Abstract
The protein kinase ribosomal S6 kinase 2 (RSK2) has been implicated in phosphorylation of transcription factor CREB and histone H3 in response to mitogenic stimulation by epidermal growth factor. Binding of phospho-CREB to the coactivator CBP allows gene activation through recruitment of the basal transcriptional machinery. Acetylation of H3 by histone acetyltransferase (HAT) activities, such as the one carried by CBP, has been functionally coupled to H3 phosphorylation. While various lines of evidence indicate that coupled histone acetylation and phosphorylation may act in concert to induce chromatin remodeling events facilitating gene activation, little is known about the coupling of the two processes at the signaling level. Here we show that CBP and RSK2 are associated in a complex in quiescent cells and that they dissociate within a few minutes upon mitogenic stimulus. CBP preferentially interacts with unphosphorylated RSK2 in a complex where both RSK2 kinase activity and CBP acetylase activity are inhibited. Dissociation is dependent on phosphorylation of RSK2 on Ser227 and results in stimulation of both kinase and HAT activities. We propose a model in which dynamic formation and dissociation of the CBP-RSK2 complex in response to mitogenic stimulation allow regulated phosphorylation and acetylation of specific substrates, leading to coordinated modulation of gene expression.
The induction of immediate-early response genes is the result of a number of coordinate events operating at the levels of intracellular signaling and transcriptional activation (33). Upon physiological challenge by mitogens and hormones and the consequent triggering of corresponding transduction pathways, various molecular events elicit the activation of transcription factors and the recruiting of specific coactivators (18, 30). Some coactivators also bear histone acetylase (HAT) activity, directly linking transcriptional activation to distinct chromatin modifications (7, 29, 46).
Various signaling routes converge on transcription factor CREB (cyclic AMP-responsive binding protein) (4, 20, 44) and control its function by modulating its phosphorylation state. Phosphorylation at a single serine residue (Ser-133) works as a molecular switch, as it dictates CREB's ability to interact with the coactivator CBP (CREB-binding protein), a large protein with HAT activity (5, 13, 39) which mediates functional contacts with the basal transcriptional machinery (34). Activation of CREB by growth factors was shown to be Ras dependent and to involve the mitogen-activated protein kinases (MAPKs) (27). Although a number of kinases downstream of the MAPKs may be implicated (19), members of the p90rsk (ribosomal S6 kinase [RSK]) family have been identified as mitogen-responsive CREB kinases (21, 51). In particular, both CREB phosphorylation and c-fos transcriptional induction are drastically impaired in response to epidermal growth factor (EGF) in human fibroblasts derived from Coffin-Lowry syndrome patients (21), which carry mutations in the gene encoding the RSK2 kinase (49). RSK2 is a member of the p90rsk family, which includes four closely related isoforms (23, 52). A conserved feature of all p90rsk proteins is the presence of two nonidentical kinase catalytic domains, the N-terminal domain being responsible for the phosphorylation of several targets (6, 22). The activity of the N-terminal domain is regulated upon direct MAPK activation of the C-terminal catalytic domain by the ERKs and involvement of PDK1 (3-phosphoinositide-dependent protein kinase 1) (17, 24, 25, 31, 41, 45, 53).
A critical set of observations have directly linked the activation of mitogen-activated transduction pathways with histone modifications and the consequent remodeling of chromatin (11, 36). In particular, the rapid and transient mitogen-induced phosphorylation of a serine residue (Ser10) in the tail of histone H3 has been coupled to the transcriptional activation of the immediate-early response genes (8, 15, 16). Interestingly, this event of phosphorylation appears to be coupled to acetylation, as the efficiency of histone acetyltransferases (HATs) to subsequently acetylate the nearby Lys14 is drastically increased (12, 35). Thus, activation of gene expression through chromatin remodeling is the result of multiple, coordinated events.
As somewhat anticipated, there are indications that common signaling pathways and effector kinases are utilized in the phosphorylation of transcription factors and histone tails (11). Although the Ser10 site in the H3 tail is the likely target of various kinases (11, 16, 43), there is evidence that RSK2 is the kinase involved in response to EGF (43). We have been wondering whether the coupling of phosphorylation and acetylation at the level of the histone substrates could possibly be paralleled by a physical and functional interplay of the respective effectors, kinases and acetylases. Here we show that CBP and RSK2 associate in a complex in quiescent cells and that they dissociate within a few minutes upon mitogenic stimulus. CBP preferentially interacts with unphosphorylated RSK2 in a complex where both RSK2 kinase activity and CBP acetylase activity are inhibited. We propose a model in which dynamic formation and dissociation of the CBP-RSK2 complex in response to mitogenic stimulation allow regulated phosphorylation and acetylation of specific substrates.
MATERIALS AND METHODS
Antibodies.
Monoclonal antibodies directed against RSK2 phosphorylated at Ser227 (P-S227) and Thr577 (P-T577) have been described previously (37). Anti-CBP (A-22) and anti-RSK2 (E1 and C-19) antibodies were from Santa Cruz Biotechnology Inc., anti-HA antibody was from Boehringer-Mannheim, anti-ERK, anti-phosphorylated ERK, anti-CREB, and anti-phosphorylated CREB antibodies were from New England Biolabs, and anti-H3 and anti-phosphorylated H3 antibodies were from Upsate Biotechnology. Preimmune sera were used in control experiments.
