Abstract
Ebola Virus Disease (EVD), caused by the Ebola virus (EBOV), is a severe illness characterized by case fatality rates up to 90%. The sporadic nature of outbreaks in resource-limited areas has hindered the ability to characterize the pathogenesis of EVD at all stages of infection, but particularly during early host responses. Pathogenesis is often studied in non-human primate (NHP) models of disease that replicate major aspects of human EVD. Typically, NHP exposures utilize a large infectious dose, are carried out through intramuscular or aerosol exposure, and have a fairly uniform disease course. In contrast, here we report our analysis of the host response to EBOV following intranasal exposure. Twelve cynomolgus macaques were infected with 100 plaque forming units of EBOV/Makona through intranasal exposure, and presented with varying time to onset of EVD. We utilized RNA-Sequencing and a newly developed NanoString codeset to monitor the host response via changes in RNA transcripts over time. When individual animal gene expression data were phased based on onset of sustained fever, the first clinical sign of severe disease, mathematical models indicated that interferon stimulated genes (ISGs) appeared as early as 4 days before fever onset. This demonstrates that lethal EVD has a uniform and predictable response to infection regardless of time-to-onset. Furthermore, expression of a subset of genes could predict disease development prior to other host-based indications of infection such as fever.
One Sentence Summary:
Examination of an animal model of ebolavirus infection with variable disease onset identifies highly conserved host markers of infection that precede symptoms by 4 days. .
Introduction
Ebola virus disease (EVD) is a serious illness that can cause up to 90% case fatalities in humans and non-human primates (NHPs) (1). The causative agent, Ebola virus (EBOV), spills over into human and NHP populations on a sporadic basis and typically causes limited outbreaks. A notable exception was the 2013–2016 outbreak in West Africa, which caused approximately 28,000 cases (2). This event highlighted the need to better understand the pathogenesis and host response to EVD as a means to create better therapeutics, vaccines, and diagnostics.
The study of EVD is difficult since human clinical samples available for research purposes are rare and priority is not usually placed on preserving samples for subsequent experimentation during an outbreak. To fill this gap, animal models have been developed to study EVD in vivo. The current model most widely used for understanding pathogenesis and testing of therapeutics and vaccines is NHPs, specifically cynomolgus and rhesus macaques (3). Upon infection with EBOV, NHPs display many of the same symptoms as humans including coagulopathy, lymphocytosis, and rash (4). The NHP EVD model typically employs an infectious dose of 1000 plaque forming units (PFU) through an intramuscular injection (IM) or aerosol exposure resulting in disease symptoms within 3–5 days and 100% lethality within 6–10 days post-exposure (5, 6). Intranasal exposures at 250 or 500 PFU also result in a uniformly lethal model of EVD, but with a delay in time to death compared to 100 PFU IM exposures (7). In contrast, human EVD is thought to result from exposure to lower infectious doses via parenteral or mucosal routes (8, 9) with the incubation period ranging widely from 2–21 days post-exposure to EBOV (10). These differences highlight the need for experimental NHP models that more closely recapitulate natural human exposure and ensuing disease.
Important data regarding the host response to infection has been gained from investigating circulating immune responses in 100% lethal NHP models (11). For example, transcriptomics has been used to explore early host responses to infection in time-resolved NHP datasets (12–15) and to define important aspects of the host response that contribute to survival in mice (16), NHPs (17, 18), and humans (19–21). These analyses have pointed to a robust and early immune response following EBOV exposure largely characterized by the upregulation of interferon-stimulated genes (ISGs).
Left unaddressed in these analyses has been whether the host response to infection in a uniform disease onset model is the same as that seen in a more variable onset disease situation that is representative of human cases of EVD. We were able to approach this question through a recently developed model of EVD. This model used a mucosal exposure route that was less than 100% fatal and showed variable onset of disease. We investigated the host response to infection in this model using telemetry devices, blood chemistry analysis, and transcriptomic approaches of RNA-Sequencing (RNA-Seq) and NanoString. This allowed us to map the NHP response to infection at both symptomatic and asymptomatic phases and to relate these findings to human EVD
Results
In the context of ongoing efforts to develop NHP models that more accurately replicate EVD observed in humans, 12 cynomolgus macaques were exposed to EBOV/Mak via the intranasal route. Virus (target dose of 100 PFU) was administered either via pipette or Mucosal Atomization Device (MAD) to each NHP. Following exposure, disease onset was not uniform, with animals showing variable onset of disease symptoms and time to death. Two animals were asymptomatic for disease during the course of the experiment.
Description of 4 distinct response groups following exposure
The NHPs clustered into 4 distinct groups based on appearance of disease symptoms, onset of viremia, and time to death (Figure 1A–B) and had an 83% fatality rate. Three animals had an expected disease course (Group 1 (Normal): NHPs 1, 2, and 12), became quantifiably EBOV-positive as detected by RT-qPCR on day 6 post-exposure and had a mean time to death of 10.47 days. Four animals had a delayed onset of disease (Group 2 (Delayed): NHPs 3, 5, 8, and 10) and had a mean time to death of 13.31 days. In this group, NHPs did not develop quantifiable viremia until days 10 or 12 post-exposure, but viral RNA was qualitatively detected (above the limit of detection but below the limit of quantification for the assay) as early as day 3 in NHP 10 and on day 6 in NHPs 3 and 8. NHP 5 did not have any detectable viral RNA until day 10. Three animals had a late onset of disease (Group 3 (Late): NHPs 4, 6, and 9) and a mean time to death of 21.42 days. NHP 6 had quantifiable viremia on day 20, but viral RNA was only qualitatively detected on days 6, 10, and 14. NHP 9 first had quantifiable viremia on day 21, but was also qualitatively positive on day 14. NHP 4 had borderline quantitative viremia on day 10, was qualitatively positive on day 14, and then quantitatively positive again on day 20. Finally, two animals did not become quantitatively RT-qPCR-positive across the time course of the experiment (Group 4 (No response): NHPs 7 and 11) and survived out to 41 days which was the pre-defined experimental endpoint. However, viral RNA was qualitatively positive in these animals on days 6 and 10 post-exposure, respectively. Survival and time to death did not correlate with the method of intranasal exposure (p = 0.1292).
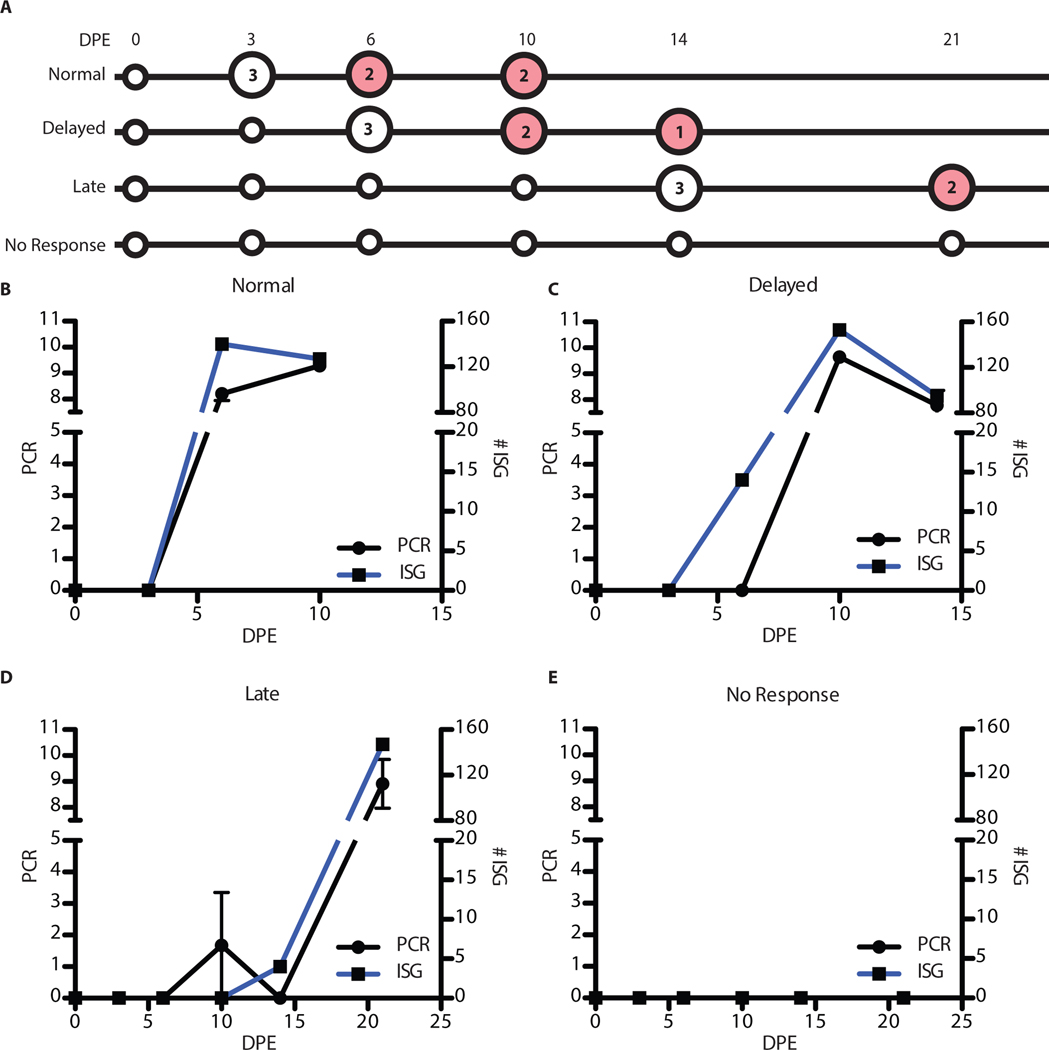
Figure 1: Overview of the separation of animals and analysis performed.
