Abstract
Background:
Abstraction of critical data from unstructured radiologic reports using natural language processing (NLP) is a powerful tool to automate the detection of important clinical features and enhance research efforts. We present a set of NLP approaches to identify critical findings in patients with acute ischemic stroke from radiology reports of computed tomography (CT) and magnetic resonance imaging (MRI).
Methods:
We trained machine learning classifiers to identify categorical outcomes of edema, midline shift (MLS), hemorrhagic transformation, and parenchymal hematoma, as well as rule-based systems (RBS) to identify intraventricular hemorrhage (IVH) and continuous MLS measurements within CT/MRI reports. Using a derivation cohort of 2289 reports from 550 individuals with acute middle cerebral artery territory ischemic strokes, we externally validated our models on reports from a separate institution as well as from patients with ischemic strokes in any vascular territory.
Results:
In all data sets, a deep neural network with pretrained biomedical word embeddings (BioClinicalBERT) achieved the highest discrimination performance for binary prediction of edema (area under precision recall curve [AUPRC] > 0.94), MLS (AUPRC > 0.98), hemorrhagic conversion (AUPRC > 0.89), and parenchymal hematoma (AUPRC > 0.76). BioClinicalBERT outperformed lasso regression (p < 0.001) for all outcomes except parenchymal hematoma (p = 0.755). Tailored RBS for IVH and continuous MLS outperformed BioClinicalBERT (p < 0.001) and linear regression, respectively (p < 0.001).
Conclusions:
Our study demonstrates robust performance and external validity of a core NLP tool kit for identifying both categorical and continuous outcomes of ischemic stroke from unstructured radiographic text data. Medically tailored NLP methods have multiple important big data applications, including scalable electronic phenotyping, augmentation of clinical risk prediction models, and facilitation of automatic alert systems in the hospital setting.
Keywords: Natural language processing, Stroke, Critical care, Diagnostic imaging
Introduction
Natural language processing (NLP) is a powerful tool to abstract and categorize critical data from unstructured radiologic reports for research and clinical purposes [1]. Assembling large cohorts for clinical and epidemiologic studies has historically required intensive manual chart review, limiting both sample size and generalizability [2-5]. Such efforts are particularly laborious in the case of critical outcomes after ischemic stroke, such as life-threatening edema and hemorrhagic transformation, for which dedicated disease classification codes may be inaccurate or nonexistent. Direct derivation of relevant data from radiology reports (electronic phenotyping) [6] represents an emerging strategy to improve the quality of retrospective data sets and develop clinical systems for triaging patients with critical findings [7].
NLP, a subfield of artificial intelligence dedicated to the study of human language [8], has emerged as a promising new approach to retrieve clinically rich data contained in medical records and diagnostic reports [2, 3, 9, 10]. NLP makes use of a variety of approaches to analyze language and classify text according to predefined outcomes of interest. Methods that identify desired text-based information include specifically tailored rule-based systems (RBSs) [11] as well as more general combinations of featurization methods (the conversion of text to numerical values) with supervised learning methodologies [12]. In the field of neurology, recent NLP studies have focused on identifying and subtyping ischemic stroke [13-15] and cerebral aneurysms [16], predicting neurologic outcomes after ischemia [17], and triaging patients with transient ischemic attack [18]. However, few studies have focused on feasibly implementable methods to classify critical and acute stroke sequelae [19].
Previously, we tested a variety of featurization and machine learning classification methods to detect the presence, acuity, and location of ischemic stroke [4]. In the present work, we sought to expand our electronic phenotyping of neuroradiology reports by applying various NLP methodologies to detect critical complications of stroke relevant to neurocritical care contexts. Specifically, we characterized the following intracranial abnormalities using both computed tomography (CT) and magnetic resonance imaging (MRI) reports: (1) edema, (2) midline shift (MLS), (3) hemorrhagic conversion, (4) parenchymal hematoma (within hemorrhagic conversion), and (5) intraventricular hemorrhage (IVH). We hypothesized that recently developed machine learning models pretrained on biomedical text would demonstrate improved classification of binary outcomes compared with traditional machine learning algorithms [20]. Moreover, we also hypothesized that customized RBSs would perform better at detecting rare and continuous outcomes of stroke than more general machine-learning-based approaches. Our work expands progress in large-scale neurocritical care research by demonstrating the application of NLP methods for electronic phenotyping of relevant emergent radiographic complications of ischemic stroke.
Methods
Study Population and Data Sources
To derive and assess the performance and generalizability of our NLP models, we accrued and labeled three distinct data sets of head CT, CT angiography, MRI, and magnetic resonance angiography radiology reports from patients 18 years or older with an International Classification of Diseases, Ninth Revision (ICD-9) (433.01, 433.11, 433.21, 433.31, 433.81, 433.91, 434.01, 434.11, 434.91) or International Classification of Diseases, 10th Revision (ICD-10) (I63) diagnosis of ischemic stroke during admission. The three data sets are described as follows:
MGB MCA Ischemic Stroke: This data set consisted of 2289 reports from 550 individuals presenting to Massachusetts General or Brigham and Women’s (MGB) hospitals with acute ischemic stroke occurring in the middle cerebral artery (MCA) territory between 2006 and 2018. Reports were identified by our previous NLP algorithm [4] from an initial Research Patient Data Registry query of 60,654 radiology reports, and the presence of MCA stroke was confirmed by expert reviewers (BB, CJO, HS).
BMC MCA Ischemic Stroke: This data set comprised 377 randomly selected reports from 154 patients out of a total of 4818 reports from 2069 patients with NLP-identified acute MCA ischemic stroke from Boston Medical Center (BMC) between 2005 and 2018. BMC, New England’s largest safety net hospital, features a population with notable socioeconomic and demographic differences relative to the MGB system despite geographic proximity. Acute MCA ischemic stroke and outcomes of interest were confirmed and labeled by trained experts (JK, MIM, OB).
MGB Any Territory Ischemic Stroke: This data set comprised 520 reports from 214 patients with ICD-9/ICD-10-identified ischemic stroke in any vascular territory (not only MCA territory) from MGB hospitals between 2003 and 2018. Trained experts (HS, KV, MT) labeled all outcomes. No reports overlapped with the MGB MCA Ischemic Stroke data set.
The MGB Human Research Committee and Boston University Medical Center Internal Review Boards (2017P002564, H-38431) approved this study.
