Abstract
Background and aim:
Gene polymorphism, coding the host proteases, which are involved in the virus entry into the cells can influence the susceptibility to and mortality from coronavirus disease 19 (COVID-19). Angiotensin-converting enzyme 2 (ACE2), transmembrane serine 2 and serine 11A proteases (TMPRSS2, TMPRSS11A), and a cell surface cluster of differentiation 147 (CD147) might be a gene candidate that exerts such influence. The aim of this study was to investigate the associations between ace2, tmprss2, tmprss11a, and cd147 polymorphic variants and the severity of COVID-19 in the Ukrainian population.
Methods:
The study population consisted of the Ukrainian population with COVID-19: patients without oxygen therapy (n=62), with non-invasive (n=92) and invasive (n=35) oxygen therapy, as well as control subjects (n=92). Allelic polymorphisms of ace2 rs4240157, tmprss2 rs12329760, and tmprss11a rs353163 were determined by real-time PCR, and cd147 rs8259 polymorphism was detected by PCR with subsequent restrictase analysis. We compared investigated polymorphisms distribution with other populations by meta-analysis.
Results:
Our study is the first to obtain data about the distribution of investigated gene polymorphisms in the Ukrainian population: tmprss2 rs12329760 – CC 60.9%, CT 35.9%, TT 3.2%; tmprss11a rs353163 – CC 46.7%, CT 40.2%, TT 13.1%; ace2 rs4240157 – CC 7.6%, C 18.5%, CT 22.8%, TT 19.6%, T 31.5%; cd147 rs8259 – TT 60.9%, AT 32.6%, AA 6.5%. This distribution was similar to the Northern, Western and Southern European populations. There was a statistically significant difference in the frequency of tmprss2 polymorphic genotypes CC 57.1%, CT 28.6%, and TT 14.3% (P<0.05) in COVID-19 patients with invasive oxygen therapy in comparison with non-invasive oxygen therapy. This tmprss2 mutation occurs in the scavenger receptor cysteine-rich (SRCR) domain and might be important for protein-protein interaction in a calcium-dependent manner.
Conclusions:
Our study indicated the presence of an association between the tmprss2 rs12329760 polymorphism and the severity of COVID-19 in the Ukrainian population. (www.actabiomedica.it)
Keywords: transmembrane serine proteases, TMPRSS2, TMPRSS11A, CD147, angiotensin-converting enzyme 2, polymorphism, COVID-19, oxygen therapy
1. Introduction
Coronavirus disease 2019 (COVID-19) is the second pandemic of the twenty-first century, with over one hundred million infections and over two million deaths to date. It is a novel strain from the Coronaviridae family, named Severe Acute Respiratory Distress Syndrome Coronavirus-2 (SARS-CoV-2). Transmission of SARS-CoV-2 is mainly via respiratory droplets, either directly from the air when an infected patient coughs or sneezes, or in the form of fomites on surfaces (1). The host genetic variability might determine individual susceptibility to SARS-CoV-2 and the course of COVID-19 (2-5). Recently, some attempts have been made to predict gene targets in polymorphism-associated studies on COVID-19 (6-9).
SARS-CoV-2 threatens target cells using SARS spike proteins (S-proteins), which bind to angiotensin (AT) – converting enzyme 2 (ACE2). It also uses transmembrane protease, serine 2 (TMPRSS2), transmembrane protease, serine 11A (TMPRSS11A), a cell surface cluster of differentiation 147 (CD147) for cell entry and for viral proliferation in host cells (10-13). The target proteins have polymorphic variants and might influence the effectivity of virus cell entry and virus proliferation.
The aim of this study was to elaborate on the associations between the polymorphic variants of ace2 (rs4240157), tmprss2 (rs12329760), tmprss11a (rs353163), cd147 (rs8259) and the severity of COVID-19 in the Ukrainian population.
2. Patients and methods
2.1 Study population
An observational analytic study with a case-control design was conducted at outpatient clinics and inpatient wards in the clinical settings of Poltava State Medical University, Ukraine, from August 2020 through February 2021. Written informed consent was obtained from all recruited patients. The study was approved by the local Ethics Committee of Poltava State Medical University, Ukraine.
The study population consisted of Ukrainian COVID-19 patients who had resided in the Poltava region (central part of Ukraine). The inclusion criteria for the study group were subjects with clinical signs and symptoms of COVID-19: i) endotracheal intubation, ii) CPAP/BiPAP ventilation, iii) oxygen therapy, iv) hospitalized without oxygen therapy, v) not hospitalized, and positive results of SARS-CoV-2 PCR tests in nasopharyngeal swabs (14).
All COVID-19 patients were divided into three clinical groups in accordance with oxygen requirement: group 1 – patients without oxygen therapy (n=62), group 2 – patients with non-invasive oxygen therapy (n=92), group 3 – patients with lung ventilation (invasive oxygen therapy) including 8 patients who died (n=35). We allocated such groups in order to be able to compare our data with previously published results on the gene polymorphisms associations with COVID-19 severity. The control group comprised 92 healthy persons, without a history of fever or respiratory symptoms who had a negative SARS-CoV-2 IgA+IgM+IgG serology and lived in Central Ukraine.
2.2 Genetic analysis
Venous blood was drawn and placed into tubes containing EDTA (BD Vacutainer® К2Е, Becton Dickinson, USA). Genomic DNA was isolated by DNeasy Blood&Tissue Kit using automatics station QIAcube Connect (QIAGEN, Germany).
The quality and integrity of DNA were checked by MAESTRONano (MAESTROGEN, Taiwan). All DNA samples were screened for purity by measuring optical density (OD) at 260 nm and 280 nm. The ratios OD 260/280 nm ranged from 1.83-1.99 indicating good quality DNA.
Allelic polymorphisms of ace2 C>T rs4240157, tmprss2 C>T rs12329760 and tmprss11a C>T rs353163 were determined by real-time polymerase chain reaction (PCR) methods and CFX96™ Real-Time PCR Detection System (Bio-Rad, USA), using Taqman® SNP Genotyping Assays (C_228018196_20, C_25622353_20 and C_1044674_10, respectively).
Allelic polymorphism of cd147 T>A rs8259 was detected on the same thermal cycler with specific primers: forward: 5’-GAGTCCACTCCCAGTGCTTG-3’; reverse: 5’-CTCGTGAAACACTTCAGAAGGAAAAGA-3’. After digesting with restriction enzyme MboI (Thermo Fisher Scientific, USA) for 2 h at 37 °C, the PCR digest was detected on the automatic capillary electrophoresis system QIAxcel Advanced (QIAGEN, Germany) with QIAxcel DNA High-Resolution Kit. The restriction enzyme digested the 162 bp PCR products into 2 fragments: 137 and 25 bp for the rs8259 A allele, whereas the uncut PCR products (162 bp) represent the T allele (15). The relevant image of representative capillary gel-electrophoresis is shown in Fig. 1.
Figure 1.
