Abstract
Mitochondrial dysfunction and Aβ accumulation are hallmarks of Alzheimer's disease (AD). Decades of research describe a relationship between mitochondrial function and Aβ production. Amyloid precursor protein (APP), of which Aβ is generated from, is found within mitochondria. Studies suggest Aβ can be generated in mitochondria and imported into mitochondria. APP and Aβ alter mitochondrial function, while mitochondrial function alters Aβ production from APP. The role these interactions contribute to AD pathology and progression are unknown. Here, we discuss prior research, the rigor of those studies, and the critical knowledge gaps of relationships between APP, Aβ, and mitochondria.
Keywords: Alzheimer's disease, amyloid beta, amyloid precursor protein, bioenergetics, γ-secretase, mitochondria
Introduction
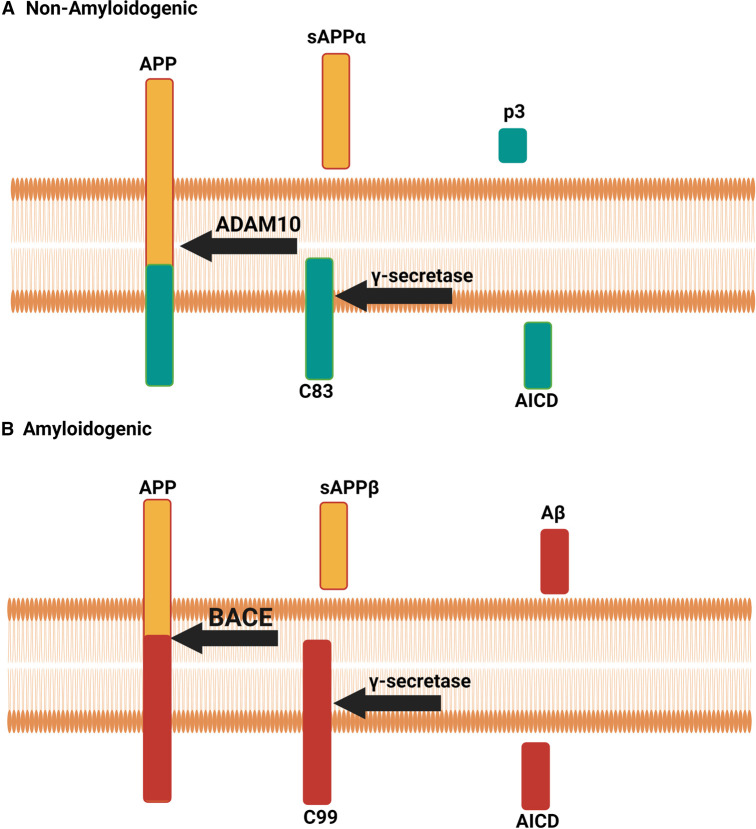
Amyloid beta (Aβ) plaques are a hallmark pathology of Alzheimer's disease (AD) and are a diagnostic requirement for AD [1,2]. Aβ is generated from cleavage of amyloid precursor protein (APP) by secretase enzymes [3]. Two APP processing pathways are currently known, the amyloidogenic and the non-amyloidogenic pathways (Figure 1). The non-amyloidogenic pathway is initiated with APP cleavage by α-secretase (ADAM10), generating soluble APPα (sAPPα) and C-terminal fragment 83 (C83) [4]. The γ-secretase enzyme complex cleaves C83 into p3 and the APP intracellular domain (AICD) [5]. In the amyloidogenic pathway, APP is first cleaved by β-secretase (BACE1), generating soluble APPβ (sAPPβ) and C-terminal fragment 99 (C99) [6]. The γ-secretase complex cleaves C99 generating Aβ and AICD. The γ-secretase complex is composed of presenilin 1 and or 2 (PS1, PS2-catalytic domain), anterior pharynx defective 1 (APH-1), presenilin enhancer 2 (Pen2), and Nicastrin. Cleavage by the γ-secretase complex occurs in a proof-reading manner, generating multiple Aβ species varying from 38 amino acids in length up to 42 amino acids [5]. Aβ42 is considered the most aggregation prone species.
Figure 1. APP processing pathways.
(A) Non-amyloidogenic APP processing. First, APP is cleaved by ADAM10 leading to the production of C83 and sAPPα. Next, C83 is cleaved γ-secretase leading to the production of AICD and p3. (B) Amyloidogenic processing. First, APP is cleaved by BACE1 leading to the production of C99 and sAPPβ. Next, C99 is cleaved by γ-secretase leading to the production of AICD and Aβ. Created using Biorender.
Mutations in APP and PS1/PS2 cause familial forms of AD [7]. Duplication of the APP gene, or chromosome 21 (in down syndrome) also lead to early onset AD [8]. Transgenic animals which express APP or PS mutations have Aβ aggregation within the brain without neuronal loss [9,10]. In many cases, cognitive changes occur in these transgenic models prior to Aβ aggregation/plaque formation which is the opposite of what occurs in human AD [11]. Overall, familial forms of AD account for <5% of all cases and data from transgenic models of familial AD do not recapitulate human disease. A major issue and critical knowledge gap is a lack of understanding of the function of APP and its processing/metabolism.
Research from the last several decades have described relationships between mitochondria, APP, Aβ, and γ-secretase. Numerous studies have established localization of APP, Aβ, and γ-secretase at or within mitochondria [12–20]. Conversely, association of APP and Aβ with mitochondria affects mitochondrial function and APP processing flux [3]. Here we will review the relationships between mitochondria, APP, Aβ, and APP processing.
Relationships between APP processing and mitochondria
Both in vitro and in vivo studies describe a strong relationship between mitochondrial function and APP processing. The first studies examined the effects of sodium azide (cytochrome oxidase or COX inhibitor) and uncoupling of mitochondrial membrane potential using FCCP [21]. Gabuzda et al. treated COS cells with both sodium azide and FCCP and noted the accumulation of APP C99 and APP accumulation within the Golgi. COX inhibition alone also reduced sAPPα levels, suggesting decreased non-amyloidogenic APP processing. Bioenergetic stress through glucose deprivation and glycolysis inhibition decreased non-amyloidogenic APP processing [22]. Separate studies verified these findings in PC12 cells and noted that glycolysis inhibition decreased APP glycosylation, an effect that was mitigated through protein kinase c (PKC) [23]. Furthermore, glycolysis inhibition was found to effect APP trafficking to ER and Golgi in HEK cells [24].