(A) Table showing the different animals (rows) and the different days post-exposure(columns). White square signifies the animal was not PCR-positive at that time point, blue indicates they were PCR positive, and greys means that the PCR results were below the limit of quantification. The number of plus signs in the grey boxes represents the number of PCR replicates that were positive. (B) Comparison of the 4 groups and their survival curves. The x-axis is the days post-exposure and the y-axis is the percent survival. The green line is the Normal group (NHP 1,2,12), blue is the Delayed group (NHP 3,5,8,10), red is the Late group (NHP 4,6,9) and black is the No Response (NHP 7,11). (C) Overview of the RNA quantification analysis performed on the animals. The x-axis is the days post-exposure and the y-axis is the different animals organized by outcome groups (Group 1: green, Group 2: blue, Group 3: red)
To determine if physiological differences that correlated to disease outcome were detectable prior to exposure, we analyzed blood chemistry data, hematology, and soluble proteins collected prior to exposure (day 0). A one-way ANOVA was performed for each of the 39 measured parameters and a regression coefficient to symptom onset (R2) value was calculated. Only one variable, mean corpuscular volume (MCV), was significantly different between the groups (p < 0.05) (Supplemental Figure 1A). The difference observed in MCV was driven by the Delayed animals which had a significantly lower value than the Normal group (p-value = 0.02)(Supplemental Figure 2) that was not significantly different from any other group. A low MCV value, which measures the average size of red blood cells, can signify blood loss or anemia. However, since the values were only significant in one of the group comparisons, we think it unlikely to have contributed to the differential disease onset.
To further investigate if there was a difference observed at the day of infection we performed PC analysis on the same parameters from day 0 (Supplemental Figure 1B). We would hypothesized that if the animals were similar across the different response groups prior to exposure, one group would not cluster differently from any of the others. There is no strong clustering within groups signifying there is no group that is distinctive from the others. Also, all the groups are centered close to the point (0,0), meaning that none of the groups had large outlier values that could have contributed to the different onset of disease. Based on this analysis, we can conclude that it was unlikely that a pre-existing physiological difference in the animals contributed to the different disease onsets.
The variable onset of disease in these animals presented a unique opportunity to identify factors that might be associated with different responses to EBOV intranasal challenge. We investigated clinical, biochemical, physiological, and transcriptomic data to identify markers of disease onset within this cohort. An overview of the transcriptomic data available can be seen in Figure 1C. We used two approaches for transcriptomic analysis – RNA-Seq and NanoString. RNA-Seq enables a hypothesis-neutral investigation of RNA abundance changes. In contrast, we also utilized a newly developed NanoString codeset that recognizes 769 NHP, and which we have previous demonstrated effective with EBOV/Mak-infected NHPs (15). Although NanoString does not provide the transcriptomic depth of RNA-seq, it is much more rapid compared to RNA-seq, and does not have as high of a requirement for RNA quality.
Clinical chemistry changes in symptomatic animals
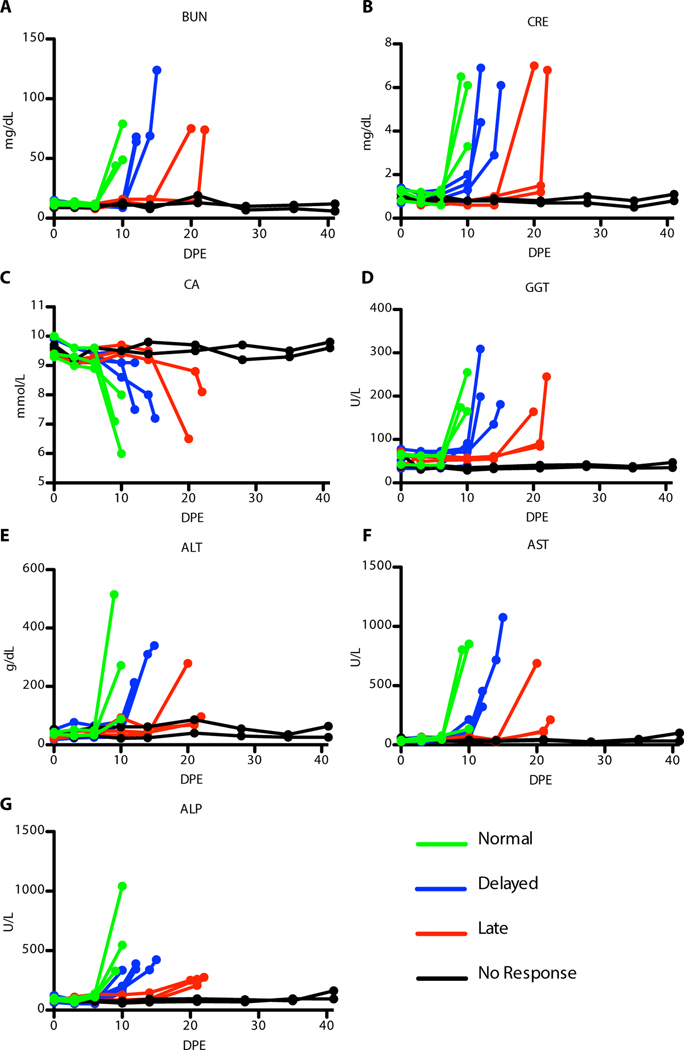
Typical manifestations of EVD were observed in all non-survivors, including maculopapular rash, lymphadenopathy, anorexia, and fever. Indicators of coagulopathy, such as bleeding or hemorrhage, were largely absent in these animals. Some animals displayed respiratory signs including cough, labored breathing, and use of accessory or abdominal muscles for breathing. Additionally, all non-survivors had alterations in hematological and serum chemistry parameters, regardless of the time-to-death. These changes mirrored previously reported EBOV infection of NHPs, including increases in the blood urea nitrogen (BUN), creatinine (CRE), calcium (CA), and gamma-glutamyl transferase (GGT) (Figure 2). Increases in liver enzymes such as alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were also noted, along with an increase in alkaline phosphatase (ALP) (Figure 2). Changes in blood chemistry coincided with the development of clinical signs and appearance of viral RNA in plasma; these changes were delayed in those animals with an extended time-to-death. The two survivors generally did not have any changes in behavior, health, or clinical pathology parameters associated with EVD.
Figure 2: Blood chemistry data for animals over time.
(A) Blood urea nitrogen (BUN) (B) creatinine (CRE) (C) calcium (CA) (D) gamma-glutamyl transferase (GGT) (E) alanine aminotransferase (ALT) (F) aspartate aminotransferase (AST) (G) alkaline phosphatase (ALP). Group 1 (green), Group 2 (blue), Group 3 (red), and Group 4 (black)
Innate immune pathway activation during acute EVD
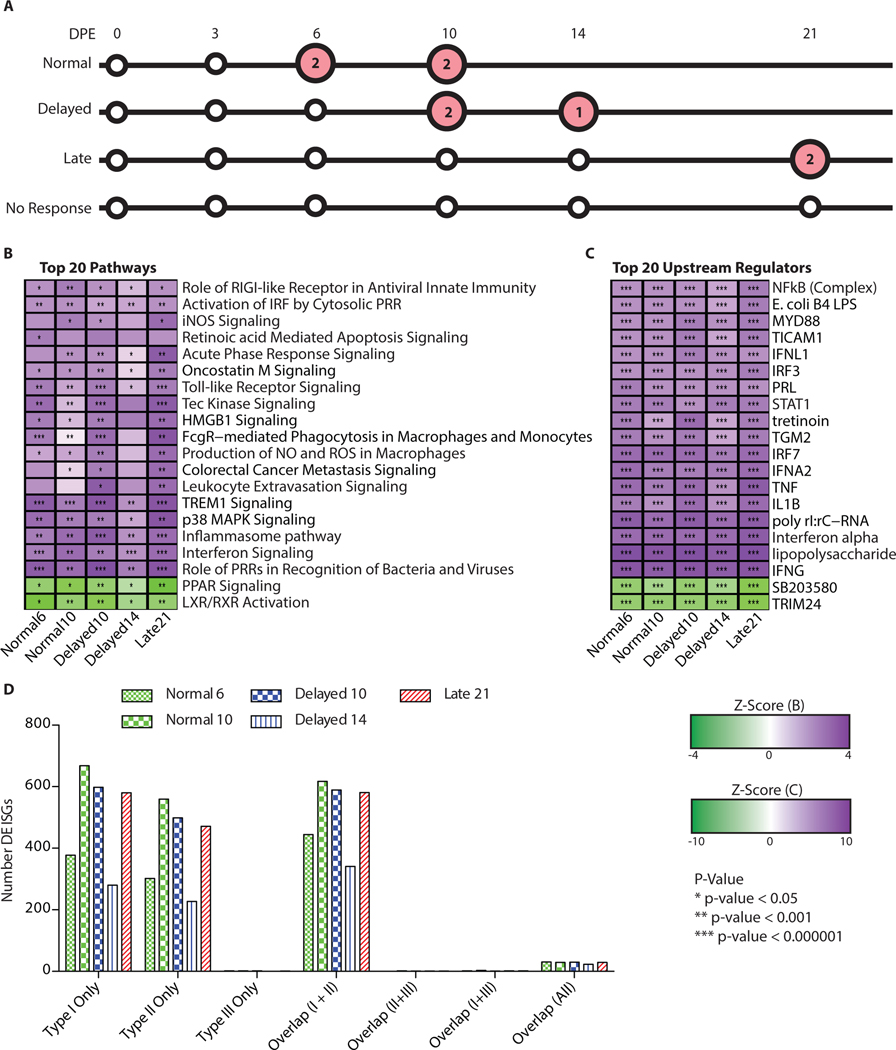
The varying times to death among the study animals led us to investigate the nature of the host immune response following onset of symptoms as determined by onset of sustained fever. We performed differential gene expression analysis in the RNA-Seq dataset and biological pathway analysis in Ingenuity Pathway Analysis (IPA) for each group relative to pre-challenge baseline for symptomatic time points (Figure 3A). All animals showed a similar host response at the end stages of disease as evidenced by upregulation of the same top 20 signaling pathways based on IPA results (Figure 3B). The highest level of conservation across the groups was the Normal and Delayed groups at 10 days post-exposure and the Late group at 21 days post-exposure. Due to loss of samples from poor RNA quality for day 14 post-exposure in the Delayed group, we only had usable samples from one animal remaining at this time point thus reducing our ability to find statistically differentially regulated genes resulting in fewer pathways with enrichment.
