unphosphorylatable (S436A) Pins fails to rescue.
- Quantification of spindle orientation phenotypes. Statistical significance in spindle orientation was determined using the Mann–Whitney test. ****P < 0.0001, ***P < 0.001.
- Pins:Tom and GFP:Mud are cortical in Dlg 1P20 /Dlg 2 mutant follicle cells.
- Pins:Tom and Pins‐myr are both laterally enriched at mitosis in Dlg 1P20 /Dlg 2 mutants. Wild‐type Pins:Tom signal quantification data as in Fig 2D (9 cells). Quantification of Pins:Tom in Dlg 1P20 /Dlg 2 is 5 cells, Pins:Myr in Dlg 1P20 /Dlg 2 is 8.
- GFP‐tagged Pins variants (control, phosphomimetic, unphosphorylatable) are observed at the mitotic cell cortex in pins p62 /pins p62 null mutant follicle cells.
- Pins‐myr‐GFP fails to rescue spindle randomization in Dlg 1P20 /Dlg 2 mutant follicle cells. Quantification in (C).
- Quantification of spindle orientation phenotypes in the Stage 9/10 embryonic ectoderm. n = 17 cells from 2 embryos in w 1118 and n = 18 cells from 2 embryos in Dlg 1P20 /Dlg 2 embryos. Statistical significance in spindle orientation was determined using the Mann–Whitney test. **P = 0.0015.
- Pins:Tom is cortical in the early embryonic ventral ectoderm from a Dlg 1P20 /Dlg 2 mother. Representative image from an embryo imaged live.
Data information: scale bars = 5 microns.
We next asked whether Dlg controlled Pins localization during mitosis. We showed previously that cortical recruitment of Pins in the FE is Dlg‐independent, but a difficulty for interpretation is that these results relied on Pins‐YFP, which is driven by a Ubiquitin promoter and therefore likely to be overexpressed (David et al, 2005; Bergstralh et al, 2013b). We repeated the experiment using Pins:Tom. We found that (i) Pins:Tom localizes to the mitotic cell cortex in Dlg 14 /Dlg 14 mutant follicle cells, which are Dlg protein null, and in Dlg 1P20 /Dlg 1P20 cells (Fig EV3A and B) and that (ii) GFP‐Mud and Pins:Tom localize to the mitotic cortex in Dlg 1P20 /Dlg 2 mutant follicle cells (Fig 4D and E). Additionally, Pins, Pins‐S436D, and Pins‐S436A are all observed at the mitotic cortex in pins null clones (Fig 4F). These results indicate that the spindle orienting function of Dlg is not through cortical anchoring of Pins. To test this even further, we expressed Pins‐myr in Dlg 1P20 /Dlg 2 mutant follicle cells. While the localization of this variant is identical to Pins‐myr in a control background, it fails to rescue spindle orientation (Fig 4C, E and G). Together, our findings indicate that while Dlg is important for spindle orientation in the FE, the effect is not through redistribution of Pins.
Figure EV3. Dlg is not required for cortical localization of Pins and explanation of the method of spindle angle measurements in the embryo—Related to Fig 4 .
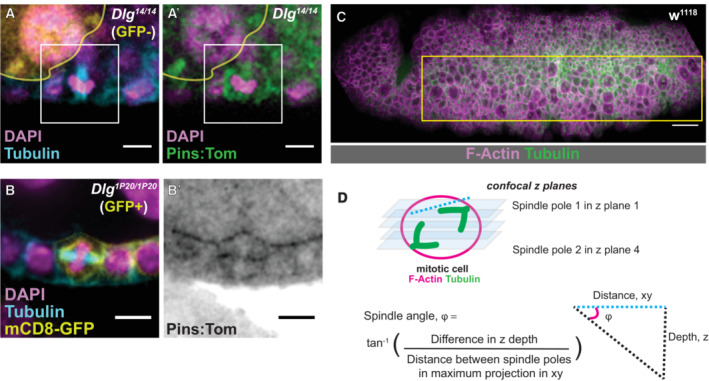
- A mitotic Dlg 14 /Dlg 14 cell and
- A mitotic Dlg 1P20 /Dlg 1P20 cell. For the former, mutant tissue is marked by the absence of RFP. For the latter, mutant tissue is marked by the presence of mCD8‐GFP. Scale bars = 5 microns (A, B).
- The region of the ventrolateral embryonic ectoderm used to quantify spindle angles from a representative fixed and immunostained embryo. Scale bar = 20 microns.
- A schematic showing the method to determine the spindle angle of mitotic cells in the early embryonic ectoderm, described in the Materials and Methods.
We also examined the relationship between Pins and Dlg in a different epithelium, the ventral ectoderm of the early (Stage 10) Drosophila embryo. Consistent with our findings in the FE, mitotic spindles in this tissue are misoriented with respect to the apical‐basal plane in embryos from Dlg 1P20 /Dlg 2 mothers (Fig 4H). As in the FE, Pins:Tom is observed at mitotic cell cortices, indicating that Pins localization in this tissue does not require direct interaction with Dlg (Fig 4I). These results contrast with the finding that cortical localization of Pins in embryonic head epithelial cells is lost when Dlg mRNA is knocked down (Chanet et al, 2017). We therefore speculate that Dlg plays an additional role in that tissue, upstream of Pins.
