Abstract
For decades research has centered on identifying the ideal balanced skin microbiome that prevents disease and on developing therapeutics to foster this balance. However, this single idealized balance may not exist. The skin microbiome changes across the lifespan. This is reflected in the dynamic shifts of the skin microbiome's diverse, inter-connected community of microorganisms with age. While there are core skin microbial taxa, the precise community composition for any individual person is determined by local skin physiology, genetics, microbe–host interactions, and microbe–microbe interactions. As a key interface with the environment, the skin surface and its appendages are also constantly exchanging microbes with close personal contacts and the environment. Hormone fluctuations and immune system maturation also drive age-dependent changes in skin physiology that support different microbial community structures over time. Here, we review recent insights into the factors that shape the skin microbiome throughout life. Collectively, the works summarized within this review highlight how, depending on where we are in lifespan, our skin supports robust microbial communities, while still maintaining microbial features unique to us. This review will also highlight how disruptions to this dynamic microbial balance can influence risk for dermatological diseases as well as impact lifelong health.
Keywords: host–microbe interactions, microbiome, skin
Introduction
The skin harbors complex microecosystems of bacteria, fungi, and viruses, each with distinct adaptions to survive on the skin. However, there is no single definition of a balanced skin microbiome. For any individual the microbial balance is dynamic, maturing with us as we grow and navigate through the environments around us. Across our lifespan, the stability and function of skin microbial communities are driven both by interactions with the host and between microorganisms.
Human skin varies in its physical characteristics across body sites [1–4], ranging from oily / sebaceous to moist or dry, resulting in distinct microenvironments that promote unique microbial populations. Sebaceous sites (e.g. face, chest, and back) have a high density of hair follicles and sebaceous glands. The lipid rich sebum produced by these glands promotes colonization by lipophilic taxa, primarily Cutibacterium bacteria and Malassezia fungi [4–6]. Moist sites (e.g. elbow crease, axilla, and groin) have high concentrations of apocrine sweat glands and are dominated by Staphylococcus spp. and Corynebacterium spp.[4]. In contrast with other sites, dry sites (e.g. forearm, abdomen, and palms) have the lowest abundance yet greatest microbial diversity, with significant populations of Cutibacterium, Corynebacterium, and Streptococcus species [2]. Thus, at a microenvironmental level, the balance of skin microbial communities is partially dictated by these physiologic and abiotic features of the skin niche.
The skin is an immunologically rich organ. To maintain the skin barrier tissue-resident immune cells collectively sample and respond to microbial products [4], produce antimicrobial peptides (AMPs) [7], and ultimately prevent penetration of skin microbes into deeper skin layers or open wounds. A hallmark of a healthy equilibrium among skin microbial communities is the maintenance of skin barrier integrity. This is accomplished by promoting skin cell maturation [8,9], training the immune system [10–12], and preventing pathogen overgrowth through niche exclusion and competitive interactions such as the production of antimicrobials [13–16]. Disruption of this equilibrium is characterized by the overgrowth of some bacterial species, such as Staphylococcus aureus, and an overall loss of community diversity [2]. This dysbiosis can lead to impaired wound healing, increased inflammation, and greater risk for infection [2,17–19].
Finally, microbe–microbe interactions within the skin microbiome can drive overall community structure. The three prominent skin taxa, Cutibacterium acnes, Corynebacterium spp., and coagulase-negative staphylococci (CoNS) are known to mediate other microbial taxa in the microenvironment and have been extensively reviewed elsewhere [5,20]. Key interactions and recent studies are summarized in Table 1.
Table 1. Key microbe–microbe interactions on the skin.
| Microbe Producing the Molecule | Anti-microbial molecule (Type) | Inhibited Microbes | Proposed Mechanism | Source |
|---|---|---|---|---|
| Cutibacterium acnes | Acnecin (peptide) | non-anecin producing C. acnes | Exact mechanism Unknown. Proposed to help C. acnes phylotypes maintain dominance within a pore. | [105,164] |
| Corynebacterium | ||||
| Cutimycin (thiopeptide) | MRSA | Unknown | [14] | |
| Staphylococcus epidermidis | ||||
| Propionic acid (SCFA) | MRSA | Lowers local pH. This can limit the growth of several pathogens while not significantly affecting growth of skin commensals. | [165-168] | |
| Escherichia coli | ||||
| Candida albicans | ||||
| Propionic acid (SCFA) | Staphylococcus epidermidis | Inhibit bacterial biofilm formation. These SCFA also influence melanocyte, keratinocyte, and sebocyte gene expression, and modulate host inflammation. | [114, 169-173] | |
| Isobutyric acid (SCFA) | ||||
| Isovaleric acid (SCFA) | ||||
| Corynebacterium accolens | Unidentified protein | Staphylococcus aureus | Inhibits biofilm formation | [13] |
| MRSA | ||||
| LipS1 (lipase) | Streptococcus pneumoniae | Metabolizes host lipids into free fatty acids that inhibit bacterial growth | [174,175] | |
| Unidentified secreted protein(s) | Staphylococcus aureus | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [176] | |
| Corynebacterium amycolatum | Unidentified secreted protein(s) | Staphylococcus aureus | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [176] |
| Corynebacterium striatum | Unidentified secreted protein(s) | Staphylococcus aureus | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [176] |
| Corynebacterium pseudodiptheriticum | Unidentified secreted protein(s) | Staphylococcus aureus | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [176] |
| Unidentified secreted factor | Staphylococcus aureus | Bactericidal against S. aureus when it expresses agr-dependent virulence factors | [177] | |
| MRSA | ||||
| Staphylococcus capitus | Capidermicin (AMP) | Lactococcus lactis | Forms pores in membranes | [178] |
| Micrococcus leuteus | ||||
| Staphylococcus aureus | ||||
| Staphylococcus intermedius | ||||
| Staphylococcus pseudointermedis | ||||
| Unidentified bacteriocin(s) | Listeria monocytogenes | Unknown | [16] | |
| Staphylococcus aureus | ||||
| MRSA | ||||
| Streptococcus alagactiae | ||||
| Streptococcus Bovis | ||||
| PSM-beta 1 to PSM-beta 6 | Micrococcus leuteus | Induces cell lysis | [179] | |
| PSM 1 to PSM 4 | Cutibacterium acnes | Act synergistically for targeted killing | [180] | |
| Staphylococcus caprae | Unidentified autoinducing peptide | Staphylococcus aureus | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [181] |
| MRSA | ||||
| Staphylococcus epidermidis | Esp (serine protease) | Staphylococcus aureus | Inhibits biofilm formation and destructs biofilms | [182] |
| Unidentified AMP | Staphylococcus aureus | Targeted killing | [183] | |
| Staphylococcus hominis | Unidentified AMP | Staphylococcus aureus | Targeted killing | [183] |
| Hominicin (AMP) | VRSA | Unknown | [184] | |
| Unidentified bacteriocin(s) | Listeria monocytogenes | Unknown | [16] | |
| Streptococcus alagactiae | ||||
| Streptococcus Bovis | ||||
| Staphylococcus mutans | Mutacin 1140 (AMP) | VRSA | Unknown | [184] |
| Staphylococcus lugdunensis | Lugdunin (thiazolidine-containing cyclic peptide, AMP) | VRSA | Unknown | [184] |
| Lugdunin (thiazolidine-containing cyclic peptide, AMP) | Enterococcus faecium | Direct killing | [185] | |
| Enterococcus faecalis | ||||
| Listeria Monocytogenes | ||||
| Staphylococcus aureus | ||||
| MRSA | ||||
| Streptococcus pneumoniae | ||||
| Lugdunin (thiazolidine-containing cyclic peptide, AMP) | Staphylococcus aureus | Direct killing and amplification of innate immune responses | [186] | |
| Staphylococcus simulans | AIP-I to AIP-III | Staphylococcus aureus & MRSA | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [187] |
| Staphylococcus warneri | AIP-I to AIP-II | Staphylococcus aureus & MRSA | Inhibits the agr quorum sensing system and prevents agr-dependent virulence factor expression | [188] |
| Unidentified bacteriocin(s) | Staphylococcus aureus | Unknown | [16] | |
| Streptococcus alagactiae |
Summary table of prominent and recently identified antimicrobial molecules produced by major skin bacterial taxa to inhibit other microbial community members and/or potential pathogens. The table is organized by the taxa that produces the antimicrobial molecule(s). Where possible the molecule's proposed mechanism of action is included.
Abbreviations: AIP, Autoinducing peptide; AMP, Antimicrobial peptide; MRSA, Methicillin resistant Staphylococcus aureus; MSSA, Methicillin sensitive Staphylococcus aureus; PSM, phenol-soluble modulins; SCFA, Short chain fatty acids; VRSA, Vancomycin resistant Staphylococcus aureus.
Changes with age
Homeostatic skin microbial community structures shift as we age and navigate the environment (Figure 1). Throughout a lifetime the skin's physiology changes as the cutaneous immune systems matures and hormones drive sweat and sebum gland development, which impacts the availability of key nutrients. As a direct interface with the environment, the skin also continuously shares microbes with the places and people around us. Below we summarize the shifts in the skin microbiome over the human lifespan and highlight where disruptions to the skin microbiome at critical age-associated stages influence the risk for disease development.
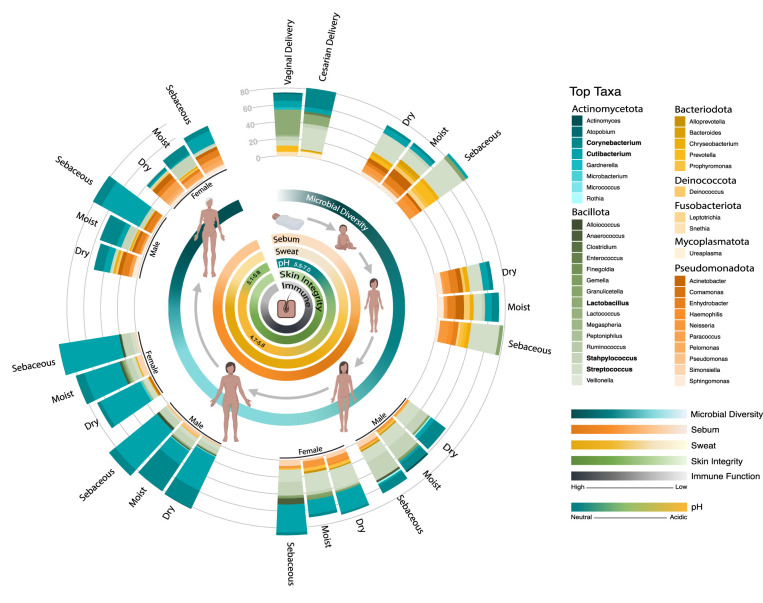
Figure 1. The dynamic balance of the skin and its microbiome over the lifespan.