Glycolysis flux and COX affect APP processing in numerous cell types. Fibroblasts from AD subjects reduced sAPPα production under conditions of glucose starvation [25]. These effects were enhanced with COX inhibition. Human fetal astrocytes showed increased Aβ production with glycolysis inhibition through both genetic knockdown of 6-phosphofructo-2-kinase/fructose-2,6-biphosphate (PFKB3) and pharmacological inhibition [26]. It is important to note that glycolysis and mitochondrial respiration are inversely linked, where decreased glycolysis can increase mitochondrial respiration. Furthermore, the downstream effects on reactive oxygen species (ROS), mitochondrial membrane potential, and ATP production were not measured in these studies. Overall, these initial studies depict a strong relationship between APP processing pathways and bioenergetic flux/mitochondrial function.
In transgenic AD mouse models altering mitochondrial function through changes to ROS decreases Aβ plaque burden. Reduced ROS production using two different methods led to a reduction in Aβ plaques with alterations in expression of secretase enzymes and full-length APP. The first study used overexpression of a mitochondrial targeted catalase in the Tg2576 AD mouse model, a method which targets H2O2 production [27]. The second study used overexpression of manganese superoxide dismutase (MnSOD, SOD2) in the Tg19959 AD mouse model, a method which targets superoxide production [28]. Overall, these studies suggest that altering redox and ROS influences APP processing and Aβ production.
Further in vivo studies have found that altering mitochondrial function through COX can also decrease Aβ production and plaque burden in AD transgenic mice. For example, reducing COX function through genetic knockdown of COX10 in AD transgenic mice decreased Aβ plaque burden [29]. In human postmortem brain, higher COX Vmax was associated with higher Aβ burden in non-demented subjects [30]. These findings are interesting because COX Vmax and function are reduced in AD subjects both within the brain (postmortem studies) and systemically (fibroblasts, platelets) [31–34]. Overall in vitro, in vivo, and postmortem studies depict a strong relationship between COX function and Aβ production.
Alterations to complex I and III activity changes Aβ production both in vitro and in vivo. Human embryonic kidney (HEK) and SH-SY5Y cells treated with complex I (rotenone) and complex III (antimycin A) inhibitors increased both ROS and Aβ production [35]. In AD transgenic mice rotenone treatment or knockout of Ndufs4, a component of complex I, also increased Aβ levels. Further studies have shown that mild reduction in complex I activity reduces Aβ production and improves cognition in AD transgenic mice [36,37]. The mechanism of benefit for mild complex I inhibition is thought to occur through activation of AMP-activated protein kinase (AMPK) using a novel compound, CP2. CP2 also altered redox pairs, including NAD/NADH, AMP/ATP, and ADP/ATP [37]. Similar effects have been observed with metformin, an anti-diabetic drug that inhibits complex I and activates AMPK [38].
Mitochondrial DNA (mtDNA) influences APP processing and Aβ production. Mice which accumulate high levels of mtDNA mutations, showed increased Aβ plaque burden [39]. A separate study created congenic transgenic AD mice with varying mtDNA sequences through breeding [40]. This study created C57Bl/6 mice with mtDNA from other mouse strains including FVB/B, NOD/LtJ, and AKR/J. Altering the mtDNA sequences changed Aβ plaque burden and plaque size, while also altering mitochondrial function. Additional work targeted an endonuclease to the mitochondria, PstI in an AD transgenic mouse model. Expression of the mitochondrial targeted endonuclease reduced mtDNA copy number and reduced Aβ plaques and Aβ42 levels [41].
Understanding the role mtDNA may contribute to altered mitochondrial function and APP processing pathways requires a unique model. Cytoplasmic hybrids (cybrids) are cells which harbor a consistent nuclear DNA background with mtDNA from donors. Cybrids are created using cells which lack mtDNA through fusion with platelets from donors. Cybrids generated using platelets from AD subjects had increased ROS, decreased ATP, and decreased COX activity [42–46]. AD cybrids also had increased Aβ deposits and Aβ40/Aβ42 ratios. Overall, mtDNA could drive changes to mitochondrial function and alter APP processing pathways.
More recent studies have examined the role of specific mitochondrial changes and their relationship with APP processing. Using SH-SY5Y cells it was found that mitochondrial membrane potential altered Aβ secretion and APP localization to mitochondria [47]. Mitochondrial depolarization reduced Aβ secretion, but increased intracellular Aβ and mitochondrial localized APP. Hyperpolarization of mitochondria increased Aβ secretion, but decreased intracellular Aβ and mitochondrial localized APP. These results were verified using induced pluripotent stem cell (iPSC) derived neurons.
Data spanning decades across in vitro, in vivo, and postmortem studies describe a strong relationship between mitochondrial function and APP processing. Further research is needed to understand how this relationship may drive AD pathology and cognitive dysfunction. Additional studies have described localization of APP, Aβ, and γ-secretase at mitochondria. There is some equipoise regarding the localization of APP and γ-secretase at mitochondria and how this affects mitochondrial function. These studies and gaps in knowledge are discussed below.
The effects of APP and APP processing on mitochondria
Full length APP
APP localizes to mitochondria and evidence of this has been documented across numerous studies and models [12,15–17,47,48]. Localization of APP at mitochondria is noted in human cell culture, mouse models, and postmortem human brain samples. APP is associated with mitochondria and in mitochondrial associated membranes (MAMs) [14,49]. MAMs are contact sites between the ER and mitochondria which regulate mitochondrial function, calcium flux, and mitophagy.
APP is synthesized and glycosylated within the ER and its currently unknown if APP localization at mitochondria is a result of direct translocation from the ER or translocation from other cell membrane components, like the plasma membrane. Studies and evidence suggest that three positive amino acids in the N-terminus of APP do impact its localization to mitochondria [12]. When those amino acids are mutated to neutral charged amino acids this reduces APP mitochondrial localization. These findings are interesting and suggest APP localizes to mitochondria based on charge-charge interactions, given the overall negative charge of the mitochondrial matrix. More detailed studies are necessary to understand the mechanisms of APP translocation to the mitochondria.
