Abstract
Structural maintenance of chromosomes (SMC) proteins fulfill pivotal roles in chromosome dynamics. In yeast, the SMC1-SMC3 heterodimer is required for meiotic sister chromatid cohesion and DNA recombination. Little is known, however, about mammalian SMC proteins in meiotic cells. We have identified a novel SMC protein (SMC1β), which—except for a unique, basic, DNA binding C-terminal motif—is highly homologous to SMC1 (which may now be called SMC1α) and is not present in the yeast genome. SMC1β is specifically expressed in testes and coimmunoprecipitates with SMC3 from testis nuclear extracts, but not from a variety of somatic cells. This establishes for mammalian cells the concept of cell-type- and tissue-specific SMC protein isoforms. Analysis of testis sections and chromosome spreads of various stages of meiosis revealed localization of SMC1β along the axial elements of synaptonemal complexes in prophase I. Most SMC1β dissociates from the chromosome arms in late-pachytene-diplotene cells. However, SMC1β, but not SMC1α, remains chromatin associated at the centromeres up to metaphase II. Thus, SMC1β and not SMC1α is likely involved in maintaining cohesion between sister centromeres until anaphase II.
The eukaryotic, evolutionarily highly conserved SMC (Structural Maintenance of Chromosomes) proteins are involved in several key DNA and chromatin dynamic processes (for recent reviews, see references 11, 21, 26, 27, 31, 48, 60, and 62). The best-documented processes are chromosome condensation and sister chromatid cohesion. Evidence is also accumulating for a function in DNA recombination and repair. A fourth role of SMC proteins is in gene dosage compensation in Caenorhabditis elegans. The phylogenetic tree comprises five subfamilies (32): SMC1 to SMC4 and an ancestral family that includes the recently defined SMC5 and SMC6 groups with the Rad18 and Spr18 proteins of Schizosaccharomyces pombe (16), which act in recombinational repair.
SMC proteins share a characteristic design. Coiled-coil domains are flanked by globular N- and C-terminal domains and are divided in the central region by a flexible hinge domain of about 150 aa. The N- and C-terminal domains of about 100 to 150 aa are highly conserved and carry important motifs. The N-terminal domain includes an NTP binding motif (Walker A box [68]), which has been shown to bind the ATP analogue azido-ATP (1). The C-terminal domain contains a DA box (68). The C-terminal and second coiled-coil domains, but not the N terminus, bind DNA (1, 2). It has been proposed that the antiparallel, heterodimeric SMC1-SMC3 protein with an N and C terminus at each end may connect two DNA molecules, such as sister chromatids, and may directly contribute to their alignment in cohesion and to recombination between sister chromatids (2, 26, 62).
In eukaryotes the SMC1-SMC3 or SMC2-SMC4 heterodimers form large multiprotein complexes. One of these complexes is condensin, which, besides the SMC2-SMC4 heterodimer, contains several non-SMC subunits. Condensin is necessary for mitotic chromosome condensation in Saccharomyces cerevisiae (61), S. pombe (64), and Xenopus laevis egg extracts (25) and has also been described in human cells (57). The MIX-1 and DPY-27 proteins of C. elegans, proteins homologous to SMC2 and SMC4, are present in a different multiprotein complex, which regulates gene dosage compensation on the X chromosomes of the hermaphrodite nematode (8, 39).
The other pair of SMC proteins, SMC1 and SMC3, is present in at least two protein complexes with distinct, albeit partially connected, functions. Genetic studies of S. cerevisiae revealed a requirement for Smc1p and Smc3p in mitotic sister chromatid cohesion (19, 45). The respective protein complex is called cohesin, contains two other polypeptides besides Smc1p and Smc3p, and interacts with several other factors required for sister chromatid cohesion and its release (reviewed in references 11, 48, and 62). One of the non-SMC cohesin subunits is the S. cerevisiae Scc1p (Mcd1p) protein, homologous to Rad21 in S. pombe (4, 19, 45). The rad21-45 mutation (in S. pombe) also causes X-ray sensitivity and a mitotic hyperrecombination phenotype (4, 18). A similar cohesin complex was identified from X. laevis cell extracts, extensively characterized, and found to be required for sister chromatid cohesion in this system (41, 42). We have identified the SMC1 and SMC3 proteins as constituents of the mammalian recombination complex RC-1, which is present in a variety of somatic cells (29, 30, 63). This complex catalyzes SMC protein-dependent cell-free transfer of duplex DNA molecules, which mimics recombinational repair of gaps and deletions (29, 30). The presence of the SMC1 and SMC3 proteins in these multiprotein complexes furthered speculations about an SMC-mediated functional link between sister chromatid cohesion and recombinational repair (26, 31, 62). Recent evidence from studies of yeast supports this concept. Klein et al. (36) reported that S. cerevisae Smc3p is required not only for meiotic sister chromatid cohesion, but also for meiotic DNA recombination.
Sister chromatid cohesion and DNA recombination are both essential for meiosis (for reviews, see references 35, 54, and 66). In mitotic cells, DNA recombination is primarily a means to repair DNA damage, and the role of cohesins may include the direction of recombinational repair towards the sister chromatid rather than the homologous chromosome (if there is one) (33, 58). In meiosis, recombination and sister chromatid cohesion are essential, but the relationship between the two processes has been modified. Whereas meiotic recombination has to be directed towards the homologue rather than the sister chromatid, cohesion between sister chromatids has to be maintained to ensure the proper orientation and disjunction of homologues at meiosis I (66). During the meiotic prophase, a characteristic, zipperlike protein structure, the synaptonemal complex (SC), is formed between homologues and likely plays an important but not entirely clarified role in adapting recombination and cohesion for meiosis (20, 23). SCs consist of two axial elements (AEs), which are connected by numerous transverse filaments along their lengths. Each AE structurally supports the two sister chromatids of one homologue.
In budding yeast, Smc3p colocalizes with an AE component during the meiotic prophase (36). In meiotic yeast cells, the cohesin protein Scc1p (Mcd1p) is largely replaced by its meiosis-specific homologue, Rec8p, or its homologues in other organisms (36, 47, 69). In S. cerevisiae, Rec8p localizes along the AEs of SCs (36).
These observations also suggest for mammalian meiotic cells an association of cohesion proteins with the SC. In mammals, two AE components have been identified, SCP2 (49) and SCP3 (37), which are specifically expressed in the meiotic prophase. Recently, we have shown that mammalian SMC1 is present in meiotic nuclei throughout prophase I. Upon permeabilization of spermatocytes in the presence of Triton X-100, SMC1 is specifically retained in a dotlike pattern along the AEs of SCs. We also showed that mammalian SMC1 and SMC3 proteins associate with AE components, e.g., SCP2 and SCP3 (13). Thus, it is intriguing to hypothesize about an essential role of mammalian SMC proteins in meiotic sister chromatid cohesion—its establishment, maintenance, and resolution—and in meiotic DNA recombination. Differences in mitotic sister chromatid cohesion, however, extend beyond meiotic prophase I. Sister chromatids separate only in the chromosome arms, but not at the centromeres, during meiosis I, leaving centromeric cohesion intact until anaphase II, when all cohesion is finally removed. An important question is whether there is specific adaptation of SMC proteins and their complexes to their specific meiotic functions. The differences mentioned above in the interplay of sister chromatid cohesion and DNA recombination between meiosis and mitosis render such adaptation likely. One way to achieve this adaptation is through expression of a meiosis-specific homologue or isoform of a somatic protein.
Here, we demonstrate the existence of a meiosis-specific isoform of mammalian SMC1, named SMC1β, which we describe in this report.
MATERIALS AND METHODS
Cloning of SMC1β cDNA.
The 150-kDa protein was isolated from testis nuclear extracts by immunoprecipitation with anti-SMC3 antibody (63) and separation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Sequences from seven peptides, generated by tryptic digest of the protein, were determined. Based on this sequence information, oligonucleotides were designed for the screening of a mouse testis 5′-STRETCH cDNA library (Clontech Inc.). Several overlapping clones were isolated, which covered a 1,735-bp fragment at the 5′ end of the cDNA. To recover the missing 3′ end, rapid amplification of cDNA ends (RACE) (15) was performed using a SMART RACE cDNA amplification kit (Clontech Inc.). Several independent clones have been obtained and sequenced. The sequence of the C terminus was further verified by performing nested PCR on 3′ RACE PCR products from testis RNA. The PCR used a gene-specific primer located at the positions corresponding to aa 1171 to 1179 of SMC1β, and the Nested Universal Primer (Clontech Inc.), located downstream of poly(A) within the region defined by the RACE universal primer. Twenty cloned PCR products were analyzed by restriction analysis, and five clones were sequenced. All belonged to the same gene and encoded identical C termini. The 4,056-bp full-length cDNA was assembled using standard cloning protocols.