Figure 3: Comparison of leading pathways and upstream regulators.
Overview of the most strongly differentially regulated pathways in the viremic animals. (A) Overview of the samples analyzed (large circles) which were symptomatic (red circles) with the number of biological replicates denoted in the circles. (B) Significantly differentially regulated pathways. Heatmap is clustered on pathways (rows) (dendogram of clustering no shown). The darker purple indicates a strong up-regulation of the pathways (positive z-score) and the green means down-regulation of the pathways (negative z-score). The p-values represent if there is a significant number of genes in the pathway that are differentially expressed as determined by IPA though a Fischer’s Exact test. * < 0.05, ** < 10−3, and *** < 10−6. (C) Up-stream regulators from IPA. Purple denotes up-regulation and green denotes down-regulation (log fold change). (D) Breakdown of the interferon response to the type of interferon responsive genes. The x-axis is the specific types (Uniquely Type I, II, III) and overlap between the different responses. The red bars are samples from the Normal group, green are from Delayed, and blue is from Late. The different patterns correspond to different days post-exposure.
Enriched pathways included activation of innate immune pathways (role of pattern recognition receptors, interferon signaling, role of RIG-I in antiviral response) as well as acute phase response signaling and pathways associated with nitric oxide (NO) and reactive oxygen species (ROS). These pathways were consistent within each group and also were consistent with host gene expression seen during viremic stages of disease following higher-dose exposure to EBOV/Kikwit (22) and other EBOV/Mak challenges (13). Interestingly we identified two pathways that were among the top 20 that were down-regulated (PPAR signaling and LXR/RXR activation) during infection. These pathways are important in lipogenesis, cell survival, and can down-regulate the inflammatory response through inhibition of NF-kB and TLR-4 (23, 24). The top 20 regulators of downstream targets were also consistent with an over-activation of the innate immune response and show a strongly conserved host response during the symptomatic stage of disease (Figure 3B and C).
To further characterize the interferon response during the symptomatic stage of infection, we used the interferome database (25) to compare the type I, II, and III interferon responses based on the number of interferon responsive genes differentially regulated. Across all the symptomatic animals, there is a strong type I and type II interferon response but minimal type III response (Figure 3D). This is consistent with the upstream regulators where interferon alpha (IFNA) and interferon gamma (IFNG) are significantly enriched (p < 10−6) across all the groups.
Cytokine gene expression during acute EVD
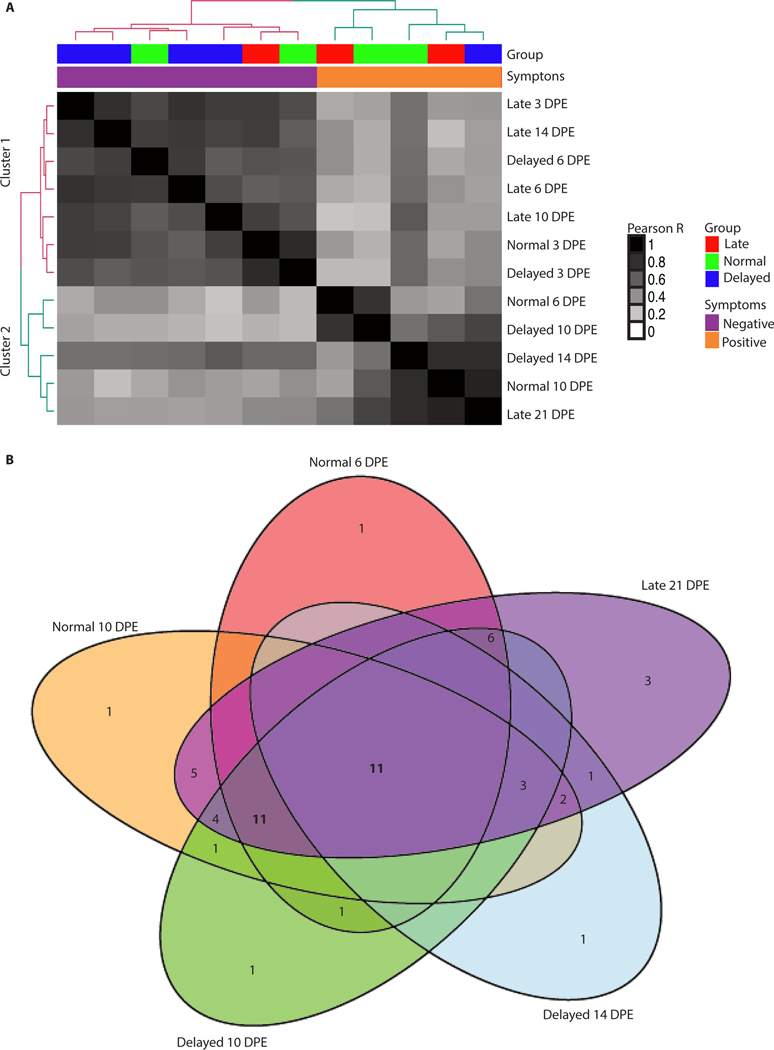
Using differentially expressed cytokine genes identified through RNA-seq, we examined whether animals that showed symptoms at different times of infection had differences in their cytokine response during the course of disease. We identified 57 differentially expressed cytokine mRNAs (Supplemental Table 1) and then determined whether the animals showed similar expression based on the groups or on the distinction between symptomatic or asymptomatic. We found that the dominant driver of clustering in this approach was symptom onset (Figure 4A). In contrast, there was little clustering based on time post EBOV exposure further suggesting the symptom onset is a more important indicator of disease course.
Figure 4: Late state cytokine response conservation.
Analysis of the conservation of the cytokine response. A) Correlation plot using only the cytokine genes. The rows and columns are the same and represent a single group at a single time point. The dendogram is colored based on a k-means clustering with 2 centers. The first color bar at the top shows the groups (green = Normal, blue = Delayed, red = Late). The second color bar indicates when the groups become symptomatic with orange for symptomatic and purple for those animals still in the incubation period. (B) Venn diagram of cluster 2 samples with cytokine genes that are differentially expressed. Spots lacking a value indicate 0 genes.
Of the cytokines in the RNA-Seq dataset that were differentially regulated, 11 were common to all groups and time points (Figure 4B). Again, due to sampling size being reduced to one animal in the Delayed group at day 14 post-exposure, fewer differentially regulated genes were identified. Comparing only the Normal group at day 6 and 10 post-exposure, Delayed group at day 10 post-exposure and the Late group at day 21 post-exposure, the number of conserved cytokines increases to 22. This suggests that the cytokine response is similar in the symptomatic animals. To confirm the host response profiles that we observed with NGS, we also used a NanoString codeset to profile the expression of 769 NHP genes that has been previously shown to be beneficial as a secondary means for RNA-quantification in NHPs infected with EBOV (15). This analysis confirmed the upregulation of the cytokine genes identified by NGS (Supplemental Figure 3).
