Abstract
Cell proliferation requires precise control to prevent mutations from replication of (unrepaired) damaged DNA in cells exposed spontaneously to mutagens. Here we show that the modified human DNA repair enzyme O6-methylguanine-DNA methyltransferase (R-MGMT), formed from the suicidal repair of the mutagenic O6-alkylguanine (6RG) lesions by MGMT in the cells exposed to alkylating carcinogens, functions in such control by preventing the estrogen receptor (ER) from transcription activation that mediates cell proliferation. This function is in contrast to the phosphotriester repair domain of bacterial ADA protein, which acts merely as a transcription activator for its own synthesis upon repair of phosphotriester lesions. First, MGMT, which is constitutively present at active transcription sites, coprecipitates with the transcription integrator CREB-binding protein CBP/p300 but not R-MGMT. Second, R-MGMT, which adopts an altered conformation, utilizes its exposed VLWKLLKVV peptide domain (codons 98 to 106) to bind ER. This binding blocks ER from association with the LXXLL motif of its coactivator, steroid receptor coactivator-1, and thus represses ER effectively from carrying out transcription that regulates cell growth. Thus, through a change in conformation upon repair of the 6RG lesion, MGMT switches from a DNA repair factor to a transcription regulator (R-MGMT), enabling the cell to sense as well as respond to mutagens. These results have implications in chemotherapy and provide insights into the mechanisms for linking transcription suppression with transcription-coupled DNA repair.
Exposure to environmental mutagens, such as UV irradiation and N-nitroso compounds, accounts for 80% of the human cancer incidence (30). The effectiveness of our cells' attempts to repair the DNA lesions inflicted by mutagens on our DNA before DNA replication is fundamentally linked to manifestation of the disease through this etiological pathway. The p53 protein is critical here for maintaining genomic integrity, since its induction upon DNA damage enables the cell to acquire sufficient time to repair the damaged DNA by halting cell cycle progression through its effector, the cell cycle-dependent kinase inhibitor p21WAFI (11, 15). However, p53 appears to be only a downstream effector of this DNA damage response pathway in the cell, since cellular factors, such as the hChk1 and hChk2 (human homologs of the yeast RAD53 and CDS1 proteins), are shown to stabilize p53 through phosphorylation upon exposure to mutagens (6, 14, 35). While much is known about cell regulation where external stimuli are transduced via the membrane receptors and kinase cascades to activate the nuclear DNA (8), knowledge of reciprocal pathways through which DNA, when it is damaged, signals cellular response through immediate factors remains circumstantial.
The high-fidelity property of DNA and RNA polymerases enables them to serve as important signaling factors for the integrity of the DNA as they are arrested at the bulky DNA lesions inflicted by mutagens (33) while processing along the DNA to carry out their functions. However, what could be an effective signaling factor for the DNA containing subtle DNA lesions that do not arrest the polymerases? DNA repair enzymes are molecular sensors of damaged DNA in the cell, since they recognize and repair damaged DNA. It would be a very effective survival strategy if the same DNA repair enzyme could also be a signaling molecule as well as a regulator for the presence of damaged DNA in the cells. The Escherichia coli ADA protein is a unique example, exhibiting these properties in protecting the bacteria from the cytotoxic effects of the phosphotriester lesions in the DNA that are induced by alkylating agents. Upon repairing the phosphotriester lesions by transferring the alkyl group from the phosphotriester lesion to the active site of the phosphotriester repair domain at its N terminus, the alkylated protein becomes a transcription activator for its own synthesis. This increases the amount of the ADA protein in the bacterial cell for protection against further damage from alkylating agents (38, 39). Unfortunately, such an elegant DNA repair and response pathway appears to be limited to the prokaryotes, since homologs of the ADA protein are not found in the eukaryotes (44).
Nevertheless, the O6-methylguanine-DNA methyltransferase (MGMT), which has an alkyl transfer repair mechanism similar to that of the E. coli ADA protein, is present in all organisms. It protects cells from the mutagenic and cytotoxic effects of alkylating carcinogens (10) by transferring the alkyl group of O6-alkylguanine (6RG) formed in the DNA by alkylating carcinogens (20) to the cysteine residue at its active site (28, 29). This repair mechanism, however, depletes instantly the MGMT activity in the cell as MGMT is converted to the active-site alkylated and inactive MGMT, R-MGMT (19). Even though the presence of unrepaired 6RG lesions in the cellular DNA is detrimental, producing point mutations upon DNA replication (1) or mutated mRNA that can be instantly translated into an altered protein (42) upon transcription (12), the MGMT suicidal repair is preserved through evolution (28, 29). Could R-MGMT serve as a unique molecular memory of exposure to alkylating carcinogens (3), similar to the alkylated ADA protein in bacteria, and therefore provide some important cellular functions?
Several observations suggested that human MGMT and R-MGMT could regulate estrogen receptor (ER)-dependent activities. First, the ligand-bound nuclear receptor activates cell proliferation (31, 37), and therefore, its activity must be controlled upon DNA damage. Second, active MGMT localizes at the active transcription sites of RNA polymerase II-dependent genes (2) (including ER-regulated genes). Third, biochemical analyses show that human R-MGMT adopts an altered conformation in exposing the VLWKLLKVV domain (26) containing an LXXLL motif that also is found in transcription coactivators for their binding to nuclear receptors (13). Fourth, the LXXLL motifs of R-MGMT and the coactivator glucocorticoid receptor interacting protein binding to the ligand binding domain of ER-α adopt similar amphipathic α-helix structures (7, 34). Finally, this LXXLL motif of human and mammalian MGMTs is not found in lower organisms that do not have ER.