Radiographic Report Labeling
We sought to identify a core set of outcomes covering major inflammatory and hemorrhagic sequelae of acute ischemic stroke. Therefore, all reports included in this study were manually labeled for the presence of edema, MLS, hemorrhagic conversion, parenchymal hematoma (as inferred according to ECASS-II criteria [21]), and IVH. Full labeling criteria are included in the Supplementary Methods. Notably, we further subdivided MLS detection into two separate prediction tasks: a categorical task, in which we labeled reports for the binary presence of any MLS, and a continuous task, in which we recorded the precise degree of MLS in millimeters. Research staff received training from three board-certified neurologists (BB, CJO, SMS) who performed regular audits of data collection to ensure accuracy and consistency. Deidentified reports were randomly distributed among study team members, and data sets were managed using a Research Electronic Data Capture electronic database [22]. Additionally, a board-certified neuroradiologist (AM) independently and in a blinded fashion assessed the accuracy of all labels in a random sample of 75 reports (25 per data set) to provide expert-level benchmarking of study labels.
Text Preprocessing
To reduce heterogeneities that impact classification, we processed raw radiology reports across all data sets in the following ways:
We converted all text to lowercase and removed excess white space.
We removed all text within the report header, deidentified the names of providers and patients, and removed any other instances of nonnarrative text (e.g., characters representing divisions between report sections).
We “tokenized” terms to quantify common word patterns describing a single entity by condensing commonly used groups of words to their corresponding “n-grams” (e.g., “subdural hematoma” → “subdural-hematoma”, “altered mental status” → “altermental-status”). The full token list is provided in the Supplementary Methods.
Featurization Methods
Following preprocessing, we used three methods to convert (“featurize”) unstructured text reports into numerical features capable of being used as direct input for statistical learning [8].
The first and simplest of these approaches, known as “Bag of Words” (BOW), counts each unique word or n-gram within a radiology report as a unique feature known as a “token.” The value of each token is determined by the number of times each given word occurs in a unique report.
Term frequency-inverse document frequency (TF-IDF) is similar to BOW but penalizes the value of each token according to the total number of reports in which that token occurs so that common words and n-grams, such as “and” and “the,” are assigned lower values.
Word-embedding strategies are more sophisticated NLP featurization methods that assign values to tokens on the basis of their relationship to other words [23]. We used an embedding method called the Bidirectional Encoder Representations from Transformers (BERT) model for our binary classification tasks [23]. Specifically, we used a domain-specific version of BERT embeddings (“BioClinicalBERT”) pretrained on 18 billion words worth of PubMed articles [24] and approximately 880 million words worth of critical care notes from the Medical Information Mart for Intensive Care-III v1.4 data set [25]. These embeddings, originally developed by Alsentzer and colleagues [26], acted as a starting point for training of our stroke-specific classifiers.
Classifier Training
For each binary task, we trained lasso regression [27], K-nearest neighbor [28], random forest [29], and multilayer perceptron [30] algorithms using the BOW and TF-IDF featurization techniques. We also further developed a BioClinicalBERT transformer network using pre-trained BERT embeddings, as described above. This type of network is a specialized type of sequential deep learning model that could be further fine-tuned for detection of poststroke outcomes. For the task of continuous MLS estimation, we employed linear regression with both BOW and TF-IDF. Further details regarding the merits and hyperparameter tuning of each algorithm are included in our Supplementary Methods. We employed a standard desktop central processing unit (CPU) for training of all models, with the exception of our BioClinicalBERT transformer, in which we used graphics processing units (GPU) for training and testing.
We trained our models on 80% of the original MGB MCA Ischemic Stroke cohort and tested it on a withheld set of 20% for internal validation. For our derivation cohort, we used bootstrapping to randomly split the data five times into training and testing sets. Both external validation cohorts were tested in full across all five splits of the data. For the parenchymal hematoma outcome, training and testing were conducted only on reports with a ground truth label of hemorrhagic conversion. Given that there were multiple tasks with differing prevalence and clinical relevance, we used 0.5 as the initial decision threshold for all classifiers.
We performed hyperparameter tuning with a grid search and tenfold cross-validation during each round of bootstrapping. BioClinicalBERT models were trained for ten epochs with a maximum sequence length of 512 and a learning rate of 4 × 10−5, which corresponds closely with hyperparameters reported in previous studies [26]. All models were developed and tested in Python 2.7 using the scikit-learn [31] package for non-BERT classifiers and the “simpletransformers” library for the BioClinicalBERT transformer. The detailed codebase is available within our Github repository (https://github.com/matthewmiller22/STROKE_NLP_2022).
RBS
We hypothesized that the previously described approaches would be ill suited for rare and continuous outcomes. Therefore, we employed alternative RBSs to identify IVH (a relatively infrequent complication of ischemic stroke [32]) and continuous MLS measurements. For IVH, we replaced phrases with periods that led to misclassification, including other hemorrhage subtypes (e.g., “subdural hemorrhage”), phrases associated with ventricular effacement (e.g., “compression [of the ventricles]”), and terms associated with neurosurgical procedures (e.g., “ventriculoperitoneal [shunt]”). We identified regular expression rules capturing both negative (e.g., “no ventricular blood”) and positive (e.g., “layering of blood”) phraseology for IVH. To identify continuous MLS, we extended our preprocessing pipeline by removing superfluous numerical characters, dates, and times. We assigned a value of 0 to cases in which there was an absence of “shift,” “deviation,” or “herniation,” as well as those in which there was explicit mention of “no midline shift” or “without midline shift.” We then split individual text reports by sentence and abstracted numeric values in proximity to words such as “shift,” “deviation,” or “herniation.” Regular expressions were constructed with the Regex library in Python 2.7. Further details of the design and implementation of each RBS are included in the Supplementary Data.
Evaluation and Statistical Analysis
To assess interrater reliability between labelers and a board-certified neuroradiologist, we calculated Cohen’s κ statistic. We report discrimination using the area under precision recall curve (AUPRC), sensitivity, specificity, and precision (positive predictive value). Similar to other work in machine learning for neurocritical care [33], we chose AUPRC to be our main performance metric given that raw accuracy and area under receiver operating characteristic curve (AUROC) may overestimate performance in imbalanced data sets [34]. We do, however, demonstrate receiver operating characteristic curves for all classifiers in Supplementary Fig. S1 and supply full performance metrics, including accuracy and AUROC, within Supplementary Tables S1-S5. Calibration curves for classifier probabilities are also provided in Supplementary Fig. S2.
