Abstract
Hpr1 forms, together with Tho2, Mft1, and Thp2, the THO complex, which controls transcription elongation and genome stability in Saccharomyces cerevisiae. Mutations in genes encoding the THO complex confer strong transcription-impairment and hyperrecombination phenotypes in the bacterial lacZ gene. In this work we demonstrate that Hpr1 is a factor required for transcription of long as well as G+C-rich DNA sequences. Using different lacZ segments fused to the GAL1 promoter, we show that the negative effect of lacZ sequences on transcription depends on their distance from the promoter. In parallel, we show that transcription of either a long LYS2 fragment or the S. cerevisiae YAT1 G+C-rich open reading frame fused to the GAL1 promoter is severely impaired in hpr1 mutants, whereas transcription of LAC4, the Kluyveromyces lactis ortholog of lacZ but with a lower G+C content, is only slightly affected. The hyperrecombination behavior of the DNA sequences studied is consistent with the transcriptional defects observed in hpr1 cells. These results indicate that both length and G+C content are important elements influencing transcription in vivo. We discuss their relevance for the understanding of the functional role of Hpr1 and, by extension, the THO complex.
The control of genome stability is essential to ensure maintenance of genetic information in all cells of a living organism. Dysfunction of this control causes mutations and chromosomal aberrations that can give rise to loss of gene function, cell death, or irreversible changes in the cell program.
Genetic recombination is required for mitotic DNA repair and for proper meiotic chromosome segregation. In addition, it may also be responsible for processes of genetic instability. A number of animal diseases, including cancer, originate by events of mitotic recombination between repeats that lead to chromosomal aberrations (34). Several elements have been described to enhance mitotic recombination, including DNA damage, replication defects, alteration of chromatin structure, and transcriptional activity (reviewed in reference 3). Ikeda and Matsumoto (26) first described the influence of transcription on recombination showing that recombination of phage λ was stimulated by transcription. In yeast, the first example of transcription-associated recombination was the finding that a hotspot of ribosomal DNA (rDNA) recombination, HOT1, was dependent on RNA polymerase I-driven transcription (55, 60). Thomas and Rothstein (56) extended transcription-induced recombination to sequences transcribed by RNA polymerase II (RNAPII). Additional examples of RNAPII-dependent recombination have been subsequently described in yeast (21, 36, 50) and mammalian cells (37, 57). Special mention must be made of the modulation of recombination at the immunoglobulin loci, as both V(D)J recombination (7, 31, 38) and class switching (15) are positively controlled by transcription.
A gene linking transcription and genome instability in Saccharomyces cerevisae is HPR1, as hpr1 mutants show both increased levels of recombination between direct repeats and chromosome loss (2, 49) as well as strong transcriptional defects (11, 44, 67). Detailed characterization of these defects has shown that the absence of Hpr1 causes impairment of transcription elongation. The intensity of such a transcriptional impairment depends on the transcribed DNA sequence (11). There is a close correlation between the reluctance of a DNA sequence to be transcribed in hpr1 cells and the ability of such a sequence to promote recombination when inserted between direct repeats (11, 44).
Biochemical and genetic analyses have contributed to identifying several factors that participate in RNAPII-mediated transcription elongation (reviewed in reference 14). According to their function in transcriptional elongation, these factors can be classified in different groups. TFIIS prevents RNAPII arrest and induces nascent transcript cleavage (reviewed in reference 64). Some other factors, like TFIIF, CSB, ELL, and elongin, influence elongation by suppressing the pausing of RNAPII (5, 46, 52, 53). P-TEFb stimulates transcription elongation in response to transactivators (reviewed in reference 45) by antagonizing negative factors like DSIF and NELF (22, 61, 65). Finally, some transcription elongation factors like FACT and Elongator play a role in facilitating RNAPII-driven transcription on chromatin templates (39, 40).
Although hpr1Δ cells are affected in transcription elongation in vivo, Hpr1 does not seem to be physically associated with any of the known elongation factors. It has been demonstrated that Hpr1 is physically present in a new form of RNAPII holoenzyme that has been proposed to respond to protein kinase C-mediated signal transduction (10). Hpr1 forms the THO complex in vivo together with the products of the THO2, MFT1, and THP2 genes (12). The absence of any of the four proteins confers similar phenotypes of transcriptional elongation impairment and hyperrecombination, indicating that the THO complex is a functional unit in gene expression and genome stability (12). However, the way THO controls these processes remains obscure.
As relevance of the THO complex in transcription depends on the transcribed DNA sequence, investigation of the features that make transcription of a particular DNA sequence dependent on Hpr1 can provide some clues to understanding its precise function. We have found that transcriptional impairment in hpr1 occurs primarily in long transcription units as well as in DNA sequences with a high G+C content fused to the GAL1 promoter. The relevance of these results for understanding the functional role of the THO complex is discussed.
MATERIALS AND METHODS
Yeast strains and plasmids.
The two isogenic yeast strains used in this study were W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) and U768-4C (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hpr1Δ::HIS3). All plasmids used are monocopy CEN-based plasmids and are listed in Table 1.
TABLE 1.
Plasmids
| Plasmid | Description | Source or reference |
|---|---|---|
| pRS416 | YCp vector based on the URA3 gene | 54 |
| p416GALI-lacZ | pRS416 containing the lacZ region fused to the GAL1 promoter | 35 |
| pSCh202 | pRS416 containing the PHO5 region fused to the GAL1 promoter | 11 |
| pSCh212 | pSCh202 with lacZ transcriptionally fused to the 3′ end UTRa of PHO5 | 11 |
| pRS314-L | YCp vector pRS314, based on the TRP1 gene and containing two 598-bp LEU2 internal fragments repeated in direct orientation, separated by a polylinker | 43 |
| pSCh211 | pSCh212 with lacZ in opposite orientation | This study |
| pSCh211Δ17-1 | pSCh202 with a ∼0.4-kb fragment of the 3′ end of lacZ fused to the 3′ end UTR of PHO5 | This study |
| pSCh229 | pSCh202 with the first 439 bp of lacZ ORF fused to the 3′ end UTR of PHO5 | This study |
| pSCh226 | pSCh202 with the HpaI-BssHII 447-bp fragment of lacZ inserted in the 3′ end UTR of PHO5 | This study |
| pSCh251 | pSCh202 with a 393-bp fragment of GAL1 (from position 843 to 1236 of the ORF) inserted in the 3′ end UTR of PHO5 | This study |
| pSCh215 | pRS416 containing a 439-bp fragment of the 5′ end of lacZ fused to the GAL1 promoter | This study |
| pSCh213 | pRS416 containing the 447-bp HpaI-BssHII fragment of lacZ fused to the GAL1 promoter | This study |
| pSCh216 | pRS416 containing the 352-bp PvuII-EcoRI fragment of lacZ fused to the GAL1 promoter | This study |
| pSCh218 | pSCh202 containing the first 439-bp fragment of the 5′ end of lacZ inserted between the GAL1 promoter and the PHO5 ORF | This study |
| pSCh219 | pSCh202 containing the 447-bp HpaI-BssHII fragment of lacZ inserted between the GAL1 promoter and the PHO5 ORF | This study |
| pSCh220 | pSCh202 containing the 352-bp PvuII-EcoRI fragment of lacZ inserted between the GAL1 promoter and the PHO5 ORF | This study |
| pSCh221 | pRS314-L containing the first 439 bp of lacZ ORF inserted between the repeats | This study |
| pSCh222 | pRS314-L containing the 447-bp HpaI-BssHII fragment of lacZ inserted between the repeats | This study |
| pSCh223 | pRS314-L containing the 352-bp PvuII-EcoRI fragment of lacZ inserted between the repeats | This study |
| pSCh205 | pRS314-L containing the entire lacZ gene inserted between the repeats | 11 |
| pSCh227 | pRS416 containing the 3.7-kbp EcoRV fragment of LYS2 fused to the GAL1 promoter | This study |
| pSCh230 | pRS314-L containing the 3.7-kbp EcoRV fragment of LYS2 inserted between the repeats | This study |
| pSCh255 | pRS416 containing the entire LAC4 coding region fused to the GAL1 promoter | This study |
| pSCh254 | pRS314-L containing the entire LAC4 coding region inserted between the repeats | This study |
| pSCh247 | pRS416 containing the entire YAT1 coding region fused to the GAL1 promoter | This study |
| pSCh248 | pRS314-L containing the entire YAT1 coding region inserted between the repeats | This study |
UTR, untranslated region.
