Abstract
Y-box proteins are major constituents of ribonucleoprotein particles (RNPs) which contain translationally silent mRNAs in gametic cells. We have recently shown that a sequence-specific RNA binding activity present in spermatogenic cells contains the two Y-box proteins MSY2 and MSY4. We show here that MSY2 and MSY4 bind a sequence, 5′-UCCAUCA-3′, present in the 3′ untranslated region of the translationally repressed protamine 1 (Prm1) mRNA. Using pre- and post-RNase T1-digested substrate RNAs, it was determined that MSY2 and MSY4 can bind an RNA of eight nucleotides containing the MSY2 and MSY4 binding site. Single nucleotide mutations in the sequence eliminated the binding of MSY2 and MSY4 in an electrophoretic mobility shift assay, and the resulting mutants failed to compete for binding in a competition assay. A consensus site of UACCACAUCCACU (subscripts indicate nucleotides which do not disrupt YRS binding by MSY2 and MSY4), denoted the Y-box recognition site (YRS), was defined from this mutational analysis. These mutations in the YRS were further characterized in vivo using a novel application of the yeast three-hybrid system. Experiments with transgenic mice show that disruption of the YRS in vivo relieves Prm1-like repression of a reporter gene. The conservation of the RNA binding motifs among Y-box protein family members raises the possibility that other Y-box proteins may have previously unrecognized sequence-specific RNA binding activities.
The mouse Y-box proteins MSY2 and MSY4 are members of a protein family found in prokaryotes and eukaryotes that contain the highly conserved cold-shock domain (CSD). This 67- to 80-amino-acid (aa) nucleic acid binding domain is 43% identical from Escherichia coli to humans, contains the RNP1 and RNP2 RNA binding motifs, and forms a five-stranded antiparallel β-barrel structure (36). The prokaryotic members of this family, such as major cold-shock protein CspA in E. coli, are about 70 aa in length and are involved in the cold-shock response in various bacteria, an adaptive response to sudden temperature downshifts (14, 19). CspA negatively regulates its own expression by acting as an RNA chaperone that destabilizes secondary structures in mRNA (1, 17).
The eukaryotic branch of the CSD family are Y-box proteins and have been characterized in all eukaryotes investigated, including fruit flies, planaria, goldfish, chickens, frogs, mice, and humans, with the exception of the yeast Saccharomyces cerevisiae (25, 30). Y-box proteins are approximately 250 to 350 aa in length, have a CSD located in the amino-terminal half of the protein, and possess amino termini which are highly divergent in sequence and length (36). The carboxy tails of invertebrate Y-box proteins are quite variable in structure, with Drosophila melanogaster YPS containing RGG repeats, Caenorhabditis elegans LIN-28 containing zinc fingers, and Schistosoma mansoni SMYB1 containing a fibroin-like domain. On the other hand, the nucleic acid binding carboxy tails of vertebrate Y-box proteins are more conserved and contain four sets of alternating basic and acidic regions, each approximately 30 aa in length (25).
Y-box proteins were originally isolated based on their ability to bind the double-stranded (dsDNA) sequence 5′-CCAAT-3′ (29). Later work defined the Y-box element as 5′-CTGATTGG(C/T)(C/T)AA-3′, which contains a reverse CCAAT box (in boldface). This regulatory element is found in the promoter regions of many vertebrate gamete-specific genes, including the Xenopus laevis oocyte-specific hsp70 gene, the rat testis-specific histone H2B gene, and the murine testis-specific Prm1 gene (37). Subsequent work has shown that some Y-box family members have specificity for binding both pyrimidine-rich dsDNA and single-stranded DNA (16). These observations have led to several models for Y-box protein function involving DNA interactions, such as roles in transcriptional regulation, chromatin modification, and DNA repair.
Many Y-box proteins have also been identified as components of messenger ribonucleoprotein particles (mRNPs). Y-box protein p50 is the major core protein of cytoplasmic mRNPs of somatic cells in rabbits (10). p50 and the poly(A) binding protein are the two most abundant proteins in these mRNPs. In Xenopus oocytes, Y-box proteins are also abundant components of mRNP3+4s containing masked mRNAs (26). Proteins homologous to mRNP3+4s are expressed during murine spermatogenesis and form complexes with stored mRNAs (21). Murine Y-box protein MSY1 has also been shown to be associated with germ cell mRNPs during spermiogenesis (34).
MSY2 and MSY4 are components of a 48- and 50-kDa RNA binding activity present in murine testis extracts (8). This activity is a component of testis mRNPs containing protamine mRNAs. The protamines are small arginine-rich proteins involved in condensation of DNA in the nuclei of mature spermatids. The protamine mRNAs are synthesized in round and early-elongating spermatids, transported to the cytoplasm, and stored as translationally repressed mRNPs until their translation from 2 to 8 days later in elongated spermatids (2, 20). The MSY2 and MSY4 proteins bind a 22-nucleotide (nt) region of the Prm1 3′ untranslated region (UTR) and a 20-nt region of the Prm2 3′ UTR (11). The 22-nt Prm1 region lies within the first 37 nt of the Prm1 3′ UTR and can delay the translation of an hGH transgene in vivo (12).
MSY2 is the murine orthologue of Xenopus protein FRGY2 (mRNP3+4) and was cloned from an expression library screen with anti-FRGY2 antibodies (15). FRGY2 was originally cloned by its ability to bind the CCAAT element (35), but as mentioned above is also found associated with germ line mRNPs. FRGY2 is therefore considered to have a dual function: a role in transcriptional activation of oogenic genes and a second role in masking gametogenic mRNAs. Msy4 was cloned from a mouse testis cDNA library using the yeast three-hybrid system with the first 37 nt of the protamine 1 (Prm1) 3′ UTR (Prm11–37wt) as bait (8). The spatial and temporal patterns of both MSY2 and MSY4 are consistent with these proteins playing roles in Prm1 mRNA storage.
In this study, we have delineated a site present in the 3′ UTR of the Prm1 3′ UTR that murine Y-box proteins MSY2 and MSY4 bind specifically. Single-nucleotide mutations within the conserved Y-box recognition site (YRS) eliminate binding of MSY2 and MSY4 in vitro and in the yeast three-hybrid system. Furthermore, transgenic experiments with mice suggest that the YRS will also function in vivo.
MATERIALS AND METHODS
Mice.
C57BL/6J male mice were purchased from Jackson Laboratory (Bar Harbor, Maine) and were sacrificed by carbon dioxide asphyxiation.
Transgenic mice were generated by microinjecting a purified DNA fragment at a concentration of 2 ng/μl in 10 mM Tris (pH 7.5)–0.25 mM EDTA into pronuclei of fertilized eggs derived from FVB/N × FVB/N (Taconic Labs, Germantown, N.Y.) matings (6). Pseudopregnant B6 CBA F1/ (Taconic Labs) foster females were used for oviduct implantation of eggs that survived microinjection. Transgenic animals were identified by PCR. Three lines of mice (3505, 3514, and 3515) were generated and analyzed. The expression of each transgenic line was analyzed by Northern analysis. Line 3505 expressed the transgene at a lower level than lines 3514 and 3515, but all three lines expressed the transgene at high levels.
Protein extracts.