Here we provide the experimental findings of how human R-MGMT serves as a DNA damage-induced transcription suppressor for regulating ER-mediated cell proliferation upon exposure to alkylating agents. These findings serve as an important example of how a DNA repair enzyme interplays with transcription factors and integrators to regulate cell proliferation upon DNA damage, and they provide a good reason for the suicidal repair of the 6RG lesions by MGMT to form R-MGMT. This appears to be a highly specific protein modification in the cell that enables the DNA to regulate itself upon alkylation damage by transducing the signal from the 6RG lesions in the damaged DNA via R-MGMT to block ER from activating the DNA directly to transcribe or indirectly to replicate by ER-transcribed gene products.
MATERIALS AND METHODS
Cell extracts and drug treatments.
Cells were from the American Type Culture Collection. Nuclear and total cell extracts were prepared as described previously (3, 19), with added cocktail protease inhibitors (1:500; Sigma) and 0.1% Triton (required for ER extraction, though it causes minor cleavage of MGMT by V8; see Fig. 2C and F). Cdex media were made from serum treated with charcoal-coated dextran (Hyclone) and RPMI medium (Gibco) without phenol red. Stock solutions of O6-benzylguanine (6BG) (200 μM) and iodomethane (MeI) (10 mM) were made in serum-free medium, and a solution of 17-β estradiol (E2) (Sigma) was made in ethanol. For experiments with Cdex medium, cells were first grown in normal medium to 60% confluence. After washing with phosphate-buffered saline (PBS), they were grown in Cdex medium for 48 h and replaced with fresh Cdex medium before experiments with E2.
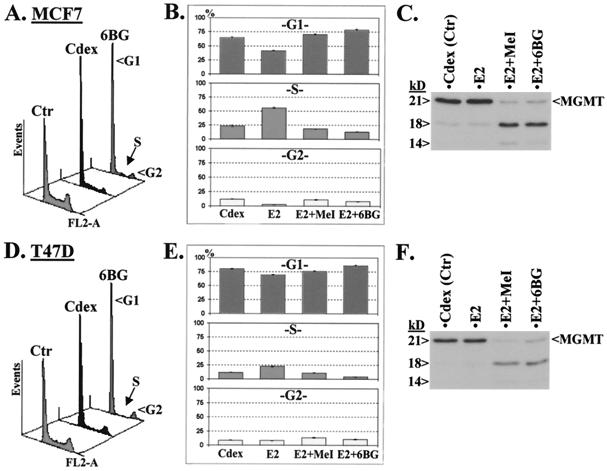
FIG. 2.
6BG and MeI block 17-β estradiol (E2)-stimulated growth of ER-positive cells. (A and D) Histograms from FC analysis of the populations of MCF7 and T47D cells, respectively, at G1, S, and G2 phases after culture in full medium alone (Ctr), Cdex medium, and full medium with 6BG for 15 h. (B and E) Summary of the percentages of cells at G1, S, and G2 phases for cells grown in Cdex medium alone (Cdex), treated with E2 for 15 h (E2), treated with MeI (1 mM) followed by E2 (E2+MeI), and treated with 6BG (50 μM) followed by E2 (E2+6BG). (C and F) R-MGMT analysis by protease V8 and Pab.MGMT in extracts from the cells analyzed in panels B and E after 2 h of E2 treatment.
Immunochemistry.
The Mab.677-F11 monoclonal antibody to steroid receptor coactivator 1 (SRC-1) was a gift from B. W. O'Malley. Antibodies for CBP/p300 (rabbit polyclonal Pab.451), ER-α (Pab.HC20, Pab.H184, and Mab.F10) and SRC-1 (goat Pab.N19 and Pab.M20) were from Santa Cruz. Mab.CBP/p300 (Power Clone) was from Upstate. They were used at a concentration of 2 μg/ml. For immunoprecipitation, antibodies (2 μg of each) were added to the nuclear extracts (800 μg by Lowry assay) in buffer A (1 ml containing 150 mM NaCl, 50 mM Tris [pH 8.0], 1 mM dithiothreitol, and 1 mM EDTA). After incubation for 12 h at 4°C followed by centrifugation (2,000 × g for 2 min), the supernatants were transferred to respective anti-immunoglobulin (Ig) agarose beads (20 μl of a 50% suspension; Sigma) for 2 h with gentle rolling. Following washing with buffer A (three times; 1 ml), the proteins were debound from the beads by boiling them in Laemmli buffer (30 μl) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis.
Flow cytometry.
A Becton Dickinson FACScan machine was used to acquire and analyze 50,000 events (using Cellquest and Modfit). DNA analysis was carried out by propidium iodide (PI) staining (50 μg/ml at 20°C, 30 min) of ethanol (80%; 12 h at −20°C)-fixed cells digested with RNase (20 μg/ml for 30 min). For dual analysis of expressed MGMT and DNA, cells transfected with wild-type (wt) and K107L MGMT cDNAs (19) were fixed in 1% paraformaldehyde (30 min on ice) followed by permeabilization with 0.2% Tween 20 and blocking with 10% sheep serum (20°C for 30 min for both steps). Mab.3B8 (100 μl of a 3 μg/ml concentration in buffer C [1% Tween 20 and 10% sheep serum in PBS]) was added to the cells (1 h at 37°C) followed by washing two times with PBS (500 μl). After treatment with fluorescein isothiocyanate anti-mouse Igs (200 μl of 1:50 dilution in buffer C) for 30 min at 37°C followed by washing with PBS, the cells were further processed for PI staining as described above.
In vitro binding.
GST-(wt) and MGMT (K107L) were prepared as described previously (26). Recombinant ER-α (50 ng; Oncogene Science) was added to buffer B (100 μl of 50 mM HEPES [pH 7.3], 0.1% Triton X, 10% glycerol, 100 mM KCl, and 1 mM dithiothreitol) with glutathione-Sepharose (20 μl of a 50% suspension)-bound fusion proteins (∼100 ng) which were first treated with bovine serum albumin (100 μl of 20% bovine serum albumin in buffer B) at 4°C for 1 h. After shaking at 4°C for 15 min, the beads were recovered and washed three times with 500 μl of buffer B before boiling in Laemmli buffer for immunoblot analysis. In peptide competition assays, high-pressure liquid chromatography-purified peptides (7 μg from a 1-mg/ml stock solution in argon-treated water; Research Genetics) were added to the recombinant ER-α for 10 min before binding.