Transient transfections and stimulation of cells.
Expression vectors encoding hemagglutinin (HA)-tagged human RSK2 or mutated versions of RSK2, S227A and T577A, were described previously (37), as well as the expression vector encoding murine wild-type CBP (1). The expression vector encoding PDK1 was a gift from G. Thomas (Basel, Switzerland). Transient transfections were performed by the phosphate calcium method. COS-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 5% fetal calf serum (FCS) and plated at 30% confluence before transfection. After transfection (24 h), cells were serum deprived for an additional 48 h and, when required, treated with PD98059, EGF, tetradecanoyl phorbol acetate (TPA), or UV light as previously described (37).
Proteins.
Cells were washed once with ice-cold phosphate-buffered saline (PBS) and resuspended in hypotonic buffer (HB; 20 mM HEPES, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], 0.2% NP-40, a cocktail of protease inhibitors, 20 mM sodium fluorure, and 1 mM sodium orthovanadate) (42). Cells were centrifuged for 20 s, giving supernatant (S0) and pellet (P0). S0 was collected and supplemented to 120 mM NaCl and 10% glycerol. Lysates were centrifuged at 13,000 × g for 20 min at 4°C, giving the cytoplasmic extract. P0 was resuspended with HB supplemented to 420 mM NaCl and 20% glycerol, rocked for 30 min at 4°C, and centrifuged at 13,000 × g for 30 min at 4°C. This fraction represented the nuclear extract. Whole-cell extracts were prepared as described (53).
Western analysis.
Protein extracts were resolved by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were electroblotted onto Protan nitrocellulose (Schleicher and Schuell). Membranes were incubated in Tris-buffered saline–1% low-fat milk overnight at 4°C with specific antibodies. Immunocomplexes were revealed by chemiluminescence with anti-mouse, anti-rabbit, or anti-goat immunoglobulin antibodies.
Kinase and HAT assays.
Equal amounts of total nuclear extracts adjusted to 120 mM NaCl were incubated with anti-RSK2 or anti-CBP antibodies overnight at 4°C and with protein G- or A-Sepharose for an additional 30 min. Immunoprecipitates were washed three times in HB supplemented to 120 mM NaCl, protease inhibitors, 1 mM sodium orthovanadate, and 0.5% NP-40, followed by one wash with kinase assay buffer (20 mM MOPS [morpholinepropanesulfonic acid, pH 7.2], 25 mM β-glycerol phosphate, 5 mM EGTA, 1 mM sodium orthovanadate, and 1 mM DTT) or one wash with acetylation buffer (50 mM Tris [pH 8.0], 10% glycerol, 10 mM sodium butyrate, 1 mM DTT, 100 mM Pefabloc), depending on whether immunoprecipitates were subjected to kinase or HAT assays. Kinase assays were performed using the standard S6 kinase assay (Upstate Biotechnology), while HAT assays were done as described (5), using a mix of histones (Sigma) or an H3 tail peptide corresponding to the first 24 amino acids.
Protein-protein association studies.
Production in bacteria of glutathione S-transferase (GST)–CBP(1–1098) and GST-CBP(1098–1877) proteins has been described (2). For purification, bacteria were centrifuged and lysed in 150 mM NaCl–1 mM DTT–5 mM EDTA–25% sucrose–50 mM Tris [pH 7.5] supplemented with protease inhibitors. Lysates were sonicated for 3 min at 4°C, and bacterial debris was removed by centrifugation at 16,000 × g for 30 min. Lysates were loaded onto glutathione-Sepharose beads overnight with COS protein whole-cell extracts containing ectopic RSK2 or not. This incubation was followed by three washes with buffer I (5 mM EDTA, 250 mM NaCl, 50 mM Tris, pH 7.5), three washes with buffer II (5 mM EDTA, 120 mM NaCl, 50 mM Tris, pH 7.5), and one wash with acetylation buffer.
RESULTS
Mitogen-regulated association of RSK2 and CBP.
