Abstract
Purpose:
Reactive oxygen species (ROS) are oxygen-containing molecules that have high reactivity and play roles in protection or harm the cancer cells. We aimed to clarify the clinical relevance of ROS in breast cancer (BC) tumor microenvironment (TME). We hypothesized that it is associated with worse BC patient outcomes.
Methods:
ROS score was generated by Gene Set Variation Analysis of Hallmark ROS pathway gene set and a total of 6,245 BC patients were analyzed.
Results:
High ROS BC significantly enriched cell proliferation-related gene sets (MYC targets v1 and v2, G2M checkpoint, E2F targets), pro-cancer-related gene sets (DNA repair, unfolded protein response, MTORC1 signaling, PI3K/AKT/MTOR signaling, glycolysis, and oxidative phosphorylation), immune-related gene sets (inflammatory response, allograft rejection, interferon-α and γ responses, complement, and IL6/JAK/STAT3 signaling), and infiltrated immune cells (CD4+ memory and CD8+ T cells, Th1 and Th2, dendritic cells, Tregs, M1 and M2 macrophages) and B-cells, as well as elevated cytolytic activity consistently in both METABRIC and GSE96058 cohorts. Cancer cells were the major source of ROS in BC TME of single cell sequence (GSE75688) cohort. High ROS was associated with intratumor heterogeneity, homologous recombination defects, mutation rates and neoantigens, and with clinical aggressiveness in AJCC stage, Nottingham grade and Ki67 expression, as well as worse overall survival in both GSE96058 and METABRIC, and with worse disease-specific survival in METABRIC.
Conclusion:
Abundant ROS in BC patients is associated with abundant mutations, aggressive cancer biology, immune response and worse survival.
Keywords: biomarker, GSVA, reactive oxygen species, survival analysis, tumor immune microenvironment
Introduction
The body maintains its homeostasis and health by balancing oxidative system that produce energy and antioxidant system that prevent the negative effects of excess oxidation, also known as oxidative stress [1, 2]. Recently, it has become clear that excessive oxidation and oxidative stress directly damages DNA and are deeply involved not only in carcinogenesis but also in cancer progression, including cancer growth, invasion, and metastatic spread.
Reactive oxygen species (ROS) are oxygen-containing molecules that have high reactivity. Hydroxyl (OH*) and superoxide (O2*) free radicals both comprise ROS along with hydrogen peroxide (H2O2). ROS are physiologically produced in the mitochondrial electron-transfer system, peroxisomes and phagosomes in the cells, where they contribute to energy production and phagocytosis [3]. ROS is also produced in small amounts by the NADPH oxidase (Nox) family at the plasma membrane and is involved in a number of intracellular signaling pathways, such as Ras, c-jun N-terminal kinase (JNK), p38 and mitogen-activated protein kinase (MAPK), and PI3K/AKT/mTOR pathways. Recently, it has been shown that ROS is involved in cell proliferation [4, 5] as well as autophagy [6], apoptosis [7], and inflammation via the NLRP3 inflammasome and nuclear factor-kB (NF-kB) pathway [8, 9]. Cells experience oxidative stress if either the ROS production increases or the number of scavenged ROS decreases [10]. ROS have been reported to be increased in breast cancer cells as well as several immune cells in tumor microenvironment (TME), and therefore, is an important factor in the TME [11]. It likely plays a multifaceted role in tumor progression and metastasis by ensuring bidirectional communication among various components [12]. Therefore, it has been suggested that ROS may be a potential therapeutic target in breast cancer [13]. While ROS have physiological functions as described, they can be harmful by directly damaging DNA during oxidative stress. Our group and others have shown that abundant DNA damage cause higher mutation load that leads to carcinogenesis and aggravation of cancer [14]. Ongoing aerobic glycolysis followed by pyruvate oxidation in the mitochondria (the Warburg effect) generates ROS which in turn increases the activity of oncogenes and receptors. This results in stimulation of oxidizing enzymes and/or growth factor-dependent pathways, thus inducing instability of genes [10]. For this reason, cells are equipped with antioxidant system that either take in antioxidants or produce them by enzymes. Although excessive induction of oxidative stress is expected to prevent and/or treat cancer since cancer cells have higher ROS than normal cells as a base line, ROS may aggravate cancer by promoting cell proliferation. To this end, the clinical relevance of abundant ROS in breast cancer remains unclear.
In this study, we generated a ROS score using ROS-related gene expressions in human breast cancer to quantify the abundance of ROS and investigated its clinical relevance. Gene Set Variation Analysis (GSVA) with 200 ROS-related genes defined by Molecular Signatures Database (MSigDB) Hallmark collection was used to generate the ROS score [15]. This approach is commonly used in genetic research because pathways involve various genes, and a single gene expression may fail to grasp the whole picture. For instance, we found the association between G2M checkpoint pathway and pancreatic cancer survival and drug response [16, 17], angiogenesis and aggressiveness of gastric cancer and worse survival [18], and DNA repair signaling [19] as well as unfolded protein response [20] and worse survival in liver cancer. We have demonstrated in breast tumor that pathways such as early estrogen response [21], KRAS signaling [22], angiogenesis [23], inflammation [24], thermogenesis [25], adipogenesis [26], and apoptosis [27] are associated with survival, and G2M checkpoint pathway [28] and E2F pathway [29] predict response to neoadjuvant chemotherapy in Luminal A subtype. In this study, we hypothesized that abundant ROS is significantly associated with cancer aggressiveness and tumor microenvironment, as well as patient survival.
Materials and Methods
Data acquisition
Clinical information and gene expression were obtained on 1,903 breast cancer patients from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort, as we previously reported [30, 31]. The GSE96058 cohort consists of full transcriptome profile of resected tumors from 3,273 breast cancer patients whose clinical data was obtained from the Swedish Breast Cancer Analysis Network (SCAN-B) [32]. Transcriptomic data was also obtained on 1,069 female breast cancer patients from The Cancer Genome Atlas (TCGA) cohort [33]. The GSE75688 cohort has single-cell RNA-sequencing data of cancer cells, myeloid cells, immune cells, and stromal cells in breast cancer [34], which was obtained from Gene Expression Omnibus.
ROS signaling score
Liberzon et al. reported the Molecular Signatures Database (MSigDB) hallmark gene set collection, one of the most comprehensive and widely used databases of gene sets for performing gene set enrichment analysis [15]. We used “HALLMARK REACTIVE OXYGENE SPECIES” gene sets, that was calculated by Gene Set Variation Analysis (GSVA) algorithm [35], in the similar manner as our previous reports on KRAS signaling [22], G2M checkpoint [16, 17, 28], E2F targets [29], angiogenesis [18], estrogen response [21], inflammation [24], thermogenesis [25], adipogenesis [26], and apoptosis [27].
Other scores
Cytolytic activity (CYT) score estimates the overall immune activity by expression pattern of granzyme A (GZMA) and perforin (PRF1) genes [36–40]. xCell scores were used as immune cells score calculating by the xCell algorithm [41], as we previously reported [42–44]. Score results provided by Thorsson et al. [45] in the TCGA cohort were utilized to calculate homologous recombination defects (HRD), intratumor heterogeneity, single nucleotide variation (SNV), silent and non-silent mutation rate, and fraction altered.
Gene set enrichment analysis
The difference in activated signaling pathways between low- and high-ROS score groups were investigated by Gene Set Enrichment Analysis (GSEA) [46] using GSEA Java software (version 4.2) with MSigDB Hallmark gene sets [15]. False discovery rate (FDR) < 25% was defined as statistically significant, as recommended by GSEA.