To compare the performance of our two best performing classifiers for each binary task, we used McNemar’s test [35], a statistical procedure used in previous NLP studies to assess the equivalence of two models tested on the same data. We also used McNemar’s test for comparison of our IVH RBS to the best performing machine learning classifier. For each clinical outcome, we report the average χ2 value across all five data partitions with 95% confidence intervals and p values. Given that McNemar’s test assumes independent observations, we performed this procedure using a dedicated data set containing only one randomly selected report patient. To assess the performance of our RBS for continuous MLS, we compared the absolute errors of RBS MLS estimates with linear regression estimates using a one-sided Student’s t-test. To account for our six total hypotheses, we used a Bonferroni correction [36] and set the significance level for all hypotheses to α = 0.008. All statistical tests were performed using the scipy.stats library within Python 2.7.
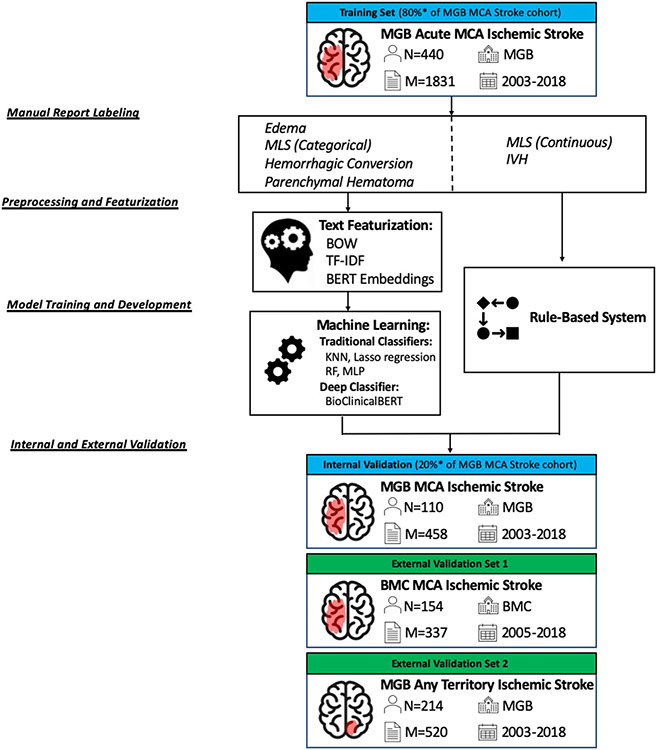
A graphical overview of our methods, including cohort selection and model development, is illustrated within Fig. 1.
Fig. 1.
Model testing and development framework. A graphic display of data labeling, preprocessing, featurization, and model training and development is illustrated. BMC Boston Medical Center, BOW Bag of Words, IVH intraventricular hemorrhage, KNN K-nearest neighbor, M number of reports, MCA middle cerebral artery, MGB Massachusetts General Brigham and Women’s, MLS midline shift, MLP multilayer perceptron, N number of patients, RF random forest, TF-IDF term frequency-inverse document frequency
Results
In our MGB MCA Ischemic Stroke data set, we labeled 2289 CT and MRI radiology reports for all stroke outcomes (Fig. 1). For external validation, we labeled 377 reports in the BMC MCA Ischemic Stroke set and 520 reports from the MGB Any Territory Ischemic Stroke data set. The baseline characteristics of the overall cohort, including demographics and clinical outcomes, are described in Tables 1 and 2. Across all three data sets, we calculated the Cohen’s κ values for interrater reliability in categorical outcomes, finding strong consistency for labeling of edema (κ = 0.70–1.00), MLS (κ = 0.74–1.00), hemorrhagic conversion (κ = 0.82–1.0), and IVH (κ = 1.00) and moderate reliability for parenchymal hematoma (κ = 0.46–0.623) [37]. Full Cohen’s κ results may be found within Supplementary Table S6.
Table 1.
Cohort characteristic by individual
| MGB MCA ischemic stroke (n = 550) |
BMC MCA ischemic stroke (n = 154) |
MGB any territory ischemic stroke (n = 214) |
|
|---|---|---|---|
| Demographics | |||
| Age (years), mean (SD) | 68.3 (14.2) | 63.6 (17.1) | 68.6 (15.0) |
| Female sex, n (%) | 265 (48.2) | 75 (48.7) | 86 (40.2) |
| Clinical outcomes, n (%) | |||
| Edema | 420 (76.3) | 101 (65.6) | 88 (41.1) |
| MLS (binary) | 223 (40.5) | 47 (30.5) | 24 (11.2) |
| Hemorrhagic conversiona | 260 (47.2) | 45 (29.2) | 63 (29.4) |
| Parenchymal hematoma | 81 (31.2) | 11 (24.4) | 11 (17.5) |
| Petechial hemorrhage | 164 (63.1) | 34 (75.6) | 47 (74.6) |
| Indeterminate | 15 (5.8) | 0 (0.0) | 5 (7.9) |
| Intraventricular hemorrhage | 39 (7.1) | 6 (3.9) | 10 (4.7) |
For unique patients, we report demographics and characteristics, and clinical outcomes are illustrated in tabular form for the MGB MCA Ischemic Stroke, BMC MCA Ischemic Stroke, and MGB Any Territory Ischemic Stroke data sets
BMC, Boston Medical Center, MCA, middle cerebral artery, MGB, Massachusetts General or Brigham and Women’s, MLS, midline shift
Hemorrhagic subtypes calculated as a percentage of patients/reports with hemorrhagic conversion. For patients with varying severities of hemorrhagic conversion, we report the most severe subtype observed
Table 2.