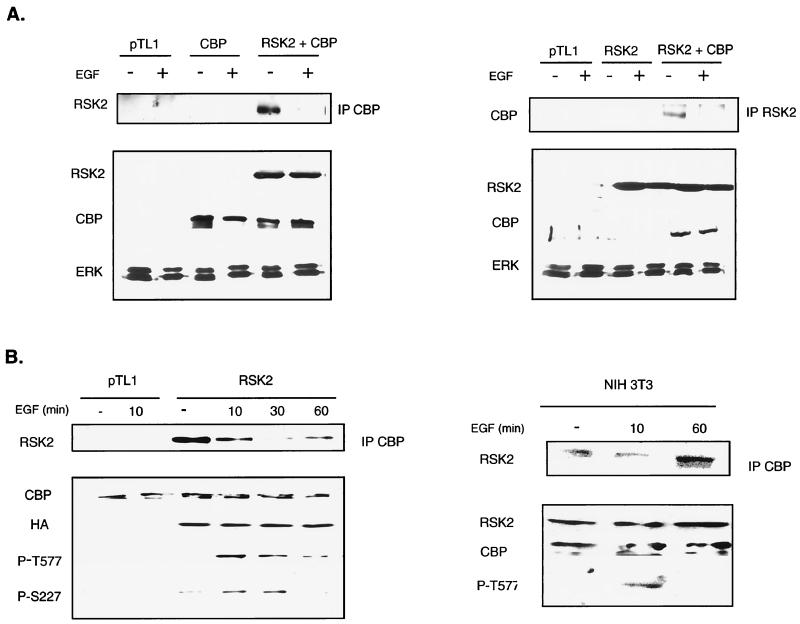
To investigate the possible interplay between phosphorylation and acetylation, we tested whether CBP could associate with RSK2. COS cells were cotransfected with expression vectors encoding RSK2 and CBP proteins, and immunoprecipitations were performed on nuclear extracts using either anti-CBP or anti-RSK2 antibodies. In both cases, RSK2 and CBP were readily coprecipitated (Fig. 1A). We tested whether stimulation by EGF could modulate the CBP-RSK2 interaction. Interestingly, association of RSK2 with CBP was detected when cells remained in a quiescent state, i.e., in a medium deprived of serum. In contrast, nearly no association of RSK2 and CBP was detected immediately (10 min) following stimulation of cells with EGF (Fig. 1A). This indicated that dissociation of the RSK2-CBP complex could be phosphorylation dependent.
FIG. 1.
Stimulation of cells with EGF induces RSK2-CBP complex dissociation. (A) Coimmunoprecipitation of RSK2 and CBP ectopically expressed in COS-1 cells. Transfected cells were serum deprived for 48 h following transfection and then treated or not for 10 min with EGF. Nuclear extracts were prepared and immunoprecipitated with anti-CBP A-22 (left) and anti-RSK2 C-19 (right) antibodies. Detection of RSK2 in anti-CBP immunoprecipitates was performed using anti-RSK2 E1 antibody (left), whereas anti-CBP A-22 antibody was used to reveal the presence of CBP in anti-RSK2 immunoprecipitates. Similar levels of ectopic CBP and RSK2 were detected in nuclear extracts prepared from cells transfected with CBP or with both CBP and RSK2 expression vectors. Detection of ERKs with anti-ERK antibody shows that equivalent amounts of total proteins were present in nuclear extracts immunoprecipitated with anti-CBP (left) or anti-RSK2 (right) antibodies. (B) The kinetics of association of RSK2 and CBP inversely correlates to the phosphorylation status of RSK2. COS-1 cells were stimulated with EGF for various times (10, 30, and 60 min). Association of RSK2 with CBP was followed by detecting the presence of RSK2 in nuclear proteins immunoprecipitated with the anti-CBP antibody. In parallel, the phosphorylation status of RSK2 was evaluated from nuclear extracts, using anti-phospho-RSK2 antibodies P-TS227 and P-T577. On the left panel, RSK2 was ectopically expressed, while on the right panel endogenous RSK2 and CBP were subjected to coimmunoprecipitation and Western analysis.
Maximal phosphorylation and activation of RSK proteins are known to occur 5 to 30 min following mitogenic stimulation (10, 21). To test the kinetics of RSK2 phosphorylation and association with CBP, we determined a time course after EGF stimulation. CBP-RSK2 association was tested in parallel with the levels of RSK2 phosphorylation using two anti-phospho-RSK monoclonal antibodies (Fig. 1B). These antibodies, P-S227 and P-T577, are directed against two critical activation sites of RSK2 located within the activation loops of the N-terminal and C-terminal kinase domains, respectively,(37). Dissociation of RSK2 from CBP was maximal 10 to 30 min after stimulation, concomitant with a drastic increase in RSK2 phosphorylation. As soon as RSK2 phosphorylation decreased, association with CBP increased (compare 30- and 60-min points in Fig. 1B). In conclusion, association of RSK2 with CBP correlates inversely with RSK2 phosphorylation.
Analogous results were obtained by analyzing the endogenous RSK2 and CBP proteins. RSK2 was present in anti-CBP immunoprecipitates prepared from NIH 3T3 (Fig. 1B, right panel), COS, and HEK 293 cells (not shown) following the same kinetics after EGF stimulation as in transfected cells. In additional experiments we have found that the other forms of p90rsk, RSK1 and RSK3, as well as the related kinase MSK1 are also found in anti-CBP immunoprecipitates after ectopic expression. In particular, paralleling what we observed here with RSK2, dissociation of the MSK1-CBP complex is concomitant with an increase in MSK1 phosphorylation (not shown). Our results confirm and extend previous observations showing CBP interaction with p90rsk (38), and reveal that phosphorylation of RSK2 is a critical event regulating RSK2-CBP complex formation.
Phorbol esters and UV light regulate RSK2-CBP association.