ERE reporter and mammalian two-hybrid assays.
The manufacturer's protocols (Qiagen) for transfection with Superfect were followed. The luciferase estrogen-responsive element (ERE) and ER reporter assay was described previously (23). First, the human ER-α cDNA was cloned into the pXJ41 vector similar to the full-length wt and mutant MGMT DNAs (19). For the mammalian two-hybrid assay (kit from Clontech), the cDNAs of Wt-MGMT, K107L-MGMT, and the nuclear receptor binding domain of SRC-1 (NR) were cloned into the V vector (with the VP-16 insert), whereas the ER cDNA was cloned into the M vector (with the GAL4 DNA insert). Besides the M and V vectors (1 μg of each), pCMVβ-gal (0.5 μg as an internal control) and 5× Gal4luc3 (1 μg as the luciferase reporter) were cotranfected to the ER− MGMT+ HeLa CCL2 cells grown on 6-well plates in Cdex medium for 24 h and stimulated with E2 (100 nM) for 24 h. For experiments with 6BG, the ER− MGMT− SV40-virus-transformed human MRC5 fibroblast (MRC5.SV40) cells were used, and the drug (25 μM) was added together with E2. The luciferase activities were normalized to beta-galactosidase (β-Gal) activities from triplicate experiments.
Labeling of RNA by [3H]uridine and RT-PCR analysis of mRNAs.
Cells were grown in six-well culture dishes and labeled with 50 μCi of [5,6-3H]uridine (Amersham, Little Chalfont, England) for 6 h. Total RNA was isolated using the Qiagen (Hilden, Germany) RNeasy-Kit. Two micrograms of the labeled RNAs were either subjected to scintillation counting or resolved on a 1% agarose gel for analyzing RNA. The resolved labeled RNAs were then transferred to a nitrocellulose membrane. The filter was then air dried and cross-linked with UV for 2 min. The amplifier-treated filter was autoradiographed for 48 h. For reverse transcription PCR (RT-PCR) (21) (kit from Promega), mRNA (2 μg) was reverse transcribed with d(T)17 (0.5 μg) followed by PCR with primers (0.05 μg of each) for β-actin (5′-AGCGGGAAATGCTGCGTG-3′ and 5′-CAGGGTACATGGTGGTGCC-3′), porphobilinogen deaminase (PBGD) (5′-TCTGGTAACGGCAATGCGGC-3′ and 5′-CCAGGGCATGTTCAAGCTCC-3′), progesterone receptor (PR) (5′-GATTCAGAAGCCAGCCAGAG-3′ and 5′-TGCCTCTCGCCTAGTTGATT-3′), and pS2 (5′-GGAGAACAAGGTGATCTGCG-3′ and 5′-CACACTCCTCTTCTGGAGGG) under the following conditions: 5 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C, and a final elongation of 5 min at 72°C. PCR products were analyzed on a 1.8% agarose gel.
RESULTS
6BG inhibits the growth of breast cells expressing ER and MGMT (mer+).
To investigate human R-MGMT function, we treated some MGMT-positive (mer+) and -deficient (mer−) cells with 6BG, which alkylates selectively the cysteine C145 residue of human MGMT without causing DNA damage (26). Flow cytometry (FC) analysis of these 6BG-treated cells in Fig. 1A shows that the populations of S-phase cells decreased significantly (i.e., growth retardation) in the MCF7 and T47D breast cell lines expressing MGMT (see the immunoblot in Fig. 1B), ER (Fig. 1C), and SRC-1 (Fig. 1D) but not the pairs of mer+ (BT549) and mer− (MDA-231) ER-negative breast cells and mer+ (HeLa CCL2) and mer− (MRC5.SV40) virus-infected cells. While the levels of ER and SRC-1 proteins were not themselves affected by 6BG, the majority of MGMT in 6BG-treated mer+ cells was cleaved by protease V8 into the 14- and 18-kDa polypeptides, the fingerprint of R-MGMT (3, 36) (Fig. 1B, bottom panel).
FIG. 1.
6BG inhibits the growth of breast cells expressing ER and MGMT (mer+). (A) Flow cytometry analysis of breast (MCF7 [panel 1], T47D [panel 2], BT549 [panel 3], and MDA-231 [panel 4]) and virus-infected (HeLa CCL2 [panel 5] and MRC5.SV40 [panel 6]) cells after 6BG treatment (60 μM) for 15 h (26). The histograms and tables represent the gated cell populations (events, y axis) and percentages, respectively, with different DNA contents stained by PI (FL2-A on the x axis defines the G1, S, or G2 cells). Ctr, control. (B) Immunoblot of MGMT and R-MGMT by Pab.MGMT in total cell extracts. Cell lines are numbered as in panel A. Control indicates untreated samples, whereas 6BG indicates 6BG-treated (2 h) samples. The 6BG+V8 panel shows the 6BG-treated samples digested with protease V8 for R-MGMT analysis (3). Two hundred micrograms of cell extract/lane was used. (C) Immunoblot of ER as for panel B by Pab.HC20. (D) Immunoblot of SRC-1 as for panel B by the antibody Pab.N19.
6BG and MeI (a DNA-damaging agent) block E2-stimulated growth of ER-positive cells.