Early and transient transcriptional responses in NanoString data
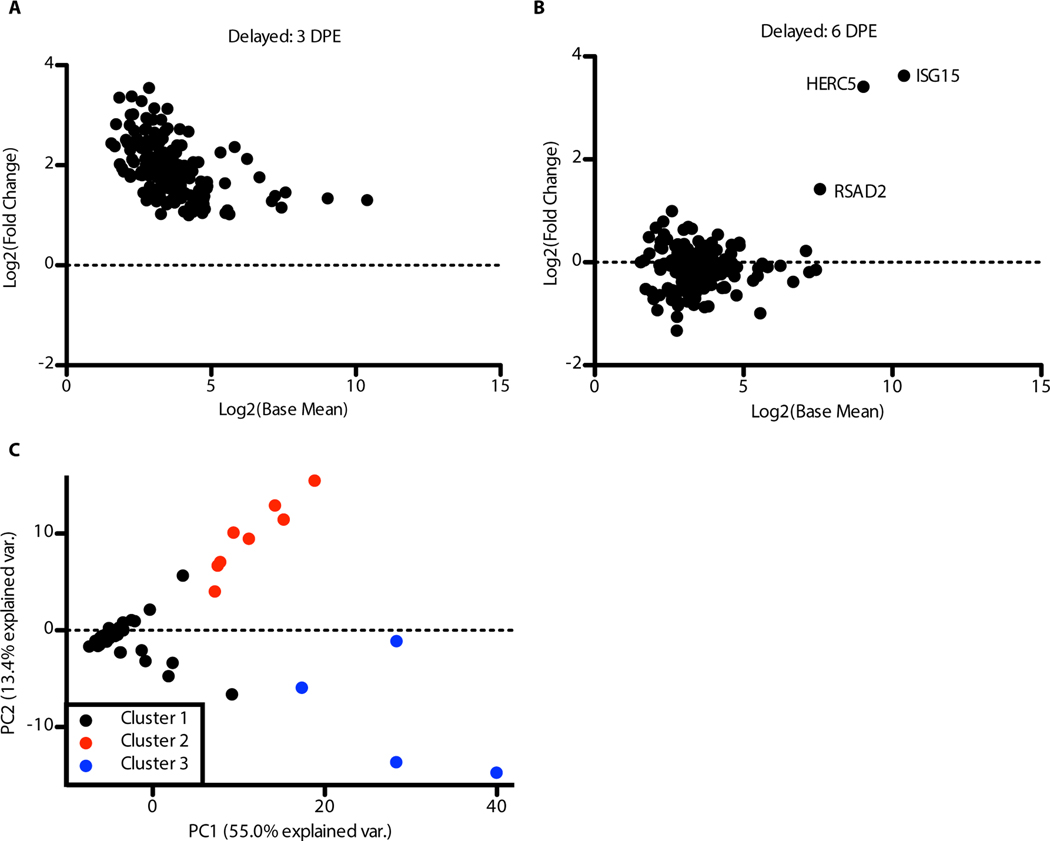
The NanoString NHP gene expression codeset has the potential for increased sensitivity vs RNA-Seq for targeted gene expression profiling (15). Notably in this dataset, we identified a subset of cytokines that were upregulated at day 3 post-exposure in NHPs that showed a delayed course of disease (NHPs 3, 5, 8, and 10).
This up-regulation was consistent across all four animals in this cohort and was not identified as up-regulated in NGS datasets. This finding led us to broaden our analysis of the NanoString data to see if there were other genes upregulated at this early time point. Subsequently, we identified 178 genes that were found to be up-regulated on day 3 post-exposure in the Delayed animals, but had returned to near baseline by day 6 post-exposure with the exception of three interferon stimulated genes (Figure 5A–B, Supplemental Figure 4, Supplemental Table 2). We performed k-means clustering (k = 3) on the NanoString expression data for all NHPs at all time points (Figure 5C) using these 178 genes. In the PCA plot, the 3 clusters are shown. Cluster 1 is made up of most samples. Cluster 2 represents the Delayed animals at day 3 post-exposure. One animal from the Normal group (NHP 2), one from the Late (NHP 9), and one from the No response group (NHP 11) at day 3 post-exposure are also included in this cluster showing a similar up-regulation at this time point. Three points formed Cluster 3. All three were end-stage samples that were close to death.
Figure 5: Early and modest immune response at day 3 post-exposure in Delayed animals.
Analysis of the NanoString samples with only the genes identified as being up-regulated at an early time point (day 3 post-exposure Delayed). (A) MA plot of the 178 up-regulated genes at day 3 post-exposure in Delayed animals. Each point represents a gene. The x-axis is the log2 of the base mean counts and the y-axis is the log2 fold change relative to day 0. (B) MA plot for the same genes at day 6 post-exposure. (C) PCA of the 178 up-regulated genes across all samples. K-means clustering was performed on all principal components with k = 3 to determine the clustering. The PCA plot is showing the first two principal components. The colors represent the different clusters.
We utilized PANTHER (26) to determine if any one set of genes was significantly enriched in these 178 genes. As a reference, we used only those genes included in the NanoString codeset to account for potential bias. We found that GO terms associated with cytokines, cytokine response (GO:0005125) and cytokine receptor binding (GO:0005126), were significantly enriched (adjusted p-value = 8.91 × 10−5 and 6.05 × 10−3 respectively). Cytokine response genes that were up-regulated at day 3 post-exposure in Delayed animals included interferons (e.g. IFNB1, IFNL2, IFNL3), pro-inflammatory cytokines (e.g. IL17F, IL6, IL10) and many T-cell recruitment/activation genes (e.g. CCL18, IL2, CCL17). Though there was detection of type I and type III interferons, there was no significant up-regulation of interferon-stimulated genes at this point. This may suggest a modest immune response to the infection in the nasal cavity and that day 3 post-exposure was optimal for detection with the Delayed animals. Transcriptional analysis of early events in the nasal cavity would be needed to support this hypothesis.
Interferon Simulated Gene activation immediately prior to onset of EVD symptoms
As the response to infection in the symptomatic stage offered few differences to explain disease onset, we moved to studying events after infection at the time point directly preceding the symptomatic stage (Figure 6A). Previous studies have suggested that transcriptional analysis can identify host innate immune response genes are induced following hemorrhagic fever virus infection and that some of these genes can be detected prior to the onset of symptoms (12, 27–29). To determine whether there was any evidence of host-response gene expression changes at early times post-exposure, we examined the RNA-Seq data for evidence of transcripts that were induced prior to the display of overt symptoms (fever as determined by telemetry) or quantifiable viremia (determined by RT-qPCR) and that were then sustained through the end of infection. Our analysis identified a number of genes that were induced at or prior to induction of fever. In challenged animals that did not develop disease we did not find these genes to be significantly differentially regulated. Many of these “sentinel” genes are classified as interferon stimulated genes, or ISGs.
Figure 6: Analysis of the pre-viremic ISG response.
(A) Analysis of the onset of the ISG response. Large circles represent samples that were used in the analysis with red denoting symptomatic samples. The numbers indicate how many samples were used. (B) Comparison of the onset of viremia to induction of the group of ISGs. The days post-exposure are on the x-axis. The left y-axis is the PCR values (black line) and the right is the number of genes annotated as ISGs that are up-regulated (blue line). (C) Similar plot for the Delayed animals, (D) Late animals, and (E) No response animals.
The transcriptome from animals in the Normal group showed increased accumulation of approximately 100 ISGs concurrent with the onset of quantifiable viremia and symptoms (Figure 6B). In animals in the Delayed group, 14 ISG transcripts were significantly up-regulated (log2(fold change) > 1, adjusted p-value < 0.05) by day 6 post-exposure. This represented a gene expression spike that occurred 4 days before the onset of quantifiable viremia and fever (Figure 6C). The pre-symptomatic expression of host innate immune genes was even more remarkable in animals that showed a late onset of disease. ISG transcripts were upregulated at 14 days post-exposure, which was 7 days before the onset of quantifiable viremia (Figure 6D).
These data showed that ISG mRNA up-regulation is a pre-symptomatic indicator of the development of EVD. This is summarized in Table 1, which shows that the ISG markers are highly expressed by day 6 in the Normal group, coincident with fever and quantifiable viremia, but appear markedly in advance of both fever and quantitative viremia in cases of delayed disease onset. In the Delayed and Late groups, the interferon response preceded a sustained fever by 4 and 5 days respectively. This pre-symptomatic response was a conserved response. Of the 4 ISGs upregulated at day 14 post-exposure in the Late animals (NHPs 4, 6, and 9), 3 of the 4 were identical to ISGs upregulated at day 6 post-exposure in the Delayed animals. All were upregulated in animals that showed the expected timing of symptom development.
Table 1.
Comparison of the ISG response, quantifiable viremia, and fever.
| Group | ISG+ | PCR+ | Fever (DPI) |
|---|---|---|---|
| Normal | 6 DPI | 6 DPI | 7.1 ± 0.99 |
| Delayed | 6 DPI | 10 DPI | 9.46 ± 0.06 |
| Late | 14 DPI | 21 DPI | 19 ± 0.74 |
| No Response | N/A | N/A | N/A |
The early upregulation of these ISGs was also seen using the NanoString codeset, which contained 80 ISGs. Differential expression analysis of genes in the NanoString codeset showed that all symptomatic animals had upregulated ISGs while asymptomatic animals (Group 4, No response) did not (Supplemental Figure 5). As was seen in our NGS datasets, at day 6 post-exposure in the Delayed group and day 14 post-exposure in the Late group, there was up-regulation of IFIT2/3/5, OASL, ISG15 and DDX58. Expression of these genes persisted through to the late stage of disease and was conserved between animals in each group (Supplemental Figure 5).