APP localization at mitochondria alters mitochondrial function. In vitro experiments utilizing a wide variety of transformed cell types (PC12, HCN1A, SH-SY5Y, N2a cells) have shown that wild-type (WT) APP expression reduced bioenergetic function of cells, including reduced ATP production, COX activity, and altered calcium homeostasis [12,48,50,51]. Other studies have shown that loss of APP at mitochondria also reduces mitochondrial function, with reduced ATP production, increased ROS, and changes to mitochondrial membrane potential [52–54]. APP null mice further support that APP is important for mitochondrial function and cognition. APP knockout (KO) mice have reduced motor performance with increased astrogliosis [55,56]. Astrocytes isolated from APP KO mice have altered calcium homeostasis with reduced mitochondrial calcium and altered mitochondrial structure.
A highly cited study suggests APP interacts with TOM and TIM complexes [17]. The conclusion of the study was that this interaction between APP/TOM/TIM blocks import of nuclear encoded mitochondrial proteins. Using blue native gel electrophoresis high molecular weight complexes positive for APP, TOMM40, and TIMM23 were observed in mitochondrial fractions from postmortem human AD brain [17]. It should be noted that mitochondrial localized APP and these high molecular weight complexes were not observed in postmortem human brain tissue from normal control subjects. This finding is inconsistent with other studies, which depict APP localization at mitochondria in normal control human postmortem brain [57,58]. The AD postmortem brain mitochondria had reduced COX expression and reduced import of COX. This study did not directly show that APP blocks import channels of mitochondria and did not directly show that APP mitochondrial localization blocks import of nuclear encoded proteins into the mitochondria. This highlights a major knowledge gap in the field.
Soluble APP
The association of sAPP species, either sAPPα or sAPPβ with mitochondria have not been established. However, through signaling mechanisms sAPPα indirectly modulates mitochondrial function. sAPPα exerts neurotropic effects through phosphatidylinositol-3-kinase-Akt kinase (PI3K) and p42/p44 mitogen-activated protein kinase (ERK1/ERK2) pathways [59]. In additional studies examining myotubes, PI3K mediated changes to glucose uptake through altered APP processing, including sAPPα levels [60]. Furthermore, sAPPα is protective against complex I inhibition through rotenone toxicity in mice expressing WT APP. The protective mechanism is likely mediated by PI3K signaling [61]. The overall neuroprotective effects of sAPPα have been reviewed elsewhere [62]. There is a lack of research examining the direct effects of sAPP species on mitochondria and vice versa. Fulfilling this knowledge gap is critical for the AD field.
C-terminal fragments of APP
The role of APP versus APP fragments in mitigating effects on mitochondria is not clearly understood. Transgenic AD mouse models, including the 5X FAD model, accumulate C99 within mitochondria [16,58]. Accumulation of C99 in mitochondria resulted in reduced complex II function, increased DNA damage, and cytochrome c release from mitochondria. When BACE1 expression was reduced mitochondrial function was improved in the 5X FAD mouse model. In mouse embryonic fibroblasts (MEF) with PS knockout, C99 accumulates in MAMs and this leads to an overall increase in MAM content [20]. C99 has also been shown to accumulate in MAMs, and this was associated with decreased mitochondrial respiration and altered lipid content. Overall, the effects of C83 on mitochondria have not been investigated directly but is found within mitochondria [58].
AICD
AICD is a 57 to 59 amino acid fragment generated from APP CTF upon γ-secretase cleavage. Studies show that AICD can translocate to the nucleus with its coactivator, Fe65 where they modulate gene transcription. Recently AICD has been found within mitochondria [58]. Increased expression of AICD and Fe65 reduce mitochondrial membrane potential, ATP production, and superoxide production [63]. The mechanism was linked to altered actin dynamics and downstream mitochondrial morphology/distribution changes. A separate study suggests that AICD alters PINK1 control of mitophagy and mitochondrial dynamics through altered forkhead box O3a (FOXO3a) activity [64]. Furthermore, AICD can inhibit the transcription of coiled-coil-helix-coiled-coil-helix domain containing 6 (CHCHD6). CHCHD6 regulates mitochondrial contact site and cristae organization [65]. When AICD is specifically targeted to mitochondria it induces apoptosis in a hippocampal cell line, and this effect could be rescued with antioxidants and caspase inhibitors [66]. The effects of AICD on mitochondria are likely due to its ability to change gene transcription in the nucleus and its localization within mitochondria. Its currently not known how AICD affects mitochondrial function, and whether it alters mitochondrial gene transcription.
Amyloid beta (Aβ)
Aβ is also found within mitochondria across multiple model types including cell cultures, mouse models, and postmortem human brain. The origin of mitochondrial Aβ could be a consequence of local APP processing or through import into the mitochondria. Several studies report a functional γ-secretase complex within mitochondria capable of generating Aβ [13,15,19,67]. Thus far limited studies have reported BACE1 co-localization with mitochondria in vitro [68]. Other studies have shown that Aβ is directly imported into mitochondria [18].
The first study to show that Aβ was imported into mitochondria leveraged isolated rat brain mitochondria with exogenous Aβ [18]. Mitochondrial localized Aβ was also shown in human brain tissue using immunogold labeling with EM. The data from this study supports a role for TOM and TIM in the import of Aβ into mitochondria. Additional studies have shown Aβ import into mitochondria and subsequent degradation by the mitochondrial presequence peptidase (PreP) [69]. The interaction between Aβ and PreP can inhibit mitochondrial presequence processing of other mitochondrial proteins, leading to a reduction in mitochondrial peptide turnover in the matrix [70].
Aβ affects mitochondrial function in silico, in vitro, and in vivo. Incubation of isolated mitochondria with Aβ42 and copper lead to a dose dependent reduction in COX activity, this was associated with increased high-molecular weight Aβ42 species [71]. In AD transgenic J20 mice, Aβ is found within mitochondria and is associated with mitochondrial dysfunction at complex III and COX [72]. Aβ was shown to interact with Aβ-binding alcohol dehydrogenase (ABAD) in yeast two-hybrid screens, in transgenic AD mice (J20), and postmortem human brain. The interaction between ABAD and Aβ reduced nicotinamide adenine dinucleotide (NAD) binding and altered cognition in transgenic AD mice [73]. Primary neurons cultured from transgenic AD mice overexpressing ABAD had increased ROS production, reduced ATP, and reduced COX activity [74]. Using a separate AD transgenic mouse model with mutant APP and PS1, Aβ accumulation was associated with early mitochondrial dysfunction with increased mitophagy and autophagy [75]. Tg2576 AD mice and neuroblastoma cells expressing mutations in APP have Aβ localized to mitochondria [76]. These models also had increased H2O2 production which correlated with the levels of Aβ. It was further found that COX activity was reduced [76]. Accumulation of H2O2 and decreased COX activity were noted before Aβ plaque deposition. Further studies have shown that Aβ can disrupt mitochondrial fission and fusion dynamics in vitro especially through direct interactions with Drp1 [77,78].