Analysis of gene expression and recombination.
For the analysis of GAL1-driven expression, mid-log phase cells were inoculated with 3% glycerol–2% lactate synthetic medium at an optical density at 600 nm (OD600) of 0.1. After 16 h of incubation at 30°C, 2% galactose was added and incubation was continued for another 8 h at 30°C. Acid phosphatase activity and mRNA levels were determined as described previously (11). It is important that all transcription analyses shown in this study, with few exceptions, were made in monocopy CEN-based plasmids, which are lost at higher frequencies in hpr1 versus wild-type strains (11). However, since in all experiments cells were grown under the selection conditions for the plasmid, more than 90% of the hpr1 cells still contained plasmids. Therefore, all observed transcriptional effects are not caused by plasmid loss.
For expression analysis of EGT2, CDC48, KAR2, OLE1, and GOG5 cells were grown in yeast extract-peptone-dextrose (YEPD)-rich medium to an OD600 of 1.0 and subsequently sampled. DNA probes for Northern experiments were obtained by PCR amplification using the following pairs of primers: TCATTTCGATACTCGGCCTAG and GCAGCATCAGAGCTAGTTGTG for EGT2; AAACCACTTTTGGACGCCTC and TCTTGTCTCTCTTTGGAGCT for CDC48; TTCAACAGACTAAGCGCTGG and CAATTTCAATACGGGTGGACA for KAR2; ATGCCAACTTCTGGAACTAC and CCGAAAGTAACAATGGCAGT for OLE1; and TTGAAAACAGGTCATGCAGG and TGGGCTTGTTGCTTCTTTTG for GOG5.
Recombination frequencies were calculated as the median of six independent cultures as previously published (43).
Mapping of MNase cleavage sites.
Yeast spheroplasts and micrococcal nuclease (MNase) digestions were performed according to Fedor and Kornberg (19) with the modifications of Chávez et al. (13). Spheroplasts prepared from mid-log phase cultures transformed with p416GAL1lacZ and grown in the appropriate selective medium containing 2% glucose or 2% galactose were lysed and immediately digested with 6.25 to 800 mU of MNase. For naked DNA controls, genomic DNA was extracted as previously described (28) and digested with 0.003 to 1.6 mU of MNase under the same conditions.
MNase-cleaved genomic DNA was digested with either EcoRI (for the endogenous GAL1 gene) or ClaI (for the GAL::lacZ fusion) and resolved in 1.5% agarose. As internal size markers, we used genomic DNA digested with SacI or XbaI (for the GAL1 promoter fused to lacZ). For the analysis of the endogenous GAL1 gene, the probe used was the 196-bp GAL1 fragment located immediately downstream of the EcoRI site and obtained by PCR with the oligonucleotides ATTCGACAGGTTATCAGCAAC and TTAAACTTCTTTGCGTCCATC. For the analysis of GAL1::lacZ the probe used was the 202-bp lacZ fragment immediately upstream of the ClaI site and obtained by PCR with the oligonucleotides TCGTTGCTGCATAAACCG and TCGATAATTTCACCGCCG.
Miscellaneous.
Serial deletions of the PHO5::lacZ fusion constructs were constructed using a double-stranded nested deletion kit from Amersham Pharmacia. Published methods were used for RNA and DNA hybridizations (13, 44).
RESULTS
Transcription impairment through lacZ caused by hpr1Δ is not dependent on particular lacZ sequences but on their distance from the promoter.
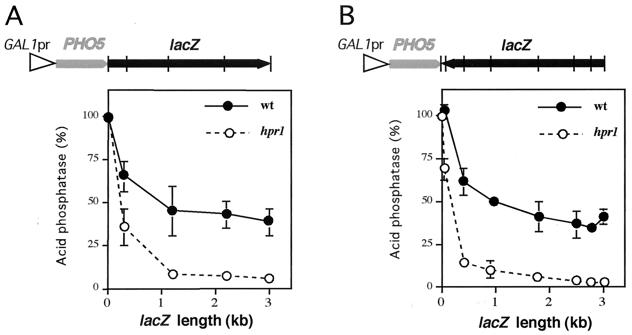
Transcription of the Escherichia coli lacZ gene in S. cerevisiae is severely impaired in hpr1 mutants at the elongation level. Transcription through PHO5 is not appreciably affected in hpr1 cells, but it becomes sensitive to hpr1 when lacZ is fused to PHO5 in a single transcription unit (11). In order to find out which structural elements or sequence motifs present in lacZ are responsible for this transcriptional elongation impairment, we constructed serial deletions of the lacZ gene in the PHO5::lacZ transcriptional fusion under control of the GAL1-regulated promoter. The resulting deletions were introduced into both wild-type and hpr1 cells. Yeast transformants grown in galactose-containing medium were then assayed for acid phosphatase activity. Expression was clearly lower in the transformants harboring PHO5-lacZ fusions than in those containing only PHO5, in both wild-type and hpr1 cells (Fig. 1A). The presence of a lacZ fragment as short as 1 kb downstream of PHO5 reduced PHO5 expression to 40% in wild-type cells. However, the reduction was considerably stronger in hpr1, reaching transcription values below 10% of those of PHO5 alone (Fig. 1A). Even the shortest fusion, encompassing ≈0.4 kb of the 5′ end of lacZ, showed reduced levels of phosphatase activity in the wild-type (65%) and, to a greater degree, hpr1 (35%) cells (Fig. 1A).
FIG. 1.
Expression patterns of serial deletions of GAL1pr::PHO5-lacZ fusion constructs. Acid phosphatase activities under induced conditions of wild-type (W303-1A) and hpr1 (U768-4C) strains transformed with lacZ-deleted variants of plasmids pSCh212 (A) or pSCh211 (B) that contain the entire lacZ coding sequence fused to PHO5 in the same and opposite orientations, respectively, under the GAL1 promoter. The average value and standard deviation of four different transformants is shown for each strain. Vertical lines across the lacZ sequences indicate the end points of the deletion constructs analyzed.