Over a lifetime the skin's physiology changes as an individual's cutaneous immune systems matures and hormones drive sweat and sebum gland development. These changes are associated with shifts in the relative abundance of prominent skin microbial taxa and shifts in the overall microbial community diversity. Microbiome data displays the average relative abundance of the top ten microbial taxa for each group as assessed by high-throughput sequencing of the bacterial 16S ribosomal RNA gene. Taxa with a relative abundance >20% in at least one group are bolded. Groups include newborns born either through vaginal delivery or cesarian section [22] as well as dry, moist, and sebaceous sites for infants (1 year old) [40], children (5 years old) [40], adolescents (Tanner Stage III) [46], adults (20–40 years old) [3], and the elderly (60 and older) [137]. Since sexual differences in skin microbial composition become more pronounced over the course of puberty [46], relative abundance plots for adolescent, adult, and elderly males and females are displayed. Inner circles represent relative microbial diversity, sebum production, sweat production, surface pH, skin integrity, and immune function throughout life [33,46,87,144,152,153].
Birth
The skin's first substantial introduction to microbes occurs at birth, where the skin is quickly colonized from the immediate environment. Mode of delivery has been shown to influence the initial community composition [21–23]. For example, the skin microbiome of vaginally delivered newborns is dominated by vaginal-associated flora, primarily Lactobacillus and Prevotella, and contains a higher abundance of Candida albicans [21,22,24]. Newborns delivered via cesarean section have microbiomes containing maternal skin-associated microbes, including Staphylococcus, Streptococcus, Corynebacterium, and Cutibacterium. Cesarian-delivered newborns intentionally exposed to their mother's vaginal flora at birth have microbiomes containing both skin- and vaginal-associated flora [25]. Although these initial communities are transient [22], the order and timing of species colonization influences how strains subsequently interact with one another [26,27]. These priority effects can shape future community structures and have long term implications for the skin, its microbiome, and overall health.
Infancy and childhood: initial microbial exposures
Skin microbial compositions gradually shift throughout infancy and childhood [22]. These communities are shaped by the skin's functional maturation, the microbiomes of close care givers, and their environment [28–30]. Particularly in infancy initial microbial exposures prime immune development [26,31] and strengthen the skin barrier through promoting proper keratinocyte differentiation and epidermal repair [8].
Newborn and infant skin has a greater water content, higher pH, suppressed sebum production, faster epidermal turnover, and greater antimicrobial properties [32,33]. Within hours after birth the skin surface begins to acidify [29] and initially homogenous microbial communities begin to diverge into body site-specific communities [26,34]. Continued site-specific skin maturation promotes reorganization of the infant microbiota across body habitats over the first few months [22]. Within 3–6 months, associations between microbial taxa and skin metabolic function (e.g. lipid production and pH) are established [35]. Compared with adults, reduced sebum production in early life is associated with lower Corynebacterium, Cutibacterium and Malassezia abundance, increased staphylococci and streptococci, and a mycobiome dominated by Candida species [22,24,34,36–39]. As children age, skin further acidifies and produces more sebum lipids, which promotes a gradual decline in acid-sensitive streptococci and increase in overall community diversity [32,33,40].
Commensal colonization also stimulates immune cell maturation, particularly regulatory T-cell (Treg) localization to developing hair follicles shortly after birth [41–43]. Specific training of commensal antigen-specific Tregs is required for establishing immune tolerance [41] that prevents inflammation against this commensal later in life [10]. However early exposure does not guarantee future tolerance. The simultaneous recognition of multiple pathogen-associated molecules and toxins (e.g. S. aureus alpha-toxin) during early colonization events limits pathogen-specific Treg formation [44]. Without microbial-specific Tregs, S. aureus exposures in later life leads to inflammation.
In early infancy, the skin microbiome of close caregivers further contributes to shaping the infant skin microbiome. At 6 weeks of age, infant and maternal skin microbiomes display very similar community structures [22], and throughout childhood the skin will continue to harbor distinct microbial taxa from caregivers [22,40,45]. However with age, older infants have greater skin microbial diversity and more microbes derived from their rural or urban environments [30] and the similarity between a mother and infant microbiome gradually declines [30,45]. Cutaneous fungal populations are also more diverse in childhood with greater interpersonal variation [37,46,47]. These early life variations and interventions in environmental microbial exposures influence the establishment of the skin microbiome and can modulate lifelong immune responses [48–51]. Disruption to the establishment of this equilibrium [26,52] is associated with greater inflammation and can increase a child's risk for atopic dermatitis and allergy development.
Microbial imbalance in pediatric atopic dermatitis
Atopic dermatitis (eczema; AD) is an inflammatory skin disorder, characterized by dry, inflamed, itchy skin patches. The typical age of onset is between 3–6 months old and it affects roughly 20% of infants and children, 15% of adolescents, and 10% of adults [53,54]. This early age of onset and evidence of skin microbiome dysbiosis preceding AD onset [52], underscore how early life microbial exposures may influence AD risk. Additionally, many children grow out of AD and experience partial or full resolution of symptoms as they enter adolescence or adulthood [54]. This showcases the important influence of the maturation of skin microbiome in AD's natural course.
Skin barrier dysfunction and immune imbalance contribute to AD's pathogenesis. In AD patients, both affected and non-affected areas of skin display increased permeability [55], reduced water retention, high pH [56], and altered lipid composition [57–59]. Subsequently external microorganisms, particularly Staphylococcus aureus, can penetrate deeper skin layers. Stressed keratinocytes and microbial antigens trigger type-2 helper T-cell-driven immune responses [60,61], which further exacerbates barrier defects through down-regulation of filaggrin [62,63], disruption of tight junctions, and reduction in stratum corneum lipids [59]. High levels of type-2 cytokines also reduce antimicrobial peptide production, further increasing the skin's susceptivity to S. aureus colonization [63]. Thus, a perpetual cycle of poor barrier function, increased microbial and irritant penetration into deeper skin layers, and increased inflammation then itching and further skin damage is developed.
Elevated S. aureus abundance can precede AD development [26,52] and several S. aureus virulence factors propagate AD related epidermal damage and inflammation [64–66]. Additional microbiome changes in AD include a reduction in Cutibacterium acnes, Corynebacterium, Dermacoccus, Micrococcus, and CoNS and an increase in Streptococcus and some Malassezia species [58,61,67–72]. These microbial shifts appear to be temporal, with a loss of community diversity and greater S. aureus dominance preceding and during AD flares followed by a gradual return to baseline following resolution [73,74]. Finally, early colonization with commensal CoNS (e.g. S. epidermidis and S. hominis) [26] and Gram-negative bacteria reduces the risk of developing AD [75].
Current AD treatment revolves around eliminating exacerbating factors (e.g. contact with allergens and strong soaps), maintaining skin moisture with emollients and reducing skin inflammation through topical corticosteroids or calcineurin inhibiters [54,76]. Many of these interventions effectively resolve the skin dysbiosis [68,73,74,77–79]. Investigations into potential therapies are increasingly centered around modulating the microbiome with oral prebiotics, probiotics and symbiotics [80,81], topical emollients containing probiotics [82], or microbial transplantation with commensal skin bacteria [75,83–85]. One promising avenue is harnessing CoNS anti-S. aureus activity. The transplantation of S. epidermidis and S. hominis can diminish S. aureus virulence, prevent epithelial damage, and reduce inflammation in murine models and clinical trials of atopic dermatitis (AD) patients [84,86].
Adolescence and puberty: hormonally driven changes
Puberty marks the next major shift in our skin microbial communities. The hormones that drive physical and sexual development also directly promote structural and functional changes in the skin such as sebum and apocrine sweat production. This leads to subsequent shifts in microbial composition (Figure 2) [87]. Due to interpersonal variability in puberty onset and progression, the Tanner staging system measures the degree of sexual maturation [88,89]. Tanner stage I corresponds to the pre-pubertal stage and stage V corresponds to adult sexual characteristics. Both cross-sectional and longitudinal studies demonstrate clear shifts in skin microbiome composition across Tanner stages [36,37,46]. Children at stage I have higher relative abundances of Streptococcus, Bacteroidota and Pseudomonadota, as well as higher bacterial and fungal diversity compared with young adults at stage V [36,46]. Furthermore, the young adult skin microbiome is dominated by lipophilic microbes, including Corynebacterium, Cutibacterium acnes, and Malassezia [36,37,46], a composition associated with higher sebum production and serum concentrations of hormones that promote sebum production [46].
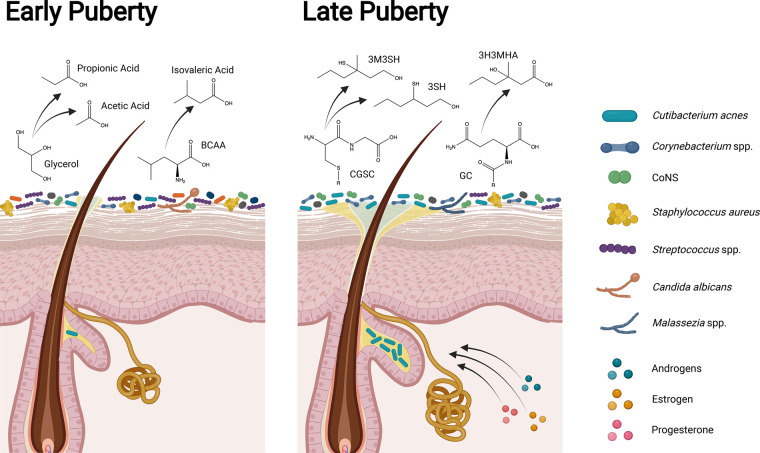
Figure 2. Differences in the skin, microbiome, and body odor production in early and late puberty.