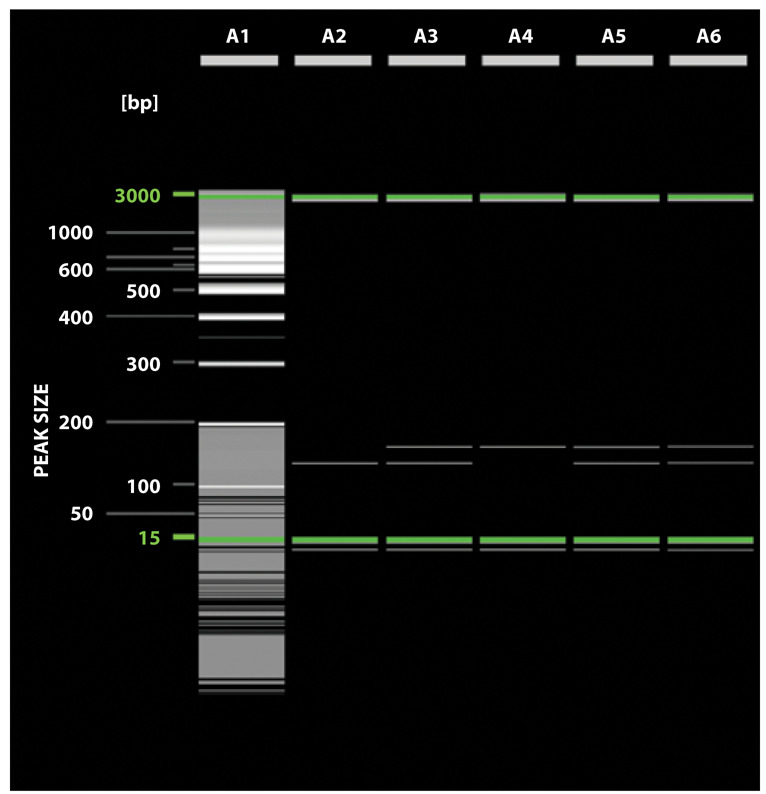
The electrophoresis result of PCR Mbol products of rs8259 cd147 polymorphism: line 1 – molecular marker 100 bp + 50 bp SibEnzyme VSLLC, line 2 – the AA genotype (137 bp, 25 bp), line 3, 5, 6 – the AT genotype (162 bp, 137 bp, and 25 bp), line 4 – the TT genotype (162 bp).
2.3 Methodology of meta-analysis
Because the recent data on the association between gene polymorphisms of our interest and COVID-19 are limited, meta-analysis was provided to: i) reveal the distribution of investigated gene polymorphisms between the Ukrainian population and other populations with these known polymorphisms, ii) to search for investigated genes polymorphisms in populations with already described gene polymorphisms’ associations with COVID-19, iii) to define the scales of COVID-19 severity used in published data of studies on the association of gene polymorphisms of our interest.
We searched PubMed, Embase, Scopus, Cochrane Library as well as Google Scholar for relevant articles. Searches of electronic databases were conducted using the terms “ACE2”, “TMPRSS2”, “TMPRSS11A”, “CD147”, “COVID-19”, “SARS-CoV-2”, and “polymorphism”. The selected articles were analyzed to contain primary data, inclusion and exclusion criteria, number of patients, demography, the proportion of gene alleles, and genotypes. The extracted data included the study design, patient data, polymorphism frequencies, and statistical analysis. Three reviewers independently determined the study eligibility and extracted the data. Since we included fewer than 10 studies for each gene, we did not use a funnel plot to assess the publication bias.
2.4 Statistical analysis
The tmprss2, tmprss11a, and cd147 (rs8259) polymorphisms were analyzed for deviation of Hardy-Weinberg equilibrium by Pearson’s χ2. Because ace2 is located on the X chromosome, this equilibrium was assessed only for females. The expected and observed heterozygosity was calculated by POPGENE (Version 1.32). Fisher’s exact test or χ2 test with Yates’s correction was used to compare the genotypic and allelic frequencies between controls and patients. The association between polymorphic gene variants and disease susceptibility / severity was assessed by calculating the odds ratio with a 95% confidential interval (CI). The analysis and calculation of descriptive statistics indicators were carried out using One-Way ANOVA & post-hoc Bonferroni test for unpaired samples using the GraphPad PRISM® (Version 5.03) statistical software package (GraphPad Software, San Diego, USA). P-values of less than 0.05 were considered to be significant.
3. Results
3.1 Characteristics of the studied population
Demographic characteristics of the entire studied population are shown in Table 1.
Table 1.
Characteristics of the studied population.
| Variables | Control subjects, n=92 | COVID-19 patients | ||
|---|---|---|---|---|
| Without oxygen therapy, n=62 | Non-invasive oxygen therapy, n=92 | Invasive oxygen therapy, n=35 | ||
|
Sex, n (%)
- Male - Female |
46 (50.0) 46 (50.0) |
25 (40.3) 37 (59.7) |
36 (39.1) 56 (60.9) |
15 (42.9) 20 (57.1) |
|
Age, years
mean (SD) |
40.36 (6.84) | 53.34 (12.79) P<0.05 |
62.55 (12.39) P<0.05 P1<0.05 |
63.71 (10.76) P<0.05 P1<0.05 |
|
BMI, kg/m2
mean (SD) |
29.25 (1.61) | 29.81 (2.08) | 30.07 (2.04) P<0.05 |
31.06 (2.12) P<0.05 P1<0.05 |
Note: P – comparison with control subjects; P1 – comparison with COVID-19 patients without oxygen therapy.
There were no significant differences in terms of sex between all study groups. The COVID-19 patients had significantly higher ages in comparison with control subjects, and COVID-19 patients with non-invasive and invasive oxygen therapy had higher ages than those without oxygen therapy. They also tend to have significantly higher body mass index (BMI) than control subjects. In addition, patients with invasive oxygen therapy had a higher BMI in contrast to patients without oxygen therapy (P<0.05).
3.2 Frequencies of tmprss2 rs12329760, tmprss11a rs353163, ace2 rs4240157, and cd147 rs8259 polymorphisms in the studied populations
The observed tmprss2, tmprss11a, ace2, and cd147 genotype distributions were not significantly different from those in the Hardy-Weinberg equilibrium in all studied populations.
There was a statistically significant difference in the frequency of the tmprss2 polymorphism, i.e., CC, CT, and TT genotypes, in the group of COVID-19 patients with invasive oxygen therapy (P=0.03) in contrast to the group with non-invasive oxygen therapy. There were no significant differences in the frequencies of tmprss11a, ace2, and cd147 polymorphisms between the groups of COVID-19 patients and control subjects (Table 2).
Table 2.
Allele frequency and genotype distribution of the tmprss2 rs12329760, tmprss11a rs353163, ace2 rs4240157, and cd147 rs8259 polymorphisms, n (%).