Aβ can be generated in MAMs and oligomers of Aβ increase MAM content and mitochondrial calcium content [79]. An active γ-secretase localizes to MAMs and reducing γ-secretase expression/activity alters MAM number [80]. MAM numbers and function are increased in AD, which is hypothesized to increase Aβ production and alter mitochondrial function [49]. MAMs resemble lipid rafts which would facilitate APP processing pathways by secretase enzymes. Aβ can increase MAM number and alter calcium uptake into mitochondria [49,81]. Post-translational modifications of APP, such as lipidation by palmitoleic acid, can alter its localization to MAMs. Reductions in MAM content and activity was shown to reduce APP processing into Aβ, particularly palmitoylated APP within synapses [14].
The relationship between Aβ and mitochondrial function/MAMs has been a large focus of research within the AD field. Despite this, most studies have focused on models which overexpress or expose cells animal models to large amounts of exogenous Aβ. These types of models are difficult to draw firm conclusions from regarding the endogenous function of Aβ within mitochondria. Furthermore, additional studies are required to understand the role of mitochondrial function in Aβ production.
Concluding remarks
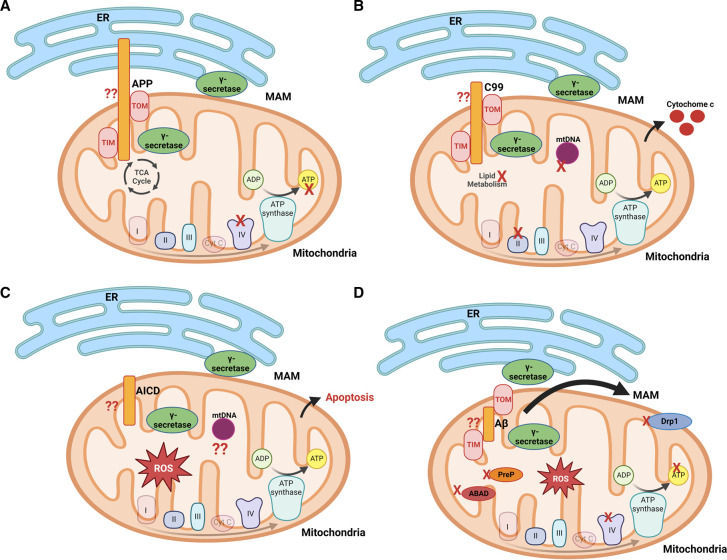
This review focused on studies which examined the relationship between APP, APP processing, and mitochondria. The data supported by current models is outlined in Figure 2. Caution should be taken when drawing conclusions from these current studies as discussed below and highlighted in Figure 2.
Figure 2. APP, APP fragments, and mitochondria.
Current knowledge and knowledge gaps. (A) Full-length APP and mitochondria. It is unknown if APP translocates into the mitochondria from the ER through MAMs. It is also unknown if APP modulates MAM function. Studies suggest APP inhibits COX (complex IV) and ATP production. Some studies suggest APP associates with TOM and TIM. These findings have not been directly confirmed. Its also unclear the role of full-length APP versus APP fragments. (B) CTF of APP (C99) and mitochondria. It is unknown if C99 translocates into the mitochondria or is generated within mitochondria. Studies suggest C99 inhibits complex II, lipid metabolism, damages mtDNA, and induces cytochrome c release. The role of C99 has not been confirmed as independent from Aβ. (C) AICD and mitochondria. It is unknown if AICD translocates into the mitochondria or is generated within mitochondria. Studies suggest AICD induces ROS and apoptosis when it is localized to mitochondria. Its unknown how it affects mtDNA or transcription of mitochondrial genes. (D) Aβ and mitochondria. It is unknown if Aβ translocates into the mitochondria, is generated within mitochondria, or both. Studies suggest Aβ inhibits COX, ATP production, PreP, ABAD, Drp1, induces ROS, and increases MAM content. Created using Biorender.
Despite decades of research there is still no consensus on the function of APP, why APP is processed to fragments including Aβ, and what causes higher rates of Aβ production in AD. Mitochondrial dysfunction is highly prevalent across AD studies and models. Mitochondrial dysfunction has been shown to alter APP processing and Aβ production. Conversely, the production of Aβ (and other APP fragments) directly alter mitochondrial function. However, the role of full-length APP versus APP proteolytic fragments in altering mitochondrial function is unknown.
A functional γ-secretase is localized within mitochondria and full-length APP and its fragments are also found within mitochondria. Localization of APP, APP fragments, and γ-secretase to mitochondria is observed in both normal control and AD postmortem human tissue. However, one highly cited study depicted localization of APP within mitochondria only in AD human postmortem tissue. This study further suggested that APP blocks the import channels of mitochondria. Many conclusions have been made based on this single study which did not directly show that APP blocks mitochondrial protein import. It has been widely assumed that APP localization at mitochondria is ‘bad', but this is based on studies with APP overexpression models. Therefore, studies with endogenous APP protein expression are required to understand the role of APP and its fragments at mitochondria.
Perspectives
APP processing to Aβ and mitochondrial dysfunction are observed in AD subjects and experimental models. There are clear relationships between mitochondrial function, APP processing, and Aβ.
Currently the field has focused on Aβ causing mitochondrial dysfunction, with studies suggesting APP inhibits protein import into mitochondria. Further evidence supports the processing of APP to Aβ within mitochondria.
Future research should focus on directly assessing the function of APP and Aβ at mitochondria. The field should leverage models which do not overexpress or expose model systems to high levels of exogenous proteins.