Conservation of ISG response to Human EVD
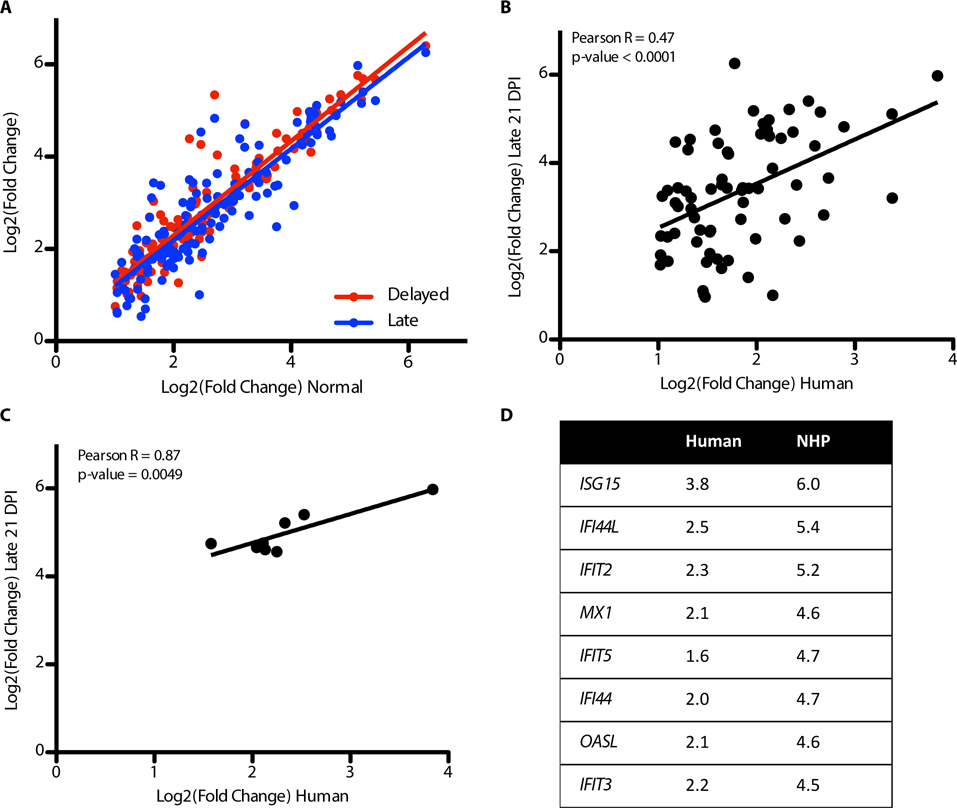
The ISG response during the acute phase of EVD in NHPs was highly conserved among the groups. Figure 7A shows the fold change of ISGs in Normal animals at day 10 post-exposure (X-axis) compared to the Delayed animals at day 10 post-exposure and the Late animals on day 21 post-exposure (Y-axis). The lines represent linear fits. As can be seen, the ISGs are highly similar in their expression values across the different groups suggesting that mRNAs that make up the response were highly conserved despite different times to the onset of disease. The conservation of the late stage animals (Delayed and Late groups) and the early response suggests that the ISG response is up-regulated at similar times and in a similar orders. There is a significant correlation of both the Delayed group ISG expression to the Early group ISG expression (Pearson r = 0.93, p < 0.0001) and the Late group ISG expression to the Early group ISG expression (Pearson r = 0.89, p < 0.0001) demonstrating a high conservation of ISG expression.
Figure 7: Correlation of NHP and human ISGs.
Analysis of the fold induction correlation with regard to the ISG response in NHPs to human fatalities. (A) Correlation plot of all ISGs that are identified as being differentially regulated at any time point in the NHP dataset. The x-axis is the log2 fold change for the Normal animals at day 6 post-exposure and the y axis is the log to fold change for the Delayed animals at day 10 post-exposure (red points and line) or the Late animals at day 21 post-exposure (blue points and line). The lines are linear fits. (B) Correlation of the human fatal cases (log2 fold change compared to controls) on the x-axis and the fold change of the Late group (log2 fold change compared to controls) at day 21 post-exposure. The line represents a linear fit of the data. (C) Correlation plot similar to B but with only the early ISGs shown. D) table of the genes in C with their comparison of the log2 fold change in humans and NHPs. Reported p-values are from a correlation test.
To determine whether the ISGs up-regulated in NHPs were also up-regulated in clinical patients with EBOV/Mak, we compared the fold induction of the ISGs that were identified in this study to ISGs that showed increased expression in a transcriptional analysis of EBOV-infected patients in Guinea (19) who would go on to succumb to disease. These patients were all symptomatic for disease at the time of sampling and had a mean time from self reported symptom onset of 5.9 days (19). A correlation plot shows that many of the same ISGs were up-regulated in both NHPs and in human host responses to EBOV infection (Figure 7B). From this analysis we identified a small subset of ISGs that could act as early indicators of disease in NHPs and humans: IFI44, IFI44L, IFIT2, IFIT3, IFIT5, ISG15, MX1, and OASL (Figure 7C and D). Based on their profile of expression in the NHPs and their conservation into the human dataset, we suggest that at a pre-symptomatic time point, these genes could act as early indicators of viral infection.
Modeling Host Dynamics of Infection
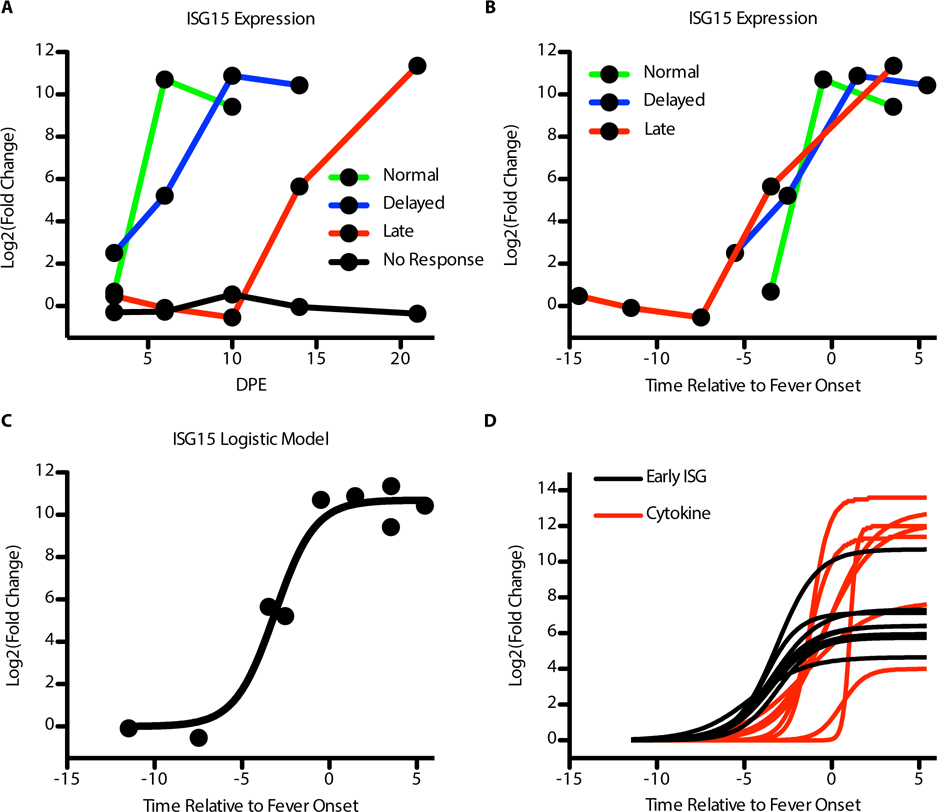
This dataset provided a chance to look at modeling the host response relative to the end of the incubation period to determine if 1) there is a way to align the samples to generally analyze the host response across the three symptomatic groups, and 2) to see if we can model the different gene groups to more robustly determine which will be up-regulated first and how we expect these genes to be up-regulated. The gene expression changes across the four groups did not align well when comparing the fold changes observed in the NanoString codeset to the days post-exposure (Figure 8A). However, using the telemetry data, we were able to determine a mean time to onset of fever for each of the days post-exposure for each group. We assigned 0 to be the mean day of onset of sustained fever (>1.5 °C above mean temperature). All the sampling days were then either negative (meaning they happened before the onset of a fever) or positive (meaning they happened after the onset of a fever). When we looked at the gene expression data using time relative to onset of sustained fever, we found that the samples aligned very well to each other (Figure 8B). We concluded that despite differential onset of symptoms and gene expression, the pattern of gene expression relative to fever onset is conserved.
Figure 8: Host response to infection modeled relative to fever onset.
Alignment and modeling of gene expression across the four NHP groups. (A) Example gene (ISG15) expression (log2 fold change, y-axis) in the NanoString dataset relative to day post-exposure (x-axis). (B) Expression of the same example gene (log2 fold change, y-axis) relative to the onset of a fever (0) on the y-axis. Green line is the Normal animals, blue is the Delayed animals, and red is the Late animals. (C) Logistic model fit for the expression of ISG15 relative to the onset of fever. The points are the mean log2 fold change for a group at its time relative to fever onset and the solid line is the model fit for the fold change. (D) Comparison of the early ISG expression (black lines) logistic model fit relative to the onset of fever (x-axis) to many cytokine genes (red lines). The dashed line is for a reference of when the genes expression crosses the threshold of a log2 fold change > 2.
Next, we used a logistic model to fit the data to curves. This type of model does limit our analysis to genes that show a pattern of expression consistent with a logistic curve. We fit a single model to a single gene at a time (Figure 8C). We did not include the time point of day 3 post-exposure since we wanted to examine gene expression relative to acute disease and the day 3 post-exposure spike of cytokines is eliminated by day 6 post-exposure. We found that the ISGs identified as early up-regulators preceded expression of cytokine genes in all cases (Figure 8D). Also, their predicted mean time to detectable expression (log2 fold change > 2) was found to be 4.6 days before the onset of fever (± 0.46 days). This means that the early interferon genes are strongly up-regulated 4.5 days before the onset of symptoms. Finally, this response is followed by the cytokine response which reached higher expression overall than the interferon response (Figure 8D) but was not significantly expressed earlier than the end of the incubation period. This shows that the cytokine response is concurrent with the onset of fever.