| Control subjects, n=92 | COVID-19 patients | |||
|---|---|---|---|---|
| Without oxygen therapy, n=62 | Non-invasive oxygen therapy, n=92 | Invasive oxygen therapy, n=35 | ||
| tmprss2 rs12329760 | ||||
| Genotype Distribution, n (%) | ||||
| CC | 56 (60.9) | 38 (61.3) | 52 (56.5) | 20 (57.1) |
| CT | 33 (35.9) | 20 (32.3) | 38 (41.3) | 10 (28.6) |
| TT | 3 (3.2) | 4 (6.4) | 2 (2.2) | 5 (14.3) |
| P<0.05 | ||||
| CT+TT | 36 (39.1) | 24 (38.7) | 40 (43.5) | 15 (42.9) |
| Allele Frequency, n (%) | 50 (71.4) | |||
| C | 145 (78.8) | 96 (77.4) | 142 (77.2) | 20 (28.6) |
| T | 39 (21.2) | 28 (22.6) | 42 (22.8) | |
| tmprss11a rs353163 | ||||
| Genotype Distribution, n (%) | ||||
| CC | 43 (46.7) | 26 (41.9) | 44 (47.8) | 16 (45.7) |
| CT | 37 (40.2) | 29 (46.8) | 41 (44.6) | 16 (45.7) |
| TT | 12 (13.1) | 7 (11.3) | 7 (7.6) | 3 (8.6) |
| CT+TT | 49 (53.3) | 36 (58.1) | 48 (52.2) | 19 (54.3) |
| Allele Frequency, n (%) | ||||
| C | 123 (66.8) | 81 (65.3) | 129 (70.1) | 48 (68.6) |
| T | 61 (33.2) | 43 (34.7) | 55 (29.9) | 22 (31.4) |
| ace2 rs4240157 | ||||
| Genotype Distribution, n (%) | ||||
| CC | 7 (7.6) | 4 (6.5) | 8 (8.7) | 2 (5.7) |
| C− | 17 (18.5) | 10 (16.1) | 15 (16.3) | 5 (14.3) |
| CT | 21 (22.8) | 22 (35.5) | 22 (23.9) | 10 (28.6) |
| TT | 18 (19.6) | 11 (17.7) | 26 (28.3) | 10 (28.6) |
| T− | 29 (31.5) | 15 (24.2) | 21 (22.8) | 8 (22.8) |
| CT+TT+T− | 68 (73.9) | 48 (77.4) | 69 (75.0) | 28 (80.0) |
| Allele Frequency, n (%) | ||||
| C | 52 (37.7) | 40 (40.4) | 53 (35.8) | 19 (33.3) |
| T | 86 (62.3) | 59 (59.6) | 95 (64.2) | 38 (66.7) |
| cd147 rs8259 | ||||
| Genotype Distribution, n (%) | ||||
| TT | 56 (60.9) | 36 (58.1) | 52 (56.5) | 22 (62.9) |
| AT | 30 (32.6) | 23 (37.1) | 36 (39.1) | 11 (31.4) |
| AA | 6 (6.5) | 3 (4.8) | 4 (4.4) | 2 (5.7) |
| AT+AA | 36 (39.1) | 26 (41.9) | 40 (43.5) | 13 (37.1) |
| Allele Frequency, n (%) | ||||
| T | 142 (77.2) | 95 (76.6) | 140 (76.1) | 55 (78.6) |
| A | 42 (22.8) | 29 (23.4) | 44 (23.9) | 15 (21.4) |
Note: P – comparison with non-invasive oxygen therapy.
3.3 Meta-analysis of tmprss2 rs12329760, tmprss11a rs353163, ace2 rs4240157, and cd147 rs8259 polymorphisms in the Ukrainian and non-Ukrainian populations with COVID-19
At the first stage, our searches of the allele frequencies and the genotype distributions of tmprss2, tmprss11a, ace2, and cd147 in the Ukrainian and non-Ukrainian populations identified 24 non-duplicate citations of which 15 were deemed eligible for inclusion based on their titles and abstracts. Eight studies were subsequently excluded because they were critical appraisal articles (Fig. 2).
Figure 2.

The Preferred Reporting Items for Meta-Analysis flow diagram.
Finally, 7 studies involving 3217 subjects from the Polish, Northern and Western European, Iberian (Spain), Tuscany (Italy), North Indian, Iranian, Jiangsu Chinese, Han Chinese, and Chinese populations were included in the analysis of investigated polymorphisms distribution. The choice of populations depended on geographic proximity or the availability of population-specific COVID-polymorphism association studies. The comparison of allele frequencies and genotype distribution of tmprss2, tmprss11a, ace2, and cd147 between The Ukrainian and other populations is shown in Table 3.
Table 3.
Allele frequency and genotype distribution of the polymorphisms of tmprss2 rs12329760, tmprss11a rs353163, ace2 rs4240157, cd147 rs8259 in different populations, n (%).
| Population | Genotypes and alleles frequencies | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tmprss2 rs12329760 | ||||||||||
| CC | CT | TT | CT+TT | C | T | |||||
| Ukrainian population, n=92 |
56 (60.9) | 33 (35.9) | 3 (3.2) | 36 (39.1) | 145 (78.8) | 39 (21.2) | ||||
| Northern and Western European population, n=99 (16) |
56 (56.6) | 40 (40.4) | 3 (3.0) | 43 (43.4) | 152 (76.8) | 46 (23.2) | ||||
| Iberian population in Spain, n=107 (16) |
73 (68.2) | 30 (28.0) | 4 (3.7) | 34 (31.8) | 176 (82.2) | 38 (17.8) | ||||
| Tuscany population in Italy, n=107 (16) |
72 (67.3) | 27 (25.2) | 8 (7.5) | 35 (32.7) | 171 (79.9) | 43 (20.1) | ||||
| P1<0.05 | ||||||||||
| tmprss11a rs353163 | ||||||||||
| Ukrainian population, n=92 |
43 (46.7) | 37 (40.2) | 12 (13.1) | 49 (53.3) | 123 (66.8) | 61 (33.2) | ||||
| North Indian population, n=310 (17) |
132 (42.6) | 151 (48.7) | 27 (8.7) | 178 (57.4) | 415 (66.9) | 205 (33.1) | ||||
| Iranian population, n=250 (18) |
130 (52.0) | 101 (40.4) | 19 (7.6) | 120 (48.0) | 361 (72.2) | 139 (27.8) | ||||
| Northern and Western European population, n=99 (19) |
42 (42.4) | 43 (43.4) | 14 (14.1) | 57 (57.6) | 127 (64.1) | 71 (35.9) | ||||
| Iberian population in Spain, n=107 (19) |
44 (41.1) | 46 (43.0) | 17 (15.9) | 63 (58.9) | 134 (62.6) | 80 (37.4) | ||||
| P2<0.05 | ||||||||||
| ace2 rs4240157 | ||||||||||
| CC | C− | CT | TT | T− | CT+TT+T− | C | T | |||
| Ukrainian population, n=92 |
7 (7.6) | 17 (18.5) | 21 (22.8) | 18 (19.6) | 29 (31.5) | 68 (73.9) | 52 (37.7) | 86 (62.3) | ||
| Northern and Western European population, n=99 (20) |
11 (11.1) | 11 (11.1) | 24 (24.2) | 15 (15.2) | 38 (38.4) | 77 (77.8) | 57 (38.3) | 92 (61.7) | ||
| Iberian population in Spain, n=107 (20) |
10 (9.3) | 17 (15.9) | 19 (17.8) | 24 (22.4) | 37 (34.6) | 80 (74.8) | 56 (35.0) | 104 (65.0) | ||
| cd147 rs8259 | ||||||||||
| TT | AT | AA | AT+AA | T | A | |||||
| Ukrainian population, n=92 |
56 (60.9) | 30 (32.6) | 6 (6.5) | 36 (39.1) | 142 (77.2) | 42 (22.8) | ||||
| Polish population, n=135 (21) |
78 (57.8) | 49 (36.3) | 8 (5.9) | 57 (42.2) | 205 (75.9) | 65 (24.1) | ||||
| Jiangsu Chinese population, n=851 (15) |
109 (12.8) | 406 (47.7) | 336 (39.5) | 742 (87.2) | 624 (36.7) | 1078 (63.3) | ||||
|
P1<0.001
P3<0.001 P4<0.001 |
P1<0.001
P3<0.001 P4<0.001 |
P1<0.001
P3<0.001 P4<0.001 |
||||||||
| Chinese population, n=1107 (22) |
156 (14.1) | 529 (47.8) | 422 (38.1) | 951 (85.9) | 841 (38.0) | 1373 (62.0) | ||||
|
P1<0.001
P3<0.001 P4<0.001 |
P1<0.001
P3<0.001 P4<0.001 |
P1<0.001
P3<0.001 P4<0.001 |
||||||||
| Han Chinese population, n=251 (23) |
106 (42.2) | 114 (45.4) | 31 (12.4) | 145 (57.8) | 326 (64.9) | 176 (35.1) | ||||
|
P1<0.05 P3=0.008 P4=0.008 P5<0.001 P6<0.001 |
P1=0.008 P3<0.001 P4<0.001 P5<0.001 P6<0.001 |
P1=0.003 P3<0.001 P4<0.001 P5<0.001 P6<0.001 |
||||||||
| Northern and Western European population, n=99 (24) |
58 (58.6) | 36 (36.4) | 5 (5.0) | 41 (41.4) | 152 (76.8) | 46 (23.2) | ||||
| Iberian population in Spain, n=107 (24) |
50 (46.7) | 43 (40.2) | 14 (13.1) | 57 (53.3) | 143 (66.8) | 71 (33.2) | ||||
|
P5<0.001
P6<0.001 |
P5<0.001
P6<0.001 |
P1<0.05
P3<0.05 P4<0.05 P5<0.001 P6<0.001 |
||||||||
Note: P1 − comparison with the Northern and Western European population, P2 − comparison with the Iranian population, P3 − comparison with the Ukrainian population, P4 − comparison with the Polish population, P5 − comparison with the Jiangsu Chinese population, P6 − comparison with the Chinese population, P7 − comparison with the Han Chinese population. We used population names as given in 1000 Genomes Project Phase 3 (16).