Testes were dissected from adult mice and placed in 1 mg of buffer A (10 mM HEPES [pH 7.6], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT])/ml containing the following protease inhibitors: p-toluenesulfonyl-l-arginine methyl ester (TAME), l-1-p-tosylamino-2-phenylethyl chloromethyl ketone (TPCK), phenylmethylsulfonyl fluoride, and soy bean trypsin inhibitor. The cells were lysed with 20 strokes of a Dounce homogenizer, and cell debris was pelleted via centrifugation at 3,000 × g for 15 min at 4°C in a fixed-angle rotor. To the supernatant was added 0.11 volume of buffer B (0.3 M HEPES [pH 7.6], 1.4 M KCl, 30 mM MgCl2), followed by the addition of glycerol to 20% (vol/vol) final concentration. Extracts were stored at −70°C following quick-freezing in liquid nitrogen.
RNA probe synthesis.
dsDNA oligonucleotides with EcoRI-BamHI 5′ overhanging ends encoding the various Prm11–37 RNAs were cloned into the EcoRI-BamHI sites of the pGEM-2 plasmid, and transformants were selected on Luria-Bertani-ampicillin (100 μg/ml) medium. RNA was synthesized in vitro using SP6 RNA polymerase and 1 μg of linearized plasmid DNA. Radiolabeled in vitro transcriptions were done in 20-μl reaction mixtures that contained 1× RNA polymerase buffer (New England Biolabs, Beverly, Mass.), 0.5 mM ATP, 0.5 mM GTP, 0.5 mM UTP, 25 μM CTP, 50 μCi of [α-32P]CTP at 3,000 Ci/mmol (NEN-Dupont, Boston, Mass.), and 40 U of RNase inhibitor (Roche, Basel, Switzerland) and that were incubated at 37°C for 1 to 2 h. The full-length RNA probe was isolated by electrophoresis of the transcription reaction mixtures on a 5% 30:1 polyacrylamide gel in 1× TBE buffer (27.8 g of Tris, 160.9 g of boric acid, and 9.3 g of EDTA per liter) at 250 V for 1 h. The transcription products were visualized by autoradiography and excised with a razor blade. The RNA was eluted from the polyacrylamide gel by incubation in 400 μl of RNA elution buffer (0.5 M ammonium acetate, 0.5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]) at 37°C overnight, and contaminating pieces of acrylamide were removed via a spin through a mini-glass wool column. The RNA probes were precipitated by addition of 50 μg of total yeast RNA–1/10 volume of 3 M sodium acetate (pH 5.2)–1 ml of 100% ethanol and storage at −20°C for 1 h. The RNA was pelleted via 20 min of centrifugation at 10,000 × g in a microcentrifuge, washed in 70% ethanol, and resuspended in 50 μl of diethyl pyrocarbonate-treated water. The amount of RNA was quantified using a scintillation counter.
Unlabeled RNA was synthesized in vitro using SP6 RNA polymerase and 2 μg of linearized plasmid DNA. Transcription reactions in 50-μl reaction mixtures were performed using 1× RNA polymerase buffer (New England Biolabs), 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.5 mM UTP, 1 μCi of [5,6-3H]UTP (NEN-Dupont), and 100 U of RNase inhibitor (Roche), and reaction mixtures were incubated at 37°C for 1 to 2 h. The RNA was precipitated by addition of 400 μl of RNA elution buffer, 50 μg of total yeast RNA, 1/10 volume of 3 M sodium acetate, pH 5.2, and 1 ml of 100% ethanol and by 20 min of centrifugation at 10,000 × g in a microcentrifuge. The RNA was washed in 70% ethanol and resuspended in 50 μl of diethyl pyrocarbonate-treated water. Trichloroacetic acid precipitations were done to quantify the amount of full-length RNA synthesized.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were done in 10-μl reaction mixtures consisting of 3 × 105 to 4 × 105 cpm of RNA probe (1 μl), approximately 30 μg of testis extract (1 μl), 1 μl of 10× binding buffer (200 mM HEPES [pH 7.6], 30 mM MgCl2, 400 mM KCl, 20 mM DTT), 1 μl of 50% glycerol, and 6 μl of H2O. Reaction mixtures were incubated for 20 min at room temperature, and then sequentially treated with 1 μl of RNase T1 (Calbiochem, La Jolla, Calif.) at 2 U/μl and 2 μl of heparin (Sigma, St. Louis, Mo.) at 5 mg/ml, each for 10 min at room temperature. After addition of 5 μl of 50% glycerol, the samples were electrophoresed on a 4% nondenaturing 60:1 polyacrylamide gel for 2½ h at 180 V and 4°C in gel shift running buffer (45 mM Trizma base, 50 mM boric acid, 1 mM EDTA). Gels were vacuum dried and visualized via autoradiography.
UV cross-linking.
Reactions were set up as described above for EMSA. After heparin treatment, the samples were placed on ice in microcentrifuge tubes, with lids open, and irradiated by a UV light source from a distance of 0.3 m for 30 min. After addition of 13 μl of 2× Laemmli buffer and boiling for 5 min, samples were loaded onto an SDS-polyacrylamide gel (5% stacking gel and 10% resolving gel) and electrophoresed at 200 V for 4 h. Prestained molecular weight markers (Gibco-BRL Life Technologies, Rockville, Md.) were used as size standards. Gels were vacuum dried and visualized via autoradiography.
Pre- and postcut RNA experiments were done via UV cross-linking. For precut RNA experiments, the RNA probe (1 μl) was digested for 10 min with 1 μl of RNase T1 (2 U/μl) in 1 μl of 10× binding buffer–1 μl of 50% glycerol–6 μl of H2O prior to addition of 1 μl (30 μg) of testis extract and incubation at room temperature for 20 min. Samples were then irradiated from a UV light source, treated with heparin, and analyzed via SDS-polyacrylamide gel electrophoresis (PAGE), all as described above. In postcut RNA experiments RNase T1 digestion was done after addition of testis extract and the subsequent incubation but prior to UV irradiation. The remaining protocol was as described for the precut RNA experiments.
Mutant RNA competitions.
Competition experiments were done by EMSA. Reactions were done in a fashion similar to that described above but in 20-μl reaction mixtures with various amounts of 3H-labeled RNA, approximately 30 μg of testis extract (1 μl), 2 μl of 10× binding buffer (200 mM HEPES [pH 7.6], 30 mM MgCl2, 400 mM KCl, 20 mM DTT), 2 μl of 50% glycerol, and H2O to 19 μl. Reaction mixtures were incubated for 20 min at room temperature, and then approximately 50,000 cpm of “hot” 32P-labeled RNA probe (1 μl, 1 ng) was added and the reaction mixtures were incubated at room temperature for an additional 20 min. Either 0, 25, 50, 100, 300, or 500 ng of “cold” RNA was used in the first binding reaction. Samples were then sequentially treated with 2 μl of RNase T1 (Calbiochem) at 2 U/μl and 4 μl of heparin (Sigma) at 5 mg/ml, each for 10 min at room temperature. EMSA was completed as described above. Competition analysis was done using the modeling program Prism (GraphPad, San Diego, Calif.).
Yeast three-hybrid system binding analysis.
