Abstract
Regulation of NF-κB transactivation function is controlled at several levels, including interactions with coactivator proteins. Here we show that the transactivation function of NF-κB is also regulated through interaction of the p65 (RelA) subunit with histone deacetylase (HDAC) corepressor proteins. Our results show that inhibition of HDAC activity with trichostatin A (TSA) results in an increase in both basal and induced expression of an integrated NF-κB-dependent reporter gene. Chromatin immunoprecipitation (ChIP) assays show that TSA treatment causes hyperacetylation of the wild-type integrated NF-κB-dependent reporter but not of a mutant version in which the NF-κB binding sites were mutated. Expression of HDAC1 and HDAC2 repressed tumor necrosis factor (TNF)-induced NF-κB-dependent gene expression. Consistent with this, we show that HDAC1 and HDAC2 target NF-κB through a direct association of HDAC1 with the Rel homology domain of p65. HDAC2 does not interact with NF-κB directly but can regulate NF-κB activity through its association with HDAC1. Finally, we show that inhibition of HDAC activity with TSA causes an increase in both basal and TNF-induced expression of the NF-κB-regulated interleukin-8 (IL-8) gene. Similar to the wild-type integrated NF-κB-dependent reporter, ChIP assays showed that TSA treatment resulted in hyperacetylation of the IL-8 promoter. These data indicate that the transactivation function of NF-κB is regulated in part through its association with HDAC corepressor proteins. Moreover, it suggests that the association of NF-κB with the HDAC1 and HDAC2 corepressor proteins functions to repress expression of NF-κB-regulated genes as well as to control the induced level of expression of these genes.
NF-κB is an inducible transcription factor that plays a role in the expression of a variety of genes involved in immune and inflammatory responses and cell survival (3, 13). There are five known members of the mammalian NF-κB/Rel family: p65 (RelA), c-Rel, RelB, p50 (NF-κB1), and p52 (NF-κB2). These proteins share sequence similarity over an approximately 300-amino-acid Rel homology domain. NF-κB subunits are able to homo- or heterodimerize through the Rel homology domain, forming transcription factor complexes with a wide range of DNA-binding and activation potentials. Although all Rel members bind DNA, only p65, c-Rel, and RelB have extended carboxy termini harboring transactivation function (37). The most widely studied and most abundant form of NF-κB is a heterodimer of p50 and p65.
NF-κB has typically been thought of as residing in the cytoplasm in an inactive form bound by its inhibitory proteins, members of the IκB family (10, 37), although evidence indicates that NF-κB may shuttle between the nucleus and cytoplasm in unstimulated cells (5, 16). The IκB family members include IκBα, IκBβ, p105 (IκBγ) (precursor of p50), p100 (precursor of p52), and IκBɛ. Activation of NF-κB is induced by a number of stimuli, including inflammatory cytokines, phorbol esters, bacterial toxins (such as lipopolysaccharides), viruses, UV light, and a variety of mitogens (13). Treatment of cells with these stimuli activates the IκB kinase complex, leading to phosphorylation of serines 32 and 36 of IκBα or serines 19 and 23 of IκBβ (7, 23, 29, 43). This phosphorylation event targets the IκB proteins for ubiquitination and degradation through the 26S proteasome complex, resulting in the release and translocation of NF-κB into the nucleus (9, 13).
Coregulatory (coactivator and corepressor) proteins have been shown to be required for the regulation of gene expression by many transcription factors. These proteins likely function by facilitating or bridging the sequence-specific activators to the basal transcriptional machinery as well as by altering chromatin structure. Coactivator proteins include CREB-binding protein (CBP) and its structural homologue p300 (4, 27), steroid receptor-coactivator-1 (SRC-1) (33), and p300/CBP-associated factor (p/CAF) (42). Consistent with their role in altering chromatin structure, many coactivator proteins contain histone acetyltransferase (HAT) domains which are capable of acetylating lysine residues in the amino-terminal tails of the core histones. In contrast to coactivators, corepressor complexes include proteins that have histone deacetylase (HDAC) activity (reviewed in reference 19). In mammalian cells, HDAC1 and HDAC2 are typically found in a large complex with either the mSin3 protein (the Sin3 complex) or as part of the Mi-2–NuRD complex (19). The Sin3 complex is brought to promoters through its interaction, either directly or indirectly, with sequence-specific transcription factors. For example, the mSin3 protein contributes to Mad-Max repression by interacting directly with the Mad family of proteins (2). Alternatively, the Sin3 complex represses nuclear receptor-mediated activation by interacting indirectly with unliganded nuclear receptors through either the N-CoR or SMRT corepressor proteins (14). In this context, HDAC1 and HDAC2 are recruited to promoters through their interaction with Sin3. However, HDACs can also repress transcription through direct interaction with sequence-specific transcription factors. For example, HDAC1 has recently been shown to directly interact with the MyoD basic helix-loop-helix transcription factor to silence MyoD-dependent transcription of p21 as well as muscle-specific genes (22). HDAC2 has been shown to interact directly with the YY1 transcription factor, converting it from an activator to a repressor (41). The homeodomain protein TGIF, which associates with the Smad proteins to mediate transforming growth factor beta signaling, has been found to interact directly with HDAC proteins (40). HDAC1 has also been shown to interact with hypophosphorylated forms of the Rb family of proteins and, through the association of Rb with E2F, to mediate repression of E2F-dependent gene expression (15).
Numerous reports indicate that a basal level of NF-κB can be found in the nuclei of unstimulated cells. The function of this low level of nuclear NF-κB is not known, but one possibility is that NF-κB functions to repress and/or activate basal gene expression in these cells. This effect may be regulated by the differential association of NF-κB with coactivator and corepressor proteins. Recent work has shown that NF-κB-dependent gene expression requires the function of transcriptional coactivator proteins. The CBP and p300 coactivators interact with the p65 subunit of NF-κB to enhance its ability to activate transcription (12, 28, 45). Inducible phosphorylation of p65 by the protein kinase A (PKA) catalytic subunit stimulates NF-κB-dependent gene expression by enhancing the interaction of p65 with CBP (44, 45). Two other coactivators are also known to be involved in regulating the transactivation function of NF-κB. The HAT function of the p/CAF coactivator was shown to be required for activation of NF-κB-dependent gene expression (31) and the SRC-1 coactivator protein was shown to interact with the p50 subunit of NF-κB to potentiate NF-κB-mediated transactivation (25). Thus, interaction with transcriptional coactivators is important in mediating the transactivation potential of NF-κB. Moreover, the differential association of NF-κB with coactivator proteins and possibly with corepressor proteins in unstimulated cells as well as in cells in which NF-κB has been activated is likely to determine the level of activation or repression of NF-κB-regulated genes.
Although the requirement of coactivators for transcriptional activation by NF-κB is well established, very little work has been done to determine the importance of corepressors in regulating NF-κB transactivation. There is some previous evidence to suggest that HDAC corepressor proteins may function to negatively regulate NF-κB transcriptional activity (8, 17, 36). In this report, we investigated whether corepressor proteins play a role in regulating NF-κB transactivation function. Here we find that chemical inhibition of HDAC activity using trichostatin A (TSA) results in increased expression of an NF-κB-dependent reporter gene. We show that both HDAC1 and HDAC2 regulate NF-κB-dependent transcription and we demonstrate that HDAC1 can interact directly with the p65 subunit of NF-κB to exert its corepressor function. Finally, we show that treatment of cells with TSA results in an increased basal level of expression as well as increased induced levels of expression of the NF-κB-dependent interleukin-8 (IL-8) gene. We further show that this increase in expression is likely caused by hyperacetylation of the IL-8 promoter due to the inhibition of HDAC activity by TSA. These results suggest that regulation of NF-κB-dependent gene expression requires the function of corepressor proteins. Moreover, we show that corepressor proteins may function to repress the transactivation function of the low level of uninduced NF-κB found in the nucleus of some cell types. Therefore, in addition to inhibition of NF-κB activity by the cytoplasmic IκB family of proteins, we propose that NF-κB transactivation function is also regulated in the nucleus through interaction with HDAC corepressor proteins and that a function of this association may be to actively repress expression of NF-κB-regulated genes.