There were no differences in the frequencies of tmprss2 polymorphism between the Ukrainian and Northern and Western European populations or the Iberian population in Spain. There was a significant difference in the frequency of tmprss2 polymorphism between the Ukrainian population and the Tuscany population in Italy. Similarly, the Ukrainian population had statistically significant differences in the frequency of tmprss11a polymorphism from the Iberian population and no differences from the North Indian, Iranian, Northern and Western European populations.
For ace2 rs4240157, there were no differences between the Ukrainian population and that of the Northern and Western Europe as well as the Iberian population in Spain.
The Ukrainian population had no differences in the frequencies of cd147 polymorphisms from the Polish, Iberian in Spain, Northern and Western European populations. At the same time, there were statistically significant differences in the frequencies of cd147 polymorphisms in the Ukrainian population and Chinese populations, where the A allele was prevalent with the exception of Han Chinese populations.
At the next step of the study, we identified 8 non-duplicate citations with the data of allele frequencies and genotype distribution of tmprss2, tmprss11a, ace2, and cd147 in the Ukrainian and non-Ukrainian populations of patients with COVID-19. In recent publications, there were no data about the association of tmprss11a rs353163 and cd147 rs8259 polymorphisms with COVID-19 severity. 4 studies were subsequently excluded because they were critical appraisal articles, and 4 studies of the Indonesian, German, and Saudi Arabian populations were included in the analysis of tmprss2 rs12329760 and, ace2 rs4240157 polymorphisms distributions. We used the data of appropriate population controls where it was needed (Table 4). We also agreed on the data on the severity of COVID-19 among all the cited sources.
Table 4.
Allele frequency and genotype distribution of the polymorphisms of tmprss2 rs12329760, ace2 rs4240157 in different populations of patients with COVID-19, n (%).
| Population | Genotype, allele | Population control / Control subjects | Asymptomatic | Mild | Moderate | Severe |
|---|---|---|---|---|---|---|
| tmprss2 rs12329760 | ||||||
| Ukrainian population, n=281 |
CC CT TT C T |
56 (60.9) 33 (35.9) 3 (3.2) 145 (78.8) 39 (21.2) |
− | 38 (61.3) 20 (32.3) 4 (6.4) 96 (77.4) 28 (22.6) |
52 (56.5) 38 (41.3) 2 (2.2) 142 (77.2) 42 (22.8) |
20 (57.1) 10 (28.6) 5 (14.3) P1<0.05 50 (71.4) 20 (28.6) |
| Indonesian population, n=95 (25) |
CC CT TT C T |
49 (49.5)* 36 (36.4) 14 (14.1) P3<0.05 134 (67.7) 64 (32.3) |
8 (38.1) 7 (33.3) 6 (28.6) 23 (54.8) 19 (45.2) |
4 (33.3) 5 (41.7) 3 (25.0) 13 (54.2) 11 (45.8) P3<0.05 |
17 (53.1) 11 (34.4) 4 (12.5) 45 (70.3) 19 (29.7) |
13 (43.4) 10 (33.3) 7 (23.3) 36 (60.0) 24 (40.0) |
| German population, n=478 (26) |
CC CT TT C T |
56 (56.6)** 40 (40.4) 3 (3.0) P4<0.05 152 (76.8) 46 (23.2) |
− | 139 (58.2) 84 (35.1) 16 (6.7) P4<0.05 362 (75.7) 116 (24.3) P4<0.05 |
91 (55.5) 61 (37.2) 12 (7.3) 243 (74.1) 85 (25.9) |
48 (64.0) 23 (30.7) 4 (5.3) P4<0.05 119 (79.3) 31 (20.7) P4<0.05 |
| Italian population, n=1177 (27) |
CC CT+TT |
72 (67.3)*** 35 (32.7) P4<0.05 |
− | 313 (64.0) 176 (36.0) |
− | 482 (70.0) 206 (30.0) P2<0.05 P4<0.05 |
| ace2 rs4240157 | ||||||
| Ukrainian population, n=281 |
CC C− CT TT T− C T |
7 (7.6) 17 (18.5) 21 (22.8) 18 (19.6) 29 (31.5) 52 (37.7) 86 (62.3) |
− | 4 (6.5) 10 (16.1) 22 (35.5) 15 (24.2) 11 (17.7) 40 (38.8) 63 (61.2) |
8 (8.7) 15 (16.3) 22 (23.9) 26 (28.3) 21 (22.8) 53 (35.8) 95 (64.2) |
2 (5.7) 5 (14.3) 10 (28.6) 10 (28.6) 8 (22.8) 19 (33.3) 38 (66.7) |
| Saudi Arabian population, n=217 (28) |
CC CT TT C T |
10 (10.0) 31 (31.0) 59 (59.0) P3<0.05 51 (25.5) 149 (74.5) P3<0.05 |
− | − | − | 29 (24.8) 47 (40.2) 41 (35.0) P5=0.0007 105 (44.9) 129 (55.1) P5<0.001 |
Note: Data provided from: * – the Vietnamese population, ** – the Northern and Western European population, *** – the Tuscany population in Italy [16]; P1 – comparison with the moderate course, P2 – comparison with the mild course, P3 – comparison with the Ukrainian population, P4 – comparison with the Indonesian population, P5 – comparison with population control / control subjects.
Initially, we compared the frequencies of tmprss2 rs12329760 polymorphism between the Ukrainian, Indonesian, German and Italian populations. There were no differences in the frequencies of this polymorphism between the Ukrainian, German and Italian populations. In contrast, in the Indonesian population, the T allele was more common than in other investigated populations.
There were no statistically significant differences in the frequencies of tmprss2 rs12329760 polymorphism between the mild, moderate, and severe course of COVID-19 in the Indonesian and German populations. In the Italian population, there was a significantly higher prevalence of the CC genotype over the combined CT+TT genotype in severe COVID-19 patients as compared to mild patients.