Cohort characteristic by report
| MGB MCA ischemic stroke (n = 2289) |
BMC MCA ischemic stroke (n = 377) |
MGB any territory ischemic stroke (n = 520) |
|
|---|---|---|---|
| Imaging modality, n (%) | |||
| CT | 1846 (80.6) | 225 (59.7) | 276 (53.1) |
| CT angiography | 97 (4.2) | 63 (16.7) | 8 (1.5) |
| MRI/MRA | 346 (15.2) | 89 (23.6) | 236 (45.4) |
| Clinical outcomes | |||
| Edema, n (%) | 1534 (67.0) | 233 (61.8) | 179 (34.4) |
| MLS (binary), n (%) | 623 (27.2) | 126 (33.4) | 47 (9.0) |
| MLS (mL), mean (SD)a | 7.2 (4.1) | 6.8 (3.9) | 6.1 (3.7) |
| Hemorrhagic conversion,b n (%) | 819 (35.8) | 86 (22.8) | 119 (22.9) |
| Parenchymal hematoma | 277 (33.8) | 23 (26.7) | 15 (12.6) |
| Petechial hemorrhage | 463 (56.5) | 58 (67.4) | 89 (74.8) |
| Indeterminate | 79 (9.6) | 5 (5.8) | 15 (12.6) |
| Intraventricular hemorrhage, n (%) | 109 (4.8) | 12 (3.2) | 18 (3.5) |
Given that a single patient may have multiple radiology reports included within the analysis, we provide baseline information across all individual stroke reports. For all reports, we provide diagnostic modalities and clinical outcomes for the MGB MCA Ischemic Stroke, BMC MCA Ischemic Stroke, and MGB Any Territory Ischemic Stroke data sets
BMC, Boston Medical Center, CT, computed tomography, MCA, middle cerebral artery, MGB, Massachusetts General or Brigham and Women’s, MLS, midline shift, MRA, magnetic resonance angiography, MRI, magnetic resonance imaging
Only indicates reports with MLS present
Hemorrhagic subtypes calculated as a percentage of patients/reports with hemorrhagic conversion
For our binary outcomes, we compared performance of lasso regression, K-nearest neighbor, random forest, multilayer perceptron, and BioClinicalBERT algorithms. In our internal test set of MGB MCA Ischemic Stroke, BioClinicalBERT consistently outperformed other classifiers trained with BOW and TF-IDF featurizations. Specifically, BioClinicalBERT achieved high performance in detecting edema (AUPRC 0.98, sensitivity 0.99, specificity 0.92), MLS (AUPRC 1.00, sensitivity 0.99, specificity 0.99), and hemorrhagic conversion (AUPRC 0.95, sensitivity 0.87, specificity 0.97). BioClinicalBERT detected parenchymal hematoma slightly less well (AUPRC 0.80, sensitivity 0.79, specificity 0.78). Our RBS for IVH detection yielded respective sensitivity and specificity of 0.94 and 0.99 in the MGB MCA Ischemic Stroke data set.
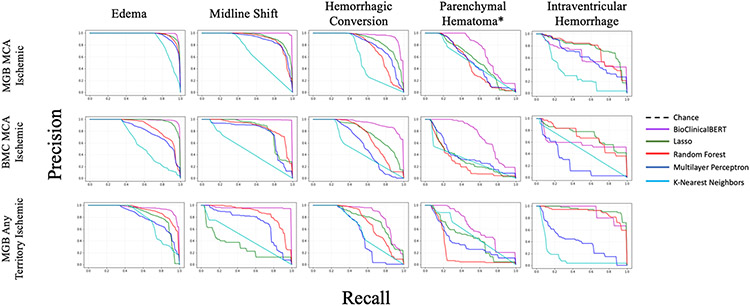
In both external validation data sets BioClinicalBERT discriminated well between reports with edema (AUPRC > 0.94, sensitivity > 0.91, specificity > 0.87), MLS (AUPRC > 0.98, sensitivity > 0.95, specificity > 0.99), hemorrhagic conversion (AUPRC > 0.89, sensitivity > 0.72, specificity > 0.94), and parenchymal hematoma (AUPRC > 0.76, sensitivity > 0.79, specificity > 0.70). Precision recall curves for all data sets, outcomes, and classifiers are included in Fig. 2.
Fig. 2.
Precision (positive predictive value) recall curves. Precision recall curves are demonstrated for binary classification of edema, midline shift, hemorrhagic conversion, and parenchymal hematoma* per ECASS-II criteria. Results are demonstrated for BioClinicalBERT, lasso regression, random forest, multilayer perceptron, and K-nearest neighbors algorithms. For all models other than BioClinicalBERT, the precision recall curve is shown for the text featurization algorithm (BOW or TF-IDF) that maximized the AUPRC value. Precision recall curves, unlike ROC curves, are influenced by class imbalance as well as performance metrics (sensitivity, specificity, and precision) that are reflective of clinical heuristics. This allows notable differences in binary classification to stand out when ROC analysis might otherwise not highlight salient differences. *Indicates parenchymal hematoma only trained using reports with ground-truth-labeled hemorrhagic conversion. AUPRC area under precision recall curve, BOW Bag of Words, European-Australasian Acute Stroke Study-II, ROC receiver operating characteristic, TF-IDF term frequency-inverse document frequency
We compared BioClinicalBERT with the next-best performing classifier (lasso regression) using McNemar testing in the MGB MCA Ischemic Stroke derivation cohort. We found statistically significant improvements in the detection of edema (χ2 = 12.07, p < 0.001), MLS (χ2 = 29.43, p < 0.001), and hemorrhagic conversion (χ2 = 18.64, p < 0.001) using BioClinicalBERT. Performance metrics for these two models are compared side-by-side within Table 3, and further details on McNemar testing are available in Supplementary Tables S7-S11.
Table 3.