In childhood and early puberty (Tanner Stages I to II) the skin microbiome is highly diverse and body odor is associated with CoNS (e.g., S. epidermidis and S. hominis) production of volatile fatty acids (e.g., propionic, acetic, and isovaleric acid; sour odors) and sulfur (rotten-egg odor) [92]. As puberty advances, steroid hormones promote sebaceous and apocrine sweat gland development [90,91], modify the types of lipids present in sebum [90,97], and enhance the skin barrier [94–96]. In later puberty (Tanner Stages IV to V), increased lipid production and altered lipid content is associated with a skin microbiome dominated by lipophilic taxa [46]. While breakdown of sweat and sebum components into volatile fatty acids still occurs, body odor in young adults becomes more associated with Corynebacterium spp. metabolism of sebum and sweat components into sulfanylalkanols (e.g., 3SH and 3M3SH; oniony odors), and volatile organic compounds (e.g., 3H3MHA; cumin like odors) [92,98–101]. BCAA: Branched chain amino acids; CGSC: Cystine-Glycine-S-conjugate; CoNS: Coagulase negative Staphylococcus spp.; GC: Glutaminyl-conjugate; 3H3MHA: 3-hydroxy-3-methylhexanoic acid; 3M3SH: 3-methyl-3-sullanylhexanol; 3SH: 3-sulfanylhexanol.
Puberty is driven by androgens (e.g. dehydroepiandrosterone and testosterone), estrogen, and progesterone [87]. On the skin androgens promote pubic and axillary hair growth and sex specific hair patterns [87], sebaceous gland development and increased sebum production [90,91], and apocrine sweat gland development with a subsequent increase in body odor [92,93]. Estrogen and progesterone enhance the skin barrier and promote wound healing through encouraging collagen synthesis and stimulating keratinocyte proliferation [94–96]. Puberty-driving hormones also modulate immune function in both pro- and anti-inflammatory ways [96]. These hormonally driven changes in skin physiology lead to changes in the skin microbial communities [46]. For instance, androgen-driven sebaceous gland development [91] and estrogen modification of the lipid types at the skin surface [97] promote the survival of lipophilic species, particularly C. acnes and Malassezia restricta [46].
Although apocrine gland secretions are initially odorless, microbial metabolism of their components (e.g. branched chain amino acids, fatty acids, and glycerol) produces odoriferous compounds (Figure 2) [92,98]. Body odor in children and young-teens is associated with microbial production of isovaleric and acetic acid (sour odor) and sulfur (rotten-egg odor) [92]. In young adults body odor, particularly from the axilla [92,98,99], is associated with corynebacterial breakdown of sebum into volatile fatty acids (cheesy odor) and sulfanylalkanols (oniony odors) [99–101].
While all puberty-driving hormones surge in both sexes, the heightened role of androgens in males and estrogens plus progesterone in females foster sexual differences in maturation. As such, sexual differences in skin microbial composition become more pronounced as puberty advances [46]. Microbial communities of females become less diverse and have greater prevalence of Cutibacterium with increasing Tanner stage [46]. Skin microbiomes of males display more inter-individual differences and greater diversity, with higher relative abundance of Corynebacterium, Staphylococcus, Streptococcus, and Haemophilus [46]. Other hormonally dependent skin physiological (lower pH, increased skin thickness) and immunological changes during puberty likely also account for many maturation-associated skin microbial shifts, but their influence is less defined [36,46].
Collectively, puberty marks major shifts in the skin microbial community structure, metabolic complexity, and function. Microbiome imbalance during this process can promote several microbe-associated skin disorders including acne vulgaris.
Cutibacterium acnes phylotype imbalance in acne vulgaris
Several microbe-associated skin disorders often begin during puberty, including acne vulgaris hidradenitis suppurativa, and psoriasis, highlighting the influence of puberty-driven skin and microbial shifts in disease pathogenesis [46]. Here we highlight acne vulgaris (acne), due to the strong support for a microbiome-driven association. Although there are microbial disturbances associated with hidradenitis suppurativa and psoriasis, no single microbe or consistent pattern of dysbiosis has been implicated [61,102,103]. Further research into the roles of the microbiome in these conditions are needed.
Acne affects ∼85% of adolescents and young adults (12–25 years old) [104]. Mild acne is characterized by clogged hair follicles while severe cases present with inflammatory, painful papules, pustules, nodules or cysts. Like how individuals experience resolution of AD as they enter adolescence, acne tends to resolve as individuals mature out of adolescence and enter adulthood. This further highlights how the natural age associated evolution of microbial community structures influence the course of a disease.
Our understanding of acne's pathogenesis recently underwent a drastic shift. Cutibacterium acnes is a lipophilic bacteria that dominates sebaceous skin sites [3,105]. Each pore is dominated by a single, nearly clonal lineage [105] and individuals have their own unique mix of C. acnes strains [3]. Earlier research suggested that increased sebum production and C. acnes over-proliferation triggered inflammation, abnormal keratinocyte proliferation, and pore duct obstruction [106]. However, healthy individuals have similar to slightly higher C. acnes bioburden [107–110]. Recent works support a dogma where hyper-proliferation of particular C. acnes phylotypes (phylotypes IA1, IA2, 1B1, and IC), reduced C. acnes phylotype diversity, and collective skin microbial dysbiosis triggers the cutaneous inflammation underlying acne development [105,108,111,112]. These acne associated phylotypes tend to induce more inflammation, display elevated porphyrin production [113], and exhibit excessive lipase activity [114]. The fatty acids produced by lipase metabolism of sebum subsequently attract neutrophils and promote hyperkeratosis [114].
Mild acne is generally treated with topical benzoyl peroxide and/or a topical retinoid [115]. Topical then oral antibiotics followed by oral isotretinoin is the mainstay for moderate-severe or refractory acne treatment [115]. Concerns for rising antibiotic resistance and isotretinoin's side effects have driven efforts to identify new therapies designed to prevent or reverse the hyperproliferation of C. acnes strains and equilibrate the microbiome. One promising avenue is selective augmentation of S. epidermidis over C. acnes [116,117] through sucrose supplementation [116] or a probiotic containing encapsulated S. epidermidis and glycerol [118]. Supplementing the skin microbiome with topical probiotics derived from Lactobacillus is another promising avenue [119]. Additional future therapeutics include; i) topical application of anti-microbial peptides [72,120], ii) bacteriophages that strategically infect C. acnes [121], and iii) using oral antibiotics to modulate the gut microbiome and indirectly alter the skin microbiome [122–124].
Homeostasis throughout adulthood
The adult skin microbiome is stable over the span of years [125]. Collectively, established microbial-microbial interaction networks (Table 1), lasting adult skin physiology, and resilient skin immunity maintain balanced adult skin microbial communities. Adult skin microbiomes are dominated by Cutibacterium, Corynebacterium, Staphylococcus and Malassezia species [4]. Each body site has a unique microenvironment and particular sweat and sebaceous gland density that dictates the prevailing microbial composition [4]. Once adulthood is reached, matured and enduring skin physiology promotes consistent sebum production, sweat composition, and surface pH, which collectively provide reliable body site microenvironments and nutrient pools [126,127]. The immune system also reaches full maturation in our early twenties [128]. Unwavering immune function [129,130] further encourages appropriate, reliable responses to our established commensal microbiome and infectious insults. These intrinsic features enable large portions of microbial communities on the skin to persist, despite daily environmental changes [125]. Microbial community stability is evidenced by the longitudinal fixation of highly abundant species (e.g. C. acnes) [125] along with the persistence of several low abundant taxa, which contribute to our unique microbial signature [125,131].
A portion of the taxa within the skin microbiome are also influenced by environmental surroundings. For example, individuals living in Egypt often have a greater abundance of bacteria within the Pseudomonadota phyla [132]; individuals in Cameroon have greater Staphylococcus and Micrococcus [133], those in South Asia tend to have greater abundance of Corynebacterium and Streptococcus [134]; while individuals in Japan have high abundance of Cutibacterium [133]; and Americans and Europeans have greater Corynebacterium species abundance [135]. Collectively this illustrates how individuals living in different geographic locations maintain slightly different skin microbial patterns. Individuals living or working in rural communities also contain more soil and agriculture associated microbes within their skin microbiome [136–138]. We also share more microbial taxa with those living in the same residence [131,139,140] and skin microbiomes become more similar the longer individuals cohabitate [141,142]. In short, the people in our lives, local environment, and geography, partially influence our skin microbial balance. The degree to which a portion of your microbiome shifts with environment changes also appears to be person and situation specific [140–142].
Advancing age
Intriguingly, of the prominent human microbiomes, the skin microbiome is the best predictor of age [143]. With advancing age, distinct skin changes occur including the decline of collagen synthesis, extracellular matrix fragmentation, and a reduction in skin cell regeneration [144]. These changes can manifest as skin wrinkles and more consequently impaired wound healing. Furthermore, these aging related changes shape microbiome composition.
As the skin barrier changes it can lose its ability to retain water, resulting in a compensatory increase in natural moisturizing factor (NFM) production [145]. NMFs both absorb water and can promote bacterial proliferation and adherence to the skin [146]. Subsequently, increased NMFs is associated with greater abundance of numerous taxa, such as Corynebacterium, Micrococcus, Streptococcus, Anaerococcus [127], and a reduction in Cutibacterium [127,144]. Skin microbial diversity also broadly increases [127,137,144,147]. Decreasing sebocyte area and sebum production after menopause in females correlates with loss of Cutibacterium and an increase in Corynebacterium, Streptococcus, Acinetobacter, and Corynebacterium abundance [127,144,148–150]. In males, sebum secretion declines significantly slower so they maintain greater Cutibacterium abundance as they age [137,151].
With age, immune system function also slowly declines [128,130,152,153]. Elderly individuals sustain a low-grade inflammatory state with increased systemic concentrations of pro-inflammatory cytokines [152]. Within the skin Langerhans cells are gradually lost from the epidermis [154]. Cutaneous dendritic cells and the remaining Langerhans also display impaired ability to migrate to lymph nodes and present antigens to T-cells [155,156]. Subsequently, this disrupted antigen presentation along with systemic and local defects in immune signaling ultimately lead to slower immune responses, reduced antimicrobial activity, and impaired wound healing [153,157]. Collectively, impaired immune defenses and increased prevalence of potentially pathogenic bacteria (e.g. beta-hemolytic Streptococci) contribute to the substantial increased risk for skin infection in the elderly and difficulty clearing the infection [153,158,159].
A myriad of dermatologic diseases are associated with advancing age, including dry skin, seborrheic dermatitis, rosacea, disrupted wound healing, and chronic wound infections. Although all these conditions are associated with skin microbiome dysbiosis, specific underlying bacterial and fungal changes remain elusive [18,160,161]. One key exception is seborrheic dermatitis, where recent works find increased abundance of Malassezia and Staphylococcus species to be potential fungal and bacterial biomarkers [162,163]. In tandem with the numerous works seeking to identify avenues that support a balanced, ‘youthful,' microbiome throughout adulthood and later life [144,150,151], greater research into the active role of the microbiome as we age and in the development of age-associated diseases are needed.