Khc‐73 is not required for spindle orientation in the follicular epithelium
Another proposed explanation for the role of Dlg in spindle orientation is that it facilitates interaction between Pins and a microtubule‐capturing activity. This has been studied an artificially polarized S2 (cultured) cell system, in which the microtubule‐capturing factor is the plus‐end directed motor Khc‐73 (Siegrist & Doe, 2005; Johnston et al, 2009; Lu & Prehoda, 2013). Khc73 is also part of a Pins/Dlg pathway that rescues division orientation in Inscuteable‐mutant neuroblasts (Siegrist & Doe, 2005). Given that epithelial cells express Dlg but not Inscuteable, the possibility that this rescue pathway is the default in epithelia is particularly attractive. We tested it in follicle cells using (i) flies transheterozygous for the null alleles Khc‐73 149 and Khc‐73 193 , (ii) expression of Khc73 shRNA in the follicular epithelium; and (iii) mitotic clones mutant for the null allele Khc73 3‐3 (Liao et al, 2018; Zajac & Horne‐Badovinac, 2022). Spindle angles were normal in all of these conditions (Fig 5A and C).
Figure 5. Putative microtubule‐capturing factors are not required for spindle orientation in the follicular epithelium.
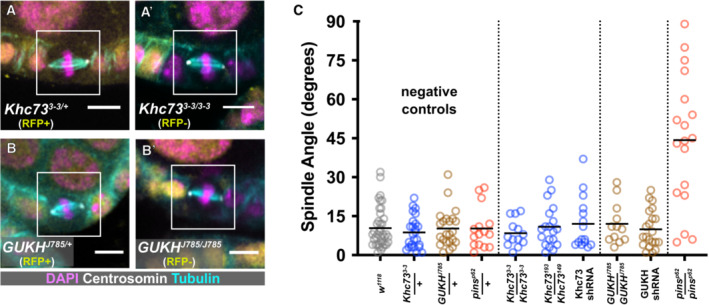
-
A–CMultiple genetic approaches were used to test a role for either Khc73 or GUKHolder in metaphase spindle orientation. Representative pictures (A, B) and quantification (C) are shown. Scale bars = 5 microns. Spindle angle randomization in pins p62 /pins p62 null mutant follicle cells is included as a positive control for comparison.
Dlg is also proposed to link the spindle orienting machinery to another microtubule capturing protein called GUKHolder (GUKH), though loss of GUKH function has a negligible effect on spindle orientation in neuroblasts (Golub et al, 2017). We examined spindle orientation in follicle cells mutant for GUKH J785 , which encodes a premature stop at Q520. (Full length isoforms are > 1700AA.) This allele causes failed projection of photoreceptor axons and lethality (Berger et al, 2008). Spindle orientation was normal in GUKH J785 cells and likewise unaffected by expression of shRNA targeting GUKH transcript, though we do not have direct evidence of knockdown (Fig 5B and C). Together, these findings show that Khc73 is dispensable for spindle orientation in the follicle epithelium and do not support a role for GUKHolder.
Two patterns of Inscuteable localization predict spindle orientation
Our findings indicate that Discs large promotes Pins‐dependent spindle orientation without acting as a localization factor or a link to microtubule‐capturing molecules. They also show that, in contradiction to the model, Pins localization is not sufficient to determine the activity of the pulling force. Another way to test this is to move Pins to a different location on the cortex.
A potential strategy for relocalizing the spindle machinery is ectopic expression of Inscuteable, which is required for apical localization of Pins in neuroblasts (Schaefer et al, 2000; Yu et al, 2000). This manipulation has a well‐established impact on the position of the pulling force; ectopic expression of Inscuteable in any of three Drosophila epithelia—the embryonic ectoderm, larval neuroepithelium, and follicular epithelium—causes reorientation of mitotic spindles such that they are parallel, rather than perpendicular, to the apical‐basal axis (Kraut et al, 1996; Bowman et al, 2006; Egger et al, 2007; Bergstralh et al, 2015). Our own earlier investigation of UAS‐Inscuteable expression in the FE was performed using the mosaic GAL4 drivers GR1‐GAL4 and T155‐GAL4 and relied on visualization of Inscuteable (by immunostaining) at the apical cell cortex to confirm expression (Bergstralh et al, 2015). We repeated the experiment using Traffic jam‐GAL4, which is less mosaic across a given egg chamber, though in our hands generally weaker than other drivers. In agreement with our previous work, cells with a strong (~ 4‐fold > median) apical enrichment of Inscuteable (InscA) demonstrate perpendicular spindle orientation (Figs 6A, B and F, and EV4A) (Bergstralh et al, 2015). However, we also identified a population of cells that demonstrates both lateral and apical localization of Inscuteable (InscB) (Fig 6C and D). In an exceptional instance, it is only lateral (Fig EV4B). Spindle orientation is normal in this group (Fig 6F). We considered whether InscA cells resolve to become InscB, but consider this possibility unlikely because we observed misoriented divisions in previous work (Bergstralh et al, 2015).
Figure 6. Inscuteable and Dlg function as distinct activators of the pulling machinery.