A derivative of the S. cerevisiae L40 strain [MATa ura3-52 leu2-3,112 hisΔ200 trp1Δ1 ade2 LYS2::(LexAop)-HIS3 ura3::(LexAop)-LacZ] with an integrated fusion gene encoding the LexA-MS2 coat protein (32) containing either plasmid pGAD10-MSY4ΔN or plasmid pGAD10-MSY2ΔN was transformed with the plasmid encoding the hybrid RNAs, pIII/MS2-2/Prm11–37. MSY4ΔN is a cDNA encoding MSY4 with an amino-terminal deletion of 76 aa, leaving only 9 amino-terminal amino acids (8). All 273 aa of the CSD and C terminus are intact. MSY2ΔN is a cDNA encoding MSY2 with a complete amino-terminal deletion. MSY2ΔN was cloned via PCR with the Matchmaker library (Clontech, Palo Alto, Calif.) as the template using primers 5′-CGCGGATCCCAAGCCGGTGCTGGCAATCC-3′ and 5′-CGCGGATCCGAATCACTCCAGTATGGTG-3′. The PCR product from this reaction was then inserted into the BamHI site of pACT (Clontech) for expression as a fusion protein with the GAL4 activation domain. These hybrid RNA constructs were generated by blunting the 5′ overhanging EcoRI-BamHI ends of the Prm11–37 oligonucleotides by Klenow filling and cloning into the SmaI site of pIII/MS2-2. Transformants were selected on synthetic medium lacking tryptophan, leucine, and uracil. Interactions between MSY4 and the various hybrid RNAs were tested in triplicate by patching single-transformant colonies onto plates of synthetic media lacking tryptophan, leucine, uracil, and histidine and containing 5 mM 3-aminotriazole. Interactions were also tested using β-galactosidase filter assays, in which filters are incubated at room temperature in Z-buffer containing 5-bromo-4-chloro-3-indolyl-β-d-galactoside as the substrate (5).
Quantitative liquid β-galactosidase assays were done on duplicate cultures for each RNA hybrid transformant. Cultures were grown in liquid synthetic medium lacking tryptophan, leucine, uracil, and histidine. Cultures were allowed to grow until log phase, approximately two doubling times (optical density at 600 nm [OD600], 0.5 to 0.8). Three 1-ml aliquots of cells were pelleted from each culture by centrifugation at 10,000 × g in a microcentrifuge. Cells were washed in 500 μl and then resuspended in 100 μl of Z-buffer and lysed by being frozen in liquid nitrogen and then thawed. Debris was pelleted by centrifugation at 10,000 × g, and 700 μl of Z-buffer was added to the supernatant. Freshly prepared o-nitrophenyl-β-d-galactopyranoside (ONPG; 160 μl; 4 mg/ml) was added to each reaction mixture. After color development, 400 μl of 1 M sodium carbonate was added to stop the reaction, and the samples were read at OD420. Statistical analysis was done using Microsoft (Redmond, Wash.) Excel, version 5.0.
RNA analysis.
Total RNA was isolated from dissected mouse tissues as previously described (7). RNA samples were electrophoresed in agarose-formaldehyde gels, transferred to nylon (Hybond-N; Pharmacia BioTech, Peapack, N.J.), and hybridized 15 to 20 h with radioactive α-32P-labeled DNA probes prepared by random oligonucleotide-primed synthesis (13). The nylon membrane was washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% SDS (final stringency) at 60°C and exposed to X-ray film.
Immunohistochemistry.
Immunohistochemistry was performed as previously described (4). Briefly, tissues were dissected from adult mice and fixed in Bouin's fixative overnight and embedded in paraffin. Sections were deparaffinized with xylene and rehydrated using standard procedures. Tissue sections were treated with a primary antibody overnight at 4°C or for 2 to 3 h at room temperature. Biotinylated goat anti-rabbit immunoglobulin G and streptavidin conjugated to horseradish peroxidase (HRP) were used as recommended by the manufacturer (Zymed Laboratories, San Francisco, Calif.). Peroxidase activity was visualized with chromogen aminoethyl carbazole. Tissue sections were counterstained with hematoxylin.
Immunoblotting.
Protein extracts were mixed with Laemmli buffer (23), boiled, and electrophoresed in SDS–8% polyacrylamide gels. The proteins were transferred to nitrocellulose (Gibco). After transfer the membrane was blocked for 30 min to several hours at room temperature in 5% nonfat dry milk and phosphate-buffered saline (PBS) and then incubated overnight at 4°C with the primary antibody at a 1:10,000 dilution. The membrane was washed once in PBS with 0.05% Tween 20 and twice in PBS and then incubated with the secondary antibody conjugated to HRP for several hours at room temperature. After the membrane was washed again as described above, the HRP activity was detected using enhanced chemiluminescence (ECL) as described previously (31). ECL reagent was prepared immediately prior to use by dissolving 40 mg of luminol (5-amino-2,3-dihydro-1,4-phthalazinedione) and 10 mg of 4-iodophenol in 1 ml of dimethyl sulfoxide. Following the addition of 10 ml of 0.1 M Tris (pH 8.5), 5 ml of 5 M NaCl,17 ml of H2O, and 125 μl of H2O2, the membrane was incubated for 2 min and exposed to X-ray film.
Transgenic constructs.
A heterologous reporter was used to evaluate translational control function in vivo as previously described (4, 12, 38). This reporter cassette contains 4.1 kb of mouse Prm1 5′ untranscribed sequence up to the transcriptional start site, a chimeric 5′ UTR of 159 bp (91 bp of Prm1 5′UTR, 7 bp of linker DNA, and 61 bp of the hGH 5′UTR), and the complete hGH coding sequence and introns (9). Oligonucleotides that contain Prm11–37mu4 were inserted into the plasmid at a BamHI site 3′ to the hGH open reading frame and 5′ to the 5′-most 23 nt of the Prm1 3′ UTR that contains the polyadenylation site. One hundred forty base pairs of sequence downstream of the polyadenylation signal is also present to ensure proper 3′ processing of the mRNA.
RESULTS
Delineation of the MSY2 and MSY4 binding site by RNA sequence homology.
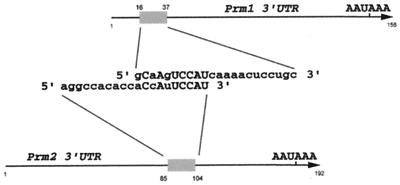
We have previously shown that murine testis extracts contain a 48- and 50-kDa RNA binding activity composed of MSY2 and MSY4 that recognizes a conserved region in the 3′ UTRs of the protamine mRNAs (8, 11). Deletion mapping of the 3′ UTRs of Prm1 and Prm2 mRNA complexes with MSY2 and MSY4 defined a binding site between nt 16 and 37 of the 156-nt Prm1 3′ UTR and between nt 85 and 104 of the 192-nt Prm2 3′ UTR. Comparison of these two binding sites revealed a region of homology, 5′-CNANUCCAU-3′ (identity at seven of nine sites) (Fig. 1); when multiple nucleotides in this region are mutated, MSY2- and MSY4-RNA complex formation is eliminated (8). Comparative sequence analysis of the Prm1 3′ UTRs from several species (Table 1) identified another two conserved nucleotides (boldface) immediately 3′ to the above binding site (5′-CNANUCCAUCA 3′), delineating a stretch with 9 of 11 conserved nucleotides. This sequence is highly conserved from mice to humans, as are both the position of the sequence within the 3′ UTR and the length of the Prm1 3′ UTR, with only slight variation. For example, within the 156-nt mouse Prm1 3′ UTR, the binding site begins at nt 16, whereas within the 148-nt human Prm1 3′ UTR this site begins at nt 13.