Abbreviations
- ABAD
Aβ-binding alcohol dehydrogenase
- AD
Alzheimer's disease
- AICD
APP intracellular domain
- AMPK
AMP-activated protein kinase
- APP
amyloid precursor protein
- CHCHD6
coiled-coil-helix-coiled-coil-helix domain containing 6
- HEK
human embryonic kidney
- KO
knockout
- MAMs
mitochondrial associated membranes
- NAD
nicotinamide adenine dinucleotide
- PI3K
phosphatidylinositol-3-kinase-Akt kinase
- ROS
reactive oxygen species
- WT
wild-type
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Funding
This study was supported by the Margaret ‘Peg' McLaughlin and Lydia A. Walker Opportunity Fund, the University of Kansas Alzheimer's Disease Center P30AG035982, and R00AG056600.
References
- 1.Braak, H. and Braak, E. (1997) Diagnostic criteria for neuropathologic assessment of Alzheimer's disease. Neurobiol. Aging 18, S85–S88 10.1016/s0197-4580(97)00062-6 [DOI] [PubMed] [Google Scholar]
- 2.Committee NS. (2020) Coding Guidebook for the NACC Neuropathology Data Form. 11
- 3.Wilkins, H.M. and Swerdlow, R.H. (2017) Amyloid precursor protein processing and bioenergetics. Brain Res. Bull. 133, 71–79 10.1016/j.brainresbull.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allinson, T.M., Parkin, E.T., Turner, A.J. and Hooper, N.M. (2003) ADAMs family members as amyloid precursor protein alpha-secretases. J. Neurosci. Res. 74, 342–352 10.1002/jnr.10737 [DOI] [PubMed] [Google Scholar]
- 5.Wolfe, M.S. and Miao, Y. (2022) Structure and mechanism of the gamma-secretase intramembrane protease complex. Curr. Opin. Struct. Biol. 74, 102373 10.1016/j.sbi.2022.102373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, H., Wang, Y., McCarthy, D., Wen, H., Borchelt, D.R., Price, D.L.et al. (2001) BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat. Neurosci. 4, 233–234 10.1038/85064 [DOI] [PubMed] [Google Scholar]
- 7. www.alzforum.org/mutations
- 8.Salehi, A., Ashford, J.W. and Mufson, E.J. (2016) The link between Alzheimer's disease and down syndrome. A historical perspective. Curr. Alzheimer Res. 13, 2–6 10.2174/1567205012999151021102914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uddin, M.S., Kabir, M.T., Rahman, M.S., Behl, T., Jeandet, P., Ashraf, G.M.et al. (2020) Revisiting the amyloid cascade hypothesis: from anti-abeta therapeutics to auspicious new ways for Alzheimer's disease. Int. J. Mol. Sci. 21, 5858 10.3390/ijms21165858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, A.M. and Roberson, E.D. (2012) Mouse models of Alzheimer's disease. Brain Res. Bull. 88, 3–12 10.1016/j.brainresbull.2011.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jack CR, J., Barrio, J.R. and Kepe, V. (2013) Cerebral amyloid PET imaging in Alzheimer's disease. Acta Neuropathol. 126, 643–657 10.1007/s00401-013-1185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anandatheerthavarada, H.K., Biswas, G., Robin, M.A. and Avadhani, N.G. (2003) Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J. Cell Biol. 161, 41–54 10.1083/jcb.200207030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behbahani, H., Pavlov, P.F., Wiehager, B., Nishimura, T., Winblad, B. and Ankarcrona, M. (2010) Association of Omi/HtrA2 with gamma-secretase in mitochondria. Neurochem. Int. 57, 668–675 10.1016/j.neuint.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya, R., Black, S.E., Lotlikar, M.S., Fenn, R.H., Jorfi, M., Kovacs, D.M.et al. (2021) Axonal generation of amyloid-beta from palmitoylated APP in mitochondria-associated endoplasmic reticulum membranes. Cell Rep. 35, 109134 10.1016/j.celrep.2021.109134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Prete, D., Suski, J.M., Oules, B., Debayle, D., Gay, A.S., Lacas-Gervais, S.et al. (2017) Localization and processing of the amyloid-beta protein precursor in mitochondria-associated membranes. J. Alzheimers Dis. 55, 1549–1570 10.3233/JAD-160953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devi, L. and Ohno, M. (2012) Mitochondrial dysfunction and accumulation of the beta-secretase-cleaved C-terminal fragment of APP in Alzheimer's disease transgenic mice. Neurobiol Dis. 45, 417–424 10.1016/j.nbd.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devi, L., Prabhu, B.M., Galati, D.F., Avadhani, N.G. and Anandatheerthavarada, H.K. (2006) Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci. 26, 9057–9068 10.1523/JNEUROSCI.1469-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansson Petersen, C.A., Alikhani, N., Behbahani, H., Wiehager, B., Pavlov, P.F., Alafuzoff, I.et al. (2008) The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl Acad. Sci. U.S.A. 105, 13145–13150 10.1073/pnas.0806192105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlov, P.F., Wiehager, B., Sakai, J., Frykman, S., Behbahani, H., Winblad, B.et al. (2011) Mitochondrial gamma-secretase participates in the metabolism of mitochondria-associated amyloid precursor protein. FASEB J. 25, 78–88 10.1096/fj.10-157230 [DOI] [PubMed] [Google Scholar]
- 20.Pera, M., Larrea, D., Guardia-Laguarta, C., Montesinos, J., Velasco, K.R., Agrawal, R.R.et al. (2017) Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in Alzheimer disease. EMBO J. 36, 3356–3371 10.15252/embj.201796797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabuzda, D., Busciglio, J., Chen, L.B., Matsudaira, P. and Yankner, B.A. (1994) Inhibition of energy metabolism alters the processing of amyloid precursor protein and induces a potentially amyloidogenic derivative. J. Biol. Chem. 269, 13623–8 10.1016/S0021-9258(17)36875-8 [DOI] [PubMed] [Google Scholar]