Conclusion
There is no single definition of a balanced skin microbiome. While there are core skin microbial members, for any individual, the precise microbial composition is dynamic and unique. This dynamic is influenced by the continuous exchange of microbes with the people we are close to and the world around us. The specific community structure in a particular skin microenvironment is also partially determined by local skin physiology, the microbe–immune interface, and complex microbe–microbe interactions. However, on the human microbiome scale, the largest and somewhat predictable shifts in our skin microbial community compositions occur as we, and our skin, age. Future investigations will continue to elucidate the active role of our dynamic skin microbiome across the lifespan, its implications for dermatologic disease risk and overall health, as well as targeted, microbiome centered therapeutic approaches.
Perspectives
The skin and its microbiome are primary interfaces with the environment. The balance of this microbial community influences our risk for various diseases and lifelong health.
There is no single definition of a balanced skin microbiome. At a microenvironment level, balance is dictated by the skin niche along with complex host immune-microbe and microbe–microbe interactions. For any individual this balance is also dynamic, shifting with us as we age and navigate the environments around us.
Future works in the field of skin microbiology will likely continue to explore i) the active role of our skin microbiome as we age, ii) the complex immune-microbe and microbe–microbe interactions and how they shift over the lifespan, iii) the cutaneous microbiomes of diverse population demographics, iv) the microbiome's influence in protection from and/or disease pathogenesis, and v) the development of novel therapeutic approaches to strategically modulate the microbial balance.
Acknowledgements
The authors gratefully acknowledge members of the Kalan laboratory for thoughtful feedback on the manuscript. RStudio, Biorender, and Adobe Illustrator were used for figure generation.
Abbreviations
- 3H3MHA
3-hydroxy-3-methylhexanoic acid
- 3M3SH
3-methyl-3-sullanylhexanol
- 3SH
3-sulfanylhexanol
- BCAA
Branched chain amino acids
- CGSC
Cystine-Glycine-S-conjugate
- CoNS
Coagulase negative Staphylococcus spp
- GC
Glutaminyl-conjugate
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by grants from the National Institutes of Health (NIAID U19AI142720, NIGMS R35 GM137828 [L.R.K]). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
E.C.T and L.R.K contributed to the conceptualization and editing of the text and figures. E. C.T. primarily contributed to the main text and figure creation.
References
- 1.Costello, E.K., Lauber, C.L., Hamady, M., Fierer, N., Gordon, J.I. and Knight, R. (2009) Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice, E.A. and Segre, J.A. (2011) The skin microbiome. Nat. Rev. Microbiol. 9, 244–253 10.1038/nrmicro2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh, J., Byrd, A.L., Deming, C., Conlan, S., Kong, H.H. and Segre, J.A. (2014) Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 10.1038/nature13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd, A.L., Belkaid, Y. and Segre, J.A. (2018) The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 5.Flowers, L. and Grice, E.A. (2020) The skin microbiota: balancing risk and reward. Cell Host Microbe 28, 190–200 10.1016/j.chom.2020.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jo, J.H., Kennedy, E.A. and Kong, H.H. (2017) Topographical and physiological differences of the skin mycobiome in health and disease. Virulence 8, 324–333 10.1080/21505594.2016.1249093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi, T. and Gallo, R.L. (2017) The critical and multifunctional roles of antimicrobial peptides in dermatology. Dermatol. Clin. 35, 39–50 10.1016/j.det.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 8.Uberoi, A., Bartow-McKenney, C., Zheng, Q., Flowers, L., Campbell, A., Knight, S.A.B.et al. (2021) Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 29, 1235–1248.e8 10.1016/j.chom.2021.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dwyer, L.R. and Scharschmidt, T.C. (2022) Early life host-microbe interactions in skin. Cell Host Microbe 30, 684–695 10.1016/j.chom.2022.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scharschmidt, T.C., Vasquez, K.S., Truong, H.A., Gearty, S.V., Pauli, M.L., Nosbaum, A.et al. (2015) A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 10.1016/j.immuni.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastar, I., O'Neill, K., Padula, L., Head, C.R., Burgess, J.L., Chen, V.et al. (2020) Staphylococcus epidermidis boosts innate immune response by activation of gamma delta T cells and induction of perforin-2 in human skin. Front. Immunol. 11, 550946 10.3389/fimmu.2020.550946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunjani, N., Ahearn-Ford, S., Dube, F.S., Hlela, C. and O'Mahony, L. (2021) Mechanisms of microbe-immune system dialogue within the skin. Genes Immun. 22, 276–288 10.1038/s41435-021-00133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menberu, M.A., Liu, S., Cooksley, C., Hayes, A.J., Psaltis, A.J., Wormald, P.J.et al. (2021) Corynebacterium accolens has antimicrobial activity against Staphylococcus aureus and methicillin-resistant S. aureus pathogens isolated from the sinonasal niche of chronic rhinosinusitis patients. Pathogens 10, 207 10.3390/pathogens10020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claesen, J., Spagnolo, J.B., Ramos, S.F., Kurita, K.L., Byrd, A.L., Aksenov, A.A.et al. (2020) A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 12, eaay5445 10.1126/scitranslmed.aay5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin, D., Goncheva, M.I., Flannagan, R.S., Deecker, S.R., Guariglia-Oropeza, V., Ensminger, A.W.et al. (2021) Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat. Commun. 12, 1887 10.1038/s41467-021-22175-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Sullivan, J.N., Rea, M.C., O'Connor, P.M., Hill, C. and Ross, R.P. (2019) Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 95, fiy241 10.1093/femsec/fiy241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanford, J.A. and Gallo, R.L. (2013) Functions of the skin microbiota in health and disease. Semin. Immunol. 25, 370–377 10.1016/j.smim.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbanic, S., Shen, Y., Lee, J., Deacon, J.M. and Chen, I.A. (2020) Microbial predictors of healing and short-term effect of debridement on the microbiome of chronic wounds. NPJ Biofilms Microbiomes 6, 21 10.1038/s41522-020-0130-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjerre, R.D., Bandier, J., Skov, L., Engstrand, L. and Johansen, J.D. (2017) The role of the skin microbiome in atopic dermatitis: a systematic review. Br. J. Dermatol. 177, 1272–1278 10.1111/bjd.15390 [DOI] [PubMed] [Google Scholar]
- 20.Swaney, M.H. and Kalan, L.R. (2021) Living in your skin: microbes, molecules, and mechanisms. Infect. Immun. 89, e00695-20 10.1128/IAI.00695-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez-Bello, M.G., Costello, E.K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N.et al. (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. U.S.A. 107, 11971–5 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu, D.M., Ma, J., Prince, A.L., Antony, K.M., Seferovic, M.D. and Aagaard, K.M. (2017) Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 10.1038/nm.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Younge, N.E., Araújo-Pérez, F., Brandon, D. and Seed, P.C. (2018) Early-life skin microbiota in hospitalized preterm and full-term infants. Microbiome 6, 98 10.1186/s40168-018-0486-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward, T.L., Dominguez-Bello, M.G., Heisel, T., Al-Ghalith, G., Knights, D. and Gale, C.A. (2018) Development of the human mycobiome over the first month of life and across body sites. mSystems 3, e00140-17 10.1128/mSystems.00140-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Bello, M.G., De Jesus-Laboy, K.M., Shen, N., Cox, L.M., Amir, A., Gonzalez, A.et al. (2016) Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253 10.1038/nm.4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy, E.A., Connolly, J., Hourihane, J.O., Fallon, P.G., McLean, W.H.I., Murray, D.et al. (2017) Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J. Allergy Clin. Immunol. 139, 166–172 10.1016/j.jaci.2016.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casterline, B.W. and Paller, A.S. (2021) Early development of the skin microbiome: therapeutic opportunities. Pediatr. Res. 90, 731–737 10.1038/s41390-020-01146-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King, A., Balaji, S. and Keswani, S.G. (2013) Biology and function of fetal and pediatric skin. Facial Plast. Surg. Clin. North Am. 21, 1–6 10.1016/j.fsc.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visscher, M.O., Adam, R., Brink, S. and Odio, M. (2015) Newborn infant skin: physiology, development, and care. Clin. Dermatol. 33, 271–280 10.1016/j.clindermatol.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 30.Manus, M.B., Kuthyar, S., Perroni-Marañón, A.G., Núñez-de la Mora, A. and Amato, K.R. (2020) Infant skin bacterial communities vary by skin site and infant age across populations in Mexico and the United States. mSystems 5, e00834-20 10.1128/mSystems.00834-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoch, J.J., Monir, R.L., Satcher, K.G., Harris, J., Triplett, E. and Neu, J. (2019) The infantile cutaneous microbiome: a review. Pediatr. Dermatol. 36, 574–580 10.1111/pde.13870 [DOI] [PubMed] [Google Scholar]