FIG. 1.
Comparison of MSY2 and MSY4 minimal binding sites in the Prm1 and Prm2 3′ UTRs, as defined by deletion mapping using EMSAs (11). Arrows, full-length Prm1 and Prm2 3′ UTRs. The 22-nt Prm1 3′ UTR site is aligned with the 20-nt Prm2 3′ UTR site, revealing a region with seven of nine conserved nucleotides. Conserved nucleotides are capitalized (11).
TABLE 1.
Comparative sequence analysis of the Prm1 3′ UTR
| Species | RNA sequencea | Organism |
|---|---|---|
| M. musculus | CAAGUCCAUCA | Mouse |
| R. novegicus | CAAGUCCACCA | Rat |
| H. sapiens | CACAUCCACCA | Human |
| S. imperator | CAAGUCCACCA | New world primate |
| H. lar | CACAUCCACCA | Gibbon |
| P. pygmaeus | CACAUCCACCA | Orangutan |
| A. seniculus | CAAGUCCACCA | Red howler monkey |
| D. marsupialis | AAACUCCAUCU | Opossum |
| I. macrouris | AAACACCAUCU | Bandicoot |
| Conserved nucleotidesb | CA UCCA CA |
Underlined nucleotides are conserved between murine Prm1 and Prm2 3′ UTRs. Boldface indicates nucleotide differences from M. musculus Prm1 3′ UTR.
Conserved in at least seven of the nine species.
Determination of minimum MSY2- and MSY4-binding RNA fragment.
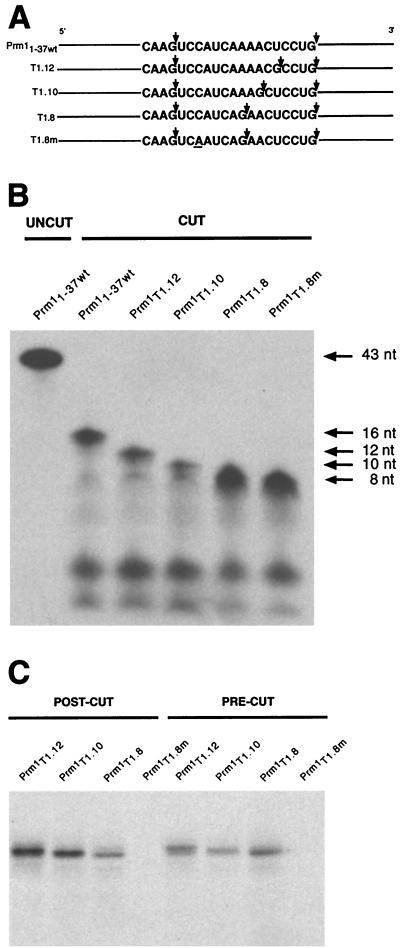
To further refine the minimal size of RNA that MSY2 and MSY4 can bind, a series of Prm1 3′UTR RNAs containing the stretch conserved at 9 of 11 nt was generated; in these RNAs guanines at various distances from a naturally occurring guanine immediately 5′ of the YRS were replaced (Fig. 2A). When digested with RNase T1, these RNAs, T1.12, T1.10, and T1.8, produce 12-, 10-, and 8-nt RNA fragments, respectively, derived from the wild-type 16-nt fragment (Fig. 2B). As a control, a mutant version of T1.8 containing a single nucleotide substitution was also generated (T1.8m).
FIG. 2.
Determination of minimum RNA fragment bound by MSY2 and MSY4. (A) Schematic depiction of Prm11–37wt RNA and four mutant RNAs engineered such that RNase T1 treatment produces different-size RNA fragments containing the YRS. Arrows, RNase T1 cleavage sites. The single nucleotide substitution in T1.8m is underlined. (B) Urea gel analysis of RNase T1 precut RNAs. Top arrow, size of the RNAs prior to cutting (43 nt). Upon treatment with RNase T1, YRS-containing RNA fragments of 12, 10, and 8 nt are released. No uncut RNA of 43 nt is seen in the cut-RNA lanes. (C) UV cross-linking analysis of MSY2 and MSY4 binding of the RNAs depicted in panels A and B. MSY2 and MSY4 were able to bind the T1.12, T1.10, and T1.8 RNA substrates before and after treatment with RNase T1.
UV cross-linking analysis was performed with the T1.12, T1.10, T1.8, and T1.8m RNAs and testis extract. The RNAs were subjected to RNase T1 digestion either before or after the addition of testis extract. After binding incubations and RNase T1 digestions, the reaction mixtures were exposed to UV irradiation and resolved by SDS-PAGE. The intensities of the UV-cross-linked complexes for all the RNAs, with either pre- or posttreatment with RNase T1, were similar (Fig. 2C). A point mutation (C23A) within T1.8m disrupts UV cross-link complex formation, indicating that the RNA bound by MSY2 and MSY4 was indeed the 8-mer fragment. Therefore, we conclude that this 8-nt RNA is sufficient for MSY2 and MSY4 binding.
Mutagenesis of the MSY2 and MSY4 binding site.
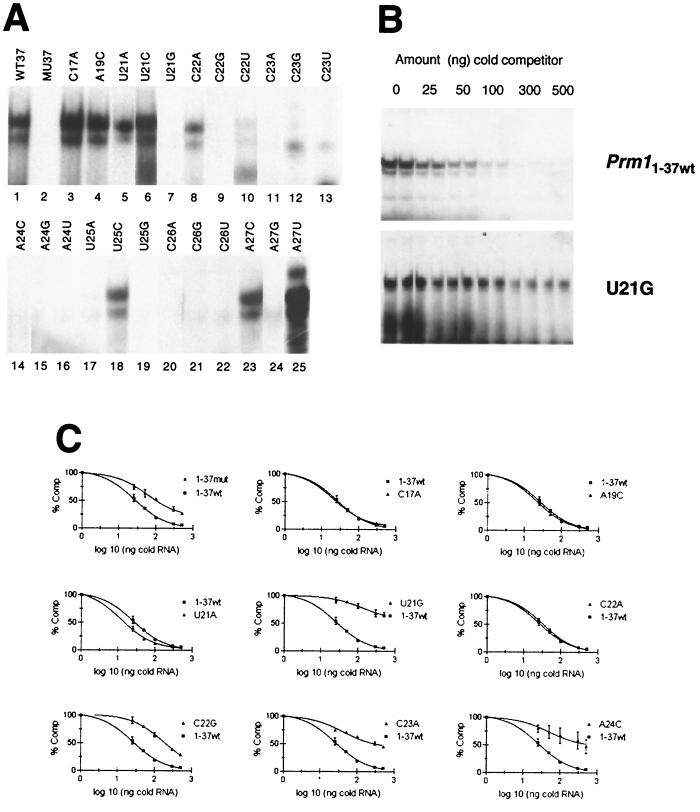
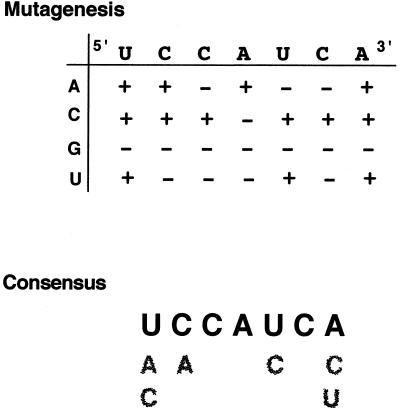
RNAs representing every possible point mutation of the seven conserved nucleotides contained within the binding site defined by T1.8 were generated and analyzed by EMSAs. Incubation of the first 37 nt of the wild-type Prm1 3′ UTR (Prm11–37wt) with testis extract, followed by RNase T1 treatment, heparin treatment, and native PAGE generated an MSY2- and MSY4-RNA EMSA complex consisting of a darker upper band and a lighter lower band (Fig. 3A, lane 1). The Prm11–37mut RNA, in which all of the nucleotides conserved between the Prm1 and Prm2 3′ UTRs within the MSY2 and MSY4 binding site are mutated, eliminated MSY2- and MSY4-RNA complex formation (Fig. 3A, lane 2). The Prm11–37 point mutations C17A, A19C, U21A, U21C, U25C, A27C, and A27U did not significantly decrease MSY2 and MSY4 binding (Fig. 3A, lanes 3 to 6, 18, 23, and 25), whereas Prm11–37 point mutations U21G, C22G, C22U, C23A, C23G, C23U, A24C, A24G, A24U, U25A, U25G, C26A, C26G, C26U, and A27G all disrupted complex formation as indicated by EMSA (Fig. 3A, lanes 7, 9 to 17, 19 to 22, and 24). The point mutation C22A (lane 8) reduced but did not eliminate binding.