MATERIALS AND METHODS
Plasmid constructs and antibodies.
The pBJ-HDAC1 expression vector was provided by S. Schrieber (Harvard Medical School, Boston, Mass.). pcDNA-HDAC1 was constructed by ligating a BamHI/EcoRI fragment from pBJ-HDAC1 into the BamHI/EcoRI sites of pcDNA3.1. The pcDNA-HDAC2 expression vector was provided by E. Seto (Moffitt Cancer Center, University of South Florida, Tampa, Fla.). The mSin3a expression plasmid was provided by D. Ayer (University of Utah, Salt Lake City, Utah). pCEP4-N-CoR was provided by C. Glass (University of California at San Diego, San Diego, Calif.). pCRX-SMRT was provided by R. Evans (The Salk Institute, San Diego, Calif.). The CMV-p65 and CMV-p50 expression plasmids have been described previously (34). The GAL4-p50 fusion was made by ligating full-length p50 in frame with the GAL4 DNA-binding domain (GAL4 1-147). GAL4-VP16 contains a fusion of the VP16 activation domain (amino acids 413 to 490) to the GAL4 DNA-binding domain and GAL4-p65 contains a fusion of amino acids 280 to 551 of p65 to the GAL4 DNA-binding domain. The plasmids used for in vitro transcription and translation were pGEM-flag-p65, pcDNA-p65(1-521), pcDNA-p65(1-313), pcDNA-p65(1-276), and pcDNA-p65(270-551). GST-HDAC1 was made by fusing the full-length HDAC1 coding sequence in frame into pGEX-4T-1 (Pharmacia). Reporter plasmids used in this study included 3XκB-Luc, which contains three copies of the NF-κB binding site from the major histocompatibility complex class I gene upstream of the luciferase reporter gene, and 5XGAL4-Luc, which contains five copies of the GAL4 DNA-binding site upstream of the luciferase reporter gene. Antibodies used in these studies were specific for p65 (Rockland), p50 (Upstate Biotechnology; 06-886), HDAC1 (Santa Cruz Biotechnology; sc-7872), and HDAC2 (Santa Cruz Biotechnology; sc-7899).
Cell culture, transfections, and luciferase assays.
HeLa and 293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. Cos-7 cells were grown in Iscove's minimal essential medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. NIH 3T3 cells with stably integrated wild-type or mutant 3XκB-Luc (provided by D. Guttridge, University of North Carolina, Chapel Hill, N.C.) and p65−/− embryonic fibroblasts expressing either Flag-tagged empty vector or Flag-p65 (38) were grown in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum, penicillin, streptomycin, and 500 μg of G418 (NIH 3T3)/ml or 450 μg of hygromycin (p65−/−)/ml. Transfections were performed using the Superfect transfection reagent (Qiagen) according to the manufacturer's recommendations. For tumor necrosis factor alpha (TNF-α) inductions, cells were treated with 10 ng of TNF-α/ml 24 h after transfection. For TSA treatments, cells were treated with 100 nM TSA (BioMol) for 16 h prior to harvesting cells for luciferase assays. Cells were harvested 16 h after induction and lysed using the mammalian protein extraction reagent (M-PER; Pierce). Luciferase assays were performed as described elsewhere (6).
Immunoprecipitations and Western blot analysis.
The indicated cell lines were harvested and lysed in IP buffer (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 0.5% NP-40, 10% glycerol, 0.1 mM EDTA, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 5 μg each of aprotinin, leupeptin, and pepstatin/ml). Whole-cell extracts were incubated with the indicated primary antibody in a total volume of 1 ml of IP buffer for 6 h at 4°C. For the final 2 h, 25 μl of protein A- and G-agarose beads (Santa Cruz Biotechnology) was added. The complexes were then centrifuged at 4°C for 30 s and the beads were washed four times with 1 ml of IP buffer. The beads were then resuspended in sodium dodecyl sulfate (SDS) sample buffer, boiled for 3 min, and analyzed on SDS–10% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell), blocked with 5% blotting grade milk (Bio-Rad) in TBST (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.5% Tween 20), and incubated with the indicated antibodies for 30 to 60 min. Membranes were then washed with TBST and incubated for 30 min with secondary antibody conjugated to horseradish peroxidase (Promega) for 30 min, washed with TBST, and incubated with ECL chemiluminescence reagent (Amersham). Membranes were exposed to X-ray film to visualize the bands.
In vitro interactions.
For in vitro interactions, the p65 full-length and deletion mutants were synthesized in vitro in the presence of [35S]methionine (NEN) using the TNT Quick Coupled transcription and translation system (Promega) according to the manufacturer's recommendations. Reactions were carried out at 30°C for 1 h. Ten microliters (of the 50-μl in vitro transcription and translation reaction) was then mixed with either glutathione S-transferase (GST) protein or GST-HDAC1 in 500 μl (total volume) of binding buffer (phosphate-buffered saline with 1% Triton X-100) in the presence of glutathione-agarose beads (Sigma) and incubated at 4°C with gentle rocking for 1 h. Beads were then washed three times with 1× phosphate-buffered saline containing 1% Trition X-100, resuspended in SDS sample buffer, boiled for 3 min, and analyzed on SDS–10% polyacrylamide gels. The gels were dried and bands were visualized by autoradiography.
Ribonuclease protection assays, Northern blot analysis, cell treatments, and RNA isolation.
A custom Riboquant Multi-probe RNase Protection Assay System (Pharmingen) with six genes known to be regulated by NF-κB was used. The genes included in the kit were those for TRAF1, TRAF2, Bfl1/A1, c-IAP-2, IL-8, and IL-2Rα. Reactions were carried out according to the manufacturer's recommendations and bands were visualized by autoradiography. Northern blot analysis was performed as previously described (1). Probes specific for human IL-8 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were labeled by nick translation and added directly to the hybridization solution. HeLa cells were treated with a final concentration of 10 ng of human recombinant TNF-α (Promega)/ml or 10 ng of mouse IL-1β (Life Technologies)/ml for the indicated times. Where indicated, TSA (Biomol) treatments were done for 18 h at a final concentration of 100 nM. Total RNA was made using Trizol reagent (Life Technologies) according to the manufacturer's recommendations.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed using the Acetyl-Histone H3 Immunoprecipitation Assay kit (Upstate Biotechnology) according to the manufacturer's recommendations. The integrated wild-type and mutant 3XκB-Luc promoter was amplified using the primer pair 5′-AGCTTGGGCTGCAGGTCG-3′ and 5′-GCGGAACCGCCGGCTCTATCC-3′. The human IL-8 promoter region from −121 to +61 was amplified with the primer pair 5′-GGGCCATCAGTTGCAAATC-3′ and 5′-TTCCTTCCGGTGGTTTCTTC-3′. The human IL-8 upstream region from −1042 to −826 was amplified with the PCR primer pair 5′-AACAGTGGCTGAACCAGAG-3′ and 5′-AGGAGGGCTTCAATAGAGG-3′. Primers were end labeled with [γ-32P]dATP. PCR was performed for 30 cycles and the products were analyzed on a 6% acrylamide gel. The gels were dried and the bands were visualized by autoradiography.
RESULTS
TSA treatment causes increased expression of an integrated NF-κB-dependent reporter gene.