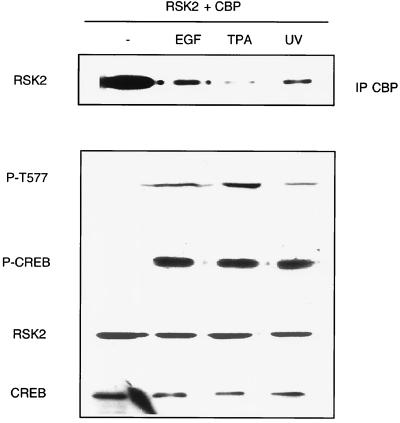
The results described above would predict that stimulation of cells with any RSK-activating factor should induce dissociation of the RSK2-CBP complex. Since RSK proteins are phosphorylated and activated not only by growth factor stimulation, but also by phorbol esters and stress factors (10, 17, 37), we tested whether treatment of cells with TPA and UV light also affected the association of RSK2 and CBP (Fig. 2). Both treatments induced dissociation of the complex within minutes following stimulation to an extent comparable to dissociation induced by EGF. Importantly, dissociation again correlated with an increase in RSK2 phosphorylation, as revealed by the P-T577 antibody.
FIG. 2.
Stimulation of cells with TPA and UV leads to RSK2-CBP complex dissociation. COS-1 cells were cotransfected with RSK2 and CBP and treated or not with TPA for 15 min, UV light for 30 min, or EGF for 10 min. Induction of RSK2 phosphorylation following treatment was determined using the P-T577 antibody. Anti-phospho-CREB antibody (P-CREB) was also used to show the effectiveness of the various treatments.
Phosphorylation of RSK2 at Ser227 controls association with CBP.
RSK proteins are composed of two unrelated kinase domains connected by a regulatory linker region (17, 31, 32). Mitogenic stimulation induces activation of the Ras-MEK-ERK cascade, which in turn leads to activation of the C-terminal kinase domain of RSK2 through direct phosphorylation of Thr577 by ERKs (14, 17, 48, 53). This results in autophosphorylation of Ser386, a residue located in the linker region, and recruitment of PDK1 to this site (24, 29). PDK1 then phosphorylates Ser227, resulting in activation of the N-terminal kinase domain, which is involved in substrate phosphorylation (6, 22, 31, 41). We wished to establish which phosphorylation event in RSK2 modulates its capacity to interact with CBP.
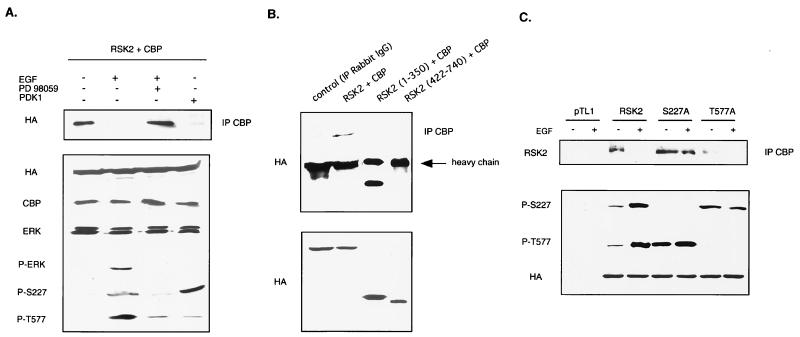
In cells stimulated with EGF but concomitantly treated with the MEK1/2-specific inhibitor PD98059 (40), the CBP-RSK2 dissociation is impaired (Fig. 3A). Thus, block of ERK signaling, which results in mostly dephosphorylated RSKs, elicits a stable RSK2-CBP complex.
FIG. 3.
Dissociation of RSK2-CBP complex is induced by phosphorylation of serine 227 within the activation loop of the N-terminal kinase domain of RSK2. (A) Unphosphorylated RSK2 binds to CBP. COS-1 cells were cotransfected with wild-type RSK2 and CBP and left untreated, treated with EGF, treated with EGF and PD98059, or cotransfected with PDK1. Phosphorylation levels of RSK2 were detected using both P-S227 and P-T577 antibodies, while activation of the MAPK/ERK pathway was evaluated using an anti-phospho-ERK antibody (P-ERK). (B) Preferential association of CBP with the N-terminal domain of RSK2 (amino acids 1 to 350). The lower panel shows that comparable levels of the truncated RSK2 proteins were used. (C) Association of CBP with the RSK2 mutant S227A is not affected by stimulation of cells with EGF. COS-1 cells were transfected with wild-type RSK2, mutant S227A, and mutant T577A and stimulated or not with EGF for 10 min. Phosphorylation levels of wild-type and mutated versions of RSK2 were detected using P-S227 and P-T577 antibodies.
In order to selectively phosphorylate the N-terminal kinase domain of RSK2, we ectopically expressed PDK1, which is a constitutively active kinase in serum-deprived cells (3) (Fig. 3A). This resulted in powerful Ser227 phosphorylation but nearly no Thr577 phosphorylation, as revealed by the anti-phospho-RSK2 specific antibodies. The effect of PDK1 on Ser227 phosphorylation disrupts CBP-RSK2 association, as it results in a threefold activation of RSK2 even in cells deprived of serum (Fig. 3A and data not shown). These results were confirmed by the use of truncated RSK2 proteins in which the two catalytic domains were separated (Fig. 3B). Only the N-terminally truncated protein RSK2(1–350) bound CBP.