- 32.Oranges, T., Dini, V. and Romanelli, M. (2015) Skin physiology of the neonate and infant: clinical implications. Adv. Wound Care 4, 587–595 10.1089/wound.2015.0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visscher, M.O., Carr, A.N., Winget, J., Huggins, T., Bascom, C.C., Isfort, R.et al. (2021) Biomarkers of neonatal skin barrier adaptation reveal substantial differences compared to adult skin. Pediatr. Res. 89, 1208–1215 10.1038/s41390-020-1035-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capone, K.A., Dowd, S.E., Stamatas, G.N. and Nikolovski, J. (2011) Diversity of the human skin microbiome early in life. J. Invest. Dermatol. 131, 2026–2032 10.1038/jid.2011.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roux, P.F., Oddos, T. and Stamatas, G. (2022) Deciphering the role of skin surface microbiome in skin health: an integrative multiomics approach reveals three distinct metabolite‒microbe clusters. J. Invest. Dermatol. 142, 469–479.e5 10.1016/j.jid.2021.07.159 [DOI] [PubMed] [Google Scholar]
- 36.Oh, J., Conlan, S., Polley, E.C., Segre, J.A. and Kong, H.H. (2012) Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 77 10.1186/gm378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo, J.H., Deming, C., Kennedy, E.A., Conlan, S., Polley, E.C., Ng, W.I.et al. (2016) Diverse human skin fungal communities in children converge in adulthood. J. Invest. Dermatol. 136, 2356–2363 10.1016/j.jid.2016.05.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, T., Duan, Y.Y., Kong, F.Q., Galzote, C. and Quan, Z.X. (2020) Dynamics of skin mycobiome in infants. Front. Microbiol. 11, 1790 10.3389/fmicb.2020.01790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul, A.A., Hoffman, K.L., Hagan, J.L., Sampath, V., Petrosino, J.F. and Pammi, M. (2020) Fungal cutaneous microbiome and host determinants in preterm andterm neonates. Pediatr. Res. 88, 225–233 10.1038/s41390-019-0719-7 [DOI] [PubMed] [Google Scholar]
- 40.Zhu, T., Liu, X., Kong, F.Q., Duan, Y.Y., Yee, A.L., Kim, M.et al. (2019) Age and mothers: potent influences of children's skin microbiota. J. Invest. Dermatol. 139, 2497–2505.e6 10.1016/j.jid.2019.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scharschmidt, T.C., Vasquez, K.S., Pauli, M.L., Leitner, E.G.. Chu, K., Truong, H.A.et al. (2017) Commensal microbes and hair follicle morphogenesis coordinately drive treg migration into neonatal skin. Cell Host Microbe 21, 467–477.e5 10.1016/j.chom.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez Rodriguez, R., Pauli, M.L., Neuhaus, I.M., Yu, S.S., Arron, S.T., Harris, H.W.et al. (2014) Memory regulatory T cells reside in human skin. J. Clin. Invest. 124, 1027–1036 10.1172/JCI72932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christoph, T., Müller-Röver, S., Audring, H., Tobin, D.J., Hermes, B., Cotsarelis, G.et al. (2000) The human hair follicle immune system: cellular composition and immune privilege. Br. J. Dermatol. 142, 862–873 10.1046/j.1365-2133.2000.03464.x [DOI] [PubMed] [Google Scholar]
- 44.Leech, J.M., Dhariwala, M.O., Lowe, M.M., Chu, K., Merana, G.R., Cornuot, C.et al. (2019) Toxin-triggered interleukin-1 receptor signaling enables early-life discrimination of pathogenic versus commensal skin bacteria. Cell Host Microbe 26, 795–809.e5 10.1016/j.chom.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaitanis, G., Tsiouri, G., Spyridonos, P., Stefos, Τ, Stamatas, G.N., Velegraki, A.et al. (2019) Variation of cultured skin microbiota in mothers and their infants during the first year postpartum. Pediatr. Dermatol. 36, 460–465 10.1111/pde.13829 [DOI] [PubMed] [Google Scholar]
- 46.Park, J., Schwardt, N.H., Jo, J.H., Zhang, Z., Pillai, V., Phang, S.et al. (2022) Shifts in the skin bacterial and fungal communities of healthy children transitioning through puberty. J. Invest. Dermatol. 142, 212–219 10.1016/j.jid.2021.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen, U.T. and Kalan, L.R. (2022) Forgotten fungi: the importance of the skin mycobiome. Curr. Opin. Microbiol. 70, 102235 10.1016/j.mib.2022.102235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roslund, M.I., Puhakka, R., Nurminen, N., Oikarinen, S., Siter, N., Grönroos, M.et al. (2021) Long-term biodiversity intervention shapes health-associated commensal microbiota among urban day-care children. Environ. Int. 157, 106811 10.1016/j.envint.2021.106811 [DOI] [PubMed] [Google Scholar]
- 49.Lehtimäki, J., Sinkko, H., Hielm-Björkman, A., Salmela, E., Tiira, K., Laatikainen, T.et al. (2018) Skin microbiota and allergic symptoms associate with exposure to environmental microbes. Proc. Natl Acad. Sci. U.S.A. 115, 4897–4902 10.1073/pnas.1719785115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein, M.M., Hrusch, C.L., Gozdz, J., Igartua, C., Pivniouk, V., Murray, S.E.et al. (2016) Innate immunity and asthma risk in amish and hutterite farm children. N. Engl. J. Med. 375, 411–421 10.1056/NEJMoa1508749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyons, S.A., Knulst, A.C., Burney, P.G.J., Fernández-Rivas, M., Ballmer-Weber, B.K., Barreales, L.et al. (2020) Predictors of food sensitization in children and adults across Europe. J. Allergy Clin. Immunol. Pract. 8, 3074–3083.e32 10.1016/j.jaip.2020.04.040 [DOI] [PubMed] [Google Scholar]
- 52.Meylan, P., Lang, C., Mermoud, S., Johannsen, A., Norrenberg, S., Hohl, D.et al. (2017) Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J. Invest. Dermatol. 137, 2497–2504 10.1016/j.jid.2017.07.834 [DOI] [PubMed] [Google Scholar]
- 53.Silverberg, J.I., Barbarot, S., Gadkari, A., Simpson, E.L., Weidinger, S., Mina-Osorio, P.et al. (2021) Atopic dermatitis in the pediatric population: a cross-sectional, international epidemiologic study. Ann. Allergy Asthma Immunol. 126, 417–428.e2 10.1016/j.anai.2020.12.020 [DOI] [PubMed] [Google Scholar]
- 54.Langan, S.M., Irvine, A.D. and Weidinger, S. (2020) Atopic dermatitis. Lancet 396, 345–360 10.1016/S0140-6736(20)31286-1 [DOI] [PubMed] [Google Scholar]
- 55.Tsakok, T., Woolf, R., Smith, C.H., Weidinger, S. and Flohr, C. (2019) Atopic dermatitis: the skin barrier and beyond. Br. J. Dermatol. 180, 464–474 10.1111/bjd.16934 [DOI] [PubMed] [Google Scholar]
- 56.Seidenari, S. and Giusti, G. (1995) Objective assessment of the skin of children affected by atopic dermatitis: a study of pH, capacitance and TEWL in eczematous and clinically uninvolved skin. Acta Derm. Venereol. 75, 429–433 10.2340/0001555575429433 [DOI] [PubMed] [Google Scholar]
- 57.Jungersted, J.M., Scheer, H., Mempel, M., Baurecht, H., Cifuentes, L., Høgh, J.K.et al. (2010) Stratum corneum lipids, skin barrier function and filaggrin mutations in patients with atopic eczema: filaggrin mutations, skin barrier and lipids. Allergy 65, 911–918 10.1111/j.1398-9995.2010.02326.x [DOI] [PubMed] [Google Scholar]
- 58.Baurecht, H., Rühlemann, M.C., Rodríguez, E., Thielking, F., Harder, I., Erkens, A.S.et al. (2018) Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J. Allergy Clin. Immunol. 141, 1668–1676.e16 10.1016/j.jaci.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 59.Berdyshev, E., Goleva, E., Bronova, I., Dyjack, N., Rios, C., Jung, J.et al. (2018) Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight 3, e98006 10.1172/jci.insight.98006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beck, L.A., Cork, M.J., Amagai, M., De Benedetto, A., Kabashima, K., Hamilton, J.D.et al. (2022) Type 2 inflammation contributes to skin barrier dysfunction in atopic dermatitis. JID Innov. 2, 100131 10.1016/j.xjidi.2022.100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fyhrquist, N., Muirhead, G., Prast-Nielsen, S., Jeanmougin, M., Olah, P., Skoog, T.et al. (2019) Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 10, 4703 10.1038/s41467-019-12253-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howell, M.D., Kim, B.E., Gao, P., Grant, A.V., Boguniewicz, M., DeBenedetto, A.et al. (2009) Cytokine modulation of atopic dermatitis filaggrin skin expression. J. Allergy Clin. Immunol. 124, R7–12 10.1016/j.jaci.2007.04.031 [DOI] [PubMed] [Google Scholar]
- 63.Hönzke, S., Wallmeyer, L., Ostrowski, A., Radbruch, M., Mundhenk, L., Schäfer-Korting, M.et al. (2016) Influence of Th2 cytokines on the cornified envelope, tight junction proteins, and β-defensins in filaggrin-deficient skin equivalents. J. Invest. Dermatol. 136, 631–639 10.1016/j.jid.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 64.Aziz, F., Hisatsune, J., Yu, L., Kajimura, J., Sato'o, Y., Ono, H.K.et al. (2020) Staphylococcus aureus isolated from skin from atopic-dermatitis patients produces Staphylococcal enterotoxin Y, which predominantly induces T-cell receptor Vα-specific expansion of T cells. Infect. Immun. 88, e00360-19 10.1128/IAI.00360-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Mierlo, M.M.F., Pasmans, S.G.M.A., Totté, J.E.E., de Wit, J., Herpers, B.L., Vos, M.C.et al. (2021) Temporal variation in Staphylococcus aureus protein A genotypes from nose and skin in atopic dermatitis patients. Dermatology 237, 506–512 10.1159/000515235 [DOI] [PubMed] [Google Scholar]
- 66.Poh, S.E., Koh, W.L.C., Lim, S.Y.D., Wang, E.C.E., Yew, Y.W., Common, J.E.A.et al. (2022) Expression of Staphylococcus aureus virulence factors in atopic dermatitis. JID Innov. 2, 100130 10.1016/j.xjidi.2022.100130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bjerre, R.D., Holm, J.B., Palleja, A., Sølberg, J., Skov, L. and Johansen, J.D. (2021) Skin dysbiosis in the microbiome in atopic dermatitis is site-specific and involves bacteria, fungus and virus. BMC Microbiol. 21, 256 10.1186/s12866-021-02302-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khadka, V.D., Key, F.M., Romo-González, C., Martínez-Gayosso, A., Campos-Cabrera, B.L., Gerónimo-Gallegos, A.et al. (2021) The skin microbiome of patients with atopic dermatitis normalizes gradually during treatment. Front. Cell Infect. Microbiol. 11, 720674 10.3389/fcimb.2021.720674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chng, K.R., Tay, A.S.L., Li, C., Ng, A.H.Q., Wang, J., Suri, B.K.et al. (2016) Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat. Microbiol. 1, 16106 10.1038/nmicrobiol.2016.106 [DOI] [PubMed] [Google Scholar]
- 70.Moosbrugger-Martinz, V., Hackl, H., Gruber, R., Pilecky, M., Knabl, L., Orth-Höller, D.et al. (2021) Initial evidence of distinguishable bacterial and fungal dysbiosis in the skin of patients with atopic dermatitis or netherton syndrome. J. Invest. Dermatol. 141, 114–123 10.1016/j.jid.2020.05.102 [DOI] [PubMed] [Google Scholar]