Northern blot analysis.
Eight-microgram aliquots of total RNA from various mouse tissues (Ambion) were separated electrophoretically and transferred to a nylon membrane (Hybond N; Amersham) using standard protocols (55). Hybridization was performed as described previously (9). The probe corresponded to the first 616 bp of the SMC1β cDNA.
Protein purification and antibody generation.
The C-terminal domain of SMC1β (SMC1β-C) was subcloned into the Escherichia coli expression vector pQE32 (Qiagen Inc.). The His6-tagged protein, with a molecular mass of 33 kDa, was overexpressed and purified on Ni-nitrilotriacetic acid agarose resin (Qiagen) and eluted in steps with increasing imidazole. The yield was 200 to 600 μg per liter of E. coli culture. About 80% of the total SMC1β-C protein was found to be insoluble under native conditions and had to be dissolved in 8 M urea. The remaining 20%, native soluble protein, was mixed with an equal amount of denatured protein and injected into mice. Monoclonal antibodies were generated by standard hybridoma techniques. Hybridoma tissue culture supernatants were screened by enzyme-linked immunosorbent assay using 96-well plates that were coated with the C-terminal protein. Subsequently, the positive supernatants were tested for their specificities by Western blotting on testis versus somatic cell extracts and by immunofluorescence on testis versus liver sections. At least six independent hybridoma clones produced antibodies specific in both procedures for SMC1β.
The rabbit anti-SMC1 serum SMC1-C1 and the anti-SMC3 serum (13, 63), the hamster anti-SCP3 serum H1 (14), the rabbit anti-SCP3 serum 175 (37), and the rabbit anti-SCP2 serum (49) have been described. For labeling of kinetochores, we used a human autoimmune serum from a patient with CREST (calcinosis, Raynaud syndrome, esophageal dismobility, sclerodactyly, and telangiectasia) syndrome; this serum reacts with kinetochore proteins and has been described by Moens et al. (46).
Immunoprecipitation.
Nuclear extracts from tissue or cells were prepared as described previously (29), with the concentration of dithiothreitol (DTT) lowered to 0.1 mM. Five micrograms of affinity-purified anti-SMC3 antibody was incubated for 4 to 16 h at 4°C with nuclear extract (50 μg of protein unless otherwise indicated), which was diluted 1:5 in buffer IP (phosphate-buffered saline plus 0.75% Brij 58, and 500 mM NaCl). A 20-μl slurry of protein A beads was added, and the mixture was further incubated for 1 h. The beads were washed six times with buffer IP or variations of it as described in the text and were boiled with SDS gel sample buffer before being loaded on reducing 5% polyacrylamide SDS gels. Detection of proteins was done by silver staining according to a published protocol (29).
Preparation of spreads and agar filtrates.
Spreads of mouse spermatocytes were prepared by the dry-down technique of Speed (59), as modified by Peters et al. (51). Agar filtrates of lysed rat spermatocytes were prepared as described by Heyting and Dietrich (22).
Immunofluorescence labeling.
Immunofluorescence labeling of dry-down preparations and agar filtrates was performed as described previously (22). The slides were mounted in Vecta Shield (Vector Laboratories Inc., Burlingame, Calif.). The monoclonal anti-SMC1β antibodies in tissue culture supernatant (from two independent hybridomas) were diluted 1:1, serum 175 (rabbit anti-SCP3) was diluted 1:500, CREST serum (anti-kinetochore) was diluted 1:1,000, and serum H1 (hamster anti-SCP3) was diluted 1:50. Goat anti-rabbit immunoglobulin G (IgG) conjugated with aminomethylcoumarin acetate (AMCA) (Vector) or with Texas red (Jackson ImmunoResearch Laboratories, West Grove, Pa.), goat anti-mouse IgG conjugated with fluorescein isothiocyanate (FITC) (Jackson), goat anti-hamster IgG conjugated with AMCA, and goat anti-human IgG conjugated with Texas red were used as secondary antibodies and were diluted according to the instructions of the suppliers.
Microscopy.
Spread preparations were examined with a Zeiss Axioplan research microscope equipped with epifluorescence illumination and Plan-Neofluar optics. Selected images were directly photographed on an ISO 400 color negative film using single-band-pass emission filters (for DAPI [4′,6′-diamidino-2-phenylindole]-AMCA, FITC, and Texas red fluorescence) with separated excitation filters. Negatives were scanned at high resolution, and their computer images were processed and combined using the CorelDraw and Corel Photopaint software packages.
Isolation of meiotic cells.
Spermatocytes were isolated from rat testes by cell elutriation and density centrifugation according to the method of Heyting and Dietrich (22), and the composition of the isolated cell fraction was analyzed by differential counts of Giemsa-stained preparations, as described previously (38). The cell fraction that was used in this study had the following composition: Sertoli cells, 0.3%; spermatogonia, 0.7%; and spermatocytes, 99%, of which 1.2% was in leptotene-zygotene, 29% was in early-mid pachytene, 54% was in late pachytene or prediffuse diplotene, and 16% was in diffuse or postdiffuse diplotene.
DNA interaction assays.
DNA concentrations are expressed as nucleotide equivalents. The assay for retention of double-stranded DNA on nitrocellulose filters through binding to protein (filter binding assay) was performed essentially as described elsewhere (3).
Linear double-stranded DNA fragments were generated by digestion of pBluescript SK(+) (Stratagene Inc.) with AluI and were labeled at their 5′ ends with 32P. Reactions were performed in 20-μl mixtures containing 25 mM Tris-HCl (pH 7.5), 10 mM KCl, 0.2 mM EDTA, 1 mM DTT, 10 ng of linear DNA, and various amounts of peptide. After 20 min at room temperature, the reaction mixtures were diluted by adding 1 ml of reaction buffer containing 10 mM sodium pyrophosphate. The reaction mixtures were filtered through prewashed 0.1-μm-pore-size nitrocellulose filters (Whatman Inc.). The filters were washed thrice with 1 ml of the reaction buffer containing sodium pyrophosphate, and the radioactivity retained on the filters was measured in a liquid scintillation counter.
The gel shift (gel retardation) assay was performed as described previously (1, 2). For a DNA substrate, we used a 200-bp DNA fragment of the 5S rRNA gene or a 230-bp DNA fragment derived from the double-stranded form of M13mp8, which we know are good binding substrates for SMC protein domains (1, 2). The reaction buffer contained 0.8 pmol of 32P 3′-end-labeled DNA (5,000 to 10,000 cpm) in 20 mM HEPES (pH 7.5), 1 mM DTT, and 0.1 mg of bovine serum albumin (ultrapure; Amersham-Pharmacia Inc.)/ml. The SMC1β peptide consisted of the C-terminal 28 aa (TEDQEGSRSHRKPRVPRVSMSPKSPQSR; theoretical pI, 11.4). The positive control peptide was derived from mouse Rad54L (34), aa 152 to 181 (KVCRPHQREGVKFLWECVTSRRIPGSHGLI; theoretical pI, 10.09). The negative control peptide was derived from human Rad21 (44), aa 612 to 631 (TQEEPYSDIIATPGPRFHII; theoretical pI, 4.65).
The assay for network formation was done as described for RecA (7) or mammalian DNA binding proteins (17), with some modifications. The assay measures coaggregation of DNA into DNA-protein complexes that sediment rapidly. The DNA substrate was AluI-digested (blunt-ended), 5′-32P-labeled plasmid DNA as used for the filter binding assay. Various amounts of peptide were incubated with the DNA (1.5 pmol) in a volume of 50 μl for 20 min at room temperature in the DNA binding buffer also used for gel shift experiments. The reaction mixture was then centrifuged for 3 min at 14,000 × g. The supernatant was transferred to a scintillation vial, and the pellet was solubilized in 100 μl of 0.1% SDS and also transferred to a scintillation vial. The supernatant (nonaggregated) and pellet (aggregated) DNAs were measured. All experiments were done in triplicate.