These results were further supported by coimmunoprecipitation assays of CBP with RSK2 mutants (Fig. 3C). To selectively prevent phosphorylation of Ser227 or Thr577, we used two modified versions of RSK2, mutants S227A and T577A, respectively. Mutant T557A only weakly associated with CBP in unstimulated cells, while EGF stimulation resulted in dissociation of the complex. In both cases, phosphorylation at Ser227 of mutant T577A was higher than that of wild-type RSK2 in basal conditions. Therefore, the importance of phosphorylation at Ser227 in decreasing the association with CBP is confirmed. In contrast to mutant T577A, mutant S227A associated with CBP when cells were both serum deprived and EGF stimulated. Interestingly, mutation of Ser227 to alanine did not preclude phosphorylation of Thr577. Indeed, Thr577 of mutant S227A was normally phosphorylated in response to EGF stimulation and, compared to wild-type RSK2, was even hyperphosphorylated in cells deprived of serum. These data indicated that dissociation of RSK2 from CBP did not require phosphorylation of Thr577, while phosphorylation at Ser227 is a prerequisite for dissociation of the complex.
Additional experiments confirmed these results. In particular, transfection of cells with truncated versions of RSK2 showed that the C-terminal kinase domain of RSK2 did not bind CBP (not shown). In contrast, the N-terminal kinase domain of RSK2 interacted with CBP when Ser227 was not phosphorylated (not shown). Collectively, these data suggested that phosphorylation of Ser227 in the N-terminal kinase domain of RSK2 is necessary and sufficient to induce dissociation of the RSK2-CBP complex. As the mechanism of activation of RSK2 by mitogens implies that phosphorylation of Ser227 depends on phosphorylation of Thr577 (17, 24), both phosphorylation events should be associated with complex dissociation. This is indeed what was observed (Fig. 1B).
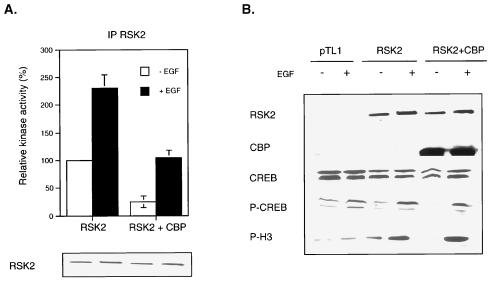
Association of CBP inhibits kinase activity of RSK2.
Our data show that RSK2 and CBP preferentially associate when RSK2 is inactive, suggesting that complex formation might constitute a mechanism to impair RSK2 kinase activity. To test this possibility, we transfected COS cells with RSK2 alone or with CBP and determined the in vitro RSK2 kinase activity on anti-RSK2 immunoprecipitates (Fig. 4A). The presence of ectopic CBP in nuclear extracts reduced RSK2 kinase activity by about fourfold when cells were deprived of serum. A weaker reduction of RSK2 kinase activity (twofold) was also observed in EGF-stimulated cells, likely due to an incomplete phosphorylation of the RSK2 pool. We also directly evaluated the phosphorylation levels of endogenous CREB and histone H3, two physiological targets of RSK2 (21, 43, 51) (Fig. 4B). Western analysis using anti-phospho-CREB and anti-phospho-H3 antibodies showed that basal phosphorylation levels of both CREB and H3 were impaired by CBP coexpression. In EGF-stimulated cells, overexpression of RSK2 induced high phosphorylation levels of CREB and H3, irrespective of the presence of ectopic CBP. Thus, altogether, these results indicated that binding of CBP to RSK2 impaired its kinase activity.
FIG. 4.
RSK2-CBP complex formation inhibits RSK2 kinase activity. (A) RSK2 kinase activity is inhibited by the presence of ectopic CBP. COS-1 cells transfected with RSK2 or with both RSK2 and CBP and stimulated or not with EGF for 10 min. Nuclear extracts were prepared and immunoprecipitated (IP) with anti-RSK2 C-19 antibody, and an in vitro kinase assay was performed using the S6 peptide as the substrate. The total levels of RSK2 protein were verified by Western analysis. (B) Basal phosphorylation levels of CREB and histone H3 are downregulated in the presence of ectopic CBP. COS-1 cells were treated as in panel A, and the phosphorylation status of endogenous CREB and histone H3, two targets of RSK2, was detected using phospho-CREB and phospho-H3 antibodies. Western analysis was performed on similar amounts of total proteins, as shown by using anti-CREB antibody. In addition, equivalent amounts of ectopic RSK2 were present in cells overexpressing CBP or not.
Association with RSK2 inhibits HAT activity of CBP.