In addition, we observed statistically significant differences in the frequencies of tmprss2 polymorphism among COVID-19 patients between the Indonesian population and German/Italian populations due to the populational distribution of the C and T alleles.
We found only one article about ace2 rs4240157 polymorphism associated with the severity of COVID-19. In the Saudi Arabian population, a high prevalence of alleles C was associated with severe COVID-19. In addition, the prevalence of the T allele in the Saudi Arabian population was higher than in the Ukrainian population (P<0.05).
4. Discussion and conclusions
Despite the fact that SARS-CoV-2 spike protein had a strong binding affinity to the ACE2 extracellular domain, the cell entry and viral proliferation might be observed throughout other membrane molecules, such as TMPRSS2, TMPRSS11A, and CD147 (6, 29).
To understand the genetic background of complex phenotypes in human populations, researchers commonly assess correlation with allele frequency (30, 31). This approach has identified a correlation between ancestral genetic composition and the case fatality rate of COVID-19 (31).
TMPRSS2 protein is composed of a small cytoplasmic region (aa 1-121), a transmembrane helix (aa 122-142), and an extracellular region (aa 143-529). In turn, the latter is composed of three domains, the LDL-receptor-like domain (aa 149-186), the SRCR-like domain (aa 187-279), and the peptidase domain (aa 293-526). It is regulated by androgenic hormones in vivo (32).
Genetic determinants of susceptibility and/or severity of COVID-19 have been sought in TMPRSS2 in Whole-Exome Sequencing studies (33, 34).
Vargas-Alarcón G. et al. (35) identified a potentially disruptive polymorphism in the tmprss2 gene (rs12329760), whose minor allele frequencies of which varied between populations (American, African, European and Asian). This mutation (V197M) occurs not in the catalytic domain but in the SRCR domain, which is probably needed for protein-protein interaction (36). Structural and energy calculation analysis of V197M amino acid change showed that it destabilizes the TMPRSS2 protein, possibly negatively affecting its ACE2 and viral spike protein processing (37).
Our study is the first to describe the genotype distribution of tmprss2 rs12329760 in the Ukrainian population: CC – 60.9%; CT – 35.9%; TT – 3.2%; CT+TT – 39.1%; C – 78.8%; and T – 21.2%. This distribution is similar in the Northern and Western European as well as the Tuscany population in Italy (16).
In contrast, in the Indonesian population, we observed statistically significant differences in the genotype distribution: CC – 49.5%; CT – 36.4%; and TT – 14.1%. This data went in parallel with the observation that the prevalence of missense mutations represented by rs12329760 varied between 10 and 65%, with the Asian population showing a significantly higher frequency than European populations (38).
At the beginning of our study, selected patients with COVID-19 were divided into three clinical groups according to the need for oxygen: patients without oxygen therapy, with non-invasive, and invasive oxygen therapy. Additionally, the sub-group of patients who died was selected from the group with invasive oxygen therapy. We used such a severity scale because previously published articles applied different scales predominantly based on the hospitalization due to COVID-19, death, need for oxygen, and the type of ventilation. This scaling enabled us to compare our data with other published results of gene polymorphisms’ associations with COVID-19 severity (as shown in our meta-analysis). All COVID-19 patients had significantly higher ages in comparison with control subjects, and patients who needed oxygen had higher ages than those without oxygen therapy. Patients with invasive oxygen therapy were also older than those with non-invasive oxygen therapy. Similarly, oxygen-dependent COVID-19 patients had elevated BMI as compared to control subjects. The patients who died of COVID-19 had the highest BMI. This data went in parallel with studies, which showed that aging and overweight were important risk factors for COVID-19 (39-42).
We observed significant differences in the frequency of CC, CT, and TT genotypes in the groups of COVID-19 patients with non-invasive (moderate severity) and invasive (severe course) oxygen therapy. The frequency of the T allele was higher in COVID-19 patients with invasive (severe course) oxygen therapy in the Ukrainian population. In the Indonesian (25) and German (26) populations, there were no differences in the frequency of CC, CT, and TT genotypes in the groups of COVID-19 patients. In contrast, Monticelli et al. (2021) found the differences in the frequency of CC and CT+TT genotypes in the groups of Italian COVID-19 patients with the mild and severe course of the disease (P<0.05). The frequency of the CC genotype was higher in COVID-19 patients with invasive (severe course) oxygen therapy as against the group of patients without oxygen therapy (mild course).
TMPRSS11A is another important molecule, which might promote virus entry into the cells. TMPRSS11A is a type II transmembrane serine protease expressed on the surface of airway epithelial cells, which has been shown to cleave and activate spike proteins of the severe acute respiratory syndrome (SARS) and MERS coronaviruses. TMPRSS11A was able to cleave the SARS-CoV-2 spike protein (43, 44). TMPRSS11A is a novel age-altered, tissue-specific regulator of migration and wound healing. Its levels increase with age in the skin and gingival tissue. Overexpression of TMPRSS11A decreases cell migration and spreading, and induces cellular senescence. TMPRSS11A interacts with integrin β1 and influences cell migration. In the coding sequences of tmprss11a, one probably-damaging polymorphism was detected. This polymorphism rs353163 C>T produced a change of amino acid and led to the nonsynonymous substitution, namely to Arg290Gln. Arg290 was predicted to undergo a translational modification (35, 45).
We are the first to inform about the genotype distribution of tmprss11a rs353163 in the Ukrainian population: CC – 46.7%, CT – 40.2%, TT – 13.1%; allele C – 66.8%, and T – 33.2%. This distribution was similar to the North Indian (17), Iranian (18), Northern and Western European, and Iberian populations in Spain (19). There were no significant differences in the frequency of CC, CT, and TT genotypes in all groups of Ukrainian COVID-19 patients. During the literature analysis, we did not find information about tmprss11a rs353163 polymorphism association with sensitivity and severity of COVID-19.
Human ACE2 has emerged as the target for SARS-CoV-2 (46). ACE2, a type I transmembrane zinc finger glycoprotein, is a monocarboxypeptidase, which converts angiotensin 1 to angiotensin 1-9. The gene for ACE2 is present on chromosome X (47). The genetic polymorphism of the ace2 gene is well-known all over the world with racial and ethnic variation having varying influences on the altered functions of the RAAS pathway (48). The polymorphism of ace2 has been associated with arterial hypertension in different populations (49-51). The ace2 polymorphism has been associated with varying degrees of disease severity and clinical outcomes of COVID-19, with the absence of the D/D genotype conferring protection against severe lung involvement (52). Controversial results were obtained in Asian patients (53).
Another ace2 polymorphism rs4240157 was also associated with arterial hypertension (54, 55), and type 2 diabetes mellitus (56). Wooster et al. (57), reported that ace2 rs4240157 polymorphism is associated with COVID-19 disease severity as it might induce higher tissue-specific expression of ace2, resulting in the hospitalization of COVID-19 patients.
Our study is the first to investigate the genotype distribution of ace2 rs4240157 in the Ukrainian population: CC – 7.6%, (C-) – 18.5%, CT – 22.8%, TT – 19.6%, (T-) – 31.5%, CT+TT+(T-) – 73.9%, allele C – 37.7%, and T – 62.3%. This data was similar to the distribution of this polymorphism in the Northern and Western European, and the Iberian populations in Spain (20). In contrast, in the Saudi Arabian population, we observed significant differences in genotype and allele frequencies (28) with a high prevalence of T alleles.