Nucleotide sequence accession numbers.
The GenBank accession number of mSMC1β is AF303827.
RESULTS
A novel SMC protein complex from testis.
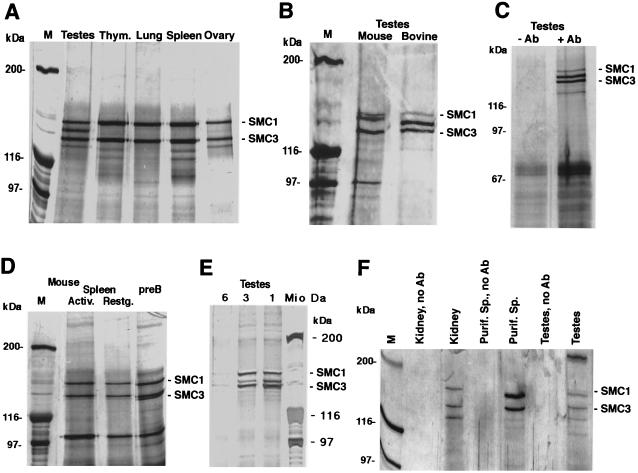
Earlier, we used affinity-purified polyclonal anti-SMC3 antibodies, raised against the C-terminal domain of bovine SMC3, in immunoprecipitation experiments with nuclear extracts of mitotically dividing cells of human, mouse, hamster, and bovine origin (reference 63 and unpublished observations). Under stringent precipitation conditions, the SMC1 and SMC3 proteins were observed as the only strong bands in silver-stained SDS polyacrylamide gels, used to analyze the precipitates. A weaker 120-kDa band was often visible and probably represented the Rad21 protein. Immunoprecipitation experiments with nuclear extracts prepared from a variety of mouse and bovine tissues, however, revealed a marked difference between extracts from testes and all other extracts. From testes, we coimmunoprecipitated a hitherto undescribed protein that migrates at an approximately 145- to 155-kDa position between SMC1 and SMC3, depending on the particular gel electrophoresis conditions (Fig. 1). The 150-kDa protein was also not observed in immunoprecipitates from various human, mouse, and hamster cell lines, nor was it present in cytoplasmic extract fractions (63; data not shown). However, a faint 150-kDa band was visible in immunoprecipitates from ovary extracts (Fig. 1A). Comparable results were obtained with mouse tissues (Fig. 1B and F). The coprecipitated protein from mouse testis extract migrates at 155 kDa, a position slightly higher than that of the bovine protein. The 150-kDa protein is present neither in resting nor in activated, proliferating somatic mouse cells (Fig. 1D).
FIG. 1.
Immunoprecipitation of SMC3 and associated SMC1 proteins with anti-SMC3 antibodies. (A) Immunoprecipitation from various bovine tissue nuclear extracts. Thym., thymus. (B) Immunoprecipitation from mouse and bovine testis nuclear extracts. (C) Control immunoprecipitation from bovine testis nuclear extract, with (+Ab) and without (−Ab) anti-SMC3 antibody included. (D) Immunoprecipitation from nuclear extracts prepared from actively proliferating (lipopolysaccharide-induced; Activ.) and resting (Restg.) mouse spleen cell cultures and from an actively growing mouse pre-B-cell line. (E) Immunoprecipitation from bovine testis nuclear extract gel filtration (BioGel A15m resin) fractions (20 μg of protein each) representing 1-, 3-, and 6-MDa molecular-mass positions. (F) Immunoprecipitation from mouse kidney, rat testis, and purified rat spermatocyte (Purif. Sp.) nuclear extracts (25 μg of protein each). Controls without antibody (no Ab) are included. All precipitates were analyzed by SDS-polyacrylamide gel electrophoresis and silver staining. M, molecular mass marker.
Incubation of the precipitates with bacterial alkaline phosphatase or the inclusion of ATP (1 mM) or of the phosphatase inihibitor o-vanadate (0.5 mM) in the extracts and all buffers did not alter the result (not shown). The characteristic band pattern also did not change upon variation of the precipitation and wash conditions, e.g., the use of different detergents, differently pretreated protein A or protein G beads, or protein G-precleared extracts (not shown). Association of the 150-kDa protein with SMC3 was found to be as resistant to stringent precipitation reaction conditions as that of SMC1 with SMC3. The 150-kDa protein was also immunoprecipitated from testis nuclear extract fractions that had been obtained by gel filtration of the extract through a large BioGel A15m chromatography column. Similar chromatography experiments were done before for either purification of RC-1 from thymus or other analysis of testis extract fractions (13, 30) (Fig. 1E). The protein was found together with SMC3 in fractions that represent molecular masses of globular proteins of around 1 MDa and—albeit in smaller amounts—at 3 MDa. At 3 MDa, more of SMC1-SMC3 was visible, possibly indicating differences in masses between complexes based on SMC1-SMC3 or the 150-kDa protein and SMC3. Thus, the 150-kDa protein coprecipitates and copurifies with SMC3 and is likely a component of a large multiprotein complex, similar but not identical to that containing the SMC1-SMC3 heterodimer (13).
In immunoblotting, neither a monoclonal nor an affinity-purified polyclonal antibody, both raised against the C-terminal domain of SMC1, recognizes any protein migrating between the SMC1 and SMC3 proteins (13, 63). Likewise, anti-SMC3 antibodies do not react with the protein in immunoblotting. Thus, the 150-kDa protein is not a degradation product of SMC1 or very homologous to SMC3.
From immunoblotting and immunoprecipitation experiments, we estimate an approximate relative abundance of SMC1α, 150-kDa protein, and SMC3 of 1:2:3 in total testis nuclear extracts. In extracts from purified spermatocyte preparations, which consist of >99% meiotic cells (70% late pachytene-diplotene [22, 38]), only a small amount of SMC1α was seen, while the 150-kDa protein and SMC3 precipitated in an approximately equimolar ratio (Fig. 1F).
Large-scale immunoprecipitation followed by SDS-polyacrylamide gel electrophoresis and amino acid sequencing of the 150-kDa protein allowed us to deduce seven peptide sequences 6 to 12 aa in length. This information was used to generate oligonucleotides with which a mouse testis library was screened. By a combination of further screening and reverse transcription-PCR from mouse testis RNA, the entire cDNA was cloned and then sequenced. This cDNA encodes a protein that has not been reported previously and that shows a high level of homology to mammalian SMC1 (Fig. 2A). The homology is highest in the conserved functional domains of SMC proteins, the N-terminal, C-terminal, and hinge domains. Lower degrees of homology were found with SMC4 and SMC3. Dendrogram analyses of the N- and C-terminal domains confirmed the close relationship to SMC1 (Fig. 2B). Therefore, we call the protein SMC1β, indicating an SMC1 variant or isoform. The “classical” SMC1 may be termed SMC1α. SMC1β bears a unique C-terminal sequence of 28 aa that has been found neither in any other SMC protein nor in the databases. Among the 28 aa are 4 proline residues and 7 arginine and lysine residues. This C-terminal peptide is very basic, with a theoretical pI of 11.4. In contrast, the entire C-terminal domain of SMC1β, with 186 aa, has a theoretical pI of 6.9. This C-terminal motif was present in all independently isolated clones and was confirmed by RT-PCR of mouse testis and subsequent sequencing of PCR products. SMC1β shows all motifs characteristic of SMC proteins, including the N-terminal Walker A box, the C-terminal Walker B box, and the signature motif typical of ABC-ATPases (28, 68), as well as the extended coiled-coil domains with heptad repeats and the hinge region.
FIG. 2.
Amino acid sequence comparison and dendrogram of SMC1β. (A) N-terminal domains of SMC1β and mammalian SMC proteins representing the four most closely related SMC subfamilies. (B) C-terminal domains of SMC1β and mammalian SMC proteins representing the most closely related SMC subfamilies. The program Megalign (DNAStar) was used. The accession numbers for the related SMC proteins are as follows: mSMC1α (mouse SMCB; AF047600), bSMC1α (bovine SMC1; AF072712), hSMC1α (human SB1.8; S78271), XSMC1 (X. laevis; AF051784), mSMC3 (mouse SMCD; AF047601), bSMC3 (bovine; AF072713), hSMC4 (human CAP-C; AB019987), and hSMC2 (human CAP-E; AF092563). mSMC1β, mouse SMC1β; hSMC1β, predicted human protein (CAB41703; partial sequence). Identical residues are shaded. Dashes indicate gaps in alignment.
