Abstract
The Saccharomyces cerevisiae mitotic exit network (MEN) is a conserved set of genes that mediate the transition from mitosis to G1 by regulating mitotic cyclin degradation and the inactivation of cyclin-dependent kinase (CDK). Here, we demonstrate that, in addition to mitotic exit, S. cerevisiae MEN gene MOB1 is required for cytokinesis and cell separation. The cytokinesis defect was evident in mob1 mutants under conditions in which there was no mitotic-exit defect. Observation of live cells showed that yeast myosin II, Myo1p, was present in the contractile ring at the bud neck but that the ring failed to contract and disassemble. The cytokinesis defect persisted for several mitotic cycles, resulting in chains of cells with correctly segregated nuclei but with uncontracted actomyosin rings. The cytokinesis proteins Cdc3p (a septin), actin, and Iqg1p/ Cyk1p (an IQGAP-like protein) appeared to correctly localize in mob1 mutants, suggesting that MOB1 functions subsequent to actomyosin ring assembly. We also examined the subcellular distribution of Mob1p during the cell cycle and found that Mob1p first localized to the spindle pole bodies during mid-anaphase and then localized to a ring at the bud neck just before and during cytokinesis. Localization of Mob1p to the bud neck required CDC3, MEN genes CDC5, CDC14, CDC15, and DBF2, and spindle pole body gene NUD1 but was independent of MYO1. The localization of Mob1p to both spindle poles was abolished in cdc15 and nud1 mutants and was perturbed in cdc5 and cdc14 mutants. These results suggest that the MEN functions during the mitosis-to-G1 transition to control cyclin-CDK inactivation and cytokinesis.
During the transition from mitosis to G1, cytokinesis, disassembly of the mitotic spindle, chromatin decondensation, and DNA licensing must be precisely coordinated to ensure the genomic stability and viability of the cellular progeny (22, 29, 31, 53, 67). A major signal that controls these events is the degradation of mitotic cyclins and the inactivation of cyclin-dependent kinase (CDK) in late mitosis (52, 68). In Saccharomyces cerevisiae, mitotic cyclin degradation and CDK inactivation are regulated by a group of genes that constitute the mitotic exit network (MEN) (45, 47). MEN genes encode four protein kinases (Cdc5p, Cdc15p, Dbf2p, and Dbf20p), Cdc14p phosphatase, a GTP binding protein (Tem1p), a GTP exchange factor (Lte1p), and Mob1p, which binds Dbf2p and Dbf20p (35, 36, 44, 57, 58, 73, 75). At the restrictive temperature, conditional alleles of the MEN genes cause cells to arrest in late mitosis with high levels of mitotic cyclin (33, 48, 58, 66, 69). The mitotic arrest of several MEN mutants can be suppressed by overexpression of CDK inhibitor SIC1 (18, 33), indicating that CDK inactivation is the major function of the MEN pathway. Indeed, a pivotal step in cyclin and CDK inactivation is mediated by the Cdc14p phosphatase, which is sequestered in the nucleolus during most of the cell cycle until it is released at the end of mitosis (3, 60, 72). Release of Cdc14p from the nucleolus requires other MEN genes (60, 72) and apparently allows access of Cdc14p to certain substrates, including Hct1p/Cdh1p (anaphase-promoting complex/cyclosome activator protein) and Sic1p, thereby facilitating CDK inactivation (32, 71).
Despite the requirement for the S. cerevisiae MEN for cyclin-CDK inactivation, defects in the homologous pathway in Schizosaccharomyces pombe, called the septation initiation network (SIN), do not result in mitotic arrest but instead cause cytokinesis and septation defects (see references 6, 25, 40, 45, and 55 for reviews). Moreover, overexpression of some SIN genes induces the synthesis of multiple septa (4, 21, 50, 56), suggesting that the SIN genes are positive regulators of cytokinesis and septum formation (6, 25, 40, 45, 55). Given the high degree of conservation between the SIN and the MEN, it is plausible that the MEN genes regulate S. cerevisiae cell separation (defined here as the sum of all the processes necessary for separating daughter cells from their mothers) in addition to regulating mitotic exit. In support of this idea, certain mutations in the S. cerevisiae CDC5 and CDC15 genes give rise to morphological phenotypes that appear consistent with a role in cell separation (2, 34, 62). However, it is not yet known whether the putative cell separation defects caused by those mutations arise as a consequence of errors in cytokinesis, septation, or another process. Nor is it known whether the putative cell separation function of CDC5 and CDC15 is genetically separable from the mitotic-exit function or if cell separation requires each of the MEN genes.
MOB1 is a MEN gene, based on the late mitotic arrest phenotype exhibited by conditional mob1 mutants and based on the genetic and biochemical interactions with other MEN genes and their products, including CDC5, CDC15, LTE1, DBF2, and DBF20 (37, 44). However, several characteristics distinguish MOB1 from other MEN genes and suggest that MOB1 has additional functions. We previously observed that many conditional mob1 mutants undergo a quantal increase in ploidy at the permissive temperature, i.e., haploid cells become diploid (44). This phenotype is characteristic of mutants that are defective in duplication of the spindle pole body (SPB; the yeast centrosome equivalent) (13) and has not been observed in other MEN mutants. In addition, Mob1p, which contains no known structural motifs, binds to and can serve as a substrate for the Mps1p protein kinase, an essential component of the SPB duplication pathway (39, 44, 76). These data have led us to propose that Mob1p has at least two functions, one for mitotic exit and another for SPB duplication (44).
We investigated whether S. cerevisiae MOB1 is required for cell cycle processes other than SPB duplication and mitotic exit, and here we present genetic and cytological evidence that MOB1 is required for cytokinesis. We identified genetic conditions that cause a cell separation defect in conditional mob1 mutants in the absence of mitotic arrest and established that this defect arises, at least in part, as a consequence of the failure of the actomyosin ring to contract. We also examined the subcellular distribution of Mob1p during the cell cycle and found that Mob1p first localizes to the SPBs and then localizes to the bud neck, consistent with multiple roles in cell division. These data support the idea that MOB1 and the MEN genes function to coordinate the execution of multiple events associated with the M-to-G1 transition.
MATERIALS AND METHODS
Strains and plasmids.
Standard yeast culture conditions, genetic procedures, and transformations were performed as described previously (28). Where indicated (see Results and the legends to Fig. 5 and 7), MATa cells were synchronized in G1 with mating pheromone (alpha factor) as described previously (74). Yeast strains and sources are listed in Table 1. Those derived from our laboratory are in the S288C strain background.
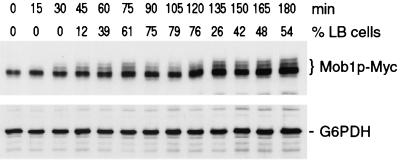
FIG. 5.
Immunoblot of 13xMyc-Mob1p from synchronized cells. Cells expressing 13xMyc-Mob1p (FLY595) were synchronized in G1 and released to fresh medium. Samples were taken at 15-min intervals and immunoblotted as described in Materials and Methods. (Top) Blot probed with anti-Myc antibody; (bottom) parallel immunoblot probed with antibody to glucose-6-phosphate dehydrogenase (G6PDH), as a protein loading control. The times (minutes) after release from G1 block and the percentages of large-budded (LB) cells (G2 and M phase cells; n = 100) are designated. At 15 and 30 min, the percentages of unbudded cells (G1 phase) were 100 and 42%, respectively.