Serial deletions were also made in a PHO5::lacZ fusion carrying lacZ in an opposite orientation. A similar profile of phosphatase activities was obtained (Fig. 1B). Although all fusions showed lower expression levels than PHO5 alone, the negative transcriptional effect was clearly stronger in hpr1 than in wild-type cells. A ≈0.4-kb lacZ fragment, for example, was enough to reduce the phosphatase activity to under 15% of the level shown by PHO5 alone in hpr1 strains (Fig. 1B). Thus, the two end fragments of lacZ were able to impair transcription in hpr1 cells.
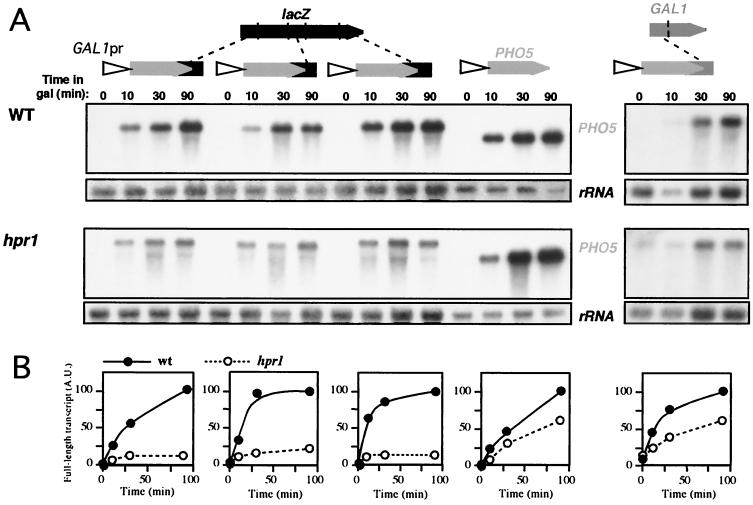
The previous results suggest that there is not a particular lacZ sequence responsible for the transcriptional elongation impairment caused by hpr1Δ. On the contrary, transcriptional impairment could occur through any lacZ region. To confirm this and to show that the negative effect of hpr1Δ on acid phosphatase expression really takes place at the transcriptional rather than posttranscriptional level, we performed Northern analyses of selected PHO5::lacZ-fragment constructs. We inserted three different ≈0.4-kb fragments of lacZ corresponding to the two ends and the center of the gene (plasmids pSCh229, pSCh226, and pSCh251; Table 1) immediately downstream of a PHO5 gene under GAL1 control. Galactose-induced transcription of the resulting fusion constructs was analyzed in wild-type and hpr1 cells. The results shown in Fig. 2 indicate a substantial decrease in the accumulation of full-length mRNA of the three fusion constructs in hpr1 cells (13 to 25% of the wild-type levels), whereas only a minor effect was observed with PHO5 alone. In addition to the full-length PHO5-lacZ mRNA, a shorter transcript exhibiting the same size as PHO5 was detected in hpr1. The presence of this shorter transcript suggests that hpr1 cells transcribe poorly through lacZ sequences, downstream of the PHO5 open reading frame (ORF). The same results were obtained when the lacZ fragments were located in the opposite orientation (data not shown). To confirm that this phenomenon was due to lacZ itself and not to the 3′ end of PHO5, we replaced the lacZ segment with a 416-bp fragment of the 5′ end of GAL1, a gene whose expression is not affected in hpr1 cells (67) (see Fig. 9A). Northern analysis of the resulting PHO5::GAL1Δ fusion was carried out in wild-type and hpr1 cells (Fig. 2). A weaker reduction in accumulation of full-length mRNA was measured in hpr1 (60% of wild-type levels), and no short transcript was detected. This confirms that the transcriptional defects of the PHO5-lacZ fusion constructs were mainly due to the presence of lacZ fragments in the transcription units.
FIG. 2.
Transcription analysis of GAL1pr::PHO5, three different GAL1pr::PHO5-lacZ fusion constructs, and a GAL1pr::PHO5-GAL1 fusion in wild-type (W303-1A) and hpr1 (U768-4C) cells. (A) Northern blot analyses of PHO5-containing mRNAs driven from the GAL1 promoter. Plasmids used were pSCh229, pSCh226, and pSCh211Δ17-1 (carrying ≈0.4 kb of the 5′ end, middle part, and 3′ end of lacZ fused to PHO5, respectively), pSCh202 (carrying the PHO5 gene), or pSCh251 (carrying ≈0.4 kb of the 5′ end of GAL1 fused to PHO5). Mid-log phase cells were cultured in 3% glycerol–2% lactate synthetic complete (SC)-Ura medium and diluted into identical fresh media to an OD600 of 0.3 and incubated for 16 h. Galactose (Gal) was then added and samples were taken for Northern analysis at different times, as specified. A 0.9-kb EcoRV PHO5 internal fragment and a 589-bp 28S rDNA internal fragment obtained by PCR (rRNA) were used as DNA probes. (B) Kinetics of induction of mRNAs as determined by quantification of Northern blots in a Fuji FLA3000. The mRNA values are given in arbitrary units (A.U.) with respects to rRNA levels. For any given construct, RNA levels are related to the wild-type (wt) levels at 90 min, which was set at 100 for each panel.
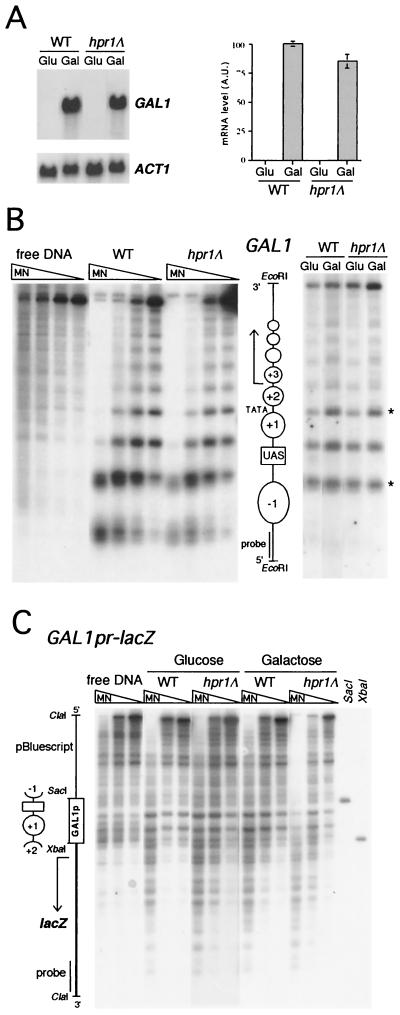
FIG. 9.