Specific expression of SMC1β in meiotic cells.
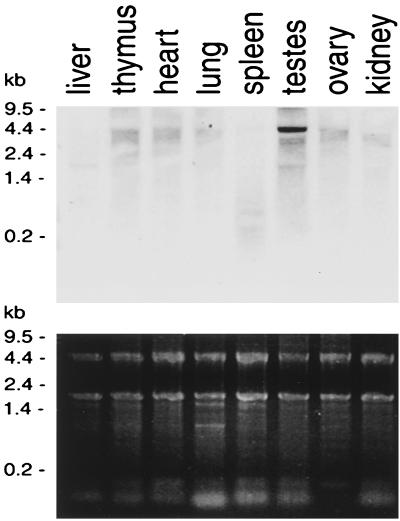
Northern blotting of RNA from a variety of mouse tissues was performed using a 616-bp 5′ DNA fragment of SMC1β as a probe (Fig. 3). This experiment confirmed testis-specific expression of the gene. The specific signal of about 4.5 kb was not seen in RNA from any of the other tissues. We also used the same probe to analyze RNA prepared from purified spermatocytes. The same 4.5-kb signal was observed, and no other band was detected (not shown).
FIG. 3.
Tissue specificity of SMC1β RNA expression. (Top) Northern blot of total RNA extracted from different mouse tissues and hybridized to an SMC1β-specific probe. (Bottom) Corresponding agarose gel stained with Radiant Red fluorescent RNA stain prior to transfer for loading control.
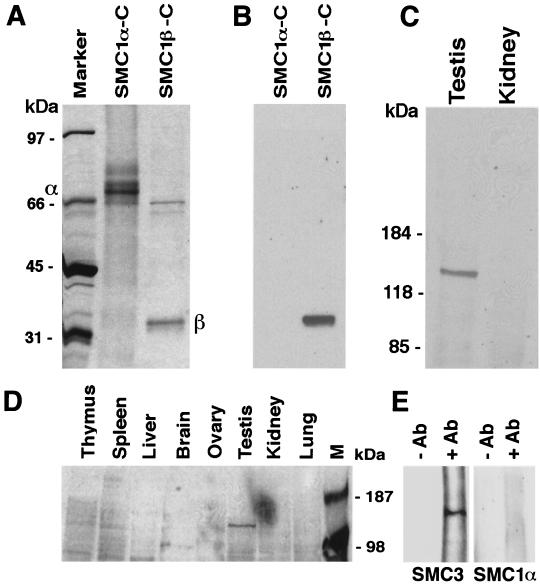
Next, we generated monoclonal antibodies for the analysis of protein expression. The antibodies were raised in mice against a C-terminal 33-kDa fragment of mouse SMC1β, expressed in E. coli, and purified by affinity chromatography on Ni-Sepharose (Fig. 4A). The C-terminal protein was partially soluble under native conditions, and the insoluble precipitates were solubilized in 8 M urea. A mixture of native and denatured protein was used for immunization. We obtained several hybridoma lines that produce antibodies that specifically recognize SMC1β but not SMC1α (Fig. 4B). No cross-reaction with even large amounts of the purified C-terminal domain of SMC1α, or with SMC proteins present in somatic cell nuclear extracts, was observed (Fig. 4B and C). The tissue specificity of SMC1β was also confirmed for a large variety of bovine tissues (Fig. 4D). We obtained several antibodies that recognize SMC1β of mouse, rat, and bovine origin. Immunoprecipitation using the anti-SMC1β antibody with testis extracts confirmed the association of SMC1β with SMC3 (Fig. 4E). We never observed SMC1α to coimmunoprecipitate with anti-SMC1β antibodies.
FIG. 4.
Generation and specificity of anti-SMC1β monoclonal antibodies. (A) Coomassie blue-stained SDS-polyacrylamide gel loaded with the C-terminal domains of SMC1α (approximately 67 kDa) and SMC1β (approximately 33 kDa), with their positions indicated by α and β. (B) Immunoblot of a gel identical to that in panel A probed with a monoclonal anti-SMC1β antibody. (C) Immunoblot of nuclear extracts from mouse testis and kidney probed with a monoclonal anti-SMC1β antibody. (D) Immunoblot of nuclear extracts from a variety of bovine tissues probed with a monoclonal anti-SMC1β antibody. M, marker. (E) Anti-SMC1β immunoprecipitates from testis nuclear extracts probed in Western blotting with anti-SMC3 or anti-SMC1α antibodies. −AB, no anti-SMC1β; +AB, with anti-SMC1β.
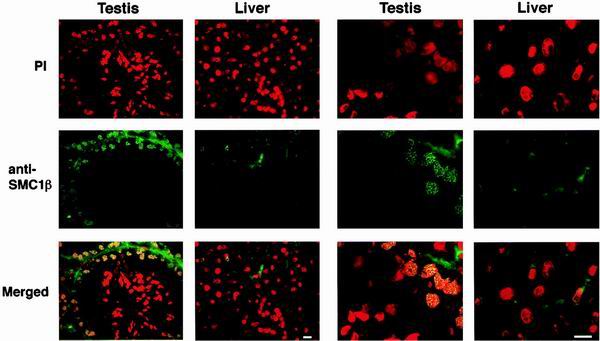
We then used these monoclonal antibodies to investigate expression and localization of SMC1β in testis. Mouse testis sections and, for control, liver sections were prepared and immunoprobed with FITC-labeled anti-SMC1β and stained with propidium iodide (after RNase treatment) to visualize nuclear DNA (Fig. 5). No specific anti-SMC1β signal was obtained with liver sections. In testis, however, strong staining of prophase I nuclei was observed. The antibodies stained the compact chromosomal axes within the meiotic nuclei, indicative of the presence of SMC1β along the SCs. In these sections, only weak staining was observed in cells of later stages. These results were confirmed with three other anti-SMC1β antibodies (not shown).
FIG. 5.
Testis and liver section stained with anti-SMC1β. Sections of mouse testis or liver were incubated with either propidium iodide (PI)-RNase or anti-SMC1β, FITC-labeled, to visualize DNA or SMC1β. The merged images are at the bottom, and two magnifications are shown as indicated. Bar = 10 μm.
By immunoblotting, SMC1β was also found in preparations of SCs from rat spermatocytes (13) (not shown). As these preparations contain only a very limited number of proteins (<20) and are prepared under stringent conditions, this indicates a close association of SMC1β with SCs.
Chromosomal localization of SMC1β throughout meiosis.