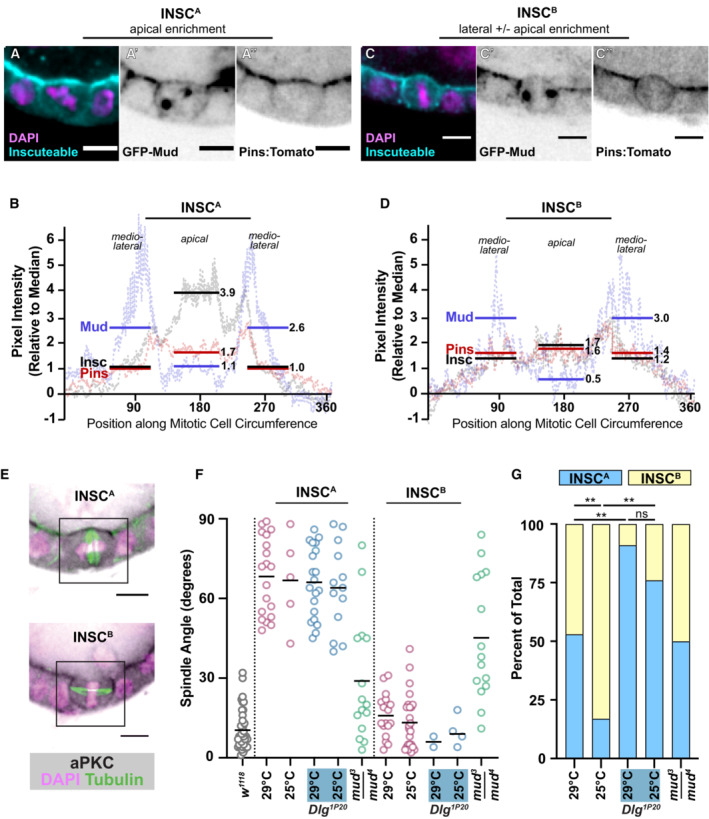
-
A–DTwo distinct patterns of Inscuteable localization are observed in the follicular epithelium when UAS‐Inscuteable is driven by Traffic jam‐GAL4. (A, C) In the first pattern (InscA), Inscuteable is highly enriched at the apical cell surface. (B, D) In the second pattern (InscB), Inscuteable is observed at both the lateral and apical cortex. Mud and Pins localizations are also shown. Representative pictures (A, B) and quantifications (InscA: average of 5 cells, InscB: average of 7 cells) (C, D) are shown.
-
EaPKC localization in InscA and InscB follicle cells.
-
FQuantification of spindle orientation phenotypes shows that mitotic spindles are reoriented in InscA cells, but not InscB cells. Reorientation relies on Mud but not on the interaction between Pins and Dlg.
-
GThe ratio of InscA to InscB is impacted by temperature and by the interaction between Pins and Dlg. Significance was determined using the chi‐square test. **P < 0.01.
Data information: scale bars = 5 microns.
Figure EV4. InscA and InscB cells are distinguished by the pattern of Inscuteable localization—Related to Fig 6 .
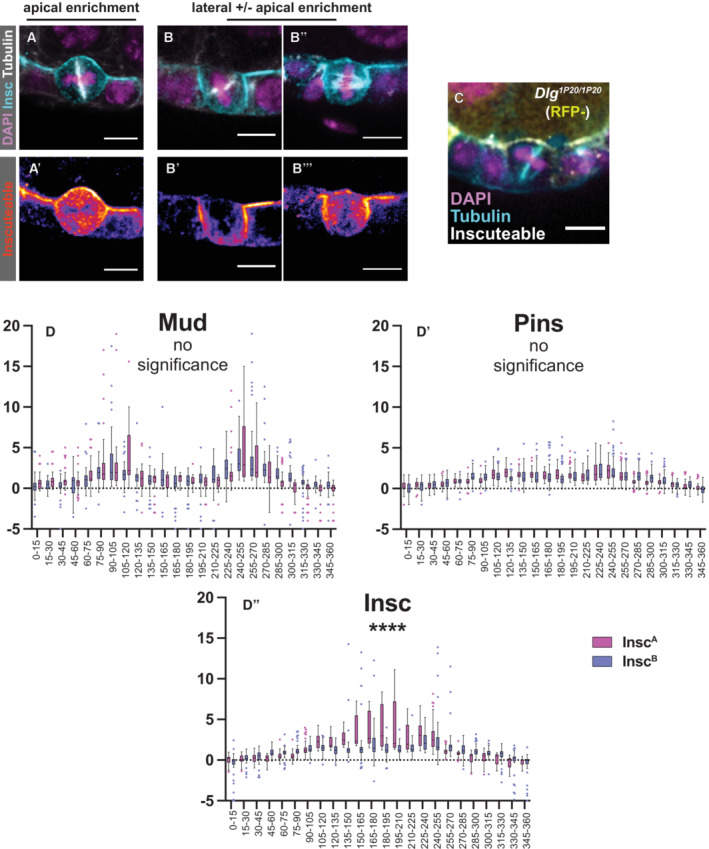
- In InscA cells, Inscuteable is strongly enriched at the apical surface.
- InscB cells show a stronger enrichment of Inscuteable at lateral surfaces. In an extreme example, Inscuteable is only lateral.
- Mitotic spindle reorientation in a Dlg1P20/Dlg1P20 cell expressing Inscuteable. Quantification in Fig 6F.
- As a complementary visualization strategy to that provided in Fig 6B and D, we divided the relative signal intensity data for Mud, Pins, and Insc in InscA and InscB cells into bins of 15 degrees each. Box plot central bands = mean, whiskers = standard deviation. These data are presented here side‐by‐side for each protein in the two conditions. Only the intensity of Inscuteable showed a statistical difference (as determined by ANOVA) between InscA and InscB cells. Quantification from InscA: 5 cells, InscB: 7 cells (biological replicates).