MNase digestion pattern of the GAL1 gene and the GAL1pr::lacZ fusion in wild-type and hpr1 strains under repression and activation conditions. (A) Northern blot analysis of GAL1 mRNAs. (B) Nucleosome positioning over the GAL1 gene. (C) MNase digestion pattern of the GAL1pr::lacZ fusion. A scheme of the analyzed regions of GAL1 and lacZ indicating the position of nucleosomes and the most relevant regulatory elements is shown. Asterisks indicate the MNase hypersensitive sites associated with the activation of transcription of GAL1.
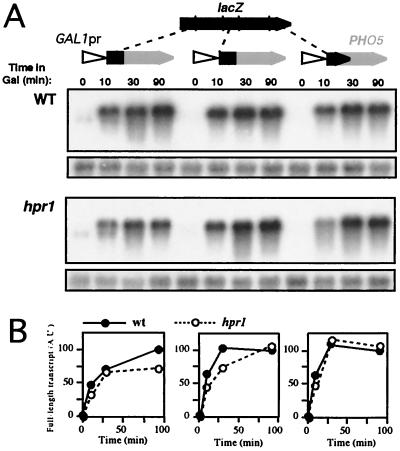
Altogether, these results suggest that the longer distance between the promoter and lacZ sequences the greater the transcriptional elongation impairment. To test this possibility, we inserted upstream of PHO5 the same three lacZ fragments used previously. The resulting transcriptional fusions exhibited similar transcription levels and patterns in wild-type and hpr1 cells (Fig. 3). Therefore, the distance between lacZ and the promoter can modulate the negative effect of the lacZ fragments on transcription in hpr1 cells.
FIG. 3.
Transcription analysis of three different GAL1pr::lacZΔ-PHO5 fusion constructs in wild-type (W303-1A) and hpr1 (U768-4C) cells. (A) Northern blot analyses of PHO5-containing mRNAs driven from the GAL1 promoter. Plasmids used were pSCh218, pSCh220, and pSCh219 (carrying ≈0.4 kb of the 5′ end, middle part, and 3′ end of lacZ fused to PHO5, respectively). As a control we used pSCh202 (carrying the PHO5 gene; data not shown), which gave identical results as those shown in Fig. 2. The transcript levels of each construct with respect to PHO5 were similar to those of the wild type shown in Fig. 2. Other details were as described for Fig. 2. (B) Quantification of Northern analyses.
Transcription through long DNA sequences is negatively affected by hpr1Δ
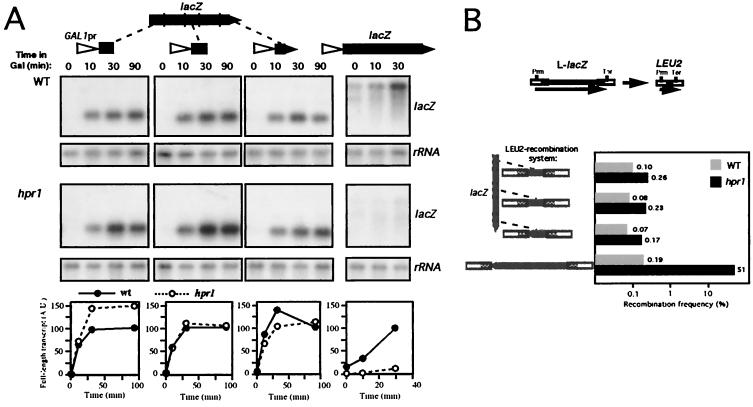
The data shown in Fig. 1 indicate that the longer the constructs containing lacZ fragments are, the lower the transcriptional yield exhibited in both wild-type and hpr1 cells. To evaluate the influence of transcript length on the transcriptional effect of hpr1, we put the same three above-mentioned lacZ fragments immediately downstream of the GAL1 promoter. The kinetics of accumulation of these three short lacZ fragments was identical in wild-type and hpr1 cells (Fig. 4A). Full-length mRNA accumulated shortly after induction in both strains and at similar levels in all three constructs, in contrast with the clear difference between the wild type and hpr1 shown by the entire lacZ (Fig. 4A). The same results were obtained when the lacZ fragments were cloned in the opposite orientation (data not shown). These results strongly suggest that short transcription units are not impaired by hpr1, even if they contain DNA fragments that hinder transcription in a different context.
FIG. 4.
Transcription and recombination analyses of several GAL1pr::lacZ fusion constructs in wild-type (W303-1A) and hpr1 (U768-4C) cells. (A) Northern analyses of lacZ-containing mRNAs transcribed from the GAL1 promoter. Plasmids used were pSCh215, pSCh213, and pSCh216 (carrying ≈0.4 kb of the 5′ end, middle part, and 3′ end of lacZ, respectively) or p416GAL1-lacZ (carrying the entire lacZ ORF). Other details were as described for Fig. 2. (B) Recombination frequencies of leu2-based direct-repeat systems containing the same short lacZ fragments used in the previous Northern experiments. Plasmids used were pSCh221, pSCh222, and pSCh223 (carrying ≈0.4 kb of the 5′ end, middle part, and 3′ end of lacZ, respectively) or pSCh205 (carrying the entire lacZ ORF). A schematic diagram of the recombination products obtained with the direct-repeat LEU2 recombination systems used is shown at the top of panel B. The LEU2 promoter (Prm) and transcriptional terminator (Ter) as well as the RNA (arrow) produced by the system are indicated. The median recombination frequency of six independent values is given in each case. All median frequencies were calculated in duplicate with two independent transformants. Recombinants were selected in SC-Leu-Trp. Data from the L-lacZ system containing the entire lacZ gene (bottom) are taken from Chávez and Aguilera (11).
We have previously shown that in hpr1 and other mutants affected in the THO complex, the ability of a given DNA segment, like lacZ, to impair transcriptional elongation correlates with hyperrecombination of a direct-repeat system containing that segment. This hyperrecombination is transcription dependent (11, 42, 44). We tested, therefore, the recombination frequency of direct-repeat systems containing either one of the three lacZ fragments used in the previous experiments flanked by two leu2 repeats. In agreement with the absence of effect of the hpr1 mutation on transcription of such lacZ fragments, we did not detect a significant stimulation of recombination when the fragments were located between the leu2 repeats (Fig. 4B). Again, in this case there was a clear difference between the short lacZ fragments and the entire lacZ, which stimulates recombination between direct repeats up to 200-fold in hpr1 cells (11) (Fig. 4B).
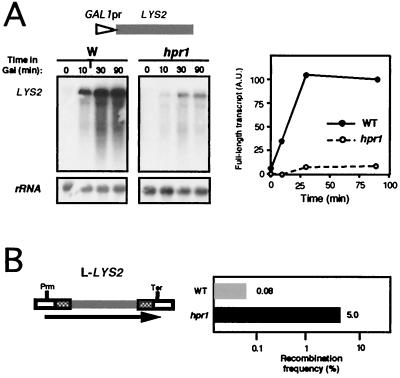
The previous results suggest that transcript length is an important feature in determining the requirement of Hpr1 in transcription. To confirm this, we constructed comparable transcription and recombination systems containing only yeast long DNA sequences. We randomly chose a fragment from a long yeast ORF, a 3.7-kb fragment of the S. cerevisiae LYS2 gene. We either fused it to the GAL1 promoter or inserted it between the leu2 repeats for transcriptional and recombinational analyses, respectively. The new constructs were introduced in wild-type and hpr1 cells (Fig. 5). Full-length mRNA from the GAL1pr::LYS2 transcriptional fusion accumulated shortly after transferring wild-type cells to galactose. However, only a smear of incomplete transcript was detected in similar Northern experiments performed with hpr1 samples (Fig. 5A), a very similar pattern to that obtained for the entire 3-kb-long lacZ in hpr1 (11) (Fig. 4A). As expected for a DNA sequence that cannot be properly transcribed in hpr1 cells, LYS2 promoted a strong hyperrecombination in hpr1 when located between leu2 repeats (L-LYS2 system). The recombination frequency reached in hpr1 (5%) is 60 times higher than the wild-type levels, but still 3- to 10-fold lower than that of analogous systems containing lacZ (11) (Fig. 4B). This result supports our hypothesis for the influence of transcript length on hpr1 sensitivity of transcription.