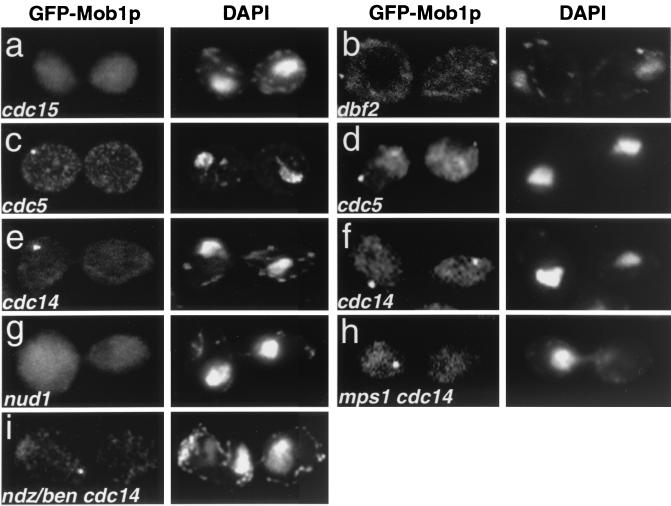
FIG. 7.
GFP-Mob1p localization in MEN mutants. Cells were synchronized in G1 with mating pheromone at 25°C and released to 37°C for 3 to 4 h. They were then fixed and treated with DAPI. (a) cdc15-2 (FLY353); (b) dbf2-1 (FLY022); (c and d) cdc5-1 (FLY343); (e and f) cdc14-1 (FLY346); (g) nud1-44 (FLY539); (h) mps1-1 cdc14-1 double mutant (FLY035); (i) cdc14-1 arrested cells treated with 15 μg of nocodazole/ml and 30 μg of benomyl/ml (ndz/ben). Cells were shifted to the restrictive temperature for 3 h prior to the addition of the ndz/ben. The image in panel i was captured 1 h after the ndz/ben treatment. The absence of microtubules in ndz/ben-treated cells was confirmed by tubulin immunofluorescence (data not shown). Note that the loss of microtubules in the ndz/ben-treated cdc14-1 mutants caused the chromatin to collapse toward the middle of the cell. The patchy cytoplasmic fluorescence of GFP-Mob1p (b and c) was not observed in living cells and thus is an artifact of fixation (data not shown).
TABLE 1.
Yeast strains
| Name | Genotype | Source or reference |
|---|---|---|
| L119-7d | MATα dbf2-1 trp1-1,2 ade1 ura3 | C. Denis |
| nud1-44 | MATanud1Δ::TRP1 nud1-44::HIS3 his3 leu2 trp1Δ1 ura3 ade2 | 1 |
| SSC166 | MATa iqg1-1 his3Δ200 leu2-3,112 trp1-1 ura3-52 ade2-1 can1-100 | 10 |
| WX257-5c | MATahis3Δ200 leu2-3,112 trp1Δ1 ura3-52 | M. Winey |
| WX257-8b | MATahis3Δ200 leu2-3,112 trp1Δ1 ura3-52 | M. Winey |
| WX255-2b | MATacdc3-1 mps1-1 his3Δ200 ura3-52 | M. Winey |
| YEF1681 | MATaMYO1-GFP::KANMX his3 leu2 trp1 ura3 lys2 | E. Bi |
| YEF2056-10a | MATamyo1Δ::HIS3 his3 leu2 trp1 ura3 lys2 [YCp50-MYO1] | E. Bi |
| FLY022 | MATadbf2-1 mob1Δ::HIS3::GFP-MOB1::URA3 leu2-3,112 trp1 | This study |
| FLY30 | MATamob1-77 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | 44 |
| FLY32 | MATamob1-95 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | 44 |
| FLY035 | MATamps1-1 cdc14-1 mob1Δ::HIS3::GFP-MOB1::URA3 leu2-3,112 trp1Δ1 | This study |
| FLY62 | MATamob1Δ::HIS3::mob1-83::LEU2 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 [pRS316-MOB1] | 44 |
| FLY258-B | MATamob1Δ::HIS3 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 [pRS314-MOB1] | This study |
| FLY329 | MATamob1Δ::HIS3::GFP-MOB1::URA3 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY330 | MATα mob1Δ::HIS3::GFP-MOB1::URA3 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY331 | MATa/α mob1Δ::HIS3::GFP-MOB1::URA3/mob1Δ::HIS3::GFP-MOB1::URA3 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 ura3-52/ura3-52 | This study |
| FLY343 | MATacdc5-1 mob1Δ::HIS3::GFP-MOB1::URA3 leu2-3,112 ura3 | This study |
| FLY346 | MATacdc14-1 mob1Δ::HIS3::GFP-MOB1::URA3 leu2 ura3 | This study |
| FLY353 | MATacdc15-2 mob1Δ::HIS3::GFP-MOB1::URA3 leu2 trp1Δ1 ura3-52 | This study |
| FLY358 | MATacdc3-1 mob1Δ::HIS3::GFP-MOB1::URA3 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY388 | MATa mob1-77 MYO1-GFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY394 | MATamob1Δ::HIS3::mob1-83::LEU2 MYO1-GFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY510 | MATadbf2-1 dbf20Δ::KANMX mob1Δ::HIS3::GFP-MOB1::URA3 leu2-3,112 trp1 | This study |
| FLY539 | MATanud1Δ::TRP1 nud1-44::HIS3 mob1Δ::HIS3::GFP-MOB1::URA3 his3 leu2 trp1 ura3 ade2 | This study |
| FLY595 | MATaMOB1-13xMYC::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY612 | MATamob1Δ::HIS3::mob1-83::LEU2 IQG1-GFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY615 | MATamob1-77 IQG1-GFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY642 | MATα IQG1-GFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY643 | MATaMOB1-YFP::HIS3 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY694 | MATα SPC42-CFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | This study |
| FLY725 | MATamob1-77 MYO1-GFP::KANMX his3Δ200 leu2-3,112 trp1Δ1 ura3-52 [YEp13-SIC1] | This study |
| FLY727 | MATa/α MOB1/MOB1-YFP::HIS3 SPC42/SPC42-CFP::KANMX his3Δ200/his3Δ200 leu2-3,112/ leu2-3,112 trp1Δ1/trp1Δ1 ura3-52/ura3-52 | This study |
| FLY732 | MATamob1-77 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 [pRS316-GFP-CDC3] | This study |
| FLY743 | MATamyo1Δ::HIS3 mob1Δ::HIS3::GFP-MOB1::URA3 his3 leu2 trp1 ura3 | This study |
| FLY744 | MATaiqg1-1 mob1Δ::HIS3::GFP-MOB1::URA3 his3Δ200 leu2-3,112 trp1 ura3-52 | This study |
| FLY800-B | MATamob1Δ::HIS3::mob1-83::LEU2 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 [pRS316-GFP-CDC3] | This study |
Strains expressing green fluorescent protein (GFP)-tagged Mob1p were constructed as follows. An integrating vector encoding N-terminally tagged GFP-Mob1p, pRS306-GFP-MOB1, was constructed by ligating three DNA fragments into the NotI and EcoRI sites of pRS306 (61). These include (i) a NotI and XbaI fragment of the 5′ upstream activation sequence of MOB1 that was amplified from yeast genomic DNA using MOB1-X (AGGGAAAAAAGCGGCCGCAAACCCTTCTTCTACGCC) and MOB1-Y (GCTCTAGAGGAAATTGAAGTCCTTAT) primers, (ii) an XbaI and BamHI fragment encoding GFP (F64L, S65T) that was amplified from pYEGFP1 (15) with GFP-1 (GCTCTAGAATGTCTAAAGGTGAAGAA) and GFP-2 (CGGGATTTGTACAATTCATCCAT) primers, and (iii) the BamHI and EcoRI fragment from pGST-MOB1 (44) containing the entire MOB1 open reading frame. pRS306-GFP-MOB1 was linearized with PmlI and integrated into the mob1Δ::HIS3 locus of FLY258-B, which contained pRS314-MOB1, a MOB1- and TRP1-containing plasmid. Transformants were grown for several days in medium containing tryptophan, and colonies that had lost pRS314-MOB1 were selected. One such isolate was designated FLY329. Proper integration of the GFP-MOB1 plasmid was confirmed by PCR. No mutant phenotypes were observed as a consequence of GFP-Mob1p expression. FLY329 was backcrossed to WX257-8b to obtain FLY330, and FLY329 was mated to FLY330 to yield FLY331. All other GFP-Mob1p strains used in this study were obtained through genetic crosses of FLY329 or FLY330 to the various cell cycle mutants listed in Table 1 or in reference 44. The absence of MYO1 in FLY743 was confirmed by PCR.