Data information: scale bars = 5 microns (A–C).
We speculate that the two populations reflect different expression levels, meaning that there is a threshold of expression over which Inscuteable causes reorientation. Two lines of evidence support this possibility. Firstly, ectopic expression of Inscuteable in the embryonic ectoderm causes a reliable reorientation of spindle angles rather than a bimodal distribution, consistent with the possibility that the threshold is met in that tissue (Kraut et al, 1996; Bergstralh et al, 2015). Secondly, we found that spindle reorientation was less common when UAS‐Inscuteable flies were maintained at 25°C rather than 29°C (Fig 6F and G). The UAS‐GAL4 system is more efficient at the higher temperature.
The spindle‐orientation machinery must be activated, not just localized
Although Inscuteable can reorient epithelial cell spindles, a careful examination of the spindle orienting machinery in epithelial cells expressing Inscuteable has not been undertaken. We first tested the influence of Inscuteable on cortical polarity. In neuroblasts, Inscuteable and the Par complex proteins aPKC and Bazooka rely on one another for apical localization (Wodarz et al, 1999, 2000; Schaefer et al, 2000; Yu et al, 2000). In wild type follicle cells aPKC is an apical polarity marker at interphase but loses its polarized enrichment during mitosis (Morais‐de‐Sá & Sunkel, 2013; Bergstralh et al, 2013b). In agreement with interdependence between Inscuteable and the Par complex, we find that aPKC is stabilized at the apical cortex in InscA cells but enriched at the lateral cortex in InscB cells (Fig 6E). This finding is consistent with an Inscuteable‐expression threshold model; below the threshold, Pins dictates lateral localization of Inscuteable and aPKC. Above the threshold, Inscuteable dictates apical localization of Pins and aPKC.
The canonical model predicts that Pins and Mud should be recruited to the apical surface in InscA cells, from which position they would draw the spindle into alignment along the apical‐basal axis, and that Pins and Mud should be primarily lateral in InscB cells, where they would promote wild type spindle orientation (perpendicular to the apical‐basal axis). These predictions are not met. Instead, we find that Pins is apically enriched in both InscA and InscB cells (Figs 6A–D and EV4D). Mud looks markedly different to Pins. Although a stronger enrichment is observed at the apical surface in InscA cells than in the InscB cells, Mud is most prominent at the mediolateral regions in both InscA and InscB cells (Figs 6A–D and EV4D).
These results conflict with the canonical model. If spindle orientation is predicted only by the location of Pins, InscA and InscB cells should both be expected to orient their spindles along the apical‐basal axis, as in neuroblasts. If spindle orientation is predicted only by the location of Mud, InscA and InscB cells should both be expected to orient their spindles along the tissue plane, as in the wild type. Our results show that the strongest predictor of spindle orientation in these cells is neither Mud nor Pins, but rather Inscuteable; if it is highly enriched at the apical surface (InscA) spindles are reoriented, but if it is not (InscB) then spindle angles are normal.
Since Mud localization is not an accurate predictor of spindle orientation, whereas Inscuteable is, a possibility to consider is that Inscuteable exerts a Mud‐independent effect. However, we find that spindle orientation is randomized in both InscA and InscB cells when Mud is genetically removed (Fig 6F). This finding indicates that in InscA cells Inscuteable impacts both the location and the activity of Mud‐dependent pulling.
We have already shown that activation of the spindle‐orientation machinery in the FE requires Dlg, and an obvious possibility is therefore that Inscuteable promotes spindle reorientation in InscA cells by changing the location of Dlg. We have been unable to test this directly because Dlg is expressed in germline cells as well as the FE, meaning that it can appear to be apical even in wild type tissue. However, to find out whether Dlg is required for Inscuteable‐mediate spindle orientation we expressed Inscuteable in Dlg 1P20 /Dlg 1P20 mutant tissue. Not only does Inscuteable reorient spindles in this tissue, but the proportion of InscA cells is significantly higher at both 29 and 25°C (Figs 6F and G, and EV4C). In other words, Dlg acts to shift the threshold towards the InscB pattern. We interpret this to mean that interaction between Pins and Dlg, which is required for spindle orientation, stabilizes the lateral spindle‐orientation machinery even if Dlg is not a direct anchor.
Taken together with the results described above, these findings show that Inscuteable and Dlg mediate distinct and competitive mechanisms for activation of the spindle‐orienting machinery in follicle cells.
Discussion
Limitations of this study
A technical challenge to undertaking this study is that current fluorescent molecules—including those introduced here—are exceedingly weak and liable to photobleaching (Pins‐myr‐GFP is an exception). This issue presents a barrier to live imaging, meaning that temporal resolution is unavailable. We are also unable to convincingly determine whether cortical Pins is decreased (as opposed to simply absent), when it is unable to interact with Dlg; a decrease is predicted by work in chick embryo neuroepithelial cells showing that a C‐terminal fragment of LGN localization is ~ 33% less cortical in the unphosphorylatable condition (Saadaoui et al, 2014). However, we find that even when Pins is anchored to the membrane by myristoylation it fails to rescue spindle orientation in Dlg 1P20 /Dlg 2 mutant tissue, meaning that a reduction of Pins is unlikely to be the only effect responsible for randomization.