Other statistical analyses
All analyses and data plots were generated using Microsoft Excel (version 16) and R software (version 4.0.1). The analysis of the comparison of groups used the Kruskal-Wallis test, the Mann-Whitney U test, or the Fisher exact test. Log-rank test and the Kaplan-Meier plot were used for survival analyses. Values of p < 0.05 generally indicate a statistically significant difference.
Results
Breast cancer with a high reactive oxygen species (ROS) score enriched cell proliferation and other pro-cancerous gene sets
The Gene set variation analysis (GSVA) algorithm on MSigDB was used to define the ROS score (Table S1), similar to how we defined the other scores in our previous publications [16–19]. We first investigated the underlying mechanism involved in the score by conducting Gene Set Enrichment Analysis (GSEA) on two independent breast cancer cohorts, GSE96058 and METABRIC. We found that tumors with a high ROS score significantly enriched expression of Hallmark cell proliferation gene sets; MYC targets v1 and v2, G2M checkpoint, and E2F targets (Fig. 1A), as well as other pro-cancerous signaling; glycolysis, MTORC1 signaling, PI3K/AKT/MTOR signaling, oxidative phosphorylation, DNA repair, and Unfolded protein response, in both the cohorts consistently (Fig. 1B). To this end, there is a significant association of high ROS score with both cell proliferation and also other pro-cancer signaling and responses in breast cancer.
Fig. 1. Cell proliferation-related and other pro-cancer signaling-related gene sets enriched to breast cancer with a high ROS score consistently in both GSE96058 and METABRIC cohorts.
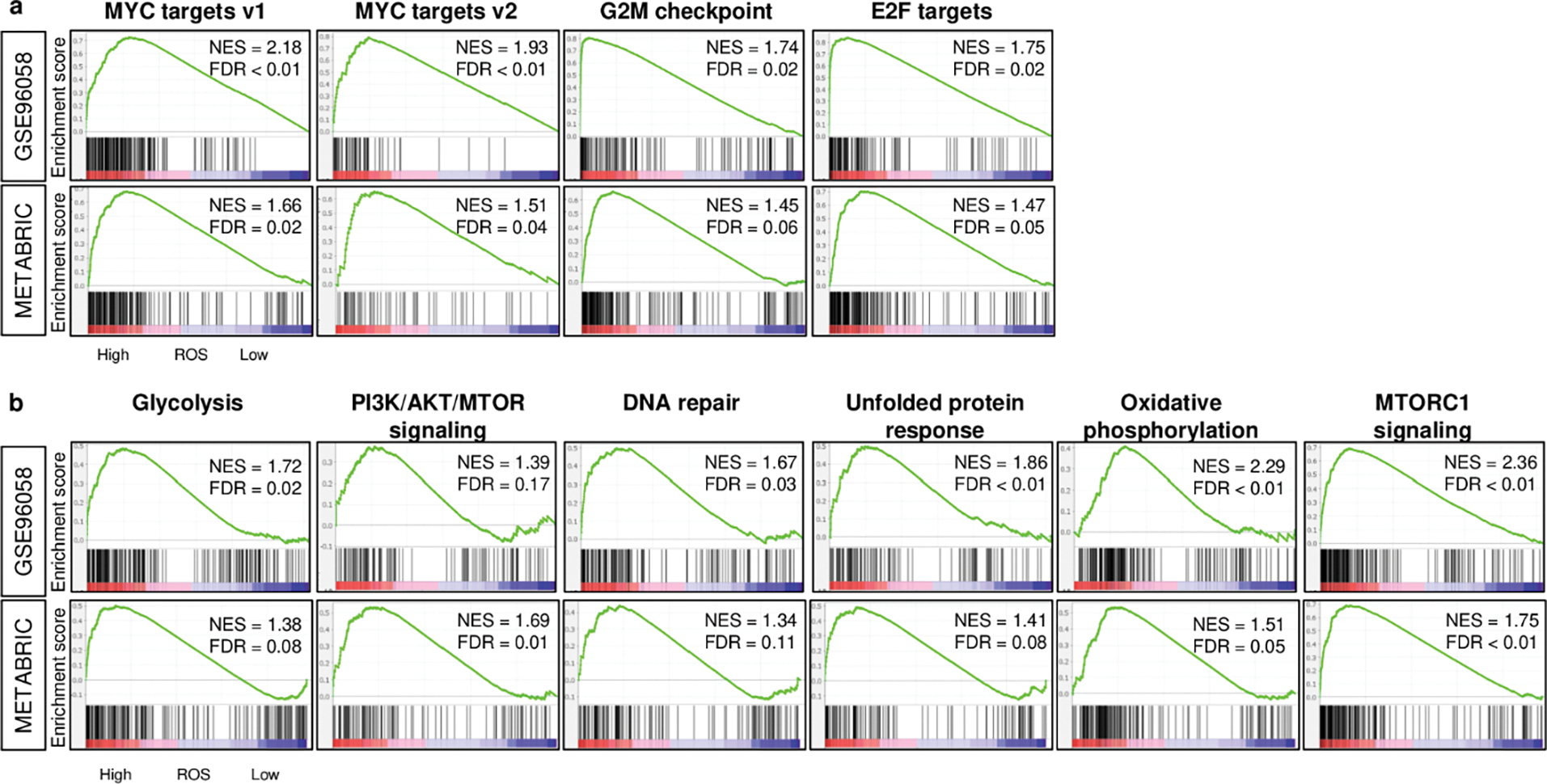
a. Enrichment plots of cell proliferation-related gene sets (MYC targets v1 and v2, G2M checkpoint, E2F targets), and b. other pro-cancerous gene sets (glycolysis, PI3K/AKT/MTOR signaling, DNA repair, Unfolded protein response, oxidative phosphorylation, and MTORC1 signaling) in the GSE96058 and METABRIC cohorts. The classical gene set enrichment analysis (GSEA) method was used to determine the NES (normalized enrichment score) and the FDR (false discovery rate) where the cut-off to divide two score groups was defined as median value
Cancer cells were the major source of ROS in tumor microenvironment (TME), and immune-related gene sets significantly enriched to high ROS breast cancer.
To identify which cell type is the major source of ROS in the tumor microenvironment (TME), the score was measured in a single-cell sequence cohort (GSE75688) that differentiate transcriptome from cancer cells, myeloid cells, T cells, B cells or stromal cells. We found that cancer cells are the major source of ROS in TME (Fig. 2A; p < 0.001). Next, we investigated the relationship between ROS and immune response in TME of breast tumor. We found that immune-related gene sets; allograft rejection, interferon (IFN)-γ response, IFN-α response, inflammatory response, complement, and IL6/JAK/STAT3 signaling, were significantly enriched to breast cancer with elevated ROS score in both METABRIC and GSE96058 cohorts consistently (Fig. 2B). To this end, immune response was significantly associated with high ROS score in breast cancer.
Fig. 2. Relationship between ROS and immune response in breast cancer TME.