Strains expressing C-terminally tagged 13xMyc-Mob1p (FLY595), yellow fluorescent protein (YFP)-Mob1p (FLY643), cyan fluorescent protein (CFP)-Spc42p (FLY694), or GFP-Iqg1p (FLY642) were constructed by the targeted integration of DNA cassettes into WX257-5c or WX257-8b, as described previously (43), using one of the following plasmids as the DNA template: pFA6a-13xMyc-kanMX6, pFA6a-GFP(S65T)-kanMX6, pDH5 (YFP), and pDH3 (CFP). The pDH5 and pDH3 plasmids were gifts from The Yeast Resource Center, University of Washington. The PCR primers used for these constructs are as follows: MOB1-Cterm-F (CCGGCTGATTTTGGTCCGCTGT TAGAAT TAGTGATGGAGT TGAGGGATAGGGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA), MOB1-Cterm-R (GTCCCATGCATGGAAGAATACA ACCTACA AGCAGAC T TATATAAATATACAATAGAAT TCGAGCTCGTTTAAAC), SPC42-Cterm-F (CCTGAAAATAATATGTCAGAAACAT TCGCAACTCCCAC TCCC A ATAATCGAGG T G G TCCCGGTGG TCGGATCCCCGGGTTAATTAA), SPC42-Cterm-R (GCCGTAATTACACAGAACGC T T TAAGAATGCGCCATACTCCT TAACTGCT T T T TAAATCAGAATTCGAGCTCGTTTAAAC), and IQG1-Cterm-F (TTACTACATTTGATTGTCAGT T T T T TCTATAAAAGGAACGCT T TGGGTGGTCCCGGTGGTCGGATCCCCGGGTTAATTAA), IQG1-Cterm-F (GGAAAATTTAGTAACAGCTTTTGCCCAATATGCTCAAAACCGAGTGAATTCGAGCTCGTTTAAAC). FLY643 was mated to FLY694 to yield FLY727. FLY642 was crossed to FLY30 and FLY62 to obtain mob1 strains expressing GFP-Iqg1p. No observable cell growth defects occurred as a consequence of expression of any of these fusions.
Strains expressing GFP-Myo1p were obtained through genetic crosses with YEF1681 (gift from Erfei Bi, University of Pennsylvania) or its derivatives (9). All GFP-Cdc3p-expressing strains contained plasmid pRS316-GFP-CDC3 (a gift from Erfei Bi) (70).
Suppression of mob1 mutants by Sic1p overproduction.
High-copy-number plasmids containing SIC1, MOB1, or WHI3 (YEp13-SIC1, YEp13-MOB1, and YEp13-WHI3, respectively) were isolated from a previously described yeast genomic DNA library (16). The plasmids were introduced into haploid mob1-77 (FLY30), mob1-95 (FLY32), and MOB1 (WX257-5c) strains, and the strains were then grown in leucine-deficient medium at 25°C. To induce cellular-chain formation, cells were grown to mid-log phase at 25°C and transferred to 34°C for 3 to 20 h. YEp13-SIC1 and YEp13-MOB1, but not YEp13-WHI3, suppressed the conditional lethality of mob1-77 and mob1-95 mutants.
Microscopy and image processing.
Where indicated (see the legends to Fig. 1, 2, and 7), cells were fixed in 3.7% formaldehyde for 1 h and stained with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml to visualize DNA or 2 U of Alexa594-phalloidin (Molecular Probes)/ml to visualize F-actin, as previously described (70). Prior to microscopic analysis, the fixed cells were briefly sonicated. Cells were observed using Leica DMR5 fluorescence microscopes illuminated with 75-W xenon or 100-W mercury arc lamps. Most images were captured as described previously (12), with some modifications. Briefly, digital images were captured using a SensiCam (Cooke Corp.) or a Roper Micromax 512BFT (Princeton Instruments) cooled charge-coupled device camera. The microscopes and cameras were controlled by SlideBook (Intelligent Imaging Innovations, Denver, Colo.) or OpenLab (Improvision, London, United Kingdom) imaging software. Typically, a series of 10 to 20 0.2-μm optical Z-sections of cells were processed for deconvolution using nearest-neighbor algorithms and then merged to a single plane. Immuno-electron microscopy (immunoEM) was performed as described previously (12, 49) using an affinity-purified rabbit polyclonal anti-GFP antibody (provided by Jason Kahana and Pam Silver) and a 5-nm gold-conjugated secondary antibody (Ted Pella, Inc.).
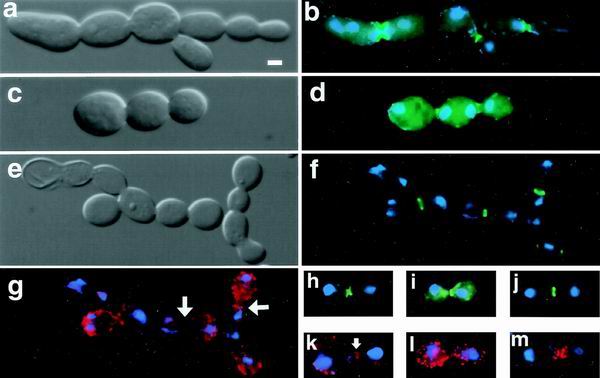
FIG. 1.
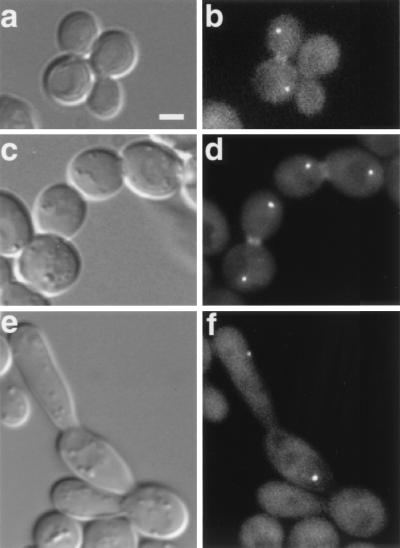
Cell separation defects are evident in mob1-83 cells. Differential interference contrast (DIC) (a, c, and e) and fluorescence microscopy (b, d, and f to i) of mob1-83 cells that were shifted to the restrictive temperature (34°C) for 3 to 4 h. Cells were fixed and stained with DAPI (blue) and/or Alexa594-phalloidin (red) to reveal nuclear DNA and F-actin, respectively. (a, b, and h) Cells expressing GFP-Cdc3p (FLY800-b); (c, d, and i) cells expressing GFP-Iqg1p (Cyk1p) (FLY612); (e, f, and j) cells expressing GFP-Myo1p (FLY394); (g, k, l, and m) F-actin (arrows, actin rings). (a to g) Typical examples of chains of mob1-83 cells; (h to l) representative images of mob1-83 cells that arrest in late mitosis; (m) example of a mob1-83 cell that was released from mitotic arrest; the image was captured 30 min after returning to the permissive temperature. Panels l and m are merges of 12 0.2-μm optical sections. The remaining panels show single optical sections. Scale bar = 2.5 μm.
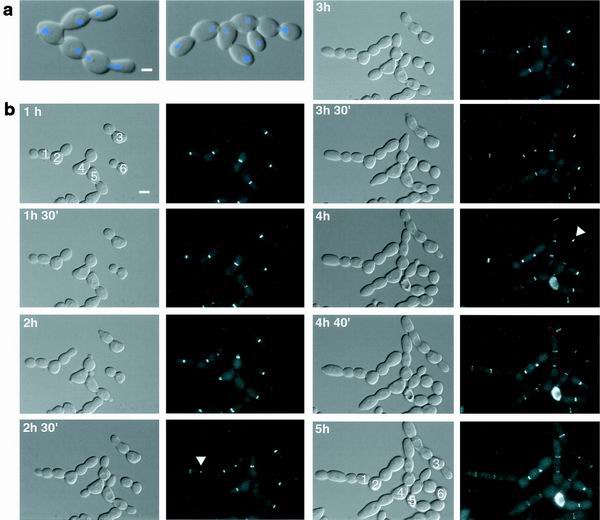
FIG. 2.