Another technical consideration is that our work makes use of transgenes under the control of Traffic jam‐GAL4. While this strategy allows us to compare our results with previous work employing the same or similar tools, a drawback is that we cannot guarantee that Traffic jam‐GAL4 drives equivalent expression to the endogenous Pins promoter (Chanet et al, 2017, Camuglia et al, 2022). However, given that Traffic jam‐GAL4 is fairly weak at the developmental stages examined, we are not especially concerned about overexpression effects.
Additional function for Pins?
While it is not a focus for our study, our work provides support for the possibility that Pins has an additional function outside of spindle orientation. This is already suggested by the fact that strong mutants of pins are lethal to the organism, whereas strong mutants of mud are viable (Schaefer et al, 2000; Yu et al, 2000, 2006). Additionally, in the developing mouse epidermis, the vertebrate ortholog of Pins (LGN) acts to promote stratification by facilitating daughter cell placement subsequent to metaphase, perhaps implicating LGN in the regulation of cell–cell adhesion (Lough et al, 2019). We show here that whereas Mud expression in the follicular epithelium ends at developmental Stage 6, coinciding with the end of follicle cell divisions, Pins continues to be detected past that point (Bergstralh et al, 2013b). This suggests that Pins function is not limited to a role in proliferation. In addition, we find that while the LGN S408D (Drosophila 436D) variant is reported to act as a phosphomimetic, this variant does not cause an obvious mutant phenotype in the follicular epithelium (Johnston et al, 2012). What then is the purpose of this modification? Since the phosphosite is highly conserved through metazoans, one possibility to consider is that the phosphorylation regulates the spindle orientation role of Pins, whereas unphosphorylated Pins plays a different role (Schiller & Bergstralh, 2021).
Dlg and Inscuteable function as distinct activators for the pulling machinery
Previous work, including our own, has concentrated on identifying mechanisms that regulate the position of the spindle‐orienting machinery. Work presented here shows that localization of either Pins or Mud is not always sufficient to predict pulling. Firstly, while Pins is required for cortical localization of Mud in the follicular epithelium, Pins and Mud localizations do not strictly overlap, meaning that Pins is not an obligate Mud anchor. We see evidence for this in wild‐type mitotic follicle cells, in which Pins appears slightly apical whereas Mud does not, and in follicle cells expressing Inscuteable, in which the pattern of Pins and Mud localizations are markedly different. Because cortical localization of Mud/NuMA has proven difficult to visualize, Pins location has been used as a proxy for the position of the entire pulling machinery (Dimitracopoulos et al, 2020). While this may very well be the case in some cell types, our work suggests that it is not a rule that can be automatically applied. Secondly, we find that even Mud/NuMA localization is not necessarily a reliable predictor for spindle orientation (Fig 6). Although elegant optogenetics work shows that cortical anchoring of NuMa is sufficient to direct pulling in unpolarized HeLa cells, we find that in the follicular epithelium at least Mud activity must also be regulated by additional factors (Okumura et al, 2018).
One of these factors is Discs large. We tested two models for the function of Dlg in Pins‐mediated spindle orientation in the follicular epithelium. The first model, based in part on our own previous work, is that Dlg acts as a cortical anchor for Pins, and is therefore important for relocalization of Pins from the apical to the lateral cortex at mitosis (Bergstralh et al, 2013b). We now show that this model is incorrect, partly because Pins relocalization at mitosis does not require a Pins‐specific mechanism, but only a membrane anchor. Gαi may serve that purpose. The second model is that Dlg links Pins with Khc73 or GUKHolder. We find that neither molecule is important for spindle orientation in the FE. Our findings indicate that instead of a localization cue or a link to microtubule‐capturing factors, Dlg acts as an activator for Pins‐mediated pulling in the follicular epithelium. This mechanism is distinct from and can compete with Inscuteable, which we show here is not only a cortical localization factor for the spindle‐orienting machinery but also an activator.
The mechanism of activation remains unclear. While the most straightforward possibility is that Dlg promotes interaction between Pins and Mud, our results show that Mud is recruited to the cortex even when Dlg is disrupted (Fig 4D). Alternatively, Discs large may promote a conformational change in the spindle‐orientation complex and/or a change in complex composition. Furthermore, the Inscuteable mechanism is not likely to work in the same way. Dlg binds to a conserved phosphosite in the central linker domain of Pins and should therefore allow for Pins to simultaneously interact with Mud (Johnston et al, 2009). Contrastingly, binding between Pins and Inscuteable is mediated by the TPR domains of Pins, meaning that Mud is excluded (Culurgioni et al, 2011, 2018). While a stable Pins‐Inscuteable complex has been suggested to promote localization of a separate Pins‐Mud‐dynein complex, our work raises the possibility that it might also or instead promote activation.
The finding that there are two distinct mechanisms for activating the spindle‐orienting machinery raises the question of how and for what purpose they evolved. Phylogenetic analysis indicates that the interaction between Pins and Dlg is evolutionarily ancient; it is predicted to have evolved in Cnidaria, predating the appearance of Inscuteable orthologs (Schiller & Bergstralh, 2021). It is therefore tempting to speculate that Inscuteable‐mediated spindle orientation evolved to promote cell type diversity, particularly in the more complex bilaterian nervous system.
Together, our results show that the canonical model that explains the mitotic spindle orientation is incomplete. Spindle orientation relies not only on the localization, but also on the activation of the pulling machinery.