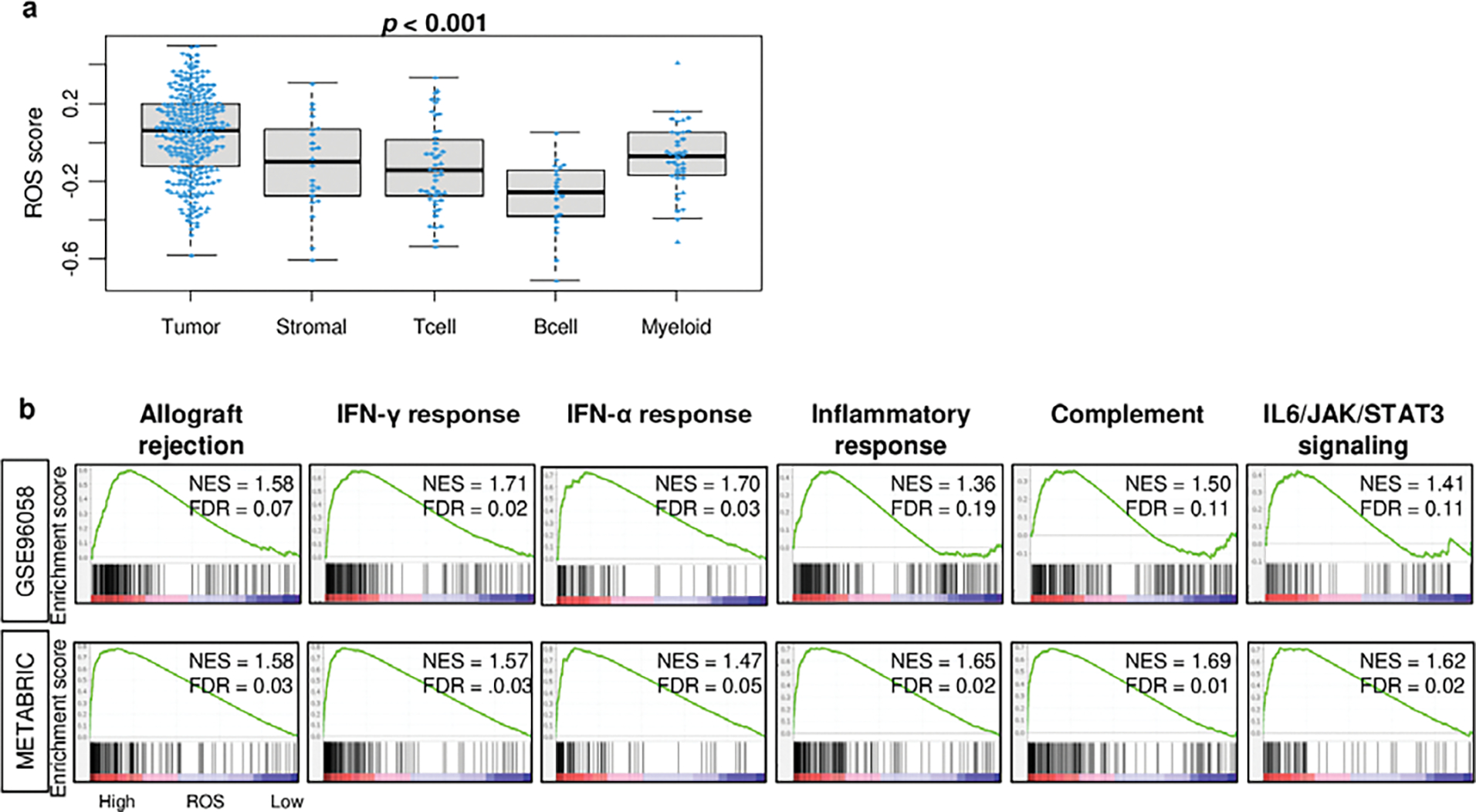
a. Boxplots showing ROS by single cells of the cancer, myeloid cells, B cells, T cells, and stromal cells in the GSE75688 cohort. P-values were calculated by the Kruskal-Wallis test. b. Gene enrichment plots of allograft rejection, interferon (IFN)-γ response, IFN-α response, inflammatory response, complement, and IL6/JAK/STAT3 signaling, that enriched to high ROS score in both cohorts. The median value was used as a cut-off to divide high vs low score groups. NES (normalized enrichment score) and FDR (false discovery rate) were determined with the classical gene set enrichment analysis (GSEA) method
Immune cells were universelly infiltrated in ROS high tumor immune microenviornment (TIME) where cytolytic activity (CYT) were elevated
Since immune response was enhanced in ROS high tumor, to investigate which type of immune cell is infiltrated in breast cancer TIME was of interest. The xCell algorithm was used to estimate immune cell infiltration, in a similar fashion as previous publications [39, 42, 47–51]. We found that in high ROS breast cancer, there was significant infiltration of not only pro-cancer immune cells; M2 macrophages, T helper type 2 (Th2) cells, and regulatory T cells, but also anti-cancer immune cells; CD4+ memory T cells, CD8+ T cells, T helper type 1 (Th1) cells, M1 macrophages, and dendritic cells (DC), and B cells, consistently in both GSE96058 and METABRIC cohorts (Fig. 3A; all p < 0.001). Furthermore, they were significantly associated with high CYT (Fig. 3B; both p < 0.001). To this end, immune cells are universally infiltrated with high cytolytic activity in high ROS breast cancer.
Fig. 3. Infiltrating immune cells by the ROS score in breast cancer.
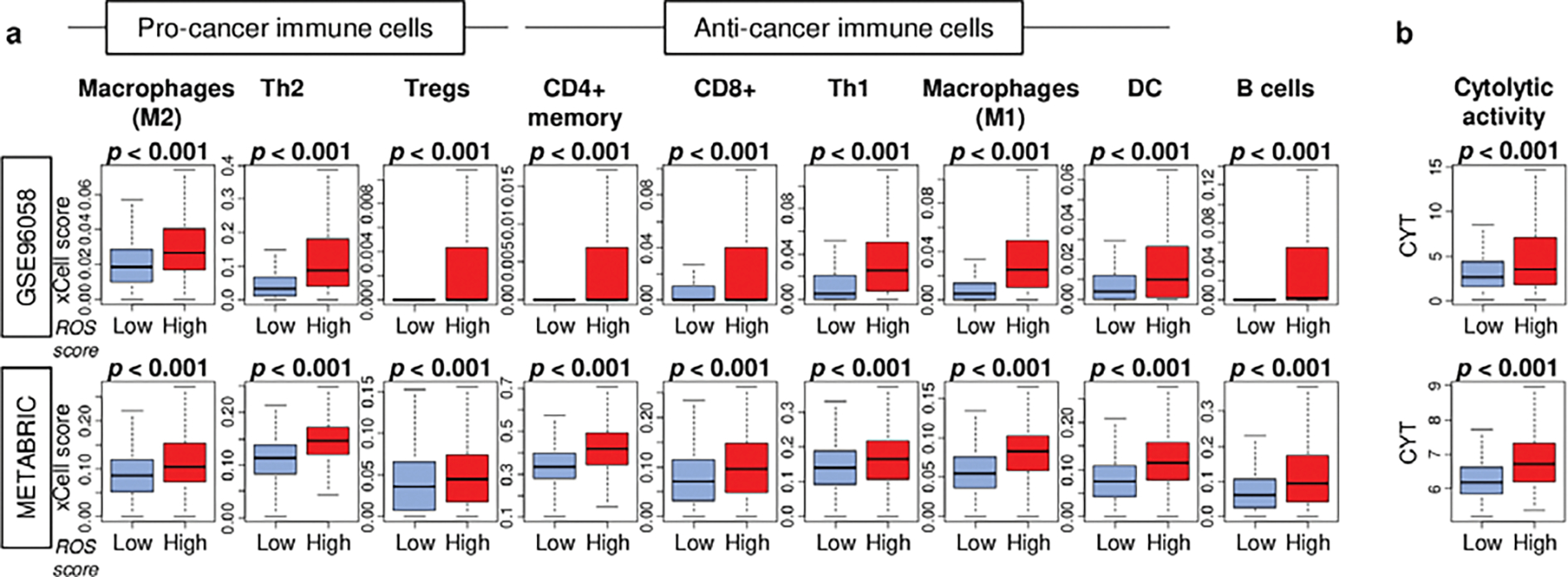
Boxplots of infiltrating fraction of a. pro-cancer immune cells; regulatory T cells (Tregs), type2 T (Th2) cells, M2 macrophages, and anti-cancer immune cells; CD4+ memory T cells, CD8+ T cells, type1 T helper (Th1) cells, M1 macrophages, and B cells by high and low ROS score groups in the GSE96058 and METABRIC cohorts. b. Box plots of cytolytic activity score (CYT) by low and high ROS score groups in both cohorts. The median value was used as a cut-off to divide two score groups. P-values were calculated by the Mann-Whitney U test
High ROS score was significantly associated with high level of intratumor heterogeneity, homologous recombination deficiency (HRD), and mutation load in breast cancer
Tumor-infiltrating immune cells are known to be attracted to TME through neoantigens generated by high tumor mutational burden [14]. To this end, investigating the relationship of the ROS score with the intratumor heterogeneity, HRD, and mutation load in breast cancer was of interest. We found that breast cancer with a high ROS score was significantly associated with HRD and intratumor heterogeneity, as well as mutation load-related score; single nucleotide variation (SNV) neoantigens, silent and non-silent mutation rates, in the TCGA cohort (Fig. 4; all p < 0.001). This result is in agreement with the notion that immune cells are attracted to cancer cells with high mutation load, and our observation that high ROS score breast cancer is associated with immune response and immune cell infiltration, commonly seen in aggressive cancer.