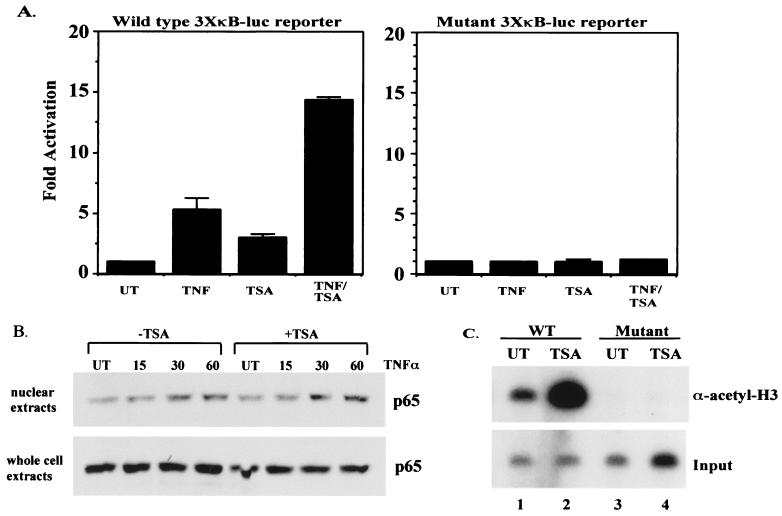
In order to begin to determine a possible role for HDAC corepressor proteins in regulating NF-κB-mediated transactivation, we utilized NIH 3T3 cell lines harboring a stably integrated NF-κB-dependent reporter gene (wild type or mutant 3XκB-Luc). This is important since HDACs function by modifying chromatin structure through deacetylation of histones and transiently transfected plasmids are not efficiently packaged into chromatin (18, 32). Treatment of the NIH 3T3 3XκB-Luc wild-type cells with TNF resulted in a five- to sixfold stimulation of the reporter gene relative to uninduced basal expression (Fig. 1A, left panel). Treatment of these cells with the HDAC inhibitor TSA caused a three- to fourfold increase over basal expression (Fig. 1A, left panel), implying that NF-κB, possibly in association with HDAC proteins, plays a role in repressing basal expression of the reporter gene. When cells were treated simultaneously with TNF and TSA, a 14-fold increase was seen (Fig. 1A, left panel), a level more than 2-fold higher than with TNF alone. The data shown are from pools of G418-resistant colonies; however, similar results were seen when four independent wild-type clones were tested (data not shown). The observed increases are dependent on an intact NF-κB binding site and not on global derepression of the repressive chromatin structure since there was no increase in expression after TNF or TSA treatment alone or simultaneous treatment with TNF and TSA in the cells with the stably integrated mutant 3XκB-Luc reporter (Fig. 1A, right panel) or with a stably integrated pGL-2 basic (promoterless vector) reporter (data not shown). Moreover, the increased expression after TSA treatment, in either untreated or TNF-induced cells, is not due to increased levels of nuclear NF-κB, since the amount of p65 in the nucleus of both untreated and TNF-stimulated cells was similar in the presence or absence of TSA treatment (Fig. 1B, upper panel). Although the amount of p65 in the nuclei of untreated cells is similar in the presence or absence of TSA, TSA treatment results in a two- to threefold increase in basal reporter gene expression, implying a role for NF-κB in regulating basal gene expression. Finally, the overall levels of p65 were not affected by treatment of cells with TSA, further indicating that the TSA effect on NF-κB transactivation is mediated through inhibition of HDAC activity and not by altering the amount of NF-κB (Fig. 1B, lower panel). These results are in agreement with a previously published report that showed that treatment of HeLa cells harboring a stably integrated human immunodeficiency virus long terminal repeat (LTR)-chloramphenicol acetyltransferase reporter gene with TSA resulted in an increase in both basal and induced expression of the reporter gene (8). In order to determine if the increased expression of the wild-type integrated reporter in response to TSA was due to hyperacetylation of the promoter region, ChIP assays were performed. Figure 1C shows that treatment of the wild-type NIH 3T3 3XκB-Luc cells with TSA results in a dramatic increase in acetylation of this promoter (Fig. 1C, upper panel, compare lanes 1 and 2). In contrast, no acetylation was seen in NIH 3T3 cells containing the mutant 3XκB-Luc reporter (Fig. 1C, upper panel, lanes 3 and 4). Taken together, these results imply that transactivation by NF-κB is regulated, at least in part, by association with HDAC corepressor proteins and that inhibition of HDAC activity results in hyperacetylation of the wild-type NIH 3T3 3XκB-Luc promoter and subsequently higher levels of reporter expression.
FIG. 1.
Inhibition of HDAC activity causes an increase in basal and induced expression of an integrated NF-κB-dependent reporter gene (3XκB-Luc). (A) NIH 3T3 cells harboring either integrated wild-type 3XκB-Luc (left) or mutant 3XκB-Luc (right) were treated in triplicate with TNF (10 ng/ml) for 8 h, TSA (100 nM) for 18 h, or with TSA for 18 h and TNF for the final 8 h. Extracts were prepared and relative luciferase activities were determined by normalizing to total protein. Activity is expressed as fold activation relative to the untreated control and is the average ± standard deviation (SD) from a representative experiment. Experiments were performed a minimum of three times, with similar results. UT, untreated cells. (B) Western blot analysis on nuclear and whole-cell extracts probing for p65 levels. (C) ChIP assay. ChIP assays were performed on NIH 3T3 3XκB-Luc wild-type (lanes 1 and 2) and mutant (lanes 3 and 4) cells. Cells were treated with either vehicle (dimethyl sulfoxide) or TSA for 18 h. DNA and protein were cross-linked with formaldehyde, and DNA was sheared and immunoprecipitated with anti-acetyl histone H3 antibody. After reversing cross-links, the DNA was amplified using end-labeled primers specific for the promoter region of the integrated 3XκB-Luc reporter gene. PCR products were analyzed by polyacrylamide gel electrophoresis and bands were visualized by autoradiography. Input, DNA prior to immunoprecipitation with the anti-acetyl histone H3 antibody.
Overexpression of HDAC1 and/or HDAC2 represses TNF-α-induced expression of an NF-κB-dependent reporter gene.
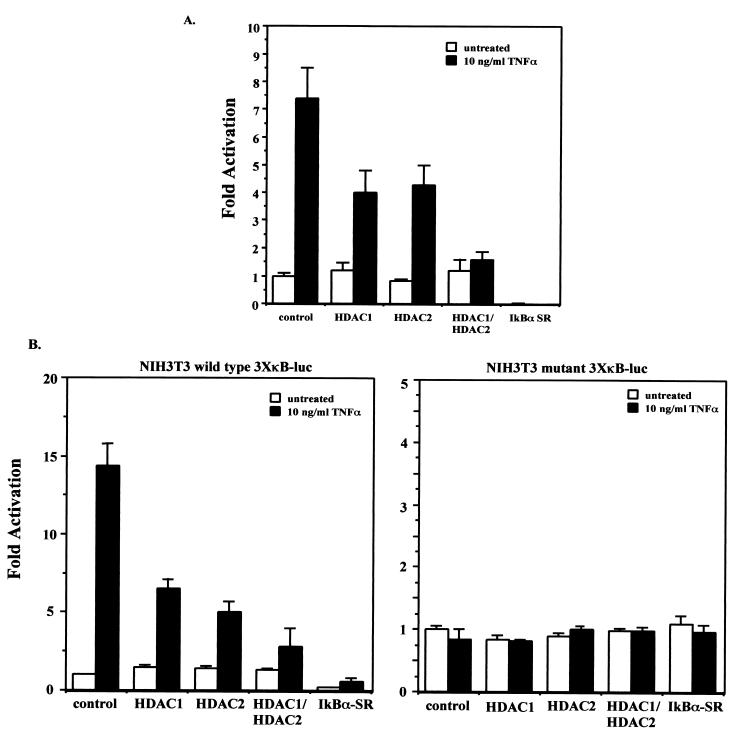
HDAC1 and HDAC2 are class I HDACs with homology to the yeast Rpd3 HDAC protein. HDAC1 and HDAC2 interact with each other and are typically found as components of large corepressor complexes such as the mSin3 and Mi-2–NURD complexes (19). We therefore decided to determine if HDAC1 and HDAC2 can regulate NF-κB-mediated transactivation of a transiently transfected NF-κB-dependent reporter gene. TNF treatment of Cos-7 cells transfected with the 3XκB-Luc reporter and a vector control yielded a sevenfold induction of 3XκB-Luc reporter gene activity (Fig. 2A). Overexpression of either HDAC1 or HDAC2 reduced TNF induction of the reporter gene to about fourfold over the basal level. Overexpression of both HDAC1 and HDAC2 together essentially blocked induction of the 3XκB-Luc reporter gene in response to TNF induction (Fig. 2A); however, expression of HDAC1 and HDAC2, either alone or together, had no effect on the basal level of expression of the reporter gene (Fig. 2A). Cells were also transfected with a mutant form of the IκBα inhibitor in which the serines at positions 32 and 36 had been changed to alanines. The resulting protein (referred to as IκBα super repressor, or IκBα-SR) blocks activation of NF-κB because it can no longer be inducibly phosphorylated and degraded by the proteasome. As expected, transfection of IκBα-SR resulted in inhibition of both basal and TNF-induced expression of the 3XκB-Luc reporter (Fig. 2A).