Materials and Methods
Drosophila genetics
A list of alleles and transgenes used in this study is found in Table EV1. Genotypes are in Table EV2. We thank the Transgenic RNAi Project at Harvard Medical School (NIH/NIGMS R01‐GM084947) for providing shRNA lines. Ectopic protein expression was accomplished using the UAS‐GAL4 system (Brand & Perrimon, 1993). Expression was driven by Traffic Jam‐GAL4 (Olivieri et al, 2010). Mitotic clones of Ubi‐Pins‐YFP were made by recombining the Ubi‐Pins‐YFP transgene onto an FRT82B chromosome.
Molecular biology
Pins:Tomato fly line was generated by cloning the entire pins gene tagged with tdTomato including 3 kb upstream and 2 kb downstream of the coding sequence into a pattB plasmid backbone construct (Bischof et al, 2013). tdTomato RFP (Shaner et al, 2004) was inserted at the C‐terminus of the Pins coding sequence separated by a linker (GGSG), using PCR and Gibson assembly. The sequence map is shown in Appendix Fig S1 and the sequence in Appendix Supplementary Methods. This plasmid was injected into attP2 line Bloomington line 25,710 (outsourced to BestGene Drosophila Embryo Injection Services) with 3rd chromosome attP docking site for phiC31 integrase‐mediated transformation.
Mitotic clones
Clones were induced by incubating larvae or pupae at 37°C for two out of every 12 h over a period of at least 2 days. Adult females were dissected at least 2 days after the last heat shock. Flies in which the Gal4‐UAS system was used were kept at 29° for 48 h before dissection unless otherwise noted. The following background stocks were used to generate mitotic clones, which were induced by heat shock at 37° for multiple periods of 2 h: RFP‐nls, hsflp, FRT19A and hsflp;;FRT82B, RFP‐nls / TM3,Sb. Mosaic Analysis with a Repressible Cell Marker was carried out using GFP‐mCD8 (under control of an actin promoter) as the marker (Lee & Luo, 1999).
Reagents
A list of reagents used in this study is found in Table EV3.
Immunostaining
Ovaries were fixed for 15 min in 10% Formaldehyde and 0.2% Tween in Phosphate Buffered Saline (PBS‐T). Embryos were fixed for 5 min at the interface of 37% Formaldehyde and 100% Heptane. Embryos were then washed in PBS and PBS‐T and the vitelline membrane was removed manually using a sharp needle. Ovaries and embryos were incubated in blocking solution (10% Bovine Serum Albumin in PBS) for about 1 h at room temperature prior to staining. Primary and secondary immunostainings lasted at least 3 h in PBS‐T. Three washes of about 10 min each in PBS‐T were carried out between stainings and after the secondary staining. Primary and secondary antibodies were used at a concentration of 1:150.
Imaging
Microscopy was performed using either a Leica SP5 point scanning confocal (63×/1.4 HCX PL Apo CS oil lens) or an Andor Dragonfly Spinning Disk confocal microscope (60× water objective). Images were collected with LAS AF or the Andor Fusion software respectively. Minor processing (Gaussian blur) was performed using FIJI. For live imaging, embryos undergoing germband extension were mounted ventro‐laterally between O2‐permeable membrane (Sartorius) and a glass coverslip in Voltalef oil (Attachem). Embryos were gently squashed such that the ventrolateral ectoderm was flattened.
Localization quantification
We used FIJI to perform our quantifications. Starting at the center of the basolateral region (which we defined as 0), a freehand line was drawn around the perimeter of the cell and pixel intensity determined at each point along the line. Background signal, measured in the cytoplasm, was subtracted from these points. Intensities were normalized by dividing the median intensity and positions were normalized to a line with a length of 360. The graphical representations show data points from multiple cells (as indicated in the legend) using a floating 10‐point average. Measurement of Mud intensity was sometimes complicated by the proximity between spindle poles, which are highly Mud‐positive, and the cortex, which is less so. In these instances, the overlapping region was excluded from the measurement. These exclusions help to explain why Mud intensity profiles are not as smooth as those measured for other proteins.
Spindle orientation measurements
Follicular epithelium
Spindle angle determination was performed as previously described (Finegan et al, 2018). Angles were determined by drawing a first line connecting either the two spindle poles (if applicable) or along the spindle and a second line along the apical surface of the tissue, then measuring the angle between them. Spindle angle values for control w 1118 are reused between Figs 1D, 3F, 4C, 5C and 6F, and for pins p62 /pins p62 between Figs 3F, 4C and 5C.
Embryonic ectoderm
Spindle angles were determined using FIJI by finding the distance between the mitotic spindle poles: (i) in the xy axis of the flattened ventrolateral ectoderm in a sum projection of the confocal z stack; (ii) in z axis by determining the z planes that the 2 poles appear. The mitotic spindle orientation angle, φ, was calculated by taking the inverse tan of the difference in z depth divided by the distance in xy between the spindle poles (Fig EV3C and D).
Statistical analyses
The Mann–Whitney test was used to determine significance when comparing spindle orientation across conditions/genotypes. No statistical method was used to predetermine sample size, the experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. The chi‐square test of independence was used to determine the significance in Fig 6G.