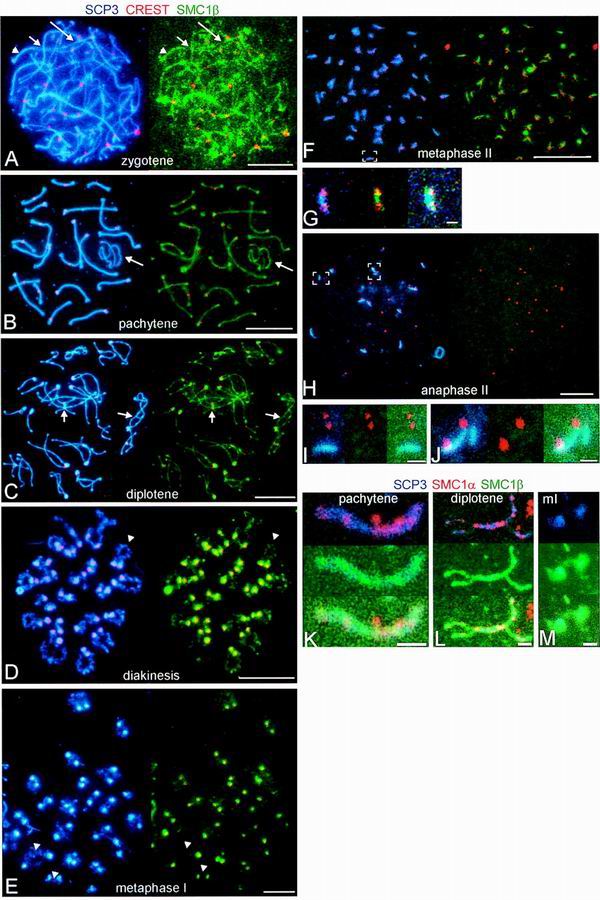
High-resolution analysis of rat spermatocyte nuclear spreads confirmed and significantly extended our initial observations (Fig. 6 and 7). Several different anti-SMC1β antibodies were used in immunofluorescence, all yielding identical results. Spreads of cells in consecutive stages of meiosis up to anaphase II were analyzed for SMC1β, the AE-specific protein SCP3, and kinetochores (Fig. 6). SMC1β tightly colocalizes with SCP3 from early prophase I (leptotene and zygotene) on along the entire AEs of the chromosomes in presynapsed (leptotene-zygotene [Fig. 6A]), synapsed (pachytene), unsynapsed (XY bivalent [Fig. 6B]), and desynapsed (diplotene [Fig. 6C]) regions. The distribution of SMC1β appears rather uniform along the AEs, with occasional more intense dots. Until diplotene, there is no concentration of SMC1β around the centromeres (Fig. 6A and B). SCP3 and SMC1β remain tightly associated with the AEs throughout pachytene. Upon desynapsis of the homologous chromosomes in diplotene, SCP3 and SMC1β start to accumulate around the centromeres and to dissociate from the chromosome arms (Fig. 7C to E). Staining for both proteins is also visible at sites of bridges between the homologues, which possibly represent sites of crossover (Fig. 6C).
FIG. 6.
Immunolocalization of SMC1β and SMC1α in successive stages of meiosis. (A to J) Images of immunofluorescence triple labeling of SCP3 (blue), SMC1β (green), and kinetochores (red) in dry-down preparations of rat spermatocytes. (A to F and H) Shown on the left are the merged images of SCP3 (blue) and kinetochores (red), and shown on the right are the merged images of the same cell of SMC1β (green) and kinetochores (red). (A) zygotene (the long arrows indicate asynapsed segments of AEs, the short arrows point to regions of presynaptic alignment, and the arrowheads designate paired segments of AEs); (B) pachytene (the XY bivalent is indicated by arrows); (C) diplotene (the arrows point to connections containing SCP3 and SMC1β between AEs of homologous chromosomes); (D) diakinesis (the arrowheads point to partial splitting of AEs); (E) metaphase I (the arrowheads indicate a weak signal for SCP3 in the chromosome arms, whereas SMC1β is hardly detectable); (F) metaphase II; (H) anaphase II. (G) Enlargement of the area indicated in panel F, with the merged images of SCP3 (blue) and kinetochores (red) (left); SMC1β (green) and kinetochores (red) (middle); and SCP3 (blue), SMC1β (green), and kinetochores (red) (right). (I and J) Enlargements of areas indicated in panel H, with the merged images of SCP3 (blue) and kinetochores (red) (left); SMC1β (green) and kinetochores (red) (middle); and SCP3 (blue), SMC1β (green), and kinetochores (red) (right). (K to M) Images of immunofluorescence triple labeling of SCP3 (blue), SMC1β (green), and SMC1α (red). Shown are a pachytene SC (K), a diplotene SC (L), and a metaphase I bivalent (M) in a dry-down preparation of rat spermatocytes. The tops of panels K to M show the merged images of SCP3 (blue), SMC1α (red), and SMC1β (green); the middles show SCP3 (blue), SMC1β (green), and SMC1α (red); and the bottoms show SCP3 (blue), SMC1β (green), and SMC1α (red). Bars = 10 μm (A to F and H) and 1 μm (G, I, J, and K to M).
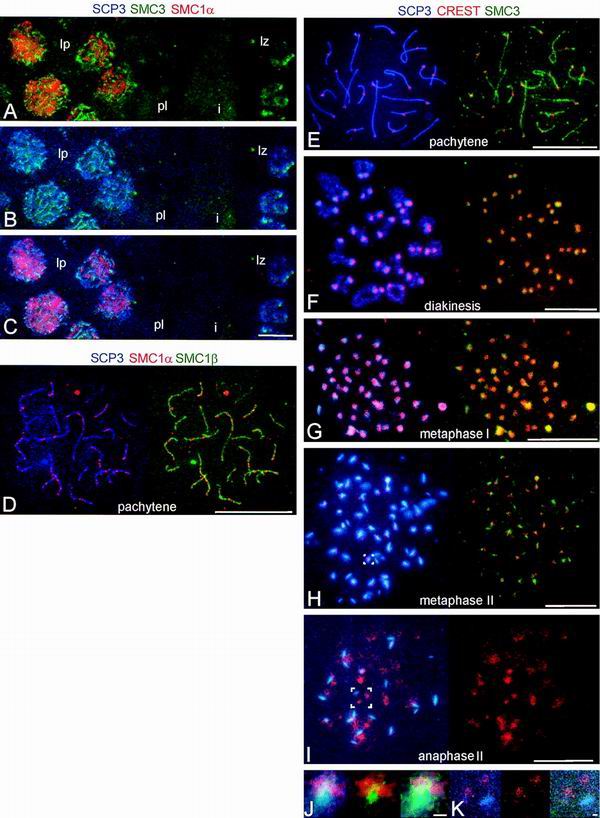
FIG. 7.
Immunolocalization of SMC3 and SMC1α in successive stages of meiosis. (A to C) Images of immunofluorescence triple labeling of SMC1α (red), SMC3 (green), and SCP3 (blue) in a frozen section of rat testis. (A) Merged images of SMC1α (red) and SMC3 (green). (B) Merged images of SCP3 (blue) and SMC1α (red). (C) Merged images of SMC1α (red), SMC3 (green), and SCP3 (blue). The pictures show parts of two testicular tubules. lp, late-pachytene spermatocytes; pl, preleptotene spermatocytes; i, interstitial zone between the two tubules; lz, late-zygotene spermatocytes. (D) Images of immunofluorescence triple labeling of SMC1α (red), SMC3 (green), and SCP3 (blue) in a rat pachytene nucleus, spread by agar filtration; on the left, the merged images of SCP3 (blue) and SMC1α (red) are shown, and on the right, the merged images of SMC3 (green) and SMC1α (red) are shown. (E to K) Images of immunofluorescence triple labeling of SCP3 (blue), SMC3 (green), and kinetochores (red) in dry-down preparations of rat spermatocytes. (E to I) On the left are the merged images of SCP3 (blue) and kinetochores (red), and on the right are SMC3 (green) and kinetochores (red). (J) Enlarged images of the area indicated in H. (K) Enlarged images of the area indicated in I. On the left are the merged images of SCP3 (blue) and kinetochores (red), in the middle are SMC3 (green) and kinetochores (red), and on the right are SCP3 (blue), SMC3 (green), and kinetochores (red). Bars = 10 μm (A to I) and 1 μm (J and K).
The further meiosis I continues, the less SCP3 and SMC1β we find in the chromosome arms, but intense staining remains present at the centromeres. This is obvious in diakinesis (Fig. 6D), metaphase I (Fig. 6E), and up to metaphase II (Fig. 6F). In anaphase II, SMC1β is not visibly associated with the chromosome anymore. Aggregates containing SCP3 can still be seen in anaphase II, but most of these have detached from the kinetochores (Fig. 6H to J). The colocalization of SCP3 and SMC1β is also not perfect in other respects. The SMC1β signal on the AEs has a more granular appearance (though far less dotty than that of SMC1α [13]), and there is a limited SMC1β but no SCP3 signal present on peripheral chromatin loops from leptotene until metaphase II. Moreover, some SCP3, but very little if any SMC1β, persists in the chromosome arms in metaphase I (Fig. 6E).