FIG. 2.
(A) HDAC1 and HDAC2 repress TNF-induced expression of a transiently transfected 3XκB-Luc reporter gene. Cos-7 cells were transfected in triplicate with the wild-type 3XκB-Luc reporter and a control vector (pcDNA3.1), pcDNA-HDAC1, pcDNA-HDAC2 alone, or HDAC1 and -2 together. Twenty-four hours after transfection the cells were either untreated or treated with 10 ng of TNF-α/ml for 8 h. Extracts were prepared and relative luciferase activities were determined by normalizing to total protein. Activity is expressed as fold activation relative to the untreated control and is the average ± SD from a representative experiment. Experiments were performed a minimum of three times, with similar results. (B) NIH 3T3 cells harboring either integrated wild-type 3XκB-Luc (left) or mutant 3XκB-Luc (right) were transfected in triplicate as described for panel A. Twenty-four hours after transfection the cells were either untreated or treated with 10 ng of TNF-α/ml for 8 h. Extracts were prepared and relative luciferase activities were determined by normalizing to total protein. Activity is expressed as fold activation relative to the untreated control and is the average ± SD from a representative experiment. Experiments were performed a minimum of three times, with similar results.
One mechanism by which the HDAC proteins likely exert their corepressor function is by modifying chromatin structure through their HDAC activity. However, since transiently transfected plasmids are not efficiently packaged into chromatin, we examined the effect of HDAC1 and HDAC2 expression on TNF induction of the integrated wild-type and mutant 3XκB luciferase reporters. TNF induction of vector control-transfected cells harboring an integrated wild-type 3XκB-Luc reporter gave a 14-fold induction of luciferase activity relative to the basal level (Fig. 2B, left panel). Transfection of HDAC1 or HDAC2 alone reduced the level of induction to about five- to sixfold over basal levels (Fig. 2B, left panel). Cotransfection of both HDAC1 and HDAC2 resulted in a further reduction of TNF-induced expression to an approximately threefold total induction relative to the basal level (Fig. 2B, left panel). Transfection of IκBα-SR almost completely blocked both basal and TNF-induced expression of the reporter (Fig. 2B, left panel), indicating a high level of transfection efficiency. In addition, the basal level of expression of the integrated reporter gene is much lower in the cells transfected with IκBα-SR, further suggesting a role for NF-κB in regulating basal, unstimulated expression of the reporter gene. However, expression of HDAC1 and HDAC2 either alone or together had no effect on the unstimulated level of expression of the reporter (Fig. 2B, left panel). The reason for this is not clear, but one explanation may be that in unstimulated cells, the amount of NF-κB in the nucleus is already limiting relative to the level of HDAC proteins. Therefore, further increasing the amount of HDACs in the cell has no effect on the basal level of expression and can only cause repression when higher levels of NF-κB are found in the nucleus after stimulation with TNF. As expected, in cells harboring an integrated mutant version of the 3XκB-Luc reporter, no increase in expression was seen after TNF induction and expression of HDAC1 and/or HDAC2, and IκBα-SR had no effect on reporter gene expression (Fig. 2B, right panel). These results indicate that the reduced levels of reporter gene expression in cells expressing HDAC1 and HDAC2 are dependent on NF-κB. In addition, these results also show that HDAC1 and HDAC2 are able to repress induction of both a transiently transfected and stably integrated NF-κB-dependent reporter equally well. This may imply that the ability of the HDAC proteins to repress NF-κB-mediated gene expression is not entirely dependent on the HDAC activity or that the chromatin structure of the transiently transfected reporter plasmid is able to affect the regulation of expression of the reporter gene and that it can be modulated by coactivator and corepressor proteins. Alternatively, it is possible that the HDACs are targeting other proteins, such as the NF-κB subunits, for modification.
mSin3a and N-CoR, but not SMRT, can repress NF-κB activation of a transiently transfected reporter gene.
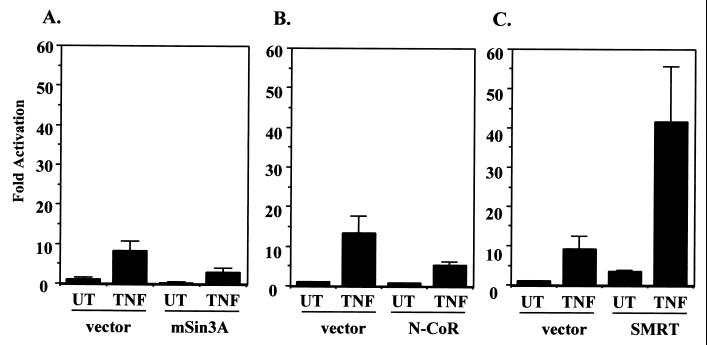
HDAC1 and HDAC2 are known to form complexes directly with the mSin3a corepressor and can interact indirectly through mSin3a with both the N-CoR and SMRT corepressor proteins (35). In addition, a recent paper showed that cotransfection of the SMRT corepressor with p65 was able to moderately repress p65-mediated transactivation (20). We therefore decided to determine if mSin3a, N-CoR, or SMRT was able to repress the ability of TNF to induce NF-κB-dependent transactivation. In order to test this, mSin3a, N-CoR, or SMRT was cotransfected into HeLa cells along with the 3XκB-Luc reporter gene. The cells were either untreated or stimulated with TNF and assayed for reporter gene activity. These data show that mSin3a was able to modestly repress TNF induction of the 3XκB-Luc reporter gene two- to threefold relative to the vector control (Fig. 3A). Expression of the N-CoR corepressor protein was also able to repress TNF-induced expression of the reporter (less than twofold reduction) (Fig. 3B), although to a slightly lesser extent than did mSin3a. Interestingly, expression of SMRT resulted in a slight increase in basal expression of the reporter gene and a 40-fold induction of expression after TNF treatment, a level about fourfold higher than TNF induction in the vector control cells (Fig. 3C). Our results suggest that mSin3a and N-CoR are capable of repressing TNF-induced transactivation mediated by NF-κB in transient transfections but that SMRT is not able to repress, and in fact, may sequester endogenous HDAC proteins to increase NF-κB-dependent transactivation.
FIG. 3.
mSin3a and N-CoR, but not SMRT, can repress TNF-induced expression of 3XκB-Luc. Cos-7 cells were transfected with wild-type 3XκB-Luc and either an appropriate control vector or pIRESHis-mSin3a (A), pCEP4–N-CoR (B), or pCMX-SMRT (C). Twenty-four hours after transfection the cells were either untreated (UT) or treated with 10 ng of TNF-α/ml for 8 h. Extracts were prepared and relative luciferase activities were determined by normalizing to total protein. Activity is expressed as fold activation relative to the untreated control and is the average ± SD from a representative experiment. Experiments were performed a minimum of three times, with similar results.
HDAC1 targets p65.