FIG. 5.
Transcription and recombination analyses of LYS2 sequences in wild-type and hpr1 cells. (A) Northern blot analyses of LYS2 mRNAs in strains transformed with plasmid pSCh227 containing a 3.7-kb fragment of the LYS2 coding sequence under the control of the GAL1 promoter. (B) Recombination frequencies of strains transformed with plasmid pSCh230 harboring a leu2-based direct repeat system containing as intervening sequence the same 3.7 kb fragment of LYS2 used for the transcription assays. Other details are as described for Fig. 4.
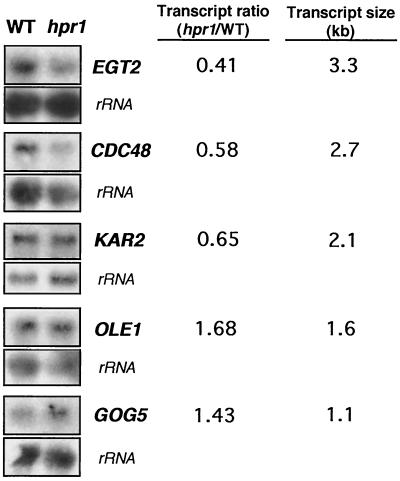
If the contribution of Hpr1 to the accumulation of long transcripts initiating at the GAL1 promoter is not restricted to the artificial construct that we have analyzed, we should expect the genome-wide effect of hpr1 to be more dramatic on long genes than on short ones in highly expressed genes. To test this idea we analyzed the effect of hpr1Δ on transcription of five endogenous chromosomal genes with sizes ranging from 0.5 to 3.1 kb. They were selected because they have high and comparable expression levels in YEPD-rich medium in wild-type cells (between 10 and 14 mRNAs per cell, according to Holstege et al. [24]). The longest genes, EGT2, CDC48, and KAR2, showed significantly lower expression levels in hpr1 than in the wild type. The shortest ones, OLE1 and GOG5, exhibited even higher expression levels in hpr1 than in the wild type (Fig. 6). As we are not controlling transcription, such as with the regulatable GAL1 promoter in these experiments, we cannot exclude the possibility that such higher expression levels are an indirect effect of hpr1. The correlation between transcript size and the hpr1:wild-type transcript ratio is not perfect. This may reflect the facts that (i) each ORF is under the control of a different promoter, (ii) the ORFs are in different chromosomal locations, and (iii) the different DNA sequence context of each gene may affect its transcription pattern. These results are consistent with transcript length being at least one feature partially responsible for impairing transcription driven from strong promoters in hpr1 cells.
FIG. 6.
Transcription analyses of five yeast endogenous genes, EGT2, CDC48, KAR2, OLE1, and GOG5, having high levels of expression and different transcript sizes, in wild-type and hpr1 cells. Total RNA was isolated from mid-log phase cells, grown in YEPD broth, and used for Northern analyses. Internal fragments of each gene and of the 23S rDNA, obtained by PCR, were used as DNA probes. The hpr1:wild-type transcript ratio was obtained from the mRNA levels that were quantified in a Fuji FLA3000 and normalized with respect to the rRNA levels.
Transcription of G+C-rich DNA sequences is severely impaired by hpr1Δ
Although the length of a gene is an important feature influencing the sensitivity of its transcription to the hpr1 mutation, there are several pieces of evidence indicating that it cannot be the only feature. First, replacement of the lacZ fragments by GAL1 in the PHO5 transcriptional fusion constructs largely suppressed the hpr1 effect (Fig. 2). In addition, the two sets of PHO5 fusions that we constructed, in which lacZ fragments were located either at the 3′ or the 5′ end, share the same length but behave differently in hpr1 cells. Finally, although both lacZ and LYS2 are hyperrecombinogenic when flanked by direct repeats, lacZ is significantly more recombinogenic than LYS2 (Fig. 3B and 4B). Therefore, we decided to explore other features of lacZ, the most hpr1-sensitive sequence detected so far, in order to identify additional elements influencing transcriptional impairment by hpr1.
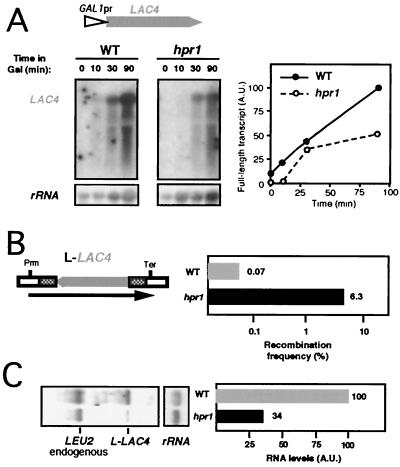
The most evident difference between lacZ and the bulk of S. cerevisiae genes is the G+C content. The majority of yeast genes show a G+C content of around 40%, whereas that of lacZ is 56.2%. In order to investigate whether the G+C content influences the transcriptional impairment of lacZ in hpr1 cells, we used the Kluyveromyces lactis LAC4 gene, a yeast homologue of lacZ with 40% G+C (1). We placed LAC4 under GAL1 control, creating a GAL1pr::LAC4 fusion similar to those previously used in this work. Transformants of wild-type and hpr1 isogenic strains were used to determine the kinetics of accumulation of mRNA. The results presented in Fig. 7A show that accumulation of LAC4 full-length mRNA was only moderately diminished in hpr1 cells (50% of the wild-type level after 90 min of induction). Nonetheless, in the same background and after an identical induction time, lacZ full-length mRNA was almost absent (Fig. 4A) (11). The overall comparison of LAC4 and lacZ transcriptional behaviors showed that transcription through LAC4 was at least fivefold more efficient than that through lacZ in hpr1 cells. Thus, two transcription units, identical in length and differing in G+C content, were differentially affected by hpr1. This indicates that, in addition to transcript length, the G+C content of a DNA sequence may be an important feature influencing transcriptional impairment by hpr1.
FIG. 7.
Transcription and recombination analyses of LAC4 in wild-type and hpr1 cells. (A) Northern analyses of LAC4 mRNAs in strains transformed with the plasmid pSCh255, which contains the entire LAC4 coding sequence under the control of the GAL1 promoter. (B) Recombination frequencies of cells transformed with plasmid pSCh254, which harbors the leu2-based direct-repeat L-LAC4 construct containing LAC4 as the intervening region. (C) Northern analyses of the L-LAC4 repeat construct in wild-type and hpr1 cells. Other details are as described for Fig. 4.