To compare the chromosomal localization of SMC1β with that of SMC1α, we performed additional immunofluorescence experiments on agar filtrates of rat spermatocytes (13), using anti-SMC1β, anti-SMC1α, and anti-SCP3 (Fig. 6K to M). As described by us earlier (13), SMC1α is present throughout spermatocyte nuclei in frozen sections (Fig. 6A). However, if spermatocytes are lysed in buffers containing Triton X-100 (as is done for agar filtration), or if they are permeabilized in such buffers (as we have done previously with frozen sections [13]), SMC1α is preferentially retained in intensely labeled dots along the AEs. Thus, the localization of SMC1α differs from that of SMC1β, which is almost uniformly distributed along the AEs. Also, the SMC1α dots appear not to be as close to the AEs as the SMC1β staining (Fig. 6K and L). Thus, SMC1β seems to localize most closely to the SC, while SMC1α also appears SC associated, albeit not centered as much towards the AEs. Strikingly, and unlike SMC1β, SMC1α did not accumulate around the centromeres in diakinesis and metaphase I (Fig. 6 M).
We also analyzed the localization of SMC3 in spermatocytes by triple labeling of SMC3, SCP3, and SMC1α, using frozen sections that had not been exposed to Triton X-100 (Fig. 7A to C) or agar filtrates (Fig. 7D), and by triple labeling of SMC3, SCP3, and kinetochores, using dry-down preparations (Fig. 7E to K). Along the AEs, SMC3, like SMC1β, occurred mainly homogeneously distributed on the AEs. From late diplotene up to metaphase II, SMC3 was concentrated at the kinetochores, very similar to the localization seen for SMC1β (Fig. 7F to H and J), whereas SMC3—like SMC1β—was not detectable anymore at the kinetochores in anaphase II (Fig. 7I and K). In SMC1α-SMC3-SCP3 triple labelings of frozen sections, SMC3 does not colocalize with SMC1α, indicating yet another difference between SMC1α and SMC1β. Unexpectedly, we could not demonstrate the tight colocalization of SMC3 with AEs if we labeled the AEs with anti-SCP2 rather than anti-SCP3 (not shown and reference 13). Apparently, the anti-SCP2 antibodies interfere with the immunolabeling of SMC3 in AEs, perhaps implying a close association of SMC3 with SCP2.
Initial analysis of the C-terminal motif.
As noted above, SMC1β carries an unusual, basic C-terminal amino acid sequence of 28 aa that has not been found in any other SMC protein. Analyzing this sequence, we found a nuclear localization signal (NLS) sequence (RKPR [24]), and therefore, the C-terminal motif may contribute to nuclear import of the protein. In addition, the basic pI of the peptide and two consecutive SP motifs (10) renders interaction with DNA likely. We tested both hypotheses.
To test for an NLS function, we cloned the 28-aa motif in frame with the enhanced green fluorescent protein (EGFP) gene into a mammalian expression vector. The construct was transfected into 293 cells, and the intracellular distribution of EGFP was monitored by fluorescence microscopy. For a positive control, we used the EGFP-Nuc protein, a variant of EGFP fused to three copies of the NLS of the simian virus 40 large T antigen (Clontech Inc.), and the unaltered EGFP was used for a negative, cytoplasmic control. Screening several thousand cells, we did not detect EGFP expression in the nucleus (not shown). Thus, the 28-aa motif does not confer NLS activity on EGFP and therefore is not likely to decisively contribute to the nuclear import of SMC1β.
Preliminary evidence for an interaction of the 28-aa peptide with DNA was obtained in filter binding and gel shift (gel retardation) assays. For a positive control in these assays, we used a peptide derived from the Rad54L protein (34); for a negative control, we used a Rad21-derived peptide from a non-DNA binding region (44).
In filter binding assays (43, 53), we used AluI-digested (blunt-ended) plasmid DNA, radioactively labeled at the 5′ ends. The DNA was efficiently retained by the peptide and the Rad54 control peptide but not by the Rad21 control peptide. The amount of DNA retained on the nitrocellulose filters was directly proportional to the amount of peptide used in the assay (not shown).
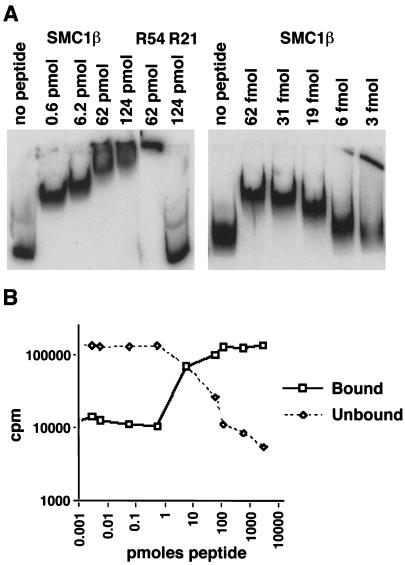
In gel shift experiments, end-labeled DNA fragments, either from M13mp18 or from the 5S rRNA gene, were used (1, 2, 40) and incubated with increasing amounts of the peptide. The results (Fig. 8A) show efficient binding of the SMC1β-C peptide to the DNA substrate. The migration distance from the start position linearly decreased with increasing amounts of peptide added, with no indication of cooperative binding. Surprisingly, almost all DNA was shifted to a higher position even with submolar amounts of peptide (the ratio of peptide to DNA was 1:12 in nucleotide equivalents or 1:2 in DNA molecules). This may indicate binding of one peptide molecule to more than one DNA molecule, i.e., network formation.
FIG. 8.
DNA interaction of the 28-aa SMC1β C-terminal motif. (A) In a gel shift assay, increasing amounts of the 28-aa peptide were incubated with 0.8 pmol of a 5′-32P-labeled 200-bp ribosomal DNA fragment as indicated. R54 and R21, Rad54 and Rad21 control peptides. (B) Assay for protein-DNA network formation. Bound (pelleted) and unbound (supernatant) DNA was measured after incubation of 1.5-pmol of DNA substrate with increasing amounts of the 28-aa peptide.
To further test this hypothesis, we used an assay for nucleoprotein network formation that was used primarily in studies of DNA recombination proteins, such as E. coli RecA or mammalian proteins (7, 17). Nucleoprotein network formation activity was found to be required for homologous DNA pairing (7). Radioactively labeled DNA is incubated with protein, and aggregates are pelleted by centrifugation. The pellet (bound DNA) and supernatant (unbound) are measured in a scintillation counter. The results (Fig. 8B) show network formation activity of the SMC1β peptide. The Rad21 peptide did not yield a signal above background, and the Rad54L peptide was active but eightfold less efficient than the SMC1β-C terminal peptide (not shown). Network formation as seen in this assay starts at a minimal molar ratio of peptide to DNA of 1:1 (in nucleotides). However, to be efficiently pelleted, DNA-protein aggregates of considerable mass have to be created. Therefore, the true minimum ratio required for one peptide molecule to bind several DNA molecules may be significantly lower. Future molecular studies will have to determine the details of the reaction requirements, specificities, and mechanism, as well as the function of the motif in the context of the entire protein or protein complex.
Together, these initial results demonstrate DNA binding activities of the unique SMC1β C-terminal motif.
DISCUSSION
In this report, we demonstrate for the first time the existence of a tissue- and cell-type-specific isoform of a mammalian SMC protein, SMC1β. Specific expression of SMC1β was found in meiotic cells and tissue from several mammalian species, including mouse, rat, and cow. This, and the high degree of evolutionary conservation generally found among SMC proteins, renders SMC1β likely to exist in humans as well. Indeed, the working draft sequence of human chromosome 22 contains a gene highly homologous to the mouse SMC1β gene (GenBank accession number NT011522). This gene was mapped to the region 22q13.31. Currently, only one locus in this region is known to be associated with human disease, i.e., methemoglobin reductase deficiency, which apparently has no direct connection to chromatin dynamics. We could not, however, identify a corresponding gene in the S. cerevisiae genome. While there exist a few open reading frames with low homology, no certain assignment to SMC1β could be made. Likewise, no C. elegans or S. pombe orthologues were detected in the database. Thus, the SMC1 gene may have diversified in higher eukaryotes throughout evolution into a universally and a meiotically expressed isoform. Swapping of SMC proteins in somatic cells has been reported for the gene dosage compensation complex in C. elegans (39), but no SMC protein isoform, and no meiosis-specific SMC protein, has been described.