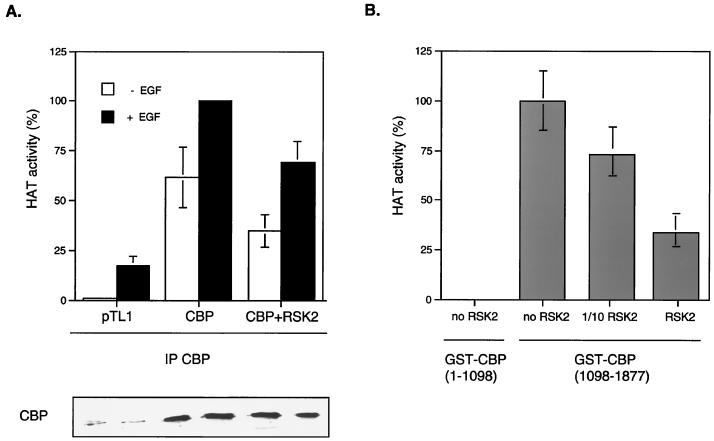
The general coactivator CBP enhances transcription of various genes by at least two molecular mechanisms: first, by bridging various transcription factors, such as CREB, with the basal transcriptional machinery, and second, CBP has a potent HAT activity which links transcription to chromatin remodeling (26). The HAT domain of CBP is adjacent to the C/H3 region (39), which corresponds to the RSK binding site (38). Interestingly, binding of the adenovirus E1A protein with this same region modulates HAT activity, probably by inducing local conformational changes (9, 28). We asked whether binding of RSK2 to CBP could also regulate its HAT activity. To test this, we transfected cells with CBP alone or with RSK2 and prepared anti-CBP immunoprecipitates from nuclear extracts. The in vitro HAT activity present in the immunoprecipitates was then assessed using an H3 peptide corresponding to the N-terminal tail of histone H3 (Fig. 5A). Ectopically expressed RSK2 impaired CBP's HAT activity, although modestly. To further support these data, a recombinant GST-CBP(1098–1877) construct, which contained both HAT and C/H3 domains, was incubated with COS cells extracts in which ectopic RSK2 was present. Reactions were then pulled down, and HAT activities were assessed using the H3 peptide. As predicted, Western analyses using anti-RSK2 antibody showed that GST-CBP(1098–1877) interacted with RSK2 (not shown). The presence of RSK2 impaired CBP(1098–1877) HAT activity in a dose-dependent manner (Fig. 5B). Thus, we concluded that binding of RSK2 to CBP inhibited its HAT activity.
FIG. 5.
RSK2-CBP complex formation inhibits CBP HAT activity. (A) CBP HAT activity is inhibited by ectopic expression of RSK2. COS-1 cells transfected with CBP or with both CBP and RSK2 were stimulated or not with EGF. Nuclear extracts were immunoprecipitated (IP) with anti-CBP antibody, and in vitro CBP HAT activity was assessed using an N-terminal H3 peptide. The total levels of CBP protein were verified by Western analysis. (B) The HAT activity of GST-CBP constructs containing HAT and E1A domains is inhibited when incubated with RSK2. The GST-CBP(1098–1877) construct was incubated with COS whole-cell extracts in which ectopic RSK2 was not (no RSK2), slightly (1/10 RSK2), or highly (RSK2) present. GST proteins were subsequently pulled down, and the HAT activity was determined as in panel A using the H3 peptide. The GST-CBP(1–1098) construct, which lacks the HAT domain, was used as the negative control.
DISCUSSION
The induction of gene transcription is the result of multiple signaling events acting in concert at the level of specific activators and coactivators, as well of selected chromatin locations where remodeling may occur following histone modifications (11, 34, 47, 50). The dynamic nature of these events is critical as it dictates the programming of gene expression. Thus, deciphering the functional interplays which operate among signaling components is an important step towards understanding all cell responses. Here we have shown that a growth factor-induced kinase, RSK2, physically associates in a dynamic fashion with a transcriptional coactivator, CBP, which has HAT activity. This finding underscores the possibility that phosphorylation and acetylation of specific substrates may be regulated by this interaction and thus that the parameters regulating the interaction itself are essential to the control of downstream events. The association between p90rsk and CBP was described previously, but the signaling events regulating the interaction were not explored in detail, also because the HAT activity of CBP was not yet characterized at the time (38). Our findings support a model in which formation of the RSK2-CBP complex is phosphorylation dependent. In contrast to the well-characterized CREB-CBP interaction, where association is induced by CREB phosphorylation at Ser133 (13), here we have shown that phosphorylation and activation of RSK2 induce dissociation from CBP (Fig. 1). Presumably, following the dissociation, RSK2 becomes available to phosphorylate CREB in response to EGF (21, 51). At the same time CBP becomes available to interact with the newly phosphorylated CREB. The dynamics of this tripartite regulation fit the kinetics of early gene transcriptional activation well (Fig. 1). In addition to CREB, other potential substrates are likely to benefit from the combined function of the activated RSK2 and CBP. Indeed, the enzymatic activities carried by CBP and RSK2 could exert a coordinate action at the level of the histone H3 tail, on which the two major sites of modification, phosphorylation (Ser10) and acetylation (Lys14), are closely spaced (12) (Fig. 6). In support of this view, it is thought that the combined phosphorylation-acetylation of the H3 tail constitutes an essential step in the local remodeling of chromatin structure (11, 12, 35).
FIG. 6.