There were no differences in the distribution of ace2 polymorphism in all groups with different severity of COVID-19 in the Ukrainian population. In contrast, in the Saudi Arabian population, there were differences between control subjects and patients with severe COVID-19. The patients with severe COVID-19 had high frequencies of the CC, CT genotypes, and the C allele. These differences might depend on the population peculiarities of special counts of genotypes and alleles in this study.
CD147 is a 58-kD cell surface glycoprotein of the immunoglobulin superfamily. It was originally described on the surface of tumor cells, whereon it can induce the synthesis of MMPs. CD147 has recently been identified as a mere marker of inflammation (58). CD147 possesses a pivotal role in the complex processes of atherogenesis, atheroprogression, and acute atherothrombosis; it is consistently associated with the risk of cardiovascular disease (15, 59).
CD147 (EMMPRIN) is the target receptor for COVID-19. It is expressed on several immune cells where it leads to induction of chemotactic cytokines (TNF-alpha, IL-10, IL-6), causes MM2, IL-9 induction, production of IFN-gamma (IL-18), T-cell activation, proliferation, invasion, adhesion, and energy activation (60, 61).
Wu et al. (62) genotyped SNPs at the CD147 locus and found that the 3’-untranslated region (3’-UTR) T/A rs8259 SNP modified the association correlation between CD147 expression and miRNA-492.
Over-expression of CD147 is involved in the pathogenesis of ACS. The cd147 3’-UTR rs8259 T allele may be a protective factor for ACS, its polymorphism can affect the CD147 protein expression in ACS patients (63).
We investigated genotype distribution of cd147 rs8259 polymorphism in the Ukrainian population: TT – 60.9%, AT – 32.6%, AA – 6.5%; (AT+AA) – 39.1%; the T allele – 77.2%, allele A – 22.8%. This data was similar to that of Polish, Northern and Western European populations. At the same time, there were significant differences in the Asian population (Jiangsu Chinese, Han Chinese, and Chinese) where the A allele had a higher distribution. There were no significant differences in the frequency of cd147 rs8259 polymorphism in all groups of Ukrainian COVID-19 patients. Unfortunately, in the literature, we did not find any information about cd147 rs8259 association with sensitivity and severity of COVID-19.
Taken together, our data showed that out of all investigated genes, only polymorphism of tmprss2 rs12329760 was associated with severe COVID-19.
We hypothesize that this association might depend on the specificity of proteolytic activation of SARS-CoV-2 S protein by TMPRSS2 as well as by other investigated proteases (TMPRSS11A, ACE2, CD147).
The S protein has two cleavage sites (S1/S2 and S2’), and the cleavage motif for furin protease at the S1/S2 site that results from a unique four-amino acid insertion, which is one of the distinguishing features of SARS-CoV-2. The viral particle incorporates the S protein, which has already undergone S1/S2 cleavage by furin, and then undergoes further cleavage at the S2 site, mediated by the TMPRSS2, after binding to the ACE2 to facilitate membrane fusion at the plasma membrane (64). The investigated tmprss2 mutation (rs12329760) occurs in the SRCR domain, which is necessary for protein-protein interaction but is not needed for catalytic cleavage.
The SRCR protein plays an important role in human diseases, such as autoimmune diseases, Alzheimer’s disease, atherosclerosis, and cancer (65). TMPRSS2 has two calcium-binding domains, the SRCR and the LDLRA (LDL receptor class A) domains, which form a binding site for calcium (66). SARS-CoV-1 employs a calcium-dependent fusion process (67), and possibly SARS-CoV-2 also uses this process (68).
Thus, tmprss2 mutation (rs12329760) might be associated with the severity of COVID-19 due to the triggering of a calcium-dependent fusion process.
Limitations of our study are the possible influence of age and BMI, as well as the limited number of subjects involved. Moreover, the distribution of tmprss2 polymorphism is different in statistical terms among the controls and COVID-19 patients, but it should be admitted that these differences have a limited extent and might not be so significant in clinical and epidemiological terms.
Prospects for future research might be in further investigation of COVID-19 susceptibility and severity associated with polymorphisms of other genes from the SRCR protein family. Experimental research is also needed to prove the hypothesis that the calcium-dependent mechanism and tmprss2 rs12329760 affect COVID-19. Additional studies will be required including a larger number of patients and a longitudinal population-based study design should be provided to receive information on COVID-19 susceptibility.
Our study indicated the presence of an association between the tmprss2 rs12329760 polymorphism and the severity of COVID-19 in the Ukrainian population. It seems that patients with severe COVID-19 had the TT genotype more often than those with moderate COVID-19.
Conflicts of Interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article.
Ethics Approval:
This study was performed in line with the principles of the Declaration of Helsinki. Ethical approval and consent to participate in the study were approved by the Committee on Bioethics and Ethical Issues of Poltava State Medical University.
Funding:
The study was a part of research project No. 0121U107440 “Genetic variants and their potential link with COVID-19 in the Ukrainian population” funded by the Ministry of Public Health of Ukraine.
References
- Mallah SI, Ghorab OK, Al-Salmi S, et al. COVID-19: breaking down a global health crisis. Ann Clin Microbiol Antimicrob. 2021;20(1):35. doi: 10.1186/s12941-021-00438-7. Published 2021 May 18. doi:10.1186/s12941-021-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Zhao J, Martin W, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):216. doi: 10.1186/s12916-020-01673-z. Published 2020 Jul 15. doi:10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicessi JR, Roden DM, Wilde AAM, Ackerman MJ. Genetic susceptibility for COVID-19-associated sudden cardiac death in African Americans. Heart Rhythm. 2020;17(9):1487–1492. doi: 10.1016/j.hrthm.2020.04.045. doi:10.1016/j.hrthm.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and Individual Genetic Susceptibility/Receptivity: Role of ACE1/ACE2 Genes, Immunity, Inflammation and Coagulation. Might the Double X-chromosome in Females Be Protective against SARSCoV-2 Compared to the Single X-Chromosome in Males? Int J Mol Sci. 2020;21(10):3474. doi: 10.3390/ijms21103474. Published 2020 May 14. doi:10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izmailova O, Shlykova O, Vatsenko A, et al. Allele С (rs5186) of at1r is associated with the severity of COVID-19 in the Ukrainian population. Infect Genet Evol. 2022;98:105227. doi: 10.1016/j.meegid.2022.105227. doi:10.1016/j.meegid.2022.105227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidashev I, Shlykova O, Izmailova O, et al. Host gene variability and SARS-CoV-2 infection: A review article. Heliyon. 