While up to 70% homologous to SMC1α in the globular domains, the new SMC1β displays a characteristic feature that distinguishes it from SMC1α: the highly basic C-terminal domain of 28 aa. Distinct C-terminal protein sequences, albeit not of such unusual composition, that are specific for meiotic isoforms of somatic proteins have been described, for example, for mammalian DNA ligase III (6). We predicted that this unique C-terminal sequence would contribute important functions to SMC1β. It may, for example, affect interactions of the protein with DNA, or it may act as an NLS. The latter proved to be unlikely, since the peptide on its own did not confer nuclear localization on EGFP. An NLS function may also not be necessary: in SMC1β, there are seven more predicted NLSs distributed all over the protein. The C-terminal peptide, however, interacts with DNA. The basic pI and the presence of two SP motifs, known to constitute DNA binding motifs (e.g., in histone H1 [10]), rendered this likely. Indeed, initial evidence from three independent assays, filter binding, gel shift, and protein-DNA network formation, suggests that the peptide efficiently binds DNA. The gel shift experiments indicated binding of one peptide molecule to more than one DNA molecule, i.e., possible network formation. Preliminary experiments with network formation confirmed the capacity of the peptide molecules to link several DNA molecules, i.e., to form DNA-peptide aggregates. The ability of short peptides to promote networking-related reactions, such as homologous DNA pairing, has been reported, for example, for a 20-aa peptide derived from the E. coli RecA protein (67). A detailed molecular analysis of these DNA interactions, and the demonstration of their significance in vivo, are important subjects for future studies. Together, the unique C-terminal sequence of SMC1β is likely to codetermine the DNA binding properties of the protein.
The new evidence for specialization of mammalian SMC proteins is reminiscent of what has been reported for a limited number of other proteins that are also involved in DNA dynamics. Examples in yeast are the Scc1-type proteins—also collectively called gordin proteins (48)—and Rec8p proteins, as well as the Rad51p and Dmc1p proteins (5, 47, 49, 65, 69). Recently, a meiosis-specific homologue of the Scc3 protein, STAG 3, has been described (52). Whereas Rad51p and MCD1p-Scc1p-RAD21 exist in both somatic and meiotic cells, Dmc1p and Rec8p of yeast and of higher eukaryotes are restricted to meiotic cells (although human Rec8p transcripts were also found in spermatids and the thymus [50]) and postmeiotic cells (47). In meiosis, the relationship between cohesion and recombination is modified, and apparently this is accompanied by replacement of components of the protein complexes involved, like the gordins. Our results suggest that in mammals one of the SMC components of cohesin, SMC1α, is replaced by a meiosis-specific isoform, SMC1β. As had been found for the gordins and Rad51 in yeast, SMC1α is only partially replaced in mammals: SMC1α is present in both somatic and meiotic cells, whereas SMC1β exists only in meiotic cells.
Earlier, we demonstrated the association of SMC1α with meiotic chromatin in rat spermatocytes, and we have shown its presence in preparations of SCs (13). Although SMC1α and SMC1β are coimmunoprecipitated from total testis extracts by anti-SMC3 antibodies, a fraction of SMC1α in spermatocytes does not colocalize with SMC3. Thus, two different SMC3-containing complexes exist in testis. The slightly different behavior of SMC1α-SMC3 and SMC1β-SMC3 in gel filtration, with the latter eluting predominantly at a lower-molecular-mass position around 1 MDa, further indicates two different higher-order complexes of different masses or stabilities that share SMC3 but contain either one of the two SMC1 isoforms. Furthermore, very little SMC1α, but an equimolar amount of SMC1β, is coimmunoprecipitated with SMC3 from late-prophase I spermatocytes. More evidence for this hypothesis originated from chromosomal localization studies.
As for SMC1α, expression of SMC1β is regulated during meiotic cell development, i.e., most protein is observed in prophase I of meiosis. There are, however, important differences in chromosomal localization between SMC1α and SMC1β. While SMC1α is distributed throughout the meiotic prophase nucleus and—upon permeabilization or lysis of cells in the presence of Triton X-100—is preferentially retained in a dotlike pattern along AEs of SCs (reference 13 and this paper), SMC1β is more tightly associated and more uniformly distributed along the AEs. In late prophase I (pachytene-diplotene), SMC1β also associates with bridges between the AEs of homologues, which possibly represent the sites of crossover. This has not been observed for SMC1α. In addition, the association of SMC1β with the AEs of SCs seems to be even closer than that of SMC1α with the SC. Thus, there may be a structural organization in layers, with SMC1β constituting the inner and SMC1α an outer SMC-containing layer at the SC. Finally, in diplotene and diakinesis, SMC1β and SMC3 accumulate around the centromeres, where the proteins persist until anaphase II, whereas SMC1α does not concentrate at the centromeres in any stage of meiosis. This strongly suggests that SMC1β, and not SMC1α, is involved with SMC3 in the maintenance of centromere cohesion during the first meiotic division in mammals. In S. cerevisiae, a similar conclusion was reached for another meiosis-specific component of cohesin, Rec8 (36). SCP3 (reference 12 and this paper) and SCP2 (56) also accumulate at the centromeres from late diplotene until anaphase II. It remains to be investigated whether these SC proteins contribute directly to maintenance of centromere cohesion. Most likely, the dissociation of SMC1α and SMC1β from the chromosome arms in late prophase contributes to the release of sister chromatid arm cohesion, while centromeric cohesion is further supported by SMC1β.
In addition, we observed that preparations of nuclear extracts and total cell lysates, especially those from purified pachytene-diplotene spermatocytes (22), contained a characteristic 85-kDa degradation product of SMC1β but no SMC1α or SMC3 degradation products (not shown). Northern blotting showed the same 4.5-kb transcript in RNA from purified spermatocytes as in testis RNA, rendering alternative splicing unlikely. Thus, SMC1β appears to be more sensitive to proteolysis than the other SMC proteins. One may speculate that such proteolysis may be required for the release of arm cohesion, similar to degradation of Rec8. The SMC1β degradation product does not coprecipitate with SMC3, indicating that the 85-kDa fragment of SMC1β is not present in a complex with SMC3. Alternatively, it may also be cleaved quickly after synthesis and thus prevented from associating with SMC3. The nature of the protease that cleaves SMC1β, and whether such cleavage is necessary for meiotic progression, are among the questions now to be addressed.
In summary, we propose the existence of two multiprotein complexes in meiotic cells that are based on two different SMC1-SMC3 cores: SMC1α-SMC3 and SMC1β-SMC3. Both complexes associate with meiotic chromatin and should contribute to meiotic sister chromatid cohesion. The “α-complex,” however, appears more loosely chromosome associated, in a punctate pattern, and also in the chromatin loops. This complex dissociates and is released from the chromatin in late prophase I. The “β-complex” closely localizes to the SC and remains chromosome associated at the centromeres beyond prophase I until metaphase-anaphase II. Therefore, the β-complex, and not the α-complex, is likely responsible for centromeric cohesion until anaphase II. The interplay among meiotic sister chromatid cohesion, DNA recombination, and the new meiosis-specific SMC1β is now amenable to future studies.
ACKNOWLEDGMENTS
We thank Mirjam van Aalderen for expert technical assistance and David Avila for protein sequencing. E.R. was supported by a grant from the Human Frontier of Science Foundation and by NIH grant GM62517. M.E. was financially supported by grant no. 901-01-097 of The Netherlands Society for Scientific Research (NWO).
An initial part of this work was done at the Basel Institute for Immunology, Basel, Switzerland.