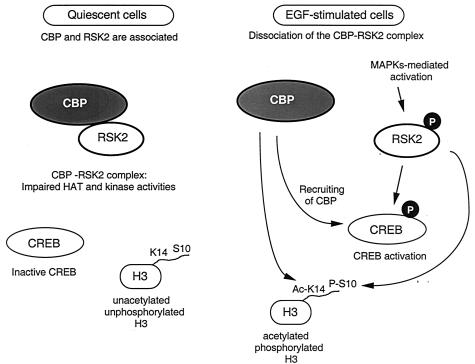
Schematic representation of the phosphorylation-regulated association of CBP and RSK2. In quiescent cells, the two proteins are found associated in a complex with low kinase and HAT activities. In serum-starved cells, the basal phosphorylation levels of two RSK2 substrates, CREB and histone H3, are also low. Upon EGF mitogenic stimulation and consequent activation by phosphorylation of RSK2 at Ser227, CBP and RSK2 dissociate. This results in increased Ser133 CREB and Ser10 H3 phosphorylation (21, 43, 51). The newly phosphorylated CREB associates with CBP, whose HAT activity is also increased. The coordinated activation of both kinase and HAT activities may converge at the histone H3 tail, where concerted modifications of Ser10 and Lys14 have been reported previously (12, 35). This possible scenario does not take into account additional signaling routes which could influence the function of the various components. While it is likely that physical interactions among other kinases and HAT molecules will be found, deciphering the regulatory pathways controlling their functions is essential to the understanding of the molecular events involved in activation of gene expression. Ac-K14, acetylated residue K14; P-S10, phosphorylated residue S10.
Earlier studies have reported that CBP and not specifically identified members of the p90rsk family interact upon activation of the Ras-dependent signaling pathway in PC12 cells (38). The difference in the mitogen-regulated CBP-RSK2 association reported here may be related to the choice of the cell type in which the analyses were performed. We have readily reproduced our results in at least three different proliferating cells (NIH 3T3, COS-1, and HEK 293), while PC12 cells were not used in our study. In addition, the use of specific anti-phospho-RSK2 antibodies and of single-amino-acid mutations (Ser227) powerfully validates the regulatory scenario reported here (Fig. 6).
An important outcome of this study concerns the reciprocal regulation of kinase and HAT activities exerted, respectively, by CBP and RSK2. Indeed, at the time the two proteins are associated in a complex, i.e., before mitogenic stimulation, both RSK2 kinase and CBP HAT activities are downregulated. Within a very few minutes after EGF stimulation and consequent dissociation of the complex, both kinase and HAT activities increase significantly. Therefore, these observations are crucial for the understanding of how the two processes of phosphorylation and acetylation are connected. A possible view of our findings is related to the antagonistic function that the inactive RSK2 may exert by associating with CBP, as we have shown that the interaction decreases the HAT activity. We favor a model in which activation of the MAPK signaling pathway and the consequent phosphorylation of RSK2 constitute a switch, as they allow dissociation of the CBP-RSK2 complex and activation of HAT function. Thus, activation of RSK2 by growth factors may result in coordinated transcription activation and chromatin remodeling.
Our findings hint at the possibility that similar scenarios may exist for other kinases and HAT molecules. It is indeed conceivable that situations like the one described here may operate under the control of various signaling pathways, specificity being provided by either the distinct combinations in the association or the dynamic modifications at various regulatory sites.
ACKNOWLEDGMENTS
We thank for help, discussions, and generous gifts of reagents the following colleagues: A. Hanauer, C. D. Allis, P. Cheung, G. Thomas, M. Frödin, G. M. Fimia, S. Jacquot, M. Zéniou, and J. L. Mandel.
This work was supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Fondation de la Recherche Médicale, Association Française contre les Myopathies, Université Louis Pasteur, Human Frontier Science Program, Organon (Akzo/Nobel), and Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali S, Ramirez S, Robin P, Trouche D, Harel-Bellan A. A rapid and sensitive assay for histone acetyl-transferase activity. Nucleic Acids Res. 1998;26:3869–3870. doi: 10.1093/nar/26.16.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 4.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Bjorbaek C, Zhao Y, Moller D E. Divergent functional roles for p90rsk kinase domains. J Biol Chem. 1995b;270:18848–18852. doi: 10.1074/jbc.270.32.18848. [DOI] [PubMed] [Google Scholar]
- 7.Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 8.Chadee D N, Hendzel M J, Tylipski C P, Allis C D, Bazett-Jones D P, Wright J A, Davie J R. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen R H, Chung J, Blenis J. Regulation of pp90rsk phosphorylation and S6 phosphotransferase activity in Swiss 3T3 cells by growth factor-, phorbol ester-, and cyclic AMP-mediated signal transduction. Mol Cell Biol. 1991;11:1861–1867. doi: 10.1128/mcb.11.4.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung P, Allis C D, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 12.Cheung P, Tanner K G, Cheung W L, Sassone-Corsi P, Denu J M, Allis C D. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 13.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 14.Chung J, Pelech S L, Blenis J. Mitogen-activated Swiss mouse 3T3 RSK kinases I and II are related to pp44mpk from sea star oocytes and participate in the regulation of pp90rsk activity. Proc Natl Acad Sci USA. 1991;88:4981–4985. doi: 10.1073/pnas.88.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton A L, Rose S, Barratt M J, Mahadevan L C. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosio C, Cermakian N, Allis C D, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 17.Dalby K N, Morrice N, Caudwell F B, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 18.Davie J R, Spencer V A. Signal transduction pathways and the modification of chromatin structure. Prog Nucleic Acid Res Mol Biol. 2000;65:299–340. doi: 10.1016/s0079-6603(00)65008-0. [DOI] [PubMed] [Google Scholar]
