Dear editor,
We read with great interest the recent study by Cailard et al.1 who reported the evolution of SARS-CoV-2 Omicron BA.2 variants in kidney transplant recipients (KTRs). They found that some KTRs could shed SARS-CoV-2 for a prolonged time and the viruses in these KTRs accumulated rare mutations which were associated with immune escape and symptom deterioration. Here, we describe a potential mechanism for the emergence of SARS-CoV-2 variants of concern (VOCs) that can rapidly spread globally, change clinical presentations, or decrease effectiveness of vaccines. Some of these VOCs, such as Alpha, Delta and Omicron, rapidly outcompeted previous variants with increased transmissibility.2, 3, 4
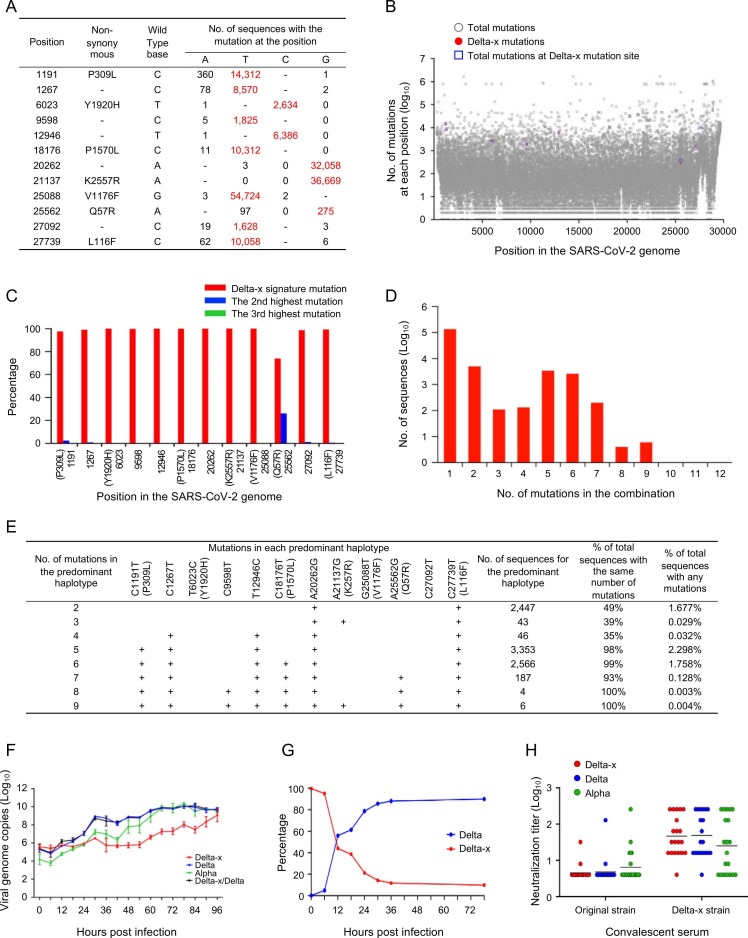
There was one epidemic spike of SARS-CoV-2 infection in Guangzhou city in China between May 21 and June 19 in 2021 (Fig. S1). Whole genome sequence analysis showed that these viruses were highly homogenous, with the majority of them (66%) identical to each other, suggesting that they were derived from a single source. Since these sequences formed a distinct cluster within the large Delta variant group, they were named Delta-x (Figs. S2-S3). Delta-x acquired 12 new signature mutations not commonly found in other variants ( Fig. 1A). The mutations were randomly scattered across the whole viral genome (Fig. 1B). We analyzed all available 1,698,654 good quality sequences available in the Global Initiative on Sharing All Influenza Data (GISAID) database by May 21, 2021. All 12 mutations were at the high variable sites (Fig. 1B). These Delta-x signature mutations were among the top 12% of mutations that occurred more than 1,000 times except one at position 25,562 (Fig. 1B).
Fig. 1.