BioClinicalBERT versus lasso regression
| BioClinicalBERT | Lasso | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUPRC | Sensitivity | Specificity | Precision (PPV) |
AUPRC | Sensitivity | Specificity | Precision (PPV) |
Featurization | ||
| MGB MCA Ischemic Stroke | Edema | 0.98 (0.97, 1.00) | 0.99 (0.98, 0.99) | 0.92 (0.89,0.96) | 0.98 (0.96,0.98) | 0.98 (0.98, 0.99) | 0.87 (0.84, 0.89) | 0.84 (0.89, 0.98) | 0.97 (0.95, 0.99) | TF-IDF |
| MLS | 1.00 (1.00, 1.00) | 0.99 (0.98, 1.00) | 0.99 (0.98, 1.00) | 0.97 (0.95, 0.99) | 0.95 (0.92, 0.98) | 0.81 (0.77, 0.86) | 0.96 (0.94, 0.98) | 0.90 (0.86, 0.95) | BOW | |
| Hemorrhagic Conversion | 0.95 (0.93, 0.97) | 0.89 (0.85, 0.93) | 0.97 (0.95, 0.99) | 0.94 (0.91, 0.97) | 0.89 (0.86, 0.91) | 0.80 (0.75, 0.84) | 0.89 (0.85, 0.92) | 0.81 (0.75, 0.87) | BOW | |
| Parenchymal Hematoma | 0.80 (0.77, 0.84) | 0.79 (0.75, 0.83) | 0.78 (0.76, 0.79) | 0.62 (0.52, 0.65) | 0.64 (0.58, 0.70) | 0.75 (0.59, 0.90) | 0.75 (0.60, 0.89) | 0.70 (0.50, 0.89) | BOW | |
| IVH | 0.66 (0.48, 0.83) | 0.57 (0.41, 0.73) | 0.99 (0.98, 0.99) | 0.71 (0.53, 0.89) | 0.76 (0.67, 0.84) | 0.37 (0.13, 0.60) | 0.99 (0.99, 1.00) | 0.94 (0.80, 1.00) | BOW | |
| BMC MCA Ischemic Stroke | Edema | 0.99 (0.99, 1.00) | 0.98 (0.97, 0.99) | 0.88 (0.84, 0.92) | 0.93 (0.91, 0.95) | 0.98 (0.97, 0.98) | 0.76 (0.74, 0.78) | 0.96 (0.95, 0.97) | 0.97 (0.97, 0.98) | TF-IDF |
| MLS | 1.00 (1.00, 1.00) | 0.99 (0.97, 1.00) | 1.00 (1.00, 1.00) | 1.00 (1.00, 1.00) | 0.80 (0.80, 0.81) | 0.81 (0.79, 0.83) | 0.84 (0.83, 0.85) | 0.69 (0.68, 0.71) | TF-IDF | |
| Hemorrhagic Conversion | 0.89 (0.85, 0.93) | 0.72 (0.69, 0.76) | 0.99 (0.98, 1.00) | 0.96 (0.93, 0.98) | 0.75 (0.74, 0.76) | 0.61 (0.58, 0.63) | 0.96 (0.95, 0.96) | 0.82 (0.79, 0.84) | TF-IDF | |
| Parenchymal Hematoma | 0.78 (0.75, 0.82) | 0.79 (0.75, 0.84) | 0.74 (0.70, 0.78) | 0.55 (0.49, 0.61) | 0.43 (0.41, 0.45) | 0.24 (0.20, 0.29) | 0.97 (0.96, 0.97) | 0.53 (0.47, 0.59) | TF-IDF | |
| IVH | 0.59 (0.23, 0.96) | 0.54 (0.09, 1.00) | 0.99 (0.97, 1.00) | 0.59 (0.14, 1.00) | 0.82 (0.76, 0.87) | 0.15 (0.06, 0.24) | 1.00 (1.00, 1.00) | 1.00 (1.00, 1.00) | TF-IDF | |
| MGB Any Territory Ischemic Stroke | Edema | 0.94 (0.91, 0.97) | 0.91 (0.88, 0.94) | 0.97 (0.95, 0.98) | 0.93 (0.90, 0.96) | 0.89 (0.89, 0.90) | 0.64 (0.62, 0.66) | 0.97 (0.97, 0.98) | 0.93 (0.91, 0.93) | TF-IDF |
| MLS | 0.98 (0.95, 1.00) | 0.96 (0.92, 1.00) | 1.00 (1.00, 1.00) | 0.99 (0.97, 1.00) | 0.82 (0.80, 083) | 0.89 (0.88, 0.90) | 0.96 (0.95, 0.96) | 0.67 (0.64, 0.71) | TF-IDF | |
| Hemorrhagic Conversion | 0.90 (0.86, 0.93) | 0.78 (0.73, 0.83) | 0.94 (0.92, 0.96) | 0.81 (0.75, 0.87) | 0.77 (0.76, 0.79) | 0.66 (0.64, 0.68) | 0.92 (0.92, 0.93) | 0.73 (0.71, 0.75) | TF-IDF | |
| Parenchymal Hematoma | 0.76 (0.69, 0.84) | 0.87 (0.86, 0.89) | 0.71 (0.70, 0.72) | 0.33 (0.31, 0.35) | 0.37 (0.34, 0.40) | 0.14 (0.11, 0.17) | 0.94 (0.94, 0.94) | 0.52 (0.48, 0.56) | TF-IDF | |
| IVH | 0.89 (0.67, 1.00) | 1.00 (1.00, 1.00) | 0.97 (0.95, 1.00) | 0.57 (0.40, 0.74) | 0.94 (0.89, 0.99) | 0.94 (0.94, 0.94) | 0.99 (0.96, 1.00) | 0.74 (0.36, 1.00) | TF-IDF | |
We demonstrate comparative performance statistics (AUPRC, sensitivity, specificity, and precision) for BioClinicalBERT and lasso regression for binary classification tasks. Across three data sets and four outcomes (edema, hemorrhagic conversion, MLS, and parenchymal hematoma), BioClinicalBERT demonstrated the greatest balanced performance, whereas lasso regression generally outperformed other machine learning classifiers. For each data set and outcome, lasso performance is shown according to the text featurization algorithm, which yielded the highest AUPRC value. Full model results across all classifiers and featurizations may be found in Supplemental Tables S2-S6
AUPRC, area under precision recall curve, BMC, Boston Medical Center, BOW, Bag of Words, IVH, intraventricular hemorrhage, MCA, middle cerebral artery, MGB, Massachusetts General Brigham and Women’s, MLS, midline shift, PPV, positive predictive value, TF-IDF, term frequency-inverse document frequency
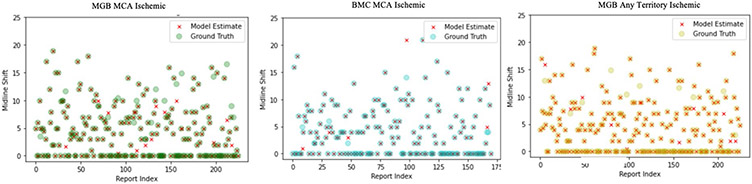
Additionally, we observed strong external performance from the RBS for IVH and continuous MLS. The IVH RBS achieved sensitivities > 90% and specificities > 99% across all data sets. McNemar’s testing demonstrated this system to outperform BioClinicalBERT (average χ2 = 427.0, p < 0.001; Supplementary Table S12). For continuous MLS estimation, the mean absolute error of our RBS was less than 0.2 mm for all data sets (Table 4, Fig. 3). In comparing absolute errors of this system with those of standard regression models, one-sided t-testing revealed that the RBS provided substantial improvements in absolute error over linear regressions trained with both BOW (t = 21.14, p < 0.001) and TF-IDF (t = 22.06, p < 0.001). (Supplementary Tables S13-S14).