FIG. 3.
Mutagenesis of the MSY2 and MSY4 binding site within the Prm1 3′ UTR (Prm11–37) and competition between Prm11–37wt and Prm11–37 mutants. (A) EMSA showing presence or absence of MSY2 and MSY4 binding Prm11–37 variants. WT37, RNA representing the first 37 nt of the Prm1 3′ UTR; MU37, mutant version of WT37 in which 10 nt in the conserved region are mutated. The remaining RNAs are WT37 point mutants. (B) EMSA showing the ability of cold Prm11–37wt to compete hot Prm11–37wt binding, while U21G, which is not bound by MSY2 and MSY4, does not compete Prm11–37wt binding. (C) Curves for competition between wild-type Prm11–37 RNA and mutant Prm11–37 RNAs. Each plot contains the same Prm11–37wt control curve. Percentages of wild-type EMSA complex formation versus the amount of mutant Prm11–37 RNA competitor are plotted.
To determine the relative binding affinity of MSY2 and MSY4 for a subset of the mutant RNAs generated, competition experiments were performed. Cold [5,6-3H-UTP]-labeled Prm11–37 mutants were synthesized by in vitro transcription and quantified by trichloroacetic acid precipitation. These RNAs were preincubated with testis extract to allow binding of the RNA by MSY2 and MSY4, followed by addition of hot [α-32P]CTP-labeled Prm11–37wt RNA, and the binding reaction mixtures were subjected to EMSA. Competition experiments were performed with either 25-, 50-, 100-, 300-, or 500-fold more cold competitor RNA than hot wild-type RNA (1 ng or approximately 50,000 cpm). In addition, control reactions with no competitor present were set up. All reactions were done in duplicate, and the intensity of the EMSA complex was measured by phosphorimaging (example in Fig. 3B). The duplicates were averaged, and percentages of competition were calculated from comparisons to the hot-RNA-only control. These percent competitions were plotted as competition curves (Fig. 3C). The relative binding affinities of RNAs that compete the wild type are shown in Table 2. In general, Prm11–37 mutants which did not disrupt band shift complex formation, such as the C17A and A19C mutants, were competent to compete Prm11–37wt RNA, whereas Prm11–37 mutant RNAs which did disrupt band shift complex formation, for example the U21G and C23A mutants, did not effectively compete Prm11–37wt.
TABLE 2.
Relative binding affinity of MSY2 and MSY4 for Prm11–37wt RNA and Prm11–37 mutant RNAs by EMSAs
| RNA | Relative binding affinitya (fold) |
|---|---|
| Prm11–37wt | 26 |
| Prm11–37mut | 59 |
| C17A | 21 |
| A19C | 22 |
| U21A | 12 |
| U21G | 134 |
| C22A | 32 |
| C22G | 174 |
| C23A | 42 |
| A24C | 50 |
Relative binding affinity is defined as the ratio of unlabeled RNA to Prm11–37wt radiolabeled RNA required to decrease binding activity to 50%.
Another RNA construct in which the 7-nt MSY2 and MSY4 binding site was placed in the context of the human growth hormone gene (hGH) 3′ UTR was generated. Previous work has shown that MSY2 and MSY4 do not bind the hGH 3′ UTR RNA (11). The inclusion of this site within the context of the hGH 3′ UTR (hGH-YRS) was sufficient to permit binding of MSY2 and MSY4 (data not shown). These results indicate that the YRS is both necessary and sufficient for MSY2 and MSY4 binding.
Analysis of MSY2 and MSY4 RNA binding using the yeast three-hybrid system.
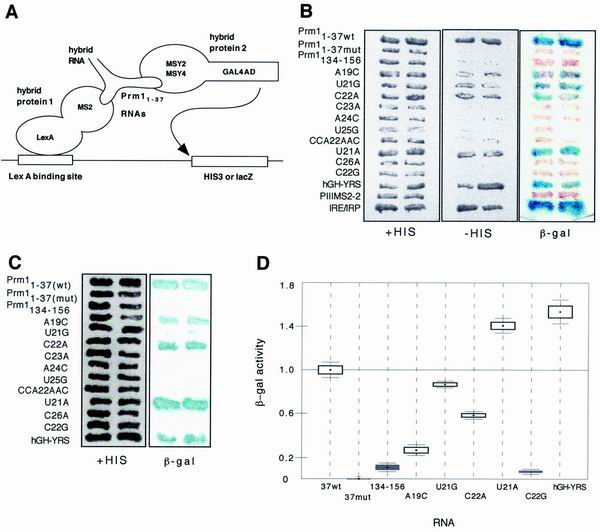
The RNA binding profiles of MSY2 and MSY4 were also tested using the yeast three-hybrid system (Fig. 4A). The yeast three-hybrid system detects RNA-protein interactions by transcriptional activation of a reporter gene (32). In our experiments, the second construct encoded an RNA hybrid of Prm11–37wt or one of the Prm11–37 mutants fused to two copies of the MS2 coat protein recognition site. The substitution of these constructs in the three-hybrid system allowed analysis of MSY2 and MSY4 interactions with several of the RNA mutants analyzed by EMSAs. In this system MSY4 and MSY2 interacted strongly with Prm11–37wt and thus produced high levels of β-galactosidase (Fig. 4B and C). The U21A, U21G, and C22A point mutants, as well as the hGH-YRS RNA, also interacted strongly with MSY2 and MSY4, causing strong activation of the lacZ reporter gene. The A19C point mutant interacted with MSY2 and MSY4 in the three-hybrid system, though weakly. On the other hand, C22G, C23A, A24C, U25G, and C26A RNA point mutants disrupted interactions with MSY2 and MSY4, as did Prm11–37mut.
FIG. 4.