Suppression of mob1 mutations by SIC1 overexpression reveals cytokinesis defects. (a) Merged and DIC fluorescence images are shown for cellular chains of mob1-77 cells (FLY30) that expressed YEp13-SIC1. Scale bar = 2.5 μm. Cells were grown for 4 h at 34°C, fixed in formaldehyde, sonicated, and treated with DAPI. (b) Series of time-lapse images of mob1-77 cells expressing GFP-Myo1p and YEp13-SIC1 (FLY725). Cells were synchronized in G1 at 25°C and released to 34°C. The time relative to the shift to 34°C is denoted. Scale bar = 5 μm. Cells are numbered 1 to 6 as guides. Arrowheads, disappearance (cell 1) or maintenance (cell 3) of the myosin ring from the first bud cycle. Myosin rings from all subsequent bud cycles were maintained. Cell 6 arrested in late mitosis and thus probably did not contain YEp13-SIC1. The presented images are a subset of those taken every 10 min for 5 h. Each fluorescence image is a merge of 10 0.2-μm optical Z-sections.
Time-lapse microscopy of cells expressing GFP-Myo1p was performed as follows. Cells were synchronized in G1 at 25°C and released to synthetic complete medium at 34°C. After 1 h at 34°C, cells were placed on a thin sheet of 2% agarose (in synthetic medium) on a prewarmed microscope slide and sealed under a coverslip with nail polish. The microscope slide was maintained at 34°C on the microscope using an objective heater (Bioptics). Cells were illuminated by the 100-W mercury arc lamp through neutral-density filters, and 15 0.2-μm optical Z-sections were captured every 10 min using the 512BFT charge-coupled device camera and OpenLab imaging software. Captured images were processed as described above.
For time-lapse microscopy of GFP-Mob1p, homozygous diploid cells expressing GFP-Mob1p were used rather than haploid cells because diploid cells were larger and exhibited greater fluorescence. Cells from a mid-log-phase culture were placed in perfusion chambers that were fashioned by affixing 2% phenylethylene amine-coated coverslips to microscope slides with two thin strips of double-stick tape. Fresh medium was added periodically to prevent the cells from drying out. Cells were monitored, and eight 0.2- or 0.5-μm optical Z-sections were captured every 1 or 2 min, as described above. The time-lapse images presented in Fig. 4 were not processed for deconvolution.
FIG. 4.
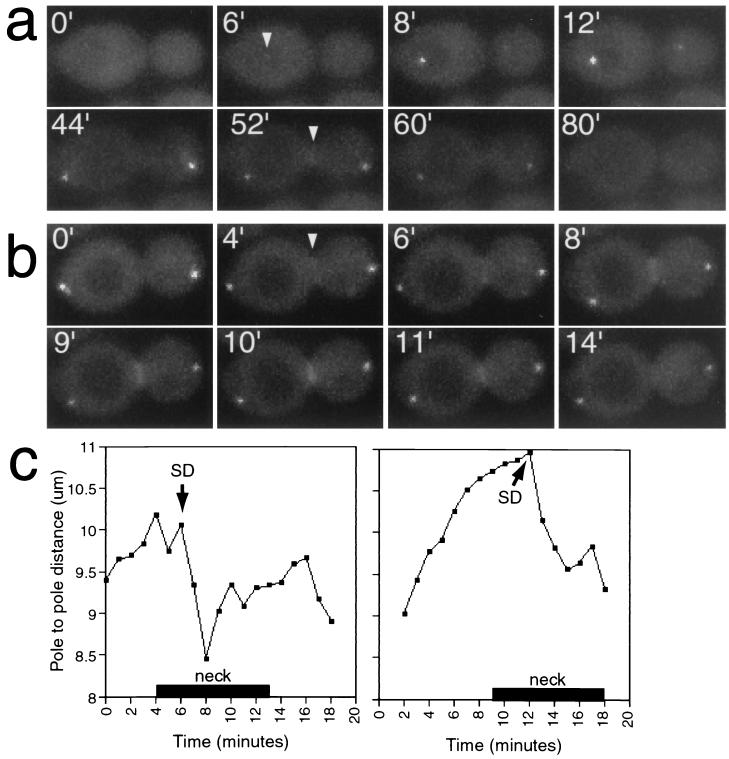
Localization of GFP-Mob1p during mitosis. (a) Time-lapse series of GFP-Mob1p in FLY331. Arrowheads, first detection of GFP-Mob1p at the SPB and the bud neck. (b) Time-lapse series of GFP-Mob1p in a late-anaphase cell. Arrowhead, first detection of GFP-Mob1p at the bud neck. The images are a subset of eight 0.5-μm optical sections captured every 2 min and merged to a single plane (a) or a subset of eight 0.2-μm optical sections captured every 1 min and merged to a single plane (b). (c) SPB-to-SPB distance versus time (see Materials and Methods) for the cell in panel b and another late-mitosis cell. Bars (“neck”) indicate the duration of detectable GFP-Mob1p at the bud neck. SD, time of spindle disassembly as inferred from the decrease in SPB-to-SPB distance.
Spindle lengths were obtained by measuring the three-dimensional distance between the GFP-Mob1p-labeled SPBs using OpenLab imaging software. The percentages of cellular chains of mob1-83 mutants were determined by counting 200 cells each from six different cultures that were grown at 25°C or shifted to 37°C for 3 to 20 h. Samples were fixed in 70% ethanol or 3.7% formaldehyde for 1 h and briefly sonicated prior to counting. Cells with three or more buds were counted as chains.
Immunoblot analysis.
Immunoblotting and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were performed as previously described (44). The blots were probed with either a mouse monoclonal anti-Myc antibody (9E10; Sigma) or rabbit polyclonal anti-glucose-6-phosphate dehydrogenase (Sigma) as the primary antibodies. Peroxidase-conjugated secondary antibodies were obtained from Pierce, and the blots were processed using the Renaissance ECL kit (NEN) in accordance with the manufacturer's protocols.
RESULTS
Mob1p performs separable mitotic-exit and cytokinesis functions.
We examined a collection of conditional mob1 mutants (44) to identify the full range of mutant defects. All alleles caused a late-mitosis arrest at the restrictive temperature as previously described for mob1-77, consistent with the essential role of MOB1 in mitotic exit. In addition, we found that some alleles, including mob1-83, caused cell separation defects at the permissive temperature. In six different mob1-83 cultures grown at 22°C, 18 to 25% of the cells persisted as unseparated chains of cells, even after brief sonication. The remaining cells were similar in morphology to wild-type cells. When shifted to 34°C for 2 to 4 h, mob1-83 cultures contained both cellular chains (Fig. 1a to g) and individual large-budded cells (Fig. 1h to m) with separated chromatin, consistent with a late mitotic arrest. The cell separation defect was allele specific, as similarly treated mob1-77 cultures did not contain cellular chains. The formation of cellular chains in mob1-83 mutants at the permissive temperature suggested that the cell separation defect is independent of mitotic arrest. Indeed, >80% of the segments within the chains contained a nucleus (Fig. 1b, d, and f), indicating that the nuclear-division and budding cycles remained coordinated and that the cell separation defect did not arise as a downstream consequence of a mitotic-exit defect.