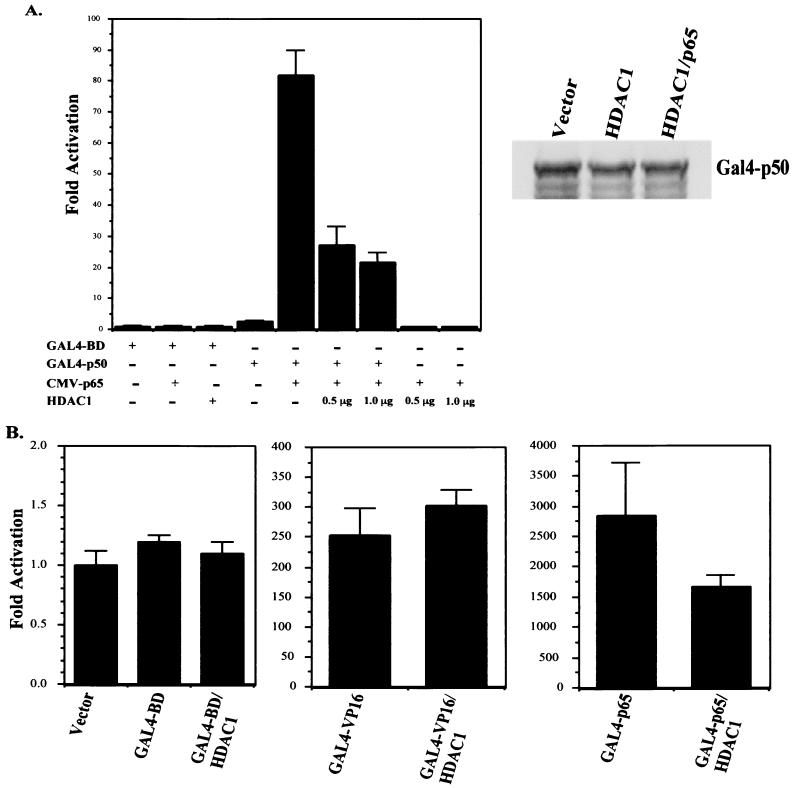
In order to determine if HDAC1 can repress p65-mediated transactivation, we used a fusion of full-length p50 to the GAL4 DNA-binding domain in cotransfection experiments with p65. The p50 protein does not have a transactivation domain and therefore the GAL4-p50 fusion cannot efficiently activate expression of the GAL4-Luc reporter gene. However, when p65 is cotransfected with GAL4-p50, a high level of expression is seen since p65 has a potent transactivation domain and can dimerize with p50. When transfected into HeLa cells, the GAL4-p50 fusion can weakly activate a 5XGAL4-Luc reporter gene, resulting in about a fourfold induction relative to expression of the GAL4 DNA-binding domain alone (Fig. 4A). Cotransfection of a plasmid expressing p65 resulted in a greater than 80-fold stimulation of reporter gene expression, due to dimerization of p65 with the GAL4-p50 fusion. This stimulation was dependent on the presence of GAL4-p50, since expression of p65 with just the GAL4 DNA-binding domain did not induce expression of the reporter gene (Fig. 4A). When HDAC1 was expressed along with GAL4-p50 and p65, the level of induction was reduced to about 20- to 25-fold (Fig. 4A). Expression of HDAC1 with the GAL4 DNA-binding domain alone or with p65 alone did not have any effect on reporter gene activity (Fig. 4A). Also, HDAC1 had no effect on the low level of expression seen with the GAL4-p50 fusion (data not shown), indicating that HDAC1 is not functioning through p50. Expression of HDAC1 or HDAC1 and p65 had no effect on the level of expression of the GAL4-p50 fusion (Fig. 4A, right panel). Therefore, the observed repression caused by HDAC1 is not due to reduced GAL4-p50 expression. As a control for specificity, the effect of HDAC1 expression on the ability of the GAL4 DNA-binding domain and fusions of the VP16 and p65 transactivation domains to the GAL4 DNA-binding domain to activate reporter gene expression was also tested. As expected, no effect was observed when HDAC1 was expressed with the GAL4 DNA-binding domain (Fig. 4B). HDAC1 expression also had no effect on the high level of activity of the GAL4-VP16 fusion (Fig 4B). A modest level of repression (less than twofold) was seen when HDAC1 was expressed with the GAL4-p65 fusion (Fig. 4B), possibly due to weak interactions between HDAC1 and the p65 C-terminal region. These results provide further evidence that the transactivation function of NF-κB is negatively regulated by the activity of HDAC corepressor proteins and that this repression is mediated through an interaction with p65.
FIG. 4.
(A) HDAC1 targets the p65 subunit of NF-κB. HeLa cells were transfected with a 5XGAL4-Luc reporter plasmid along with either the GAL4 DNA-binding domain (GAL4-BD) as a control or a full-length fusion of p50 to the GAL4 DNA-binding domain (GAL4-p50). Where indicated, cells were also transfected with pCMV-p65 and/or pcDNA-HDAC1 (either 0.5 or 1.0 μg). Forty-eight hours after transfection extracts were prepared and relative luciferase activities were determined by normalizing to total protein. Activity is expressed as fold activation relative to the untreated control and is the average ± SD from a representative experiment. Experiments were performed a minimum of three times, with similar results. Right panel, Western blot probing for GAL4-p50 to show that its expression was not affected by expression of HDAC1. (B) HeLa cells were transfected with the GAL4 DNA-binding domain, GAL4-VP16, or GAL4-p65 and either control vector or pcDNA-HDAC1 along with 5XGAL4-Luc reporter plasmid. Forty-eight hours after transfection extracts were prepared and relative luciferase activities were determined by normalizing to total protein. Activity is expressed as fold activation relative to the untreated control and is the average ± SD from a representative experiment. Experiments were performed a minimum of three times, with similar results.
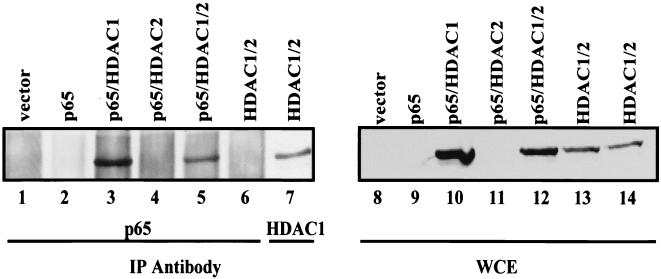
p65 coimmunoprecipitates with HDAC1.
In order to determine if p65 and HDAC1 and/or HDAC2 can interact in vivo, 293 cells were transfected with expression plasmids for p65, HDAC1, and HDAC2 and coimmunoprecipitations were performed. As shown in Fig. 5, HDAC1 was coimmunoprecipitated with p65 in cells transfected with p65 and HDAC1 alone or with HDAC1 and HDAC2 (Fig. 5, lanes 3 and 5). This interaction was dependent on the presence of both p65 and HDAC1, since HDAC1 was not coimmunoprecipitated with p65 in vector control cells or in cells transfected with just p65 or just HDAC1 and HDAC2 (Fig. 5, lanes 1, 2, and 6). No direct interaction was seen between p65 and HDAC2 (data not shown), although HDAC2 was coimmunoprecipitated with p65 in the presence of HDAC1 (data not shown). As a control, an immunoprecipitation and Western blot for HDAC1 from cells transfected with HDAC1 and HDAC2 were performed. These data indicate that HDAC1 interacts with p65 and that HDAC2, probably based on its interaction with HDAC1, is indirectly associated with p65.
FIG. 5.
HDAC1 interacts with p65 in transient transfections. 293 cells were transfected with the indicated plasmids and extracts were made and used for immunoprecipitations (IP) with either a p65-specific antibody (lanes 1 to 6) or an HDAC1-specific antibody (lane 7). Immunoprecipitates were then used in Western blot analysis to probe for the presence of HDAC1. Lanes 8 to 14, Western blot probed for HDAC1 on the whole-cell extracts (WCE) used for the immunoprecipitation reactions.