Discussion
A striking aspect of our results is that despite the different times to onset of disease, the host response was similar across the three symptomatic groups (Group 1 (Normal), 2 (Delayed), 3 (Late)). The Delayed and Late groups each had a longer mean time to death than the standard 6–10 days. By shifting the sampling days to align the different response groups by time to symptom onset instead of days post exposure, we observed that the host response follows a conserved and predictable pattern of gene expression. We were able to then generate mathematical models of gene expression to show that pre-symptomatic gene expression can be detected up to 4 days prior to fever
An important implication of this study is the potential use of host markers as pre-symptomatic indicators of disease. We found that through modeling the gene data relative to the onset of a fever, the interferon response was detectable up to 4 days earlier. When we corrected for sampling days, ISGs expression was still detectable up to 3 days before the onset of fever. Earlier studies have shown that gene expression changes are detectable in vivo following EBOV exposure (12, 30, 31), but our results here greatly expand the utility of these markers. The appearance of host-based markers up to 4 days prior to fever suggests that they could be used to track individuals at high risk for infection including hospital personnel caring for EBOV-infected individuals, close-contacts of the same, or individuals who have been in an environment where EBOV is present. Based on our work here, NanoString based assays appear to be a feasible approach as a host based diagnostic, but other multiplexed nucleic acid detection platforms could be equally useful.
The interferon response in symptomatic animals was followed closely by a cytokine response that was more concurrent with the onset of a fever. This cytokine response was similar to what has been seen previously in NHP infections with a 100% lethal model of infection including cytokines such as IL-6, CCL8, and CXCL10 (12). This cytokine response developed after the interferon response and generally reached higher maximum levels of expression than ISGs. Similar to our modeling with the expression patterns of interferon-responsive genes, we found that we could also model the timing of cytokine expression. The cytokines found to be up-regulated are consistent with the cytokine storm that is characteristic of dysregulation of the immune system (32). This is consistent with a strong pro-inflammatory response in innate immune cells circulating in the blood.
The response observed during the symptomatic phase is consistent with transcriptional observation in human infections. In a patient treated at the NIH Clinical Center, the earliest response to viral infection involved many interferon stimulated genes and cytokine expression (20). Since this patient ultimately survived, these responses eventually subsided. In a Guinea cohort of patients from the 2013–2016 EBOV outbreak in West Africa, one of the gene sets strongly enriched in the fatal cases was interferon-stimulated genes (19). Interestingly, when we compare the expression of the early genes to human fatal cases, a correlation between the genes during the acute phase in both the humans and NHPs was appearent. This correlation and the conservation of interferon genes at the earliest stages of disease suggest that these genes may be up-regulated early in infection in humans. This introduces the possibility of using the host response as an early diagnostic, and preliminary work to do so has been performed (30). Though the interferon response is not unique to EBOV, identifying other genes with a similar expression pattern may help to isolate potential early markers of infection.
Smaller changes in RNA abundance were detected in our NanoString assays, allowing for the characterization of a modest immune response very early in infection. This response mostly contained pro-inflammatory cytokines and well as general immune response genes. This response could be the animals responding to the infection in the nasal cavity, which was the site of initial exposure. The presence of interferon lambda (IFNL) may suggest an immune response in epithelial cells consistent with a modest immune response occurring in the nasal cavity. IFNL has been previously shown to be an important factor in protection of respiratory epithelial cells against viral infections (33).
The early immune response detected by NanoString was present in all the Delayed animals, one Late animal, one Normal animal, and one No response animal. The No response animal that had a detectable early response at day 3 post-exposure did have more instances of elevated body temperature than the other No response animal as determined by implanted telemetry devices. The combination of increased instances of fever over the course of the study and the presence of the early response suggests that the two No response animals had different host responses to the initial infection. This early host response was no longer detectable by day 6 post-exposure in all the animals except the one Normal animal as that animal had viremia and fever at this time. During this initial immune response it is likely that myeloid cells such as dendritic cells and macrophages become infected as they are suggested as initial targets for EBOV (34) and found to be major contributors to the host response (35). This would contribute to viral dissemination and later transcriptional changes.
One potential limitation of this study is the small sampling size that is a byproduct arising from the challenges associated with performing NHP studies in high containment. Though large changes could be detected and described, a more complete analysis of the changes would require additional studies with a similar outcome of disease. Furthermore, we could not make any claims about the route of infection and its relationship to the differential onset of disease. Though there was a separation of animals based on incubation time, we cannot rule out that the Normal or Delayed groups caused a secondary infection in the Late animals that eventually caused them to spike a fever. Nevertheless, it is important to emphasize that the host response to infection after all infection events (including possible secondary events) was highly conserved in the experiment following a predictable pattern. We were unable to find an association between exposure route and survival or exposure route and time to onset of disease. This may become clearer as additional NHP studies with similar outcomes are performed.
Overall, this animal model provided a unique opportunity to study the host response to infection with differential onsets of disease and time to death. Also, the combination of telemetry, blood chemistry data, and gene expression allowed for an in-depth analysis of the exact timings of the host response relative to clinical presentation. This study highlighted that once the disease has started, it follows a predictable pattern over time with interferon genes being up-regulated early, followed by fever and cytokines, and ultimately the end stage of disease with have excessive immune dysregulation. Despite the different onset of symptoms between animals, the host response, once started, occurred in approximately the same time frame and can be modeled to create predictions of disease progression in the host.
Materials and Methods
Study Design
Twelve healthy adult cynomolgus macaques (Macaca fascicularis), equally distributed by sex, were used for this study. All macaques were surgically implanted with T27F-1B radiotelemetry devices (Konigsberg Instruments) at least 12 months prior to the start of the experiment, and were allowed to fully recover. Surgeries were done under anesthesia and, if necessary, with post-operational buprenorphine analgesia to minimize pain.
Animals were randomly assigned to receive a target dose of 100 PFU EBOV/Mak by intranasal exposure either by pipette (n=6, Animals 1–6) or by mucosal atomization device (MAD, n=6, Animals 7–12). Predefined endpoint euthanasia criteria were used to determine endpoints with the total experiment planned to run for 41 days.
Research was conducted under an IACUC-approved animal protocol at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID). This protocol complied with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011. All experiments were conducted in an ABSL-4 laboratory at USAMRIID. Technicians were not blinded to method of intranasal delivery
Virus
The virus stock used has been previously described in (15). Briefly, a master stock was made from an isolate from serum of a 2014 Sierra Leone fatal human case (SL 3864.1) (Gen Bank accession #KR013754.1). The sequence obtained from the clinical specimen SL-3864 was identical to 17 other P0 sequences from the outbreak. Eight of those come from non-survivors. Thus, the sequence of SL-3864 is identical to the sequence of lethal viruses. The stock did not have detectable amounts of contaminates such as mycoplasma, endotoxin or adventitious agents based on the assays and techniques used. Particles were examined through electron microscopy and determined to have characteristic shapes of filoviruses with a uniform diameter of approximately 80 nm.
Challenge
Animals were anesthetized with an intramuscular injection of ketamine prior to challenge. The pipette group received 250 μl of virus inoculum in each nare dropwise by pipette. For the MAD exposure, the device (Teleflex Medical) was attached to a 1 mL syringe containing the virus inoculum. The animals’ heads were tilted so that the nostrils were angled upward. Each animal received 0.5 mL of virus inoculum in each nare delivered as a single puff. The head remained tilted upwards for 1–2 min following challenge to ensure proper delivery of the challenge material. The challenge dose for all animals was determined by neutral red plaque assay as previously described (36) was 65 PFU.
In-life study activities
Animals were monitored at least once daily for general health and well-being. Additional checks were performed when animals began to display signs of illness. Physical examinations and blood collection were performed under anesthesia on day 0 (pre-challenge), 3, 6, 10, 14, 21, 28, 35, 41 (end of study), and terminally. Euthanasia was performed via intracardiac administration of a pentobarbital-based euthanasia solution under deep anesthesia when an animal was deemed moribund (37).
Clinical Pathology
Chemistry parameters were evaluated in serum samples using a Piccolo Point-Of-Care Analyzer (Abaxis) and Piccolo General Chemistry 13 panel. Hematology was assessed on K3 EDTA-treated whole blood using an ADVIA 120 Hematology System (Siemens).
Telemetry Monitoring and Pathophysiological Analysis
The implanted radiotelemetry devices allowed for the continuous monitoring of body temperature, aortic pressure (AOP), left ventricular pressure (LVP), heart rate (HR), and respiratory rate (RR) throughout the baseline and study periods. The telemetry data were captured and analyzed using the Notocord-hem Evolution software platform (Version 4.3, Notocord Inc.). Thirty-minute averages of the above listed physiologic parameters were calculated for each subject. Telemetry data obtained during the five days prior to challenge were used to calculate baseline values, and provided the average and standard deviation (SD) of each 30-min time period of a 24-h day. Using the average and SD determinations from the baseline physiologic data, values greater or less than 3 SD of the corresponding baseline value were considered significant. Fever was defined as > 1.5°C above and hyperpyrexia was defined as > 3.0°C above their corresponding baseline values. The phases of infection were defined as the incubation period (time from challenge to onset of sustained fever response for more than 2 h), the clinical period (time from onset of sustained fever until onset of sustained hypotension, decline in blood pressure for more than 2 h), and terminal period (time from onset of sustained hypotension until death or euthanasia).
Viral RNA isolation and RT-qPCR
Freshly collected plasma samples (100 μl) were combined with 300 μl TRIzol LS (ThermoFisher Scientific) and then RNA was extracted manually using either the QIAamp Viral RNA Mini Kit (Qiagen) or with an EZ1 Advanced XL system using EZ1 Virus Mini Kit (Qiagen). Viral genome equivalents per mL (ge/mL) were quantified using an EBOV-specific RT-qPCR assay and a synthetic RNA standard curve as previously described (38). Alternatively, PFU equivalents per mL (PFUe/mL) were extrapolated from the data using an RNA standard curve derived from a viral stock of known infectivity.