2021;7(8):e07863. doi: 10.1016/j.heliyon.2021.e07863. doi:10.1016/j.heliyon.2021.e07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe Covid-19 GWAS Group. Ellinghaus D, Degenhardt F, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. doi:10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Xiao Y, Kang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019 [published correction appears in Clin Infect Dis. 2021 Dec 16;73(12):2374] Clin Infect Dis. 2020;71(15):713–720. doi: 10.1093/cid/ciaa203. doi:10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabibzadeh A, Zamani F, Laali A, et al. SARS-CoV-2 Molecular and Phylogenetic analysis in COVID-19 patients: A preliminary report from Iran. Infect Genet Evol. 2020;84:104387. doi: 10.1016/j.meegid.2020.104387. doi:10.1016/j.meegid.2020.104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. doi:10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J Virol. 2019;93(6):e01815–18. doi: 10.1128/JVI.01815-18. Published 2019 Mar 5. doi:10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Mi L, Xu J, et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis. 2005;191(5):755–760. doi: 10.1086/427811. doi:10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella F, Wang W, Moreno I, Roson B, Quake SR, Simon C. Single-cell RNA sequencing of SARS-CoV-2 cell entry factors in the preconceptional human endometrium. Hum Reprod. 2021;36(10):2709–2719. doi: 10.1093/humrep/deab183. doi:10.1093/humrep/deab183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleghani N, Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: A review of the state-of-the-art. Biosens Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. doi:10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Mao Y, Wang C, Wang Z. Association Study between an SNP in CD147 and Its Expression With Acute Coronary Syndrome in a Jiangsu Chinese Population. Medicine (Baltimore) 2015;94(42):e1537. doi: 10.1097/MD.0000000000001537. doi:10.1097/MD.0000000000001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=21:41480070-41481070;v=rs12329760;vdb=variation;vf=722896905#ncbialfa_anchor. [Google Scholar]
- Umar M, Upadhyay R, Kumar S, Ghoshal UC, Mittal B. Modification of risk, but not survival of esophageal cancer patients by esophageal cancer-related gene 1 Arg290Gln polymorphism: a case-control study and meta-analysis. J Gastroenterol Hepatol. 2013;28(11):1717–1724. doi: 10.1111/jgh.12335. doi:10.1111/jgh.12335. [DOI] [PubMed] [Google Scholar]
- Akbari MR, Malekzadeh R, Shakeri R, et al. Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res. 2009;69(20):7994–8000. doi: 10.1158/0008-5472.CAN-09-1149. doi:10.1158/0008-5472.CAN-09-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=4:67918556-67919556;v=rs353163;vdb=variation;vf=90470389. [Google Scholar]
- https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=X:15568341-15569341; v=rs4240157;vdb=variation;vf=93325552. [Google Scholar]
- Łacina P, Butrym A, Mazur G, Bogunia-Kubik K. BSG and MCT1 Genetic Variants Influence Survival in Multiple Myeloma Patients. Genes (Basel) 2018;9(5):226. doi: 10.3390/genes9050226. Published 2018 Apr 24. doi:10.3390/genes9050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MP, Hu XL, Yang YL, et al. Basigin rs8259 Polymorphism Confers Decreased Risk of Chronic Heart Failure in a Chinese Population. Int J Environ Res Public Health. 2017;14(2):211. doi: 10.3390/ijerph14020211. Published 2017 Feb 21. doi:10.3390/ijerph14020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Chen M, Yang K, Shao J, Fu Y, Zhou W. Association of CD147 genetic polymorphisms with carotid atherosclerotic plaques in a Han Chinese population with cerebral infarction. Thromb Res. 2017;156:29–35. doi: 10.1016/j.thromres.2017.05.027. doi:10.1016/j.thromres.2017.05.027. [DOI] [PubMed] [Google Scholar]
- https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=19:582427-583427;v=rs8259;vdb=variation;vf=201683794. [Google Scholar]
- Wulandari L, Hamidah B, Pakpahan C, et al. Initial study on TMPRSS2 p. Val160Met genetic variant in COVID-19 patients. Hum Genomics. 2021;15(1):29. doi: 10.1186/s40246-021-00330-7. Published 2021 May 17. doi:10.1186/s40246-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfelder K, Breuckmann K, Elsner C, et al. Transmembrane serine protease 2 Polymorphisms and Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Type 2 Infection: A German Case-Control Study. Front Genet. 2021;12:667231. doi: 10.3389/fgene.2021.667231. Published 2021 Apr 21. doi:10.3389/fgene.2021.667231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli M, Hay Mele B, Benetti E, et al. Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women. Genes (Basel) 2021;12(4):596. doi: 10.3390/genes12040596. Published 2021 Apr 19. doi:10.3390/genes12040596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MM, Mir R, Alghamdi MAA, et al. Strong Association of Angiotensin Converting Enzyme-2 Gene Insertion/Deletion Polymorphism with Susceptibility to SARS-CoV-2, Hypertension, Coronary Artery Disease and COVID-19 Disease Mortality. J Pers Med. 2021;11(11):1098. doi: 10.3390/jpm11111098. Published 2021 Oct 27. doi:10.3390/jpm11111098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre J, Cracowski JL, Richard V, Bouhanick B. 'Drugs, COVID-19' working group of the French Society of Pharmacology, Therapeutics. Renin-angiotensin-aldosterone system and COVID-19 infection. Ann Endocrinol (Paris) 2020;81(2-3):63–67. doi: 10.1016/j.ando.2020.04.005. doi:10.1016/j.ando.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY) 2020;12(11):10087–10098. doi: 10.18632/aging.103415. doi:10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Ghate D. Investigating the likely association between genetic ancestry and COVID-19 manifestations. doi: https://doi.org/10.1101/2020.04.05.20054627. [Google Scholar]
- Lucas JM, Heinlein C, Kim T, et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4(11):1310–1325. doi: 10.1158/2159-8290.CD-13-1010. doi:10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla F, Mohammad A, Al Madhoun A, et al. ACE2 and FURIN variants are potential predictors of SARS-CoV-2 outcome: A time to implement precision medicine against COVID-19. Heliyon. 2021;7(2):e06133. doi: 10.1016/j.heliyon.2021.e06133. doi:10.1016/j.heliyon.2021.e06133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini A, Agolini E, Novelli A, et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes (Basel) 2020;11(9):1010. doi: 10.3390/genes11091010. Published 2020 Aug 27. doi:10.3390/genes11091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Alarcón G, Posadas-Sánchez R, Ramírez-Bello J. Variability in genes related to SARS-CoV-2 entry into host cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and its potential use in association studies. Life Sci. 2020;260:118313. doi: 10.1016/j.lfs.2020.118313. doi:10.1016/j.lfs.2020.118313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester E, Sasaki T, Timpl R. Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat Struct Biol. 1999;6(3):228–232. doi: 10.1038/6669. doi:10.1038/6669. [DOI] [PubMed] [Google Scholar]