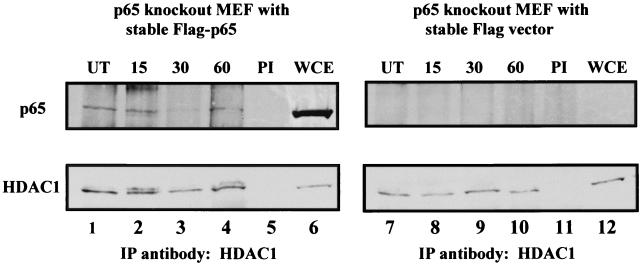
In order to determine if endogenous HDAC1 interacts with NF-κB in vivo, coimmunoprecipitation experiments were performed using immortalized mouse embryonic fibroblasts (MEFs) from a p65 knockout mouse which had been reconstituted with either Flag-tagged p65 or the Flag vector (38). The level of expression of p65 in these cells is similar to the level seen in p65+/− heterozygotes and is therefore not an overexpression system (data not shown). The Flag-p65 line and the vector control line were either untreated or treated for the indicated times with TNF. Cells were harvested and immunoprecipitations were performed on whole-cell extracts using an anti-HDAC1 antibody. Results from the vector control p65 knockout line and from the Flag-p65-reconstituted line are shown in Fig. 6. As expected, there was no detectable p65 in either the whole-cell extracts or in the HDAC1 immunoprecipitations from the vector control cells. In contrast, p65 was coimmunoprecipitated with HDAC1 in the Flag-p65-reconstituted cells. p65 was found associated with HDAC1 in untreated cells, and treatment with TNF for 15, 30, or 60 min did not diminish the amount of p65 associated with HDAC1 (Fig. 6). There appears to be a slight decrease in the amount of p65 associated with HDAC1 after 30 min of TNF treatment (Fig. 6, upper panel, lane 3); however, the amount of HDAC1 precipitated at this time point was also slightly diminished (Fig. 6, lower panel, lane 3). It therefore appears that p65 is associated with HDAC1 even after activation of NF-κB with TNF and that there is no change in the amount of HDAC1 associated with NF-κB after stimulation with TNF.
FIG. 6.
p65 can be coimmunoprecipitated with endogenous HDAC1. Immunoprecipitations (IP) were performed with an antibody specific for HDAC1 on whole-cell extracts (WCE) from p65 null MEFs that had been stably reconstituted with Flag-tagged p65 (left) or the Flag vector (right). Lower panels, Western blot probing the immunoprecipitations for HDAC1 from the Flag-p65-reconstituted cells (lanes 1 to 5) or from the Flag vector-reconstituted cells (lanes 7 to 11); upper panels, Western blot probed for p65 from the HDAC1 immunoprecipitations. Lanes 6 and 12, whole-cell extracts to show the presence of HDAC1 and p65 in the extracts. Prior to harvesting extracts, the cells were either untreated (UT) or treated with 10 ng of TNF-α/ml for the indicated times. PI, preimmune serum.
HDAC1 can interact in vitro directly with p65.
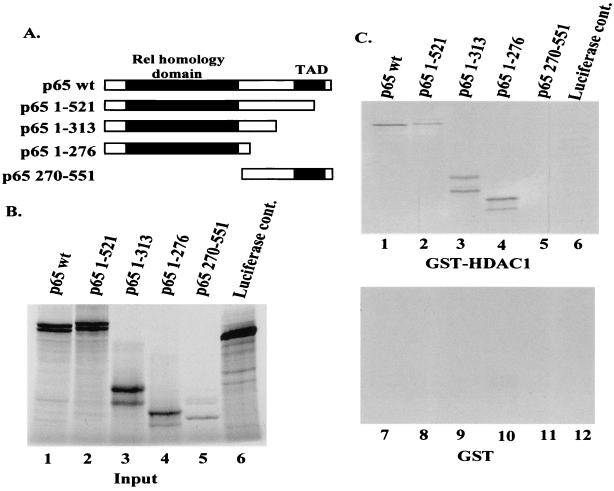
In order to determine if the interaction between p65 and HDAC1 observed in the coimmunoprecipitation experiments was direct, in vitro interaction assays using in vitro-transcribed and -translated p65 and GST-HDAC1 fusions were performed. Figure 7A shows a diagram of full-length p65 as well as three carboxyl-terminal deletions and one amino-terminal deletion of p65 that was used in these assays. The products of the in vitro transcription and translation reactions are shown in Fig. 7B (lanes 1 to 5). The luciferase expression plasmid included with the in vitro transcription and translation kit is also shown as a control (Fig. 7B, lane 6). When the in vitro transcribed and translated p65 was incubated with GST-HDAC1, interactions were seen with full-length p65 as well as with all three C-terminal deletions [p65 wt, p65(1-521), p65(1-313), and p65(1-276)] (Fig. 7C, lanes 1 to 4). However, no interaction was seen between GST-HDAC1 and the amino-terminal deletion [p65(270-551)] (Fig. 7B, lane 5), indicating that the region of p65 which interacts with HDAC1 lies within the Rel homology domain of p65 (Fig. 7C, upper panel). As expected, no interaction was seen between the luciferase control and GST-HDAC1 (Fig. 7C, lane 6) or between any of the p65 proteins and the GST control (Fig. 7C, lower panel).
FIG. 7.
HDAC1 interacts directly with p65 in in vitro binding assays. (A) Diagram of p65 full-length and deletion mutants used for in vitro interaction. (B) Lanes 1 to 6, 1/10 of the indicated input in vitro transcription and translation products used for in vitro binding assays. The products of the in vitro transcription and translation reactions were fractionated by SDS-polyacrylamide gel electrophoresis and the bands were visualized by autoradiography. (C) After the in vitro transcription and translation reactions were performed, 10 μl (of a 50-μl reaction) of the indicated products was mixed with GST-HDAC1 (upper panel) or GST (lower panel) bound to glutathione-agarose beads. After incubation for 1 h at 4°C, the beads were washed extensively and interacting proteins were visualized as for panel A.
Inhibition of HDAC activity causes increased expression of the IL-8 gene.
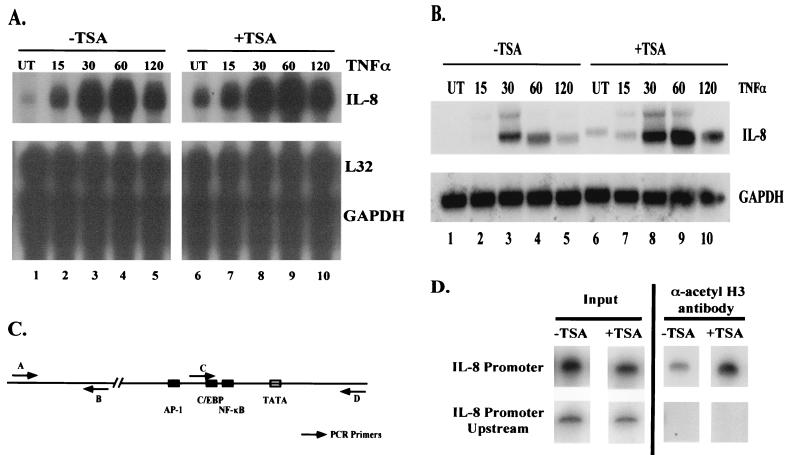
In order to determine if inhibition of deacetylase corepressor proteins had any effect on basal or TNF-induced expression levels of NF-κB-regulated genes, ribonuclease protection assays were performed on RNA harvested from HeLa cells after the indicated treatments. A custom ribonuclease protection assay template containing six NF-κB-regulated genes (TRAF1, TRAF2, Bfli/A1, c-IAP-2, IL-8, and IL-2Rα) was used. Of these genes, expression of the IL-8 gene was most dramatically affected by inhibition of HDAC activity using TSA. A very slight induction of basal and induced expression of the c-IAP-2 gene was also seen after TSA treatment (data not shown). In the absence of TSA, TNF induction caused an increase in IL-8 expression beginning at about 15 min postinduction and peaking at about 1 h, with the signal beginning to go down by 2 h postinduction (Fig. 8A, lanes 2 to 5). In the presence of TSA, a higher level of basal IL-8 expression (Fig. 8A, compare lanes 1 and 6) was seen, correlating with the increase in basal expression of the integrated 3XκB-Luc reporter gene after TSA treatment (Fig. 1A). After stimulation with TNF and TSA, the kinetics of induction of IL-8 were similar to those of TNF stimulation alone; however, the level of induction of IL-8 was greater in the presence of TSA treatment (Fig. 8A, lanes 7 to 10). To further illustrate the effect of TSA treatment on TNF-induced expression of IL-8, Northern blot analysis was performed (Fig. 8B). These results show that TSA treatment results in an increase in basal IL-8 expression (Fig. 8B, lanes 1 and 6) and causes enhanced and prolonged expression of IL-8 after TNF induction (Fig. 8B, lanes 2 to 5 versus lanes 7 to 10). Expression of two housekeeping genes, L32 (Fig. 8A) and GAPDH (Fig. 8A and B), was unaffected by treatment with TNF either in the presence or absence of TSA.