- 71.Jagielski, T., Rup, E., Ziółkowska, A., Roeske, K., Macura, A.B. and Bielecki, J. (2014) Distribution of Malassezia species on the skin of patients with atopic dermatitis, psoriasis, and healthy volunteers assessed by conventional and molecular identification methods. BMC Dermatol. 14, 3 10.1186/1471-5945-14-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han, S.H., Cheon, H.I., Hur, M.S., Kim, M.J., Jung, W.H., Lee, Y.W.et al. (2018) Analysis of the skin mycobiome in adult patients with atopic dermatitis. Exp. Dermatol. 27, 366–373 10.1111/exd.13500 [DOI] [PubMed] [Google Scholar]
- 73.Kong, H.H., Oh, J., Deming, C., Conlan, S., Grice, E.A., Beatson, M.A.et al. (2012) Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwon, S., Choi, J., Shin, J., Huh, C., Park, K., Du, M.et al. (2019) Changes in lesional and non-lesional skin microbiome during treatment of atopic dermatitis. Acta Derm. Venerol. 99, 284–290 10.2340/00015555-3089 [DOI] [PubMed] [Google Scholar]
- 75.Myles, I.A., Williams, K.W., Reckhow, J.D., Jammeh, M.L., Pincus, N.B., Sastalla, I.et al. (2016) Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight 1, e86955 10.1172/jci.insight.86955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howe, W. (2022) Treatment of atopic dermatitis (eczema) [Internet]. UpToDate. [cited 2022 Jun 10]. Available from: https://www.uptodate.com/contents/treatment-of-atopic-dermatitis-eczema#H1022550
- 77.Seite, S., Flores, G.E., Henley, J.B., Martin, R., Zelenkova, H., Aguilar, L.et al. (2014) Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J. Drugs Dermatol. 13, 1365–1372 PMID: [PubMed] [Google Scholar]
- 78.Liu, Y., Wang, S., Dai, W., Liang, Y., Shen, C., Li, Y.et al. (2020) Distinct skin microbiota imbalance and responses to clinical treatment in children with atopic dermatitis. Front. Cell Infect. Microbiol. 10, 336 10.3389/fcimb.2020.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olesen, C.M., Ingham, A.C., Thomsen, S.F., Clausen, M.L., Andersen, P.S., Edslev, S.M.et al. (2021) Changes in skin and nasal microbiome and staphylococcal species following treatment of atopic dermatitis with dupilumab. Microorganisms 9, 1487 10.3390/microorganisms9071487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makrgeorgou, A., Leonardi-Bee, J., Bath-Hextall, F.J., Murrell, D.F., Tang, M.L., Roberts, A.et al. (2018) Probiotics for treating eczema. Cochrane Database Syst. Rev. 11, CD006135 10.1002/14651858.CD006135.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang, Y.S., Trivedi, M.K., Jha, A., Lin, Y.F., Dimaano, L. and García-Romero, M.T. (2016) Synbiotics for prevention and treatment of atopic dermatitis: a meta-analysis of randomized clinical trials. JAMA Pediatr. 170, 236 10.1001/jamapediatrics.2015.3943 [DOI] [PubMed] [Google Scholar]
- 82.Ambrożej, D., Kunkiel, K., Dumycz, K. and Feleszko, W. (2021) The use of probiotics and bacteria-derived preparations in topical treatment of atopic dermatitis: a systematic review. J. Allergy Clin. Immunol. Pract. 9, 570–575.e2 10.1016/j.jaip.2020.07.051 [DOI] [PubMed] [Google Scholar]
- 83.Myles, I.A., Earland, N.J., Anderson, E.D., Moore, I.N., Kieh, M.D., Williams, K.W.et al. (2018) First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 3, 120608 10.1172/jci.insight.120608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakatsuji, T., Hata, T.R., Tong, Y., Cheng, J.Y., Shafiq, F., Butcher, A.M.et al. (2021) Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat. Med. 27, 700–709 10.1038/s41591-021-01256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakatsuji, T., Gallo, R.L., Shafiq, F., Tong, Y., Chun, K., Butcher, A.M.et al. (2021) Use of autologous bacteriotherapy to treat Staphylococcus aureus in patients with atopic dermatitis: a randomized double-blind clinical trial. JAMA Dermatol. 157, 978–982 10.1001/jamadermatol.2021.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams, M.R., Costa, S.K., Zaramela, L.S., Khalil, S., Todd, D.A., Winter, H.L.et al. (2019) Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 11, eaat8329 10.1126/scitranslmed.aat8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gratton, R., Del Vecchio, C., Zupin, L. and Crovella, S. (2022) Unraveling the role of sex hormones on keratinocyte functions in human inflammatory skin diseases. Int. J. Mol. Sci. 23, 3132 10.3390/ijms23063132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marshall, W.A. and Tanner, J.M. (1969) Variations in pattern of pubertal changes in girls. Arch. Dis. Child. 44, 291–303 10.1136/adc.44.235.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marshall, W.A. and Tanner, J.M. (1970) Variations in the pattern of pubertal changes in boys. Arch. Dis. Child. 45, 13–23 10.1136/adc.45.239.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen, F., Hu, X., He, Y. and Huang, D. (2021) Lipidomics demonstrates the association of sex hormones with sebum. J. Cosmet. Dermatol. 20, 2015–2019 10.1111/jocd.14055 [DOI] [PubMed] [Google Scholar]
- 91.Strauss, J.S., Kligman, A.M. and Pochi, P.E. (1962) The effect of androgens and estrogens on human sebaceous glands. J. Invest. Dermatol. 39, 139–155 10.1038/jid.1962.94 [DOI] [PubMed] [Google Scholar]
- 92.Lam, T.H., Verzotto, D., Brahma, P., Ng, A.H.Q., Hu, P., Schnell, D.et al. (2018) Understanding the microbial basis of body odor in pre-pubescent children and teenagers. Microbiome 6, 213 10.1186/s40168-018-0588-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beier, K., Ginez, I. and Schaller, H. (2005) Localization of steroid hormone receptors in the apocrine sweat glands of the human axilla. Histochem. Cell Biol. 123, 61–65 10.1007/s00418-004-0736-3 [DOI] [PubMed] [Google Scholar]
- 94.Raghunath, R.S., Venables, Z.C. and Millington, G.W.M. (2015) The menstrual cycle and the skin. Clin. Exp. Dermatol. 40, 111–115 10.1111/ced.12588 [DOI] [PubMed] [Google Scholar]
- 95.Wilkinson, H.N. and Hardman, M.J. (2017) The role of estrogen in cutaneous ageing and repair. Maturitas 103, 60–64 10.1016/j.maturitas.2017.06.026 [DOI] [PubMed] [Google Scholar]
- 96.Kanda, N., Hoashi, T. and Saeki, H. (2019) The roles of sex hormones in the course of atopic dermatitis. Int. J. Mol. Sci. 20, 4660 10.3390/ijms20194660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang, M., Zhou, M., Li, Y., Huang, H. and Jia, Y. (2021) Lipidomic analysis of facial skin surface lipid reveals the causes of pregnancy-related skin barrier weakness. Sci. Rep. 11, 3229 10.1038/s41598-021-82624-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.James, A.G., Austin, C.J., Cox, D.S., Taylor, D. and Calvert, R. (2013) Microbiological and biochemical origins of human axillary odour. FEMS Microbiol. Ecol. 83, 527–540 10.1111/1574-6941.12054 [DOI] [PubMed] [Google Scholar]
- 99.Troccaz, M., Gaïa, N., Beccucci, S., Schrenzel, J., Cayeux, I., Starkenmann, C.et al. (2015) Mapping axillary microbiota responsible for body odours using a culture-independent approach. Microbiome 3, 3 10.1186/s40168-014-0064-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Emter, R. and Natsch, A. (2008) The sequential action of a dipeptidase and a β-lyase is required for the release of the human body odorant 3-methyl-3-sulfanylhexan-1-ol from a secreted cys-gly-(S) conjugate by corynebacteria. J. Biol. Chem. 283, 20645–20652 10.1074/jbc.M800730200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baumann, T., Bergmann, S., Schmidt-Rose, T., Max, H., Martin, A., Enthaler, B.et al. (2014) Glutathione-conjugated sulfanylalkanols are substrates for ABCC 11 and γ -glutamyl transferase 1: a potential new pathway for the formation of odorant precursors in the apocrine sweat gland. Exp. Dermatol. 23, 247–252 10.1111/exd.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen, L., Li, J., Zhu, W., Kuang, Y., Liu, T., Zhang, W.et al. (2020) Skin and gut microbiome in psoriasis: gaining insight into the pathophysiology of it and finding novel therapeutic strategies. Front. Microbiol. 11, 589726 10.3389/fmicb.2020.589726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wark, K.J.L. and Cains, G.D. (2021) The microbiome in hidradenitis suppurativa: a review. Dermatol. Ther. (Heidelb) 11, 39–52 10.1007/s13555-020-00465-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heng, A.H.S. and Chew, F.T. (2020) Systematic review of the epidemiology of acne vulgaris. Sci. Rep. 10, 5754 10.1038/s41598-020-62715-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Conwill, A., Kuan, A.C., Damerla, R., Poret, A.J., Baker, J.S., Tripp, A.D.et al. (2022) Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 30, 171–182.e7 10.1016/j.chom.2021.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dawson, A.L. and Dellavalle, R.P. (2013) Acne vulgaris. BMJ 346, f2634 10.1136/bmj.f2634 [DOI] [PubMed] [Google Scholar]
- 107.Barnard, E., Shi, B., Kang, D., Craft, N. and Li, H. (2016) The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 6, 39491 10.1038/srep39491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fitz-Gibbon, S., Tomida, S., Chiu, B.H., Nguyen, L., Du, C., Liu, M.et al. (2013) Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 10.1038/jid.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dessinioti, C. and Katsambas, A. (2017) Propionibacterium acnes and antimicrobial resistance in acne. Clin. Dermatol. 35, 163–167 10.1016/j.clindermatol.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 110.Pécastaings, S., Roques, C., Nocera, T., Peraud, C., Mengeaud, V., Khammari, A.et al. (2018) Characterisation of Cutibacterium acnes phylotypes in acne and in vivo exploratory evaluation of Myrtacine®. J. Eur. Acad. Dermatol. Venereol. 32, 15–23 10.1111/jdv.15042 [DOI] [PubMed] [Google Scholar]
- 111.Dagnelie, M., Montassier, E., Khammari, A., Mounier, C., Corvec, S. and Dréno, B. (2019) Inflammatory skin is associated with changes in the skin microbiota composition on the back of severe acne patients. Exp. Dermatol. 28, 961–967 10.1111/exd.13988 [DOI] [PubMed] [Google Scholar]