- 22.Gasparini, L., Racchi, M., Benussi, L., Curti, D., Binetti, G., Bianchetti, A.et al. (1997) Effect of energy shortage and oxidative stress on amyloid precursor protein metabolism in COS cells. Neurosci. Lett. 231, 113–117 10.1016/s0304-3940(97)00536-3 [DOI] [PubMed] [Google Scholar]
- 23.Henriques, A.G., Domingues, S.C., Fardilha, M., da Cruz e Silva, E.F. and da Cruz e Silva, O.A. (2005) Sodium azide and 2-deoxy-D-glucose-induced cellular stress affects phosphorylation-dependent AbetaPP processing. J. Alzheimers Dis. 7, 201–212 10.3233/jad-2005-7302 [DOI] [PubMed] [Google Scholar]
- 24.Domingues, S.C., Henriques, A.G., Wu, W., da Cruz e Silva, E.F. and da Cruz e Silva, O.A. (2007) Altered subcellular distribution of the Alzheimer's amyloid precursor protein under stress conditions. Ann. N. Y. Acad. Sci. 1096, 184–195 10.1196/annals.1397.085 [DOI] [PubMed] [Google Scholar]
- 25.Gasparini, L., Benussi, L., Bianchetti, A., Binetti, G., Curti, D., Govoni, S.et al. (1999) Energy metabolism inhibition impairs amyloid precursor protein secretion from Alzheimer's fibroblasts. Neurosci. Lett. 263, 197–200 10.1016/s0304-3940(99)00155-x [DOI] [PubMed] [Google Scholar]
- 26.Fu, W., Shi, D., Westaway, D. and Jhamandas, J.H. (2015) Bioenergetic mechanisms in astrocytes may contribute to amyloid plaque deposition and toxicity. J. Biol. Chem. 290, 12504–12513 10.1074/jbc.M114.618157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao, P., Manczak, M., Calkins, M.J., Truong, Q., Reddy, T.P., Reddy, A.P.et al. (2012) Mitochondria-targeted catalase reduces abnormal APP processing, amyloid beta production and BACE1 in a mouse model of Alzheimer'sAlzheimer's disease: implications for neuroprotection and lifespan extension. Hum. Mol. Genet. 21, 2973–2990 10.1093/hmg/dds128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumont, M., Wille, E., Stack, C., Calingasan, N.Y., Beal, M.F. and Lin, M.T. (2009) Reduction of oxidative stress, amyloid deposition, and memory deficit by manganese superoxide dismutase overexpression in a transgenic mouse model of Alzheimer's disease. FASEB J. 23, 2459–2466 10.1096/fj.09-132928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukui, H., Diaz, F., Garcia, S. and Moraes, C.T. (2007) Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc. Natl Acad. Sci. U.S.A. 104, 14163–8 10.1073/pnas.0705738104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troutwine, B.R., Strope, T.A., Franczak, E., Lysaker, C.R., Hamid, L., Mansel, C.et al. (2022) Mitochondrial function and abeta in Alzheimer's disease postmortem brain. Neurobiol. Dis. 171, 105781 10.1016/j.nbd.2022.105781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosetti, F., Brizzi, F., Barogi, S., Mancuso, M., Siciliano, G., Tendi, E.A.et al. (2002) Cytochrome c oxidase and mitochondrial F1F0-ATPase (ATP synthase) activities in platelets and brain from patients with Alzheimer's disease. Neurobiol. Aging 23, 371–376 10.1016/s0197-4580(01)00314-1 [DOI] [PubMed] [Google Scholar]
- 32.Cardoso, S.M., Proenca, M.T., Santos, S., Santana, I. and Oliveira, C.R. (2004) Cytochrome c oxidase is decreased in Alzheimer's disease platelets. Neurobiol. Aging 25, 105–110 10.1016/s0197-4580(03)00033-2 [DOI] [PubMed] [Google Scholar]
- 33.Parker, Jr, W.D. and Parks, J.K. (1995) Cytochrome c oxidase in Alzheimer's disease brain: purification and characterization. Neurology 45, 482–486 10.1212/wnl.45.3.482 [DOI] [PubMed] [Google Scholar]
- 34.Wilkins, H.M., Koppel, S.J., Bothwell, R., Mahnken, J., Burns, J.M. and Swerdlow, R.H. (2017) Platelet cytochrome oxidase and citrate synthase activities in APOE epsilon4 carrier and non-carrier Alzheimer's disease patients. Redox Biol. 12, 828–832 10.1016/j.redox.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leuner, K., Schutt, T., Kurz, C., Eckert, S.H., Schiller, C., Occhipinti, A.et al. (2012) Mitochondrion-derived reactive oxygen species lead to enhanced amyloid beta formation. Antioxid. Redox Signal. 16, 1421–1433 10.1089/ars.2011.4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojakovic, A., Trushin, S., Sheu, A., Khalili, L., Chang, S.Y., Li, X.et al. (2021) Partial inhibition of mitochondrial complex I ameliorates Alzheimer's disease pathology and cognition in APP/PS1 female mice. Commun. Biol. 4, 61 10.1038/s42003-020-01584-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, L., Zhang, S., Maezawa, I., Trushin, S., Minhas, P., Pinto, M.et al. (2015) Modulation of mitochondrial complex I activity averts cognitive decline in multiple animal models of familial Alzheimer's disease. EBioMedicine 2, 294–305 10.1016/j.ebiom.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, X.Y., Huang, S., Chen, Q.B., Zhang, D., Li, W., Ao, R.et al. (2020) Metformin ameliorates abeta pathology by insulin-degrading enzyme in a transgenic mouse model of Alzheimer's disease. Oxid. Med. Cell. Longev. 2020, 2315106 10.1155/2020/2315106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trifunovic, A., Wredenberg, A., Falkenberg, M., Spelbrink, J.N., Rovio, A.T., Bruder, C.E.et al. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- 40.Scheffler, K., Krohn, M., Dunkelmann, T., Stenzel, J., Miroux, B., Ibrahim, S.et al. (2012) Mitochondrial DNA polymorphisms specifically modify cerebral beta-amyloid proteostasis. Acta Neuropathol. 124, 199–208 10.1007/s00401-012-0980-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto, M., Pickrell, A.M., Fukui, H. and Moraes, C.T. (2013) Mitochondrial DNA damage in a mouse model of Alzheimer's disease decreases amyloid beta plaque formation. Neurobiol. Aging 34, 2399–2407 10.1016/j.neurobiolaging.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardoso, S.M., Santana, I., Swerdlow, R.H. and Oliveira, C.R. (2004) Mitochondria dysfunction of Alzheimer's disease cybrids enhances abeta toxicity. J. Neurochem. 89, 1417–1426 10.1111/j.1471-4159.2004.02438.x [DOI] [PubMed] [Google Scholar]