Fig. 4. Association of the ROS score with the intratumor heterogeneity, homologous recombination deficiency (HRD), and mutation-related score in the TCGA cohort.
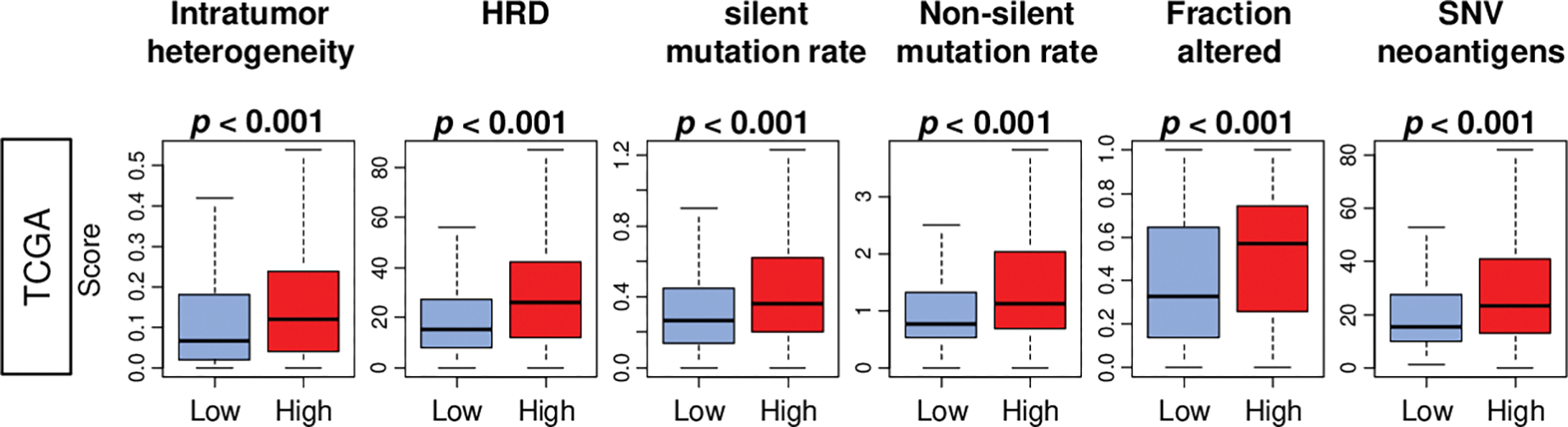
Boxplots of homologous recombination defects (HRD), and intratumor heterogeneity, and the mutation-related scores; silent and non-silent mutation rate, fraction altered, single nucleotide variation (SNV) neoantigens, by low and high ROS score groups. The median value was used as a cut-off to divide two score groups. P-values were calculated by the Mann-Whitney U test
A high ROS breast cancer was significantly associated with clinical aggressiveness
To study the clinical relevance of the ROS score in breast cancer, its association with breast cancer subtypes, AJCC (American Joint Committee on Cancer) staging including the lymph node category, the Nottingham histological grade were investigated in the GSE96058 and METABRIC cohorts. In the METABRIC cohort, the high score was significantly associated with triple-negative breast cancer (TNBC) subtype, advanced stage and lymph node metastasis, and higher grade (Fig. 5, all p < 0.001). GSE96058 cohort was used to validate these results (all p < 0.001). Therefore, ROS score was associated with clinical parameters of tumor aggressiveness.
Fig. 5. Breast cancer with a high ROS score was significantly associated with advanced stage, lymph node metastasis, higher pathological grade, and TNBC.
