Figure 1.
Gene co-expression modules for TG-GATEs rat liver dataset
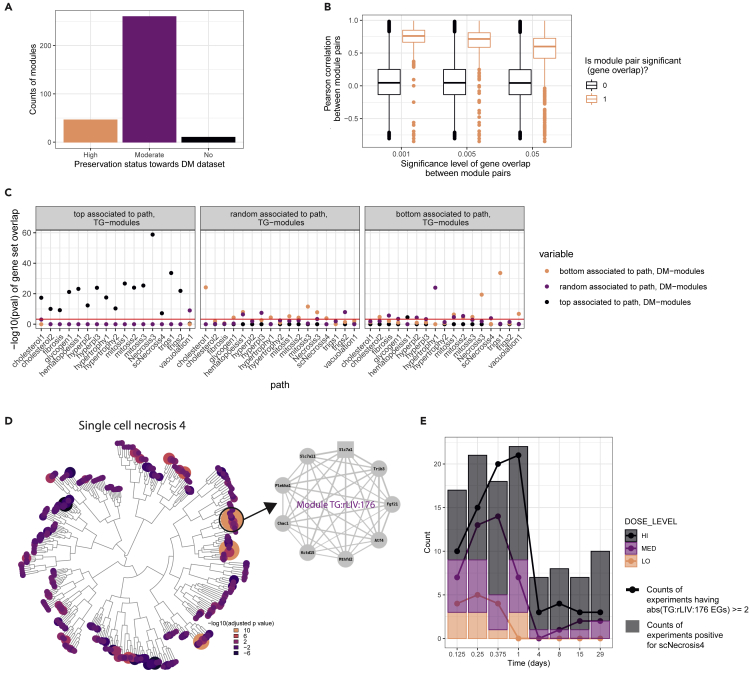
(A) Number of TG-based modules that show low (Z-summary <2), medium (2≤ Z-summary <10) and high (Z-summary ≥10) preservation toward DM rat liver dataset.
(B) Boxplot of Pearson correlation values between EGs of overlapping or non-overlapping modules pairs (color) between the 2 builds, shown by significance level of overlap (xaxis).
(C) −log10(pval) overlap between the gene sets consisting of the top 20, or bottom 20 associated modules in TG-GATEs to each toxicity phenotypes or 20 randomly selected modules in TG-GATEs and the gene sets consisting of the top 20, bottom 20 associated modules in DM to each toxicity phenotypes and 20 randomly selected modules in DM. The red solid line indicates-log10(0.0005). Toxicity phenotypes are abbreviated as specified in Table S3.
(D) Dendrogram of TG-modules, showing-log10 transformed adjusted p values for the association of each module with the occurrence of single cell necrosis (scNecrosis4). The size of each circle is proportional to the absolute log10(adjusted p value) and the color to the-log10(adjusted p value). Highlighted and zoomed out below the structure of module TG:rLIV:176. Each node corresponds to the genes included in the module, the square node corresponds to the hub gene and the edges weight indicates the correlation between each genes’ log2FC across TG-data.
(E) Count of experiments showing EGs for module TG:rLIV:176 higher than 2 or lower than −2 (points connected by lines) and showing positive score for the toxicity phenotype scNecrosis 4 (bar plots). Different colors indicated the three dosage levels.