Yeast three-hybrid analysis of MSY4 and MSY2 RNA binding. (A) Diagrammatic depiction of the three-hybrid system. The Prm11–37 RNAs were used as baits in hybrid RNAs with the bacteriophage MS2 coat protein RNA binding site to test for interactions with either MSY2 or MSY4 cDNAs fusions with the GAL4 activation domain. Binding of MSY2 or MSY4 to Prm11–37 RNA results in transcriptional activation of HIS3 and lacZ reporter genes. (B) Two assays for interactions between MSY4 and Prm11–37 RNAs. Transcriptional activation of HIS3 and lacZ indicates an interaction between MSY4 and the bait RNA and is assayed by measuring growth on media lacking histidine and β-galactosidase expression. The Prm1134–156 bait contains nt 134 to 156 of the Prm1 3′UTR, a region of the 3′ UTR which does not contain the YRS and thus serves as a negative control. CCA22AAC, mutation of 3 nt in the YRS which eliminates MSY2 and MSY4 EMSA complex formation (F. Giorgini, unpublished data). PIII/MS2-2 encodes the MS2 RNA binding site alone and serves as a negative control. The iron response element and its binding protein (IRE/IRP) were used as a positive control for the three-hybrid assay. (C) HIS3-dependent growth assay and lacZ assay for interaction between MSY2 and the Prm11–37 RNAs. (D) Quantitative liquid β-galactosidase assays for MSY4 binding of wild-type Prm11–37 RNA, various Prm11–37 mutant RNAs, and a negative-control RNA (Prm1134–156). β-Galactosidase activity is normalized to that of Prm11–37wt. β-Galactosidase assays used ONPG as the substrate. Boxes, standard errors; lines, 95% confidence intervals; black boxes, β-galactosidase activities that are not significantly different from each other.
Quantitative liquid β-galactosidase assays were performed to determine the relative affinities of MSY4 for the various Prm11–37 mutant RNAs. Prm11–37wt β-galactosidase activity was normalized to 1.0 U of β-galactosidase activity. In general, the liquid β-galactosidase assays confirmed the filter assays (Fig. 4D).
YRS binding in vivo.
Several lines of transgenic mice expressing the hGH reporter have been derived to analyze the cis elements required for Prm1 translational repression. Previous experiments showed that the 156-nt Prm1 3′ UTR was sufficient for Prm1-like translational control of the hGH reporter (4). Subsequently, two regions of the Prm1 3′ UTR, Prm11–37wt (which contains the YRS) and Prm193–156, were shown to independently confer translational control (4, 12).
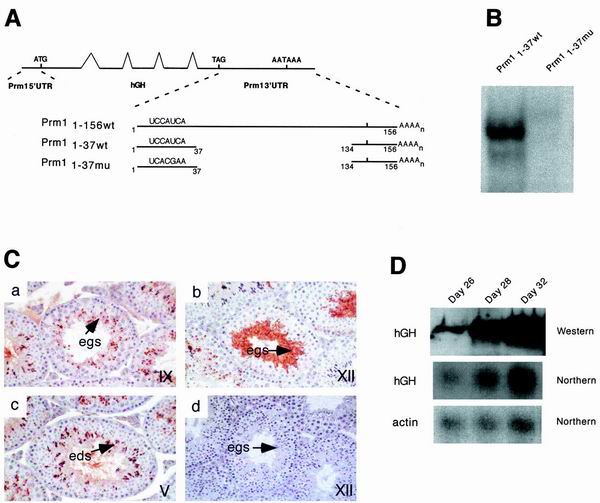
To test the necessity of the YRS for Prm11–37wt-dependent translational delay, Prm11–37mu4 transgenic lines were generated using a Prm1-hGH reporter cassette with a mutant YRS. This transgene encodes a chimeric reporter mRNA consisting of the Prm1 5′ UTR, hGH coding sequence, and a 3′ UTR containing Prm11–37 with a 4-nt mutation (CAUC23–26 to ACGA23–26) in the YRS fused to the 3′-most 23 nt, which contain the nuclear polyadenylation signal (Fig. 5A). This mutant RNA is not bound by MSY2 or MSY4 in an EMSA (Fig. 5B).
FIG. 5.
YRS binding in vivo. (A) Structure and design of the Prm1-hGH-Prm1 1–37mu4 transgene. Top, schematic of a transgenic mRNA including the Prm1 5′ UTR, hGH genomic region, and full-length Prm1 3′ UTR. The first of the three transgenic mRNAs shown below is derived from this transgenic mRNA. The second and third transgenic mRNAs are Prm1 3′ UTR deletion variants missing nt 38 to 133. The wild-type version of the transgene, reported in reference 12, is shown second, with mutant version Prm11–37mu4 shown third. (B) EMSA with testis extracts showing the binding of MSY2 and MSY4 to Prm11–37wt but not to the mutant version used in the transgene. (C) Immunohistochemistry of Prm1-hGH-Prm11–37mu4 transgene. Immunohistochemistry detected the hGH protein in a stage IX tubule (a), a stage XII tubule (b), a stage V tubule (c), and a control stage XII tubule section with no primary antibody (d). Sections were counterstained with hematoxylin. The germ cells are indicated as elongating spermatids (egs) and elongated spermatids (eds). (D) Northern and Western blot analyses of extracts from prepubertal Prm11–37mu4 transgenic animals. RNA was isolated from the testes of mice 26, 28, and 32 days old for Northern blotting analysis. The Northern blot membrane was hybridized with a 32P-labeled probe specific for hGH coding sequences. Total SDS-soluble protein extracts from testes were prepared from the same prepubertal animals. Western analysis was performed with these protein extracts using an anti-hGH antibody.
Three lines of mice were generated and analyzed. The developmental regulation of a transgene can be studied in the testis from a single adult mouse since spermatogenesis is ongoing in the adult testis. Germ cells at different stages of development can be identified histologically by their morphological characteristics and predictable association with cells at other stages of development. To determine if the transgene is regulated like the endogenous protamines, adult testes were analyzed by immunohistochemistry. A minimum of two mice were analyzed from each line. Immunohistochemistry with an hGH antibody showed expression of the hGH protein in the acrosomes of elongating and elongated spermatids (Fig. 5C, stages IX, XII, and V). The hGH protein was also detected in the cytoplasm of stage XII elongating spermatids (Fig. 5C) and continued to be detected in elongated spermatids. The early expression and accumulation of hGH in the acrosomes of spermatids are the hallmark of Prm1-hGH transgenes that are not under translational control (4).
Translational regulation in the mouse can also be studied in prepubertal animals by monitoring the time course of the first wave of spermatogenesis, which is synchronous. This first round of spermatogenesis starts at birth and is complete by day 35. The time after birth at which an mRNA appears can be used to determine in which cell type the gene is first expressed. At day 26, cells at the leading edge of the first wave of spermatogenesis are well into the round spermatid stage, and by day 28 these cells have become elongating spermatids. By day 32, elongated spermatids are found in the testes. Transgenic mice with the hGH reporter fused to Prm11–37wt express transgenic mRNA when 26, 28, and 32 days old and hGH protein by day 32, a pattern indicative of Prm1-like translational delay (12). However, the transgenic mice described here containing Prm11–37mu4 do not have a pattern indicative of Prm1-like translational control. Expression of both the transgenic mRNA and the hGH protein is seen in 26-, 28-, and 32-day-old mice, indicating that repression is relieved by mutation of the YRS (Fig. 5D). These results suggest that a factor, likely MSY2 and MSY4, binds this site in vivo in a functional manner.