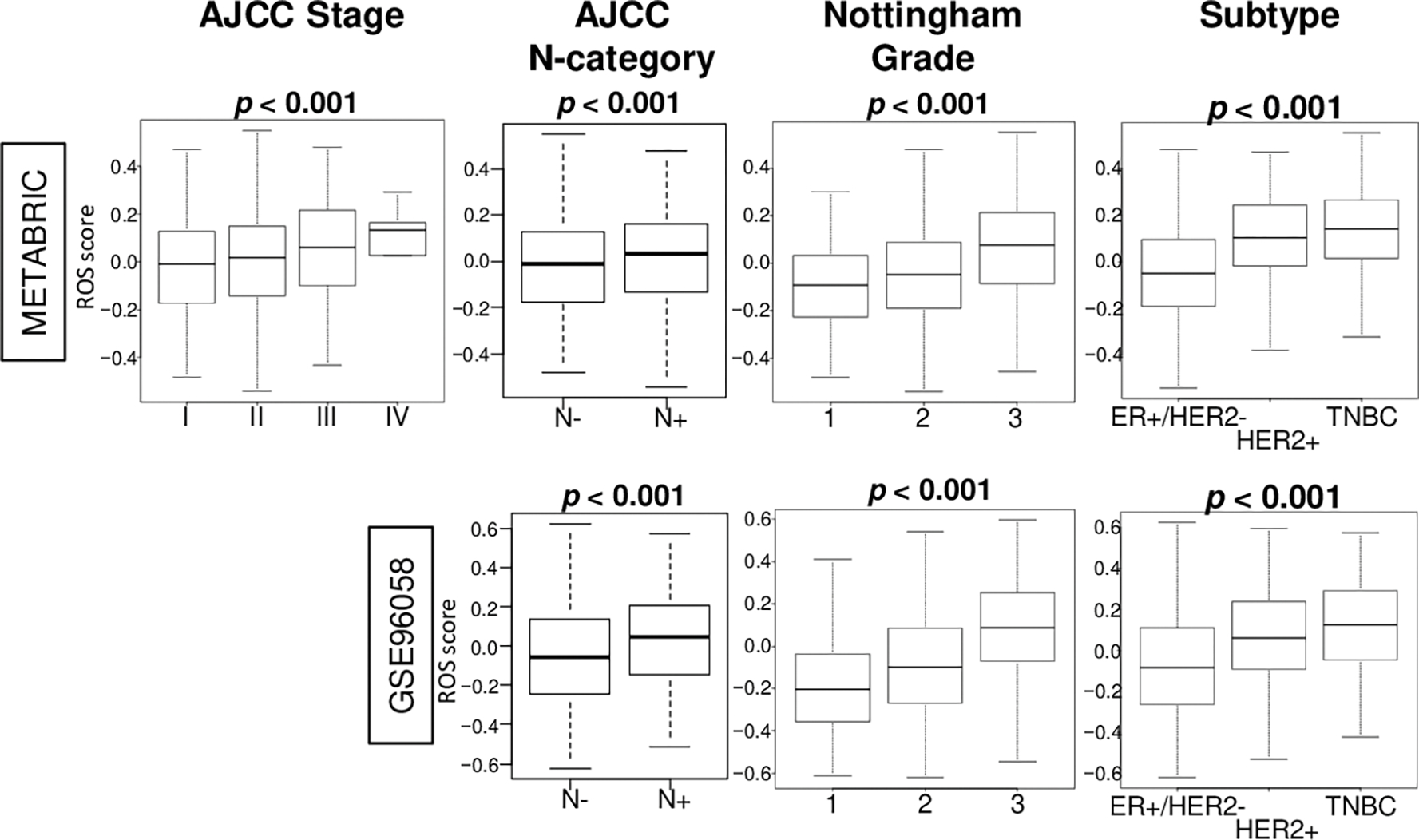
Box plots of the score by American Joint Committee on Cancer (AJCC) stage in the METABRIC, and lymph node metastasis status, Nottingham pathological grade, and breast cancer subtypes in the METABRIC and GSE96058 cohorts. P values were calculated by Kruskal-Wallis and Mann-Whitney U test.
High ROS ER-positive/HER2-negative breast cancer was significantly associated with worse survival, but not the other subtypes
Given that ROS score was significantly different by breast cancer subtypes, we hypothesized that there may be a different relationship of each subtype with ROS. Therefore, overall survival (OS) in the GSE96058 and METABRIC cohorts and disease specific survival (DSS) in the METABRIC by subtypes were analyzed. High ROS score was significantly associated with worse OS in ER-positive/HER2-negative subtype and the whole group, but not in the other subtypes in the GSE96058 cohort (Fig. 6). OS and DSS in METABRIC cohort validated these results. To this end, high ROS was associated with worse survival outcome only in ER-positive/HER2-negative breast cancer subtype. Furthermore, we investigated the association of ROS score with infiltrating immune cells in ER-positive/HER2-negative breast cancer. We found that high ROS ER-positive/HER2-negative breast cancer was also associated with high infiltration of Th1 and Th2 cells, CD4+ memory T cells, and M1 macrophages, consistently in both cohorts (Fig. S1).
Fig. 6. ER-positive/HER2-negative breast cancer with a high score is significantly associated with worse survival in the GSE96058 and METABRIC cohorts.
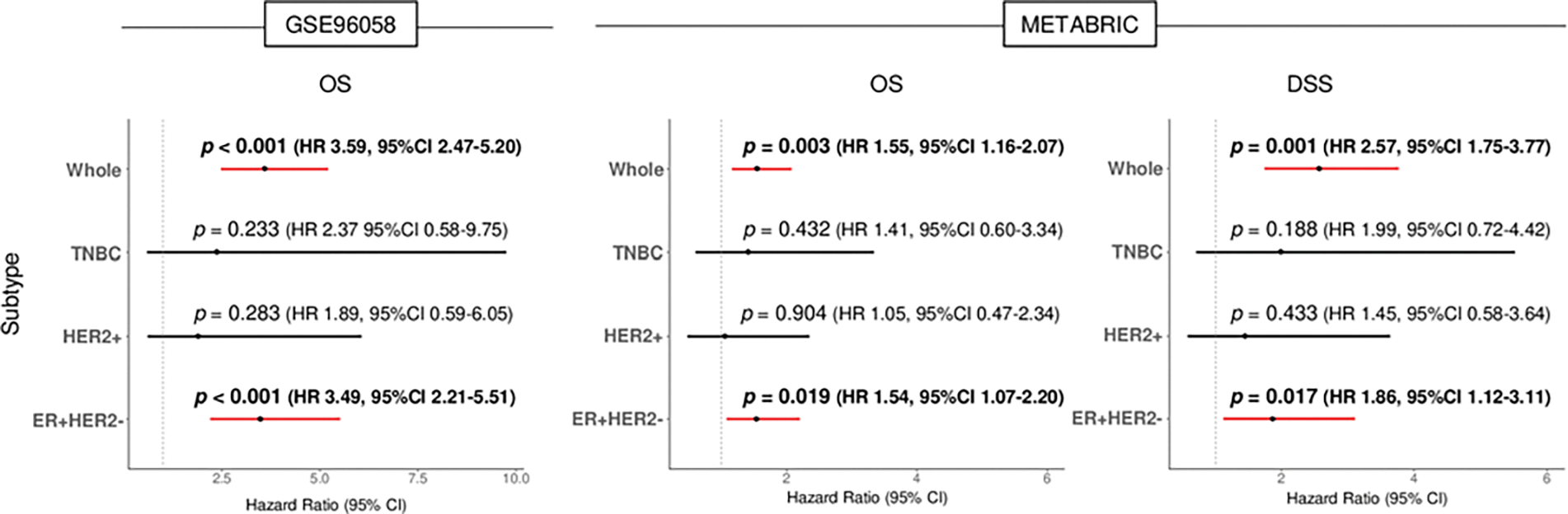
Forest plots with log-rank p-values and hazard ratio (HR) of OS in the GSE96058 cohort, and OS and DSS in the METABRIC cohort between low and high ROS score in the whole, ER+/HER2−, HER2+ and TNBC subgroups. Log-rank test was used to calculate p values. DSS, disease-specific survival; ER, estrogen receptor; HER2, human epidermal growth factor 2; OS, overall survival; TNBC, triple-negative breast cancer
Discussion
In this study, we generated a ROS score utilizing Hallmark ROS pathway gene sets by GSVA algorithm and quantified the abundance of ROS in total of 6,245 breast cancer patients. We found that high ROS breast cancer enriched cell proliferation gene sets; MYC targets v1 and v2, G2M checkpoint, E2F targets, as well as other pro-cancerous gene sets; glycolysis, PI3K/AKT/MTOR signaling, DNA repair, unfolded protein response, MTORC1 signaling, and oxidative phosphorylation, consistently in both GSE96058 and METABRIC cohorts. Cancer cells were the major source of ROS among the cells in TIME, including myeloid cells, T cells, B cells, and stromal cells, based on single cell sequence cohort. High ROS breast cancer enriched immune gene sets; inflammatory response, IL6/JAK/STAT3 signaling, IFN-γ response, IFN-α response, allograft rejection, and complement, in both the cohorts consistently. High ROS breast cancer was significantly associated with high infiltration of pro-cancer immune cells; Tregs, Th2 cells, and M2 macrophages, as well as anti-cancer immune cells; M1 macrophages, CD4+ memory T cells, CD8+ T cells, dendritic cells, Th1 cells, and B-cells in both cohorts. Furthermore, high ROS breast cancer was significantly associated with enhanced CYT. High ROS score was significantly associated with high levels of homologous recombination defects and intratumor heterogeneity, along with mutation-related score, including single nucleotide variant and indel neoantigens, fraction altered, silent and non-silent mutation rate in the TCGA cohort. Clinically, high ROS breast cancer was significantly associated with advanced cancer by AJCC stage, lymph node metastasis, and Nottingham grade. Furthermore, high ROS was significantly associated with worse OS in both cohorts, and DSS in the METABRIC cohort in ER-positive/HER2-negative subtype.