- 112.Dagnelie, M.A., Corvec, S., Saint-Jean, M., Nguyen, J.M., Khammari, A. and Dréno, B. (2019) Cutibacterium acnes phylotypes diversity loss: a trigger for skin inflammatory process. J. Eur. Acad. Dermatol. Venereol. 33, 2340–2348 10.1111/jdv.15795 [DOI] [PubMed] [Google Scholar]
- 113.Barnard, E., Johnson, T., Ngo, T., Arora, U., Leuterio, G., McDowell, A.et al. (2020) Porphyrin production and regulation in cutaneous propionibacteria. mSphere 5, e00793-19 10.1128/mSphere.00793-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sanford, J.A., O'Neill, A.M., Zouboulis, C.C. and Gallo, R.L. (2019) Short-chain fatty acids from Cutibacterium acnes activate both a canonical and epigenetic inflammatory response in human sebocytes. J. Immunol. 202, 1767–1776 10.4049/jimmunol.1800893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zaenglein, A.L. (2018) Acne vulgaris. N. Engl. J. Med. 379, 1343–1352 10.1056/NEJMcp1702493 [DOI] [PubMed] [Google Scholar]
- 116.Wang, Y., Kao, M.S., Yu, J., Huang, S., Marito, S., Gallo, R.et al. (2016) A precision microbiome approach using sucrose for selective augmentation of staphylococcus epidermidis fermentation against Propionibacterium acnes. Int. J. Mol. Sci. 17, 1870 10.3390/ijms17111870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marito, S., Keshari, S., Traisaeng, S., My, D.T.T., Balasubramaniam, A., Adi, P.et al. (2021) Electricity-producing Staphylococcus epidermidis counteracts Cutibacterium acnes. Sci. Rep. 11, 12001 10.1038/s41598-021-91398-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang, A.J., Marito, S., Yang, J.J., Keshari, S., Chew, C.H., Chen, C.C.et al. (2018) A microtube array membrane (MTAM) encapsulated live fermenting Staphylococcus epidermidis as a skin probiotic patch against Cutibacterium acnes. Int. J. Mol. Sci. 20, 14 10.3390/ijms20010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sathikulpakdee, S., Kanokrungsee, S., Vitheejongjaroen, P., Kamanamool, N., Udompataikul, M. and Taweechotipatr, M. (2022) Efficacy of probiotic-derived lotion from Lactobacillus paracasei MSMC 39-1 in mild to moderate acne vulgaris, randomized controlled trial. J. Cosmet. Dermatol. 21, 5092–5097 10.1111/jocd.14971 [DOI] [PubMed] [Google Scholar]
- 120.Woodburn, K.W., Jaynes, J. and Clemens, L.E. (2020) Designed antimicrobial peptides for topical treatment of antibiotic resistant acne vulgaris. Antibiotics 9, 23 10.3390/antibiotics9010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Castillo, D.E., Nanda, S. and Keri, J.E. (2019) Propionibacterium (Cutibacterium) acnes bacteriophage therapy in acne: current evidence and future perspectives. Dermatol. Ther. (Heidelb) 9, 19–31 10.1007/s13555-018-0275-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jung, G.W., Tse, J.E., Guiha, I. and Rao, J. (2013) Prospective, randomized, open-label trial comparing the safety, efficacy, and tolerability of an acne treatment regimen with and without a probiotic supplement and minocycline in subjects with mild to moderate acne. J. Cutan. Med. Surg. 17, 114–122 10.2310/7750.2012.12026 [DOI] [PubMed] [Google Scholar]
- 123.Fabbrocini, G., Bertona, M., Picazo, Ó., Pareja-Galeano, H., Monfrecola, G. and Emanuele, E. (2016) Supplementation with Lactobacillus rhamnosus SP1 normalises skin expression of genes implicated in insulin signalling and improves adult acne. Benef. Microbes 7, 625–630 10.3920/BM2016.0089 [DOI] [PubMed] [Google Scholar]
- 124.Thompson, K.G., Rainer, B.M., Antonescu, C., Florea, L., Mongodin, E.F., Kang, S.et al. (2020) Minocycline and its impact on microbial dysbiosis in the skin and gastrointestinal tract of acne patients. Ann. Dermatol. 32, 21–30 10.5021/ad.2020.32.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Oh, J., Byrd, A.L., Park, M., Kong, H.H. and Segre, J.A. (2016) Temporal stability of the human skin microbiome. Cell 165, 854–866 10.1016/j.cell.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luebberding, S., Krueger, N. and Kerscher, M. (2013) Skin physiology in men and women: in vivo evaluation of 300 people including TEWL, SC hydration, sebum content and skin surface pH. Int. J. Cosmet. Sci. 35, 477–483 10.1111/ics.12068 [DOI] [PubMed] [Google Scholar]
- 127.Howard, B., Bascom, C.C., Hu, P., Binder, R.L., Fadayel, G., Huggins, T.G.et al. (2022) Aging-associated changes in the adult human skin microbiome and the host factors that affect skin microbiome composition. J. Invest. Dermatol. 142, 1934–1946.e21 10.1016/j.jid.2021.11.029 [DOI] [PubMed] [Google Scholar]
- 128.Simon, A.K., Hollander, G.A. and McMichael, A. (2015) Evolution of the immune system in humans from infancy to old age. Proc. R. Soc. B. 282, 20143085 10.1098/rspb.2014.3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weitzmann, A., Naumann, R., Dudeck, A., Zerjatke, T., Gerbaulet, A. and Roers, A. (2020) Mast cells occupy stable clonal territories in adult steady-state skin. J. Invest. Dermatol. 140, 2433–2441.e5 10.1016/j.jid.2020.03.963 [DOI] [PubMed] [Google Scholar]
- 130.Tokura, Y., Phadungsaksawasdi, P., Kurihara, K., Fujiyama, T. and Honda, T. (2021) Pathophysiology of skin resident memory T cells. Front. Immunol. 11, 618897 10.3389/fimmu.2020.618897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Richardson, M., Gottel, N., Gilbert, J.A. and Lax, S. (2019) Microbial similarity between students in a common dormitory environment reveals the forensic potential of individual microbial signatures. mBio 10, e01054-19 10.1128/mBio.01054-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramadan, M., Solyman, S., Taha, M. and Hanora, A. (2016) Preliminary characterization of human skin microbiome in healthy Egyptian individuals. Cell. Mol. Biol. 62, 21–27 PMID: [PubMed] [Google Scholar]
- 133.Ogai, K., Nana, B.C., Lloyd, Y.M., Arios, J.P., Jiyarom, B., Awanakam, H.et al. (2022) Skin microbiome profile of healthy Cameroonians and Japanese. Sci. Rep. 12, 1364 10.1038/s41598-022-05244-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chaudhari, D.S., Dhotre, D.P., Agarwal, D.M., Gaike, A.H., Bhalerao, D., Jadhav, P.et al. (2020) Gut, oral and skin microbiome of Indian patrilineal families reveal perceptible association with age. Sci. Rep. 10, 5685 10.1038/s41598-020-62195-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cho, H.W. and Eom, Y.B. (2021) Forensic analysis of human microbiome in skin and body fluids based on geographic location. Front. Cell Infect. Microbiol. 11, 695191 10.3389/fcimb.2021.695191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peng, M. and Biswas, D. (2020) Environmental influences of high-density agricultural animal operation on human forearm skin microflora. Microorganisms 8, E1481 10.3390/microorganisms8101481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ying, S., Zeng, D.N., Chi, L., Tan, Y., Galzote, C., Cardona, C.et al. (2015) The influence of age and gender on skin-associated microbial communities in urban and rural human populations. PLoS ONE 10, e0141842 10.1371/journal.pone.0141842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hospodsky, D., Pickering, A.J., Julian, T.R., Miller, D., Gorthala, S., Boehm, A.B.et al. (2014) Hand bacterial communities vary across two different human populations. Microbiology 160, 1144–1152 10.1099/mic.0.075390-0 [DOI] [PubMed] [Google Scholar]
- 139.Lax, S., Smith, D.P., Hampton-Marcell, J., Owens, S.M., Handley, K.M., Scott, N.M.et al. (2014) Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 10.1126/science.1254529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Leung, M.H.Y., Tong, X., Wilkins, D., Cheung, H.H.L. and Lee, P.K.H. (2018) Individual and household attributes influence the dynamics of the personal skin microbiota and its association network. Microbiome 6, 26 10.1186/s40168-018-0412-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sharma, A., Richardson, M., Cralle, L., Stamper, C.E., Maestre, J.P., Stearns-Yoder, K.A.et al. (2019) Longitudinal homogenization of the microbiome between both occupants and the built environment in a cohort of United States Air Force Cadets. Microbiome 7, 70 10.1186/s40168-019-0686-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Voorhies, A.A., Mark Ott, C., Mehta, S., Pierson, D.L., Crucian, B.E., Feiveson, A.et al. (2019) Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep. 9, 9911 10.1038/s41598-019-46303-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang, S., Haiminen, N., Carrieri, A.P., Hu, R., Jiang, L., Parida, L.et al. (2020) Human skin, oral, and gut microbiomes predict chronological age. mSystems 5, e00630-19 10.1128/mSystems.00630-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim, H.J., Kim, J.J., Myeong, N.R., Kim, T., Kim, D., An, S.et al. (2019) Segregation of age-related skin microbiome characteristics by functionality. Sci. Rep. 9, 16748 10.1038/s41598-019-53266-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Boireau-Adamezyk, E., Baillet-Guffroy, A. and Stamatas, G.N. (2021) The stratum corneum water content and natural moisturization factor composition evolve with age and depend on body site. Int. J. Dermatol. 60, 834–839 10.1111/ijd.15417 [DOI] [PubMed] [Google Scholar]
- 146.Feuillie, C., Vitry, P., McAleer, M.A., Kezic, S., Irvine, A.D., Geoghegan, J.A.et al. (2018) Adhesion of Staphylococcus aureus to corneocytes from atopic dermatitis patients is controlled by natural moisturizing factor levels. mBio 9, e01184-18 10.1128/mBio.01184-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dimitriu, P.A., Iker, B., Malik, K., Leung, H., Mohn, W.W. and Hillebrand, G.G. (2019) New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. mBio 10, e00839-19 10.1128/mBio.00839-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shibagaki, N., Suda, W., Clavaud, C., Bastien, P., Takayasu, L., Iioka, E.et al. (2017) Aging-related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci. Rep. 7, 10567 10.1038/s41598-017-10834-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jugé, R., Rouaud-Tinguely, P., Breugnot, J., Servaes, K., Grimaldi, C., Roth, M.P.et al. (2018) Shift in skin microbiota of Western European women across aging. J. Appl. Microbiol. 125, 907–916 10.1111/jam.13929 [DOI] [PubMed] [Google Scholar]