To determine the effect of LAC4 in recombination, we placed the entire LAC4 ORF between the leu2 direct repeats. The resulting L-LAC4 system exhibited a high frequency of recombination in hpr1 cells (90-fold above wild-type levels [Fig. 7B]). This frequency was eightfold lower than that shown by L-lacZ but was comparable to the frequency reached by L-LYS2 (Fig. 3B and 4B). An increase in transcription efficiency is accompanied therefore by a lower recombination frequency. It is important that the L-LAC4 system shows a hyperrecombination phenotype, because the size of the full transcript in this system is approximately 5 kb. Indeed, and consistent with our hypothesis, we have shown by Northern analysis (Fig. 7C) that transcription of the L-LAC4 system is impaired in hpr1 cells.
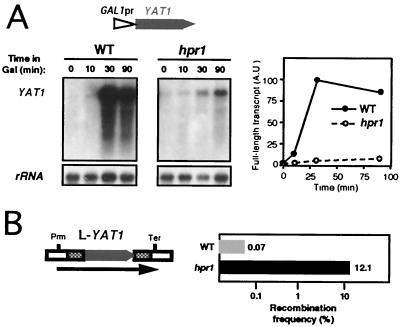
To further investigate the influence of G+C content on transcription of a DNA sequence, regardless of whether coming from bacteria or yeast, we decided to analyze a yeast gene with a G+C content comparable to that of lacZ. YAT1, a 2-kb long ORF, is the gene in the S. cerevisiae genome with the highest G+C content (58%). We placed YAT1 under GAL1 control in a transcriptional system similar to those used in the previous experiments. Northern analysis showed that high levels of YAT1 full-length mRNA were reached after galactose induction in the wild type, but only a minimal accumulation was detected in hpr1 cells (Fig. 8A). In agreement with this transcriptional impairment, a recombination system bearing YAT1 as intervening sequence (L-YAT1) displayed an extremely high frequency of recombination in hpr1 (12%) that was 173 times the frequency reached in the wild type (Fig. 8B). This level of hyperrecombination is comparable to that of L-lacZ (Fig. 4B) and higher than the levels of L-LYS2 (Fig. 5B) and L-LAC4 (Fig. 7B). Thus, transcription through a medium-size G+C-rich gene is clearly hpr1 sensitive, indicating that G+C content can modulate the Hpr1 dependency of gene transcription.
FIG. 8.
Transcription and recombination analyses of YAT1 in wild-type and hpr1 cells. (A) Northern blot analyses of YAT1 mRNAs in cells transformed with plasmid pSCh247 containing the entire YAT1 coding sequence under the control of the GAL1 promoter. (B) Recombination analyses of cells transformed with plasmid pSCh248, which harbors the leu2-based direct-repeat system containing the entire YAT1 gene as intervening sequence. Other details are as described for Fig. 4.
Nucleosome positioning is lacking in lacZ sequences.
The organization of DNA in a proper nucleosome-positioned chromatin structure has been shown to be favored by A+T-rich motifs (27) and prevented by G+C-rich sequences (62). In order to test whether there is a relationship between chromatin structure and transcriptional efficiency in hpr1 cells, we determined whether the chromatin structure of G+C-rich sequences such as lacZ was different from that of low-G+C-content sequences. We performed MNase sensitivity assays of the GAL1pr::lacZ fusion construct and the GAL1 endogenous genes, in which transcription was strongly and poorly impaired in hpr1 cells, respectively (11, 17, 67) (Fig. 9A). As previously shown (19), clear and specific nucleosome positioning along the endogenous GAL1 gene was observed (Fig. 9B). Such a pattern of MNase sensitivity was identical for both wild-type and hpr1Δ cells. The more diffuse pattern of MNase digestion under induced conditions in both wild-type and hpr1 cells reflects the destabilization of chromatin structure caused by transcription (9). Interestingly, the pattern of MNase sensitivity of lacZ shows no nucleosome organization in either wild-type or hpr1Δ cells under both induced and repressed conditions of transcription. Nucleosome positioning is only limited to the GAL1 promoter (Fig. 9C). Indeed, a lack of nucleosome positioning is also observed over the bacterial sequences upstream of the GAL1 promoter, through which transcription has also been shown to be impaired in hpr1 mutants (44). Our results, therefore, show that lacZ adopts a random nucleosomal organization in yeast and that hpr1 has no effect on nucleosome positioning.
DISCUSSION
In this work we have investigated why transcription of DNA sequences like E. coli lacZ is especially sensitive to hpr1. We have shown that 0.4-kb lacZ fragments fused to PHO5 under the GAL1 promoter are sufficient to increase the Hpr1 dependency of transcription, but not when they are transcribed alone. Such an effect is position dependent: the longer the distance between the lacZ sequence and the GAL1 promoter, the stronger the impairment of transcription caused by hpr1. In addition, we see transcription of long yeast DNA sequences like LYS2 fused to the GAL1 promoter is negatively affected by hpr1. We have also shown that transcription of K. lactis LAC4, a eukaryotic homologue of lacZ that is equal in length but with a much lower G+C content, exhibits a milder Hpr1 dependency in S. cerevisiae, whereas YAT1, an S. cerevisiae G+C-rich gene shorter than lacZ, is dramatically affected by hpr1. Taken together, these results indicate that both length and G+C content are important elements influencing gene transcription in vivo and that Hpr1 is an important factor controlling transcription of either long or G+C-rich DNA sequences fused to a strong promoter such as GAL1pr.
Hpr1 is required for proper transcription of long DNA sequences.
We have shown that transcription of long DNA sequences is compromised in hpr1 cells, whereas shorter sequences are either unaffected or mildly influenced. This conclusion is supported by the inappreciable effect of hpr1 on transcription of short lacZ fragments directly fused to the GAL1 promoter, whether or not upstream of PHO5, whereas the same lacZ fragments do confer hpr1 dependency when fused downstream of PHO5 (Fig. 2 and 3). This conclusion is also supported by the marked negative effect of hpr1 on transcription of the 3.7-kb-long LYS2 fragment under control of the GAL1 promoter (Fig. 5). The transcriptional analysis of five highly transcribed chromosomal genes showed a negative effect of hpr1 on transcription of the three longest ones, EGT2 (3.3 kb), CDC48 (2.7 kb), and KAR2 (2.1 kb), but no negative effect on the other two, OLE1 (1.6 kb) and GOG5 (1.1 kb) (Fig. 6). Further experiments would be required to know whether these results are also valid for poorly expressed genes.