Table 4.
RBS performance
| Sensitivity | Specificity | Precision | Accuracy | MAE (mm) | ||
|---|---|---|---|---|---|---|
| MGB MCA Ischemic Stroke | MLS | 0.95 (0.94, 0.96) | 0.95 (0.95, 0.97) | 0.92 (0.91, 0.93) | 0.97 (0.96, 0.97) | 0.18 (0.16, 0.20) |
| IVH | 0.94 (0.92, 0.95) | 0.99 (0.99, 0.99) | 0.85 (0.82, 0.88) | 0.99 (0.98, 0.99) | -- | |
| BMC MCA Ischemic Stroke | MLS | 0.96 (0.95, 0.96) | 0.96 (0.95, 098) | 0.95 (0.94, 0.95) | 0.98 (0.97, 0.98) | 0.14 (0.13, 0.16) |
| IVH | 0.91 (0.89, 0.93) | 0.99 (0.99, 1.00) | 0.83 (0.80, 0.85) | 0.99 (0.99, 1.00) | -- | |
| MGB Any Territory Ischemic Stroke | MLS | 0.95 (0.94, 0.96) | 0.95 (0.93, 0.99) | 0.95 (0.94, 0.96) | 0.99 (0.98, 1.00) | 0.03 (0.03, 0.04) |
| IVH | 0.94 (0.92, 0.96) | 0.99 (0.99, 1.00) | 0.85 (0.83, 0,87) | 0.99 (0.99, 1.00) | -- |
RBSs were developed for the detection of IVH as well as the quantification of MLS. In the case of our quantitative RBS for MLS, correct numerical detection was converted to true and false positives and negatives. Thus, for both outcomes, we report sensitivity, specificity, precision, and accuracy. Mean absolute error in millimeters is reported in the final column for the quantitative MLS system
BMC, Boston Medical Center, IVH, intraventricular hemorrhage, MAE, Mean average error, MCA, middle cerebral artery, MGB, Massachusetts General Brigham and Women’s, MLS, midline shift, RBS, rule-based system
Fig. 3.
RBS for quantitative midline shift. Scatterplots are demonstrated with overlap of RBS-detected midline shift (“model estimates”) and ground truths from computed tomography and magnetic resonance imaging reports in three separate cohorts. Points representing individual radiology reports are randomly selected from each cohort for visualization and arranged according to their order (“index”) within the overall sample. Significant concordance is seen between RBS-detected midline shift and ground truths. Full metrics for RBSs, including midline shift, are found in Table 3. BMC Boston Medical Center, MCA middle cerebral artery, MGB Massachusetts General Brigham and Women’s, RBS rule-based system
Lastly, among traditional machine learning classifiers, we observed the fastest CPU runtimes in lasso regression and K-nearest neighbor models, which each took < 60 s per round of bootstrapping. Runtimes for BioClinicalBERT on a GPU similarly took < 60 s per epoch. Full runtime statistics may be found in Supplementary Tables S15-S16.
Discussion
NLP is a promising approach to classify important unstructured data for research and clinical purposes [6, 38, 39]. In the present study, we analyzed a variety of NLP methods tailored to specific tasks in neurocritical care. Overall, we observed that a BioClinicalBERT transformer network pretrained on large critical care corpora outperformed other binary classifiers trained with both BOW and TF-IDF featurizations for detection of edema, MLS, hemorrhagic conversion, and parenchymal hematoma after acute ischemic stroke. Specifically, our fine-tuned BioClinicalBERT network discriminated well between reports with and without edema (AUPRC > 0.94), MLS (AUPRC > 0.98), and hemorrhagic conversion (AUPRC > 0.89) across three separate data sets that differed by hospital and ischemic stroke location. We found that performance was not quite as good for parenchymal hematoma (AUPRC > 0.76), which we speculate may be related to variations in how hemorrhage is described and the difficulty in distinguishing between confluent petechiae versus parenchymal hematoma from report alone. Notably, BioClinicalBERT ran efficiently on GPU hardware, with runtimes of < 60 s per epoch across all outcomes. We observed similar computational efficiency when training our second-highest performing model (lasso regression) on a standard desktop CPU. Finally, for IVH and continuous MLS detection, we found that customized RBS outperformed more generalized machine learning methods.
The field of neurocritical care is increasingly recognizing the potential for clinical and research applications that leverage the ability of NLP to organize information from unstructured radiologic data sets. In our own work, we previously reported improved detection of ischemic stroke, stroke location, and acuity using a modality-agnostic approach with word embeddings adapted from neurologic texts [4], which notably demonstrated significant mismatches between patient outcomes and officially documented diagnostic codes. Our current study expands the scope of these efforts by classifying sequelae of stroke and identifying which features are better suited to machine learning approaches versus expert-tailored RBS. Importantly, given variations in radiologist documentation [13], we have also shown that our current tool kit achieved robust external performance across differing hospital systems and patient populations.
Although our results demonstrate successful electronic phenotyping of neuroradiology reports, the full scope of applications for such NLP tools remains to be fully explored. From a research perspective, we believe that our methods can help expedite, scale, and control quality in clinical investigative endeavors by automating or semiautomating the identification of important outcomes in large retrospective data sets. Past work has highlighted the potential for inaccuracy when relying on administrative records, such as ICD codes, for classifying ground truth hospital outcomes [40, 41]. This mismatch is of direct consequence to stroke research, with Mandava and colleagues [42] notably arguing that the heterogeneous nature of stroke subtypes contributes to mistaken statistical inferences on clinical trial data. Consequently, further development of NLP methodologies such as ours could enable appropriate stratification of retrospective patient information according to critical stroke sequelae, thereby furthering efforts already underway in the field to automatically characterize ischemic stroke phenotypes [14, 15]. Additionally, NLP tools could potentially facilitate timely identification of patients for prospective studies. Improved characterization of stroke from radiologic reports may be used to screen for inclusion or exclusion criteria in clinical trials, which may in turn optimize administrative efficiency [43]. Applying NLP to identify or flag potentially eligible patients for research studies could decrease labor and costs associated with prospective investigation and lower the barrier for these research endeavors [44]. Although automated text mining for trial recruitment has been shown in other disciplines, such as oncology [45, 46], our work may help to bring similar NLP strategies into prospective trials for stroke.