The above-described results raise the question of whether the specific conversion of active MGMT to R-MGMT by 6BG inhibits the growth of MGMT+ ER+ cells. To address this, we compared 6BG treatment with the effect of withdrawal of estrogen-like stimuli on these ER+ cells (using medium without phenol red and steroid-depleted serum, Cdex medium [37]), since some ER-responsive genes regulate cell proliferation (31). The FC analysis in Fig. 2A and D shows, respectively, that the MCF7 and T47D cultures grown in Cdex medium contain fewer S-phase cells than those grown in full medium with 6BG. These results suggest that 6BG may antagonize ER. Thus, we analyzed cells grown in Cdex medium treated with 6BG or MeI (because MGMT is a DNA repair enzyme, we tested whether generation of R-MGMT by an alternative means involving DNA damage by MeI will behave similarly) followed by stimulation with E2, an activating ligand for ER. The FC analysis shows that 6BG and MeI behave similarly in blocking E2-stimulated growth on cells grown in Cdex medium; see the S-phase cells of E2, E2 plus MeI, and E2 plus 6BG in Fig. 2B and E. Again, most MGMT in the drug-treated cells was converted to R-MGMT (Fig. 2C and F). Thus, modification of MGMT may be causal in blocking the activation of ER by E2, because 6BG and MeI convert MGMT to R-MGMT via independent pathways (26).
The inhibition of ER activity by 6BG and MeI is independent of p53.
To ensure that the failure of E2 in activating the ER responses in the ER+ MGMT+ cells treated with the DNA-damaging agent MeI is indeed related to the conversion of MGMT to R-MGMT but not p53, we compare the levels of the p53 protein in these treated cells with those in the γ-irradiated cells (11, 15). The Western blots in Fig. 3A show that p53 levels in the MCF7 cells exposed to the dosage of γ irradiation that is sufficient to induce growth arrest in these cells (Fig. 3D) are significantly higher than in those cells undergoing growth arrest induced by 6BG or MeI treatment. This is not due to a low dosage of MeI being used, since the majority of MGMT in the MeI-treated cells is converted to the protease V8-sensitive R-MGMT (compare Fig. 3B and C) through the repair of the 6RG lesions in the DNA induced by MeI or the direct alkylation of MGMT by MeI (26). Furthermore, the T47D cells, which undergo growth arrest induced by 6BG or MeI treatment (Fig. 1A and 2E), express high levels of the mutated p53 protein (4).
FIG. 3.
Inhibition of ER response by 6BG and MeI is independent of p53. MCF7 cells grown in normal medium were treated with 2 Gy of γ irradiation (from a cobalt 65 source), 60 μM 6BG, or 1 mM MeI. Total cell extracts were prepared at 1, 3, and 6 h after treatment for Western blot analysis using Mab.DO1 for p53 and Mab.3B8 for MGMT as shown in panels A and B, respectively. The cell extracts were also treated with protease V8 to confirm the presence of R-MGMT as shown in panel C. Panel D shows the FC analysis of the cell population profiles 24 h after treatment with 6BG, Mel, and γ irradiation. Ctr, untreated control cells. The inserted table gives the summary of the average percentage of cells at G1, S, and G2 from three independent experiments.
R-MGMT disrupts the ER–SRC-1 interaction and fails to associate with the CBP/p300 complex.
The above-described results indicate that the growth arrest induced by 6BG and MeI in ER+ MGMT+ cells is independent of p53, since the protein is poorly induced under these drug treatments. What molecular mechanism might lie behind the above observations? Since R-MGMT adopts an altered conformation to expose the VLWKLLKVV domain (26) containing the LXXLL motif found in steroid hormone coactivators for their binding to nuclear hormone receptors (13), we investigated whether R-MGMT can regulate ER. First, we quantified the levels of the two proteins in the cell context using purified recombinant proteins. Figure 4A shows that MCF7 or T47D cell extracts contain ∼6-fold more MGMT than the ER proteins (∼80 ng of MGMT [21 kDa] and ∼40 ng of ER [67 kDa] are present in 200 μg of cell extract). Thus, there is potentially sufficient R-MGMT generated by drug treatment to mediate the function of ER, since >90% of cellular MGMT can be converted to R-MGMT (Fig. 2C and F). We then studied the ER–SRC-1 complex (27) by analyzing the amount of ER coimmunoprecipitated with SRC-1 from various nuclear extracts. The immunoblot in Fig. 4B shows that ER coprecipitates with SRC-1 from normal cell extracts but coprecipitates poorly with SRC-1 from those treated with 6BG and MeI. This explains the overriding effect of the drugs on E2-driven cell proliferation (Fig. 2B and E), since SRC-1 coactivates transcription by ER (27). However, does R-MGMT promote the dissociation of ER from SRC-1? We therefore tested whether R-MGMT interacts with ER. Interestingly, MGMT was present at high levels in the ER immunoprecipitates from the drug-treated cells containing R-MGMT but not in those from the untreated cells containing active MGMT (Fig. 4C), suggesting that ER preferentially binds the R-MGMT. This is not the case for the ER+ MGMT− SV-ER cells (see Fig. 6E, lane 2), which are ER− MGMT− MRC5.SV40 cells stably expressing exogenous ER (interestingly, we were unable to obtain “native” ER+ MGMT− cells) (Fig. 4D). Unlike the case with ER+ MGMT+ MCF7 cells, as shown in Fig. 4B, the immunoprecipitable ER–SRC-1 complex remained intact in the ER+ MGMT− SV-ER cells treated with 6BG or MeI (Fig. 4E). These results establish a unique role for R-MGMT in these events.
FIG. 4.