Processivity defects of an RNA polymerase can be more easily detected with transcription of long rather than short DNA templates. Comparison between long and short transcripts is in fact a common method to quantify the effect of transcription factors on RNAPII-mediated elongation (66). Consequently, a length-dependency effect of hpr1 on transcription is expected if Hpr1 controls elongation. The longer the transcription unit, the higher the probability of RNAPII reaching a DNA region requiring the function of Hpr1. Even in the wild-type strain, long transcription units are less efficiently expressed than short ones, as we have observed by comparing the expression levels of several PHO5::lacZ fusion constructs that differ in the size of the lacZ fragment (Fig. 1). Thus, it is possible that the absence of Hpr1 enhances elongation defects already present in the wild type, the occurrence of such defects being more likely as the DNA sequence to be transcribed becomes longer.
Transcription of G+C-rich DNA sequences is Hpr1 dependent.
Our results support a correlation between G+C content and HPR1 function. This conclusion is based on three sets of data. First, although most tested genes are either slightly or not affected by hpr1 (Fig. 7) (11, 67), transcription of lacZ (56% G+C) in Saccharomyces is strongly affected and direct-repeat systems containing lacZ sequences that are transcribed exhibit hyperrecombination in hpr1 cells (11). Second, transcription of LAC4, a 40% G+C-rich lacZ orthologue of K. lactis, is at least five times more efficient than lacZ in hpr1 cells (Fig. 7A), and direct repeats flanking LAC4 recombine at frequencies eightfold lower than those containing lacZ (Fig. 7B). Finally, YAT1, a 2-kb-long gene from Saccharomyces with a 58% G+C content, is strongly affected by hpr1 both in transcription and in recombination (Fig. 8).
As far as we know, neither a direct influence of the G+C content of the template on transcription elongation efficiency nor the requirement of auxiliary factors for transcription of G+C-rich genes has been described. The only sequence features that have been shown to affect elongation are those of specific transcriptional pausing sites (59). Some pause sites identified in bacteria are G+C rich, like the ops signals in E. coli (4), but others are not. It is, therefore, unlikely that G+C-rich genes exhibit, in general, a higher probability of containing a pause signal. Nonetheless, a high G+C content might affect transcriptional elongation by stabilizing secondary structures in the nascent RNA that can function as pausing signals. At least in the case of T7 phage, a lower number of hydrogen bonds in the RNA hairpins eliminates some pauses (32), suggesting that a high G+C content might contribute to stronger RNA-mediated elongation impairments.
A G+C-rich nascent RNA might also form more stable RNA-DNA hybrids within the template. Twelve-nucleotide-long RNA-DNA hybrids negatively affect RNAPII processivity in vitro (29). Another class of RNA-DNA hybrids are R-loops, produced by the association of nascent mRNA with upstream template DNA. It has been proposed that these R-loops are formed during transcription of the G+C-rich human immunoglobulin switch region in vivo (15), and they have been detected after in vitro transcription (58). The cleavage of R-loops by specific nucleases might initiate class-switch recombination (58), providing a mechanism to explain transcription-associated recombination. Nothing is known about the influence of RNA-DNA structures on RNAPII-dependent transcription (20). However, it has been proposed that R-loops formed during rRNA transcription elongation in E. coli constitute roadblocks for the next transcribing RNA polymerase (25).
Molecular features making transcription elongation Hpr1 dependent.
Unless Hpr1 plays more than one function, we should expect a common mechanistic requirement during transcription of long versus G+C-rich genes requiring its action. It is not evident which kind of transcription-impairing signal might link long and G+C-rich DNA sequences. One possibility would be the existence of shorter G+C-rich regions hindering transcription within long genes. However, we can exclude this possibility with the results of this work. Considering a 300-bp-long window (the minimal lacZ fragment conferring an effect of hpr1 in transcription), the maximum G+C content is 46% for LYS2, 45.5% for LAC4, and 59% for lacZ. Shorter or longer windows show similar results. In addition, no difference in the G or C content is found on the cDNA strands. As LYS2 is no more G+C-rich than LAC4, length is the most likely reason the two genes behave differently in hpr1 mutants. Consistently, when LAC4 is located between the leu2 repeats in the L-LAC4 construct, the transcription unit containing LAC4 becomes longer and transcription becomes significantly affected in hpr1 cells (Fig. 7).
Alternatively, the link between long and G+C-rich sequences might be unrelated to the DNA sequence itself. Transcription of long and G+C-rich sequences may produce some kind of transcriptional event that would be overcome by the action of Hpr1. Genetic analyses have provided some hints as to the nature of this kind of event. Several mutants have been described that display synthetic phenotypes with hpr1 (2, 67). The fact that topoisomerase mutants (top1, top2, and top3) become sick in an hpr1 background may establish a link with DNA topology (2, 48). For example, the accumulation of negative supercoiling impairs transcriptional elongation of bacterial genes in vitro (30), and positive supercoiling diminishes RNAPII-dependent transcription in yeast cells. R-loops are formed in the absence of DNA topoisomerase I in E. coli (16), and they have been proposed to impair transcription elongation (25). Elongation by RNAPII alters template topology (8), producing an accumulation of positive and negative supercoiling ahead of and behind the RNAPII, respectively (33). It is, therefore, expected that positive supercoiling will be stronger at the 3′ region of long transcription units. Examples of DNA sequences that impair transcription elongation more efficiently in distal locations have been reported (63). The strong effect of hpr1 on transcription of long DNA sequences agrees with this view.
Chromatin structure is another source of stress affecting RNAPII-mediated transcription elongation that requires the action of specific auxiliary factors (reviewed in reference 39). Some mutations resulting in a poor growth phenotype with hpr1 are in fact related to chromatin structure, like SIN1-2 or those causing histone imbalance (67). The organization of DNA in a proper, nucleosome-positioned chromatin structure is favored by some A+T-rich motifs (27) and is prevented by some G+C-rich sequences (62). A G+C-rich sequence might, therefore, be biased against a proper chromatin structure. The lacZ gene is in fact unable to support stably organized chromatin in S. cerevisiae (Fig. 9). Transcription of G+C-rich genes might then be impaired in hpr1 due to an aberrant chromatin structure.
If these hypotheses are true, the location of a G+C-rich region in the 3′ region of a gene should produce a stronger effect, since it would combine two sources of transcriptional stress in the same place: superhelicity and aberrant chromatin structure. Indeed, negative supercoiling can induce changes of DNA structure in CG sequences, with this altered structure being able to produce transcriptional elongation blocks (41). Our results with the PHO5::lacZ fusion constructs support this view, since the location of the G+C-rich lacZ fragments at the 3′ region produced a clear transcriptional effect in hpr1 mutants (Fig. 2), whereas their location at the 5′ region had no effect (Fig. 2 and 3).
Finally, we cannot exclude the possibility that the size and G+C content of the nascent RNA, and not of its DNA template, are what determines the requirement of Hpr1 in transcription. This would imply that Hpr1, and by extension the THO complex, might also control RNA metabolism beyond transcription elongation. Such a possibility is consistent with the observation that transcription and RNA processing are coupled (reviewed in references 6, 23, 47) and could explain the observed RNA export defects of hpr1 mutants (51). In this respect, the recent observations that hpr1 is suppressed by overexpression of the putative RNA helicase SUB2 and that sub2 mutants are also hyperrecombinant (18) are noteworthy.