- Jeon S, Blazyte A, Yoon C, et al. Regional TMPRSS2 V197M Allele Frequencies Are Correlated with COVID-19 Case Fatality Rates. Mol Cells. 2021;44(9):680–687. doi: 10.14348/molcells.2021.2249. doi:10.14348/molcells.2021.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin A, Stepanov V, Markov A, et al. Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy. Genes (Basel) 2020;12(1):19. doi: 10.3390/genes12010019. Published 2020 Dec 25. doi:10.3390/genes12010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw AJM, Oude Luttikhuis MAM, Wellen AC, Müller C, Calkhoven CF. Obesity and its impact on COVID-19. J Mol Med (Berl) 2021;99(7):899–915. doi: 10.1007/s00109-021-02072-4. doi:10.1007/s00109-021-02072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamara A, Tahapary DL. Obesity as a predictor for a poor prognosis of COVID-19: A systematic review. Diabetes Metab Syndr. 2020;14(4):655–659. doi: 10.1016/j.dsx.2020.05.020. doi:10.1016/j.dsx.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klein SL, Garibaldi BT, et al. Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res Rev. 2021;65:101205. doi: 10.1016/j.arr.2020.101205. doi:10.1016/j.arr.2020.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parohan M, Yaghoubi S, Seraji A, Javanbakht MH, Sarraf P, Djalali M. Risk factors for mortality in patients with Coronavirus disease 2019 (COVID-19) infection: a systematic review and meta-analysis of observational studies. Aging Male. 2020;23(5):1416–1424. doi: 10.1080/13685538.2020.1774748. doi:10.1080/13685538.2020.1774748. [DOI] [PubMed] [Google Scholar]
- Qian W, Kallergi M, Clarke LP, et al. Tree structured wavelet transform segmentation of microcalcifications in digital mammography. Med Phys. 1995;22(8):1247–1254. doi: 10.1118/1.597562. doi:10.1118/1.597562. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang Y, Zhang S, et al. Intracellular autoactivation of TMPRSS11A, an airway epithelial transmembrane serine protease. J Biol Chem. 2020;295(36):12686–12696. doi: 10.1074/jbc.RA120.014525. doi:10.1074/jbc.RA120.014525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C, Burgos A, Morales D, et al. TMPRSS11a is a novel age-altered, tissue specific regulator of migration and wound healing. FASEB J. 2021;35(5):e21597. doi: 10.1096/fj.202002253RRR. doi:10.1096/fj.202002253RRR. [DOI] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875–879. doi: 10.1038/nm1267. doi:10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275(43):33238–33243. doi: 10.1074/jbc.M002615200. doi:10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Yi L, Gu YH, Wang XL, et al. Association of ACE, ACE2 and UTS2 polymorphisms with essential hypertension in Han and Dongxiang populations from north-western China. J Int Med Res. 2006;34(3):272–283. doi: 10.1177/147323000603400306. doi:10.1177/147323000603400306. [DOI] [PubMed] [Google Scholar]
- Patnaik M, Pati P, Swain SN, et al. Association of angiotensin-converting enzyme and angiotensin-converting enzyme-2 gene polymorphisms with essential hypertension in the population of Odisha, India. Ann Hum Biol. 2014;41(2):145–152. doi: 10.3109/03014460.2013.837195. doi:10.3109/03014460.2013.837195. [DOI] [PubMed] [Google Scholar]
- Pinheiro DS, Santos RS, Jardim PCBV, et al. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: A genetic association study in Brazilian patients. PLoS One. 2019;14(8):e0221248. doi: 10.1371/journal.pone.0221248. Published 2019 Aug 20. doi:10.1371/journal.pone.0221248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Cao JJ. Angiotensin-Converting Enzyme Gene Polymorphism and Severe Lung Injury in Patients with Coronavirus Disease 2019. Am J Pathol. 2020;190(10):2013–2017. doi: 10.1016/j.ajpath.2020.07.009. doi:10.1016/j.ajpath.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancy CW. COVID-19 and African Americans. JAMA. 2020;323(19):1891–1892. doi: 10.1001/jama.2020.6548. doi:10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
- Aung AK, Aitken T, Teh BM, et al. Angiotensin converting enzyme genotypes and mortality from COVID-19: An ecological study. J Infect. 2020;81(6):961–965. doi: 10.1016/j.jinf.2020.11.012. doi:10.1016/j.jinf.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Wang T, Li Y, et al. Association of ACE2 polymorphisms with susceptibility to essential hypertension and dyslipidemia in Xinjiang, China. Lipids Health Dis. 2018;17(1):241. doi: 10.1186/s12944-018-0890-6. Published 2018 Oct 20. doi:10.1186/s12944-018-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, Yang Y, Wang Y, et al. ACE2 gene polymorphism and essential hypertension: an updated meta-analysis involving 11,051 subjects. Mol Biol Rep. 2012;39(6):6581–6589. doi: 10.1007/s11033-012-1487-1. doi:10.1007/s11033-012-1487-1. [DOI] [PubMed] [Google Scholar]
- Patel SK, Wai B, Ord M, et al. Association of ACE2 genetic variants with blood pressure, left ventricular mass, and cardiac function in Caucasians with type 2 diabetes. Am J Hypertens. 2012;25(2):216–222. doi: 10.1038/ajh.2011.188. doi:10.1038/ajh.2011.188. [DOI] [PubMed] [Google Scholar]
- Wooster L, Nicholson CJ, Sigurslid HH, Cardenas CLL, Malhotra R. Polymorphisms in the ace2 locus associate with severity of COVID-19 infection [preprint] MedRxiv. 2020.06.18.20135152; doi: https://doi.org/10.1101/2020.06.18.20135152. [Google Scholar]
- Gwinn WM, Damsker JM, Falahati R, et al. Novel approach to inhibit asthma-mediated lung inflammation using anti-CD147 intervention. J Immunol. 2006;177(7):4870–4879. doi: 10.4049/jimmunol.177.7.4870. doi:10.4049/jimmunol.177.7.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, von Brühl ML, Barocke V, et al. EMMPRIN (CD147/basigin) mediates platelet-monocyte interactions in vivo and augments monocyte recruitment to the vascular wall. J Thromb Haemost. 2011;9(5):1007–1019. doi: 10.1111/j.1538-7836.2011.04235.x. doi:10.1111/j.1538-7836.2011.04235.x. [DOI] [PubMed] [Google Scholar]
- Venkatesan B, Valente AJ, Prabhu SD, Shanmugam P, Delafontaine P, Chandrasekar B. EMMPRIN activates multiple transcription factors in cardiomyocytes, and induces interleukin-18 expression via Rac1-dependent PI3K/Akt/IKK/NF-kappaB andMKK7/JNK/AP-1 signaling. J Mol Cell Cardiol. 2010;49(4):655–663. doi: 10.1016/j.yjmcc.2010.05.007. doi:10.1016/j.yjmcc.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad AR, Bashir I, Ijaz F, et al. Is COVID-19 Fatality Rate Associated with Malaria Endemicity? Discoveries (Craiova) 2020;8(4):e120. doi: 10.15190/d.2020.17. Published 2020 Dec 11. doi:10.15190/d.2020.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LS, Li FF, Sun LD, et al. A miRNA-492 binding-site polymorphism in BSG (basigin) confers risk to psoriasis in central south Chinese population. Hum Genet. 2011;130(6):749–757. doi: 10.1007/s00439-011-1026-5. doi:10.1007/s00439-011-1026-5. [DOI] [PubMed] [Google Scholar]
- Mao Y, Yan J, Wang C, Wang Z, Liu P, Yuan W. Zhonghua Xin Xue Guan Bing Za Zhi. 2014;42(7):566–570. [PubMed] [Google Scholar]
- Takeda M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol Immunol. 2022;66(1):15–23. doi: 10.1111/1348-0421.12945. doi:10.1111/1348-0421.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez VG, Moestrup SK, Holmskov U, Mollenhauer J, Lozano F. The conserved scavenger receptor cysteine-rich superfamily in therapy and diagnosis. Pharmacol Rev. 2011;63(4):967–1000. doi: 10.1124/pr.111.004523. doi:10.1124/pr.111.004523. [DOI] [PubMed] [Google Scholar]
- Paoloni-Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3 [published correction appears in Genomics 2001 Sep;77(1-2):114] Genomics. 1997;44(3):309–320. doi: 10.1006/geno.1997.4845. doi:10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- Lai AL, Millet JK, Daniel S, Freed JH, Whittaker GR. The SARS-CoV Fusion Peptide Forms an Extended Bipartite Fusion Platform that Perturbs Membrane Order in a Calcium-Dependent Manner. J Mol Biol. 2017;429(24):3875–3892. doi: 10.1016/j.jmb.2017.10.017. doi:10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman DP. Why the lower reported prevalence of asthma in patients diagnosed with COVID-19 validates repurposing EDTA solutions to prevent and manage treat COVID-19 disease. Med Hypotheses. 2020;144:110027. doi: 10.1016/j.mehy.2020.110027. doi:10.1016/j.mehy.2020.110027. [DOI] [PMC free article] [PubMed] [Google Scholar]