ROS attack DNA, thus inducing DNA damage which, could either inhibit or induce transcription, replication errors, signal transduction pathways and genomic instability, resulting in carcinogenesis [52]. Several risk factors of breast cancer are related to ROS production and oxidative stress, thus, supporting the role of ROS involvement in breast cancer pathogenesis. Aging is associated with the production of ROS, like H2O2 and HOCl, that induce mutations in mitochondrial DNA [53]. Mutations in BRCA1 and BRCA2, the breast cancer susceptibility genes, are also implicated in breast cancer. BRCA1 upregulates the expression of multiple genes that are involved in the cytoprotective antioxidant response, such as, glutathione-S-transferase (GST), oxidoreductase, glutathione peroxidase, and other antioxidant genes. A potential source of ROS, ionizing radiation, can induce irreversible DNA damage, thus initiating carcinogenesis. Female breast tissue is particularly susceptible to the carcinogenic effects of ionizing radiation, especially before it is fully differentiated before first child birth [54]. Estrogens are known key player in all stages of breast cancer, where estrogen receptor (ER) independent effects result from oxidative metabolism of estradiol to genotoxic metabolites, which then cause direct damage to DNA [55]. This supports our observed finding that ROS was associated with aggressive breast cancer, as it was associated with higher stage and pathological grade.
We also observed that ROS was associated with higher mutation load in tumors. It is known that ROS can cause unpaired or misrepaired DNA damage in cellular division leading to mutations. ROS induce mutations that commonly cause G to T transversions [56], which are the most frequent mutations in the human tumors in the p53 suppressor gene [57]. The presence of oxidative DNA damage in cancer supports the fact that initiation of cancer may at least partly be triggered by ROS [58]. In our study, tumors with high ROS were shown to be enriched for cell proliferation-related gene sets. This is consistent with previous reports that ROS in high amounts is associated with uncontrolled cell proliferation [4, 5].
Our study showed that all the immune cells were higher in ROS high breast cancers. Proper T cell activation, proliferation and differentiation depends on low levels of ROS [59, 60]. Tregs are able to persist in the oxidant environment, and therefore, a greater number of Tregs have been detected at tumor sites. Tregs have high antioxidant capacity, and are therefore, less sensitive to oxidant stress-induced cell death [61]. In breast cancer models, ROS were essential for tumor associated macrophages (TAMs) to invade the TIME and acquire a M2 pro-tumorigenic phenotype [62]. ROS also results in impairment of antigen presentation by dendritic cells [63–65]. However, we observed higher M1 macrophages as well. This could be explained by a general inflammatory state induced by the ROS in the TIME [66]. Our result that cytolytic activity was elevated in ROS high tumor agrees with a recent report that suggested that ROS contribute to immunogenic cell death and T-cell mediated immunity [67], and it was shown that immune cells infiltrated in breast cancer with high ROS, not because of the characteristics of TNBC which have high ROS, as it has been shown that even ER-positive/HER2-negative breast cancer with high ROS attract high level of immune cells. Furthermore, we found that not only tumor cells but also myeloid cells have high ROS score in breast cancer. Since there are several types of myeloid cells with different functions, it is of interest to investigate which type of myeloid cells have high ROS in breast cancer in the future. It is well known that breast cancer with high infiltration of immune cells have better prognoses, especially in TNBC [68]. On the other hand, the type of infiltrating immune cells is the key in relationship to patient prognoses [69]. We have previously reported that breast cancers with high CD8+ T cell infiltrations have better prognoses [51]. On the other hand, breast cancer with high Treg infiltration was not associated with patient survival, although they showed significant association with NAC response [42]. In the case of high ROS breast cancer, we found that overall pro-cancerous immune cells appear to outperform the anti-cancer immune cell infiltration. The same trend was seen in immune response. IFN-γresponse and Inflammatory response are known to be associated with better outcome, particularly in TNBC [24], whereas IL-6 is known to negatively affect anti-immune response [70] and we observed that both are elevated in high ROS tumor. Based upon our observation in high mutation load breast cancer, where aggressive biology and high immune response are both counterbalanced [14], we cannot help but speculate that IL-6 is outperforming the favorable effects of IFN-γresponse and Inflammatory response in high ROS tumor.
Unlike the ER-positive subtype, a high ROS score did not correlate with worse survival in either TNBC or HER2-positive subtype. Given that ROS score was significantly high in TNBC as well as HER2-positive breast cancer, and the fact that both subtypes are biologically aggressive and initially respond to chemotherapy better than ER-positive subtype, we speculate that the ROS signaling alone was not strong enough to predict clinical outcomes of both tumor subtypes. We speculate that ROS score may have the potential to be used as a predictive biomarker, and is a useful tool to investigate new drug treatment for breast cancer.
There were several limitations in our study. The degree of ROS signaling was determined by GSVA algorithm with hallmark collection in MSigDB, that may or may not include all the genes related to ROS pathway. Since this is a retrospective study that utilized multiple publicly available large patient cohorts, we did not have access to some of the pertinent clinical data including the details on systemic treatments received and it is assumed that all the patients received “Standard of Care”. Our results are based on tumor transcriptomics alone and we are incapable of generating any data on mechanistic role of ROS activity. The novelty of our study is to assess the role of ROS in the TIME using a score combining multiple genes. A prospective trial is needed to confirm the utility of ROS score as a predictive biomarker in breast cancer.
Conclusions
In conclusion, ROS abundance is associated with cancer aggressiveness, enhanced immune response, and with worse survival in ER-positive/HER2-negative breast cancer.
Supplementary Material
Funding:
This research was supported by National Institutes of Health, USA grant number R37CA248018, R01CA250412, R01CA251545, R01EB029596, as well as US Department of Defense BCRP grant number W81XWH-19-1-0674 and W81XWH-19-1-0111 to K.T. National Cancer Institute, cancer center support grant P30CA016056 supports Roswell Park Comprehensive Cancer Center. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award numbers KL2TR001413 and UL1TR001412. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- AJCC
American Joint Committee on Cancer
- DFS
disease-free survival
- DSS
disease-specific survival
- ER
estrogen receptor
- FDR
false discovery rate
- GSEA
Gene set enrichment analysis
- GSVA
Gene set variation analysis
- HER2
human epidermal growth factor receptor 2
- METABRIC
Molecular Taxonomy of Breast Cancer International Consortium
- NES
normalized enrichment score
- OS
overall survival
- TNBC
triple negative breast cancer
Footnotes
Competing Interests: The authors have no relevant financial or non-financial interests to disclose.
Statements & Declarations
Ethics approval: Institutional review board (IRB) approval at Roswell Park Comprehensive Cancer Center (Buffalo, New York, United States of America) was waived as publicly available deidentified databases were used.