Hpr1 is stably associated in the cell nucleus with Tho2, Mft1, and Thp2, forming the THO complex (12). Mutations affecting any of the four proteins cause the same phenotypes in transcription and transcription-associated recombination, although the quantitative effect of each mutation is different (12). Since the THO complex is a functional unit, the information obtained studying the gene spectrum affected by hpr1 sheds light on the in vivo functional role of the complex. Characterization of the biochemical and functional properties of the THO complex in vitro will help in understanding why transcription of long as well as G+C-rich genes preferentially requires a specific cellular function. It remains to be seen whether these conclusions can be extended to endogenous yeast genes or to DNA sequences transcribed from poorly expressed promoters. The existence of Tho2 homologues as well as of long genes and G+C-rich DNA sequences in Drosophila, mice, and humans opens the possibility that this might be a general phenomenon in all eukaryotes.
ACKNOWLEDGMENTS
We thank J. Polaina for providing the LAC4 gene, M. Beato for providing his facilities (IMT, Philipps Universitat, Marburg) for part of the chromatin analyses presented in this study, E. Santero for critical reading of the manuscript, and D. Haun for style supervision.
This project was supported by grants from the Spanish Ministry of Science and Culture (PB96-1350) and the Human Frontier Science Program (RG0075/1999-M).
REFERENCES
- 1.Adam A C, Prieto J A, Rubio-Texeira M, Polaina J. Construction of a lactose-assimilating strain of baker's yeast. Yeast. 1999;15:1299–1305. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1299::AID-YEA454>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera A, Klein H L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera A, Chávez S S, Malagón F. Mitotic recombination in yeast: elements controlling its incidence. Yeast. 2000;16:731–754. doi: 10.1002/1097-0061(20000615)16:8<731::AID-YEA586>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aso T, Lane W S, Conaway J W, Conaway R C. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 6.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell T K, Moore M W, Yancopoulos G D, Suh H, Lutzker S, Selsing E E, Alt F W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986;324:585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- 8.Brill S J, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 9.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang M, French-Cornay D, Fan H Y, Klein H L, Denis C L, Jaehning J A. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chávez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chávez S, Beilharz T, Rondón A G, Erdjument-Bromage H, Tempst P, Svejstrup J Q, Lithgow T, Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez S, Candau R, Truss M, Beato M. Constitutive repression and nuclear factor I-dependent hormone activation of the mouse mammary tumor virus promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6987–6998. doi: 10.1128/mcb.15.12.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaway J W, Shilatifard A, Dvir A, Conaway R C. Control of elongation by RNA polymerase II. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- 15.Daniels G A, Lieber M R. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drolet M, Phoenix P, Menzel R, Massé E, Liu L F, Crouch R. Overexpression of RNase H partially complements the growth defect of an Escherichia coli ΔtopA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan H Y, Klein H L. Characterization of mutations that suppress the temperature-sensitive growth of the hpr1Δ mutant of Saccharomyces cerevisiae. Genetics. 1994;137:945–956. doi: 10.1093/genetics/137.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan H-Y, Merker R J, Klein H L. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol Cell Biol. 2001;21:5459–5470. doi: 10.1128/MCB.21.16.5459-5470.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedor M J, Kornberg R D. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol Cell Biol. 1989;9:1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gartenberg M R, Wang J C. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci USA. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 24.Holstege F C, Jennings E G, Wyrick J J, Lee T L, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 25.Hraiky C, Raymond M-A, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem. 1995;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda H, Matsumoto T. Transcription promotes recA-independent recombination mediated by DNA-dependent RNA polymerase in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:4571–4575. doi: 10.1073/pnas.76.9.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ioshikhes I, Bolshoy A, Derenshteyn K, Borodovsky M, Trifonov E N. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J Mol Biol. 1996;262:129–139. doi: 10.1006/jmbi.1996.0503. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser C, Michaelis M, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 29.Kireeva M L, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J Mol Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 30.Krohn M, Pardon B, Wagner R. Effects of template topology on RNA polymerase pausing during in vitro transcription of the Escherichia coli rrnB leader region. Mol Microbiol. 1992;6:581–589. doi: 10.1111/j.1365-2958.1992.tb01504.x. [DOI] [PubMed] [Google Scholar]
- 31.Lauster R, Reynaud C A, Martensson I L, Peter A, Bucchini D, Jami J, Weill J C. Promoter, enhancer and silencer elements regulate rearrangement of an immunoglobulin transgene. EMBO J. 1993;12:4615–4623. doi: 10.1002/j.1460-2075.1993.tb06150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin J R, Chamberlin J M. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- 33.Liu L F, Wang J C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupski J R. Charcot-Marie-Tooth disease: lessons in genetic mechanisms. Mol Med. 1998;4:3–11. [PMC free article] [PubMed] [Google Scholar]
- 35.Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevo-Caspi Y, Kupiec M. Transcriptional induction of Ty recombination in yeast. Proc Natl Acad Sci USA. 1994;91:12711–12715. doi: 10.1073/pnas.91.26.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickoloff J A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oltz E M, Alt F W, Lin W C, Chen J, Taccioli G, Desiderio S, Rathbun G. A V(D)J recombinase-inducible B-cell line: role of transcriptional enhancer elements in directing V(D)J recombination. Mol Cell Biol. 1993;13:6223–6230. doi: 10.1128/mcb.13.10.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 40.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M, Gustafsson C M, Erdjument-Bromage H, Tempst P P, Svejstrup J Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 41.Peck L J, Wang J C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985;40:129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- 42.Piruat J I, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prado F, Piruat J I, Aguilera A. Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price D H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proudfoot N. Connecting transcription to messenger RNA processing. Trends Biochem Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 48.Sadoff B U, Heath-Pagliuso S, Castano I B, Zhu Y, Kieff F S, Christman M F. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics. 1995;141:465–479. doi: 10.1093/genetics/141.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos-Rosa H, Aguilera A. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1Δ strains. Mol Gen Genet. 1994;245:224–236. doi: 10.1007/BF00283271. [DOI] [PubMed] [Google Scholar]
- 50.Saxe D, Datta A, Jinks-Robertson S. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol Cell Biol. 2000;20:5404–5414. doi: 10.1128/mcb.20.15.5404-5414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneiter R, Guerra C E, Lampl M, Gogg G, Kohlwein S D, Klein H L. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol Cell Biol. 1999;19:3415–3422. doi: 10.1128/mcb.19.5.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selby C P, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart S E, Roeder G S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 57.Thyagarajan B, Johnson B L, Campbell C. The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res. 1995;23:2784–2790. doi: 10.1093/nar/23.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian M, Alt F W. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 59.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 60.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 61.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y H, Griffith J D. The [(G/C)3NN]n motif: a common DNA repeat that excludes nucleosomes. Proc Natl Acad Sci USA. 1996;93:8863–8867. doi: 10.1073/pnas.93.17.8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiest D K, Hawley D K. In vitro analysis of a transcription termination site for RNA polymerase II. Mol Cell Biol. 1990;10:5782–5795. doi: 10.1128/mcb.10.11.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 66.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y, Peterson C L, Christman M F. HPR1 encodes a global positive regulator of transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1698–1708. doi: 10.1128/mcb.15.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]