R-MGMT disrupts the ER–SRC-1 interaction and fails to associate with the CBP/p300 complex. (A) Quantification of MGMT and ER proteins. Lanes labeled MCF7 and T47D contain 200 μg of total cell extracts, whereas lanes with numbers contain the purified recombinant MGMT and ER proteins, with numbers indicating amounts in nanograms (ng). Western blotting was performed with Mab.3B8 for MGMT and with Pab.HC20 for ER. (B) Immunoblot analysis of SRC-1 and ER in SRC-1 immunoprecipitates from MCF7 nuclear extracts. The lanes labeled Ctr, Mel, and 6BG are untreated (control), Mel (1 mM)-treated, and 6BG (50 μM)-treated cells grown in full medium for 2 h, respectively. The top panel is the immunoblot of the SRC-1 protein visualized by Pab.N19 in the SRC-1 Mab.677-F11 immunoprecipitate, and the bottom panel is the coimmunoprecipitated ER shown by Pab.HC20. (C) Immunoblot analysis of ER (Mab.F10) and MGMT or R-MGMT (Mab.3B8) in ER immunoprecipitates (Pab.H184), similar to panel B. (D) Survey of breast cells. Immunoblots of ER and MGMT in reported breast cell lines are shown: note the absence of the dual ER-positive and MGMT-deficient phenotypes. (E) Immunoblot analysis of SRC-1 and ER in SRC-1 immunoprecipitates from SV-ER (ER+ MGMT−) nuclear extracts, similar to panel B: note that the levels of ER protein were not affected by MeI and 6BG treatments compared to the MCF7 (ER+ MGMT+) cells in panel B. (F) Immunoblot analysis of CBP/p300 (Pab.451) and MGMT (Pab.MGMT) and in CBP/p300 immunoprecipitates (Mab.CBP/p300), similar to panel B.
FIG. 6.
Effect of R-MGMT on the cell cycle and transcription. (A) Growth arrest. MCF7 cells were transfected with expression vector (Ctr) or vectors fused with wt or K107L mutant MGMT cDNAs for 24 h (19) before analysis by FC using dual channels to quantify the fluorescence in the cells from the stainings of MGMT by Mab.3B8 and of DNA by PI. Panels a, b, and c show the distributions of transfected cells stained by MGMT Mab.3B8 (FL1-A, y axis) versus their DNA contents (FL2-A, x axis). Cells with greater MGMT stainings than the control (Ctr), due to the expression of the MGMT cDNAs, are gated (the region R) to generate the histograms (d, e, and f) where the cell populations (events, y axis) were plotted against their DNA contents (x axis) representing G1, S, and G2 phases. The arrow in panel c indicates that a large population of cells expressing the K107L-MGMT proteins are at G1 phase. (B) Inhibition of RNA synthesis. Cells were labeled for 6 h with [3H]uridine after being cultured under full medium (FM) (lane 2), FM with 6BG (lane 1, pretreatment with 60 μM 6BG for 2 h before labeling), Cdex medium (lane 3), Cdex with E2 (lane 4) (pretreatment with 100 nM E2 for 2 h before labeling), or Cdex with E2 and 6BG together (lane 5) (pretreatment with 6BG and E2 together for 2 h before labeling). The labeled total RNAs isolated (2 μg/lane) were resolved on a 1% agarose gel. The top panel is the autoradiograph of the membrane with the labeled RNAs resolved by the agarose gel. The histogram is a summary of the 3H counts from 2 μg of labeled RNAs (averaged from three independent experiments). The bottom panel is the description of the samples. (C) RT-PCR analysis of the levels of PR, pS2, PBGD, and β-actin mRNAs obtained from cells cultured for 24 h, similar to panel B without [3H]uridine labeling. (D) Abbreviations of the wt and Mu (K107L mutant) MGMT full-length (FL) cDNAs used for transfection into the SV-ER cells (19). M is the first methionine. (E) Reporter assay. Top panel, immunoblot of the expressed ER (Pab.HC20) and MGMT (Pab.MGMT) proteins in SV-ER cells grown in Cdex medium 24 h after cotransfection with ERE-Luc and MGMT (Wt, Mu, Wt-2A, and Mu-2A) constructs followed by 6 h of treatment with 1 μM E2. SV indicates the parental ER- and MGMT-negative MRC5.SV40 cells. Lower panel, the corresponding luciferase activities in the cell extracts.
To assess its proximity to transcriptionally active ER–SRC-1 complexes in the cell, we analyzed MGMT in the immunoprecipitates of CBP/p300, which are transcription integrators for various nuclear receptor-coactivator complexes (40, 43). Figure 4F shows that MGMT was recovered at high levels from normal cell extracts but not from drug-treated extracts, suggesting that after MGMT was converted to R-MGMT in cells exposed to alkylating agents, R-MGMT might migrate from the CPB/p300-containing complex to bind proteins such as ER.
The LXXLL motif of R-MGMT interacts with ER in vitro and in vivo.
Since results of the above-described immunoprecipitation experiments strongly suggest that there is a close relationship between ER and R-MGMT, we tested whether the exposed LXXLL motif of R-MGMT can bind to ER directly by using synthetic peptides containing the LXXLL motifs of MGMT and CBP (as a control) in competition experiments (13). These peptides were added to the recombinant ER-α before binding to the immobilized glutathione S-transferase fusion protein of the unique K107L-MGMT mutant (see the scheme in Fig. 5A, which adopts an altered conformation [3] identical to that of R-MGMT with an exposed VLWKLLKVV domain [26]). The immunoblot in Fig. 5B confirms that the K107L mutant preferentially binds to ER-α compared to the wt MGMT. This binding is sensitive to both Wt-CBP and Wt-MGMT peptides (peptides 1 and 3 in Fig. 5C) but not mutant peptides (2 and 4) substituted in a conserved leucine residue, indicating that the intact LXXLL motif is critical for binding of R-MGMT to ER-α. Similar results were obtained when R-MGMT was generated via the TATAC6MGTATA oligonucleotide substrate (18) (data not shown).
FIG. 5.