- 43.Khan, S.M., Cassarino, D.S., Abramova, N.N., Keeney, P.M., Borland, M.K., Trimmer, P.A.et al. (2000) Alzheimer's disease cybrids replicate beta-amyloid abnormalities through cell death pathways. Ann. Neurol. 48, 148–155 [DOI] [PubMed] [Google Scholar]
- 44.Silva, D.F., Selfridge, J.E., Lu J, E.L., Roy, N., Hutfles, L., Burns, J.M.et al. (2013) Bioenergetic flux, mitochondrial mass and mitochondrial morphology dynamics in AD and MCI cybrid cell lines. Hum. Mol. Genet. 22, 3931–3946 10.1093/hmg/ddt247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swerdlow, R.H., Koppel, S., Weidling, I., Hayley, C., Ji, Y. and Wilkins, H.M. (2017) Mitochondria, cybrids, aging, and Alzheimer's disease. Prog. Mol. Biol. Transl. Sci. 146, 259–302 10.1016/bs.pmbts.2016.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trimmer, P.A., Keeney, P.M., Borland, M.K., Simon, F.A., Almeida, J., Swerdlow, R.H.et al. (2004) Mitochondrial abnormalities in cybrid cell models of sporadic Alzheimer's disease worsen with passage in culture. Neurobiol. Dis. 15, 29–39 10.1016/j.nbd.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 47.Wilkins, H.M., Troutwine, B.R., Menta, B.W., Manley, S.J., Strope, T.A., Lysaker, C.R.et al. (2022) Mitochondrial membrane potential influences amyloid-beta protein precursor localization and amyloid-beta secretion. J. Alzheimers Dis. 85, 381–394 10.3233/JAD-215280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, T.T., Hsu, C.T. and Kuo, Y.M. (2009) Amyloid precursor protein, heat-shock proteins, and Bcl-2 form a complex in mitochondria and modulate mitochondria function and apoptosis in N2a cells. Mech. Ageing Dev. 130, 592–601 10.1016/j.mad.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 49.Area-Gomez, E., Del Carmen Lara Castillo, M., Tambini, M.D., Guardia-Laguarta, C., de Groof, A.J., Madra, M.et al. (2012) Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31, 4106–4123 10.1038/emboj.2012.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez Sanchez, M.I.G., Waugh, H.S., Tsatsanis, A., Wong, B.X., Crowston, J.G., Duce, J.A.et al. (2017) Amyloid precursor protein drives down-regulation of mitochondrial oxidative phosphorylation independent of amyloid beta. Sci. Rep. 7, 9835 10.1038/s41598-017-10233-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keil, U., Bonert, A., Marques, C.A., Scherping, I., Weyermann, J., Strosznajder, J.B.et al. (2004) Amyloid beta-induced changes in nitric oxide production and mitochondrial activity lead to apoptosis. J. Biol. Chem. 279, 50310–50320 10.1074/jbc.M405600200 [DOI] [PubMed] [Google Scholar]
- 52.Wang, Y., Wu, F., Pan, H., Zheng, W., Feng, C., Wang, Y.et al. (2016) Lost region in amyloid precursor protein (APP) through TALEN-mediated genome editing alters mitochondrial morphology. Sci. Rep. 6, 22244 10.1038/srep22244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Del Prete, D., Rice, R.C., Rajadhyaksha, A.M. and D'Adamio, L. (2016) Amyloid precursor protein (APP) may act as a substrate and a recognition unit for CRL4CRBN and Stub1 E3 ligases facilitating ubiquitination of proteins involved in presynaptic functions and neurodegeneration. J. Biol. Chem. 291, 17209–17227 10.1074/jbc.M116.733626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaefer, P.M., von Einem, B., Walther, P., Calzia, E. and von Arnim, C.A. (2016) Metabolic characterization of intact cells reveals intracellular amyloid beta but not its precursor protein to reduce mitochondrial respiration. PLoS ONE 11, e0168157 10.1371/journal.pone.0168157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, H., Jiang, M., Trumbauer, M.E., Sirinathsinghji, D.J., Hopkins, R., Smith, D.W.et al. (1995) beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81, 525–531 10.1016/0092-8674(95)90073-x [DOI] [PubMed] [Google Scholar]
- 56.Montagna, E., Crux, S., Luckner, M., Herber, J., Colombo, A.V., Marinkovic, P.et al. (2019) In vivo Ca2+ imaging of astrocytic microdomains reveals a critical role of the amyloid precursor protein for mitochondria. Glia 67, 985–998 10.1002/glia.23584 [DOI] [PubMed] [Google Scholar]
- 57.Walls, K.C., Coskun, P., Gallegos-Perez, J.L., Zadourian, N., Freude, K., Rasool, S.et al. (2012) Swedish Alzheimer mutation induces mitochondrial dysfunction mediated by HSP60 mislocalization of amyloid precursor protein (APP) and beta-amyloid. J. Biol. Chem. 287, 30317–30327 10.1074/jbc.M112.365890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaillant-Beuchot, L., Mary, A., Pardossi-Piquard, R., Bourgeois, A., Lauritzen, I., Eysert, F.et al. (2021) Accumulation of amyloid precursor protein C-terminal fragments triggers mitochondrial structure, function, and mitophagy defects in Alzheimer's disease models and human brains. Acta Neuropathol. 141, 39–65 10.1007/s00401-020-02234-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng, G., Yu, Z., Zhou, D. and Mattson, M.P. (2002) Phosphatidylinositol-3-kinase-Akt kinase and p42/p44 mitogen-activated protein kinases mediate neurotrophic and excitoprotective actions of a secreted form of amyloid precursor protein. Exp. Neurol. 175, 407–414 10.1006/exnr.2002.7920 [DOI] [PubMed] [Google Scholar]