REFERENCES
- 1.Akhmedov A T, Frei M, Tsai-Pflugfelder C, Kemper B, Gasser S M, Jessberger R. Structural maintenance of chromosomes: protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- 2.Akhmedov A T, Gross B, Jessberger R. Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends, and prevent DNA bending. J Biol Chem. 1999;274:38216–38224. doi: 10.1074/jbc.274.53.38216. [DOI] [PubMed] [Google Scholar]
- 3.Bayne M L, Alexander R F, Benbow R M. DNA binding protein from ovaries of the frog Xenopus laevis which promotes concatenation of linear DNA. J Mol Biol. 1984;172:87–108. doi: 10.1016/0022-2836(84)90416-9. [DOI] [PubMed] [Google Scholar]
- 4.Birkenbihl R P, Subramani S. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Tomkinson A E, Ramos W, Mackey Z R, Danehower S, Walter C A, Schultz R A, Besterman J M, Husain I. Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol Cell Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow S A, Radding C M. Ionic inhibition of formation of RecA nucleoprotein networks blocks homologous pairing. Proc Natl Acad Sci USA. 1985;82:5646–5650. doi: 10.1073/pnas.82.17.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang P T, Lieb J D, Meyer B. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–1738. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- 9.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churchill M E, Suzuki M. SPKK motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989;8:4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobbe N, Heck M M S. SMCs in the world of chromosome biology—from prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- 12.Dobson M, Pearlman R E, Karaiskakis A, Spyropoulos B, Moens P B. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- 13.Eijpe M, Heyting C, Gross B, Jessberger R. Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J Cell Sci. 2000;113:673–682. doi: 10.1242/jcs.113.4.673. [DOI] [PubMed] [Google Scholar]
- 14.Eijpe M, Offenberg H, Goedecke W, Heyting C. Localisation of RAD50 and MRE11 in spermatocyte nuclei of mouse and rat. Chromosoma. 2000;109:123–132. doi: 10.1007/s004120050420. [DOI] [PubMed] [Google Scholar]
- 15.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fousteri M I, Lehmann A R. A novel SMC protein complex in Schizosaccharomyces pombecontains the Rad18 DNA repair protein. EMBO J. 2000;19:1691–1702. doi: 10.1093/emboj/19.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganea D, Moore P, Chekuri L, Kucherlapati R. Characterization of an ATP-dependent DNA strand transferase from human cells. Mol Cell Biol. 1987;7:3124–3130. doi: 10.1128/mcb.7.9.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossenbacher-Grunder A M, Thuriaux O. Spontaneous and UV-induced recombination in radiation-sensitive mutants of Schizosaccharomyces pombe. Mutat Res. 1981;81:37–48. doi: 10.1016/0027-5107(81)90085-3. [DOI] [PubMed] [Google Scholar]
- 19.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–58. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawley R S, Arbel T. Yeast genetics and the fall of the classical view of meiosis. Cell. 1993;72:301–303. doi: 10.1016/0092-8674(93)90108-3. [DOI] [PubMed] [Google Scholar]
- 21.Heck M M S. Condensins, cohesins, and chromosome architecture: how to make and break a mitotic chromosome. Cell. 1997;91:5–8. doi: 10.1016/s0092-8674(01)80002-7. [DOI] [PubMed] [Google Scholar]
- 22.Heyting C, Dietrich A J J. Meiotic chromosome preparation and labeling. Methods Cell Biol. 1991;35:177–202. doi: 10.1016/s0091-679x(08)60573-7. [DOI] [PubMed] [Google Scholar]
- 23.Heyting C. Synaptonemal complexes: structure and function. Curr Opin Cell Biol. 1996;8:389–396. doi: 10.1016/s0955-0674(96)80015-9. [DOI] [PubMed] [Google Scholar]
- 24.Hicks G R, Raikhel N V. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 25.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophilabarren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 26.Hirano T. SMC protein complexes and higher-order chromosome dynamics. Curr Opin Cell Biol. 1998;10:317–322. doi: 10.1016/s0955-0674(98)80006-9. [DOI] [PubMed] [Google Scholar]
- 27.Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 28.Hopfner K P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 29.Jessberger R, Podust V, Hübscher U, Berg P. A mammalian protein complex that repairs double-strand breaks and deletions by recombination. J Biol Chem. 1993;268:15070–15079. [PubMed] [Google Scholar]
- 30.Jessberger R, Riwar B, Baechtold H, Akhmedov A T. SMC proteins constitute two subunits of the mammalian recombination protein complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- 31.Jessberger R, Frei C, Gasser S M. Chromosome dynamics: the SMC protein family. Curr Opin Genet Dev. 1998;8:254–259. doi: 10.1016/s0959-437x(98)80149-4. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Sgouros J. The cohesin complex: sequence homologies, interaction networks and shared motifs. Genome Biol. 2001;2:0009.1–0009.12. doi: 10.1186/gb-2001-2-3-research0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadyk L C, Hartwell L H. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics. 1993;133:469–489. doi: 10.1093/genetics/133.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanaar R, Troelstra C, Swagemakers S M, Essers J, Smit B, Franssen J H, Pastink A, Bezzubova O Y, Buerstedde J M, Clever B, Heyer W D, Hoeijmakers J H J. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr Biol. 1996;6:828–830. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- 35.Kleckner N. Meiosis: how could it work? Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein F, Mahr P, Galova M, Buonomo S B C, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 37.Lammers J H, Offenberg H H, van Aalderen M, Vink A C R, Dietrich A J J, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lammers J H M, van Aalderen M, Peters A H F M, van Pelt A A M, Gaemers I C, de Rooij D G, de Boer P, Offenberg H H, Dietrich A J J, Heyting C. A change in the phosphorylation pattern of the 30,000–33,000 Mr synaptonemal complex proteins of the rat between early and mid-pachytene. Chromosoma. 1995;104:154–163. doi: 10.1007/BF00352179. [DOI] [PubMed] [Google Scholar]
- 39.Lieb J D, Albrecht M R, Chuang P T, Meyer B J. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:1–20. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 40.Logie C, Peterson C L. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopusand human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McEntee K, Weinstock G M, Lehman I R. RecA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci USA. 1998;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKay M J, Troelstra C, van der Spek P, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers J H. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- 45.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–46. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 46.Moens P B, Heyting C, Dietrich A J, van Raamsdonk W, Chen Q. Synaptonemal complex antigen location and conservation. J Cell Biol. 1987;105:93–103. doi: 10.1083/jcb.105.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molnar M, Bähler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:167–187. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nasmyth K, Peters J-M, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1384. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 49.Offenberg H H, Schalk J A C, Meuwissen R L J, van Aalderen M, Kester H A, Dietrich A J J, Heyting C. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 1998;26:2572–2579. doi: 10.1093/nar/26.11.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parisi S, McKay M J, Molnar M, Thompson M A, van der Spek P J, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers J H, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters A H, Plug A W, van Vugt M J, de Boer P A. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 52.Pezzi N, Prieto I, Kremer L, Perez Jurado L A, Valero C, del Mazo J, Martinez C, Barbero J L. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking Williams-Beuren syndrome deletion. FASEB J. 2000;14:581–592. doi: 10.1096/fasebj.14.3.581. [DOI] [PubMed] [Google Scholar]
- 53.Potter H, Dressler D. Biochemical assay designed to detect formation of recombination intermediates in vitro. Proc Natl Acad Sci USA. 1979;76:1084–1088. doi: 10.1073/pnas.76.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Schalk J A C, Dietrich A J J, Vink A C G, Offenberg H H, van Aalderen M, Heyting C. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma. 1998;107:540–548. doi: 10.1007/s004120050340. [DOI] [PubMed] [Google Scholar]
- 57.Schmiesing J A, Gregson H C, Zhou S, Yokomori K. A human condensin complex containing hCAP-C–hCAP-E and CNAP1: a homolog of XenopusXCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol Cell Biol. 2000;20:6696–6700. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 59.Speed R M. Meiosis in the foetal mouse ovary. I. An analysis at the light microscope level using surface-spreading. Chromosoma. 1982;85:427–437. doi: 10.1007/BF00330366. [DOI] [PubMed] [Google Scholar]
- 60.Strunnikov A V. SMC proteins and chromosome structure. Trends Cell Biol. 1998;8:454–459. doi: 10.1016/s0962-8924(98)01370-1. [DOI] [PubMed] [Google Scholar]
- 61.Strunnikov A V, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 62.Strunnikov A V, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 63.Stursberg S, Riwar B, Jessberger R. Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complex RC-1. Gene. 1999;228:1–12. doi: 10.1016/s0378-1119(99)00021-9. [DOI] [PubMed] [Google Scholar]
- 64.Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- 65.Tarsounas M, Morita T, Pearlman R E, Moens P. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol. 1999;147:207–219. doi: 10.1083/jcb.147.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Heemst D, Heyting C. Sister chromatid cohesion and recombination in meiosis. Chromosoma. 2000;109:10–26. doi: 10.1007/s004120050408. [DOI] [PubMed] [Google Scholar]
- 67.Voloshin O, Wang L, Camerini-Otero R D. Homologous DNA pairing promoted by a 20-amino acid peptide derived from RecA. Science. 1996;272:868–872. doi: 10.1126/science.272.5263.868. [DOI] [PubMed] [Google Scholar]
- 68.Walker J E, Sarasate M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]