We also envision a range of clinical applications for this work in the future. Notably, we feel that NLP tools such as these may be used to safeguard the timely communication of critical radiologic results in hospital settings. Although standardized systems for reporting are available in many tertiary care centers, delays in communication between radiologists and bedside providers remain a potential source of error in patient care [47], especially during periods of understaffing, including nights, weekends, and holidays. Moreover, given that radiologists in high-income nations are increasingly participating in global health delivery via teleradiology, our work also has implications for low-resource settings where critical results reporting may be underused [48]. Beyond improvements in communication, we also suggest that automated flagging of stroke sequelae could facilitate automated quality improvement initiatives. Escobar and colleagues [49] demonstrated better 30-day outcomes after implementing an automated model identifying concerning data trends that triggered a series of response protocols. Conceivably, our tools could be similarly used to develop quality improvement initiatives, including electronic-health-record-based warnings, such as flagging anticoagulation use in patients with documented hemorrhage on radiology reports or suggested stepwise protocols for level of care escalation in response to cerebral edema. Finally, additional work has shown how medical NLP systems could be potentially integrated with evidence-based guidelines to manage specific conditions identified by imaging reports [50]. Automatic retrieval of such information may allow for immediate review of evidence-based guidelines for providers’ convenience rather than relying on the clinician to initiate database searches de novo.
Although the potential applications of NLP may be intriguing, we acknowledge that the necessary technological infrastructure is lacking for immediate clinical translation of our findings. Although other studies have suggested pathways toward implementation of NLP systems in neurocritical care settings, including improved prehospital stroke diagnosis [7], prediction of stroke outcome from radiology reports [17], and automated phenotyping of stroke subclass [15] for treatment recommendations, only a fraction of medical NLP applications reach the level of in-hospital deployment [3]. It is important to recognize that fundamental proofs of principle in machine learning often fall short of effectively providing clinicians with the kind of clinical support systems that they often suggest to be possible [51]. With this perspective, we must state that our present NLP models, although well performing overall, still require the development of user-friendly interfaces, external validation, and well-designed implementation trials before they can be regarded as ready for patient care [47]. We concede that our work is foundational and hope that it can serve as a guide toward clinician-led directives that leverage technology to serve patients and improve outcomes. At the present time, we advocate that our NLP methods are best viewed as a promising analytic strategy for others conducting big data research in neurocritical care rather than a ready tool for clinical use.
We also recognize several methodological limitations within our study. Although our manually labeled data set of neurologic complications after ischemic stroke is one of the largest in literature to date, we acknowledge that we had a modest sample size and number of unique patients. We also note that our gold standard labeling of radiology reports reflects a professional interpretation of raw imaging data and may not represent the clinical ground truth of a patient’s condition. Additionally, we found lower performance of parenchymal hematoma detection relative to other outcomes, which may be, in part, due to our annotation strategy. Our labelers assigned parenchymal hematoma occurrence on the basis of interpretation of ECASS-II criteria and not direct specific radiology phrases, leading to more heterogeneity and lower interrater reliability. It is also possible that we overestimated the degree of external validity of our methods. Given that our clinical data sets contain some patients with multiple reports, models might have been trained with linguistically correlated data, which could potentially limit generalizability to other unseen data sets. Because the two hospital systems used for comparison are located within the same city in the United States, a substantial number of staff radiologists between the two institutions have received training or experience between both hospitals during their careers. Therefore, it is possible that reporting styles in the MGB and BMC data sets may be more similar than comparators derived from other regions.
Nevertheless, our results suggest that a new generation of NLP models holds promise for characterizing and subtyping large amounts of radiologic text data in ischemic stroke. Future studies may apply our methodology and models to identify cohorts, outcomes of interest, and/or covariates for epidemiologic and clinical research in neurocritical care. Further work in this area is necessary to ensure generalizability of these methods for future research and clinical applications.
Conclusions
Automated NLP approaches detect critical complications of ischemic stroke, including edema, MLS, hemorrhagic conversion, parenchymal hematoma, and IVH. A pre-trained BioClinicalBERT model yielded the highest overall performance in detecting common binary outcomes and outperformed traditional machine learning classifiers trained with BOW and TF-IDF featurizations. Our fine-tuned BioClinicalBERT also demonstrated strong external validity, generalizing well from our derivation cohort of MCA territory ischemic strokes to external data sets from a separate institution and to patients presenting with a variety of locations of cerebral infarctions. Our results reinforce the utility of deep attention neural networks in clinical NLP tasks. The presented approach has multiple important clinical and research applications, including scalable electronic phenotyping, augmentation of clinical risk prediction models, and facilitation of automatic alert systems in the hospital setting. Additional research in this area will continue the development of novel strategies for electronic phenotyping in neurocritical care.
Supplementary Material
Source of Support
CJO receives support from National Institutes of Health National Institute of Neurological Disorders and Stroke grant K23NS116033 and the Peter Paul Young Career Development Foundation at Boston University. EJB is supported in part by 2R01 HL092577, 1R01 HL141434 01A1, 1R01AG066010, 1R01AG066914, and American Heart Association grant AHA_18SFRN34110082. DMG receives support from R01 NS102574. SMS receives support from RO1 EY024019 and Veterans Affairs Merit award I01RX002981.
Footnotes
Conflicts of interest
The authors do not have any relevant conflicts of interest that may impact this work.
Ethical Approval/Informed Consent
This work adheres to ethical guidelines and was approved by the Institutional Review Boards of Massachusetts General and Brigham & Women’s Hospitals, as well as Boston Medical Center.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.1007/s12028-022-01513-3.