DISCUSSION
Comparative sequence analysis and mutagenesis were used to define a consensus sequence, 5′-UACCACAUCCACU-3′ (the YRS), that is present in the Prm1 3′ UTR and that is bound by the murine Y-box proteins MSY2 and MSY4 (Fig. 6). The sequence-specific binding of MSY2 and MSY4 to the YRS in vitro and the presence of the YRS within the Prm1 3′ UTR, suggest that the YRS specifically recruits MSY2 and MSY4 into the Prm1 mRNP and is important for its function. In addition, mutation of the YRS in vivo relieved Prm1-like repression of a reporter construct, further suggesting that this site is functionally bound by MSY2 and MSY4.
FIG. 6.
Consensus YRS sequence and summary of mutational analysis. A schematic diagram depicting Prm1 3′ UTR nt 21 to 27 along the x axis and all possible base substitutions along the y axis is shown. +, mutations that do not disrupt MSY2 and MSY4 binding; −, mutations that eliminate MSY2 and MSY4 binding. A consensus sequence for the YRS is also shown.
In a novel application of the yeast three-hybrid system, mutational analysis of the YRS binding site was performed using both MSY2 and MSY4. Comparison of these results to those by EMSA showed a very similar spectrum of RNAs bound by MSY2 and MSY4 (Table 3). For example, mutants with point mutations of a single nucleotide which either retain binding (C22A) or abolish binding (C22G) as indicated by EMSAs behave in the same manner in the three-hybrid system. In most cases there was agreement between the in vitro EMSA and yeast three-hybrid data; however, the measures of relative affinity calculated by these assays did not always agree. For example, in the competition experiments, the A19C mutant was bound by MSY2 and MSY4 with an affinity equal to that for Prm11–37wt. On the other hand, the A19C mutant was tested in the three-hybrid system with MSY4 and produced less β-galactosidase activity than Prm11–37wt.
TABLE 3.
Comparison of MSY2 and MSY4 binding for Prm11–37wt and various Prm11–37 mutants by the yeast three-hybrid system and EMSAs
| RNA | MSY2 and MSY4 binding by three-hybrid system | MSY2 and MSY4 binding by EMSA |
|---|---|---|
| Prm11–37wt | + | + |
| Prm11–37mut | − | − |
| Prm1134–156 | − | − |
| A19C | + | + |
| U21A | + | + |
| U21G | + | − |
| C22A | + | + |
| C22G | − | − |
| C23A | − | − |
| A24C | − | − |
| U25G | − | − |
| C26A | − | − |
| hGH-YRS | + | + |
EMSAs and the three-hybrid assay define binding profiles and substrate affinities for MSY2 and MSY4 which are overlapping, but different. These variations could be due to a difference in the sensitivities of the two assays. The activation of the lacZ reporter gene in the three-hybrid screen requires only a relatively weak transient interaction between protein and RNA, whereas EMSA requires that protein-RNA interactions withstand migration in an electric field through a polyacrylamide gel. It is also possible that these variations reflect differences in the binding behavior of MSY2 and MSY4 in vivo versus in vitro. Another possibility is that both MSY2 and MSY4 behave differently in isolation than when in the presence of the other. Nonetheless, these data highlight the potential of the three-hybrid system as a valuable tool for binding site analysis of RNA binding proteins.
Several lines of transgenic mice carrying fusions of the hGH reporter and Prm1 cis elements have been generated. Two regions of the Prm1 3′ UTR, one containing the YRS and another containing a conserved sequence in the 3′-most region of the 3′ UTR, have been shown to confer Prm1-like translational control of the hGH reporter (4, 12). The transgenic experiments herein indicate that mutating the YRS in the context of Prm11–37 relieves this translational control. However, further mutational analysis has shown that while the 3′-most conserved region is required for translational control in the context of the full-length Prm1 3′ UTR, the YRS alone is not sufficient (39). It is possible, however, that the binding of MSY2 and MSY4 to the YRS is an important event in repression, which functions in concert with the conserved downstream element. MSY2 and MSY4 may also have roles other than translational repression. They may function in stabilizing the Prm1 mRNA and protecting it from RNases by its sequestration in an mRNP. A secondary effect of this packaging may be to keep the repressed mRNA unavailable to the translational machinery. Finally, it is also possible that interactions between MSY2 and MSY4 and other factors may be important for activation of translation of the Prm1 messages contained in these mRNPs.
Binding sites similar to the YRS have been defined for other Y-box proteins, including FRGY2 and chkYB-1b and chkYB-2. RNA binding protein FRGY2 has been shown to bind the FRGY2 YRS 5′-AACAUC-3′ using the Selex methodology (3). The spectra of RNA sequences that MSY2 and MSY4 and that FRGY2 bind are similar but different. MSY2 and MSY4 can bind the FRGY2 YRS in the context of the Prm1 3′ UTR (F. Giorgini, unpublished data). Recently, chk-YB-1b and chk-YB-2 have been shown by RNA EMSAs to specifically bind an RNA sequence, 5′-GUAACAAC-3′, which is present in Rous sarcoma virus long-terminal repeats present in avian cells and which is also similar to the MSY2 and MSY4 YRS (33). Despite variations in binding patterns of the MSY2 and MSY4 YRS, FRGY2 YRS, and chk-YB YRS, it seems that Y-box family members can bind a conserved subset of sequences.
Work with FRGY2 has shown that the CSD is required for sequence-specific RNA binding, while nonspecific RNA binding interactions of the C-terminal tail are required for stable association of FRGY-2 into mRNPs (24). Preliminary domain mapping using recombinant MSY4 supports the role of the CSD in sequence-specific RNA binding (8). In addition, both MSY2 and MSY4 bind the YRS specifically in the three-hybrid system (Fig. 4B and C). It is interesting that the domain of Y-box proteins likely to be responsible for sequence-specific binding, the CSD, is also the most highly conserved region of the protein. How can different Y-box proteins with highly similar CSDs bind specific RNAs? One model is that certain Y-box proteins will be preferentially recruited to specific RNAs based on proximity to the nascent transcript in the nucleus. For example, the Prm1 promoter contains two Y-box DNA elements that MSY2 and MSY4 could potentially bind (18). MSY2 present in mouse testis nuclear extracts has been shown to interact with the Prm2 promoter (27). Thus, it is possible that MSY2 and MSY4 first bind Y-box DNA elements in the Prm1 promoter and then bind to the YRS RNA element in the 3′ UTR; they are then exported as a complex from the nucleus to the cytoplasm. Such dual functionality has been seen with other nucleic acid binding proteins. The best studied is likely Xenopus protein TFIIIA, which forms complexes with both 5S rRNA gene box C DNA and with 5S rRNA in cytoplasmic 7S particles (22).
There are at least two other examples of Y-box proteins where specific RNA binding is likely to be important to their in vivo function. Y-box proteins chk-YB-1b and chk-YB-2 have been implicated in both transcription from the Rous sarcoma virus promoter and translation repression by sequence-specific RNA binding (33). A mitochondrial Y-box protein in Trypanosoma brucei, RBP16, which binds guide RNAs (gRNAs) specifically in vitro, has been shown to interact with gRNAs in vivo (28). It is likely that RPB16 is involved in kinetoplastid RNA editing. These two examples show that many Y-box proteins may function in vivo by specific interactions with RNAs.