FIG. 8.
Inhibition of HDAC activity causes an increase in IL-8 expression. (A) Ribonuclease protection assay showing effect of TSA treatment on IL-8 expression. Total cellular RNA was harvested from HeLa cells after the indicated treatments and used in a ribonuclease protection assay. TNF-α was used at a final concentration of 10 ng/ml for the indicated times. TSA treatment was for 18 h at a final concentration of 100 nM. L32 and GAPDH are shown as loading controls. Other genes used in the assay are not shown. (B) Same as panel A, but RNA was used in Northern blot analysis to analyze the IL-8 expression pattern. (C) Diagram of IL-8 promoter and IL-8 upstream region. Arrows indicate primer pairs used in PCR for ChIP assays. (D) ChIP assay on IL-8 promoter and IL-8 upstream region. Cells were treated with either vehicle (dimethyl sulfoxide) or TSA for 18 h. DNA and protein were cross-linked with formaldehyde, and DNA was sheared and immunoprecipitated with anti-acetyl histone H3 antibody. After reversing cross-links, the DNA was amplified using end-labeled primers specific for the promoter region of the IL-8 gene or for a region upstream of the IL-8 promoter devoid of any known promoter elements. PCR products were analyzed by polyacrylamide gel electrophoresis and bands were visualized by autoradiography. Input, DNA prior to immunoprecipitation with the anti-acetyl histone H3 antibody. UT, untreated cells.
Treatment of cells with TSA blocks the activity of HDAC proteins, resulting in hyperacetylation of histones and subsequently a higher level of gene expression. In order to determine if TSA treatment caused hyperacetylation of the IL-8 promoter around the NF-κB binding site similar to that seen in the NIH 3T3 cells with the wild-type 3XκB-Luc reporter, ChIP assays were performed. Figure 8C shows a diagram of the IL-8 promoter, including the NF-κB binding site and the primers used for PCR amplification of the IL-8 promoter region and a region upstream of the IL-8 promoter which does not contain any known transcription factor binding sites. ChIP assays were performed on either untreated cells or cells treated with TSA using an anti-acetyl histone H3 antibody. In Fig. 8D, amplification of the input DNA from the IL-8 promoter and the upstream region used in the ChIP assay and of the DNA after immunoprecipitation with the anti-acetyl histone H3 antibody are shown. Treatment of cells with TSA resulted in hyperacetylation of the IL-8 promoter region (Fig. 8D, upper right panel), whereas no acetylation and no TSA effect was seen in the upstream region (Fig. 8D, lower right panel). Based on this data, it is likely that the increased level of expression of the IL-8 gene observed in the ribonuclease protection assay and Northern blot analyses is due to a localized hyperacetylation of the IL-8 promoter.
DISCUSSION
Activation of transcription by NF-κB has been shown to require a number of different coactivator proteins including CBP (p300), p/CAF, and SRC-1 (12, 25, 28, 31, 44). This study was undertaken to determine the potential role of corepressor proteins in regulating the transactivation potential of NF-κB. Our results show that treatment of cells harboring an integrated NF-κB-dependent reporter gene (NIH 3T3 3XκB-Luc) with TSA, a chemical inhibitor of HDAC corepressor proteins, leads to an increase in both basal and TNF-induced activation of the reporter gene. These results indicate that HDAC corepressor proteins are involved in regulating the transactivation function of NF-κB. In support of this, we show that transient transfection of HDAC1 and HDAC2, either alone or together, is able to repress TNF-induced activation of both a transiently transfected 3XκB-Luc reporter and the integrated 3XκB-Luc reporter. Using both in vitro binding assays and coimmunoprecipitation and Western blot analysis, we show that HDAC1 can interact directly with the p65 subunit of NF-κB and likely exerts its corepressor function through this interaction. Importantly, we show that inhibition of HDAC activity using TSA results in an increase in basal and inducible expression of the NF-κB-regulated IL-8 gene. The inducible expression of IL-8, as well as the 3XκB-Luc reporter gene, requires activation of NF-κB. However, it is not known if expression of these genes is also repressed in part by NF-κB. The increase in basal expression of the 3XκB-Luc reporter as well as the IL-8 gene in the presence of an HDAC inhibitor indicates that HDAC proteins are functioning to repress their expression; however, it is not yet known if this repression is mediated through NF-κB. Infection of HeLa cells with an adenovirus expressing IκBα-SR (Ad-IκBα-SR) results in a block in activation of IL-8 expression (Brian P. Ashburner and Albert S. Baldwin, Jr., unpublished results). However, a low basal level of expression is also seen after treatment of Ad-IκBα-SR-infected cells with TSA, indicating that repression of IL-8 expression may require the presence of NF-κB and the activity of HDAC corepressor proteins (Ashburner and Baldwin, unpublished results). Based on these results, we conclude that an important component regulating NF-κB-dependent transactivation is mediated through the interaction of NF-κB with HDAC corepressor proteins. At present, we do not know if the presumed ability of NF-κB to repress basal, unstimulated IL-8 gene expression is unique to IL-8 or whether other genes will exhibit this phenomenon. Overall, these findings add yet another level to the already complex regulation of gene expression controlled by NF-κB.
In addition to an increase in basal expression of the 3XκB-Luc reporter and the IL-8 gene, TSA treatment also causes an increase in the inducible level of expression of these genes. Treatment of the NIH 3T3 3XκB-Luc cells with TNF and TSA resulted in a 15-fold increase in reporter gene expression compared to a 5-fold increase with TNF alone. The level of induced expression of IL-8 was also higher after TNF and TSA treatment than treatment with TNF alone. Interestingly, only the level of expression, not the kinetics of IL-8 expression, was altered by TSA treatment. That is, expression can begin to be seen 15 min after TNF induction, peaks at 30 to 60 min after induction, and begins to decrease 2 h after TNF induction either with or without TSA treatment. The only difference is that the level of induction in the presence of TSA is greater than in the absence of TSA. These data indicate that HDAC proteins may play a role in regulating the inducible level of expression of certain NF-κB-regulated genes in addition to repressing expression of these genes.
The primary level of regulation of NF-κB activity is through its retention in the cytoplasm through interactions with IκB and its subsequent release and transport into the nucleus upon activation by a variety of stimuli. Although the majority of cellular NF-κB in unstimulated cells is cytoplasmic, many cell types appear to have a constitutively low basal level of nuclear NF-κB. The function of this nuclear NF-κB in unstimulated cells is not clear, although it may be involved in regulating basal gene expression as well as in repressing expression of certain genes. Evidence from our lab and others has shown that another mechanism controlling NF-κB activity is the regulation of the p65 transactivation domain. For example, oncogenic forms of H-Ras activate the transactivation function of p65 in a manner that is independent of nuclear translocation (11, 26). In addition, the serine/threonine protein kinase Akt has also been shown to stimulate the transactivation function of p65 (21). The mechanism by which these proteins stimulate the transactivation function of p65 is not yet known, but one possibility is by modulating the interaction of NF-κB with coactivator and corepressor proteins. The p65 subunit of NF-κB is known to be inducibly phosphorylated (30, 38, 39, 44, 45) in response to a number of different stimuli which may regulate the interaction of p65 with coactivator and corepressor proteins. In support of this, inducible phosphorylation of p65 on serine 276 by the catalytic subunit of PKA enhances the interaction of p65 with CBP, thus enhancing the ability of NF-κB to activate transcription (44, 45).