Genetic and biological characterization of the new Delta-x variant. (A) Delta-x signature mutations (red) were the highest among all three possible mutations (black) at each position. (B) Frequencies of all mutations in the SARS-CoV-2 genomes. All mutations and Delta-x signature mutations are shown in blue squares and red dots, respectively, while all others are shown as gray open circles. (C) Percentage of the highly mutable Delta-x signature mutations is shown in red, while other two mutations are shown in blue and green, respectively. A total of 1,698,654 sequences (collected before May 21, 2021) were used for analysis. (D) Sequences with different numbers of the combined Delta-x signature mutations. (E) Analysis of the predominant haplotypes with different numbers of mutations among total sequences with the same number of mutations and total sequences with any mutations. (F) Viral replication kinetics was monitored by measuring viral RNA copy numbers in the cell culture supernatants using RT-qPCR. (G) Viral fitness was compared between the Delta-x and Delta viruses by determining the proportion of each virus in the same culture using NGS. (H) Neutralization of the Delta-x virus. The neutralization susceptibility of the Delta-x to convalescent sera from individuals infected with original viruses (n = 23) or Delta-x (n = 20) was compared to those of the Delta and Alpha viruses.
At each site, there are three possible mutations. Interestingly, the frequencies of all 12 mutations were the highest among them (Fig. 1C). This suggests that Delta-x signature mutations are a unique combination of 12 highly mutable nucleotides. Examining the frequencies of all possible combinations of mutations showed that the vast majority (91.40%) of the sequences did not contain any of the Delta-x unique mutations. Among the 145,929 sequences carrying any Delta-x mutations, the sequences with one mutation were predominant (92.15%), while the sequences with 2, 5, or 6 mutations were more frequent than others (Fig. 1D). No sequences with more than nine signature mutations were detected.
All predominant haplotypes for the sequences with the same number of mutations accounted for at least 35% of the population (Fig. 1E). The predominant haplotypes for the combinations of higher numbers of mutations (5−9) were nearly exclusive (93%−100%). These results suggest that the predominant haplotypes present at high frequency or exclusively are more viable than others. Interestingly, each predominant haplotype generally differed from that with one fewer mutation by gaining an additional mutation (Fig. 1E). The predominant haplotypes with the most detectable mutations (8 or 9) were rare, but each was the only haplotype detected in that population. The sequences containing eight or nine Delta-x mutations were found in different countries in four continents (Fig. S4), suggesting that the ancestors of Delta-x had been widely presented in different human populations.
When compared to Delta and Alpha in cell culture, the Delta-x replicated at a delayed rate (Fig. 1F, S5). To more accurately compare their fitness, both Delta and Delta-x were cultured together. The Delta virus quickly outcompeted (88.2%) the Delta-x virus, demonstrating that Delta-x was less fit than Delta (Fig. 1G). When tested with the sera from the donors infected with original and Delta-x strains, all three viruses were similarly neutralized (Fig. 1H), indicating that both Delta-x and Delta have a similar neutralization profile.
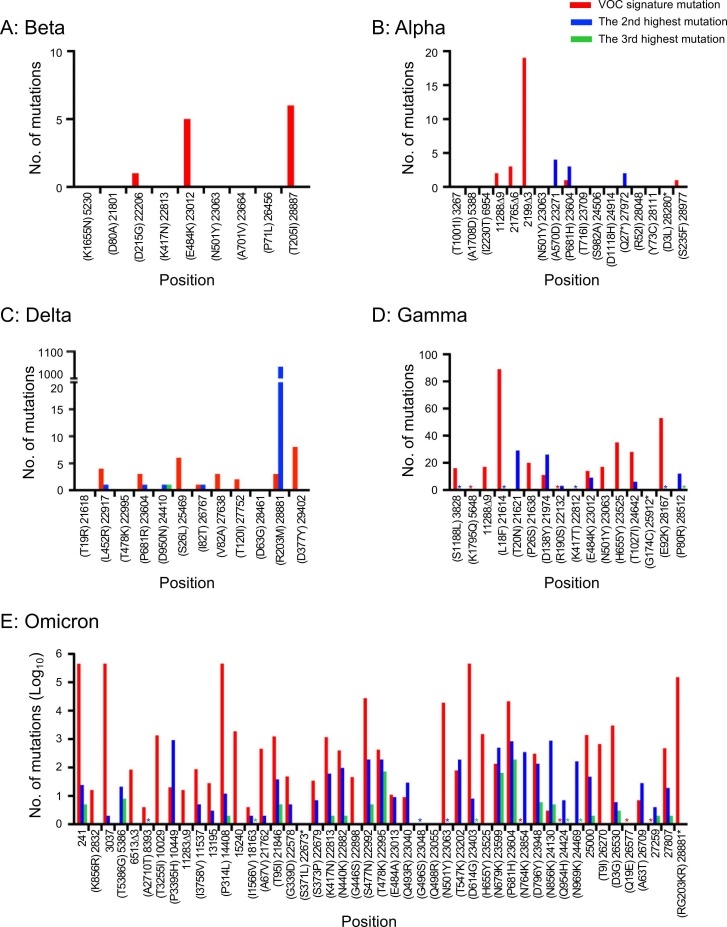
To investigate if signature mutations in other VOCs were also generated through the similar mechanism, we obtained good quality SARS-CoV-2 whole genome sequences (8,846,680) from GISAID by March 5, 2022. As seen with Delta-x, the majority of VOC signature mutations in all five VOCs (Alpha, Beta, Delta, Gamma, and Omicron) were also those with the highest mutation rates at their positions ( Fig. 2). Thus, they were all generated through acquiring a set of the highest mutable nucleotides at the majority of signature mutation sites. For the earlier VOCs, combinations of signature mutations in the same viral genome were either not found (Beta and Alpha) or very rare (Delta and Gamma). However, many sequences with 9 or fewer Omicron signature mutations were found among 473,719 sequences. Examination of the predominant haplotypes for sequences with 2–9 mutations showed that those with higher numbers of mutations were also generated by gaining additional mutations, as observed for Delta-x (Fig. S6).