In conclusion, it is clear that the image of Y-box proteins as either dsDNA-binding transcription factors or nonspecific RNA masking proteins is changing. Several Y-box proteins are now known to bind RNA specifically in a variety of biological roles. The discovery of MSY2 and MSY4 as sequence-specific RNA binding proteins and likely factors involved in Prm1 and Prm2 metabolism suggests an additional role of Y-box proteins as factors important for targeting specific mRNAs to mRNPs.
ACKNOWLEDGMENTS
We thank Mark A. Fajardo for insight into the experimental design and for many lively discussions on this research. We are also indebted to many members of the Braun laboratory for critical discussions about this work and help assembling the manuscript.
This work was supported by National Institutes of Health grant HD27215 to R.E.B.
REFERENCES
- 1.Bae W, Jones P G, Inouye M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol. 1997;179:7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balhorn R, Weston S, Thomas C, Wyrobek A J. DNA packaging in mouse spermatids. Synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150:298–308. doi: 10.1016/0014-4827(84)90572-x. [DOI] [PubMed] [Google Scholar]
- 3.Bouvet P, Matsumoto K, Wolffe A P. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 4.Braun R E, Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Protamine 3′-untranslated sequences regulate temporal translational control and subcellular localization of growth hormone in spermatids of transgenic mice. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 5.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 6.Brinster R L, Chen H Y, Trumbauer M E, Yagle M K, Palmiter R D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cathala G, Savouret J F, Mendez B, West B L, Karin M, Martial J A, Baxter J D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2:329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- 8.Davies H G, Giorgini F, Fajardo M A, Braun R E. A sequence-specific RNA binding complex expressed in murine germ cells contains MSY2 and MSY4. Dev Biol. 2000;221:87–100. doi: 10.1006/dbio.2000.9658. [DOI] [PubMed] [Google Scholar]
- 9.DeNoto F M, Moore D D, Goodman H M. Human growth hormone DNA sequence and mRNA structure: possible alternative splicing. Nucleic Acids Res. 1981;9:3719–3730. doi: 10.1093/nar/9.15.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evdokimova V M, Wei C L, Sitikov A S, Simonenko P N, Lazarev O A, Vasilenko K S, Ustinov V A, Hershey J W, Ovchinnikov L P. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem. 1995;270:3186–3192. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- 11.Fajardo M A, Butner K A, Lee K, Braun R E. Germ cell-specific proteins interact with the 3′ untranslated regions of Prm-1 and Prm-2 mRNA. Dev Biol. 1994;166:643–653. doi: 10.1006/dbio.1994.1344. [DOI] [PubMed] [Google Scholar]
- 12.Fajardo M A, Haugen H S, Clegg C H, Braun R E. Separate elements in the 3′ untranslated region of the mouse protamine 1 mRNA regulate translational repression and activation during murine spermatogenesis. Dev Biol. 1997;191:42–52. doi: 10.1006/dbio.1997.8705. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein J, Pollitt N S, Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Tekur S, Reinbold R, Eppig J J, Choi Y C, Zheng J Z, Murray M T, Hecht N B. Mammalian male and female germ cells express a germ cell-specific Y-box protein, MSY2. Biol Reprod. 1998;59:1266–1274. doi: 10.1095/biolreprod59.5.1266. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa S L, Doetsch P W, Hamilton K K, Martin A M, Okenquist S A, Lenz J, Boss J M. DNA binding properties of YB-1 and dbpA: binding to double-stranded, single-stranded, and abasic site containing DNAs. Nucleic Acids Res. 1991;19:4915–4920. doi: 10.1093/nar/19.18.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 18.Johnson P A, Peschon J J, Yelick P C, Palmiter R D, Hecht N B. Sequence homologies in the mouse protamine 1 and 2 genes. Biochim Biophys Acta. 1988;950:45–53. doi: 10.1016/0167-4781(88)90071-1. [DOI] [PubMed] [Google Scholar]
- 19.Jones P G, VanBogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleene K C, Distel R J, Hecht N B. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol. 1984;105:71–79. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- 21.Kwon Y K, Murray M T, Hecht N B. Proteins homologous to the Xenopus germ cell-specific RNA-binding proteins p54/p56 are temporally expressed in mouse male germ cells. Dev Biol. 1993;158:99–100. doi: 10.1006/dbio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 22.Ladomery M. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto K, Meric F, Wolffe A P. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. J Biol Chem. 1996;271:22706–22712. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto K, Wolffe A P. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 1998;8:318–323. doi: 10.1016/s0962-8924(98)01300-2. [DOI] [PubMed] [Google Scholar]
- 26.Murray M T, Schiller D L, Franke W W. Sequence analysis of cytoplasmic mRNA-binding proteins of Xenopus oocytes identifies a family of RNA-binding proteins. Proc Natl Acad Sci USA. 1992;89:11–15. doi: 10.1073/pnas.89.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolajczyk B S, Murray M T, Hecht N B. A mouse homologue of the Xenopus germ cell-specific ribonucleic acid/deoxyribonucleic acid-binding proteins p54/p56 interacts with the protamine 2 promoter. Biol Reprod. 1995;52:524–530. doi: 10.1095/biolreprod52.3.524. [DOI] [PubMed] [Google Scholar]
- 28.Pelletier M, Miller M M, Read L K. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 2000;28:1266–1275. doi: 10.1093/nar/28.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakura H, Maekawa T, Imamoto F, Yasuda K, Ishii S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene. 1988;73:499–507. doi: 10.1016/0378-1119(88)90514-8. [DOI] [PubMed] [Google Scholar]
- 30.Salvetti A, Batistoni R, Deri P, Rossi L, Sommerville J. Expression of DjY1, a protein containing a cold shock domain and RG repeat motifs, is targeted to sites of regeneration in planarians. Dev Biol. 1998;201:217–229. doi: 10.1006/dbio.1998.8996. [DOI] [PubMed] [Google Scholar]
- 31.Schneppenheim R, Rautenberg P. A luminescence Western blot with enhanced sensitivity for antibodies to human immunodeficiency virus. Eur J Clin Microbiol. 1987;6:49–51. doi: 10.1007/BF02097190. [DOI] [PubMed] [Google Scholar]
- 32.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swamynathan S K, Nambiar A, Guntaka R V. Chicken Y-box proteins chk-YB-1b and chk-YB-2 repress translation by sequence-specific interaction with single-stranded RNA. Biochem J. 2000;348:297–305. [PMC free article] [PubMed] [Google Scholar]
- 34.Tafuri S R, Wolffe A P. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4) J Biol Chem. 1993;268:24255–24261. [PubMed] [Google Scholar]
- 35.Tafuri S R, Wolffe A P. Xenopus Y-box transcription factors: molecular cloning, functional analysis and developmental regulation. Proc Natl Acad Sci USA. 1990;87:9028–9032. doi: 10.1073/pnas.87.22.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolffe A P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 37.Wolffe A P, Tafuri S, Ranjan M, Familari M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 1992;4:290–298. [PubMed] [Google Scholar]
- 38.Zhong J, Peters A H, Lee K, Braun R E. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 39.Zhong J, Peters A H F M, Kafer K, Braun R E. A highly conserved sequence that is essential for translational repression of the protamine 1 mRNA in spermatids. Biol Reprod. 2001;64:1784–1789. doi: 10.1095/biolreprod64.6.1784. [DOI] [PubMed] [Google Scholar]