If the mitotic-exit and cell separation defects reflect separate MOB1 functions, then it might be possible to suppress one phenotype and not the other. We identified this type of suppressor in a screen for dosage suppressors of the mob1-77 mutation. A high-copy-number plasmid containing SIC1 (YEp13-SIC1) allowed mob1-77 cells to grow at the restrictive temperature (34°C), but the cells exhibited a dramatic cell separation defect (Fig. 2). After 4 h at 34°C, greater than 50% of cells were in chains of four or more buds, and after 20 h at 34°C nearly 70% of the cells were in chains (Table 2). The mitotic defect of the mob1-95 mutant was also suppressed by the YEp13-SIC1 plasmid, resulting in a similar cell separation defect. The cellular-chain phenotype was not observed in mob1-77 cells that contained YEp13-WHI3, a randomly chosen plasmid from the yeast DNA library that was used as a negative control, nor was the phenotype observed in MOB1 cells containing the YEp13-SIC1 plasmid (Table 2). These data suggest that the mob1-77 and mob1-95 mutants are defective in both mitotic exit and cell separation at the restrictive temperature and that overexpression of SIC1 specifically suppresses the mitotic-exit defect. Thus, the mitotic-exit and cell separation functions of MOB1 are genetically separable.
TABLE 2.
Suppression of mitotic-exit defect of mob1-77 mutantsa
| Cell type | % LB/% chains at:
|
||
|---|---|---|---|
| 0 h | 4 h | 20 h | |
| WT + pWHI3 | 37/0 | 29/0 | 22/0 |
| WT + pSIC1 | 34/0 | 30/0 | 17/0 |
| mob1-77 + pMOB1 | 39/0 | 37/0 | 46/0 |
| mob1-77 + pWH13 | 38.5/0 | 86.5/2.5 | 92/0 |
| mob1-77 + pSIC1 | 40/0 | 47/51 | 26.5/69.5 |
Wild-type (WT; WX257-5c) and mob1-77 cells (FLY30) containing YEp13-WH13 (pWH13), YEp13-SIC1 (pSIC1), or YEp13-MOB1 (pMOB1) were grown in selective medium at 25°C to mid-log phase and shifted to 34°C in rich medium. Samples were taken 0, 4, and 20 h after the 34°C shift, sonicated, and observed by microscopy. The percentages of large-budded cells (% LB) and chains of cells (% chains) were determined by counting at least 200 cells.
Recruitment of contractile-ring proteins to the bud neck is independent of MOB1.
One possible function for MOB1 in cell separation is the recruitment of actomyosin ring proteins or other cell separation components to the bud neck. If so, the distribution of cytokinesis or septation proteins might be altered in mob1 mutants. To test this, we assayed the distribution of GFP-tagged Cdc3p (a septin protein), Myo1p (type II myosin), and Iqg1p/Cyk1p (IQGAP protein) in mob1-83 mutants at the restrictive temperature. We also assayed F-actin distribution by treating cells with Alexa594-phalloidin. In the mob1-83 cells that persisted as chains, we found that Cdc3p, Iqg1p, Myo1p, and F-actin localized to rings at approximately 50% of the bud neck regions (Fig. 1b, d, f, and g). We observed similar distributions for these proteins in mob1-83 chains at the permissive temperature (data not shown). For the majority of cells, which arrested in late mitosis as large-budded cells, we found that GFP-Cdc3p localized to a single band across the bud neck and that GFP-Myo1p and GFP-Iqg1p each localized to a single ring at the bud neck (Fig. 1h to j). These are normal distributions for these proteins during late mitosis (9, 16, 20, 29, 42). Close inspection revealed that approximately 50% of the arrested cells contained discernible actin rings at the bud neck (Fig. 1k) and that nearly all of the cells arrested with randomly dispersed cortical actin patches at the restrictive temperature (Fig. 1l). It is probable that the percentage of actin rings is an underestimate because the rings are frequently obscured by the cortical actin patches. Upon release from mitotic arrest, the cortical patches redistribute to the bud neck during the completion of mitosis (Fig. 1m). We observed similar subcellular distributions for these proteins in mob1-77 and mob1-95 cells when shifted to the restrictive temperature (data not shown). These data suggest that the recruitment and formation of septin and contractile rings are not inhibited in mob1-77, mob1-95, and mob1-83 mutants.
Cellular chains arise due to a defect in cytokinesis.
The cellular-chain phenotype of mob1 mutants could arise due to a defect in either septum formation or cytokinesis. To investigate which process was affected, we used GFP-Myo1p and time-lapse microscopy to observe ring contraction. The SIC1-suppressed mob1-77 strain was used because the cellular-chain phenotype was inducible and occurred in a high percentage of cells. At 22°C, all cells initiated and completed cytokinesis, as indicated by the contraction of the myosin ring and the eventual disappearance of GFP-Myo1p from the bud necks (data not shown) and as previously described for wild-type cells (9). To monitor the formation of cellular chains from single cells, we synchronized cells in G1 with mating pheromone and released them into fresh medium at 34°C. GFP-Myo1p localized to the bud sites prior to bud emergence and then remained localized to single rings at the base of growing buds, as previously described for wild-type cells (9). The buds grew with normal morphology during the first cell cycle, but the myosin ring failed to contract or disappear in 30% (n = 50) of the cells (Fig. 2b, cell 3). Subsequently, new buds emerged, often at both poles of the large-budded cells (Fig. 2b, cell 4 at 2 h). New bud growth was accompanied by the development of myosin rings. The myosin rings that formed after the first bud cycle almost never contracted or disappeared for the duration of the experiment, even as additional buds emerged from the tips of the previous buds (Fig. 2b, cells 1 to 5). Similar results were obtained using asynchronous cells (data not shown). In the absence of the YEp13-SIC1 plasmid, new buds never emerged from the arrested mob1-77 cells at the restrictive temperature. These data indicate that cytokinesis is impaired in the cellular chains of the Sic1p-overexpressing mob1-77 cells and suggest that the chains arise as a consequence of this impairment.
Mob1p localizes to SPBs during anaphase and to the bud neck during cytokinesis.
To determine the subcellular localization of Mob1p, we tagged Mob1p at the N terminus with GFP. The GFP-Mob1p-tagged strains contained a single copy of the gene fusion expressed from the MOB1 promoter in a mob1Δ background. The GFP-Mob1p fusion complemented the mob1 null mutation with no detectable defects in cell cycle progression. During late mitosis, GFP-Mob1p localized to two spots at the poles of the cell, reminiscent of spindle pole localization (Fig. 3a). GFP-Mob1p also localized to a single ring at the bud neck in late mitotic cells (Fig. 3a and 4). In G1, S, and G2 phases GFP-Mob1p localized diffusely to the cytoplasm (data not shown).
FIG. 3.
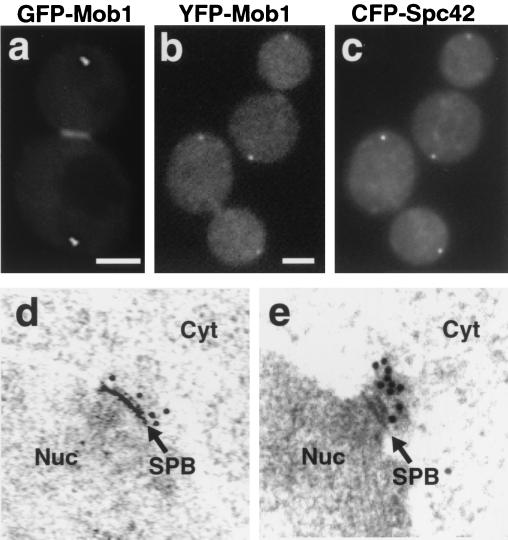
Mob1p localizes to SPBs and the bud neck during late mitosis. (a) GFP-Mob1p distribution in a living late-mitosis cell (FLY331). Scale bar = 2.5 μm. (b and c) Colocalization of YFP-Mob1p (b) and CFP-Spc42p (c) in two fixed late-mitosis cells (FLY727). Scale bar = 2.5 μm. There was no detectable bleed-through of the YFP or CFP fluorescence in the single-tagged control strains, FLY643 and FLY694 (data not shown). The patchy cytoplasmic fluorescence of YFP-Mob1p and CFP-Spc42p was not observed in living cells and thus is an artifact of fixation. (d and e) Cells expressing GFP-Mob1p were fixed and processed for immunoEM, as described in Materials and Methods. (d) Localization of GFP-Mob1p to the cytoplasmic side of the SPB in a late-mitosis cell (FLY331). The second SPB was also labeled (data not shown). (e) Localization of GFP-Mob1p to an SPB in a cell cycle-arrested cdc14-1 cell (FLY346). Nuc, nucleus; Cyt, cytoplasm; arrows, SPBs.