Fig. 2.
Number of all three possible mutations at each signature mutation site. The numbers of the highly mutable signature mutations in each VOC are shown in red, while other two mutations are shown in blue and green, respectively. The numbers of sequences used for Beta (A), Alpha (B), Delta (C), Gamma (D), and Omicron (E) are 1152, 6969, 6969, 73733, and 473719, respectively. An asterisk indicates one sequence at the position.
Among 12 Delta-x mutations, 5 were synonymous mutations and only 1 (V1176F) was found in the end of the S gene (Table S1). Therefore, all those mutations should not be driven by neutralizing antibodies. This was in good agreement with the observation that no significant differences were observed between Delta and Delta-x. However, four mutations (Y1920H, V1176F, Q57R and L116F) in NSP3, S, ORF3a, and ORF7a, respectively, were found in the known CD8 restricted T cell epitopes.5 Thus, escaping from T cell immune responses may play a critical role in the generation of the Delta-x variant.
The majority of Delta-x signature mutations (11 of 12) were transition mutations (A:G or T:C), which are favored by the ExoN defective viruses.6, 7 Similar results were also observed for other VOCs. Therefore, accumulation of these mutable nucleotides is likely an intrinsic property of SARS-CoV-2 when the function of the proofreading enzyme ExoN is affected. Our study also confirmed that the high transmissibility of VOCs is not necessarily associated with their replication capacity and pathogenicity.8, 9, 10
Understanding of the mechanisms of generation of new VOCs can have important implications. Since the accumulation of highly mutable nucleotides in the SARS-CoV-2 genome may be the intrinsic property of its replication-associated enzymes, the new combinations of these mutable nucleotides are more likely to produce viable variants with higher transmissibility. This may explain why new VOCs constantly emerge and rapidly replace the previous variants.2, 3, 4 Highly mutable nucleotides, especially those at known biological function domains or epitopes targeted by immune responses, should be closely monitored. Furthermore, modeling of such highly mutable nucleotides may be used to predict new VOCs in the future.
Funding
This work was supported by the National Key Research and Development Program of China (Grant Nos. 2021YFC2301500 and 2021YFC0863300), the Project of Medicine Discipline of Guangzhou (2021-2023-11).
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgment
We thank all those who have contributed SARS-CoV-2 genome sequences to the GISAID database.
Addition information
Supplementary information contains figures, table, methods, and accession number for the sequences used in this study.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jinf.2023.03.003.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Caillard S., Laugel E., Benotmane I., Kremer S.F. Molecular evolution of the SARS-CoV-2 omicron BA.2 variant in kidney transplant recipients with prolonged viral shedding. J Infect. 2023 doi: 10.1016/j.jinf.2023.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo P., de Benito N. Alpha variant SARS-CoV-2 infection: how it all starts. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595(7865):17–18. doi: 10.1038/d41586-021-01696-3. [DOI] [PubMed] [Google Scholar]
- 4.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398(10317):2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., et al. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29(7):1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLOS Pathog. 2013;9(8) doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian D., Sun Y., Xu H., Ye Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J Med Virol. 2022;94(6):2376–2383. doi: 10.1002/jmv.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karim F., Moosa M., Gosnell B., Cele S., Giandhari J., Pillay S., et al. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv. 2021 doi: 10.1101/2021.06.03.21258228. [DOI] [Google Scholar]
- 9.Shuai H., Chan J.F., Hu B., Chai Y., Yuen T.T., Yin F., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603(7902):693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material