The LXXLL motif of R-MGMT binds to ER in vitro and in vivo. (A) The scheme for binding of recombinant ER-α to immobilized GST-(K107L)MGMT. (B) Immunoblot of recombinant ER-α (by Mab.F10) bound to immobilized GST-(wt)MGMT and GST-(K107L)MGMT (visualized by Mab.3B8). (C) Peptide competition. The top panel is the sequence of the peptide used. Peptides (7 μg) were added to the ER-α before binding to the immobilized GST-(K107L)MGMT. The associated ER-α was analyzed by immunoblot using Mab.F10. (D) Mammalian two-hybrid assay. The top panel shows the region of SRC-1 (NR) used, similar to MGMT in size and in positioning of the LXXLL motif. Codons are numbered. The bottom panel shows the average luciferase activity obtained from three independent experiments in the ER− MGMT+ HeLa CCL2 cells normalized to the control β-Gal activities obtained. V represents the VP-16-containing vector, and M represents the GAL4 DNA-binding domain-containing vector. The fusion constructs are V-K107L-MGMT, V-SRC-1 (NR), and M-ER. The luciferase activities were normalized to β-Gal activities from triplicate experiments. (E) Effect of 6BG on mammalian two-hybrid assay. The experiment is similar to that shown in panel D except that the ER− MGMT MRC5.SV40 cells were used and 6BG (25 μM) was added together with E2.
Furthermore, in vivo mammalian two-hybrid analysis comparing K107L-MGMT to a similar-size SRC-1 fragment containing the nuclear receptor-interating domain (NR) in binding to ER-α showed equivalent binding in the ER− MGMT+ HeLa CCL2 cells (Fig. 5D). Similar results were obtained when the ER− MGMT− MRC5.SV40 cells were used (Fig. 5E). In addition, when R-MGMT was generated in situ by using 6BG (added together with E2) to convert the expressed Wt-MGMT to R-MGMT, a positive signal was observed (see the increase in luciferase activity of the Wt-MGMT sample treated with 6BG over that of the untreated sample in Fig. 5E). However, we are unclear as to why generation of R-MGMT via this procedure is less efficient than the K107L mutant MGMT in initiating the two-hybrid response. Nevertheless, together these in vitro and in vivo data suggest that R-MGMT mimics the NR box of SRC-1 in binding to ER through sequences containing the LXXLL motif.
The R-MGMT-equivalent K107L-MGMT mutant induces the growth arrest of ER-positive cells.
The above observation that R-MGMT can behave similarly to the NR box of SRC-1 in binding to ER (Fig. 5D) provides a reason for the disruption of the ER–SRC-1 interaction observed upon treatment with 6BG or MeI (Fig. 4B). This raises the question of whether sequestering ER from the SRC-1-associated transcriptional machinery by R-MGMT is the molecular event behind the growth arrest of ER+ MGMT+ cells induced by 6BG or MeI treatments (which generate R-MGMT [26]) (Fig. 1A and 2B and E). To establish such a link, we analyzed the MCF7 cells by FC after transient transfection with the cDNAs of wt and mutant (K107L) MGMT. Figure 6A shows that cells expressing the K107L-MGMT (i.e., R-MGMT) proteins (26) are predominantly at the G1 phase, but the Wt-MGMT-expressing cells are not (see the cell distribution histograms of cells stained by MGMT antibody in the gated region R in panels d, e, and f). Thus, generation of R-MGMT by expression of a conformational equivalent mutant (K107L) MGMT protein alone is sufficient to inhibit the growth of ER-driven cells, similar to the 6BG or MeI treatments, which generate R-MGMT (Fig. 2).
The exposed LXXLL motif in R-MGMT suppresses ER-mediated transcription.
Together these results demonstrate a direct relationship between growth inhibition (Fig. 6A) and disruption of the ER–SRC-1 complex (Fig. 4B) by R-MGMT generated in the ER+ MGMT+ cells. To understand the mechanism behind these observations, we investigated the effect of R-MGMT on ER function (i.e., ER-mediated transcription activities) in vivo by first studying the effect of 6BG treatment on the incorporation of [3H]uridine. Figure 6B shows that addition of 6BG to MCF7 cells grown in full medium indeed inhibits the incorporation of [3H]uridine into the RNA; compare the labeled RNA in samples 2 (control/FM) and 1 (with 6BG) in the autoradiogram (top panel) and histogram. Further pulse-labeling of MCF7 cells cultured in estrogen-depleted Cdex medium and then stimulated with E2 showed that E2 activates transcription of these cells, as shown by the increased [3H]uridine incorporation into the RNA, but fails to do so in the presence of 6BG (compare samples 3 [control/Cdex], 4 [with E2], and 5 [with E2 and 6BG] in Fig. 6B). These results indicate that R-MGMT inhibits E2-mediated ER transcription. Consistent with this observation, RT-PCR analysis of the mRNAs of the known ER-regulated progesterone receptor (PR) and pS2 genes (Fig. 6C, compare lane 1 with lane 2 and lane 5 with lane 4) shows that their mRNA levels are significantly lower in the 6BG-treated cells, but the β-actin and PBGD mRNAs (21) are not.
To show that the sequences containing the LXXLL motif of R-MGMT (or K107L-MGMT itself) is involved in blocking ER-mediated transcription, we used the ER+ MGMT− stable SV-ER cell line and transfected full-length wt and mutant MGMT cDNAs (see Fig. 6D). The results portrayed in Fig. 6E show that the Mu (K107L) construct produced significantly less luciferase activity (i.e., transcription inhibition) in the luciferase reporter assay for ERE in SV-ER cells (23). The residual transcription activity observed for the Mu sample probably arises from the background ER activity of the untransfected SV-ER cells, since immunostaining showed that 60% of the transfected cells express the MGMT protein (data not shown). Significantly, both the Wt-2A and Mu-2A constructs, which contain mutations in the LXXLL domain, show luciferase activities comparable to the control's. These results show that R-MGMT, which binds to ER (Fig. 4C and 5B to E), blocks ER from transcription that is activated through the binding of the LXXLL motif of coactivators, presumably to the activation function 2 domain of ER (25, 34). This explains the effects of 6BG or MeI treatment on the inability of SRC-1 antibodies to immunoprecipitate ER (Fig. 4B) and failure of growth stimulation by E2 (Fig. 2B and E). Together, these results thus link transcription suppression of ER-responsive genes and growth inhibition mediated by R-MGMT.