To ascertain whether Mob1p localizes to SPBs, we constructed a strain that expressed both YFP-tagged Mob1p and CFP-tagged Spc42p, an SPB protein (17). YFP-Mob1p colocalized with CFP-Spc42p (Fig. 3b and c), confirming that Mob1p localizes to SPBs. To determine the topology of Mob1p's SPB localization, we performed immunoEM on serial sections of asynchronous GFP-Mob1p-expressing cells. Sections were probed with an anti-GFP antibody followed by a gold-conjugated secondary antibody. Electron-microscopic analysis revealed that the immunogold decorated the cytoplasmic side of SPBs in late mitosis (Fig. 3d; n = 20 SPBs). We did not detect any immunogold labeling on the SPBs of cells from other cell cycle stages or in untagged cells (data not shown). We performed a similar analysis for cells that were synchronized in late mitosis, using cdc14-1 mutants. Immunogold was found decorating at least one SPB in all cdc14-1 cells expressing GFP-Mob1p that were arrested at the restrictive temperature (Fig. 3e; n = 10 cells). Moreover, the cdc14-1 cells usually displayed a greater amount of immunogold labeling than comparably treated wild-type cells (Fig. 3e).
We performed time-lapse microscopy to determine the relative timing of Mob1p localization to the SPBs and bud neck. Cells were photographed at five to eight focal planes for each time point, and the sections were later projected into a single composite image to ensure equal detection of both SPBs and the bud neck. Specific Mob1p localization was not detected until cells entered mitosis, when GFP-Mob1p was first detectable on the SPB in the mother cell (Fig. 4a). After a short delay, GFP-Mob1p was observed on the second SPB, which was located in the bud. The intensity of the fluorescence signals of GFP-Mob1p on the SPBs was initially asymmetric, but the fluorescence signal of the second SPB gradually increased to that of the first. The distance between spindle poles at the time of earliest detection of the bipolar localization for Mob1p (6.3 μm for the example shown in Fig. 4a) indicates that Mob1p is recruited to the SPBs during mid-anaphase. After the spindle reached its maximal length, the SPB-to-SPB distance rapidly decreased, signaling the disassembly of the mitotic spindle and the end of mitosis (65). GFP-Mob1p remained on both SPBs throughout mitotic exit, and the signal gradually diminished in intensity until it became undetectable during early G1 phase.
Subsequent to the SPB localization, Mob1p localized to the bud neck (Fig. 4b) in an arrangement similar to that observed for contractile-ring proteins, such as Iqg1p (20, 42). Mob1p was first detected on the bud neck 2 to 4 min prior to mitotic-spindle disassembly, yet the intensity of the Mob1p signal at that location peaked 3 to 4 min after spindle disassembly. The total duration of Mob1p localization at the neck was 6 to 10 min, and its disappearance was rapid (within 2 min). The timing of Mob1p localization at the neck relative to spindle length is shown (Fig. 4c). The pattern of Mob1p localization suggests that Mob1p first performs a function at the SPBs and subsequently performs a function at the bud neck during cytokinesis and cell separation.
Mob1p is present throughout the cell cycle.
It is possible that changes in MOB1 expression contribute to the dramatic cell cycle-dependent redistribution of Mob1p. Indeed, previous studies have suggested that MOB1 mRNA levels are cell cycle regulated, with peak expression occurring in mitosis (14, 37, 64). To determine whether the levels of Mob1p change significantly during cell cycle progression, we analyzed the expression of Myc-tagged Mob1p on immunoblots of synchronized cells (Fig. 5). The degree of synchrony and cell cycle stages were inferred from fluorescence-activated cell sorter analysis and by assaying the percentage of unbudded (G1), small-budded (S), and large-budded cells (G2 and M) (Fig. 5 and data not shown). We found that Mob1p-Myc was present at fairly constant levels throughout the cell cycle. In addition, slower electrophoretic variants of Mob1p were detectable in S, G2, and M phases. Similar electrophoretic variants of Mob1p were previously determined to be the result of phosphorylation (44). The Mob1p levels were also constant in cells arrested in G1, S, and M phases and in cells expressing other epitope-tagged versions of Mob1p, including GFP-Mob1p (data not shown). These data indicate that fluctuations in cellular Mob1p levels do not account for the transient nature of Mob1p localization.
The bud neck localization of Mob1p is dependent on septin but not cytokinesis proteins.
We asked if the localization of Mob1p is dependent on contractile-ring or septin proteins by monitoring GFP-Mob1p in live cells containing iqg1-1, myo1Δ, or cdc3-1 mutations. In iqg1-1 and myo1Δ mutants, GFP-Mob1p transiently localized to the SPBs and the bud neck, as in wild-type cells (Fig. 6a to d). In contrast, GFP-Mob1p could not be detected on the bud neck in cdc3-1 cells (n > 500 cells) that were grown at the restrictive temperature. Nevertheless, GFP-Mob1p localization was observed on one or both SPBs in cdc3-1 mutants at the restrictive temperature, suggesting the presence of anaphase spindles (Fig. 6e and f). At the permissive temperature, the subcellular distribution of GFP-Mob1p in cdc3-1 mutants was similar to that in wild-type cells (data not shown). From these results, we conclude that at least one septin protein is essential for the localization of Mob1p to the bud neck.
FIG. 6.
GFP-Mob1p localization in contractile ring and septin mutants. DIC (a, c, and e) and fluorescence microscopy (b, d, and f) of live cells expressing GFP-Mob1p are shown. (a and b) iqg1-1 (FLY744); (c and d) myo1Δ (FLY743); (e and f) cdc3-1 (FLY358). iqg1-1 and cdc3-1 mutants were shifted to the restrictive temperature (37°C) for 3 to 4 h prior to analysis. In panels b, d, and f, 10 of the 16 0.2-μm optical sections were merged into a single plane, as described in Materials and Methods. Scale bar = 2.5 μm.
Normal Mob1p localization requires MEN genes but not MPS1 or microtubules.
To determine whether Mob1p localization to the bud neck or SPB requires MEN function, we examined the GFP-Mob1p distribution in conditional MEN mutants. GFP-Mob1p was not detectable on bud necks in any living or fixed cdc5-1, cdc14-1, cdc15-2, or dbf2-1 cells when shifted to the restrictive temperature. However, when the cells were released from the restrictive temperature, GFP-Mob1p localized to both SPBs (if not already there) and to the bud neck prior to completion of mitosis (data not shown). In cdc15-2 cells that were arrested at the restrictive temperature, GFP-Mob1p remained localized to the cytoplasm (Fig. 7a). In contrast, in dbf2-1 mutants, Mob1p localized to both SPBs (Fig. 7b). The distribution of Mob1p in dbf2-1 dbf20Δ double mutants was indistinguishable from that in dbf2-1 mutants (data not shown). In cdc5-1 and cdc14-1 mutants that were arrested at the restrictive temperature, GFP-Mob1p localized more strongly to the SPB in the mother cell (Fig. 7c to f). However, 68% (n = 40) of cdc5-1 cells and 38% (n = 27) of cdc14-1 cells arrested with GFP-Mob1p at both spindle poles (Fig. 7d and f). In contrast to what was found for wild-type late mitotic cells, the SPBs in the buds often exhibited a weaker fluorescence than those in the mother cells. The percentages of cdc5-1 and cdc14-1 cells that displayed the bipolar distribution of GFP-Mob1p localization increased with time at the restrictive temperature, which suggested that these alleles are “leaky” at the restrictive temperature (data not shown).