RNA isolation from whole blood
Freshly-collected whole blood samples (500 μl) were diluted with an equal volume of molecular biology grade water (500 μl), and then combined with 3 ml TRIzol LS. For RNA-Sequencing (RNA-Seq) analyses, RNA was isolated from whole blood in TRIzol LS using the PureLink RNA Mini Kit (ThermoFisher Scientific) and RNA was evaluated for quality on the Agilent 2200 TapeStation. For NanoString analyses, total RNA was isolated using the miRNeasy Mini Kit (Qiagen) and then quantified using the Qubit RNA HS Assay Kit (ThermoFisher Scientific).
RNA-Sequencing
Libraries were generated on the Sciclone G3 Liquid Handling Robot (PerkinElmer) using the TruSeq Stranded Total RNA Library Prep Kit (Illumina). Library quality was evaluated on the Agilent 2200 TapeStation 2200 and quantified by qPCR using the KAPA Complete (Universal) qPCR kit (Kapa Biosystems) for Illumina libraries. Libraries were diluted to 12 pM and cluster generation was performed on the Illumina cBot. Libraries were sequenced on the HiSeq 2500 using the paired end 2×100 bp, dual-index format.
Nanostring RNA collection and run
Total RNA samples were analyzed using a custom-designed NanoString nCounter gene expression codeset targeting 769 NHP genes. For each gene, a single probeset was designed with predicted cross-hybridization to homologous genes for cynomolgus macaque and rhesus macaque (Macaca mulatta). Probeset-target RNA hybridization reactions were performed according to the manufacturer’s protocol. For each hybridization reaction, 100 ng total RNA was used or any quantity that was present in a 5-μl aliquot of purified RNA if less than 100 ng. Purified probeset-target RNA complexes from each reaction were processed and immobilized on nCounter Cartridges using an nCounter MAX Prep Station and transcripts were quantified on the Digital Analyzer (GEN 2).
RNA-Seq data processing
Raw sequencing reads were trimmed on the edges for low quality reads and filtered using the FASTX-Toolkit v0.0.14. Paired reads were then aligned to the cynomolgus macaque genome v5.0 (39) using Bowtie2 v2.2.6 (40) and Tophat v2.1.0 (41) with default alignment parameters. Finally, count tables were generated using HTSeq Count v0.5.4 (42) and Python v2.7.3. Counts were generated using an alignment to a gene and parameter --stranded set to no.
Count tables were then read into R v3.2.2. Data normalization was performed with the rlog function of DESeq2 (43). Genes were filtered to only include genes whose median count was greater than 1. RNA-Seq samples were filtered if the number of aligned reads was less than one million mapped reads to genes or if they were determined to be outliers by principal components analysis (PCA). In total, 6 samples were classified as outliers and were not included in the analysis. Finally, differential gene expression analysis was done through DESeq2 and statistical significance was determined if the absolute log fold-change was greater than 1 and the Benjamini-Hochberg (BH) corrected p-value was less than 0.05.
Human datasets were collected from BioProject PRJNA352396 and processed according to (19). The data was read into R using DESeq2 to generate a fold-change table. Gene conversion between human Ensembl IDs and macaque gene names was done with Ensembl Biomart (44).
NanoString Data Processing
NanoString raw counts were compiled into a single counts table. Normalization of the counts was done using a standard method. First a global background was calculated using the mean plus 2 standard deviations of the negative controls. Then normalization to internal positive controls was performed to adjust for system variation between lanes. This was achieved by first calculating the positive control factor that was the mean of the geometric means of the positive controls of the different lanes. A normalization factor was calculated for each lane by dividing the positive control factor by the geometric mean of each lane. This value was used as a multiplier for each lane’s count values.
To determine genes that would perform as good input normalization factors, we first filtered all the genes to include only those whose count values were above the global background in all samples. We then applied the NormFinder R package (45) on these genes to determine which were most stable in the data set. The top 5 most stable genes were utilized for reference gene normalization across the samples to control for input variation of the amount of RNA. This was done similarly to the positive control normalization.
Differential expression analysis of the NanoString data was performed using a one-way ANOVA test on each gene for the different conditions. This was achieved through the lm function in R. P-values were adjusted with a BH correction.
Statistical Analysis and Gene Set Analysis
PCA was performed using the stats package prcomp function in R. Clustering was determined by k-means clustering with the cluster package (46) pam function in R. Significant blood chemistry data was determined with a one-way ANOVA test. Significance was determined by a p-value < 0.05. All p-value adjustments were done with a BH correction using the p.adjust function in the stats package. Gene set analysis was done using a Fisher’s exact test on a pre-defined gene set using standard cutoffs for the differential expression analysis (p-adjusted < 0.05 and absolute log fold-change > 1). Pathway analysis was done using IPA (Ingenuity Pathways Analysis) knowledge base. Interferon type determination was done using the interferome knowledge base (25). Statistical significance was determined with a hypergeometric test to determine if the overlap of interferon response type was significant.
Clustering of the samples based on time to death was determined by k-means clustering in R. To determine if clustering was significant, 10000 iterations of k-means clustering was performed on the time-to-death data. This was compared to 10000 iterations of randomly shuffling the animals’ times-to-death.
To determine a logistic model fit to the data, we first converted all the days post-infection into a time point relative to the group mean incubation time. Thus, 0 represents when that group ended the incubation time, which was characterized by a sustained fever of greater than 1.5 °C change. For the fitting, the day 3 post-exposure time point was not included since it exhibited early cytokine expression. Also, the non-responder animals were not included since they did not develop disease. The R stats package was used with the function nls and the following formula for the fit.
Starting parameters were determined as follows: a = log fold change at the end of the timecourse, b = slope of a linear model fit, c = intercept of a linear model fit. A deviance cutoff of 20 was used to determine if the data fit the model well (Supplemental Figure 6).
Plotting of the data was done in GraphPad Prism v.5 and ggplot2.
Supplementary Material
Supplemental Table 1: Cytokine Expression in RNA-Seq
Supplemental Table 2: Early expressed genes in Delayed Group
Supplemental Table 3: Normalized NanoString Counts
Supplemental Figure 1. Comparison of the pre-infection animals
Supplemental Figure 2: Comparison of MCV between groups.
Supplemental Figure 3: Correlation of Cytokines in Symptomatic Animals
Supplemental Figure 4: Example genes from NanoString in Delayed
Supplemental Figure 5: Interferon Response in NanoString Codeset.
Supplemental Figure 6: Histogram of Deviances from Curve Fitting
Acknowledgments:
We would like to thank Jeremy J. Bearss for work as a pathologist on this study.
Funding:
This research was performed with a National Science Foundation Graduate Research Fellowship under Grant No. DGE-1247312 to ES. JHC was supported through BPS/STP-15-015 and RO1AI1096159. LA, TM, CTH, CB, and WA were supported through the Defense Threat Reduction Agency Project # CB-102245. SB was supported through a Chemical and Biological Defense Research Associateship award from the National Research Council of the National Academies and the Defense Threat Reduction Agency. This research was performed while CEB held an NRC Research Associateship award at Center for Genome Sciences, U.S. Army Medical Research Institute of Infectious Diseases. The research described herein was sponsored by the Defense Threat Reduction Agency (DTRA), Program number CB3713 to SB, AG, WP, FR, JS, SW, and JZ.
Footnotes
Competing interests: The authors declare no competing interest. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Data and materials availability:
The normalized NanoString count data is available in Supplemental Table 3. The RNA-Sequencing data can be accessed through GEO accession number GSE103825.
References and Notes:
- 1.Feldman H, Geisbert T, Ebola haemorrhagic fever., Lancet 377, 849–862 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Center for Disease Control (CDC), 2014. Ebola Outbreak in West Africa - Case Counts | Ebola Hemorrhagic Fever | CDC (available at https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html).