DISCUSSION
Unlike histone deacetylase-mediated transcription suppression (via the mSIN3 [45] or MeCP proteins [24]), which cannot operate immediately, R-MGMT functions as a DNA damage-induced transcription suppressor (Fig. 6E), providing timely suppression of ER-mediated cell proliferation when cells are exposed spontaneously to alkylating agents (Fig. 2). This serves two immediate functions. First, cells that proliferate under the control of ER will not enter S phase upon exposure to alkylating agents (Fig. 2B and E), thus avoiding 6RG-directed mutations during DNA replication (1). Secondly, even though alkylating agents can inflict damage on the transcribing DNA of ER-regulated genes, this DNA will be blocked from transcription (Fig. 6E) to prevent the instant formation of mutated RNA (12). Thus, the “suicide” of MGMT upon repair of the 6RG lesions immediately converts the DNA repair enzyme to a transcription suppressor (R-MGMT), allowing it to directly couple detection and the subsequent response specifically to the insult of alkylating carcinogens. This mechanism should ensure that ER would not activate proliferation of cells with damaged DNA, so that genomic integrity could be maintained. Furthermore, it would be important to establish how the activities of ER could be regulated when cells were exposed to DNA-damaging agents other than alkylating agents given the observed potency of ER in activating cell growth (see the significant increases in the S-phase cells upon E2 stimulation in Fig. 2B and E) and transcription activities (compare the [3H]uridine incorporated into the RNA in lanes 3 and 4 of Fig. 6B) in the ER+ cells.
One might also predict from the prevalent MGMT and ER dual positive phenotypes (Fig. 4D) that in patients treated with 6BG (used in clinical trials for brain tumors) (28, 29), breast-derived tumors would be arrested (Fig. 1). Perhaps R-MGMT, as an endogenous ER modulator that blocks ER-driven cell proliferation even in the presence of its ligand E2 (see Fig. 2B and E and Fig. 6B, lane 5), would be as effective as the exogenous agent tamoxifen, a selective estrogen receptor modulator that functions by competing with the ligand of ER (9), which has been shown to reduce the breast cancer incidence in high-risk women by 45% (36). In retrospect, monitoring the MGMT levels in the ER+ cells within the respective tissues of those women who received the drug estrogen, which activates ER, during hormone replacement therapy may be necessary, since sufficient MGMT levels must be maintained to control the rapid growth of ER+ cells, due to stimulation by estrogen, when they are exposed to environmental alkylating agents. Failure to control cell proliferation upon DNA damage, thus allowing damaged DNA to be replicated, could be a putative factor responsible for the elevated cancer incidence in this group (5, 32). Nevertheless, exposure to environmental alkylating agents is of concern because these agents may also affect ER-dependent homeostatsis of neural, skeletal, cardiovascular, and reproductive tissues (16), since they rapidly convert MGMT in the cell to R-MGMT (26).
Although a DNA repair pathway is shown here to directly impinge on cell regulation, it remains unclear how different DNA lesions in the transcribing DNA are perceived by the cell. The RNA polymerase, which encounters every transcribing base residue, serves as an important checkpoint protein for the presence of bulky lesions in the transcribing DNA, since it cannot transcribe across these lesions. Although it does not possess DNA repair activity, this stalled RNA polymerase orchestrates the effective removal of the bulky DNA lesion in transcribing DNA by recruiting the required DNA repair factors (22, 33). Thus, the high-fidelity RNA polymerase enables the cell to regulate transcription as well as repair when bulky DNA lesions are formed in the transcribing DNA. However, what could be the mechanism behind the repair of those DNA lesions (i.e., 6RG) that escape the editing mechanism of RNA polymerase? Such a mechanism would demand a constant surveillance at the sites of active transcription by DNA repair factors that are also capable of overseeing the transcription activities on the DNA. MGMT fulfills some of these requirements. First, MGMT as a DNA repair factor is constitutively present at active transcription sites, whereas R-MGMT, as a transcription suppressor (Fig. 6E), appears instantly upon exposure to alkylating agents (2). Second, active MGMT is a component of the CBP/p300-containing complex (Fig. 4F) that integrates external signals to activate the transcription activities of nuclear receptors (40, 43). Furthermore, CBP/p300 is a histone acetylase (17), which can modify the histone octamer in the nucleosome to poise the DNA for transcription (41) and also expose the DNA to undesirable damage by mutagens (2). Thus, it is strategic for MGMT to be a component of the CBP/p300-containing complex (Fig. 4F) in linking DNA repair events and transcription regulation to deal with the 6RG lesions in the damaged transcription-active DNA prior to their transcription by RNA polymerases.
ACKNOWLEDGMENTS
Alvin K. C. Teo and Hue Kian Oh contributed equally to this work.
We thank E. Manser for critical reading of the manuscript, R. Moschel (National Cancer Institute) and D. B. Yarosh (Applied Genetic Inc.) for 6BG, B. W. O'Malley (Baylor College of Medicine) for SRC-1 antibody, V. Yu for the ER and ERE constructs, R. Kaushik for SV-ER cells, W. Stuenkel for advice, L. S. H. Chuang and K. S. W. Li for discussions, and Y. H. Tan and NSTB Singapore for support.