Recent data suggest that Nud1p, an SPB protein, is a component of the MEN pathway, since at restrictive temperature nud1 mutants arrest with phenotypes similar to those of MEN mutants (1, 26). It is possible that Nud1p is required, at least indirectly, to tether or recruit Mob1p to SPBs. In support, in nud1-44 mutants at the restrictive temperature, GFP-Mob1p did not localize to SPBs or the bud neck but rather remained in the cytoplasm (Fig. 7g). These data indicate that proper Mob1p localization requires Nud1p and MEN proteins.
To elucidate a possible role for Mps1p, a Mob1p binding protein (44), in regulating the subcellular distribution of Mob1p, we localized GFP-Mob1p in an mps1-1 cdc14-1 double mutant at the restrictive temperature. The double mutant was used because mps1-1 single mutants do not undergo cell cycle arrest due to a defect in the spindle assembly checkpoint (74, 76). The cdc14-1 mutation causes cells to arrest in late mitosis at the restrictive temperature and thereby facilitates observation of GFP-Mob1p at the SPB. In these cells, GFP-Mob1p localized to the sole SPB (Fig. 7h).
To test whether microtubules are required for Mob1p localization, we added microtubule-destabilizing drugs to cdc5-1 and cdc14-1 mutants that had been arrested at the restrictive temperature for 2 to 3 h. In these cells, microtubules were eliminated (data not shown), causing the separated chromatin to collapse toward the middle of the cell, but GFP-Mob1p remained on the SPB (Fig. 7i). Thus, the SPB localization of Mob1p is independent of MPS1 and does not require microtubules for its maintenance.
DISCUSSION
MOB1 and cytokinesis.
In S. cerevisiae, cell separation comprises at least three processes: cytokinesis, septum formation, and the digestion of the chitin and cell wall material that connect the daughter cells (29). Cytokinesis results in the separation of the mother and daughter cell cytoplasm with plasma membranes and involves the function of the actomyosin contractile ring. Septum formation involves the localized deposition of chitin and cell wall material at the division site and can occur independently of actomyosin contractile-ring function (9). Chitin digestion occurs after cell wall deposition has been completed and is needed to fully separate the daughter cells (38). Intriguingly, the completion of cell separation is not essential in S. cerevisiae, since defects in components of cell separation often result in the accumulation of cellular chains in the absence of a mitotic arrest (8, 9, 29, 38, 41, 70). Both cytokinesis and septum formation are dependent on some events that occur early in the cell cycle, such as septin assembly. Septin proteins assemble into a band at the bud neck during bud emergence in late G1 and persist there until cell division has been completed (11, 23). They serve as structural scaffolds that are necessary to recruit cytokinesis and regulatory proteins to the bud neck (9, 10, 23, 42).
In this study, we demonstrated that MOB1 is required for cytokinesis in addition to mitotic exit. An indication of a cytokinesis role was provided by the presence of cellular chains in cultures of the mob1-83 mutant. The cytokinesis function of MOB1 was further supported by the isolation of suppressors of the mitotic-arrest phenotype. The mitotic-arrest phenotype of conditional mob1 mutants was suppressed with high-copy-number SIC1 plasmids, but the suppressed cells exhibited a cellular-chain phenotype at 34°C. Apparently, lowering CDK activity by overexpressing the CDK inhibitor protein Sic1p suppressed the mitotic-exit defect but not the cytokinesis defect of mob1 mutants. SIC1 suppression of the mitotic-exit defect allowed us to efficiently induce cellular-chain formation in mob1 mutants and monitor contractile-ring function by time-lapse microscopy using a GFP-Myo1 fusion. We established that the cellular chains arose by repeated rounds of budding in the absence of myosin ring contraction. Thus, the mitotic-exit and cytokinesis functions of MOB1 could be genetically separated.
The failure of mob1 mutants to undergo cytokinesis could be caused by defects either in the contractile process or in its regulation. Because the Mob1p gene belongs to the MEN, a complex regulatory network, we suspect that the role for Mob1p is regulatory. It is possible that Mob1p might mediate the recruitment or assembly of critical components of the contractile ring. Although we cannot rule out this possibility, we observed no defect in mob1 mutants in the recruitment of F-actin, Myo1p, and Iqg1p to the bud neck. Alternatively, Mob1p might initiate actomyosin ring contraction. Indeed, the timing of Mob1p localization to the bud neck just prior to actomyosin ring contraction and septum formation is consistent with such a regulatory role. In addition, the timing of maximal Mob1p concentration at the bud neck (inferred from the relative intensity of GFP-Mob1p fluorescence) suggests that Mob1p continues to function after cytokinesis is initiated, perhaps to regulate septum formation.
Coordination of mitotic exit and cytokinesis.
The tight regulatory coupling of mitotic exit and cytokinesis is also evident in the phenotypes of some other MEN mutants. For instance, overexpression of truncated Cdc5p or Cdc15p can induce cellular chains (46, 62), and cell separation defects have been reported in a certain cdc15 mutant (34). Although it has not yet been established what aspect of cell separation is defective in these instances, these results suggest that the MEN plays a critical role in controlling cell separation. The role in cell separation for Mob1p and other MEN proteins is also supported by their localization. In addition to Mob1p, Cdc5p, Cdc15p, and Dbf2p localize to the bud neck (24, 46, 62, 77).
The SIN genes of S. pombe, including the MOB1 homolog, show extensive sequence homology with the S. cerevisiae MEN genes, but, instead of exhibiting defects in mitotic exit, SIN mutants fail to undergo cytokinesis or synthesize septa (4, 5, 19, 21, 27, 30, 40, 50, 54, 56, 63). Moreover, overexpression of many SIN genes induces the formation of multiple septa, suggesting that these genes are positive regulators of cytokinesis and septum formation (4, 21, 50, 56). The cytokinesis role of Mob1p described here and the cell separation defect of cdc15 mutants (34) help to resolve the disparity between the essential functions of the S. cerevisiae MEN and S. pombe SIN genes. Moreover, the cell separation functions of the S. pombe SIN genes underscore the connection between mitotic exit and cell separation.
An essential role for the MEN in cytokinesis is supported by the interdependence of localization of Mob1p and other MEN proteins to the bud neck. We demonstrated that the bud neck localization of Mob1p requires DBF2, CDC5, CDC14, and CDC15. Similarly, bud neck localization of Mob1p requires at least one septin but not Myo1p or Iqg1p. Dbf2p and Cdc5p may also closely associate with septins, since they appear to partially colocalize with septin proteins (24, 62). Thus, binding to septin (at least indirectly) may be compulsory for Mob1p and other MEN proteins to regulate cytokinesis or septum formation.
MOB1 function at the SPBs.
In addition to the bud neck localization, we report here that Mob1p localizes to the cytoplasmic surfaces of SPBs from mid-anaphase through mitotic exit (Fig. 4). Based on interactions of Mob1p with Mps1p, which is required for SPB duplication, we previously speculated that Mob1p performs an SPB duplication function (44, 76). However the transient SPB localization of some MEN proteins, Tem1p, Cdc5p, Cdc15p, and Dbf2p (7, 24, 46, 51, 59, 62, 77), and S. pombe SIN proteins suggests that SPB localization is an important prerequisite for MEN function. In agreement, mutants defective in SPB gene NUD1 fail to exit mitosis (1, 26) and fail to recruit or maintain Mob1p and other MEN proteins at SPBs (Fig. 7) (7, 26). These data suggest that Nud1p tethers MEN proteins to SPBs and support the idea that MEN proteins must localize to SPBs in order to perform their essential mitotic-exit functions.
The functional significance of the brief asymmetry in Mob1p localization at spindle poles is not known. However, mechanisms for analogous asymmetries in S. cerevisiae Tem1p (7, 51) and Cdc15p localization (46) have been proposed. Tem1p was reported to appear first on the mother SPB and later on the daughter SPB (7, 51). Its localization to the second SPB requires a modification in the bud and is proposed to be a cellular signal for proper spindle orientation (7). Cdc15p may also appear first on the mother SPB (46). Its localization to the daughter SPB at the end of mitosis is reportedly mediated by Cdc14p-dependent dephosphorylation (46). Mob1p's localization to the second SPB is not likely to be regulated by either of these mechanisms, because Mob1p localizes to both SPBs during mid-anaphase (Fig. 4), which occurs after the spindle is properly oriented and before Cdc14p phosphatase is released from the nucleolus.