The mechanism by which HDAC proteins regulate the activity of NF-κB is presumably through their deacetylase activity since TSA inhibits this activity. However, the mechanisms regulating the interactions between NF-κB and HDAC proteins are not yet known. It is possible that the strength of the signal activating NF-κB may determine the activity of NF-κB by regulating the interaction of NF-κB with coactivator proteins such as CBP and corepressor proteins such as HDAC1 and HDAC2. In this case, a stronger activation signal may result in a greater affinity of CBP for p65 and potentially weaker interactions with HDAC proteins, resulting in higher levels of expression. Reduced signaling (presumably resulting in reduced phosphorylation of p65) would result in less binding of CBP (or other coactivators) to p65 and a greater amount of HDAC associated with p65, giving a lower level of expression. However, since TSA inhibits the deacetylase activity of the HDAC proteins, we do not yet know if this may also cause dissociation of HDACs from binding to p65, allowing more CBP (or other transcriptional coactivators) to bind. Interestingly, we have found that treatment of HeLa cells with TSA results in enhanced phosphorylation of p65 after a 30-min and 2-h TNF stimulation (Julie L. Hansen, Brian P. Ashburner, and Albert S. Baldwin, Jr., unpublished data). These results are similar to that observed for the CREB transcription factor, which showed that HDAC inhibition potentiated gene expression via cyclic AMP by enhancing phosphorylation of CREB in a chromatin-dependent manner (24). It is therefore possible that by inhibiting HDAC activity, the resulting prolonged phosphorylation of p65 may serve to enhance association of CBP (p300) with p65 and thus enhance gene expression.
We are presently attempting to determine the importance of other corepressor proteins in regulating NF-κB activity. Our data indicate that both mSin3a and N-CoR can repress TNF-induced NF-κB activation of a transiently transfected reporter. However, we were unable to detect an interaction between p65 and either mSin3a or N-CoR by coimmunoprecipitation. Interestingly, transfection of the SMRT corepressor resulted in a higher level of basal and TNF-induced reporter gene activity. The reason for this is not clear; however, it is possible that SMRT is acting in a manner to sequester the endogenous HDAC proteins, thus causing the higher levels of expression. Our data are in contrast to another report which showed that SMRT as well as mSin3a is able to repress p65-mediated transactivation when cotransfected with p65 (20). At this time we do not understand this discrepancy, but it is possible that there are cell type effects since different cell lines were used. In addition, although we transfected SMRT, we looked at the effect of SMRT on TNF induction of endogenous NF-κB instead of on transfected p65. The fact that expression of SMRT resulted in a higher level of NF-κB-dependent expression whereas mSin3a and N-CoR repressed expression may further imply that mSin3a and N-CoR, but not SMRT, are involved in regulating NF-κB-dependent gene expression. In conclusion, we have identified a novel mechanism regulating the transactivation function of NF-κB. Furthermore, our work shows that NF-κB, in association with HDAC1 and HDAC2, plays an important role in repressing gene expression. Since blocking the activity of NF-κB has become a primary target for drug therapy in a number of different diseases, it is important to understand what effect this may have on genes negatively regulated by NF-κB and on the efficacy of the therapy itself.
ACKNOWLEDGMENTS
We thank D. Ayer, S. Schrieber, E. Seto, C. Glass, and R. Evans for providing plasmids used in this work and D. Guttridge for providing the NIH 3T3 cells with the stable wild-type and mutant 3XκB-Luc reporters. We also thank D. Guttridge for critical review of the manuscript and the members of the Baldwin lab for many helpful discussions.
This work was supported by Public Health Service grants to A.S.B. (AI35098 and CA 73756) from the National Cancer Institute. B.P.A. was supported by a postdoctoral fellowship from the Cancer Research Institute. S.D.W. was supported by American Cancer Society grant PF-00-023-01-MGO.
REFERENCES
- 1.Ashburner B P, Shackelford R E, Baldwin A S, Jr, Paules R S. Lack of involvement of ataxia telangiectasia mutated (ATM) in regulation of nuclear factor-kappaB (NF-kappaB) in human diploid fibroblasts. Cancer Res. 1999;59:5456–5460. [PubMed] [Google Scholar]
- 2.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin A S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Carlotti F, Dower S K, Qwarnstrom E E. Dynamic shuttling of nuclear factor kappa B between the nucleus and cytoplasm as a consequence of inhibitor dissociation. J Biol Chem. 2000;275:41028–41034. doi: 10.1074/jbc.M006179200. [DOI] [PubMed] [Google Scholar]
- 6.Cogswell P C, Mayo M W, Baldwin A S., Jr Involvement of Egr-1/RelA synergy in distinguishing T cell activation from tumor necrosis factor-alpha-induced NF-kappa B1 transcription. J Exp Med. 1997;185:491–497. doi: 10.1084/jem.185.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 8.El Kharroubi A, Piras G, Zensen R, Martin M A. Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol. 1998;18:2535–2544. doi: 10.1128/mcb.18.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finco T S, Baldwin A S. Mechanistic aspects of NF-kappa B regulation: the emerging role of phosphorylation and proteolysis. Immunity. 1995;3:263–272. doi: 10.1016/1074-7613(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 10.Finco T S, Beg A A, Baldwin A S., Jr Inducible phosphorylation of I kappa B alpha is not sufficient for its dissociation from NF-kappa B and is inhibited by protease inhibitors. Proc Natl Acad Sci USA. 1994;91:11884–11888. doi: 10.1073/pnas.91.25.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finco T S, Westwick J K, Norris J L, Beg A A, Der C J, Baldwin A S., Jr Oncogenic Ha-Ras-induced signaling activates NF-kappaB transcriptional activity, which is required for cellular transformation. J Biol Chem. 1997;272:24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 12.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh S, May M J, Kopp E B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 14.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 15.Harbour J W, Luo R X, Dei Santi A, Postigo A A, Dean D C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 16.Huang T T, Kudo N, Yoshida M, Miyamoto S. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc Natl Acad Sci USA. 2000;97:1014–1019. doi: 10.1073/pnas.97.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito K, Barnes P J, Adcock I M. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol Cell Biol. 2000;20:6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 20.Lee S K, Kim J H, Lee Y C, Cheong J, Lee J W. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factor. J Biol Chem. 2000;275:12470–12474. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- 21.Madrid L V, Wang C Y, Guttridge D C, Schottelius A J, Baldwin A S, Jr, Mayo M W. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-kappaB. Mol Cell Biol. 2000;20:1626–1638. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mal A, Sturniolo M, Schiltz R L, Ghosh M K, Harter M L. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 24.Michael L F, Asahara H, Shulman A I, Kraus W L, Montminy M. The phosphorylation status of a cyclic AMP-responsive activator is modulated via a chromatin-dependent mechanism. Mol Cell Biol. 2000;20:1596–1603. doi: 10.1128/mcb.20.5.1596-1603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na S Y, Lee S K, Han S J, Choi H S, Im S Y, Lee J W. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappaB-mediated transactivations. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 26.Norris J L, Baldwin A S., Jr Oncogenic Ras enhances NF-kappaB transcriptional activity through Raf-dependent and Raf-independent mitogen-activated protein kinase signaling pathways. J Biol Chem. 1999;274:13841–13846. doi: 10.1074/jbc.274.20.13841. [DOI] [PubMed] [Google Scholar]
- 27.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 28.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 29.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard K A, Rose D W, Haque Z K, Kurokawa R, McInerney E, Westin S, Thanos D, Rosenfeld M G, Glass C K, Collins T. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol Cell Biol. 1999;19:6367–6378. doi: 10.1128/mcb.19.9.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C, Hager G. Transcriptional regulation of mammalian genes in vivo: a tale of two templates. J Biol Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 33.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 34.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 36.Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 37.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 38.Wang D, Baldwin A S., Jr Activation of nuclear factor-kappaB-dependent transcription by tumor necrosis factor-alpha is mediated through phosphorylation of RelA/p65 on serine 529. J Biol Chem. 1998;273:29411–29416. doi: 10.1074/jbc.273.45.29411. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Westerheide S D, Hanson J L, Baldwin A S., Jr Tumor necrosis factor alpha-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J Biol Chem. 2000;275:32592–32597. doi: 10.1074/jbc.M001358200. [DOI] [PubMed] [Google Scholar]
- 40.Wotton D, Lo R S, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 41.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 43.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 44.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 45.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]