- 60.Hamilton, D.L., Findlay, J.A., Montagut, G., Meakin, P.J., Bestow, D., Jalicy, S.M.et al. (2014) Altered amyloid precursor protein processing regulates glucose uptake and oxidation in cultured rodent myotubes. Diabetologia 57, 1684–1692 10.1007/s00125-014-3269-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimdins, K., Waugh, H.S., Chrysostomou, V., Lopez Sanchez, M.I.G., Johannsen, V.A., Cook, M.J.et al. (2019) Amyloid precursor protein mediates neuronal protection from rotenone toxicity. Mol. Neurobiol. 56, 5471–5482 10.1007/s12035-018-1460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dar, N.J. and Glazner, G.W. (2020) Deciphering the neuroprotective and neurogenic potential of soluble amyloid precursor protein alpha (sAPPalpha). Cell. Mol. Life Sci. 77, 2315–2330 10.1007/s00018-019-03404-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward, M.W., Concannon, C.G., Whyte, J., Walsh, C.M., Corley, B. and Prehn, J.H. (2010) The amyloid precursor protein intracellular domain(AICD) disrupts actin dynamics and mitochondrial bioenergetics. J. Neurochem. 113, 275–284 10.1111/j.1471-4159.2010.06615.x [DOI] [PubMed] [Google Scholar]
- 64.Goiran, T., Duplan, E., Chami, M., Bourgeois, A., El Manaa, W., Rouland, L.et al. (2018) beta-amyloid precursor protein intracellular domain controls mitochondrial function by modulating phosphatase and tensin homolog-induced kinase 1 transcription in cells and in Alzheimer mice models. Biol. Psychiatry 83, 416–427 10.1016/j.biopsych.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 65.Shang, Y., Sun, X., Chen, X., Wang, Q., Wang, E.J., Miller, E.et al. (2022) A CHCHD6-APP axis connects amyloid and mitochondrial pathology in Alzheimer's disease. Acta Neuropathol. 144, 911–938 10.1007/s00401-022-02499-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandberg, A.A., Manning, E., Wilkins, H.M., Mazzarino, R., Minckley, T., Swerdlow, R.H.et al. (2022) Mitochondrial targeting of amyloid-beta protein precursor intracellular domain induces hippocampal cell death via a mechanism distinct from amyloid-beta. J. Alzheimers Dis. 86, 1727–1744 10.3233/JAD-215108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hansson, C.A., Frykman, S., Farmery, M.R., Tjernberg, L.O., Nilsberth, C., Pursglove, S.E.et al. (2004) Nicastrin, presenilin, APH-1, and PEN-2 form active gamma-secretase complexes in mitochondria. J. Biol. Chem. 279, 51654–51660 10.1074/jbc.M404500200 [DOI] [PubMed] [Google Scholar]
- 68.Francelin, C., Mitter, S.K., Qian, Q., Barodia, S.K., Ip, C., Qi, X.et al. (2021) BACE1 inhibition increases susceptibility to oxidative stress by promoting mitochondrial damage. Antioxidants (Basel) 10, 1539 10.3390/antiox10101539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falkevall, A., Alikhani, N., Bhushan, S., Pavlov, P.F., Busch, K., Johnson, K.A.et al. (2006) Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J. Biol. Chem. 281, 29096–29104 10.1074/jbc.M602532200 [DOI] [PubMed] [Google Scholar]
- 70.Mossmann, D., Vogtle, F.N., Taskin, A.A., Teixeira, P.F., Ring, J., Burkhart, J.M.et al. (2014) Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab. 20, 662–669 10.1016/j.cmet.2014.07.024 [DOI] [PubMed] [Google Scholar]
- 71.Crouch, P.J., Blake, R., Duce, J.A., Ciccotosto, G.D., Li, Q.X., Barnham, K.J.et al. (2005) Copper-dependent inhibition of human cytochrome c oxidase by a dimeric conformer of amyloid-beta1-42. J. Neurosci. 25, 672–679 10.1523/JNEUROSCI.4276-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caspersen, C., Wang, N., Yao, J., Sosunov, A., Chen, X., Lustbader, J.W.et al. (2005) Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. FASEB J. 19, 2040–2041 10.1096/fj.05-3735fje [DOI] [PubMed] [Google Scholar]
- 73.Lustbader, J.W., Cirilli, M., Lin, C., Xu, H.W., Takuma, K., Wang, N.et al. (2004) ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science 304, 448–452 10.1126/science.1091230 [DOI] [PubMed] [Google Scholar]
- 74.Takuma, K., Yao, J., Huang, J., Xu, H., Chen, X., Luddy, J.et al. (2005) ABAD enhances Abeta-induced cell stress via mitochondrial dysfunction. FASEB J. 19, 597–598 10.1096/fj.04-2582fje [DOI] [PubMed] [Google Scholar]
- 75.de la Cueva, M., Antequera, D., Ordonez-Gutierrez, L., Wandosell, F., Camins, A., Carro, E.et al. (2022) Amyloid-beta impairs mitochondrial dynamics and autophagy in Alzheimer's disease experimental models. Sci. Rep. 12, 10092 10.1038/s41598-022-13683-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manczak, M., Anekonda, T.S., Henson, E., Park, B.S., Quinn, J. and Reddy, P.H. (2006) Mitochondria are a direct site of A beta accumulation in Alzheimer's disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum. Mol. Genet. 15, 1437–1449 10.1093/hmg/ddl066 [DOI] [PubMed] [Google Scholar]
- 77.Wang, X., Su, B., Siedlak, S.L., Moreira, P.I., Fujioka, H., Wang, Y.et al. (2008) Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl Acad. Sci. U.S.A. 105, 19318–19323 10.1073/pnas.0804871105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manczak, M., Calkins, M.J. and Reddy, P.H. (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet. 20, 2495–2509 10.1093/hmg/ddr139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calvo-Rodriguez, M., Hernando-Perez, E., Nunez, L. and Villalobos, C. (2019) Amyloid beta oligomers increase ER-mitochondria Ca2+ cross talk in young hippocampal neurons and exacerbate aging-induced intracellular Ca2+ remodeling. Front. Cell Neurosci. 13, 22 10.3389/fncel.2019.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Area-Gomez, E., de Groof, A.J., Boldogh, I., Bird, T.D., Gibson, G.E., Koehler, C.M.et al. (2009) Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 175, 1810–1816 10.2353/ajpath.2009.090219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hedskog, L., Pinho, C.M., Filadi, R., Ronnback, A., Hertwig, L., Wiehager, B.et al. (2013) Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer's disease and related models. Proc. Natl Acad. Sci. U.S.A. 110, 7916–7921 10.1073/pnas.1300677110 [DOI] [PMC free article] [PubMed] [Google Scholar]