Author contributions
Kathryn E Neville: Resources; data curation; methodology. Tara M Finegan: Data curation; formal analysis; investigation; visualization; methodology; writing—review and editing. Nicholas Lowe: Resources. Philip M Bellomio: Resources. Daxiang Na: Resources. Dan T Bergstralh: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; investigation; visualization; methodology; writing—original draft; project administration; writing—review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
PDF+
Acknowledgements
We are grateful to the Rochester‐Manchester‐Carolina Meeting Group, the Rochester Invertebrate Group, Holly Lovegrove, and to Bergstralh lab members for comments. Rebecca (Becky) Bastock suggested clonal expression of Ubi‐Pins‐YFP. We thank Allison Zajac (Horne‐Badovinac lab), Adam Martin, and Pejmun Haghighi for sharing flies. We extend very warm thanks to Mark Khoury and David Bilder, who provided emergency flies shortly after our lab's 18 degree incubator broke. This work was supported by National Science Foundation CAREER award 2042280 (PI: Bergstralh) and by National Institutes of Health Grants R01GM125839 (PI: Bergstralh) and S10 RR024577‐01.
EMBO reports (2023) 24: e56074
Data availability
This study includes no data deposited in external repositories.
References
- Berger J, Senti KA, Senti G, Newsome TP, Asling B, Dickson BJ, Suzuki T (2008) Systematic identification of genes that regulate neuronal wiring in the Drosophila visual system. PLoS Genet 4: e1000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Haack T, St Johnston D (2013a) Epithelial polarity and spindle orientation: intersecting pathways. Philos Trans R Soc Lond B Biol Sci 368: 20130291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Lovegrove HE, St Johnston D (2013b) Discs large links spindle orientation to apical‐basal polarity in Drosophila epithelia. Curr Biol 23: 1707–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Lovegrove HE, St Johnston D (2015) Lateral adhesion drives reintegration of misplaced cells into epithelial monolayers. Nat Cell Biol 17: 1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Lovegrove HE, Kujawiak I, Dawney NS, Zhu J, Cooper S, Zhang R, St Johnston D (2016) Pins is not required for spindle orientation in the Drosophila wing disc. Development 143: 2573–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Dawney NS, St Johnston D (2017) Spindle orientation: a question of complex positioning. Development 144: 1137–1145 [DOI] [PubMed] [Google Scholar]
- Bischof J, Björklund M, Furger E, Schertel C, Taipale J, Basler K (2013) A versatile platform for creating a comprehensive UAS‐ORFeome library in Drosophila . Development 140: 2434–2442 [DOI] [PubMed] [Google Scholar]
- Bosveld F, Markova O, Guirao B, Martin C, Wang Z, Pierre A, Balakireva M, Gauge I, Ainslie A, Christophorou N et al (2016) Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature 530: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Neumüller RA, Novatchkova M, Du Q, Knoblich JA (2006) The Drosophila NuMA homolog mud regulates spindle orientation in asymmetric cell division. Dev Cell 10: 731–742 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Camuglia J, Chanet S, Martin AC (2022) Morphogenetic forces planar polarize LGN/Pins in the embryonic head during Drosophila gastrulation. Elife 11: e78779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati M, Gallini S, Pirovano L, Alfieri A, Bisi S, Mapelli M (2016) Concomitant binding of Afadin to LGN and F‐Actin directs planar spindle orientation. Nat Struct Mol Biol 23: 155–163 [DOI] [PubMed] [Google Scholar]
- Chanet S, Sharan R, Khan Z, Martin AC (2017) Myosin 2‐induced mitotic rounding enables columnar epithelial cells to interpret cortical spindle positioning cues. Curr Biol 27: 3350–3358.e3 [DOI] [PubMed] [Google Scholar]
- Chugh P, Paluch EK (2018) The actin cortex at a glance. J Cell Sci 131: jcs.186254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culurgioni S, Alfieri A, Pendolino V, Laddomada F, Mapelli M (2011) Inscuteable and NuMA proteins bind competitively to Leu‐Gly‐Asn repeat‐enriched protein (LGN) during asymmetric cell divisions. Proc Natl Acad Sci U S A 108: 20998–21003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culurgioni S, Mari S, Bonetti P, Gallini S, Bonetto G, Brennich M, Round A, Nicassio F, Mapelli M (2018) Insc:LGN tetramers promote asymmetric divisions of mammary stem cells. Nat Commun 9: 1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaïche Y (2005) Drosophila Ric‐8 regulates Galphai cortical localization to promote Galphai‐dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol 7: 1083–1090 [DOI] [PubMed] [Google Scholar]
- Dimitracopoulos A, Srivastava P, Chaigne A, Win Z, Schlomovitz R, Lancaster OM, Le Berre M, Piel M, Franze K, Salbreux G et al (2020) Mechanochemical crosstalk produces cell‐intrinsic patterning of the cortex to orient the mitotic spindle. Curr Biol 30: 3687–3696.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG (2001) A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol 3: 1069–1075 [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ (2007) Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev 2: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan TM, Na D, Cammarota C, Skeeters AV, Nádasi TJ, Dawney NS, Fletcher AG, Oakes PW, Bergstralh DT (2018) Tissue tension and not interphase cell shape determines cell division orientation in the Drosophila follicular epithelium. EMBO J 38: e100072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloerich M, Bianchini JM, Siemers KA, Cohen DJ, Nelson WJ (2017) Cell division orientation is coupled to cell‐cell adhesion by the E‐cadherin/LGN complex. Nat Commun 8: 13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub O, Wee B, Newman RA, Paterson NM, Prehoda KE (2017) Activation of discs large by aPKC aligns the mitotic spindle to the polarity axis during asymmetric cell division. Elife 6: e32137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG (2010) Par3 controls epithelial spindle orientation by aPKC‐mediated phosphorylation of apical pins. Curr Biol 20: 1809–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F (2006) Drosophila pins‐binding protein mud regulates spindle‐polarity coupling and centrosome organization. Nat Cell Biol 8: 586–593 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, Doe CQ (2009) Identification of an Aurora‐a/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 138: 1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Doe CQ, Prehoda KE (2012) Structure of an enzyme‐derived phosphoprotein recognition domain. PloS One 7: e36014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM (2013) Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gönczy P (2013) NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J 32: 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA (1996) Role of inscuteable in orienting asymmetric cell divisions in Drosophila . Nature 383: 50–55 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461 [DOI] [PubMed] [Google Scholar]