- 150.Kim, H.J., Oh, H.N., Park, T., Kim, H., Lee, H.G., An, S.et al. (2022) Aged related human skin microbiome and mycobiome in Korean women. Sci. Rep. 12, 2351 10.1038/s41598-022-06189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alkema, W., Boekhorst, J., Eijlander, R.T., Schnittger, S., De Gruyter, F., Lukovac, S.et al. (2021) Charting host-microbe co-metabolism in skin aging and application to metagenomics data. PLoS ONE 16, e0258960 10.1371/journal.pone.0258960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brodin, P. and Davis, M.M. (2017) Human immune system variation. Nat. Rev. Immunol. 17, 21–29 10.1038/nri.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chambers, E.S. and Vukmanovic-Stejic, M. (2020) Skin barrier immunity and ageing. Immunology 160, 116–125 10.1111/imm.13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hasegawa, T., Feng, Z., Yan, Z., Ngo, K.H., Hosoi, J. and Demehri, S. (2020) Reduction in human epidermal langerhans cells with age is associated with decline in CXCL14-mediated recruitment of CD14+ monocytes. J. Invest. Dermatol. 140, 1327–1334 10.1016/j.jid.2019.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pilkington, S.M., Ogden, S., Eaton, L.H., Dearman, R.J., Kimber, I. and Griffiths, C.E.M. (2018) Lower levels of interleukin-1 β gene expression are associated with impaired Langerhans’ cell migration in aged human skin. Immunology 153, 60–70 10.1111/imm.12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Grolleau-Julius, A., Harning, E.K., Abernathy, L.M. and Yung, R.L. (2008) Impaired dendritic cell function in aging leads to defective antitumor immunity. Cancer Res. 68, 6341–6349 10.1158/0008-5472.CAN-07-5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Choi, E.H. (2019) Aging of the skin barrier. Clin. Dermatol. 37, 336–345 10.1016/j.clindermatol.2019.04.009 [DOI] [PubMed] [Google Scholar]
- 158.Cranendonk, D.R., Lavrijsen, A.P.M., Prins, J.M. and Wiersinga, W.J. (2017) Cellulitis: current insights into pathophysiology and clinical management. Neth. J. Med. 75, 366–378 PMID: [PubMed] [Google Scholar]
- 159.Haran, J.P., Wilsterman, E., Zeoli, T., Beaudoin, F.L., Tjia, J. and Hibberd, P.L. (2017) Elderly patients are at increased risk for treatment failure in outpatient management of purulent skin infections. Am. J. Emerg. Med. 35, 249–254 10.1016/j.ajem.2016.10.060 [DOI] [PubMed] [Google Scholar]
- 160.Daou, H., Paradiso, M., Hennessy, K. and Seminario-Vidal, L. (2021) Rosacea and the microbiome: a systematic review. Dermatol. Ther. (Heidelb) 11, 1–12 10.1007/s13555-020-00460-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu, C., Ponsero, A.J., Armstrong, D.G., Lipsky, B.A. and Hurwitz, B.L. (2020) The dynamic wound microbiome. BMC Med. 18, 358 10.1186/s12916-020-01820-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin, Q., Panchamukhi, A., Li, P., Shan, W., Zhou, H., Hou, L.et al. (2021) Malassezia and Staphylococcus dominate scalp microbiome for seborrheic dermatitis. Bioprocess Biosyst. Eng. 44, 965–975 10.1007/s00449-020-02333-5 [DOI] [PubMed] [Google Scholar]
- 163.Tao, R., Li, R. and Wang, R. (2021) Skin microbiome alterations in seborrheic dermatitis and dandruff: a systematic review. Exp. Dermatol. 30, 1546–1553 10.1111/exd.14450 [DOI] [PubMed] [Google Scholar]
- 164.Fujimura, S. and Nakamura, T. (1978) Purification and properties of a bacteriocin-like substance (Acnecin) of oral Propionibacterium acnes. Antimicrob. Agents Chemother. 14, 893–898 10.1128/AAC.14.6.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang, Y., Dai, A., Huang, S., Kuo, S., Shu, M., Tapia, C.P.et al. (2014) Propionic acid and its esterified derivative suppress the growth of methicillin-resistant Staphylococcus aureus USA300. Benef. Microbes 5, 161–168 10.3920/BM2013.0031 [DOI] [PubMed] [Google Scholar]
- 166.Shu, M., Wang, Y., Yu, J., Kuo, S., Coda, A., Jiang, Y.et al. (2013) Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 8, e55380 10.1371/journal.pone.0055380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gribbon, E.M., Cunliffe, W.J. and Holland, K.T. (1993) Interaction of Propionibacterium acnes with skin lipids in vitro. J. Gen. Microbiol. 139, 1745–1751 10.1099/00221287-139-8-1745 [DOI] [PubMed] [Google Scholar]
- 168.Brüggemann, H., Salar-Vidal, L., Gollnick, H.P.M. and Lood, R. (2021) A Janus-faced bacterium: host-beneficial and -detrimental roles of Cutibacterium acnes. Front. Microbiol. 12, 673845 10.3389/fmicb.2021.673845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Megyeri, K., Orosz, L., Bolla, S., Erdei, L., Rázga, Z., Seprényi, G.et al. (2018) Propionibacterium acnes induces autophagy in keratinocytes: involvement of multiple mechanisms. J. Invest. Dermatol. 138, 750–759 10.1016/j.jid.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 170.Kao, H.J., Wang, Y.H., Keshari, S., Yang, J.J., Simbolon, S., Chen, C.C.et al. (2021) Propionic acid produced by Cutibacterium acnes fermentation ameliorates ultraviolet B-induced melanin synthesis. Sci. Rep. 11, 11980 10.1038/s41598-021-91386-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schwarz, A., Bruhs, A. and Schwarz, T. (2017) The short-chain fatty acid sodium butyrate functions as a regulator of the skin immune system. J. Invest. Dermatol. 137, 855–864 10.1016/j.jid.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 172.Sanford, J.A., Zhang, L.J., Williams, M.R., Gangoiti, J.A., Huang, C.M. and Gallo, R.L. (2016) Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci. Immunol. 1, eaah4609 10.1126/sciimmunol.aah4609 [DOI] [PubMed] [Google Scholar]
- 173.Keshari, S., Balasubramaniam, A., Myagmardoloonjin, B., Herr, D.R., Negari, I.P. and Huang, C.M. (2019) Butyric acid from probiotic staphylococcus epidermidis in the skin microbiome down-regulates the ultraviolet-induced pro-inflammatory IL-6 cytokine via short-chain fatty acid receptor. Int. J. Mol. Sci. 20, 4477 10.3390/ijms20184477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Horn, K.J., Jaberi Vivar, A.C., Arenas, V., Andani, S., Janoff, E.N. and Clark, S.E. (2022) Corynebacterium species inhibit Streptococcus pneumoniae colonization and infection of the mouse airway. Front. Microbiol. 12, 804935 10.3389/fmicb.2021.804935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bomar, L., Brugger, S.D., Yost, B.H., Davies, S.S. and Lemon, K.P. (2016) Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7, e01725-15 10.1128/mBio.01725-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ramsey, M.M., Freire, M.O., Gabrilska, R.A., Rumbaugh, K.P. and Lemon, K.P. (2016) Staphylococcus aureus shifts toward commensalism in response to corynebacterium species. Front. Microbiol. 7, 1230 10.3389/fmicb.2016.01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hardy, B.L., Dickey, S.W., Plaut, R.D., Riggins, D.P., Stibitz, S., Otto, M.et al. (2019) Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. mBio 10, e02491-18 10.1128/mBio.02491-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lynch, D., O'Connor, P.M., Cotter, P.D., Hill, C., Field, D. and Begley, M. (2019) Identification and characterisation of capidermicin, a novel bacteriocin produced by Staphylococcus capitis. PLoS ONE 14, e0223541 10.1371/journal.pone.0223541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kumar, R., Jangir, P.K., Das, J., Taneja, B. and Sharma, R. (2017) Genome analysis of Staphylococcus capitis TE8 reveals repertoire of antimicrobial peptides and adaptation strategies for growth on human skin. Sci. Rep. 7, 10447 10.1038/s41598-017-11020-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.O'Neill, A.M., Nakatsuji, T., Hayachi, A., Williams, M.R., Mills, R.H., Gonzalez, D.J.et al. (2020) Identification of a human skin commensal bacterium that selectively kills Cutibacterium acnes. J. Invest. Dermatol. 140, 1619–1628.e2 10.1016/j.jid.2019.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Paharik, A.E., Parlet, C.P., Chung, N., Todd, D.A., Rodriguez, E.I., Van Dyke, M.J.et al. (2017) Coagulase-negative staphylococcal strain prevents staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22, 746–756.e5 10.1016/j.chom.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Iwase, T., Uehara, Y., Shinji, H., Tajima, A., Seo, H., Takada, K.et al. (2010) Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- 183.Nakatsuji, T., Chen, T.H., Narala, S., Chun, K.A., Two, A.M., Yun, T.et al. (2017) Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680 10.1126/scitranslmed.aah4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hernández-Aristizábal, I. and Ocampo-Ibáñez, I.D. (2021) Antimicrobial peptides with antibacterial activity against vancomycin-resistant Staphylococcus aureus strains: classification, structures, and mechanisms of action. Int. J. Mol. Sci. 22, 7927 10.3390/ijms22157927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zipperer, A., Konnerth, M.C., Laux, C., Berscheid, A., Janek, D., Weidenmaier, C.et al. (2016) Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 10.1038/nature18634 [DOI] [PubMed] [Google Scholar]
- 186.Bitschar, K., Sauer, B., Focken, J., Dehmer, H., Moos, S., Konnerth, M.et al. (2019) Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun. 10, 2730 10.1038/s41467-019-10646-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Brown, M.M., Kwiecinski, J.M., Cruz, L.M., Shahbandi, A., Todd, D.A., Cech, N.B.et al. (2020) Novel peptide from commensal Staphylococcus simulans blocks methicillin-resistant Staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob. Agents Chemother. 64, e00172-20 10.1128/AAC.00172-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Severn, M., Cho, Y.S.K., Manzer, H., Bunch, Z., Shahbandi, A., Todd, D.et al. (2022) The commensal Staphylococcus warneri makes peptide inhibitors of MRSA quorum sensing that protect skin from atopic or necrotic damage. J. Invest. Dermatol. 142, 3349–3352.e5 10.1016/j.jid.2022.05.1092 [DOI] [PMC free article] [PubMed] [Google Scholar]