Our data suggest that Mob1p functions in concert with other MEN proteins to coordinate several critical cell cycle processes at the end of mitosis, including cyclin degradation, CDK inactivation, cytokinesis, and septation. Moreover, the interactions between Mob1p and Mps1p may suggest a link between the SPB duplication or spindle assembly checkpoint pathways and mitotic exit. Future work may reveal that the “cell cycle-coordinating” functions may be the most fundamentally conserved elements of the MEN pathway.
ACKNOWLEDGMENTS
We thank members of the Winey laboratory and Erfei Bi for many helpful discussions and Shelly Q. Jones, Michael Atchison, and Erika Holzbaur for critical reading of the manuscript. We also thank Jason Kahana and Pamela Silver for providing anti-GFP antibodies and Erfei Bi, Clyde Denis, Clive Price, and John Kilmartin for yeast strains and reagents.
This work was supported by grants from the Leukemia Society of America (F.C.L.), American Cancer Society IRG-78-002-22 (F.C.L.), and National Institutes of Health R01 GM60575 (F.C.L.) and R01 GM51312 (M.W.).
REFERENCES
- 1.Adams I R, Kilmartin J V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asakawa K, Yoshida S, Otake F, Tohe A. A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics. 2001;157:1437–1450. doi: 10.1093/genetics/157.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachant J B, Elledge S J. Mitotic treasures in the nucleolus. Nature. 1999;398:757–758. doi: 10.1038/19641. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian M K, McCollum D, Chang L, Wong K C, Naqvi N I, He X, Sazer S, Gould K L. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian M K, McCollum D, Gould K L. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- 6.Balasubramanian M K, McCollum D, Surana U. Tying the knot: linking cytokinesis to the nuclear cycle. J Cell Sci. 2000;113:1503–1513. doi: 10.1242/jcs.113.9.1503. [DOI] [PubMed] [Google Scholar]
- 7.Bardin A J, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 8.Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi E, Maddox P, Lew D J, Salmon E D, McMillan J N, Yeh E, Pringle J R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyne J R, Yosuf H M, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113:4533–4543. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- 11.Chant J. Septin scaffolds and cleavage planes in Saccharomyces. Cell. 1996;84:187–190. doi: 10.1016/s0092-8674(00)80972-1. [DOI] [PubMed] [Google Scholar]
- 12.Chial H J, Rout M P, Giddings T H, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chial H J, Winey M. Mechanisms of genetic instability revealed by analysis of yeast spindle pole body duplication. Biol Cell. 1999;91:439–450. [PubMed] [Google Scholar]
- 14.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 15.Cormack B P, Bertram G, Egerton M, Gow N A, Falkow S, Brown A J. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 16.DeMarini D J, Adams A E, Fares H, De Virgilio C, Valle G, Chuang J S, Pringle J R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson A D, Kilmartin J V. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donovan J D, Toyn J H, Johnson A L, Johnston L H. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- 19.Eng K, Naqvi N I, Wong K C, Balasubramanian M K. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- 20.Epp J A, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 21.Fankhauser C, Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 1994;13:3011–3019. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field C, Li R, Oegema K. Cytokinesis in eukaryotes: a mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- 23.Field C M, Kellogg D. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- 24.Frenz L M, Lee S E, Fesquet D, Johnston L H. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- 25.Gould K L, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- 26.Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guertin D A, Chang L, Irshad F, Gould K L, McCollum D. The role of the Sid1p kinase and Cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- 29.Hales K G, Bi E, Wu J Q, Adam J C, Yu I C, Pringle J R. Cytokinesis: an emerging unified theory for eukaryotes? Curr Opin Cell Biol. 1999;11:717–725. doi: 10.1016/s0955-0674(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 30.Hou M C, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- 31.Hoyt M A. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- 32.Jaspersen S L, Charles J F, Morgan D O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 33.Jaspersen S L, Charles J F, Tinker-Kulberg R L, Morgan D O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiménez J, Cid V J, Cenamor R, Yuste M, Molero G, Nombela C, Sánchez M. Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J Cell Biol. 1998;143:1617–1634. doi: 10.1083/jcb.143.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keng T, Clark M W, Storms R K, Fortin N, Zhong W, Ouellette B F, Barton A B, Kaback D B, Bussey H. LTE1 of Saccharomyces cerevisiae is a 1435 codon open reading frame that has sequence similarities to guanine nucleotide releasing factors. Yeast. 1994;10:953–958. doi: 10.1002/yea.320100710. [DOI] [PubMed] [Google Scholar]
- 36.Kitada K, Johnson A L, Johnston L H, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komarnitsky S I, Chiang Y C, Luca F C, Chen J, Toyn J H, Winey M, Johnston L H, Denis C L. Dbf2 protein kinase binds to and acts through the cell cycle-regulated Mob1 protein. Mol Cell Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 39.Lauze E, Stoelcker B, Luca F C, Weiss E, Schutz A R, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Goff X, Utzig S, Simanis V. Controlling septation in fission yeast: finding the middle, and timing it right. Curr Genet. 1999;35:571–584. doi: 10.1007/s002940050455. [DOI] [PubMed] [Google Scholar]
- 41.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longtine M S, McKenzie III A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Luca F C, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol Biol Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCollum D, Gould K L. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- 46.Menssen R, Neutzner A, Seufert W. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol. 2001;11:345–350. doi: 10.1016/s0960-9822(01)00095-1. [DOI] [PubMed] [Google Scholar]
- 47.Morgan D O. Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- 48.Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 1995;270:1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- 49.Munoz-Centeno M C, McBratney S, Monterrosa A, Byers B, Mann C, Winey M. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol Biol Cell. 1999;10:2393–2406. doi: 10.1091/mbc.10.7.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohkura H, Hagan I M, Glover D M. The conserved Schizosaccharomyces pombe kinase Plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 51.Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 52.Peters J M. Subunits and substrates of the anaphase-promoting complex. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- 53.Rao P N, Adlakha R C. Chromosome condensation and decondensation factors in the life cycle of eukaryotic cells. Symp Fundam Cancer Res. 1984;37:45–69. [PubMed] [Google Scholar]
- 54.Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae MOB1 gene is essential and functions in signaling the onset of septum formation. J Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- 55.Sawin K E. Cytokinesis: Sid signals septation. Curr Biol. 2000;10:547–550. doi: 10.1016/s0960-9822(00)00613-8. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- 57.Schweitzer B, Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991;7:265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- 58.Shirayama M, Matsui Y, Toh-e A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Charbonneau H, Deshaies R J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 61.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song S, Grenfell T Z, Garfield S, Erikson R L, Lee K S. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sparks C A, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straight A F, Sedat J W, Murray A W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the Cdc28/Clb mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tada S, Blow J J. The replication licensing system. Biol Chem. 1998;379:941–949. [PMC free article] [PubMed] [Google Scholar]
- 68.Townsley F M, Ruderman J V. Proteolytic ratchets that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- 69.Toyn J H, Johnston L H. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vallen E A, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Visintin R, Craig K, Hwang E S, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 72.Visintin R, Hwang E S, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 73.Wan J, Xu H, Grunstein M. CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis, and transcription during the cell cycle. J Biol Chem. 1992;267:11274–11280. [PubMed] [Google Scholar]
- 74.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickner R B, Koh T J, Crowley J C, O'Neil J, Kaback D B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the MAK16 gene and analysis of an adjacent gene essential for growth at low temperatures. Yeast. 1987;3:51–57. doi: 10.1002/yea.320030108. [DOI] [PubMed] [Google Scholar]
- 76.Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu S, Huang H K, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10:329–332. doi: 10.1016/s0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]