Consent to participate: Not applicable.
Consent for publication: Not applicable.
Data availability:
The datasets generated during and analyzed during the current study are available from the original source as they are publicly available deidentified databases.
References
- [1].Zhou Z, Ni K, Deng H, Chen X, Dancing with reactive oxygen species generation and elimination in nanotheranostics for disease treatment, Adv Drug Deliv Rev, 158 (2020) 73–90. [DOI] [PubMed] [Google Scholar]
- [2].Schieber M, Chandel NS, ROS function in redox signaling and oxidative stress, Curr Biol, 24 (2014) R453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Circu ML, Aw TY, Reactive oxygen species, cellular redox systems, and apoptosis, Free Radic Biol Med, 48 (2010) 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ray PD, Huang BW, Tsuji Y, Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling, Cell Signal, 24 (2012) 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jenkins T, Gouge J, Nrf2 in Cancer, Detoxifying Enzymes and Cell Death Programs, Antioxidants (Basel), 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kimmelman AC, The dynamic nature of autophagy in cancer, Genes Dev, 25 (2011) 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD, The role of nuclear lamin B1 in cell proliferation and senescence, Genes Dev, 25 (2011) 2579–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martinon F, Signaling by ROS drives inflammasome activation, Eur J Immunol, 40 (2010) 616–619. [DOI] [PubMed] [Google Scholar]
- [9].Tschopp J, Schroder K, NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production?, Nat Rev Immunol, 10 (2010) 210–215. [DOI] [PubMed] [Google Scholar]
- [10].Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation, Science (New York, N.Y.), 324 (2009) 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Malla R, Surepalli N, Farran B, Malhotra SV, Nagaraju GP, Reactive oxygen species (ROS): Critical roles in breast tumor microenvironment, Crit Rev Oncol Hematol, 160 (2021) 103285. [DOI] [PubMed] [Google Scholar]
- [12].Kalyanaraman B, Cheng G, Hardy M, Ouari O, Bennett B, Zielonka J, Teaching the basics of reactive oxygen species and their relevance to cancer biology: Mitochondrial reactive oxygen species detection, redox signaling, and targeted therapies, Redox Biol, 15 (2018) 347–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sajadimajd S, Yazdanparast R, Differential behaviors of trastuzumab-sensitive and -resistant SKBR3 cells treated with menadione reveal the involvement of Notch1/Akt/FOXO1 signaling elements, Mol Cell Biochem, 408 (2015) 89–102. [DOI] [PubMed] [Google Scholar]
- [14].Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K, Biologically Aggressive Phenotype and Anti-cancer Immunity Counterbalance in Breast Cancer with High Mutation Rate, Scientific reports, 10 (2020) 1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection, Cell systems, 1 (2015) 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K, High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection, Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oshi M, Patel A, Le L, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K, G2M checkpoint pathway alone is associated with drug response and survival among cell proliferation-related pathways in pancreatic cancer, American journal of cancer research, 11 (2021) 3070–3084. [PMC free article] [PubMed] [Google Scholar]
- [18].Oshi M, Satyananda V, Angarita FA, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K, Angiogenesis is associated with an attenuated tumor microenvironment, aggressive biology, and worse survival in gastric cancer patients, American journal of cancer research, 11 (2021) 1659–1671. [PMC free article] [PubMed] [Google Scholar]
- [19].Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K, Enhanced DNA Repair Pathway is Associated with Cell Proliferation and Worse Survival in Hepatocellular Carcinoma (HCC), Cancers, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Patel A, Oshi M, Yan L, Matsuyama R, Endo I, Takabe K, The Unfolded Protein Response Is Associated with Cancer Proliferation and Worse Survival in Hepatocellular Carcinoma, Cancers, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K, Degree of Early Estrogen Response Predict Survival after Endocrine Therapy in Primary and Metastatic ER-Positive Breast Cancer, Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K, KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival, American journal of cancer research, 10 (2020) 897–907. [PMC free article] [PubMed] [Google Scholar]
- [23].Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K, Intra-Tumoral Angiogenesis Is Associated with Inflammation, Immune Reaction and Metastatic Recurrence in Breast Cancer, International journal of molecular sciences, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K, Inflammation Is Associated with Worse Outcome in the Whole Cohort but with Better Outcome in Triple-Negative Subtype of Breast Cancer Patients, J Immunol Res, 2020 (2020) 5618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gandhi S, Oshi M, Murthy V, Repasky EA, Takabe K, Enhanced Thermogenesis in Triple-Negative Breast Cancer Is Associated with Pro-Tumor Immune Microenvironment, Cancers, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Oshi M, Tokumaru Y, Angarita FA, Lee L, Yan L, Matsuyama R, Endo I, Takabe K, Adipogenesis in triple-negative breast cancer is associated with unfavorable tumor immune microenvironment and with worse survival, Scientific reports, 11 (2021) 12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Murthy V, Oshi M, Tokumaru Y, Endo I, Takabe K, Increased apoptosis is associated with robust immune cell infiltration and cytolytic activity in breast cancer, American journal of cancer research, 11 (2021) 3674–3687. [PMC free article] [PubMed] [Google Scholar]
- [28].Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K, G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer, International journal of molecular sciences, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K, The E2F Pathway Score as a Predictive Biomarker of Response to Neoadjuvant Therapy in ER+/HER2− Breast Cancer, Cells, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rueda OM, Sammut SJ, Seoane JA, Chin SF, Caswell-Jin JL, Callari M, Batra R, Pereira B, Bruna A, Ali HR, Provenzano E, Liu B, Parisien M, Gillett C, McKinney S, Green AR, Murphy L, Purushotham A, Ellis IO, Pharoah PD, Rueda C, Aparicio S, Caldas C, Curtis C, Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups, Nature, 567 (2019) 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups, Nature, 486 (2012) 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brueffer C, Gladchuk S, Winter C, Vallon-Christersson J, Hegardt C, Häkkinen J, George AM, Chen Y, Ehinger A, Larsson C, Loman N, Malmberg M, Rydén L, Borg Å, Saal LH, The mutational landscape of the SCAN-B real-world primary breast cancer transcriptome, EMBO molecular medicine, 12 (2020) e12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H, An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics, Cell, 173 (2018) 400–416.e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH, Kan Z, Han W, Park WY, Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer, Nature communications, 8 (2017) 15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hänzelmann S, Castelo R, Guinney J, GSVA: gene set variation analysis for microarray and RNA-seq data, BMC bioinformatics, 14 (2013) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K, Cytolytic Activity Score to Assess Anticancer Immunity in Colorectal Cancer, Annals of surgical oncology, 25 (2018) 2323–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takeshita T, Torigoe T, Yan L, Huang JL, Yamashita H, Takabe K, The Impact of Immunofunctional Phenotyping on the Malfunction of the Cancer Immunity Cycle in Breast Cancer, Cancers, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Oshi M, Kawaguchi T, Yan L, Peng X, Qi Q, Tian W, Schulze A, McDonald KA, Narayanan S, Young J, Liu S, Morris LG, Chan TA, Kalinski P, Matsuyama R, Otsuji E, Endo I, Takabe K, Immune cytolytic activity is associated with reduced intra-tumoral genetic heterogeneity and with better clinical outcomes in triple negative breast cancer, American journal of cancer research, 11 (2021) 3628–3644. [PMC free article] [PubMed] [Google Scholar]