- Liao EH, Gray L, Tsurudome K, El‐Mounzer W, Elazzouzi F, Baim C, Farzin S, Calderon MR, Kauwe G, Haghighi AP (2018) Kinesin Khc‐73/KIF13B modulates retrograde BMP signaling by influencing endosomal dynamics at the Drosophila neuromuscular junction. PLoS Genet 14: e1007184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough KJ, Byrd KM, Descovich CP, Spitzer DC, Bergman AJ, Beaudoin GMJ, Reichardt LF, Williams SE (2019) Telophase correction refines division orientation in stratified epithelia. eLife 8: e49249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove HE, Bergstralh DT, St Johnston D (2019) The role of integrins in Drosophila egg chamber morphogenesis. Development 146: dev182774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MS, Prehoda KE (2013) A NudE/14‐3‐3 pathway coordinates dynein and the kinesin Khc73 to position the mitotic spindle. Dev Cell 26: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas EP, Raff JW (2007) Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol 178: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais‐de‐Sá E, Sunkel C (2013) Adherens junctions determine the apical position of the midbody during follicular epithelial cell division. EMBO Rep 14: 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Bellaïche Y (2011) Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell 21: 102–119 [DOI] [PubMed] [Google Scholar]
- Nakajima YI, Lee ZT, McKinney SA, Swanson SK, Florens L, Gibson MC (2019) Junctional tumor suppressors interact with 14‐3‐3 proteins to control planar spindle alignment. J Cell Biol 218: 1824–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M, Natsume T, Kanemaki MT, Kiyomitsu T (2018) Dynein–dynactin–NuMA clusters generate cortical spindle‐pulling forces as a multi‐arm ensemble. Elife 7: e36559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J (2010) An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila . EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R (2015) The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N (1988) The maternal effect of lethal(1)discs‐large‐1: a recessive oncogene of Drosophila melanogaster . Dev Biol 127: 392–407 [DOI] [PubMed] [Google Scholar]
- di Pietro F, Echard A, Morin X (2016) Regulation of mitotic spindle orientation: an integrated view. EMBO Rep 17: 1106–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadaoui M, Machicoane M, di Pietro F, Etoc F, Echard A, Morin X (2014) Dlg1 controls planar spindle orientation in the neuroepithelium through direct interaction with LGN. J Cell Biol 206: 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Knoblich JA (2000) A protein complex containing Inscuteable and the Galpha‐binding protein pins orients asymmetric cell divisions in Drosophila . Curr Biol 10: 353–362 [DOI] [PubMed] [Google Scholar]
- Schiller EA, Bergstralh DT (2021) Interaction between discs large and pins/LGN/GPSM2: a comparison across species. Biol Open 10: bio058982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalen M, Johnston CA, Martin CA, Dumortier JG, Prehoda KE, David NB, Doe CQ, Bellaïche Y (2010) The Fz‐Dsh planar cell polarity pathway induces oriented cell division via mud/NuMA in Drosophila and zebrafish. Dev Cell 19: 740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L, Poulson ND, Foote HP, Lechler T (2013) NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol Biol Cell 24: 3651–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ (2005) Microtubule‐induced pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123: 1323–1335 [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ (2009) Spindle orientation during asymmetric cell division. Nat Cell Biol 11: 365–374 [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ (2006) The NuMA‐related mud protein binds pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol 8: 594–600 [DOI] [PubMed] [Google Scholar]
- Werts AD, Goldstein B (2011) How signaling between cells can orient a mitotic spindle. Semin Cell Dev Biol 22: 842–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E (1999) Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402: 544–547 [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E (2000) Drosophila atypical protein kinase C associates with bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol 150: 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ (1989) Molecular cloning of the lethal(1)discs large‐1 oncogene of Drosophila . Dev Biol 134: 222–235 [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W (2000) Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100: 399–409 [DOI] [PubMed] [Google Scholar]
- Yu JX, Guan Z, Nash HA (2006) The mushroom body defect gene product is an essential component of the meiosis II spindle apparatus in Drosophila oocytes. Genetics 173: 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AL, Horne‐Badovinac S (2022) Kinesin‐directed secretion of basement membrane proteins to a subdomain of the basolateral surface in Drosophila epithelial cells. Curr Biol 32: 735–748.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Graham OS, Raposo A, St Johnston D (2012) Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal‐ventral axis. Science 336: 999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Wan Q, Meixiong G, Du Q (2014) Cell cycle‐regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol Biol Cell 25: 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shang Y, Xia C, Wang W, Wen W, Zhang M (2011) Guanylate kinase domains of the MAGUK family scaffold proteins as specific phospho‐protein‐binding modules. EMBO J 30: 4986–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]