References
- 1.Foreman B. Neurocritical care: bench to bedside (Eds. Claude Hemphill, Michael James) integrating and using big data in neurocritical care. Neurotherapeutics. 2020;17(2):593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casey A, et al. A systematic review of natural language processing applied to radiology reports. BMC Med Inform Decis Mak. 2021;21(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pons E, et al. Natural language processing in radiology: a systematic review. Radiology. 2016;279(2):329–43. [DOI] [PubMed] [Google Scholar]
- 4.Ong CJ, et al. Machine learning and natural language processing methods to identify ischemic stroke, acuity and location from radiology reports. PLoS ONE. 2020;15(6): e0234908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkins JS, et al. Coding neuroradiology reports for the Northern Manhattan Stroke Study: a comparison of natural language processing and manual review. Comput Biomed Res. 2000;33(1):1–10. [DOI] [PubMed] [Google Scholar]
- 6.Langlotz CP, et al. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology. 2019;291(3):781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayampurath A, et al. Improving prehospital stroke diagnosis using natural language processing of paramedic reports. Stroke. 2021;52(8):2676–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manning C, Schutze H. Foundations of statistical natural language processing. MIT Press; 1999. [Google Scholar]
- 9.Arsava EM, et al. Assessment of the predictive validity of etiologic stroke classification. JAMA Neurol. 2017;74(4):419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mozayan A, et al. Practical guide to natural language processing for radiology. Radiographics. 2021;41(5):1446–53. [DOI] [PubMed] [Google Scholar]
- 11.Friedlin J, McDonald CJ. A natural language processing system to extract and code concepts relating to congestive heart failure from chest radiology reports. In: AMIA annual symposium proceedings. American Medical Informatics Association; 2006. [PMC free article] [PubMed] [Google Scholar]
- 12.Hinton G. Deep learning—a technology with the potential to transform health care. JAMA. 2018;320(11):1101–2. [DOI] [PubMed] [Google Scholar]
- 13.Li M, et al. Analysis of stroke detection during the COVID-19 pandemic using natural language processing of radiology reports. Am J Neuroradiol. 2021;42(3):429–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg R, et al. Automating ischemic stroke subtype classification using machine learning and natural language processing. J Stroke Cerebrovasc Dis. 2019;28(7):2045–51. [DOI] [PubMed] [Google Scholar]
- 15.Guan W, et al. Automated electronic phenotyping of cardioembolic stroke. Stroke. 2021;52(1):181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro VM, et al. Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology. 2017;88(2):164–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heo TS, et al. Prediction of stroke outcome using natural language processing-based machine learning of radiology report of brain MRI. J Pers Med. 2020;10(4):286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacchi S, et al. Deep learning natural language processing successfully predicts the cerebrovascular cause of transient ischemic attack-like presentations. Stroke. 2019;50(3):758–60. [DOI] [PubMed] [Google Scholar]
- 19.Zech J, et al. Natural language-based machine learning models for the annotation of clinical radiology reports. Radiology. 2018;287(2):570–80. [DOI] [PubMed] [Google Scholar]
- 20.Devlin J, et al. Bert: pre-training of deep bidirectional transformers for language understanding. 2018. arXiv preprint arXiv:1810.04805. [Google Scholar]
- 21.Larrue V, et al. Risk factors for severe hemorrhagic transformation in ischemic stroke patients treated with recombinant tissue plasminogen activator: a secondary analysis of the European-Australasian Acute Stroke Study (ECASS II). Stroke. 2001;32(2):438–41. [DOI] [PubMed] [Google Scholar]
- 22.Harris PA, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaswani A, et al. Attention is all you need. In: Advances in neural information processing systems. 2017. [Google Scholar]
- 24.Lee J, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AE, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alsentzer E, et al. , Publicly available clinical BERT embeddings. 2019. arXiv preprint arXiv:1904.03323. [Google Scholar]
- 27.Franklin J. The elements of statistical learning: data mining, inference and prediction. Math Intell. 2005;27(2):83–5. [Google Scholar]
- 28.Cover T, Hart P. Nearest neighbor pattern classification. IEEE Trans Inf Theory. 1967;13(1):21–7. [Google Scholar]
- 29.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 30.Chaudhuri B, Bhattacharya U. Efficient training and improved performance of multilayer perceptron in pattern classification. Neurocomputing. 2000;34(1–4):11–27. [Google Scholar]
- 31.Pedregosa F, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 32.Darby DG, et al. Primary intraventricular hemorrhage: clinical and neuropsychological findings in a prospective stroke series. Neurology. 1988;38(1):68–68. [DOI] [PubMed] [Google Scholar]
- 33.Foroushani HM, et al. Accelerating prediction of malignant cerebral edema after ischemic stroke with automated image analysis and explainable neural networks. Neurocrit Care. 2021;36:471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. 2015;10(3):e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 1998;10(7):1895–923. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong RA. When to use the Bonferroni corrrection. Ophthalmic Physiol Opt. 2014;34:502–8. [DOI] [PubMed] [Google Scholar]
- 37.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 38.Cai T, et al. Natural language processing technologies in radiology research and clinical applications. Radiographics. 2016;36(1):176–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorin V, et al. Deep learning for natural language processing in radiology—fundamentals and a systematic review. J Am Coll Radiol. 2020;17(5):639–48. [DOI] [PubMed] [Google Scholar]
- 40.Jetté N, et al. How accurate is ICD coding for epilepsy? Epilepsia. 2010;51(1):62–9. [DOI] [PubMed] [Google Scholar]
- 41.Quan H, et al. Assessing validity of ICD-9-CM and ICD-10 administrative data in recording clinical conditions in a unique dually coded database. Health Serv Res. 2008;43(4):1424–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandava P, et al. A critical review of stroke trial analytical methodology: outcome measures, study design, and correction for imbalances. Transl Stroke Res. 2012. 10.1007/978-1-4419-9530-8_40. [DOI] [Google Scholar]
- 43.Elkins JS, et al. Recruiting subjects for acute stroke trials: a meta-analysis. Stroke. 2006;37(1):123–8. [DOI] [PubMed] [Google Scholar]
- 44.Woo M. An AI boost for clinical trials. Nature. 2019;573(7775):S100–S100. [DOI] [PubMed] [Google Scholar]
- 45.Liu R, et al. Evaluating eligibility criteria of oncology trials using real-world data and AI. Nature. 2021;592(7855):629–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander M, et al. Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients. JAMIA Open. 2020;3(2):209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy DR, Singh H, Berlin L. Communication breakdowns and diagnostic errors: a radiology perspective. Diagnosis. 2014;1(4):253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mollura DJ, et al. 2016 RAD-AID conference on international radiology for developing countries: gaps, growth, and United Nations sustainable development goals. J Am Coll Radiol. 2017;14(6):841–7. [DOI] [PubMed] [Google Scholar]
- 49.Escobar GJ, et al. Automated identification of adults at risk for in-hospital clinical deterioration. N Engl J Med. 2020;383(20):1951–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Do BH, et al. Automatic retrieval of bone fracture knowledge using natural language processing. J Digit Imaging. 2013;26(4):709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Citerio G. Big data and artificial intelligence for precision medicine in the Neuro-ICU: Bla, Bla, Bla. Neurocrit Care. 2022. 10.1007/s12028-021-01427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.