- 19.Deak M, Clifton A D, Lucocq L M, Alessi D R. Mitogen-and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Cesare D, Fimia G M, Sassone-Corsi P. Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem Sci. 1999;24:281–285. doi: 10.1016/s0968-0004(99)01414-0. [DOI] [PubMed] [Google Scholar]
- 21.De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Natl Acad Sci USA. 1998;95:12202–12207. doi: 10.1073/pnas.95.21.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher T L, Blenis J. Evidence for two catalytically active kinase domains in pp90. Mol Cell Biol. 1996;16:1212–1219. doi: 10.1128/mcb.16.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frodin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 24.Frodin M, Jensen C J, Merienne K, Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavin A C, Nebreda A R. A MAP kinase docking site is required for phosphorylation and activation of p90rsk/MAPK/AP kinase-1. Curr Biol. 1999;9:281–284. doi: 10.1016/s0960-9822(99)80120-1. [DOI] [PubMed] [Google Scholar]
- 26.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 27.Ginty D D, Bonni A, Greenberg M E. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 28.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 29.Imhof A, Wolffe A P. Transcription: gene control by targeted histone acetylation. Curr Biol. 1998;8:R422–R424. doi: 10.1016/s0960-9822(98)70268-4. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R, Hunter T. Transcription: a growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 31.Jensen C J, Buch M B, Krag T O, Hemmings B A, Gammeltoft S, Frodin M. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J Biol Chem. 1999;274:27168–27176. doi: 10.1074/jbc.274.38.27168. [DOI] [PubMed] [Google Scholar]
- 32.Jones S W, Erikson E, Blenis J, Maller J L, Erikson R L. A Xenopus ribosomal protein S6 kinase has two apparent kinase domains that are each similar to distinct protein kinases. Proc Natl Acad Sci USA. 1988;85:3377–3381. doi: 10.1073/pnas.85.10.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 34.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 35.Lo W S, Trievel R C, Rojas J R, Duggan L, Hsu J Y, Allis C D, Marmorstein R, Berger S L. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 36.Mahadevan L C, Willis A C, Barratt M J. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- 37.Merienne K, Jacquot S, Zeniou M, Pannetier S, Sassone-Corsi P, Hanauer A. Activation of RSK by UV-light: phosphorylation dynamics and involvement of the MAPK pathway. Oncogene. 2000;19:4221–4229. doi: 10.1038/sj.onc.1203712. [DOI] [PubMed] [Google Scholar]
- 38.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 39.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 40.Pang L, Sawada T, Decker S J, Saltiel A R. Inhibition of MAP kinase kinase blocks the differentiation of PC-12 cells induced by nerve growth factor. J Biol Chem. 1995;270:13585–13588. doi: 10.1074/jbc.270.23.13585. [DOI] [PubMed] [Google Scholar]
- 41.Richards S A, Fu J, Romanelli A, Shimamura A, Blenis J. Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr Biol. 1999;9:810–820. doi: 10.1016/s0960-9822(99)80364-9. [DOI] [PubMed] [Google Scholar]
- 42.Sadowski H B, Gilman M Z. Cell-free activation of a DNA-binding protein by epidermal growth factor. Nature. 1993;362:79–83. doi: 10.1038/362079a0. [DOI] [PubMed] [Google Scholar]
- 43.Sassone-Corsi P, Mizzen C A, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis C D. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 44.Shaywitz A J, Greenberg M E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 45.Smith J A, Poteet-Smith C E, Malarkey K, Sturgill T W. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J Biol Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 46.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 47.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 48.Sturgill T W, Ray L B, Erikson E, Maller J L. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 49.Trivier E, De Cesare D, Jacquot S, Pannetier S, Zackai E, Young I, Mandel J L, Sassone-Corsi P, Hanauer A. Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature. 1996;384:567–570. doi: 10.1038/384567a0. [DOI] [PubMed] [Google Scholar]
- 50.Wolffe A P, Pruss D. Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 51.Xing J, Ginty D D, Greenberg M E. Coupling of the RAS-MAPK pathway to gene activation by RSK2: a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 52.Yntema H G, van den Helm B, Kissing J, van Duijnhoven G, Poppelaars F, Chelly J, Moraine C, Fryns J P, Hamel B C, Heilbronner H, Pander H J, Brunner H G, Ropers H H, Cremers F P, van Bokhoven H. A novel ribosomal S6-kinase (RSK4; RPS6KA6) is commonly deleted in patients with complex X-linked mental retardation. Genomics. 1999;62:332–343. doi: 10.1006/geno.1999.6004. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y, Bjorbaek C, Moller D E. Regulation and interaction of pp90(rsk) isoforms with mitogen-activated protein kinases. J Biol Chem. 1996;271:29773–29779. doi: 10.1074/jbc.271.47.29773. [DOI] [PubMed] [Google Scholar]