REFERENCES
- 1.Abott P, Saffhill R. DNA synthesis with methylated poly (dC · dG) templates; evidence for a competitive nature of miscoding by O6-methylguanine. Biochim Biophys Acta. 1979;567:51–61. doi: 10.1016/0005-2787(79)90125-4. [DOI] [PubMed] [Google Scholar]
- 2.Ali R B, Teo A K C, Oh H K, Chuang L S H, Ayi T C, Li B F L. Implication of localization of human DNA repair enzyme O6-methylguanine-DNA methyltransferase at active transcription sites in transcription-repair coupling of the mutagenic O6-methylguanine lesion. Mol Cell Biol. 1998;8:1660–1669. doi: 10.1128/mcb.18.3.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayi T C, Oh H K, Lee T K Y, Li B F L. A method for the simultaneous identification of human active and active-site alkylated DNA repair enzyme, O6-methylguanine-DNA methyltransferase, and its application for monitoring human exposure to alkylating carcinogens. Cancer Res. 1994;54:3726–3731. [PubMed] [Google Scholar]
- 4.Bartek J, Iggo R, Gannon J, Lane D P. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990;6:893–899. [PubMed] [Google Scholar]
- 5.Beral V, Banks E, Reeves G, Appleby P. Use of HRT and the subsequent risk of cancer. J Epidemiol Biostat. 1999;4:191–210. [PubMed] [Google Scholar]
- 6.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels D S, Mol C D, Arvai A S, Kanugula S, Pegg A E, Tainer J A. Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding. EMBO J. 2000;19:1719–1730. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell J E. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 9.Dhingra K. Antiestrogens-tamoxifen, SERMs and beyond. Investig New Drugs. 1999;17:285–311. doi: 10.1023/a:1006348907994. [DOI] [PubMed] [Google Scholar]
- 10.Dumenco L L, Allay E, Norton K, Gerson S L. The prevention of thymic lymphomas in transgenic mice by human O6-alkylguanine-DNA alkyltransferase. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W S. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 12.Gerchman L L, Ludlum D B. The properties of O6-methylguanine in template for RNA polymerase. Biochim Biophys Acta. 1973;308:310–316. doi: 10.1016/0005-2787(73)90160-3. [DOI] [PubMed] [Google Scholar]
- 13.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 14.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 15.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 16.Korach K S. Insights from the study of animals lacking functional estrogen receptor. Science. 1994;266:1524–1527. doi: 10.1126/science.7985022. [DOI] [PubMed] [Google Scholar]
- 17.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 18.Liem L K, Wong C W, Lim A, Li B F L. Factors influencing the repair of the mutagenic lesion O6-methylguanine in DNA by human O6-methylguanine-DNA methyltransferase. J Mol Biol. 1993;231:950–959. doi: 10.1006/jmbi.1993.1344. [DOI] [PubMed] [Google Scholar]
- 19.Lim A, Li B F L. The nuclear targeting and nuclear retention properties of a human DNA repair protein MGMT are both required for its nuclear localization: the possible implications. EMBO J. 1996;15:4050–4060. [PMC free article] [PubMed] [Google Scholar]
- 20.Loveless A. Possible relevance of O6-alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 21.Mass R A, Bruning P F, Top B, Breedijk A J, Peterse H L. Growth arrest associated changes of mRNA levels in breast cancer cells measured by semi-quantitative RT-PCR: potential early indicators of treatment response. Cancer Lett. 1995;97:107–116. doi: 10.1016/0304-3835(95)03959-z. [DOI] [PubMed] [Google Scholar]
- 22.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 23.Naar A M, Boutin J M, Lipkin S M, Yu V C, Holloway J M, Glass C K, Rosenfeld M G. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 24.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 25.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 26.Oh H K, Teo A K C, Ali R B, Lim A, Ayi T C, Yarosh D B, Li B F L. Conformational change in human DNA repair enzyme O6-methylguanine-DNA methyltransferase upon alkylation of its active site by DNA damage-related SN1 and direct-acting SN2 alkylating agents: breaking a “salt-link”? Biochemistry. 1996;35:12259–12266. doi: 10.1021/bi9603635. [DOI] [PubMed] [Google Scholar]
- 27.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 28.Pegg A E. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogens and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 29.Pegg A E. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 30.Perera F P, Weinstein I B. Molecular epidemiology: recent advances and future directions. Carcinogenesis. 2000;21:517–524. doi: 10.1093/carcin/21.3.517. [DOI] [PubMed] [Google Scholar]
- 31.Prall O W J, Rogan E M, Sutherland R L. Estrogen regulation of cell cycle progression in breast cancer cells. J Steroid Biochem Mol Biol. 1998;65:169–174. doi: 10.1016/s0960-0760(98)00021-1. [DOI] [PubMed] [Google Scholar]
- 32.Ross R K, Paganini-Hill A, Wan P C, Pike M C. Effect of hormone replacement therapy on breast cancer risk: estrogen versus estrogen plus progestin. J Natl Cancer Inst. 2000;92:328–332. doi: 10.1093/jnci/92.4.328. [DOI] [PubMed] [Google Scholar]
- 33.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 34.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 35.Shieh S Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 36.Smigel K. Breast cancer prevention trial shows major benefit, some risk. J Natl Cancer Inst. 1998;90:647–648. doi: 10.1093/jnci/90.9.647. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland R L, Hall R E, Taylor I W. Cell proliferation kinetics of MCF-7 human mammary carcinoma cells in culture and effects of tamoxifen on exponentially growing and plateau-phase cells. Cancer Res. 1983;43:3998–4006. [PubMed] [Google Scholar]
- 38.Teo I, Sedgwick B, Demple B, Li B, Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984;9:2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teo I, Sedgwick B, Kilpatrick M W, McCarthy T V, Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986;45:315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 40.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 41.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 42.Viswanathan A, You H J, Doetsch P W. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 43.Wadgaonkar R, Phelps K M, Haque Z, Williams A J, Silverman E S, Collins T. CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J Biol Chem. 1999;274:1879–1882. doi: 10.1074/jbc.274.4.1879. [DOI] [PubMed] [Google Scholar]
- 44.Wood R D, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 45.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]