- 3.Nakayama E, Saijo M, Animal models for Ebola and Marburg virus infectionsFront. Microbiol. 4 (2013), doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bente D, Gren J, Strong JE, Feldmann H, Disease modeling for Ebola and Marburg viruses, Dis. Model. Mech 2, 12–17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson E, Jaax N, White J, Jahrling P, Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus., Int. J. Exp. Pathol 76, 227–36 (1995). [PMC free article] [PubMed] [Google Scholar]
- 6.Jaax NK, Davis KJ, Geisbert TJ, Vogel P, Jaax GP, Topper M, Jahrling PB, Lethal experimental infection of rhesus monkeys with Ebola-Zaire (Mayinga) virus by the oral and conjunctival route of exposure, Arch. Pathol. Lab. Med 120, 140–155 (1996). [PubMed] [Google Scholar]
- 7.Alfson K, Avena L, Worwa G, Carrion R, Griffiths A, Development of a Lethal Intranasal Exposure Model of Ebola Virus in the Cynomolgus Macaque, Viruses 9, 319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rewar S, Mirdha D, Transmission of Ebola Virus Disease: An Overview, Ann. Glob. Heal 80, 444–451 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Lawrence P, Danet N, Reynard O, Volchkova V, Volchkov V, Human transmission of Ebola virus, Curr. Opin. Virol 22, 51–58 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Laupland KB, Valiquette L, Ebola virus disease., Can. J. Infect. Dis. Med. Microbiol. = J. Can. des Mal. Infect. la Microbiol. medicale 25, 128–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speranza E, Connor JH, Host Transcriptional Response to Ebola Virus Infection, Vaccines 5, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caballero IS, Honko AN, Gire SK, Winnicki SM, Melé M, Gerhardinger C, Lin AE, Rinn JL, Sabeti PC, Hensley LE, Connor JH, In vivo Ebola virus infection leads to a strong innate response in circulating immune cells., BMC Genomics 17, 707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Versteeg K, Menicucci AR, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jeske D, Messaoudi I, Geisbert TW, Infection with the Makona variant results in a delayed and distinct host immune response compared to previous Ebola virus variants, Sci. Rep 7, 9730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cilloniz C, Ebihara H, Ni C, Neumann G, Korth MJ, Kelly SM, Kawaoka Y, Feldmann H, Katze MG, Functional genomics reveals the induction of inflammatory response and metalloproteinase gene expression during lethal Ebola virus infection., J. Virol 85, 9060–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speranza E, Altamura LA, Kulcsar K, Bixler SL, Rossi CA, Schoepp RJ, Nagle E, Aguilar W, Douglas CE, Delp KL, Minogue TD, Palacios G, Goff AJ, Connor JH, Comparison of Transcriptomic Platforms for Analysis of Whole Blood from Ebola-Infected Cynomolgus Macaques, Sci. Rep 7, 14756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta M, Robertson SJ, Okumura A, Scott DP, Chang J, Weiss JM, Sturdevant GL, Feldmann F, Haddock E, Chiramel AI, Ponia SS, Dougherty JD, Katze MG, Rasmussen AL, Best SM, A Systems Approach Reveals MAVS Signaling in Myeloid Cells as Critical for Resistance to Ebola Virus in Murine Models of Infection, Cell Rep. 18, 816–829 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garamszegi S, Yen JY, Honko AN, Geisbert JB, Rubins KH, Geisbert TW, Xia Y, Hensley LE, Connor JH, Transcriptional correlates of disease outcome in anticoagulant-treated non-human primates infected with ebolavirus, PLoS Negl Trop Dis 8, e3061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yen JY, Garamszegi S, Geisbert JB, Rubins KH, Geisbert TW, Honko A, Xia Y, Connor JH, Hensley LE, Therapeutics of Ebola hemorrhagic fever: whole-genome transcriptional analysis of successful disease mitigation, J Infect Dis 204 Suppl, S1043–52 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Speranza E, Muñoz-Fontela C, Haldenby S, Rickett NY, Garcia-Dorival I, Fang Y, Hall Y, Zekeng E-G, Lüdtke A, Xia D, Kerber R, Krumkamp R, Duraffour S, Sissoko D, Kenny J, Rockliffe N, Williamson ED, Laws TR, N’Faly M, Matthews DA, Günther S, Cossins AR, Sprecher A, Connor JH, Carroll MW, Hiscox JA, Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus, Genome Biol. 18, 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kash JC, Walters K-A, Kindrachuk J, Baxter D, Scherler K, Janosko KB, Adams RD, Herbert AS, James RM, Stonier SW, Memoli MJ, Dye JM, Davey RT, Chertow DS, Taubenberger JK, Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease, Sci. Transl. Med 9 (2017) (available at http://stm.sciencemag.org/content/9/385/eaai9321). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisfeld AJ, Halfmann PJ, Wendler JP, Kyle JE, Burnum-Johnson KE, Peralta Z, Maemura T, Walters KB, Watanabe T, Fukuyama S, Yamashita M, Jacobs JM, Kim Y-M, Casey CP, Stratton KG, Webb-Robertson B-JM, Gritsenko MA, Monroe ME, Weitz KK, Shukla AK, Tian M, Neumann G, Reed JL, van Bakel H, Metz TO, Smith RD, Waters KM, N’jai A, Sahr F, Kawaoka Y, Multi-platform ‘Omics Analysis of Human Ebola Virus Disease Pathogenesis., Cell Host Microbe 0 (2017), doi: 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrenas F, Green RR, Thomas MJ, Law GL, Proll SC, Engelmann F, Messaoudi I, Marzi A, Feldmann H, Katze MG, Next generation sequencing reveals a controlled immune response to Zaire Ebola virus challenge in cynomolgus macaques immunized with VSVΔG/EBOVgp, Clin. Vaccine Immunol 22, CVI.00733–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tall AR, Yvan-Charvet L, Cholesterol, inflammation and innate immunity, Nat. Publ. Gr 15 (2015), doi: 10.1038/nri3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong C, Tontonoz P, Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors, Curr. Opin. Genet. Dev 18, 461–467 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ, INTERFEROME v2.0: an updated database of annotated interferon-regulated genes, Nucleic Acids Res. 41, D1040–D1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A, PANTHER: a library of protein families and subfamilies indexed by function., Genome Res. 13, 2129–41 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caballero IS, Yen JY, Hensley LE, Honko AN, Goff AJ, Connor JH, Lassa and Marburg viruses elicit distinct host transcriptional responses early after infection, BMC Genomics 15, 960 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malhotra S, Yen JY, Honko AN, Garamszegi S, Caballero IS, Johnson JC, Mucker EM, Trefry JC, Hensley LE, Connor JH, Transcriptional profiling of the circulating immune response to lassa virus in an aerosol model of exposure, PLoS Negl Trop Dis 7, e2171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connor JH, Yen J, Caballero IS, Garamszegi S, Malhotra S, Lin K, Hensley L, Goff AJ, Transcriptional Profiling of the Immune Response to Marburg Virus Infection., J. Virol 89, 9865–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navalkar K, Kim T-K, Gelinas R, Pre-Symptomatic Diagnosis of Ebola Virus Infection, J Emerg Virol Dis 3 (2013), doi: 10.16966/2473-1846.129. [DOI] [Google Scholar]
- 31.Rubins KH, Hensley LE, Wahl-Jensen V, Daddario DiCaprio KM, Young HA, Reed DS, Jahrling PB, Brown PO, Relman DA, Geisbert TW, The temporal program of peripheral blood gene expression in the response of nonhuman primates to Ebola hemorrhagic fever, Genome Biol. 8, R174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bixler SL, Goff AJ, The Role of Cytokines and Chemokines in Filovirus Infection., Viruses 7, 5489–507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Günther S, Drosten C, Michiels T, Staeheli P, Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections., J. Virol 84, 5670–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüdtke A, Ruibal P, Wozniak DM, Pallasch E, Wurr S, Bockholt S, Gómez-Medina S, Qiu X, Kobinger GP, Rodríguez E, Günther S, Krasemann S, Idoyaga J, Oestereich L, Muñoz-Fontela C, Ebola virus infection kinetics in chimeric mice reveal a key role of T cells as barriers for virus dissemination, (2017), doi: 10.1038/srep43776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bray M, Geisbert TW, Ebola virus: The role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever, Int. J. Biochem. Cell Biol 37, 1560–1566 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Shurtleff A, Biggins J, Keeney A, Zumbrun E, Bloomfield H, Kuehne A, Audet J, Alfson K, Griffiths A, Olinger G, Bavari S, Filovirus Animal Nonclinical Group (FANG) Assay Working Group, Standardization of the Filovirus Plaque Assay for Use in Preclinical Studies, Viruses 4, 3511–3530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren T, Trefry J, Marko S, Chance T, Wells J, Pratt W, Johnson J, Mucker E, Norris S, Chappell M, Dye J, Honko A, Euthanasia Assessment in Ebola Virus Infected Nonhuman Primates, Viruses 6, 4666–4682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trombley AR, Wachter L, Garrison J, Buckley-Beason VA, Jahrling J, Hensley LE, Schoepp RJ, Norwood DA, Goba A, Fair JN, Kulesh DA, Comprehensive Panel of Real-Time TaqManTM Polymerase Chain Reaction Assays for Detection and Absolute Quantification of Filoviruses, Arenaviruses, and New World Hantaviruses, Am. J. Trop. Med. Hyg 82, 954–960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee A, Khiabanian H, Kugelman J, Elliott O, Nagle E, Yu G-Y, Warren T, Palacios G, Rabadan R, Transcriptome reconstruction and annotation of cynomolgus and African green monkey, BMC Genomics 15, 846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2., Nat. Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Pachter L, Salzberg SL, TopHat: discovering splice junctions with RNA-Seq., Bioinformatics 25, 1105–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Pyl PT, Huber W, HTSeq A Python framework to work with high-throughput sequencing data, Bioinformatics 31, 166–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durinck S, Spellman PT, Birney E, Huber W, Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt, Nat. Protoc 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen CL, Jensen JL, Ørntoft TF, Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets., Cancer Res. 64, 5245–50 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K, cluster: Cluster Analysis Basics and Extensions (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Cytokine Expression in RNA-Seq
Supplemental Table 2: Early expressed genes in Delayed Group
Supplemental Table 3: Normalized NanoString Counts
Supplemental Figure 1. Comparison of the pre-infection animals
Supplemental Figure 2: Comparison of MCV between groups.
Supplemental Figure 3: Correlation of Cytokines in Symptomatic Animals
Supplemental Figure 4: Example genes from NanoString in Delayed
Supplemental Figure 5: Interferon Response in NanoString Codeset.
Supplemental Figure 6: Histogram of Deviances from Curve Fitting
Data Availability Statement
The normalized NanoString count data is available in Supplemental Table 3. The RNA-Sequencing data can be accessed through GEO accession number GSE103825.