- [39].Oshi M, Gandhi S, Huyser MR, Tokumaru Y, Yan L, Yamada A, Matsuyama R, Endo I, Takabe K, MELK expression in breast cancer is associated with infiltration of immune cell and pathological compete response (pCR) after neoadjuvant chemotherapy, American journal of cancer research, 11 (2021) 4421–4437. [PMC free article] [PubMed] [Google Scholar]
- [40].Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N, Molecular and genetic properties of tumors associated with local immune cytolytic activity, Cell, 160 (2015) 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Aran D, Hu Z, Butte AJ, xCell: digitally portraying the tissue cellular heterogeneity landscape, Genome biology, 18 (2017) 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K, Abundance of Regulatory T Cell (Treg) as a Predictive Biomarker for Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer, Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Oshi M, Newman S, Murthy V, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K, ITPKC as a Prognostic and Predictive Biomarker of Neoadjuvant Chemotherapy for Triple Negative Breast Cancer, Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Oshi M, Tokumaru Y, Patel A, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K, A Novel Four-Gene Score to Predict Pathologically Complete (R0) Resection and Survival in Pancreatic Cancer, Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I, The Immune Landscape of Cancer, Immunity, 51 (2019) 411–412. [DOI] [PubMed] [Google Scholar]
- [46].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proceedings of the National Academy of Sciences of the United States of America, 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Satyananda V, Oshi M, Tokumaru Y, Maiti A, Hait N, Matsuyama R, Endo I, Takabe K, Sphingosine 1-phosphate (S1P) produced by sphingosine kinase 1 (SphK1) and exported via ABCC1 is related to hepatocellular carcinoma (HCC) progression, American journal of cancer research, 11 (2021) 4394–4407. [PMC free article] [PubMed] [Google Scholar]
- [48].Katsuta E, Huyser M, Yan L, Takabe K, A prognostic score based on long-term survivor unique transcriptomic signatures predicts patient survival in pancreatic ductal adenocarcinoma, American journal of cancer research, 11 (2021) 4294–4307. [PMC free article] [PubMed] [Google Scholar]
- [49].Oshi M, Gandhi S, Angarita FA, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K, A novel five-gene score to predict complete pathological response to neoadjuvant chemotherapy in ER-positive/HER2-negative breast cancer, American journal of cancer research, 11 (2021) 3611–3627. [PMC free article] [PubMed] [Google Scholar]
- [50].Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K, Plasmacytoid Dendritic Cell (pDC) Infiltration Correlate with Tumor Infiltrating Lymphocytes, Cancer Immunity, and Better Survival in Triple Negative Breast Cancer (TNBC) More Strongly than Conventional Dendritic Cell (cDC), Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K, CD8 T Cell Score as a Prognostic Biomarker for Triple Negative Breast Cancer, International journal of molecular sciences, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M, Free radicals, metals and antioxidants in oxidative stress-induced cancer, Chem Biol Interact, 160 (2006) 1–40. [DOI] [PubMed] [Google Scholar]
- [53].Harman D, Free radical theory of aging: an update: increasing the functional life span, Ann N Y Acad Sci, 1067 (2006) 10–21. [DOI] [PubMed] [Google Scholar]
- [54].Preston DL, Mattsson A, Holmberg E, Shore R, Hildreth NG, Boice JD Jr., Radiation effects on breast cancer risk: a pooled analysis of eight cohorts, Radiat Res, 158 (2002) 220–235. [DOI] [PubMed] [Google Scholar]
- [55].Yue W, Santen RJ, Wang JP, Li Y, Verderame MF, Bocchinfuso WP, Korach KS, Devanesan P, Todorovic R, Rogan EG, Cavalieri EL, Genotoxic metabolites of estradiol in breast: potential mechanism of estradiol induced carcinogenesis, J Steroid Biochem Mol Biol, 86 (2003) 477–486. [DOI] [PubMed] [Google Scholar]
- [56].Higinbotham KG, Rice JM, Diwan BA, Kasprzak KS, Reed CD, Perantoni AO, GGT to GTT transversions in codon 12 of the K-ras oncogene in rat renal sarcomas induced with nickel subsulfide or nickel subsulfide/iron are consistent with oxidative damage to DNA, Cancer research, 52 (1992) 4747–4751. [PubMed] [Google Scholar]
- [57].Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Pontén J, A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma, Proceedings of the National Academy of Sciences of the United States of America, 88 (1991) 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Poulsen HE, Prieme H, Loft S, Role of oxidative DNA damage in cancer initiation and promotion, Eur J Cancer Prev, 7 (1998) 9–16. [PubMed] [Google Scholar]
- [59].Weinberg SE, Sena LA, Chandel NS, Mitochondria in the regulation of innate and adaptive immunity, Immunity, 42 (2015) 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kirtonia A, Sethi G, Garg M, The multifaceted role of reactive oxygen species in tumorigenesis, Cell Mol Life Sci, 77 (2020) 4459–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mougiakakos D, Johansson CC, Kiessling R, Naturally occurring regulatory T cells show reduced sensitivity toward oxidative stress-induced cell death, Blood, 113 (2009) 3542–3545. [DOI] [PubMed] [Google Scholar]
- [62].Zhang Y, Choksi S, Chen K, Pobezinskaya Y, Linnoila I, Liu ZG, ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages, Cell Res, 23 (2013) 898–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Giovanelli P, Sandoval TA, Cubillos-Ruiz JR, Dendritic Cell Metabolism and Function in Tumors, Trends Immunol, 40 (2019) 699–718. [DOI] [PubMed] [Google Scholar]
- [64].Chougnet CA, Thacker RI, Shehata HM, Hennies CM, Lehn MA, Lages CS, Janssen EM, Loss of Phagocytic and Antigen Cross-Presenting Capacity in Aging Dendritic Cells Is Associated with Mitochondrial Dysfunction, J Immunol, 195 (2015) 2624–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Veglia F, Tyurin VA, Mohammadyani D, Blasi M, Duperret EK, Donthireddy L, Hashimoto A, Kapralov A, Amoscato A, Angelini R, Patel S, Alicea-Torres K, Weiner D, Murphy ME, Klein-Seetharaman J, Celis E, Kagan VE, Gabrilovich DI, Lipid bodies containing oxidatively truncated lipids block antigen cross-presentation by dendritic cells in cancer, Nature communications, 8 (2017) 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kotsafti A, Scarpa M, Castagliuolo I, Scarpa M, Reactive Oxygen Species and Antitumor Immunity-From Surveillance to Evasion, Cancers, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Guo S, Burcus NI, Scott M, Jing Y, Semenov I, The role of reactive oxygen species in the immunity induced by nano-pulse stimulation, Scientific reports, 11 (2021) 23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, Piccart MJ, Loibl S, Denkert C, Smyth MJ, Joensuu H, Sotiriou C, Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial, Ann Oncol, 25 (2014) 1544–1550. [DOI] [PubMed] [Google Scholar]
- [69].Wang Q, Xiang Q, Yu L, Hu T, Chen Y, Wang J, Nie X, Cheng J, Changes in Tumor-Infiltrating Lymphocytes and Vascular Normalization in Breast Cancer Patients After Neoadjuvant Chemotherapy and Their Correlations With DFS, Front Oncol, 9 (2019) 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Nagahashi M, Hait NC, Maceyka M, Avni D, Takabe K, Milstien S, Spiegel S, Sphingosine-1-phosphate in chronic intestinal inflammation and cancer, Adv Biol Regul, 54 (2014) 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the original source as they are publicly available deidentified databases.


